Abstract
Programmed death-ligand 1 (PD-L1) is critical for cancer cells ability to evade attacks from the host immune system. However, the molecular mechanisms controlling the PD-L1 expression have not been fully understood. Here, we demonstrate that Sorting Nexin 6 (SNX6) is a novel regulator of PD-L1 expression. Knockdown of SNX6 in cancer cells significantly decreases PD-L1 protein levels. In contrast, loss of SNX6 does not reduce the PD-L1 mRNA levels. Instead, SNX6 interacts with Cullin3, an E3 ubiquitin ligase responsible for PD-L1 ubiquitination and subsequent degradation. By binding with Cullin3, SNX6 decreases the interaction between the adaptor protein, Speckle-type POZ protein (SPOP), and Cullin3, which in turn downregulates Cullin3-mediated PD-L1 ubiquitination. This research reveals a novel molecular nexus in modulating PD-L1.
Keywords: SNX6, PD-L1, Cullin3, SPOP, immune checkpoint, immunotherapy, ubiquitination
Introduction
Programmed death-ligand 1 (PD-L1) is a transmembrane protein that plays critical roles as a negative regulator in the T-cell immune response [1, 2]. PD-L1 is the ligand of the Programed cell death 1 (PD-1), another membrane protein expressed in T-cells [3]. By binding with PD-1, PD-L1 provides negative signals to suppress T-cell activation, which is important to prevent an autoimmune response [4]. A previous study showed that both PD-1- and PD-L1-knockout mice are prone to autoimmune diseases as severe symptoms develop when they are immunized with autoantigens [5]. Cancer cells can use the PD-1/PD-L1 pathway to escape from a host’s anti-tumor immune attack [6, 7]. Therefore, reactivating the antitumor immunity by blockading the function of PD-1/PD-L1 has been verified to be a promising strategy to treat cancer [8-10].
PD-L1 expression is regulated by a multitude of feedback signaling pathways. For example, PD-L1 expression in cancer cells can be stimulated by secreted IFN-γ from immune cells within the tumor microenvironment [11-14]. Furthermore, expression of PD-L1 can also be driven by intrinsic mechanisms, such as amplification of the PD-L1 gene in tumors [15]. PD-L1 can be downregulated via proteasome-dependent or lysosome-dependent degradation pathways. Speckle-type POZ protein (SPOP) interacts with PD-L1 to mediate PD-L1 ubiquitination through the E3 ubiquitin ligase, Cullin3, which is required for the proteasome-dependent degradation of PD-L1 [16]. By stabilizing SPOP expression, the cell cycle regulators, Cyclin D1 and CDK4, downregulate PD-L1 levels [16]. Huntingtin-interacting protein 1-related protein (HIP1R) modulates PD-L1 levels in a lysosome-dependent manner. By interacting with PD-L1, HIP1R promotes PD-L1 sorting to lysosomes for degradation [17]. Further exploration of the molecular mechanisms controlling PD-L1 expression could lead to the discovery of novel targets to improve current immune checkpoint blockade therapies to treat cancer.
Here, we identified Sorting Nexin 6 (SNX6) as a novel regulator of PD-L1 expression. As a member of the Sorting Nexin family proteins, SNX6 contains a phosphoinositide-binding SNX-phox homology domain and a membrane-binding carboxy-terminal Bin/amphiphysin/Rvs domain [18, 19]. SNX6 is an important component of the retromer complex and plays critical roles in the endosome-to-Trans Golgi Network (TGN) transport [20, 21]. Our current research indicates that SNX6 modulates PD-L1 levels in a retromer function-independent manner. SNX6 can bind to Cullin3 and block the Cullin3-SPOP interaction. Hence, SNX6 decreases PD-L1 ubiquitination and modulates PD-L1 expression. These results suggest a novel function of SNX6 in modulating immune response by regulating PD-L1 stability.
Materials and Methods
Cell cultures and transfection.
HEK-293 cells, the head and neck cancer cell lines, UMSCC22B and UMSCC1, and the breast cancer cell line, MDA-MB-231, were cultured using Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Jurkat T-cells were cultured using RPMI 1640 Medium supplemented with 10% fetal bovine serum (FBS). For siRNA transfection, cells were transfected with Oligofectamine (Invitrogen, Carlsbad, CA, USA) for 72 hours following the manufacturer’s instructions.
Reagents.
IFN-γ (cat#: Z02915) was purchased from GenScipt (Piscataway, NJ). Antibodies to SNX1 (cat#: 611482) and SNX2 (cat#: 611308) were from BD Bioscience (San Jose, CA). Antibody to SNX5 (cat#: ab180520) was from Abcam (Cambridge, MA). Antibodies to SNX6 (cat#: sc-8679) and tubulin (cat#: sc-398103) were from Santa Cruz Biotechnology (Santa Cruz, California). Antibodies to PD-L1 (cat#: 13684), NEDD4-1 (cat#: 2740), Cullin3 (cat#: 2759), and Myc (cat#: 2778) were from Cell Signaling (Danvers, MA). Anti-SPOP antibody (cat#: 16750-1-AP) was from Proteintech (Rosemont, IL). Anti-Flag antibody (cat#: F3165) was from Sigma-Aldrich (St. Louis, MO). Secondary antibodies were obtained from Jackson Immuno Research Laboratories (West Grove, PA). Human recombinant SNX6 protein (GST tag) and recombinant Cullin3 protein (Flag tag) were purchased from OriGene (Rockville, MD).
Constructs.
SNX6 ORF clone (Genbank accession number: NM_152233), PD-L1 ORF clone (Genbank accession number: NM_014143), and SPOP ORF clone (Genbank accession number: NM_003563) were purchased from GenScript and were sub-cloned into pcDNA3.1 vector with a Flag-tag at the C-terminus. SNX6 truncation mutants were generated using PCR primer overlap extension with primers containing the desired mutations. Cullin3 ORF clone (Genbank accession number: NM_003590) was purchased from GenScript and was sub-cloned into pcDNA3.1 vector with a Myc-tag at the C-terminus.
siRNA.
The sequence of control scrambled siRNA is 5’-GUACCUGUACUUCAUGCAG-3’. SNX6 siRNA_1 sequence is 5’-UAAAUCAGCAGAUGGAGUA-3’. SNX6 siRNA_2 sequence is 5’-GAGUUGCUGCAUUCAGAAA-3’. SNX5 siRNA sequence is 5’-CUACGAAGCCCGACUUUGA-3’. SNX1 siRNA sequence is 5’-CCACGUGAUCAAGUACCUU-3’. SNX2 siRNA sequence is 5’-GAUAGACCAGUUACAUCAA-3’. Cullin3 siRNA (ID 139187) and SPOP siRNA (ID 13286) were purchased from ThermoFisher.
RNA extraction and Real-Time PCR.
UMSCC1 cells were harvested, and total RNA was extracted using ISOLATE RNA mini kit (Bioline, cat#: BIO-52072). 2ug total RNA was used for the cDNA reverse transcription with the kit (Applied Biosystems, cat#: 4368814). SYBR Green master mix (Applied Biosystems, cat#: A25742) was used for Real-Time PCR and the results was analyzed by QUANTSTUDIO 3 software. The PD-L1 mRNA abundance was normalized to the expression of GAPDH. Primers used for the PCR were: 5’- TCCTGAGGAAAACCATACAGC-3’ (forward) and 5’-GATGGCTCCCAGAATTACCA-3’ (reverse) for PD-L1; 5’- AATCCCATCACCATCTTCCA-3’ (forward) and 5’-TGGACTCCACGACGTACTCA-3’ (reverse) for GAPDH.
Cycloheximide chase assay.
Control or SNX6 siRNAs were transfected into UMSCC22B cells using Oligofectamine following the manufacturer’s instruction. 72 h after transfection, cells were treated with 50 μg/ml Cycloheximide (CHX) for the indicated times. The cells were then harvested and processed for western blotting analysis.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed as described [22]. Briefly, cells were harvested and lysed in 25 mM HEPES, pH 7.2, 150 mM NaCl, 0.5% NP-40, 1 mM MgCl2, and protease inhibitor cocktail, then centrifuged and incubated with Myc-Trap® magnetic agarose beads (Chromotek) at 4°C for 4 hours. The immunocomplexes were separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and analyzed as indicated.
In vivo ubiquitination assay.
The ubiquitination of PD-L1 was evaluated as described previously [23]. His6-Ubiquitin-conjugated Flag-PD-L1 in HEK-293 cells was purified by Ni2+-nitrilotriacetic acid (NTA) beads. HEK-293 cell was lysed in IP buffer (25 mM HEPES, pH 7.2, 150 mM NaCl, 0.5% NP-40, 1 mM MgCl2, and protease inhibitor cocktail) and incubated with Ni2+-NTA beads (Qiagen) for 2 hours at 4 °C. The beads were washed with IP buffer, buffer A (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-HCl, pH 8.0, 10 mM β-mercaptoethanol), and buffer B (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-HCl, pH 5.3, 10 mM β-mercaptoethanol), and bound proteins were eluted with buffer C (200 mM imidazole, 0.15 M Tris-HCl, pH 6.7, 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS). The eluted proteins were analyzed by Western blotting for the presence of His6-Ubiquitin-conjugated PD-L1 via using anti-Flag antibody.
T-cell IL-2 expression measurement.
Jurkat T-cells were first activated with Dynabeads Human T-Activator CD3/CD28 (Life Technologies). Then the Jurkat T-cells were co-cultured with UMSCC1 cells at 5:1 (Jurkat: UMSCC1 cell) ratio for 24 h. Secreted IL-2 level in the medium was measured by Human IL-2 ELISA Kits as described by the manufacturer (Thermo Scientific).
Tumor cell-killing assay.
T-cell-mediated tumor-cell-killing assay was performed according to the manufacturer’s instructions (Essen Bioscience) [24]. To activate the primary T-cells, human Primary Peripheral Blood Mononuclear cells (ATCC) were incubated with anti-CD3 antibody (100 ng/ml) and IL-2 (10 ng/ml) for 96 h. Then nuclear-restricted red fluorescent protein (RFP)-expressing MDA-MB-231 cells were co-cultured with activated T-cells in the presence of caspase 3/7 substrate (Essen Bioscience). RFP and green fluorescent (NucView 488 Caspase 3/7 substrate) signals were then monitored. The green-fluorescent cells were counted as dead cells.
Statistics.
All data analysis was performed using SigmaPlot. Bar graphs represent means ± SEM., as indicated. Statistical significance was assessed using Student’s t-test.
Results
Loss of SNX6 decreases the expression of PD-L1.
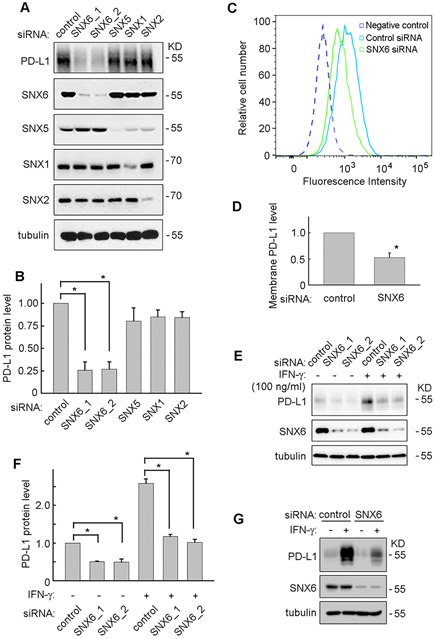
SNX5 and SNX6 are important components of the retromer complex [20]. By modulating the endosomal trafficking, SNX5 and SNX6 control the trafficking and degradation of many membrane receptors such as Epidermal Growth Factor Receptor, Insulin-like Growth Factor 1 Receptor and TSH Receptor [25-27]. To test whether SNX5 and SNX6 can modulate the PD-L1 expression, siRNAs targeting SNX5 or SNX6 were used. UMSCC22B is a head and neck squamous carcinoma cell (HNSCC) line that expresses high basal levels of PD-L1. As shown in Figure 1A and 1B, loss of SNX6 significantly decreased PD-L1 proteins levels in UMSCC22B cells. In contrast, knockdown of SNX5 did not have significant effect on PD-L1 levels. In addition, knockdown of SNX1 or SNX2, other two components of retromer complex, did not affect PD-L1 levels (Fig. 1A and 1B). This suggests SNX6 specifically modulates PD-L1 expression. To rule out siRNA off-target effects, two different SNX6 siRNAs, SNX6_1 and SNX6_2, were used and both knocked down SNX6 with similar depletion of PD-L1 expression (Fig. 1A and 1B).
Figure 1. Loss of SNX6 decreases PD-L1 expression.
(A) UMSCC22B cells were transfected with control siRNA or siRNAs targeting SNX6, SNX5, SNX1, or SNX2, and then the indicated protein levels were detected by Western Blot. Tubulin levels were used as a loading control. (B) Quantification of PD-L1 protein levels in UMSCC22B cells transfected with different siRNA. (C) UMSCC22B cells were transfected with control or SNX6 siRNA, and then the cell surface PD-L1 levels were detected via flow cytometry. (D) Quantification of cell surface PD-L1 levels in control or SNX6-knockdown UMSCC22B cells. (E) UMSCC1 cells were transfected with control or two different SNX6 siRNAs (SNX6_1 and SNX6_2), and treated with or without IFNγ (100 ng/ml) for 24 hours. Then the PD-L1 and SNX6 protein levels were detected by Western Blot. (F) Quantification of PD-L1 protein levels in control or SNX6-knockdown UMSCC1 cells treated with or without IFNγ. (G) MDA-MB-231 cells were transfected with control or SNX6 siRNA, and then treated with or without IFNγ (100 ng/ml) for 24 hours. The PD-L1 and SNX6 protein levels were detected by Western Blot. Error bars indicate mean ± SEM from three independent experiments. *, P < 0.05.
PD-L1 is a transmembrane protein and has functional roles at the cell surface [9]. Therefore, a flow cytometry method was used to determine the change of PD-L1 levels on the plasma membrane. As shown in Figure 1C and 1D, the PD-L1 levels on the cell surface were significantly decreased in the SNX6-knockdown UMSCC22B cells as compared to the control cells. This result is consistent with the downregulation of total PD-L1 protein levels when SNX6 is knocked down (Fig. 1A and 1B).
The expression of PD-L1 in cancer cells can be stimulated by IFN-γ [28-30]. Therefore, the effects of SNX6 on IFN-γ-induced PD-L1 expression was further examined. UMSCC1 is a HNSCC cell line that expresses low basal levels of PD-L1. IFN-γ treatment strongly enhanced PD-L1 expression in control cells. Furthermore, the IFN-γ-induced PD-L1 expression was significantly decreased in SNX6-knockdown cells compared with in control cells (Fig. 1E, and 1F). This result indicates that SNX6 controls both basal and IFN-γ-induced PD-L1 expression.
To further determine whether the effect of SNX6 is cell line specific, a breast cancer cell line, MDA-MB-231, was also tested. Similarly as shown in UMSCC1 cells, loss of SNX6 in MDA-MB-231 cells dramatically decreased IFN-γ-induced PD-L1 expression, which suggests the function of SNX6 in modulating PD-L1 is not limited to HNSCC (Fig. 1G).
SNX6 modulates PD-L1 protein degradation.
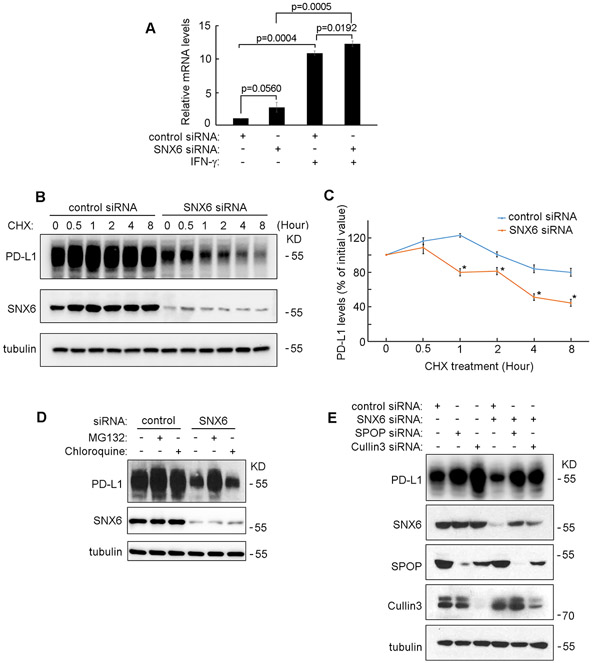
To determine whether the mRNA levels of PD-L1 were affected by SNX6-knockdown, real-time PCR was used. As shown in Figure 2A, IFN-γ treatment strongly induced PD-L1 mRNA transcription in UMSCC1 cells. Loss of SNX6 did not decrease IFN-γ-stimulated PD-L1 mRNA levels when compared to the control cells, which indicates that the function of SNX6 in modulating PD-L1 expression is not due to a change in the PD-L1 mRNA levels (Fig. 2A). Therefore, the effect of SNX6 on PD-L1 protein stability was further detected by Cycloheximide (CHX) chasing assay. CHX is an inhibitor of translation that arrests protein synthesis [31]. As shown in Figure 2B and 2C, loss of SNX6 increased the degradation rates of PD-L1. These results suggest that SNX6 may regulate PD-L1 expression levels by modulating PD-L1 protein stability instead of regulating the mRNA levels.
Figure 2. SNX6 modulates the protein degradation of PD-L1.
(A) Control siRNA or SNX6 siRNA was transfected into UMSCC1 cells. 48 hours after siRNAs transfection, the cell were treated with or without IFNγ (100 ng/ml) for 12 hours and the PD-L1 mRNA levels in these cells were quantified by Real-Time PCR. (B) UMSCC22B cells were transfected with control or SNX6 siRNA, the cells were then treated with CHX (50 μg/ml) CHX for the indicated times. Then the levels of PD-L1 and SNX6 were detected by western Blot. Levels of tubulin were used as loading control. (C) Quantification of PD-L1 levels in control or SNX6-knockdown UMSCC22B cells treated with CHX (relative to time point 0). (D) UMSCC22B cells were transfected with control or SNX6 siRNA, and then treated with proteasome inhibitor MG132 (50 μM) or lysosome inhibitor chloroquine (80 μM) for 6 hours. The PD-L1 and SNX6 protein levels were then detected by Western Blot. (E) SPOP siRNA or Cullin3 siRNA was transfected alone or co-transfected with SNX6 siRNA into UMSCC22B cells as indicated. 72 hours after siRNAs transfection, the PD-L1, SNX6, SPOP, and Cullin3 expression levels were detected by Western Blot. Error bars indicate mean ± SEM from three independent experiments. *, P < 0.05.
PD-L1 degradation can be mediated by a proteasome-dependent pathway or a lysosome-dependent pathway [17, 32, 33]. To determine whether the loss of PD-L1 protein in SNX6-knockdown cells is dependent on the proteasome or lysosome, specific inhibitors were used. As shown in Figure 2D, treatment of the proteasome inhibitor MG132 can rescue the PD-L1 protein levels in SNX6-knockdown cells. In contrast, treatment of the lysosome inhibitor, chloroquine, could not rescue PD-L1 protein levels in SNX6-knockdown cells (Fig. 2D). These results suggest that SNX6 may exert it function in modulating PD-L1 protein expression through the proteasome-dependent degradation pathway.
Proteasome-dependent degradation of PD-L1 requires SPOP and Cullin3, which mediate the ubiquitination of PD-L1 [16]. Consistently, knockdown of SPOP or Cullin3 dramatically enhanced PD-L1 levels (Fig. 2E). Loss of either SPOP or Cullin3 could rescue PD-L1 expression in SNX6-knockdown cells, which suggests that the function of SNX6 in regulating PD-L1 may be dependent on SPOP/Cullin3-mediated PD-L1 ubiquitination (Fig. 2E).
SNX6 interacts with Cullin3.
To explore the possibility that SNX6 may modulate Cullin3 function, the ability of SNX6 to interact with Cullin3 was tested. Cullin3 was immunoprecipitated (IP) from cell lysates and examined by Western blot (IB) to detect association with SNX6. As shown in Figure 3A and 3B, SNX6 was detected with the Cullin3 complex, indicating that SNX6 can bind to Cullin3. To determine which region on SNX6 is responsible for interacting with Cullin3, a series of SNX6 mutants expressing different lengths of SNX6 were constructed. As shown in Figure 3A and 3B, SNX6_225 (truncation mutant expressing 225 amino acids of the N-terminus) and SNX6_215 (truncation mutant expressing 215 amino acids of the N-terminus) still can bind to Cullin3, whereas SNX6_205 (truncation mutant expressing 205 amino acids of the N-terminus) lost the ability to interact with Cullin3. This suggest the amino acids 206-215 of SNX6 may be required for SNX6 to bind Cullin3. The sequence of SNX6 amino acids 206-215 and its alignment with SNX1, SNX2, and SNX5 are shown in Figure 3C. SNX5, another Sorting Nexin family member sharing about 70% similarity with SNX6 [20, 34], cannot be co-immunoprecipitated with Cullin3 (Fig. 3D and 3E). This result indicates the specificity of SNX6 to interact with Cullin3. Furthermore, recombinant SNX6 can bind with Cullin3 in the in vitro binding assay (Fig. 3F), which suggests that the SNX6-Cullin3 binding is direct.
Figure 3. SNX6 interacts with Cullin3.
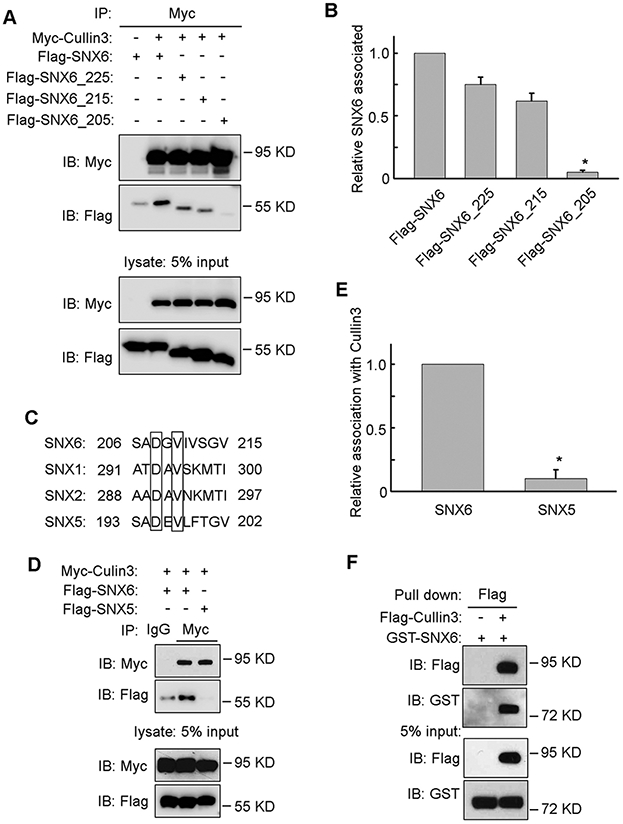
(A) Myc-Cullin3 was co-transfected with Flag-SNX6, Flag-SNX6_225 (truncation mutant expressing the N-terminus 225 amino acids), Flag-SNX6_215 (truncation mutant expressing the N-terminus 215 amino acids), or Flag-SNX6_205 (truncation mutant expressing the N-terminus 205 amino acids) separately into HEK-293T cells, subjected to immunoprecipitation with anti-Myc antibody and then immunoblotted with antibodies as indicated. (B) The amount of Cullin3 co-immunoprecipitated with wild type SNX6 or SNX6 truncation mutants was quantified. (C) Alignment of SNX6 amino acids 206-215 with SNX1, SNX2, and SNX5. (D) Myc-Cullin3 was co-transfected with Flag-SNX6 or Flag-SNX5 separately into HEK-293T cells, subjected to immunoprecipitation with anti-Myc antibody and then immunoblotted with antibodies as indicated. (E) The amount of Cullin3 co-immunoprecipitated with SNX6 or SNX5 was quantified. (F) 1 μg recombinant GST-SNX6 was mixed with or without 1 μg recombinant Flag-Cullin3 and then subjected to pulldown assay with anti-Flag Magnetic beads. Error bars indicate mean ± SEM from three independent experiments. *, P<0.05.
SNX6 modulates PD-L1 ubiquitination.
As SNX6 interacts with Cullin3 and regulates PD-L1 expression, the effects of SNX6 on PD-L1 ubiquitination was detected by an in vivo ubiquitination assay. As shown in Figure 4A and 4B, the expression of SNX6 significantly decreased PD-L1 ubiquitination levels. This result suggests that SNX6 can modulate PD-L1 expression by regulating PD-L1 ubiquitination.
Figure 4. SNX6 regulates PD-L1 ubiquitination.
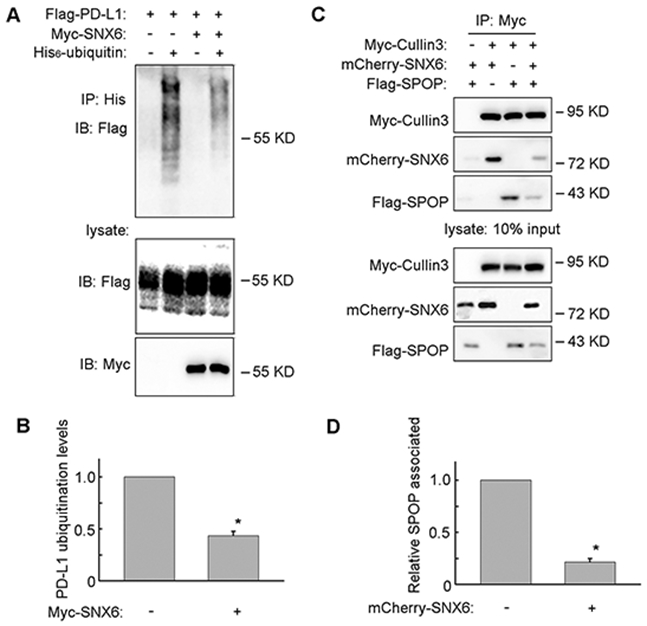
(A) Flag-PD-L1 was co-transfected with His6-Ubiquitin, with or without Myc-SNX6 into HEK-293T cells as indicated, and the Flag-PD-L1 ubiquitination was detected by an in vivo ubiquitination assay. (B) Quantification of PD-L1 ubiquitination levels. (C) Myc-Cullin3 was co-transfected with mCherry-SNX6 and/or Flag-SPOP into HEK-293T cells, subjected to immunoprecipitation with anti-Myc antibody and then immunoblotted with antibodies as indicated. (D) The amount of Cullin3 co-immunoprecipitated with SPOP was quantified. Error bars indicate mean ± SEM from three independent experiments. *, P<0.05.
SPOP functions as a substrate adaptor for Cullin3 and recruits PD-L1 for ubiquitination by Cullin3 [16]. Co-expression of SNX6 with SPOP significantly decreased SPOP binding with Cullin3 (Fig. 4C and 4D). The expression of SPOP also decreased SNX6 interaction with Cullin3 (Fig. 4C). These results suggest that SNX6 and SPOP compete for Cullin3 binding.
SNX6 modulates tumor-T-cell response.
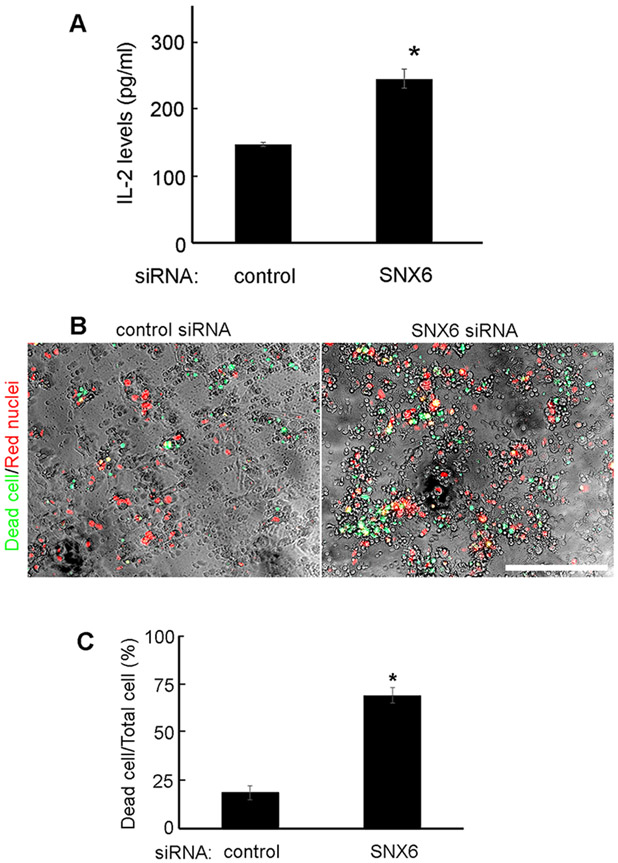
PD-L1 expressed in cancer cells can bind to PD-1 on T-cells to suppress T-cell activation. Since SNX6-knockdown decreases PD-L1 expression in cancer, the effects on T-cell suppression were further explored. Jurkat T-cells were first activated with Dynabeads Human T-Activator CD3/CD28, and then co-cultured with UMSCC1 cells transfected with control or SNX6 siRNA. The activation levels of Jurkat T-cells were evaluated by monitoring the secreted IL-2 levels [24]. As shown in Figure 5A, the Jurkat T-cells co-cultured with SNX6-knockdown UMSCC1 cells produced higher levels of IL-2 as compared with the Jurkat T-cells co-cultured with control UMSCC1 cells. This is consistent with the finding that loss of SNX6 decreased PD-L1 expression in UMSCC1 cells (Fig. 1). To further confirm the effect of SNX6 on tumor-T-cell response, a T-cell-mediated tumor cell-killing assay was performed. Control or SNX6-knockdown MDA-MB-231 cells were co-cultured with activated T-cells. As shown in Figure 5B and 5C, the loss of SNX6 enhanced T-cell-mediated killing of MDA-MB-231 cells. These results suggest that SNX6 can modulate tumor-T-cell response by controlling tumor cell PD-L1 levels.
Figure 5. SNX6 modulates tumor-T-cell response.
(A) UMSCC1 cells was transfected with control or SNX6 siRNA, and then co-cultured with activated Jurkat T-cells. The production of IL-2 by Jurkat T-cells was detected by IL-2 ELISA Kits. (B) Nuclear-restricted RFP-expressing MDA-MB-231 cells were transfected with control or SNX6 siRNA and then co-cultured with activated T-cells in the presence of caspase 3/7 substrate (Essen Bioscience). RFP and green fluorescent (NucView 488 Caspase 3/7 substrate) signals were then monitored. The green-fluorescent cells were counted as dead cells. (C) Quantification of dead cells in control or SNX6-knockdown MDA-MB-231 cells co-cultured with activated T-cells.
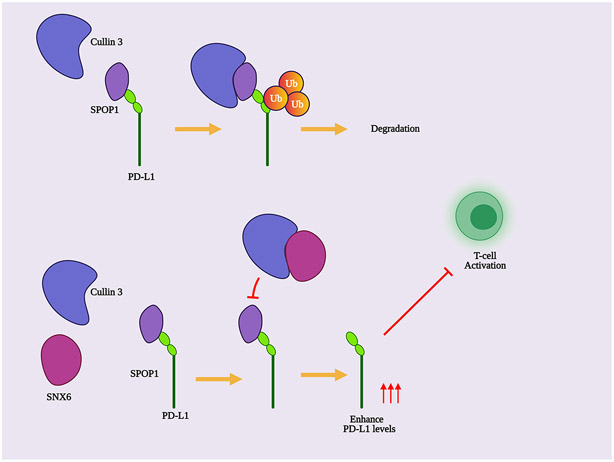
Our current research suggests a model illustrating the novel role of SNX6 in modulating PD-L1 expression and tumor-immune response (Fig. 6). By interacting with Cullin3, SNX6 blocks SPOP binding to Cullin3, which impairs Cullin3-mediated PD-L1 ubiquitination. This event protects PD-L1 from being degraded by proteasomes and increases PD-L1 expression levels. The loss of SNX6 enhances SPOP-Cullin3 interaction and promotes PD-L1 ubiquitination, which leads to PD-L1 degradation.
Figure 6. Model for SNX6 regulation of PD-L1 expression.
SPOP functions as an adaptor to link the E3 ubiquitin ligase Cullin3 with the substrate PD-L1, which is required for PD-L1 ubiquitination and degradation. By interacting with Cullin3, SNX6 competes with SPOP for binding with Cullin3. Thus, SNX6 decreases SPOP interaction with Cullin3, which blocks PD-L1 ubiquitination and protects PD-L1 from being degraded. The change of PD-L1 levels on tumor cells regulates tumor-T-cell response.
Discussion
Immune checkpoints such as PD-1/PD-L1 negatively modulate immune response to maintain self-tolerance and prevent autoimmunity [35]. Cancer cells can hijack immune checkpoint regulatory circuits to escape from the attack from host immune systems [36, 37]. Blocking the PD-1-PD-L1 interaction or decreasing PD-L1 expression in cancer cells releases the suppression of T-cell activation and enhances T-cell-mediated cancer cell killing [2, 38, 39]. Therefore, the identification of novel molecular mechanisms modulating PD-L1 expression in cancer cells may lead to discovery of new targets for cancer treatment.
Previous research works of SNX6 are mostly focused on its function in membrane trafficking. SNX6 functions as a critical component of the retromer complex to modulate the retrograde trafficking between endosomes and Trans-Golgi Networks (TGN) by forming dimers with other Sorting Nexin proteins, such as SNX1, SNX2, and SNX5 [21]. SNX6 interacts with the p150glued component of dynactin, which is required for the formation and movement of tubular retrograde intermediates between endosomes and TGN [21, 40]. Knockdown of the retromer components SNX1, SNX2, or SNX5 cannot affect PD-L1 expression (Fig. 1A and 1B), which suggests the role of SNX6 in modulating PD-L1 is not dependent on the retromer function. Our data indicate that knockdown of SNX1 or SNX2 can lead to loss of SNX5 expression (Fig. 1A). This effect was also reported by the publication of other colleagues [41]. SNX1, SNX2, SNX5, and SNX6 can form a network of interactions [21]. It is possible that the interaction of SNX5 with SNX1 or SNX2 can increase the stability of SNX5 protein. Therefore, loss of SNX1 or SNX2 leads to the decrease of SNX5 levels.
The function of SNX6 in cancer progression has been reported. In pancreatic cancer, SNX6 expression modulates TGF-β-induced epithelial to mesenchymal transition [42]. SNX6 interacts with tumor suppressor p27 (Kip1) to mediate the lysosomal sorting and degradation of p27 [43]. Breast cancer metastasis suppressor 1 (BRMS1) significantly reduce breast cancer and melanoma metastasis by modulating genes expression and phosphoinositide signaling [44-47]. SNX6 can interact with BRMS1 and promote BRMS1-dependent transcriptional repression [48]. Our current work reveals a novel function of SNX6 in cancer. By modulating PD-L1 ubiquitination and degradation, SNX6 controls the cancer immune response. Therefore, SNX6 has the potential to be a novel target to improve current immunotherapy to treat cancer.
Although it has been reported that both proteasomes and lysosomes can mediate PD-L1 degradation, we only found that the treatment of a proteasome inhibitor could rescue PD-L1 protein levels (Fig. 2B). Blocking the lysosome function did not change PD-L1 levels in our cell line system (Fig. 2B). This may suggest that the PD-L1 degradation pathways are cell context specific and should be explored further.
Cullin3 is an E3 ubiquitin ligase that plays important roles in mammalian cell differentiation, response to cellular stress, and cell cycle regulation [49, 50]. Cullin3 interacts with a wide range of BTB-proteins, which function as adaptors to recruit specific substrates for ubiquitination. SPOP serves as a substrate adaptor for Cullin3 and has tumor-suppressive functions. A number of SPOP substrates have been identified to control cancer progression, including PD-L1, androgen receptor, steroid receptor coactivator (SRC)-3, DEK, ERG, and SENP7 [51]. SNX6 can compete with SPOP for cullin3 binding, which decreases Cullin3-mediated PD-L1 ubiquitination and degradation (Fig. 4). By blocking SPOP-Cullin3 interaction, SNX6 may also modulate other SPOP-mediated substrates ubiquitination and then plays other biological functions. Our results suggest that the amino acids 206-215 of SNX6 may be required for SNX6 to bind Cullin3 (Fig. 3). Although SNX5 shares 60% similarity in this region with SNX6, SNX5 cannot bind with Cullin3 (Fig. 3).
Our study identified a novel function of SNX6. The ability of SNX6 to regulate PD-L1 protein expression levels through competitively binding to cullin3 and thus inhibiting PD-L1 ubiquitination is a new insight into PD-L1 regulation.
Acknowledgments:
This study was supported by grant R01DE029496 from the National Institute of Dental & Craniofacial Research of the National Institutes of Health and the Research Scholar Grant RSG-17-064-01-TBE from the American Cancer Society. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work is also supported by Virginia Commonwealth University Presidential Research Quest Fund. This study is also supported by Key Medical Specialty Construction project of shanghai (Grant No. ZK2019C06).
Abbreviations
- PD-L1
Programmed death-ligand 1
- PD-1
Programed cell death 1
- SNX6
Sorting Nexin 6
- SPOP
Speckle-type POZ protein
- HIP1R
Huntingtin-interacting protein 1-related protein
- TGN
Trans Golgi Network
- CHX
Cycloheximide
- BRMS1
Breast cancer metastasis suppressor 1
- HNSCC
Head and neck squamous carcinoma cell
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Data accessibility: The data that support the findings of this study are available from the corresponding author [ysun4@vcu.edu] upon reasonable request.
References
- 1.Latchman YE, Liang SC, Wu Y, Chernova T, Sobel RA, Klemm M, Kuchroo VK, Freeman GJ & Sharpe AH (2004) PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells, Proc Natl Acad Sci U S A. 101, 10691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Q, Qiu X, Zhang Z, Zhang S, Zhang Y, Liang Y, Guo J, Peng H, Chen M, Fu YX & Tang H (2020) PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade, Nat Commun. 11, 4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR & Honjo T (2000) Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation, J Exp Med. 192, 1027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurieva RI, Liu X & Dong C (2009) Yin-Yang of costimulation: crucial controls of immune tolerance and function, Immunol Rev. 229, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong H, Zhu G, Tamada K, Flies DB, van Deursen JM & Chen L (2004) B7-H1 determines accumulation and deletion of intrahepatic CD8(+) T lymphocytes, Immunity. 20, 327–36. [DOI] [PubMed] [Google Scholar]
- 6.Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S & Lou Y (2018) Next generation of immune checkpoint therapy in cancer: new developments and challenges, J Hematol Oncol. 11, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leemans CR, Snijders PJF & Brakenhoff RH (2018) The molecular landscape of head and neck cancer, Nat Rev Cancer. 18, 269–282. [DOI] [PubMed] [Google Scholar]
- 8.Chen L & Han X (2015) Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future, J Clin Invest. 125, 3384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C, Mezzadra R & Schumacher TN (2018) Regulation and Function of the PD-L1 Checkpoint, Immunity. 48, 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh C, Luong G & Sun Y (2021) A snapshot of the PD-1/PD-L1 pathway, J Cancer. 12, 2735–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L (2004) Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity, Nat Rev Immunol. 4, 336–47. [DOI] [PubMed] [Google Scholar]
- 12.Flies DB, Sandler BJ, Sznol M & Chen L (2011) Blockade of the B7-H1/PD-1 pathway for cancer immunotherapy, Yale J Biol Med. 84, 409–21. [PMC free article] [PubMed] [Google Scholar]
- 13.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL & Chen L (2012) Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape, Sci Transl Med. 4, 127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou W & Chen L (2008) Inhibitory B7-family molecules in the tumour microenvironment, Nat Rev Immunol. 8, 467–77. [DOI] [PubMed] [Google Scholar]
- 15.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O'Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL & Shipp MA (2010) Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma, Blood. 116, 3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P & Wei W (2018) Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance, Nature. 553, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Yao H, Li C, Shi H, Lan J, Li Z, Zhang Y, Liang L, Fang JY & Xu J (2019) HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity, Nat Chem Biol. 15, 42–50. [DOI] [PubMed] [Google Scholar]
- 18.Cullen PJ (2008) Endosomal sorting and signalling: an emerging role for sorting nexins, Nat Rev Mol Cell Biol. 9, 574–82. [DOI] [PubMed] [Google Scholar]
- 19.Seet LF & Hong W (2006) The Phox (PX) domain proteins and membrane traffic, Biochim Biophys Acta. 1761, 878–96. [DOI] [PubMed] [Google Scholar]
- 20.Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ & Cullen PJ (2007) A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer, J Cell Sci. 120, 45–54. [DOI] [PubMed] [Google Scholar]
- 21.Wassmer T, Attar N, Harterink M, van Weering JR, Traer CJ, Oakley J, Goud B, Stephens DJ, Verkade P, Korswagen HC & Cullen PJ (2009) The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network, Dev Cell. 17, 110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling K, Doughman RL, Iyer VV, Firestone AJ, Bairstow SF, Mosher DF, Schaller MD & Anderson RA (2003) Tyrosine phosphorylation of type Igamma phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch, J Cell Biol. 163, 1339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y & Chen J (2003) MDM2 promotes ubiquitination and degradation of MDMX, Molecular and cellular biology. 23, 5113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN & Hung MC (2016) Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity, Nat Commun. 7, 12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Hedman AC, Tan X, Schill NJ & Anderson RA (2013) Endosomal type Igamma PIP 5-kinase controls EGF receptor lysosomal sorting, Dev Cell. 25, 144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jitsukawa S, Kamekura R, Kawata K, Ito F, Sato A, Matsumiya H, Nagaya T, Yamashita K, Kubo T, Kikuchi T, Sato N, Hasegawa T, Kiyonari H, Mukumoto Y, Takano KI, Himi T & Ichimiya S (2017) Loss of sorting nexin 5 stabilizes internalized growth factor receptors to promote thyroid cancer progression, J Pathol. 243, 342–353. [DOI] [PubMed] [Google Scholar]
- 27.Simonetti B, Paul B, Chaudhari K, Weeratunga S, Steinberg F, Gorla M, Heesom KJ, Bashaw GJ, Collins BM & Cullen PJ (2019) Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins, Nat Cell Biol. 21, 1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mimura K, Teh JL, Okayama H, Shiraishi K, Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, Fazreen Z, Asuncion BR, Shabbir A, Yong WP, So J, Soong R & Kono K (2018) PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer, Cancer Sci. 109, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loke P & Allison JP (2003) PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells, Proc Natl Acad Sci U S A. 100, 5336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellucci R, Martin A, Bommarito D, Wang K, Hansen SH, Freeman GJ & Ritz J (2015) Interferon-gamma-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression, Oncoimmunology. 4, e1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B & Liu JO (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin, Nat Chem Biol. 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gou Q, Dong C, Xu H, Khan B, Jin J, Liu Q, Shi J & Hou Y (2020) PD-L1 degradation pathway and immunotherapy for cancer, Cell Death Dis. 11, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Sun Q, Mu N, Sun X, Wang Y, Fan S, Su L & Liu X (2020) The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells, Cell Commun Signal. 18, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seaman MN (2012) The retromer complex - endosomal protein recycling and beyond, J Cell Sci. 125, 4693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He X & Xu C (2020) Immune checkpoint signaling and cancer immunotherapy, Cell Res. 30, 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messerschmidt JL, Prendergast GC & Messerschmidt GL (2016) How Cancers Escape Immune Destruction and Mechanisms of Action for the New Significantly Active Immune Therapies: Helping Nonimmunologists Decipher Recent Advances, Oncologist. 21, 233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ & Sharpe AH (2017) PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity, J Exp Med. 214, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei SC, Duffy CR & Allison JP (2018) Fundamental Mechanisms of Immune Checkpoint Blockade Therapy, Cancer Discov. 8, 1069–1086. [DOI] [PubMed] [Google Scholar]
- 39.Akinleye A & Rasool Z (2019) Immune checkpoint inhibitors of PD-L1 as cancer therapeutics, J Hematol Oncol. 12, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong Z, Yang Y, Zhang C, Niu Y, Li K, Zhao X & Liu JJ (2009) The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport, Cell Res. 19, 1334–49. [DOI] [PubMed] [Google Scholar]
- 41.Simonetti B, Danson CM, Heesom KJ & Cullen PJ (2017) Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR, J Cell Biol. 216, 3695–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu P, Liang Y, Hu Q, Wang H, Cai Z, He J, Cai J, Liu M, Qin Y, Yu X, Jiang C, Zhang B & Wang W (2018) SNX6 predicts poor prognosis and contributes to the metastasis of pancreatic cancer cells via activating epithelial-mesenchymal transition, Acta Biochim Biophys Sin (Shanghai). 50, 1075–1084. [DOI] [PubMed] [Google Scholar]
- 43.Fuster JJ, Gonzalez JM, Edo MD, Viana R, Boya P, Cervera J, Verges M, Rivera J & Andres V (2010) Tumor suppressor p27(Kip1) undergoes endolysosomal degradation through its interaction with sorting nexin 6, FASEB J. 24, 2998–3009. [DOI] [PubMed] [Google Scholar]
- 44.Seraj MJ, Samant RS, Verderame MF & Welch DR (2000) Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13, Cancer Res. 60, 2764–9. [PubMed] [Google Scholar]
- 45.Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT & Welch DR (2002) Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1, Exp Cell Res. 273, 229–39. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Zhang B, Lin Y, Yang Y, Liu X & Lu F (2008) Breast cancer metastasis suppressor 1 inhibits SDF-1alpha-induced migration of non-small cell lung cancer by decreasing CXCR4 expression, Cancer Lett. 269, 46–56. [DOI] [PubMed] [Google Scholar]
- 47.DeWald DB, Torabinejad J, Samant RS, Johnston D, Erin N, Shope JC, Xie Y & Welch DR (2005) Metastasis suppression by breast cancer metastasis suppressor 1 involves reduction of phosphoinositide signaling in MDA-MB-435 breast carcinoma cells, Cancer Res. 65, 713–7. [PubMed] [Google Scholar]
- 48.Rivera J, Megias D & Bravo J (2010) Sorting nexin 6 interacts with breast cancer metastasis suppressor-1 and promotes transcriptional repression, J Cell Biochem. 111, 1464–72. [DOI] [PubMed] [Google Scholar]
- 49.Dubiel W, Dubiel D, Wolf DA & Naumann M (2018) Cullin 3-Based Ubiquitin Ligases as Master Regulators of Mammalian Cell Differentiation, Trends Biochem Sci. 43, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderica-Romero AC, Gonzalez-Herrera IG, Santamaria A & Pedraza-Chaverri J (2013) Cullin 3 as a novel target in diverse pathologies, Redox Biol. 1, 366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen HY & Chen RH (2016) Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications, Front Oncol. 6, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]