Abstract
Pre-existing conditions modulate sensitivity to numerous xenobiotic exposures such as air pollution. Specifically, individuals suffering from metabolic syndrome (MetS) demonstrate enhanced acute inflammatory responses following particulate matter inhalation. The mechanisms associated with these exacerbated inflammatory responses are unknown, impairing interventional strategies and our understanding of susceptible populations. We hypothesize MetS-associated lipid dysregulation influences mediators of inflammatory resolution signaling contributing to increased acute pulmonary toxicity. To evaluate this hypothesis, healthy and MetS mouse models were treated with either 18-hydroxy eicosapentaenoic acid (18-HEPE), 14-hydroxy docosahexaenoic acid (14-HDHA), 17-hydroxy docosahexaenoic acid (17-HDHA), or saline (control) via intraperitoneal injection prior to oropharyngeal aspiration of silver nanoparticles (AgNP). In mice receiving saline treatment, AgNP exposure resulted in an acute pulmonary inflammatory response that was exacerbated in MetS mice. A targeted lipid assessment demonstrated 18-HEPE, 14-HDHA, and 17-HDHA treatments altered lung levels of specialized pro-resolving lipid mediators (SPMs). 14-HDHA and 17-HDHA treatments more efficiently reduced the exacerbated acute inflammatory response in AgNP exposed MetS mice as compared to 18-HEPE. This included decreased neutrophilic influx, diminished induction of inflammatory cytokines/chemokines, and reduced alterations in SPMs. Examination of SPM receptors determined baseline reductions in MetS mice compared to healthy as well as decreases due to AgNP exposure. Overall, these results demonstrate AgNP exposure disrupts inflammatory resolution, specifically 14-HDHA and 17-HDHA derived SPMs, in MetS contributing to exacerbated acute inflammatory responses. Our findings identify a potential mechanism responsible for enhanced susceptibility in MetS that can be targeted for interventional therapeutic approaches.
Keywords: Nanoparticles, Inflammatory Resolution, Specialized Pro-Resolving Mediators, Metabolic Syndrome, Susceptibility, Omega-3 polyunsaturated fatty acids, Lipid Supplementation, Nanotoxicity
Introduction
Inhaled exposures can induce pulmonary inflammation, promoting the development of diseases such as asthma, fibrosis, chronic obstructive pulmonary disease (COPD), and others (Agarwal et al., 2020; Arias-Pérez et al., 2020; Klomp et al., 2021; Lotfi et al., 2020; Schett and Neurath, 2018; Utell and Samet, 1990). Inflammation is a coordinated process involving initiation and resolution allowing the immune system to respond to a variety of insults and return to homeostasis (Chen et al., 2018). Appropriate regulation of inflammation is necessary for the lung, due to constant interactions with the atmosphere and its sensitive alveolar structure. Inappropriate resolution can exacerbate and prolong the inflammatory processes and thus, may contribute to many exposure-related pulmonary diseases. Pulmonary inflammation is characterized by infiltration of immune cells (macrophages and neutrophils), goblet cell hyperplasia, and tissue damage resulting in increased epithelial permeability and edema (Leitch et al., 2008). Both initiation and resolution of inflammation are active processes regulated by lipid mediators (Serhan, 2010; Serhan and Petasis, 2011). Specifically, resolution is mediated via specialized pro-resolving mediators (SPMs) produced by metabolism of the ω−6 polyunsaturated fatty acid (PUFA) arachidonic acid to lipoxins and ω−3 PUFAs eicosatetraenoic acid (EPA) and docosahexaenoic acid (DHA) to resolvins, maresins, and protectins (Chiang and Serhan, 2020). SPMs promote resolution in a time-dependent manner and are highly potent functioning at the pico- to nanomolar level (Chiang and Serhan, 2020). Specifically, SPMs induce anti-inflammatory signaling, neutrophilic apoptosis, and macrophage clearance of apoptotic neutrophils, as well as initiate repair processes (Chiang and Serhan, 2020). Although not the focus of most toxicity assessments, exposures may impair the coordinated resolution of inflammation resulting in adverse effects. For instance, decreases in SPMs were determined following ozone inhalation contributing to pulmonary inflammation (Kilburg-Basnyat et al., 2018; Yaeger et al., 2021). Individual factors such as age, genetics, and underlying disease state may modulate resolution, resulting in increased susceptibility to inhaled exposures.
Pre-existing diseases can enhance susceptibility to environmental exposures, resulting in enhanced adverse effects (Sacks et al., 2011). Many diseases involve lipid dysregulation may impair the coordinated inflammation signaling following exposures. Metabolic syndrome (MetS) is a major health concern, affecting 34.2% of all U.S. adults, 50% of adults 60 years or older, and 9.8% of children (Moore et al., 2017). MetS consists of a combination of components including hypertension, hyperlipidemia, hyperglycemia, and increased waist circumference (Cornier et al., 2008). MetS predisposes individuals to serious chronic diseases such as Type 2 diabetes, cardiovascular disease, cancer, and others (Grundy et al., 2005). Evaluations of the National Health and Nutrition Examination Survey database in conjunction with the U.S. EPA’s Aerometric Information Retrieval system demonstrated an increase of circulating white blood cells, indicative of enhanced inflammation, due to particulate matter exposure in those with MetS compared to healthy individuals (Chen and Schwartz, 2008). Furthermore, individuals with MetS exposed to dust at the World Trade Center exhibited diminished lung function compared to healthy individuals (Naveed et al., 2012; Weiden et al., 2013). Together these epidemiological assessments establish that individuals suffering from MetS are increasingly sensitive to inhaled exposures. However, the mechanisms responsible for these exacerbated inflammatory effects following inhalation exposures in MetS remain unknown. To date, most toxicity assessments are performed in healthy models limiting our knowledge of distinct toxicity mechanisms in susceptible populations such as MetS. This deficiency impairs effective public health protections for this prominent susceptible subpopulation.
Nanosized particulate matter composes a significant portion of air pollution and is associated with the development of many diseases including cardiopulmonary and metabolic diseases (Brook et al., 2010; Nemmar et al., 2013; Ning et al., 2021; Zhang et al., 2021). Engineered nanoparticles (NPs) represent a technology that is increasingly incorporated into applications, products, and procedures, enhancing environmental, occupational, consumer, and biomedical exposures (Kessler, 2011). Following inhalation, NPs efficiently deposit within the respiratory tract where they can cause toxicity, including inflammation, apoptosis, oxidative stress, damage to the epithelial air interface, fibrotic changes, and impaired pulmonary function in healthy models (Sharifi et al., 2012). Recently, we demonstrated that a mouse model of MetS exhibited enhanced acute pulmonary inflammation following silver NP (AgNP) exposure via oropharyngeal aspiration compared to healthy mice (Alqahtani et al., 2020). This enhanced inflammation corresponded with decreases in pulmonary SPMs suggesting diminished inflammatory resolution may contribute to susceptibility observed in MetS. Specifically, our lipid screening approach determined AgNP exposure significantly reduced the pulmonary levels of EPA, DHA, 18-HEPE, 17-HDHA, 14-HDHA, maresin-1, and a number of resolvins in the MetS model. These lipid alterations and the enhanced inflammatory response were resolved by treatment of MetS mice with atorvastatin. Together, these results suggest lipid dysregulation in MetS impairs resolution of inflammation following NP exposure exacerbating pulmonary inflammation. Statins have a number of immune effects that cause anti-inflammatory effects beyond lipid regulation (Jain and Ridker, 2005; Ota et al., 2010). Further, the contribution of distinct SPMs and their dysregulation to MetS-associated exacerbated inflammation is unelucidated. This information is necessary to understand susceptibility and formulate targeted interventional strategies.
In our current study, we hypothesize MetS-associated dysregulation of specific SPM classes differentially contribute to increased acute pulmonary toxicity via impairment of inflammatory resolution. To evaluate this hypothesis, healthy and MetS mouse models were treated with distinct SPM precursors (18-hydroxy eicosapentaenoic acid (18-HEPE), 14-hydroxy docosahexaenoic acid (14-HDHA), 17-hydroxy docosahexaenoic acid (17-HDHA), or saline (control) via intraperitoneal injection) to evaluate their ability to modulate AgNP-induced acute pulmonary inflammation. The precursors selected are metabolized to produce distinct types and classes of SPMs. Specifically, 14-HDHA is a precursor to maresin-1 (Colas et al., 2016; Dalli et al., 2013), 17-HDHA is a precursor to protectin D1 and the resolvin D series (Hansen et al., 2019; Hong et al., 2003; Serhan et al., 2006, 2002), and 18-HEPE is a precursor to the resolvin E series (Duvall and Levy, 2016; Serhan and Levy, 2018). Following exposure, pulmonary SPM levels were assessed via a targeted mass spectrometry-based approach and alterations in inflammation and injury were assessed. Overall, this study determines the contributions of specific SPM classes that are dysregulated in MetS to enhanced inflammation and toxicity following inhalation exposures. This information can be directly applied to interventional approaches for the growing and susceptible MetS subpopulation.
Materials and Methods
AgNP Characterization.
Silver nanoparticles (AgNPs) coated with citrate and a diameter of 20 nm were purchased from NanoComposix (San Diego, CA, United States). AgNPs were characterized by assessment of hydrodynamic size, polydispersion, and ζ-potential at a concentration of 25 μg/mL in DI water (ZetaSizer Nano, Malvern) to verify manufacturer’s specifications (n = 3).
Animals Models, Diet-Induced Metabolic Syndrome, Lipid Treatment, and AgNP Exposure.
C57BL/6J male mice were obtained at 6 weeks of age from the Jackson Laboratory (Bar Harbor, ME, United States). Mice were randomly assigned to two main groups receiving either a healthy diet or high-fat western diet (HFWD) for 14 weeks. The healthy control diet had 10% of kcal coming from fat and contained 51.6 mg/kg cholesterol (D12450B, Research Diets Inc., New Brunswick, NJ, United States), while the HFWD had 60% of kcal from fat and contained 279.6 mg/kg cholesterol (D12492, Research Diets). Food was refreshed every other day and was provided with water ad libitum. This HFWD is well established to produce mouse models of MetS and has previously been utilized by our group and others for studies evaluating MetS (Alqahtani et al., 2020; Della Vedova et al., 2016; Gallou-Kabani et al., 2007; Kobos et al., 2020). This feeding plan resulted in two groups of mice: (1) Healthy and (2) MetS. After 14 weeks on either diet, subsets of mice were i.p. injected with 1μg of 18-HEPE, 14-HDHA, or 17-HDHA in 250 μL of sterile saline (Cayman Chemical, Ann Arbor, MI, United States) or vehicle (250 μL of sterile saline), 30 minutes prior to AgNP exposure. Specifically, following treatment, mice were anesthetized with isoflurane (Akorn, IL, USA) and exposed to either 50 μL of sterile water (VetOne, Boise ID, United States) as a control or 50 μL of AgNPs at a concentration of 1 mg/mL (50 μg of AgNPs in sterile water) via oropharyngeal aspiration. This experimental design resulted in sixteen final groups: (1) Healthy/Control, (2) Healthy/AgNPs, (3) MetS/Control, (4) MetS/AgNPs, (5) Healthy/14-HDHA/Control, (6) Healthy/14-HDHA/AgNPs, (7) MetS/14-HDHA/Control, (8) MetS/14-HDHA/AgNPs, (9) Healthy/17-HDHA/Control, (10) Healthy/17-HDHA/AgNPs, (11) MetS/17-HDHA/Control, (12) MetS/17-HDHA/AgNPs, (13) Healthy/18-HEPE/Control, (14) Healthy/18-HEPE/AgNPs, (15) MetS/18-HEPE/Control, and (16) MetS/18-HEPE/AgNPs. 24 h following AgNP exposure, mice were euthanized with ketamine/xylazine (90 mg/kg / 10 mg/kg), and immediately blood, bronchoalveolar lavage fluid (BALF), and organs were isolated for assessment of acute lung toxicity. A similar amount (1 μg) of SPM precursors in 250 μL of saline delivered by intraperitoneal injection has been utilized in other studies to evaluate effects of DHA- and EPA-derived SPMs in mouse models (Aoki et al., 2008; Francos-Quijorna et al., 2017; Hsiao et al., 2015; Kilburg-Basnyat et al., 2018; Kosaraju et al., 2017). Intraperitoneal injection of precursors can elevate lung levels of SPMs and reduce inflammation following an inflammatory stimulus (Kilburg-Basnyat et al., 2018; Liu et al., 2016; Nordgren et al., 2015). Specifically, intraperitoneal injection of a combination of SPM precursors was able to reduce ozone-induced pulmonary inflammation in a mouse model that corresponded with modification in lung SPM levels (Kilburg-Basnyat et al., 2018). The AgNP dosage of 50 μg/mouse was chosen based on previous studies demonstrating the induction of an acute inflammatory response that could be used to examine differences caused by MetS and/or lipid interventions (Alessandrini et al., 2017; Roda et al., 2019; Roursgaard et al., 2010). Exposure to 50 μg of AgNPs/mouse, in particular, stimulate pulmonary neutrophil recruitment and elevated mRNA expression of inflammatory markers in mouse models (Alessandrini et al., 2017; Alqahtani et al., 2020; Li et al., 2021; Roda et al., 2019). All animal procedures were conducted in accordance with the National Institutes of Health guidelines and approved by the Purdue University Animal Care and Use Committee.
Model Characterization.
Body weight was measured prior to necropsy to determine differences due to diet. During necropsy, blood was collected via cardiac puncture and serum was collected via centrifugation for quantification of circulating lipid levels. Specifically, triacylglycerides (Cayman Chemical, Ann Arbor, MI, United States), total cholesterol, high-density lipoprotein (HDL), and low-density lipoprotein (LDL) (Bioassay Systems, Hayward, CA, United States) were measured using commercially available kits via manufacturer’s protocols.
Collection of Bronchoalveolar Lavage Fluid (BALF).
BALF was collected from the right lung’s lobes immediately after euthanasia (Wang et al., 2011). Briefly, the left bronchus was tied off and the trachea was cannulated with a 20-gauge sterile syringe catheter. The right lung lobes were then gently washed four times with cold Phosphate-Buffered Saline (PBS) (17.5 mL/kg body weight). The first lavage was collected, centrifuged (1,800 rpm, 6 min, 4°C), and the protein-rich supernatant was stored for evaluation of total protein levels using the Pierce BCA Protein-Assay Kit (Thermo Scientific, Hercules, CA, United States), albumin using a Mouse Albumin ELISA Kit (ICL, Portland, OR, United States), and assessments of chemokine and cytokine levels via mouse-specific ELISAs (described below). BALF cell pellets from all four washes were resuspended, combined, and counted using a cellometer (Nexcelom, MA, United States) to determine total cell counts. An equal number of BALF cells were adhered to slides prior to staining with a three-step hematology statin (Fisher Scientific, Newington, NH, United States) using a Cytospin IV (Shandon Scientific Ltd., Cheshire, United Kingdom). Slides were viewed under a bright-field microscopy and differential cell counts were determined by examination of cellular morphology and assessment of ≥ 300 cells per slide blindly by two individuals. The average of the counts was used to produce percentages of specific cell types within each BALF sample. These assessments have been used in previous studies to examine acute lung inflammation and injury to the alveolar capillary barrier (Matute-Bello et al., 2011; Tighe et al., 2018).
Hyperspectral Imaging Assessment of AgNPs.
BALF cell slides were also viewed and examined by enhanced hyperspectral dark-field microscopy (Cytoviva, Auburn, AL, USA), similar to previous assessments (Anderson et al., 2015; Holland et al., 2015; Shannahan et al., 2015). 2 μL of AgNPs (1mg/mL) were pipetted directly onto premium clean microscope slides and a mean spectrum was created utilizing pixels with an intensity of >1,000 arbitrary units. This mean spectrum was then compared to AgNPs identified within BALF cells to determine alterations following internalization. AgNPs within cells were selected by focusing on the nucleus of the cell, and a hyperspectral image was collected at a magnification of 100x. At least 1,000 pixels of internalized AgNPs were collected to form a mean spectrum. All spectra were then normalized and compared to the original spectrum of the AgNP sample. This evaluation allowed for qualitative assessment of AgNP uptake by BALF cells as well as examination of spectral alterations due to internalization.
ELISA Assays to Evaluate BALF Cytokine Levels.
Chemokine and cytokine levels including interleukin-6 (IL-6), macrophage inflammatory protein-2 (MIP-2), monocyte chemoattractant protein-1 (MCP-1), and chemokine ligand 1 (CXCL1) were quantified from collected BALF using Mouse DuoSet ELISA kits (R&D Systems, Minneapolis, MN, United States), according to manufacturer’s instructions.
mRNA Expression Analysis.
Total RNA was extracted from the middle section of the left lung lobe using Trizol (Invitrogen, Carlsbad, CA, United States). Samples were moved to 2 mL vials with 1.4mm ceramic (zirconium oxide) beads (CK 14 soft tissue homogenizer Precellys, Bertin Technologies, Rockville, MD) and homogenized twice at a speed of 5 m/s for 30 s. Total RNA was extracted from the homogenate and purified using Direct-zolTM RNA MiniPrep Kits (Zymo Research, Irvine, CA, United States) as per the manufacturer’s instructions. RNA concentrations were quantified and quality assessed using a Nanodrop (Thermo Scientific, Hercules, CA, United States). An aliquot of 1 μg of RNA was reverse transcribed into cDNA using an iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States) as per the manufacturer’s instructions. Quantitative real time RT-PCR analysis was performed using inventoried primers (Qiagen, Hilden, Germany) to evaluate altered gene expression of interleukin-6 (IL-6), interleukin 1β (IL-1β), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein-2 (MIP-2), chemokine ligand 1 (CXCL1), arachidonate 5-lipoxygenase (ALOX-5), and arachidonate 15-lipoxygenase (ALOX15). In all cases, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control. Fold changes were calculated by comparing all sample values individually to the average of the control healthy mouse model (health mouse group not receiving a lipid treatment or exposed to AgNPs).
Lipid Extraction and Quantification of Specialized Pro-Resolving Mediators (SPMs). Mice were treated with either vehicle, 14-HDHA, 17-HDHA, or 18-HEPE prior to AgNP exposure. These lipid treatments represent precursors that are metabolized into distinct classes of specialized pro-resolving mediators (SPMs). Liquid chromatography tandem mass spectrometry (LC-MS/MS) was used to detect and quantify both SPM precursors and specific SPMs within the unlavaged left lung tissue (Table 1). Briefly, left lung samples (approximately 10 mg) were transferred to 2 mL vials containing 1.4 mm ceramic (zirconium oxide) beads (Precellys, CK14 kit, Bertin Corp. Rockville, MD, United States) with 1 mL of DDI water with 0.1% formic acid and 10 μL of internal standard mixture consistent of 25ng of PGE-d4, 2.5 ng of RvE-d4, 5ng of RvD-d5, 5ng of LtA, 5 ng of 9–10-diHOME-d4 and 15(s)-HETE-d8 from Cayman Chemical (Ann Arbor, MI, United States). Samples were homogenized three times for 30 seconds at 6,000 rpm using a Precellys24 tissue homogenizer (Bertin Technologies, Rockville, MD, United States). The total volume of the homogenate was extracted with methanol at 1:4 volume ratio (v/v). Samples were vortexed and centrifuged at 14,000 rpm for 10 min. Supernatant was transferred to a new vial, evaporated and stored at −80°C until analysis. Dried extracts were reconstituted with 100 μL of methanol/water at 1:1 volume ratio and 8μL were delivered to a Acquity UPLC BEH C18 1.7 μm particle size −2.1×100 (Waters, Milford, MA) column through an Agilent UPLC (G7120A) multisampler into a QQQ6470A triple quadrupole mass spectrometer (Agilent Technologies, San Jose, CA, United States) equipped with ESI Jet Stream ion source. The binary pump flow rate was set at 0.3 mL/min in an Agilent UPLC (G7120A) using water and 0.1% formic acid as mobile phase A and acetonitrile and 0.1% formic acid as mobile phase B. The LC column was pre-equilibrated with 20% of mobile phase B for 1 min. The binary pump was set in a linear gradient to 100% of mobile phase B for 27 min. It was then returned to 20% mobile phase B and re-equilibrated for 3 min. The mass spectrometer was operated in the negative ion mode, with capillary voltage was 3500, source gas temperature of 325°C, gas flow set at 7 l/min, nebulizer pressure 45 PSI, sheath gas heater at 250°C, and the sheath gas flow at 7 l/min. The source fragmentation was set to 100 (arbitrary units), dwell time was 30ms and the cell accelerator voltage was 4 V. To verify instrument performance, the system was flushed with pure methanol between each sample, quality control samples made of a mixture of lipids were injected, and chromatogram intensity and time were assessed. Blanks consisting of dilution solvent were injected and analyzed throughout. The raw data was processed with MassHunter Quantitative version B.10.00 (Agilent Technologies, Santa Clara, CA, United States). Quantification of SPMs was achieved defining the response ratio (RR) of the area under the curve of the endogenous lipid and their respective internal standard, then multiplying it by the concentration of the deuterated standard. For SPMs without a deuterated internal standard, calibration curves were prepared in water with 0.1% formic acid by 5 serial dilutions with ranging concentrations as follows: 250μg to 0.025 μg for AA, DHA and EPA; 500ng to 31.25 ng for 14HDHA; 500ng to 0.05 ng for RvD1; 100 ng to 0.01 ng for Mar1 and 17HDHA; 50ng to 0.005 ng for PD1. Concentrations in ng/mg of tissue were obtained via normalizing the concentration of the SPM by the weight of the sample.
Table 1.
Lipid Internal Standards and the amount Spiked into the Samples
| Internal Lipid Standards | Item No. | Spiked Amount |
|---|---|---|
| Resolvin D1-d5 | 11182 | 5 ng |
| Prostaglandin E2-d4 | 314010 | 25 ng |
| Leukotriene B4-d4 | 320110 | 5 ng |
| 15(S)-HETE-d8 | 334720 | 5 ng |
| Resolvin E1-d4 | 10009854 | 2.5 ng |
| (±)9(10)-DiHOME-d4 | 10009993 | 5 ng |
| PD1 | 10010390 | 50 to 0.005 ng |
| AA | 90010 | 250 to 0.025 μg |
| DHA | 90310 | 250 to 0.025 μg |
| EPA | 90110 | 250 to 0.025 μg |
| 14HDHA | 15253 | 500 to 31.25 ng |
| RvD1 | 10012554 | 500 to 0.05 ng |
| Mar1 | 10878 | 100 to 0.01 ng |
| 17HDHA | 10009799 | 100 to 0.01 ng |
Resolution Receptor Protein Expression.
Samples were isolated from collected left lung tissue and homogenized using three runs of a Bead Mill 4 homogenizer (Fisher Scientific, Newington, NH, United States) at a speed of 5 m/s (500 Watts) for 30 s with radioimmunoprecipitation assay buffer (RIPA) buffer containing 20mM Tris-HCL pH7. 4, 150mM NaCl, 1mM EDTA, 1% Triton-X100, 1% sodium deoxycholate, 0.1% SDS, and 0.2% protease inhibitor cocktail (Sigma). Using 10% SDS polyacrylamide gels, 10μl of precision plus protein ladder (catalog no 1610374, Bio-Rad, CA, USA), and 15 μg of protein were separated by electrophoresis and subsequently transferred to polyvinylidene difluoride (PVDF) membrane. After transfer, the PVDF membrane was washed and then blocked with 5% BSA in TBST for 1 h at room temperature. Afterward, the membrane was incubated overnight at 4°C with commercially available primary antibodies: G protein-coupled receptor 18 (GPR18) Antibody (catalog no. NBP2–24918) (Novus Biologicals, CO, USA); Anti- Chemokine like receptor 1 (ChemR23) (catalog no. sc-398769) or Anti- Leucine rich repeat containing G protein coupled receptor 6 (LGR6) Antibody (catalog no. sc-393010) (Santa Cruz Biotechnology, TX, US). Membranes were then washed with TBST and incubated with the secondary HRP-linked antibody (Catalog no 31430, Invitrogen, Carlsbad, CA, United States) for 1h at room temperature. After incubation, the membrane was washed and an enhanced chemiluminescence substrate (catalog no 1705060S, Bio-Rad, CA, USA) was used to facilitate the detection of protein bands. Blots images were captured under the ChemiDoc XRS+ System. Image Lab Software was used to analyze the protein bands, and were normalized to β-actin Loading Control Monoclonal Antibody (BA3R) (Catalog no MA5–15739, Invitrogen, Carlsbad, CA, United States). Quantification of the Western blots was carried out with Image Lab Software Version 6.1 (Bio-Rad Laboratories Inc., Hercules, California, USA).
Statistical Analyses.
Results are expressed as mean values ± S.E.M. with 6–8 animals/group. All samples were assessed in duplicate and values averaged for endpoints including serum parameters, BALF cell counts, mRNA expression, and BALF chemokines and cytokines. For statistical analysis, a three-way analysis of variance (ANOVA) was used to determine statistical differences between groups, with disease (healthy or MetS), treatment (lipid supplementation or no lipid supplementation), and exposure (control or AgNPs exposure), as the three factors. For western blots, all samples were run in duplicate and intensity band values averaged for each sample. Intensity was normalized to β-actin, and a two-way (ANOVA) was used to determine statistical differences between groups using disease (healthy or MetS) and exposure (control or AgNPs exposure) as the two factors. A Bonferroni test was utilized for multi-comparison analysis. All statistical examinations were performed using GraphPad Prism 8 software (Graph Pad, San Diego, CA, United States), and p < 0.05 was considered to be statistically significant.
Results
Silver Nanoparticle Characterization.
Citrate-coated silver nanoparticles (AgNPs) were characterized to verify manufacturer specifications. The AgNPs were determined to have a hydrodynamic size of 29.7 nm ± 1.1, polydispersion index of 0.23 ± 0.04, and a ζ-potential of −35.1 mV ± 2.6 (ZetaSizer Nano, Malvern) in DI water at a concentration of 25 μg/mL. These parameters are all reported as mean ± standard deviation (n = 3).
Mouse Model Characterization.
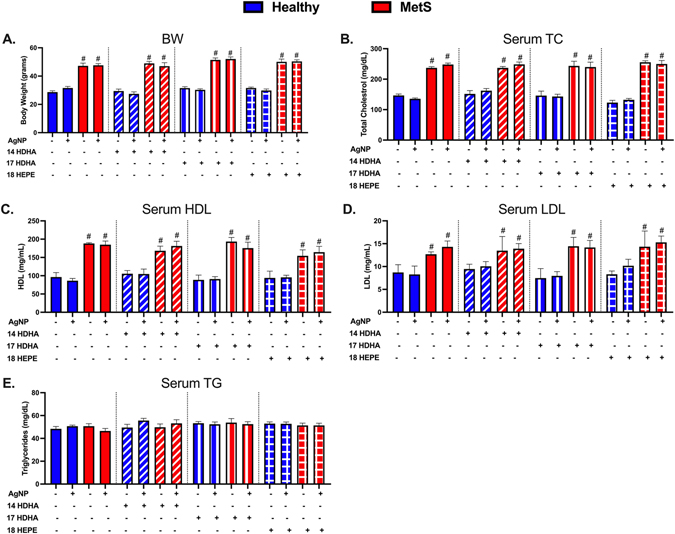
After 14 weeks on either the healthy or HFWD diet, markers of metabolic syndrome including body weight (BW), serum total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), and triacylglycerides were measured to characterize models. HFWD increased body weight in MetS mice compared to those placed on the healthy diet (Fig. 1A). Serum TC, HDL, and LDL were elevated in mice fed the HFWD compared to mice on the healthy diet (Fig. 1B–D). Serum triacylglycerides did not differ between mice receiving the healthy or HFWD after 14 weeks (Fig. 1E). Treatment with SPM precursors (14-HDHA, 17-HDHA, 18-HEPE) and AgNP exposure did not alter serum levels of TC, HDL, LDL, and triacylglycerides in either healthy or MetS mouse models (Fig. 1).
Fig. 1.
Characterization of body weight and serum lipid levels in healthy and MetS mouse models following 14 weeks on either a healthy or high-fat western diet (HFWD). Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1 μg of 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Healthy and MetS mice without or with lipid interventions were characterized by examination of (A) body weight (BW), (B) serum total cholesterol (TC), (C) serum high-density lipoprotein (HDL), (D) serum low-density lipoprotein (LDL), and (E) serum triacylglycerides (TG) levels. Values are expressed as mean ± SEM (n = 6–8/group). # denotes significant differences between healthy and MetS models receiving similar treatment and exposure, (p < 0.05).
BALF Markers of Acute Pulmonary Injury and Inflammation.
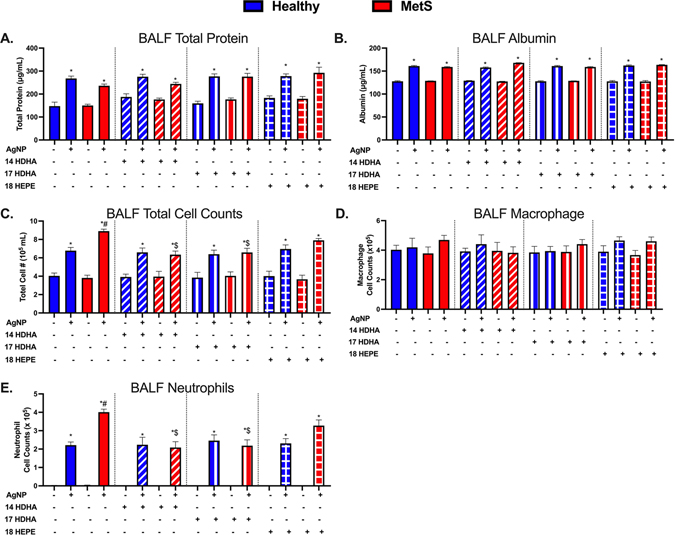
Twenty four hours following AgNP exposure, BALF parameters were evaluated to determine lung injury and the acute inflammatory response. Specifically, we examined BALF total protein and albumin levels as well as alterations in cytometry. Increases in BALF total protein concentration and albumin (a high molecular weight protein) are markers of pulmonary edema and impaired alveolar capillary barrier integrity. Alterations in cellular content of BALF such as influx of immune cells are examined to determine ongoing pulmonary inflammation. Total protein levels, albumin levels, total cell counts, macrophages counts, and neutrophils counts within the BALF where not altered at baseline between healthy and MetS controls (Fig. 2). AgNP exposure elevated BALF levels of total protein and albumin in both mouse models (Fig. 2A & B). AgNP exposure increased total cell counts in both mouse models, however, MetS mice demonstrated elevations compared to healthy (Fig. 2C). BALF macrophage counts were not altered 1 day after AgNP exposure compared to each model’s controls (Fig. 2D). AgNP exposure resulted in an influx of neutrophils into the BALF in both mouse models that was exacerbated in MetS compared to healthy (Fig. 2E).
Fig. 2.
Acute effect of AgNP exposure on BALF (A) total protein, (B) albumin, (C) total cells, (D) macrophages and (E) neutrophils from healthy and MetS mice and modulation by lipid interventions. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1 μg of 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Values are expressed as mean ± SEM (n = 6–8/group). * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) alone did not alter parameters measured in BALF samples (total protein and albumin levels, total cell counts, macrophages, and neutrophils) in healthy and MetS mice not exposed to AgNPs as compared to mice not receiving treatments (Fig. 2). 14-HDHA, 17-HDHA, and 18-HEPE treatments did not alter BALF parameters in healthy mice exposed to AgNPs in comparison to mice not receiving treatment and exposed to AgNPs (Fig. 2). Additionally, treatments were not found to affect BALF total protein or albumin levels in MetS mice following AgNP exposure (Fig. 2A & B). 14-HDHA treatment decreased total cells and neutrophil influx in AgNP exposed MetS compared to exposed MetS not receiving 14-HDHA treatment (Fig. 2C & E). 17-HDHA treatment was also determined to decrease total cells and neutrophil influx in AgNP exposed MetS compared to exposed MetS that were not treated with 17-HDHA (Fig. 2C & E). 18-HEPE lipid treatment, however, was not determined to significantly alter the total cell counts nor the neutrophilic influx due to AgNP exposure (Fig. 2C & E).
Hyperspectral Analysis of Internalized AgNPs.
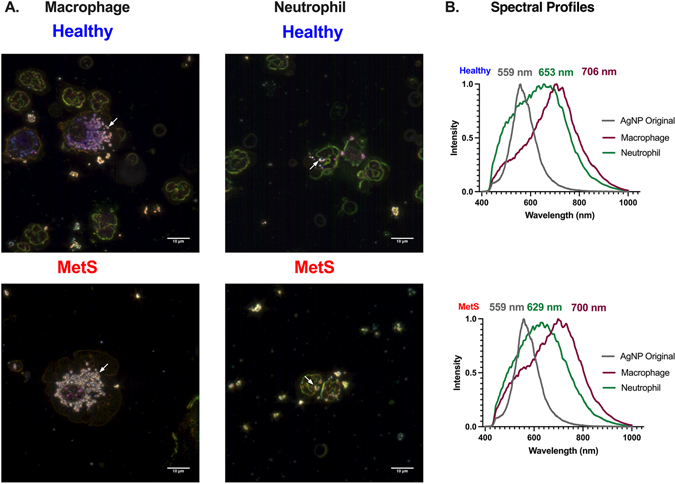
Similar to previous publications, AgNPs were identified and characterized in collected BALF immune cells via the use of hyperspectral darkfield microscopy (Anderson et al., 2015; Holland et al., 2015; Shannahan et al., 2015). AgNPs were placed onto a slide and a mean spectral profile was generated to compare to AgNPs associated with collected macrophages and neutrophils from the BALF of exposed mice. AgNPs were observed in macrophages and neutrophils in both exposed mouse models (Fig. 3A). Spectral profiles generated from internalized AgNPs demonstrated cell-specific shifts compared to the original AgNP spectrum (Fig. 3B). Specifically, internalization of AgNPs by macrophages and neutrophils in both healthy and MetS mice resulted in a red-shift in the light-scattering spectrum compared the original AgNP spectrum (Fig. 3B). AgNPs internalized by macrophages and neutrophils were demonstrated to generate distinct spectral profiles (Fig. 3B). However, no differences in AgNP spectral profiles were determined based on disease status (healthy vs MetS). Treatment with lipid precursors were not determined to alter the spectral profiles of AgNPs internalized by healthy or MetS macrophages or neutrophils (Supplemental Figs. 1–4). Spectral profiles generated are a mean of over 1,000 pixels of internalized AgNPs. These profiles were then mapped against representative images identifying only AgNPs in samples, confirming the accuracy of the spectral assessments (Supplemental Figs. 1–4).
Fig. 3.
Hyperspectral analysis of AgNPs within macrophages and neutrophils collected from BALF exposed healthy and MetS mouse models. (A) Representative enhanced darkfield images of macrophages and neutrophils 24 h post-exposure to AgNPs at a concentration of 50 μg. White arrows indicate the accumulation of AgNPs within BALF cells. White bar identifies 10 μm scaling. (B) Differences in mean spectra of AgNPs following association with BALF cells. At least 1,000 pixels of AgNPs were collected to form mean spectra. All spectra were normalized based on intensity for comparisons. Gray curve is the spectral profile of the original AgNP sample, the green curve represents AgNPs internalized by neutrophils, and the purple curve represents AgNPs internalized by macrophages. Numbers correspond to peak wavelengths. The top set of curves represent spectra of AgNPs internalized by BALF cells in the healthy model while the bottom set of curves represent spectra of AgNPs internalized by BALF cells in the MetS model. Supplemental Figure 1 includes representative image of original AgNPs as well as mapping results confirming curves represent internalized AgNPs. Representative images of internalized cells and spectral profiles of internalized AgNPs from groups receiving lipid treatments can be found in Supplemental Figure 2 (14-HDHA), Supplemental Figure 3 (17-HDHA), and Supplemental Figure 4 (18-HEPE).
Pulmonary Gene Expression Analysis of Acute Inflammatory Markers.
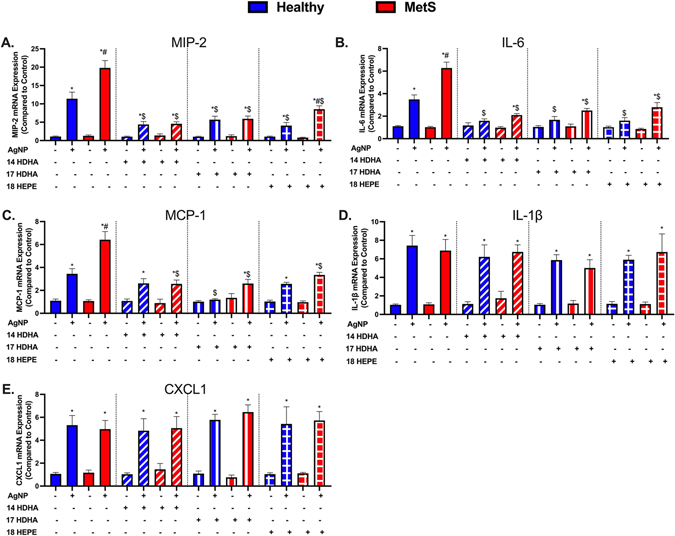
Inflammatory gene expression levels within lung tissue samples were evaluated to determine lipid treatment modifications in the AgNP-induced acute inflammatory response. Gene expression of macrophage inflammatory protein (MIP-2), interleukin-6 (IL-6), monocyte chemoattractant protein (MCP-1), interleukin 1β (IL-1β), and CXCL1 were not altered between healthy and MetS models at baseline. AgNP exposure elevated gene expression of macrophage inflammatory protein (MIP-2), interleukin-6 (IL-6), monocyte chemoattractant protein (MCP-1) in both models which was exacerbated in MetS compared to healthy (Fig. 4A–C). IL-1β and CXCL1 levels were elevated equivalently in both models following AgNP exposure (Fig. 4D & E). Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) alone did not alter the pulmonary expression of MIP-2, IL-6, MCP-1, IL-1β, and CxCL1 compared to models not receiving treatment (controls not exposed to AgNPs). Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) inhibited AgNP-induced elevations in MIP-2, IL-6, MCP-1 in both models compared to exposed mice not receiving a treatment (Fig. 4A–C). MIP-2 levels were still exacerbated in AgNP exposed MetS receiving 18-HEPE treatment compared to AgNP exposed healthy. Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) did not alter the gene expression of IL-1β, and CxCL1 in either model exposed to AgNPs compared to mice not receiving the lipid treatment (Fig. 4 D & E).
Fig. 4.
AgNP-induced pulmonary inflammatory gene expression including (A) macrophage inflammatory protein-2 (MIP-2), (B) interleukin-6 (IL-6), (C) monocyte chemoattractant protein-1 (MCP-1), (D) interleukin-1β (IL-1β), and (E) chemokine 1 (CXCL1) were evaluated in healthy and MetS lung tissue. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg of 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Values are expressed as mean ± SEM (n = 6–8/group). * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
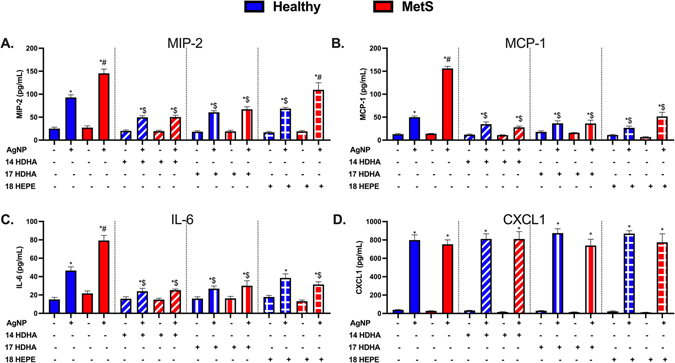
Acute Inflammatory Cytokines/Chemokines.
To further evaluate SPM precursor modifications of the AgNP-induced acute inflammatory response, cytokine/chemokine levels were measured within collected BALF samples utilizing ELISAs. Concentrations of macrophage inflammatory protein (MIP-2), interleukin-6 (IL-6), monocyte chemoattractant protein (MCP-1), and chemokine (CXCL1) were not determined to be different between BALF samples collected from healthy and MetS controls (Fig. 5). MIP-2, IL-6, and MCP-1 were elevated in both models following AgNP exposure and were exacerbated in exposed MetS mice compared to exposed healthy mice (Fig. 5A–C). CXCL1 levels were elevated similarly in AgNP exposed healthy and MetS mice (Fig. 5D). Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) did not alter BALF concentrations of MIP-2, MCP-1, IL-6, and CXCL1 at baseline in healthy or MetS mice. Treatment with either 14-HDHA or 17-HDHA decreased MIP-2, MCP-1, and IL-6 concentrations in both models exposed to AgNPs compared to groups not receiving SPM precursor treatment (Fig. 5A–C). 18-HEPE treatment reduced concentrations of MIP-2 and MCP-1 in AgNP exposed healthy mice but did not significantly affect IL-6 (Fig. 5 A–C). In AgNP exposed MetS mice, 18-HEPE treatment was not determined to reduce MIP-2 levels but did reduce MCP-1 and IL-6 (Fig. 5A–C). Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) did not alter elevations in BALF CXCL1 levels observed in either mouse model following AgNP exposure (Fig. 5D).
Fig. 5.
Alterations in BALF cytokine/chemokines proteins 24 h after AgNP exposure in healthy and MetS mice and the influence of lipid interventions. Proteins levels of (A) MIP-2, (B) MCP-1, (C) IL-6, and (D) CXCL1 were examined in BALF to determine disease-related differences in AgNP-induced inflammatory response and the influence of lipid treatments. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Values are expressed as mean ± SEM (n = 6–8/group). * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
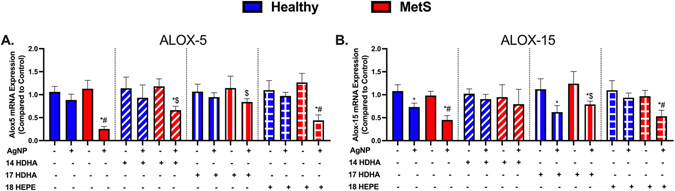
Pulmonary Gene Expression Analysis of Lipid Metabolism.
Gene expression levels of lipoxygenase arachidonate-5 lipoxygenase (ALOX-5), and arachidonate 15-lipoxygenase (ALOX-15) were measured within lung samples to evaluate modifications in lipid metabolism due to lipid treatments and AgNP exposure. ALOX-5 and ALOX-15 gene expression levels did not differ at baseline between healthy and MetS control mice (Fig. 6). Expression of ALOX-5 was decreased in only AgNP exposed MetS (Fig. 6A). However, ALOX-15 was decreased in both models due to AgNP exposure compared to model-matched controls (Fig. 6B). Treatment with individual SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) did not alter gene expression of ALOX-5 or ALOX-15 healthy and MetS controls. 14-HDHA and 17-HDHA lipid treatment inhibited AgNP exposure related reductions in ALOX-5 and ALOX-15 gene expression in MetS. 18-HEPE treatment did not modify alterations in ALOX-5 or ALOX-15 gene expression associated with AgNP exposure (Fig. 6).
Fig. 6.
Alterations in genes involved in lipid metabolism 24 h following AgNP exposure in lung tissue from healthy and MetS mice and the influence of lipid interventions. (A) arachidonate-5 lipoxygenase (ALOX-5), and (B) arachidonate 15-lipoxygenase (ALOX-15) were evaluated in lung tissue. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Values are expressed as mean ± SEM (n = 6–8/group) fold change compared with control. * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
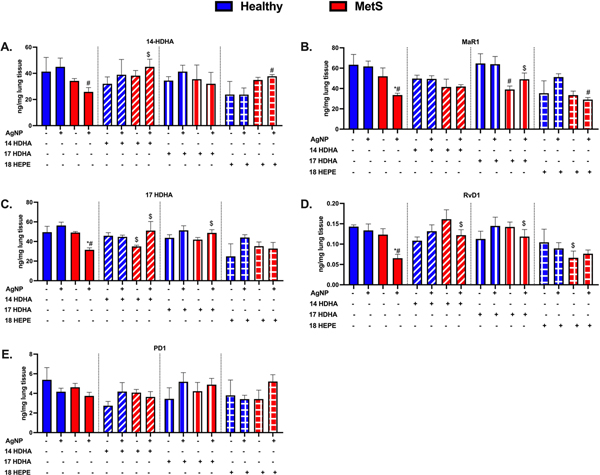
Assessment of Pulmonary SPM Levels.
A targeted LC/MS/MS approach was utilized to evaluate alterations in SPM precursors and SPMs within the lung due to AgNP exposure and treatments. Specifically, we measured arachidonic acid (AA), eicosatetraenoic acid (EPA) and docosahexaenoic acid (DHA) (Moore et al., 2017), their metabolites and the SPM precursors we treated with 14-HDHA, and 17-HDHA, as well as the SPMs MaR1 (14-HDHA derived), RvD1 (17-HDHA derived), and PD1 (17-HDHA derived). Data for AA, EPA, and DHA can be found in Supplemental Fig. 5. AA increased in MetS due to AgNP exposure while, treatment with either 14-HDHA or 17-HDHA inhibited these AgNP-induced increases in AA. 18-HEPE did not affect the increase in AA observed in MetS mice in response to AgNPs. (Supplemental Fig. 5). None of the SPM precursors or SPMs evaluated were determined to be changed at baseline between control healthy and MetS mice (Fig. 7). Pulmonary levels of 14-HDHA, MaR1, 17-HDHA, RvD1, and DHA were decreased following AgNP exposure in MetS mice while unchanged in AgNP exposed healthy mice compared to model-matched controls (Fig. 7 and Supplemental Fig. 5). 14-HDHA treatment did not alter 14-HDHA, MaR1, RvD1, AA, EPA, or DHA levels compared to control healthy and MetS mice not receiving treatments (Fig. 7 and Supplemental Fig. 5). 14-HDHA treatment increased 14-HDHA, 17-HDHA, RvD1, and DHA in AgNP exposed MetS compared to AgNP exposed MetS mice not receiving treatment (Fig. 7 and Supplemental Fig. 5). 17-HDHA treatment elevated the level of MaR1, 17-HDHA, RvD1, and DHA in AgNP exposed MetS mice compared to exposed MetS mice not receiving treatment. 18-HEPE treatment did not alter 14-HDHA, MaR1, 17-HDHA, RvD1, or PD1 levels in healthy mice compared to healthy mice not receiving treatments (Fig. 7). 18-HEPE treatment elevated 14-HDHA, decreased MaR1 levels in AgNP exposed MetS mice compared to AgNP exposed mice not receiving the treatments (Fig. 7).
Fig. 7.
Assessment of docosahexaenoic acid (DHA)-derived SPMs in the lung tissue of healthy and MetS mice 24 h following AgNP exposure and the influence of 14-HDHA intervention. A targeted LC/MS/MS approach determined quantitative differences in (A) 14-HDHA, (B) the 14-HDHA-derived SPM Maresin-1 (MaR1), (C) 17-HDHA, (D) the 17-HDHA-derived SPM resolvin D1 (RvD1), and (E) the 17-HDHA-derived SPM protectin-D1 (PD1). Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). Additional lipids (AA, DHA, EPA) evaluated in pulmonary tissue can be found in Supplemental Figure 6. * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
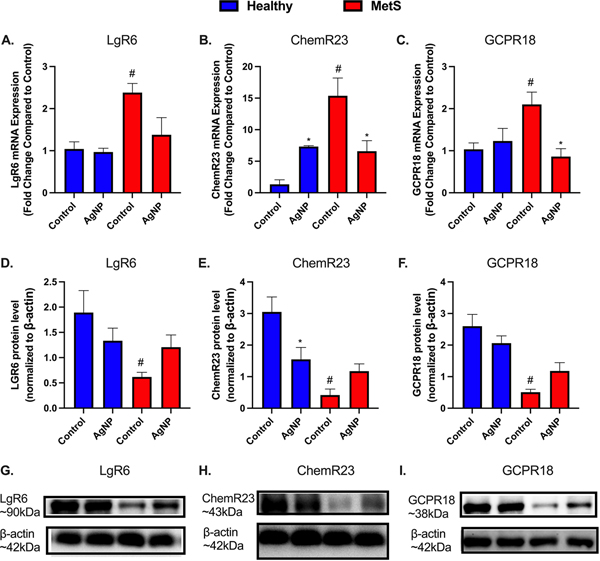
Lung Tissue SPM Receptors.
Gene expression and protein levels of SPM receptors (LgR6 a receptor for MaR1, ChemR23 a receptor for RvE1, and GCPR18 a receptor for RvD2) were evaluated within lung tissue to determine effects of disease, and AgNP exposure (Fig. 8). This assessment was performed to determine if there is reduced capacity of SPMs to signal via receptors due to MetS contributing to exacerbated inflammatory responses. LgR6, ChemR23, and GCPR18 gene expressions were elevated in control MetS mice compared to control healthy mice. LgR6 and GCPR18 gene expression were not altered in healthy mice due to AgNP exposure while ChemR23 was up-regulated (Fig. 8A–C). LgR6, ChemR23, and GCPR18 gene expression were decreased in AgNP exposed MetS mice compared to control MetS mice (Fig. 8A–C). LgR6, ChemR23, and GCPR18 protein levels were decreased in lungs of control MetS mice compared to control healthy mice (Fig. 8D–I). LgR6 and GCPR18 protein levels were not altered in AgNP exposed healthy mice compared to control healthy mice while ChemR23 protein levels were decreased (Fig. 8D–I). AgNP exposure did not significantly alter LgR6, ChemR23, or GCPR18 levels in MetS mice compared to control MetS mice (Fig. 8D–I).
Fig. 8.
Alterations in pulmonary resolution receptors 24 h following AgNP exposure in healthy and MetS mouse models. AgNP-induced alterations in the gene expression of (A) leucine-rich repeat-containing G-protein coupled receptor 6 (Lgr6), (B) Chemokine like receptor 1 (ChemR23), and (C) G protein-coupled receptor 18 (GPCR18) were evaluated in lung tissue. Alterations in receptors associated with lipid mediator of inflammatory resolution were assessed by western blot in healthy and MetS. Receptor levels of (D) LgR6, a receptor for Mar1, (E) ChemR23, a receptor for RvE1, and (F) GCPR18, a receptor for RvD2 were examined in lung tissue to determine disease-related differences in AgNP-induced inflammatory response. (G-I) Representative western blot images of LgR6, ChemR23, and GCPR18. Values are expressed as mean ± SEM (n = 6–8/group). * denotes significant differences due to AgNP exposure, # denotes significant differences between healthy and MetS mouse models (p < 0.05).
Discussion
Individuals suffering from chronic diseases are increasingly susceptible to inhaled particulate exposures, however, the toxicological mechanisms responsible remain unelucidated (Chen and Schwartz, 2008; Devlin et al., 2014; McCormack et al., 2015; Wagner et al., 2014). Many prevalent diseases such as metabolic syndrome (MetS) are characterized by lipid dysregulation which may facilitate exacerbated exposure-induced inflammatory responses. Recently we demonstrated via a lipid screening approach, specialized pro-resolving lipid mediators (SPMs) responsible for resolution of inflammation were uniquely disrupted in the lungs of MetS mice following AgNP exposure compared to healthy (Alqahtani et al., 2020). In the current assessment we hypothesized supplementation with specific SPM precursors would differentially modulate these enhanced responses observed in MetS. To test this hypothesis, healthy and MetS mouse models were treated with distinct precursors of SPMs (14-HDHA, 17-HDHA, or 18-HEPE) 30 min prior to oropharyngeal aspiration of AgNPs and the acute inflammatory response was evaluated 24h after exposure. Consistent with our previous results, MetS mice demonstrated increased AgNP-induced pulmonary inflammation compared to the healthy model. Pretreatment with either 14-HDHA or 17-HDHA was determined to reduce pro-inflammatory signaling in both models while reducing AgNP-induced neutrophilia in MetS mice to levels similar to those observed in the AgNP exposed healthy mouse model. Treatment with either 14-HDHA or 17-HDHA was determined to reduce inflammatory markers following AgNP exposure more so than 18-HEPE. This suggests deficits in 14-HDHA and 17-HDHA derived SPMs may be primarily responsible for the exacerbated inflammation observed in metabolic syndrome following AgNP exposure. Targeted assessment of pulmonary SPM precursors and SPMs demonstrated reductions in MetS mice exposed to AgNPs and increased levels in mice receiving SPM precursor treatments demonstrating treatments were effective at modulating lung levels of SPMs. Both models internalized AgNPs in collected lung inflammatory cells with macrophage and neutrophil associated AgNPs producing unique cell-specific spectral profiles. Lastly, MetS mice demonstrated reductions in pulmonary SPM receptor expression at baseline while AgNP exposure further reduced receptor expression suggesting further impairment of resolution signaling.
Inhalation is the primary route of AgNP exposure in environmental and occupational settings (Kuempel et al., 2021; Miyayama et al., 2012). Physicochemical properties such as size, charge, surface coating, etc. influence the toxicity of AgNPs (Ferdous and Nemmar, 2020). NPs can penetrate deep into the lungs and interact with alveolar macrophages, leading to pulmonary inflammation and damage (Lu et al., 2014). Many studies have demonstrated acute pulmonary toxicity associated with AgNP inhalation exposure (Braakhuis et al., 2014; Ferdous and Nemmar, 2020; Li et al., 2021). AgNPs similar to those used in our current study and at the same concentration, cause exposure-induced acute inflammation consisting of pulmonary neutrophilic influx, oxidative stress, and induction of pro-inflammatory signaling (cytokines and chemokines) (Ferdous and Nemmar, 2020; Li et al., 2021). Currently, our knowledge regarding AgNP-induced pulmonary toxicity in disease environments such as MetS is limited. Recently, we demonstrated that AgNP-induced acute inflammation was exacerbated due to MetS corresponding with reductions in a number of pulmonary SPMs (Alqahtani et al., 2020). These alterations in SPMs and the exacerbated inflammatory response were inhibited in MetS via treatment with atorvastatin. However, treatment with atorvastatin was not observed to alter the inflammatory response induced by AgNPs in the healthy model. These findings suggested that lipid dysregulation was a contributor to the enhanced susceptibility observed. Our current study confirms our previous findings and specifically elucidates the contribution of specific SPMs to the enhanced inflammatory response associated with MetS. To examine specific SPM groups, mice were pretreated with distinct SPM precursors (14-HDHA, 17-HDHA, or 18-HEPE) prior to AgNP exposure. Previous studies have utilized pretreatment of SPM precursor mixtures to evaluate the overall role of SPMs in exposure-induced responses (Kilburg-Basnyat et al., 2018; Neuhofer et al., 2013). The use of distinct SPM precursors in our study allows for the elucidation of specific SPMs deficiencies in MetS-associated susceptibility which is necessary for the development of targeted interventional and treatment strategies.
Pretreatment with either 14-HDHA or 17-HDHA reduced AgNP-induced pulmonary neutrophilic influx in the MetS mouse model to levels observed in the healthy mouse model. However, MetS mice pretreated with 18-HEPE still demonstrated exacerbated neutrophilic influx similar to MetS mice not receiving a treatment. Gene expression and protein levels of MIP-2, IL-6, and MCP-1 were enhanced in MetS mice not receiving precursor treatments. Similar alterations in CXCL1 and IL-1β were observed due to AgNP exposure in both healthy and MetS mice. Treatment with the SPM precursors, 14-HDHA and 17-HDHA, inhibited AgNP-induced increases of MIP-2, IL-6, and MCP-1 in both healthy and MetS mice without modifying induction of CXCL1 or IL-1β. Further, induction of CXCL1 and IL1β by AgNPs was not modified by any of the treatments in either model. This suggests that enhanced MIP-2, IL-6, and MCP-1 contribute to the exacerbated MetS inflammatory response and are sensitive to regulation via SPMs derived from 14-HDHA and 17-HDHA. However, CXCL1 and IL-β are regulated by other mechanisms and are not as responsive to SPMs. For example, CXCL1 is regulated by the transcription factor PARP binding to NF-κB whereas IL-6 is regulated by C/EBP binding to NF-κB (Amiri and Richmond, 2003). Previously, the SPM, resolvin D1, was found to suppress lung NF-κB and C/EBP activation, and decrease BAL fluid levels of IL-6 in inflamed lungs by IgG immune complex (Tang et al., 2014). Further, the SPM resolvin D2 has been shown to inhibit the NLRP3 inflammasome leading to reduced release IL-1β, result in resolution in murine peritonitis (Lopategi et al., 2019). This suggests that supplementation with resolvin D1 may specifically target MetS-associated exacerbations in pulmonary inflammation by inhibiting IL-6 while resolvin D2 may benefit healthy and MetS following particulate exposures by inhibiting IL-1β.
Inflammation is a highly coordinated process, and its appropriate regulation is necessary for responses to exposures. A blunted inflammatory response may enhance susceptibility to pathogens and clearance of foreign materials whereas an exacerbated response may cause tissue damage. Therefore, SPM precursor treatment appears to reestablish the healthy response to AgNP exposure via inhibition of specific pro-inflammatory signaling. Interestingly, in our previous investigation atorvastatin was determined to reduce all of these inflammatory signaling mediators we evaluated (MIP-2, MCP-1, IL-6, CXCL1, IL-1β) while also modulating pulmonary SPM levels following AgNP exposure in MetS. This suggests atorvastatin’s pulmonary protective effect functions through additional mechanisms beyond lipid regulation such as inhibition of farnesyltransferase and CXC chemokine formation leading to decreases of neutrophil recruitment (Zhang et al., 2012). 18-HEPE treatment was not determined to reduce AgNP induction of MIP-2 protein levels in MetS mice. This suggests that disruption of SPM-mediated regulation of MIP-2 may be a primary contributor to the exacerbated acute inflammatory responses observed in MetS following AgNP exposure.
SPMs are derived from dietary long chain and endogenous fatty acids via enzymatic and non-enzymatic reactions (Bannenberg and Serhan, 2010; Thatcher et al., 2019). SPMs are primarily derived from ω−3 fatty acids eicosatetraenoic acid (EPA) and docosahexaenoic acid (DHA). EPA is metabolized to the resolvins E series via the intermediate 18-HEPE, and DHA is metabolized to maresins via 14-HDHA, and/or to protectin and the resolvin D series via 17-HDHA (Chiang and Serhan, 2020; Serhan and Levy, 2018). SPMs can also be derived from ω−6 fatty acid, arachidonic acid (AA), which is metabolized to lipoxins (Buckley et al., 2014). Endogenous biosynthesis of SPMs is engaged by cell-to-cell interactions resulting in metabolism of precursors to SPMs mediated by lipoxygenases such as 5-lipoxygenase (ALOX5) and 15-lipoxygenase (ALOX15). AgNP exposure was determined to reduce ALOX5 and ALOX15 gene expression in MetS mice suggesting alterations in metabolic processes may be mediating deficits in SPM production and the exacerbated acute inflammatory response. Both 14-HDHA and 17-HDHA treatments inhibited AgNP-induced alterations in lipoxygenase gene expression. 14-HDHA was determined to be more efficient in inhibiting alterations in ALOX15 gene expression due to AgNP exposure than 17-HDHA. These treatment-induced alterations in lipoxygenase corresponded with increased pulmonary SPMs. Specifically, 14-HDHA treatment increased pulmonary levels of 14-HDHA, 17-HDHA, and RvD1, while 17-HDHA elevated MaR1, 17-HDHA, and RvD1. Similar to our findings, administration of a combination of SPM precursors via i.p. injection prevents reduction in SPM and the lipoxygenase activities in healthy mice lung tissues post-ozone exposure (Kilburg-Basnyat et al., 2018). This suggests that treatment with 14-HDHA and 17-HDHA may reestablish lipoxygenase activity in MetS mice, allowing for sustained production of SPMs following inhalation exposures. Reestablishing ALOX5 and ALOX15 within the lung by the treatments may be a mechanism to target to address MetS-associated susceptibility.
SPMs are ligands that act on resolution receptors, signaling to inhibit pro-inflammatory processes, initiate resolution, and facilitate repair (Berrueta et al., 2016; Freire and Van Dyke, 2013; Ishihara et al., 2019; Sansbury and Spite, 2016). Our study demonstrates baseline reductions in resolution receptor expression in MetS that may contribute to exacerbated inflammatory responses. Further, expression of resolution receptors were reduced 24h after AgNP exposure in both models suggesting that NP exposures can impair resolution signaling. We specifically evaluated LgR6, ChemR23, and GCPR18 as these receptors interact with SPMs we have previously determined were reduced in MetS following AgNP exposure (Alqahtani et al., 2020). LgR6 is a G-protein coupled receptor for maresins and signals to accelerate inflammatory resolution processes, including efferocytosis (Chiang et al., 2019). ChemR23 is a G-protein coupled receptor that interacts with the peptide chemerin and RvE1. Chemerin functions as a chemoattractant for monocytes and macrophages, whereas RvE1 initiates a cascade of pathways including the activation of multiple transcription factors contributing to resolution (Herová et al., 2015). RvD2 binds GCPR18, a receptor presented on the surface of the most immune cells, and signals for neutrophilic influx to cease while also enhancing efferocytosis, thereby accelerating resolution (Chiang et al., 2015). Alterations in these resolution receptors in MetS may delay resolution by impairing macrophage activity contributing to chronic inflammation. Previously, maresin-1 treatment of macrophages was determined to stimulate resolution processes via LGR6-mediated responses (Chiang et al., 2019). Interestingly, alterations in mRNA expression of SPM receptors were not observed to match receptor protein expression suggesting compensatory responses and possible post-transcriptional processes regulating receptor presentation. Together, reductions in SPMs and the diminished presence of resolution receptors suggest impaired resolution signaling contributes to exacerbated inflammation in MetS following AgNP exposure.
Our study establishes specific deficiencies in resolution signaling contributing to susceptibility to inhalation exposures in MetS which require future investigation. 18-HEPE, 17-HDHA, and 14-HDHA were selected for investigation based on findings from our previous study. The use of these precursors allowed for the determination of specific SPM classes and their impact on MetS resolution signaling. However, there are a number of SPMs generated from the metabolism of these precursors that require specific examination such as RvD1 and maresin-1. Further, there are other SPMs such as lipoxins generated by metabolism of arachidonic acid that may also contribute to exacerbated responses that should be evaluated. Additionally, our study only evaluated a single acute time point following AgNP exposure demonstrating exacerbated inflammation. To better understand SPM-mediated signaling deficiencies in MetS, investigations of sustained inflammation examining multiple time points and long-term impacts are needed. Specifically, our study suggests due to reductions in SPMs that inflammation may be sustained which may facilitate pulmonary disease development.
In conclusion, MetS is associated with enhanced susceptibility to inhalation exposure-induced inflammation and toxicity. In a mouse model of MetS, we determined through the use of SPM precursor treatments (18-HEPE, 14-HDHA, and 17-HDHA) that disruption of SPM-mediated resolution signaling contributes to susceptibility. Specifically, SPM precursors, 14-HDHA and 17-HDHA were determined to more efficiently attenuate susceptibility to AgNP exposure-induced inflammation compared to 18-HEPE. Overall, strategies to enhance SPMs specifically those produced by metabolism of 14-HDHA and 17-HDHA such as maresin-1, resolvin D series, and protectin-D1 may benefit individuals with MetS following particulate inhalation exposures.
Supplementary Material
Supplemental Figure 1. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice not receiving lipid treatments. A. Representative darkfield image of original AgNPs and spectral profile curve (gray) generated from over 1,000 pixels. Representative enhanced darkfield images of B. macrophages and C. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 2. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 14-HDHA. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 3. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 17-HDHA. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 4. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 18-HEPE. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 5. Quantitative differences in (A) AA, (B) EPA, and (C) DHA due to AgNP exposure in MetS and healthy mice and the effect of lipid interventions. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).
Highlights.
Metabolic syndrome exacerbates nanoparticle-induced acute pulmonary inflammation.
Resolution precursors modulated inflammation in exposed metabolic syndrome mice.
Docosahexaenoic acid-derived mediators more efficiently resolved inflammation.
Resolution receptors were reduced due to metabolic syndrome and exposure.
Engagement of resolution may assist susceptible groups after particulate exposure.
Acknowledgements
The authors would like to acknowledge the support of the Purdue Metabolite Profiling Facility for their assistance in generating the data presented within the manuscript.
Funding
This work was funded by the National Institute of Environmental Health Sciences (NIEHS) grant R00/ES024392.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval and consent to participate
All use of animals in this study was approved by the Purdue Animal Care and Use Committee.
Consent for Publication
All authors have read and consent for publication of this manuscript.
Availability of data and material
All data will be made available according to the National Institutes of Health policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- Agarwal AK, Raja A, Brown BD, 2020. Chronic obstructive pulmonary disease (COPD). StatPearls [Internet]. [PubMed] [Google Scholar]
- Alessandrini F, Vennemann A, Gschwendtner S, Neumann A, Rothballer M, Seher T, Wimmer M, Kublik S, Traidl-Hoffmann C, Schloter M, 2017. Pro-inflammatory versus immunomodulatory effects of silver nanoparticles in the lung: The critical role of dose, size and surface modification. Nanomaterials 7, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani SM, Kobos L, Xia L, Ferreira CR, Marmolejo JF, Du X, Shannahan J, 2020. Exacerbation of Nanoparticle-Induced Acute Pulmonary Inflammation in a Mouse Model of Metabolic Syndrome. Frontiers in Immunology 11, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri KI, Richmond A, 2003. Fine tuning the transcriptional regulation of the CXCL1 chemokine. Progress in nucleic acid research and molecular biology 74, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DS, Patchin ES, Silva RM, Uyeminami DL, Sharmah A, Guo T, Das GK, Brown JM, Shannahan J, Gordon T, 2015. Influence of particle size on persistence and clearance of aerosolized silver nanoparticles in the rat lung. Toxicological Sciences 144, 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Hisada T, Ishizuka T, Utsugi M, Kawata T, Shimizu Y, Okajima F, Dobashi K, Mori M, 2008. Resolvin E1 dampens airway inflammation and hyperresponsiveness in a murine model of asthma. Biochemical and biophysical research communications 367, 509–515. [DOI] [PubMed] [Google Scholar]
- Arias-Pérez RD, Taborda NA, Gómez DM, Narvaez JF, Porras J, Hernandez JC, 2020. Inflammatory effects of particulate matter air pollution. Environmental Science and Pollution Research 1–15. [DOI] [PubMed] [Google Scholar]
- Bannenberg G, Serhan CN, 2010. Specialized pro-resolving lipid mediators in the inflammatory response: an update. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 1801, 1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta L, Muskaj I, Olenich S, Butler T, Badger GJ, Colas RA, Spite M, Serhan CN, Langevin HM, 2016. Stretching impacts inflammation resolution in connective tissue. Journal of cellular physiology 231, 1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis HM, Gosens I, Krystek P, Boere JAF, Cassee FR, Fokkens PHB, Post JA, Van Loveren H, Park MVDZ, 2014. Particle size dependent deposition and pulmonary inflammation after short-term inhalation of silver nanoparticles. Particle and fibre toxicology 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA III, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, 2010. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation 121, 2331–2378. [DOI] [PubMed] [Google Scholar]
- Buckley CD, Gilroy DW, Serhan CN, 2014. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-C, Schwartz J, 2008. Metabolic syndrome and inflammatory responses to long-term particulate air pollutants. Environmental health perspectives 116, 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, Li Y, Wang X, Zhao L, 2018. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 9, 7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N, Dalli J, Colas RA, Serhan CN, 2015. Identification of resolvin D2 receptor mediating resolution of infections and organ protectionResolvin D2 activates pro-resolving GPCR. The Journal of experimental medicine 212, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N, Libreros S, Norris PC, de la Rosa X, Serhan CN, 2019. Maresin 1 activates LGR6 receptor promoting phagocyte immunoresolvent functions. The Journal of clinical investigation 129, 5294–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N, Serhan CN, 2020. Specialized pro-resolving mediator network: an update on production and actions. Essays in Biochemistry 64, 443–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas RA, Dalli J, Chiang N, Vlasakov I, Sanger JM, Riley IR, Serhan CN, 2016. Identification and actions of the maresin 1 metabolome in infectious inflammation. The Journal of Immunology 197, 4444–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier M-A, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, Van Pelt RE, Wang H, Eckel RH, 2008. The metabolic syndrome. Endocrine reviews 29, 777–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalli J, Zhu M, Vlasenko NA, Deng B, Haeggström JZ, Petasis NA, Serhan CN, 2013. The novel 13S, 14S-epoxy-maresin is converted by human macrophages to maresin 1 (MaR1), inhibits leukotriene A4 hydrolase (LTA4H), and shifts macrophage phenotype. The FASEB Journal 27, 2573–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Vedova MC, Muñoz MD, Santillan LD, Plateo-Pignatari MG, Germanó MJ, Tosi MER, Garcia S, Gomez NN, Fornes MW, Mejiba SEG, 2016. A mouse model of diet-induced obesity resembling most features of human metabolic syndrome. Nutrition and metabolic insights 9, NMI-S32907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RB, Smith CB, Schmitt MT, Rappold AG, Hinderliter A, Graff D, Carraway MS, 2014. Controlled exposure of humans with metabolic syndrome to concentrated ultrafine ambient particulate matter causes cardiovascular effects. Toxicological Sciences 140, 61–72. [DOI] [PubMed] [Google Scholar]
- Duvall MG, Levy BD, 2016. DHA-and EPA-derived resolvins, protectins, and maresins in airway inflammation. European journal of pharmacology 785, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous Z, Nemmar A, 2020. Health impact of silver nanoparticles: a review of the biodistribution and toxicity following various routes of exposure. International journal of molecular sciences 21, 2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francos-Quijorna I, Santos-Nogueira E, Gronert K, Sullivan AB, Kopp MA, Brommer B, David S, Schwab JM, López-Vales R, 2017. Maresin 1 promotes inflammatory resolution, neuroprotection, and functional neurological recovery after spinal cord injury. Journal of Neuroscience 37, 11731–11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire MO, Van Dyke TE, 2013. Natural resolution of inflammation. Periodontology 2000 63, 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallou-Kabani C, Vigé A, Gross M, Rabès J, Boileau C, Larue-Achagiotis C, Tomé D, Jais J, Junien C, 2007. C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity 15, 1996–2005. [DOI] [PubMed] [Google Scholar]
- Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, Smith SC Jr, 2005. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation 112, 2735–2752. [DOI] [PubMed] [Google Scholar]
- Hansen TV, Vik A, Serhan CN, 2019. The protectin family of specialized pro-resolving mediators: Potent immunoresolvents enabling innovative approaches to target obesity and diabetes. Frontiers in pharmacology 9, 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herová M, Schmid M, Gemperle C, Hersberger M, 2015. ChemR23, the receptor for chemerin and resolvin E1, is expressed and functional on M1 but not on M2 macrophages. The Journal of Immunology 194, 2330–2337. [DOI] [PubMed] [Google Scholar]
- Holland NA, Becak DP, Shannahan JH, Brown JM, Carratt SA, Winkle LSV, Pinkerton KE, Wang CM, Munusamy P, Baer DR, 2015. Cardiac ischemia reperfusion injury following instillation of 20 nm citrate-capped nanosilver. Journal of nanomedicine & nanotechnology 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Gronert K, Devchand PR, Moussignac R-L, Serhan CN, 2003. Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood, and glial cells: autacoids in anti-inflammation. Journal of Biological Chemistry 278, 14677–14687. [DOI] [PubMed] [Google Scholar]
- Hsiao H-M, Thatcher TH, Colas RA, Serhan CN, Phipps RP, Sime PJ, 2015. Resolvin D1 reduces emphysema and chronic inflammation. The American journal of pathology 185, 3189–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Yoshida M, Arita M, 2019. Omega-3 fatty acid-derived mediators that control inflammation and tissue homeostasis. International immunology 31, 559–567. [DOI] [PubMed] [Google Scholar]
- Jain MK, Ridker PM, 2005. Anti-inflammatory effects of statins: clinical evidence and basic mechanisms. Nature reviews Drug discovery 4, 977. [DOI] [PubMed] [Google Scholar]
- Kessler R, 2011. Engineered nanoparticles in consumer products: understanding a new ingredient. [DOI] [PMC free article] [PubMed]
- Kilburg-Basnyat B, Reece SW, Crouch MJ, Luo B, Boone AD, Yaeger M, Hodge M, Psaltis C, Hannan JL, Manke J, 2018. Specialized pro-resolving lipid mediators regulate ozone-induced pulmonary and systemic inflammation. Toxicological Sciences 163, 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp M, Ghosh S, Mohammed S, Nadeem Khan M, 2021. From virus to inflammation, how influenza promotes lung damage. Journal of Leukocyte Biology 110, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobos L, Alqahtani S, Xia L, Coltellino V, Kishman R, McIlrath D, Perez-Torres C, Shannahan J, 2020. Comparison of silver nanoparticle-induced inflammatory responses between healthy and metabolic syndrome mouse models. Journal of Toxicology and Environmental Health, Part A 83, 249–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaraju R, Guesdon W, Crouch MJ, Teague HL, Sullivan EM, Karlsson EA, Schultz-Cherry S, Gowdy K, Bridges LC, Reese LR, 2017. B cell activity is impaired in human and mouse obesity and is responsive to an essential fatty acid upon murine influenza infection. The Journal of Immunology 198, 4738–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel ED, Roberts JR, Roth G, Zumwalde RD, Nathan D, Hubbs AF, Trout D, Holdsworth G, 2021. Health effects of occupational exposure to silver nanomaterials.
- Leitch AE, Duffin R, Haslett C, Rossi AG, 2008. Relevance of granulocyte apoptosis to resolution of inflammation at the respiratory mucosa. Mucosal immunology 1, 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bi Z, Hu Y, Sun L, Song Y, Chen S, Mo F, Yang J, Wei Y, Wei X, 2021. Silver nanoparticles and silver ions cause inflammatory response through induction of cell necrosis and the release of mitochondria in vivo and in vitro. Cell biology and toxicology 37, 177–191. [DOI] [PubMed] [Google Scholar]
- Lim CS, Porter DW, Orandle MS, Green BJ, Barnes MA, Croston TL, Wolfarth MG, Battelli LA, Andrew ME, Beezhold DH, 2020. Resolution of pulmonary inflammation induced by carbon nanotubes and fullerenes in mice: role of macrophage polarization. Frontiers in Immunology 11, 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou D, Long F-W, Chen K-L, Yang H-W, Lv Z-Y, Zhou B, Peng Z-H, Sun X-F, Li Y, 2016. Resolvin D1 protects against inflammation in experimental acute pancreatitis and associated lung injury. American Journal of Physiology-Gastrointestinal and Liver Physiology 310, G303–G309. [DOI] [PubMed] [Google Scholar]
- Lopategi A, Flores-Costa R, Rius B, López-Vicario C, Alcaraz-Quiles J, Titos E, Clària J, 2019. Frontline science: specialized proresolving lipid mediators inhibit the priming and activation of the macrophage NLRP3 inflammasome. Journal of Leukocyte Biology 105, 25–36. [DOI] [PubMed] [Google Scholar]
- Lotfi R, Davoodi A, Mortazavi SH, Karaji AG, Tarokhian H, Rezaiemanesh A, Salari F, 2020. Imbalanced serum levels of resolvin E1 (RvE1) and leukotriene B4 (LTB4) in patients with allergic rhinitis. Molecular biology reports 47, 7745–7754. [DOI] [PubMed] [Google Scholar]
- Lu X, Zhu T, Chen C, Liu Y, 2014. Right or left: the role of nanoparticles in pulmonary diseases. International journal of molecular sciences 15, 17577–17600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, Kuebler WM, 2011. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. American journal of respiratory cell and molecular biology 44, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MC, Belli AJ, Kaji DA, Matsui EC, Brigham EP, Peng RD, Sellers C, D’Ann LW, Diette GB, Breysse PN, 2015. Obesity as a susceptibility factor to indoor particulate matter health effects in COPD. European Respiratory Journal 45, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyayama T, Arai Y, Hirano S, 2012. Environmental exposure to silver and its health effects. Nihon eiseigaku zasshi. Japanese journal of hygiene 67, 383–389. [DOI] [PubMed] [Google Scholar]
- Moore JX, Chaudhary N, Akinyemiju T, 2017. Peer reviewed: Metabolic syndrome prevalence by race/ethnicity and sex in the United States, National Health and Nutrition Examination Survey, 1988–2012. Preventing chronic disease 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed B, Weiden MD, Kwon S, Gracely EJ, Comfort AL, Ferrier N, Kasturiarachchi KJ, Cohen HW, Aldrich TK, Rom WN, 2012. Metabolic syndrome biomarkers predict lung function impairment: a nested case-control study. American journal of respiratory and critical care medicine 185, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemmar A, Holme JA, Rosas I, Schwarze PE, Alfaro-Moreno E, 2013. Recent advances in particulate matter and nanoparticle toxicology: a review of the in vivo and in vitro studies. BioMed research international 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhofer A, Zeyda M, Mascher D, Itariu BK, Murano I, Leitner L, Hochbrugger EE, Fraisl P, Cinti S, Serhan CN, 2013. Impaired local production of proresolving lipid mediators in obesity and 17-HDHA as a potential treatment for obesity-associated inflammation. Diabetes 62, 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Zhang Y, Hu H, Hu W, Li L, Pang Y, Ma S, Niu Y, Zhang R, 2021. Association between ambient particulate matter exposure and metabolic syndrome risk: A systematic review and meta-analysis. Science of The Total Environment 146855. [DOI] [PubMed] [Google Scholar]
- Nordgren TM, Bauer CD, Heires AJ, Poole JA, Wyatt TA, West WW, Romberger DJ, 2015. Maresin-1 reduces airway inflammation associated with acute and repetitive exposures to organic dust. Translational Research 166, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Eto M, Kano MR, Kahyo T, Setou M, Ogawa S, Iijima K, Akishita M, Ouchi Y, 2010. Induction of endothelial nitric oxide synthase, SIRT1, and catalase by statins inhibits endothelial senescence through the Akt pathway. Arteriosclerosis, thrombosis, and vascular biology 30, 2205–2211. [DOI] [PubMed] [Google Scholar]
- Roda E, Bottone MG, Biggiogera M, Milanesi G, Coccini T, 2019. Pulmonary and hepatic effects after low dose exposure to nanosilver: Early and long-lasting histological and ultrastructural alterations in rat. Toxicology reports 6, 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roursgaard M, Poulsen SS, Poulsen LK, Hammer M, Jensen KA, Utsunomiya S, Ewing RC, Balic-Zunic T, Nielsen GD, Larsen ST, 2010. Time-response relationship of nano and micro particle induced lung inflammation. Quartz as reference compound. Human & experimental toxicology 29, 915–933. [DOI] [PubMed] [Google Scholar]
- Sacks JD, Stanek LW, Luben TJ, Johns DO, Buckley BJ, Brown JS, Ross M, 2011. Particulate matter–induced health effects: who is susceptible? Environmental health perspectives 119, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansbury BE, Spite M, 2016. Resolution of acute inflammation and the role of resolvins in immunity, thrombosis, and vascular biology. Circulation research 119, 113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G, Neurath MF, 2018. Resolution of chronic inflammatory disease: universal and tissue-specific concepts. Nature communications 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, 2010. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? The American journal of pathology 177, 1576–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Gotlinger K, Hong S, Lu Y, Siegelman J, Baer T, Yang R, Colgan SP, Petasis NA, 2006. Anti-inflammatory actions of neuroprotectin D1/protectin D1 and its natural stereoisomers: assignments of dihydroxy-containing docosatrienes. The Journal of Immunology 176, 1848–1859. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, Moussignac R-L, 2002. Resolvins a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. The Journal of experimental medicine 196, 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Levy BD, 2018. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. The Journal of clinical investigation 128, 2657–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Petasis NA, 2011. Resolvins and protectins in inflammation resolution. Chemical reviews 111, 5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannahan JH, Podila R, Brown JM, 2015. A hyperspectral and toxicological analysis of protein corona impact on silver nanoparticle properties, intracellular modifications, and macrophage activation. International journal of nanomedicine 10, 6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi S, Behzadi S, Laurent S, Forrest ML, Stroeve P, Mahmoudi M, 2012. Toxicity of nanomaterials. Chemical Society Reviews 41, 2323–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Liu Y, Yan C, Petasis NA, Serhan CN, Gao H, 2014. Protective Actions of Aspirin-Triggered (17R) Resolvin D1 and Its Analogue, 17R-Hydroxy-19-Para-Fluorophenoxy-Resolvin D1 Methyl Ester, in C5a-Dependent IgG Immune Complex–Induced Inflammation and Lung Injury. The Journal of Immunology 193, 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher TH, Woeller CF, McCarthy CE, Sime PJ, 2019. Quenching the fires: pro-resolving mediators, air pollution, and smoking. Pharmacology & therapeutics 197, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe RM, Birukova A, Yaeger MJ, Reece SW, Gowdy KM, 2018. Euthanasia- and lavage-mediated effects on bronchoalveolar measures of lung injury and inflammation. American journal of respiratory cell and molecular biology 59, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utell MJ, Samet JM, 1990. Environmentally mediated disorders of the respiratory tract. The Medical clinics of North America 74, 291–306. [DOI] [PubMed] [Google Scholar]
- Wagner JG, Allen K, Yang H, Nan B, Morishita M, Mukherjee B, Dvonch JT, Spino C, Fink GD, Rajagopalan S, 2014. Cardiovascular depression in rats exposed to inhaled particulate matter and ozone: effects of diet-induced metabolic syndrome. Environmental health perspectives 122, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Katwa P, Podila R, Chen P, Ke PC, Rao AM, Walters DM, Wingard CJ, Brown JM, 2011. Multi-walled carbon nanotube instillation impairs pulmonary function in C57BL/6 mice. Particle and fibre toxicology 8, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiden MD, Naveed B, Kwon S, Cho SJ, Comfort AL, Prezant DJ, Rom WN, Nolan A, 2013. Cardiovascular biomarkers predict susceptibility to lung injury in World Trade Center dust-exposed firefighters. European respiratory journal 41, 1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger MJ, Reece SW, Kilburg-Basnyat B, Hodge MX, Pal A, Dunigan-Russell K, Luo B, You DJ, Bonner JC, Spangenburg EE, 2021. Sex Differences in Pulmonary Eicosanoids and Specialized Pro-Resolving Mediators in Response to Ozone Exposure. Toxicological Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-S, Gui Z-H, Zou Z-Y, Yang B-Y, Ma J, Jing J, Wang H-J, Luo J-Y, Zhang X, Luo C-Y, 2021. Long-term exposure to ambient air pollution and metabolic syndrome in children and adolescents: a national cross-sectional study in China. Environment International 148, 106383. [DOI] [PubMed] [Google Scholar]
- Zhang Songen, Rahman M, Zhang Su, Jeppsson B, Herwald H, Thorlacius H, 2012. Streptococcal m1 protein triggers farnesyltransferase-dependent formation of CXC chemokines in alveolar macrophages and neutrophil infiltration of the lungs. Infection and immunity 80, 3952–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice not receiving lipid treatments. A. Representative darkfield image of original AgNPs and spectral profile curve (gray) generated from over 1,000 pixels. Representative enhanced darkfield images of B. macrophages and C. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 2. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 14-HDHA. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 3. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 17-HDHA. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 4. Darkfield hyperspectral analysis of AgNPs in BALF cells collected from Healthy and MetS mice receiving 18-HEPE. Representative enhanced darkfield images of A. macrophages and B. neutrophils 24 h after exposure to AgNPs at a concentration of 50 μg. Spectral profiles were generated by selecting over 1,000 pixels representing internalized AgNPs from multiple macrophages and neutrophils. Purple curves show the spectral profile of AgNPs internalized by macrophages while green curves show the spectral profile of AgNPs internalized by neutrophils. Numbers correspond to peak wavelengths. Spectral profiles generated from multiple cells were then applied to representative images to map matching pixels in red to demonstrate curves accurately identify AgNPs within cells.
Supplemental Figure 5. Quantitative differences in (A) AA, (B) EPA, and (C) DHA due to AgNP exposure in MetS and healthy mice and the effect of lipid interventions. Thirty minutes prior to oropharyngeal aspiration of pharmaceutical grade sterile water (control) or AgNPs (50 μg) in sterile water, mice were i.p. injected with 1μg 14-HDHA, 17-HDHA, or 18-HEPE or vehicle (250 μl of sterile saline). * denotes significant differences due to AgNP exposure comparing to model-matched control receiving the same treatment, # denotes significant differences between healthy and MetS mouse models receiving the same treatments and exposure, $ denotes significant differences due to lipid interventions comparing to groups not receiving treatment but the same exposure (p < 0.05).