Abstract
Organ-on-a-chip platforms involve the miniaturization of cell culture systems and enable a variety of novel experimental approaches. These range from modeling the independent effects of biophysical forces on cells to the screening novel drugs in multi-organ microphysiological systems, all within microscale devices. As in living systems, the incorporation of vascular structures is a key feature common to almost all organ-on-a-chip systems. In this review we highlight recent advances in organ-on-a-chip technologies with a focus on the vasculature. We first present the developmental process of the blood vessels through which vascular cells assemble into networks and remodel to form complex vascular beds under flow. We then review self-assembled vascular models and flow systems for the study of vascular development and biology as well as pre-patterned vascular models for the generation of perfusable microvessels for modeling vascular and tissue function. We finally conclude with a perspective on developing future OOC approaches for studying different aspects of vascular biology. We highlight the fit for purpose selection of OOC models towards either simple but powerful testbeds for therapeutic development, or complex vasculature to accurately replicate human physiology for specific disease modeling and tissue regeneration.
Keywords: Vascular biology, organ-on-a-chip, endothelial cells, organoids, microfluidics, mechanotransduction
Introduction
Vasculature is the universal tissue infrastructure, known for providing nutrients, taking away waste, maintaining homeostasis, and shaping tissue and organ structure. In the developing embryo, a primitive vascular network first emerges de novo via the differentiation and assembly of a group of vascular progenitors, known as angioblasts [1–3]. As development proceeds, this primitive network remodels, presumably triggered by changes in hemodynamic forces [4,5], surrounding cell types, and microenvironment, including chemical factors and architectural features of surrounding extracellular matrix, to establish a hierarchical vascular tree with tissue-specific functionality important for the function of each organ [6,7]. The process of de novo vascular formation is known as vasculogenesis, whereas remodeling and vascular growth from pre-existing vasculature, either by sprouting or intussusception [8], has been defined as angiogenesis. Modern evidence has shown that blood vessels also provide instructive regulatory signals to surrounding non-vascular target cells [7,9]. Building vascular models and vascularized tissues has been a major thrust in biomedical research in recent decades for understanding vascular biology, regenerating tissue, modeling diseases, and testing preclinical drugs.
Among various biomedical efforts, organ-on-a-chip (OOC) technologies have gained significant research attention in recent years, with the more specific target of designing and engineering miniaturized functional units of human tissues and organs [10–13]. These miniaturized devices promise to better replicate native biological functions ex vivo and enable high-throughput techniques essential to disease modeling and drug testing. Central to this approach is the use of perfusable micro- and milli-fluidic platforms in which fluid flow both sustains the metabolic needs of OOC devices and replicates the distinct vascular compartment found in living organisms. In this review we explore recent advances in OOC technologies with a focus on this vascular component and technologies for replicating vascular structure and function in vivo. We will briefly review vascular structure and function, highlighting OOC systems that have advanced our understanding of fundamental aspects of vascular biology. We then explore OOC systems that utilize an engineered vasculature to explore tissue level functions in organ-specific models. We conclude with prospective remarks on choosing OOC systems of varying complexity and including proper cell populations to target suitable vascular biology questions.
Vasculature: Heterogenous structure and cell populations.
Blood vessels are organized in a hierarchical branching network in three-dimensional (3D) space and characterized by diameters ranging from arteries and veins (1 mm to 1 cm for large vessels, and 100 μm to 1 mm for small vessels), to arterioles and venules (20 – 100 μm) and capillaries (5 – 20 μm). The curvature of vessels ranges from 10−3 to 102 mm−1 with the largest vessel curvature found in the microvasculature of complex vascular beds like the brain [14]. Variation in size and curvature corresponds to the difference in vessel wall composition and blood flow, leading to heterogeneous biophysical forces on the vessel walls. As the innermost layer of cells on the vessel wall, endothelial cells also have distinct identities in arteries, veins, large and small vessels and different vascular beds in various organs to support diverse vascular function and tissue demands [15–18]. Oxygenated blood is carried in arteries and arterioles, whereas deoxygenated blood drains into venules and veins. A continuous endothelial cell layer in high density capillary networks supports the high metabolic demand of organs such as the heart and brain, whereas a fenestrated endothelium allows for filtration, secretion, and reabsorption of small molecules in kidney and guts. Sinusoidal or discontinuous endothelia are critical to support macromolecular and cellular trafficking functions in liver, bone marrow and spleen [19–22]. The endothelium provides critical signals to construct a specific vascular microenvironment and instruct organ development and maturation [7,9]. The organ-specific microenvironment (i.e. cytokines, metabolites, biophysical signals, and direct cell-cell contact from parenchyma cells) simultaneously interacts with ECs to induce unique EC functions [23].
Heterogeneity therefore is an important part of vascularization that needs to be considered for recapitulating specific tissue and organ function and may be used for promoting tissue regeneration.
Hemodynamics: a critical driver for vascular formation
A central process in vascular biology is sensing blood flow. Once a vascular network forms, blood flow induces mechanical stimuli that is constantly sensed by the vasculature, driving vascular growth and remodeling. As blood flows over the endothelial surface, its viscosity generates shear forces in the direction of flow. Simultaneously, pressure within the blood vessel generates a force normal to the surface of endothelial cells causing the vessel to stretch.
Vessels with different size, identity (i.e., artery, vein, or capillary), or in different organs often have different characteristic pressure and shear stress levels. Changes to shear stress level have been associated with vascular pathologies for decades, with regions of altered blood flow near vessel bends and bifurcations frequently acting as sites for atherosclerotic plaque growth or aneurysm development [24].
Endothelial cells sense these biophysical cues through a variety of mechanosensory mechanisms [25]. Numerous EC mechanosenors for wall shear stress (WSS) have been identified, including the junctional complex specific to endothelial cells consisting of VE-Cadherin, VEGFR2, and PECAM1 [26–30]. Additional mechanosensitive complexes include G proteins, ion channels, integrins, and NOTCH1 [31–36], which can be activated by WSS to induce downstream signaling to modulate vascular tone, barrier function, and proliferation. In addition to WSS magnitude, endothelial mechanosensors can also be activated by flow patterns. Physiological laminar flow activates the expression of transcription factor Krüppel-like factor (KLF2) and nuclear factor (erythroid-derived 2)-like (NRF2), and downstream antiinflammatory pathways. Disturbed flow inhibits KLF2 and NRF2 activation and activates pro-inflammatory pathways, leading to an atheroprone phenotype [31,37]. Recapitulating the diverse flow conditions found in vivo in engineered vascular models may be key to better understanding the physiologically heterogeneous vascular response to flow mediated conditions.
Blood flow plays a key role in regulating essential endothelial functions, including the release of nitric oxide for paracrine signaling [38] and endothelial barrier function [39]. These endothelial responses are often intimately tied to the function of perivascular cells and parenchymal tissues [40]. Thus, the studies of vascular biology and vascular support of parenchymal function requires fine control over biophysical and biochemical processes. OOC technologies are uniquely suited to pursuing these questions as they enable control of flow parameters that are difficult to study in vivo and let researchers specify and explore the types and interactions of endothelial cells with different parenchymal populations. In this review, we will investigate these applications of OOC devices to different aspects of vascular biology, from the discovery of fundamental endothelial mechanisms to the engineering of multi-organ models health and disease.
Self-assembled vascular models – vascular development:
Forming EC networks
The initial process of vasculogenesis (also called tubulogenesis) has been modeled with direct cell-in-gel culture systems, by embedding endothelial cells in cell-remodelable 3D matrices like collagen, fibrin, or Matrigel [41,42]. Once encased in these extracellular matrix (ECM) hydrogels, ECs can create lumens via either intracellular or extracellular mechanisms and form connective vascular tubes [41]. Vacuoles develop in ECs and then fuse with those in neighboring cells to form an intracellular lumen [43]. In addition, membrane invagination occurs at the junctions of neighboring ECs to hollow out the space between cells and create a lumen between them [44].
Extensive studies have utilized this direct cell-in-gel culture system to define factors and conditions which coax endothelial cells into luminal structures that connect and remodel following biophysical and biochemical cues [45,46]. Pericytes can be embedded in the bulk gel and recruited to support the developing vessel tube by stimulating basement membrane matrix assembly [47]. Since then, multiple OOC devices have been developed to further elucidate the role of biochemical and biophysical signals on tubulogenesis. For example, the Kamm group has developed microfluidic vasculogenesis devices with multiple culture compartments, which allowed for the inclusion of stromal cells or pericytes, and the flexibility of modifying growth factors, extracellular matrices, cellular composition, and imposing shear flow in parallel with the vasculogenic compartment [48]. Using this system, they also identified that angiopoietin-1 was a critical factor in the formation of stable and perfusable vessels with direct mural-EC cell-cell contacts (Fig. 1A) [49]. In a different microfluidic model developed by the Lee group, series of microtissue chambers containing self-assembled vascular networks, are connected between long microfluidic channels with full control of interstitial flow and mass transport. Using such an OOC device, Hsu and colleagues showed that interstitial flow across a cell-in-gel mixture could stimulate vasculogenesis [50]. This technique was then applied to create a higher throughput multi-channel platform for drug screening applications by the Hughes group [51] (Fig. 1B) and used to develop various vascularized microphysiological systems [52–54]. Self-assembled vascular networks have also been useful in identifying the role of endothelial cells and other vascular cells in supporting vascularization, tissue regeneration and OOC development [55–57].
Fig. 1.
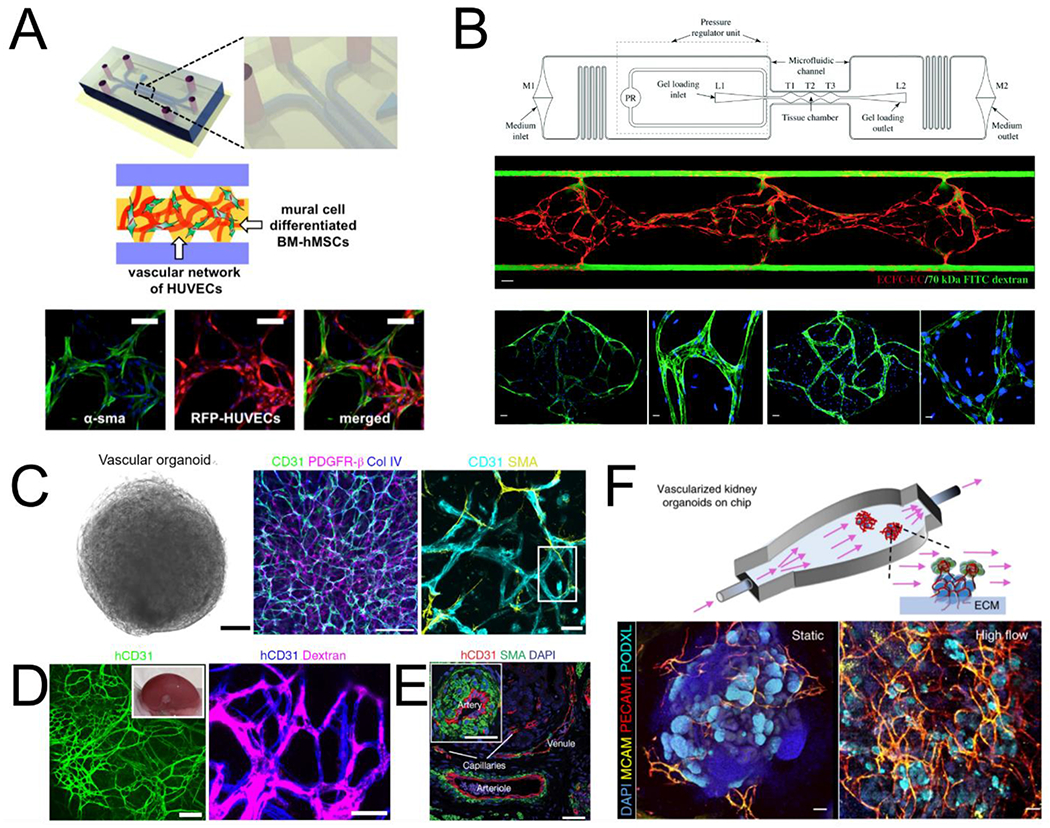
Models of vascular self-assembly. (A) Endothelial cells and mural cells embedded in a fibrin matrix and supported by perfusion self-assemble into perfusable networks with close contact between cell types (adapted from Jeon et al[49]). (B) Controlled interstitial flow and mass transport in a microfluidic design (top) across EC-laden hydrogels stimulates vasculogenesis and the formation of perfusable vascular networks (adapted from Phan et al[51]). Middle panel: three tissue chamber with red for ECs and green for perfusion of 70kDa FITC-Dextran. Bottom panels: staining of claudin-5 (left two), and VE-Cad (right two). (C) Vascular organoids after 15 days of differentiation compact into a sphere comprised of CD31 expressing endothelial cells that are enveloped with mural cells (PDGFR-β, SMA) and basement membrane (COL IV). (D) hVOs transplanted in mice survive and form robust connection with the host vasculature. (E) > 1 month after transplantation, hVOs develop hierarchical vascular structures identifiable as arteries, venules, and capillaries (C,D,E adapted from Wimmer et al[62]). (F) Using microfluidics to deliver flow through vascularized kidney organoids enhanced vascular network formation as well as maturation of kidney organoid parenchyma (adapted from Homan et al[68]).
Vascular organoids
Early vascular development starts with the assembly of vascular progenitors, which can continue to differentiate and self-renew to expand the vascular network and communicate with the parenchyma to modify endothelial cell identity. Existing model systems, however, have mostly relied on terminally differentiated endothelial cells, which lack sufficient plasticity or proliferation potential and limit the understanding and modeling of vascular development. Recent advances in pluripotent stem cell technology hold promise in deriving renewable vascular cells towards modeling blood vessels in vitro [58–60]. In particular, a recent report by Wimmer et al. demonstrated the development of human vascular organoids (hVOs) from human induced pluripotent stem cells (hiPSCs) with an organized vascular network [61]. By aggregating hiPSCs in 3D suspension culture, hVOs can be differentiated using growth factors and small molecules into organized endothelial and perivascular populations. These can be further matured and compacted in a collagen-Matrigel hydrogel to allow for sprouting and vascular network formation by VEGF and FGF stimulation. After compaction, hVOs displayed an organized vascular network with endothelial cells (CD31+) forming connective lumens and coated with COL IV+ basement membrane and PDGFR-β+ pericytes, and αSMA+ mural cells (Fig. 1C). There are also cell populations at similar differentiation stage including mesenchymal stem cells (CD90+ CD73+ CD44+) and hematopoietic cells (CD45+), suggesting simultaneous development of vascular and hematopoietic cell populations in hVOs. When transplanted in immunodeficient mice, the hVOs showed successful host integration (Fig. 1D) [62]. Notably, transplanted hVOs matured to form perfusable hierarchical vascular networks containing branching arteries and veins as well as capillary networks (Fig. 1E) enveloped with mural cells and basement membranes. This is likely the first demonstration of hierarchical vascular network formation in an implanted graft. In recent studies, this same group showed that hVOs can be directly infected with SARS-CoV-2 isolates, express viral RNA, shed progeny virus and substantially resist infection under the introduction of human recombinant soluble ACE2 [63]. The faithful recapitulation of human vasculature by hVOs promises in-depth understanding of vascular associated changes in human context and its application as a drug screening platform for treating diseases accompanying vascular complications.
Beyond hVOs, organoids have been developed to study organ development and model many organ-specific diseases [64]. The immediate milestone in these self-assembled culture models is to establish functional vascularization and perfusion. Endeavors have been made to promote vascular formation in organoids via endogenous growth factors [65,66]. and reprogramming [55,67]. One remaining but critical challenge is the lack of luminal perfusion in these self-assembled vascular structures. A recent effort led by Homan et al. managed to incorporate flow around cultured organoids, which promoted vascular network formation in the kidney organoids (Fig. 1F) [68], though luminal perfusion remains to be achieved. A perfusable vasculature is needed for the long-term culture and development of these self-assembled systems in vitro. Once that is achieved, these culture systems could be used to trace the formation of organ-specific endothelial and mural cell populations in vitro, and to model the complex organ-specific vascular physiology and pathology. It is expected that engineering tools, such as microfluidic and bioprinting techniques presented in the following sections, would be needed to help bridge these gaps and advance our understanding of vascular biology.
Flow systems – vascular development and biology
Blood flow is a key driver for the proper vascular growth and remodeling. The study of the hemodynamic response of vessels has been focused in two areas: endothelial cell sensing of flow and network expansion via angiogenesis. Different model systems have been developed in these two distinct areas.
Flow systems to model EC flow sensing
Flow chambers have been designed to model laminar or disturbed flow profiles and study the flow responses of cultured endothelial cell monolayers [26–28,69–71]. Endothelial cells respond to flow with cytoskeletal remodeling and cellular alignment in the direction of flow, changes in adherens junction complexes, and changes in gene expression and cell proliferation [28,71]. With sustained physiological shear stress under laminar flow, endothelial cells undergo elongation and alignment, strengthened adherens junctions, and lower permeability. When exposed to disturbed flow, often defined as a flow separation or recirculation with higher or lower than physiological wall shear stress, endothelial cells experience rounding, weakened adherens junctions, higher permeability, and increased proliferation and proinflammatory expression [69,71]. Similarly, in response to defined circumferential strain endothelial cells experience cytoskeletal remodeling leading to actin stress fiber orientation perpendicular to the stretch axis [72]. Physiological circumferential strain is associated with matrix remodeling, angiogenesis, growth factor secretion, and integrin engagement [37].
The state of the art of engineering models for the study of endothelial cell responses to flow have been limited in parallel plate flow chambers, 2D substrate assays, and deformable rubber membranes for many years. Parallel plate flow chambers are 2D systems which allow for a monolayer of endothelial cells to be exposed to a defined flow allowing for the effective study of WSS on the endothelium [73,74]. Additionally, endothelial cells cultured on 2D substrates and transwell plates have been used for evaluating endothelial cell function in basic mechanistic studies [24]. To investigate circumferential strain, deformable rubber membranes have been used to apply a defined strain to a monolayer of endothelial cells [37]. These simplified models have been crucial for understanding the endothelial cell response to mechanical forces. However, endothelial cells in arteries, veins, and microvasculature experience different levels of WSS, blood flow profiles, and pressure. Additionally, there are differences in metabolic demand and transport needs across different organ systems leading to specialized endothelial cell and vascular structure and function in different vascular beds [75–77]. Thus there are significant limitations in simple flow chambers due to the lack of a physiological 3D structure, lack of tissue-specific extracellular matrix and stromal support cells, and the lack of complex flow characteristics, all of which lead to a failure to fully recapitulate the microvascular niche.
In the body, the blood vessels bend and twist through 3D space with varying levels of curvature and torsion (i.e. deviation of curvature from its osculating plane). Small vessels with low flow and high curvature and torsion have been associated with heterogeneous vascular modeling, which remains not well studied. The response of ECs to flow has been binned into either laminar or disturbed flow, which however represent the two extremes of the wide spectrum of flow profiles found throughout the vasculature. To bridge the gap and understand the flow response in small vessels with 3D curvature in a laminar flow regime, our group has recently developed a spiral microvessel system with precisely controlled diameter, curvature and torsion in 3D space (Fig. 2A) [78]. We showed small spiral vessels in laminar flow regions induced distinct changes in EC phenotype and transcriptional profiles compared to conventionally defined laminar flow in straight vessels. This new engineering model expand the control over 3D geometrical parameters to better mimic vascular structure in vivo and accurately understand the effect of 3D curvature on vascular biology. The results also point to potential flow mediated mechanism of heterogeneous vascular development and remodeling.
Fig. 2.
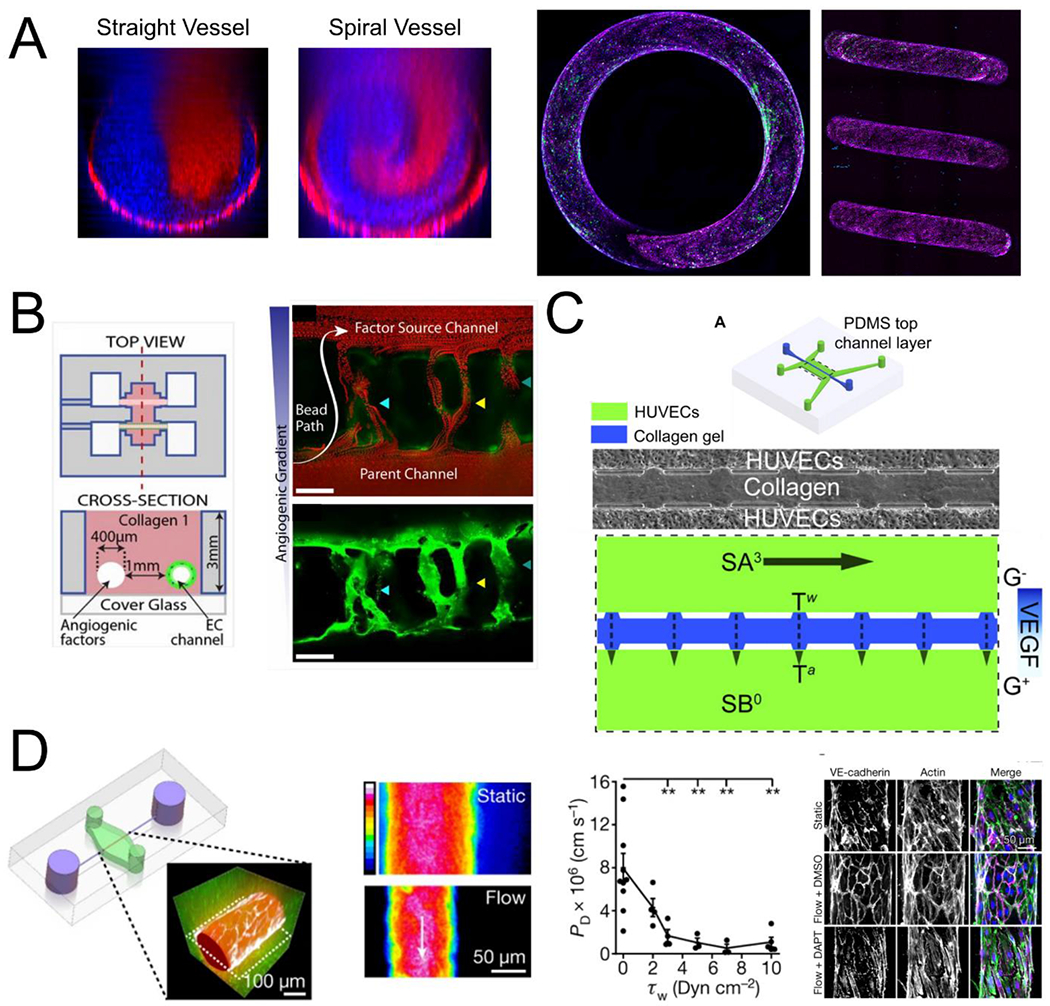
Engineering biophysical and biochemical cues in OOCs. (A) The flow in curved geometries is characterized by secondary flows and mixing not seen in straight vessels (left) and its effect can be studied in endothelialized spiral vessels (right, VECAD, magenta; VWF, green, adapted from Mandrycky et al[78]). (B) Endothelial cells cultured in channels can be stimulated by engineered biochemical gradients to stimulate angiogenesis and anastomosis to the factor source channel (adapted from Nguyen et al[86]). (C) Biochemical gradients can be combined with defined biophysical forces in opposing channels to investigate the influence of shear stress and interstitial flow differences on sprouting into collagen hydrogels (adapted from Song and Munn[88]). (D) Perfusion vascularized channels leads to a decrease in vascular permeability with increasing shear stress that can be disrupted using the γ-secretase inhibitor DAPT (adapted from Polacheck et al[90]).
Flow mediated sprouting angiogenesis and barrier function
Vascular networks expand through angiogenesis to establish hierarchical structure. This directed expansion of the vasculature is a critical process in normal development and wound healing, as well as in tumor progression [6]. Sprouting angiogenesis starts when ECs branch off of an existing blood vessel and extend into an avascular space, often motivated by the presence of specific chemical gradients in the tissue (e.g. VEGF) [79]. The sprouts are known to be led by a specialized “tip” cell EC, which migrates forward while trailing “stalk” cells proliferate to maintain a continuous lumen as the new vessel lengthens [80]. Much of this process has been observed in model organisms such as zebrafish and rodent models. Most engineering models have focused on biochemical factors that drive sprouting of an endothelial cell monolayer into ECM hydrogels [81,82]. The development of flow-mediated vascular model, however, is essential for a complete understanding of the underlying mechanisms and for the investigation of new therapeutic approaches. Given this, a wide variety of OOC systems have been developed for investigating angiogenesis [81–85]. Key to these designs is the patterning of an initial parent vessel, which is often achieved through casting of hydrogel around a removable mold (e.g. an acupuncture needle [83]) or carefully designed microfluidics devices where the hydrogel is confined to the space between two independently perfused channels [84]. For example, Nguyen and colleagues utilized needle subtraction method to generate two independent channels in a collagen hydrogel to study sprouting angiogenesis [86]. By seeding one channel with endothelial cells and perfusing angiogenic factors (HGF, bFGF, MCP-1, VEGF, S1P, and PMA) through the other channel to create a chemokine gradient they induced angiogenic sprouting (Fig. 2B). The formation and maturation of these sprouts appeared to follow the mechanics of what has been observed in vivo, representing a powerful testbed for further study of the molecular mechanisms of angiogenesis.
Other OOC models of angiogenesis have been developed to further investigate the biomechanical and biochemical regulators of angiogenesis [85,87]. Song and Munn developed microfluidic system and showed that fluid forces can regulate angiogenesis [88]. They seeded endothelial cells into two microfluidic channels separated by a block of collagen gel and established a VEGF gradient across the two channels to stimulate EC sprouting (Fig. 2C). They found that high shear stress attenuated VEGF induced sprouting through nitric oxide signaling and that interstitial flow through the collagen gel stimulated sprouting. This interstitial flow finding has also been replicated in additional OOC models [89]. Flow has been shown to regulate the production of matrix-metalloproteinase 1, which is an important factor for angiogenic sprouting [70]. In addition to sprouting, the use of cell-remodelable matrices provides important features for assessing vessel barrier function. For example, using single microvessel tubes, NOTCH signaling has been shown to be regulated by WSS, and further regulate vascular barrier function (Fig. 2D) [90], promote arterial gene expression [91], and maintain junctional integrity and suppress proliferation [92].
Together, this new generation of 3D vascular microfluidic models enables the control of input flow, vessel geometry in 3D space, vessel network structure, leading to fine control of flow magnitude, flow profiles, and pressure in individual branches and networks and better recapitulation of conditions in vivo. These advances would allow for the studies of individual contribution of flow parameters in step wise fashion, to uncover new cellular behaviors and gene regulations in in vivo like vascular microenvironment. These new models could also be used to test effects of isolated components of signaling pathways through stimulating or inhibiting (by activators, knockdowns, or cleavage methods) specific regulators and subjecting them to identical mechanical stimuli. Incorporating these new OOC technologies would provide new insights into core aspects of vascular biology at each of the stages of vessel formation and expansion, from individual cells, to lumens, and connected networks.
Patterning perfusable microvascular networks – vascular and tissue functions:
Beyond the fundamental study of vascular biology itself, introducing vasculature into OOC systems is critical for the development of functional tissue and the study of biologic processes including vascular-parenchymal and blood-vessel interactions [93]. Perfusion through the vasculature is necessary for the nutritional support of parenchymal tissue, with tissues generally needing to be located within a few hundred microns of a vascular lumen [94]. In addition to transport and hemodynamic-related phenomenon, vascular cells themselves guide the development and behavior of parenchymal and perivascular cells and are intimately involved in biological processes like inflammation, hemostasis, and wound healing [95–97]. The function of organs such as the heart, kidney, lungs, liver, or spleen is dependent on continuous perfusion, which is required for truly modeling the behavior of such organs.
An ideal vascularized OOC would include precise three-dimensional cellular arrangement, high vascular density, close apposition of vasculature and parenchyma, organ-specific function, cell-remodelable extracellular matrix materials, in vivo-like hemodynamics, hierarchical and multicellular blood vessels, and multiple organ-specific cell types. Current OOC systems generally focus on a limited number of these features and achieving them all simultaneously remains an outstanding challenge for the field. Various patterning techniques have been used to generate perfusable vasculature. The prepatterned approach involves the creation of hollow lumens within a bulk material followed by the introduction of endothelial cells. Methods for prepatterning vascular channels in OOC systems range from relatively planar microfluidic approaches, to lithographic or molding approaches within cell-remodelable hydrogels, to direct-writing strategies with full 3D control such as 3D printing or laser degradation [98,99].
PDMS based microvascular structure.
Microfluidic approaches to modeling a vascularized organ generally involve casting a biologically inert material such as polydimethylsiloxane (PDMS) over a micromachined mold with raised features, and then removing and bonding it to a glass surface or another piece of PDMS to create hollow channels. Such approaches are exemplified by the landmark design of Huh et al, consisting of two rectangular channels cast in PDMS and separated by a porous PDMS membrane [100,101]. Seeding vascular cells into one chamber and parenchymal cells into the other allows observation of transport phenomena and interactions at the tissue interface, with additional vacuum chambers on either side allowing the application of cyclical stretch/strain to mimic biological forces (Fig 3A). Initially used to model a breathing lung, organ functions including surfactant production were demonstrated, and processes occurring at the vascular-parenchymal interface such as neutrophil adherence and diapedesis in response to inflammation were replicated [100]. Since this initial report, this technology has been used to emulate specific functions of numerous organs. Notable examples include an anaerobic human intestine-on-a-chip replete with a complex bacterial microbiome [102] and a human glomerulus-on-a-chip incorporating human glomerular endothelial cells and human induced pluripotent stem cell-derived podocytes while demonstrating basic filtration function [103]. Connecting similar devices cultured with different organ-specific cells into a “body-on-a-chip” and automating cell culture and imaging was demonstrated recently, highlighting exciting future possibilities for monitoring organism-level processes [104].
Fig. 3.
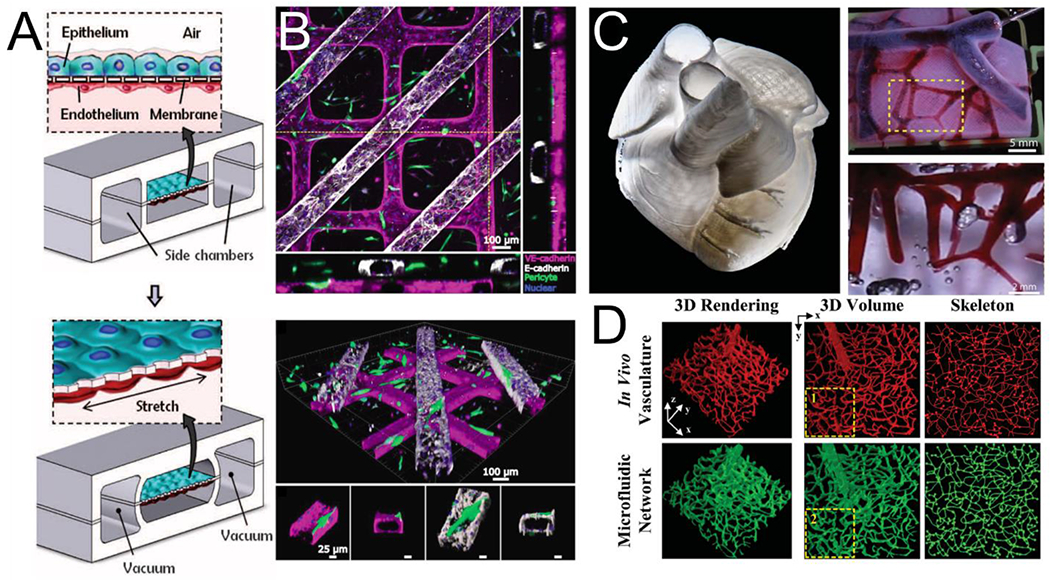
Strategies for vascularization of multicellular organ-on-a-chip platforms: (A) A schematic for microfluidic-based approach to a vascularized lung-on-a-chip (adapted from Huh et al[100]) (B) A human renal tubule on a chip created through soft lithography showing tubular channels overlying endothelial channels with flanking orthogonal views (top) and perspective views showing perivascular association with the vessel (bottom, adapted from Rayner et al[109]). (C) A 3D human heart model bioprinted in collagen (left) with vessels perfused in red (top right) and highlighted area shown magnified (bottom right, adapted from Lee et al[113]). (D) In vivo vasculature (top) was used to create a photomask for direct laser-writing of channels into a PEG-based hydrogel (bottom, adapted from Heintz et al[114]).
Hydrogel based microvascular structure.
The PDMS based microfluidic devices often succeed at replicating specific tissue function, but are limited by their essentially planar nature, the use of non-biologic materials, and difficulty supporting multiple cell types. Lithographic and/or molding-based approaches within biologic hydrogels allow greater three-dimensionality, the opportunity to include cells in the bulk matrix material, and the ability for cells to remodel the surrounding matrix. These approaches generally have in common the use of a mold or sacrificial material around which matrix material is cast, with removal of the mold or sacrificial material then leading to channels that can be seeded with endothelial cells. One simple, yet effective, iteration of this is the use of thin needle as a mold which is removed following matrix gelation, resulting in vascular lumens within a bulk hydrogel. This approach has been used to study tissue phenomenon including the vascular invasion of pancreatic cancer [105].
Our lab has developed a technique to create geometrically complex engineered vessels by lithographically patterning PDMS molds and using them to imprint channels into liquid collagen hydrogels. Following collagen gelation PDMS molds are removed and patterned slabs are assembled against a flat piece of collagen to create rectangular lumens within a bulk hydrogel [106]. This approach has been used to model the bone marrow niche [107] and create implantable cardiac vascular patches [108]. By assembling two patterned collagen slabs across a thin collagen membrane and seeding top and bottom channels with renal tubular epithelial and endothelial cells, respectively, this approach was used to emulate the human renal vascular-tubular unit (Fig 3B). Incorporated cells secreted basement membrane components, remodeled the collagen membrane into an ~1 μm exchange interface, and supported blood perfusion and albumin resorption [109]. The Radisic group has also used a stacking approach to increase vascular density and generate thick, multilayered cardiac constructs [110].
Bioprinting approaches offer the potential for full 3D control and precise cellular placement. Bioprinting can either involve the construction of sacrificial networks that leave lumens within cellularized hydrogel upon dissolution, or the direct deposition of vascular and parenchymal tissues. The intermediate step of creating sacrificial molds is appealing given the technical challenge of accurately direct-writing living cells within soft matrix materials. Using this approach, a 3D-printed dissolvable carbohydrate lattice was used to vascularize thick, cellularized Poly(ethylene glycol) (PEG)-based hydrogels, supporting the function of embedded hepatocytes [111]. Printing with a pluronic (PEO-poly(propylene)-PEO)-based ink was used to create a 3D vascularized proximal renal tubule with separate tubular and vascular channels within an engineered extracellular matrix material. This construct was shown to model both renal transport and pathophysiology related to a diseased (hyperglycemic) conditions [112]. In a recent breakthrough, direct-writing of collagen within a supportive hydrogel bath was used to bioprint a vascularized and contractile model 3D human heart with ~20 μm resolution (Fig 3C) [113]. Finally, an emerging related strategy is the use of subtractive patterning through direct laser-writing within hydrogels. Computer-controlled multiphoton manufacturing can pattern tissues with submicron resolution and offers the opportunity to create capillary-scale vascular beds in biologic hydrogels– a scale that is difficult to achieve with bioprinting approaches. This approach has been used to re-create human capillary geometries with PEG-based hydrogels (Fig 3D) [114]. While direct cell seeding cannot reliably be performed at the capillary scale, we have had success creating cellularized capillaries by ablating capillary-sized lumens (5-10 μm) extending from a larger cellularized vessels (100 - 300 μm) and applying a pressure gradient to encourage migration [115]. This technology has the potential to address current challenges in creating large scale vascularized and functional tissues, which is also an ongoing effort in our lab.
Microvasculature for studying blood-vessel interactions.
Perfusable microvessels have been useful in understanding blood vessel interactions [106,116–118], particularly in small vessels that are extremely challenging to study in vivo due to imaging constraints and structural heterogeneity. For example, we have developed microvessels with a 13 x 13 grid network in collagen hydrogel gel [106]. This large grid structure provides large range of flow velocity and wall shear stress that recapitulate both health and disease. In acute inflammation and activation, endothelia from these microvessels have been found to release von Willebrand Factor (VWF), a glycoprotein essential to hemostasis which assembles into thick fibers and 3D complex meshes under flow (Fig. 4A) [119]. These VWF fibers appear to serve as binding reservoirs for blood cells including platelets, leukocytes, and red blood cells (RBCs). In the absence of ADAMTS13, an enzyme known to cleave VWF in normal blood, this led to vascular occlusion. The same microvessel format has been adapted to study P. falciparum-infected RBC interactions with primary brain endothelium [120]. This work found that iRBCs binding to the endothelium is strongly influenced by the flow shear stress and different strands of parasite displayed different shear-dependent binding characteristics to the endothelium.
Fig. 4.
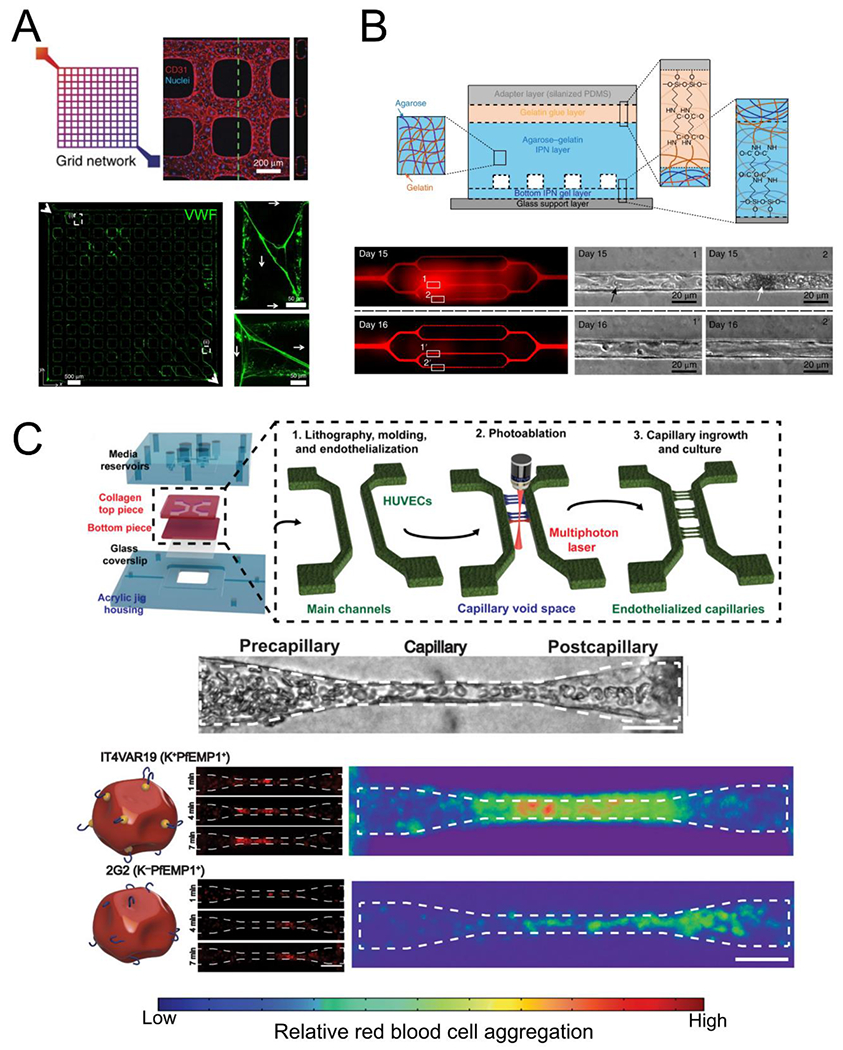
Blood-endothelial interactions in organ-on-a-chip systems. (A) Lithographically patterned endothelialized grid networks release VWF (green) when stimulated, which form complex fiber meshes under perfusion (adapted from Zheng et al[119]). (B) High concentrations of irreversibly sickled red blood cells perfused through patterned microvascular networks in agarose-gelatin hydrogels cause occlusions and increase vascular permeability (“Day 15”) that can be recovered within 24 hours with continuous perfusion (adapted from Qiu et al[116]). (C) Generation of capillary scale vessels (5 – 10 pm diameter) is possible using a combined molding and photoablation strategy (top). Malaria infected red blood cells perfused through capillaries can aggregate and different parasite variants aggregate in different regions of the capillary space (adapted from Arakawa et al[115]).
The occlusion of capillary size blood vessels, known as microvascular occlusion, is a hallmark of many vascular and blood diseases. Models at this scale have been difficult to fabricate and cellularize in biological hydrogels. The Lam group previously fabricated vessels as small as 20 μm and were able to model the effects of sickle RBCs and malaria infected RBCs on vascular barrier function (Fig 4B) [116]. While these vessels reached the scale of the post-capillary venule, no suitable models for this phenomena at the capillary scale were available until our recent development of functional capillaries with full control of size and perfusion [115]. Combining soft lithography and multiphoton ablation vessel fabrication techniques, we generated an arteriole-capillary-venule unit and demonstrated that iRBC sequestration occurs predominantly in capillary and post-capillary sized vessels (Fig 4C). These microvascular models push forward the frontier of accurately recapitulating vascular structures in vitro and enable more accurate models of microvascular diseases.
Together, these prepatterned microvessels offers full control of perfusion and vascular geometry with a large range of diameter and network complexity. These precise controls provide means to study the contribution of individual factors in a stepwise fashion and further develop potential therapies for various microvascular and blood diseases. Replicating high-density vascular networks with perfusion is crucial for the next level of tissue regeneration and the generation of functional vasculature. These pre-patterning tools could also be combined with vascular self-assembly and organoid technologies to build fully hierarchical vascular network with cellular renewal and remodeling potential.
Vascularized cardiac and organ-specific tissues.
As the most metabolically active organ in the body, the heart requires significant vascular density to support efficient nutrient delivery [121]. Hence, proper vascularization has been a key requirement towards engineering thick and functional cardiac constructs. Prior efforts to vascularize cardiac grafts have often relied on the self-assembly of ECs to form connected tubes within cardiac constructs using primary ECs (e.g. HUVECs) or ECs differentiated from pluripotent stem cells [108,122]. The presence of these vessels has been found to improve cardiomyocyte maturation and tissue function; however, the formed networks lack perfusion, preventing large scale construct fabrication and culture. Following implantation, the self-assembled vessels frequently regress and the grafts, though integrated with the host, showed insufficient perfusion. Recent developments by our group and others have shown that perfusable vessels can support cardiac tissue survival both in vitro and in vivo [108,110,123].
To support organ-specific tissue and chip designs, a variety of endothelial populations have been used. Organ-specific ECs in OOC devices are often primary cells isolated from healthy or diseased donors, but stem cell differentiation protocols have enabled the generation of these cells from hPSCs. In addition to the heart, these include brain vascular cells in models of the blood brain barrier [124], tubule and glomerular ECs in models of the kidney [125,126], pulmonary ECs in models of the lung [100], and sinusoidal ECs in models of the liver [136,137]. However, some cells such as liver sinusoidal ECs have an unstable phenotype in vitro, and HUVECs are still predominantly used in liver-specific OOC devices [134,135]. These EC populations in combination with other organ-specific cell types from both primary and hPSC-derived sources have been used to improve the function and maturation of organ-specific OOC devices (Table 1).
Table 1.
Cell types commonly used in organ specific OOC systems
| Organ System | Endothelial cell types | Secondary cell types |
|---|---|---|
| Heart | HUVECs [110,123], hiPSC-ECs [108] | Cardiomcyoytes and fibroblasts [127,128] |
| Brain | Brain microvascular endothelial cells [129,130], hiPSC-ECs [131] | Pericytes, astrocytes [129] |
| Kidney | Glomerular [112,125] and peritubular microvascular endothelial cells [132] | Podocytes [125], proximal tubule epithelial cells [112] |
| Lung | Pulmonary microvascular endothelial cells [100] | Alveolar epithelial cells [100], bronchial epithelial cells [133] |
| Liver | HUVECs [134,135], Liver sinusoidal endothelial cells [136,137] | Hepatocytes [134,136,137], stellate cells [134,136], Kupffer cells [135,136] |
Future directions – Choosing biomechanical, biochemical, and cellular complexity:
OOC devices have been applied to a wide range of research questions, spanning basic endothelial biology to disease modeling and drug discovery. Essential to these approaches is the extent to which different vascular features and compartments are modeled and the tailoring of device designs to the research question at hand. In the slow progression from simple to complex models, simplified systems targeting only some biophysical or biochemical cues have provided great insight into core aspects of endothelial biology. Laminar flow chambers, for example, have revealed fundamental mechanisms of endothelial mechanotransduction. Similarly, cell-in-gel OOCs provided new insights into the mechanisms of angiogenesis and potential strategies to induce vascular growth (or inhibit it in the case of tumorigenesis) in living or engineered tissues. In totality, however, these works as well as a variety of in vivo studies have demonstrated a wide range of factors which may influence vascular function. Recent work has increasingly reinforced the concept of vascular heterogeneity, going beyond long appreciated organ-specific or vessel-size-specific identities. Single cell transcriptomics have recently been used to identify at least 6 distinct endothelial cell types within the mouse brain [138]. Determining which populations are most important to incorporate into a given OOC device will be critical and depend on the study goals. Developing highly accurate tissue OOCs for disease modeling and drug testing, for example, may be sensitive to the presence of the full complement of native vascular phenotypes.
The question of what level of complexity to model extends beyond the varieties of endothelial cells to include. Recent work by our lab and others have highlighted additional levels of heterogeneity in the vasculature resulting from differences in hemodynamic factors. Laminar fluid flows with secondary flow characteristics can induce unique transcriptional profiles in endothelial cells [78]. This finding suggests the complex 3D geometries found in the native vasculature may lead to more diverse endothelial responses to flow than were previously thought. The functional consequences of these differences and their role in health and disease, if any, remain an open question which OOC systems are poised to provide new insight into. In addition, blood flow is pulsatile, and the pulsatility introduces temporal accelerations and decelerations which can combine with complex 3D vascular architectures to yield a continuum of complex flows. How these flows translate to vascular function is an intense area of research, particularly in hemodynamically sensitive pathologies like cerebral aneurysms [139]. On the other hand, many critical vascular phenomena occur at the smallest vessels – capillaries of 5 – 10 μm, which scale remains an engineering limit since most existing OOC technologies have resolution at the order of 100 μm or more. The development of robust capillary scale engineered vessels has enabled new investigations into the dynamics of vascular interactions like those between malaria infected red blood cells and endothelial cells when exposed to controlled flow. The inclusion of these capillary structures in OOC platforms may be crucial for studies of vascular interactions with different blood components, including altered RBCs, immune cells, platelets, and others. In these capillary vessels, the blood cells and blood vessels are of nearly the same caliber. Such close interactions may contribute to disease progression and their modulation may be core to a variety of therapeutic approaches.
There are several additional important elements in constructing appropriate organ-on-a-chip devices. One is to recapitulate the extracellular matrix (ECM) microenvironment that is found in vivo. ECM has a complex composition and function and is a regulator of vascular and tissue functions. Unlike in vivo, many OOC designs have used very simplified ECM forms. Endothelial cells are highly sensitive to different matrix molecules and the composition of the endothelial basement membrane is a mediator in the process of vascular growth and angiogenesis [140]. Hydrogels from decellularized extracellular matrices (dECM) have been developed to add back some of this biological complexity and create more organ-specific microenvironments [141]. We have previously shown that kidney dECM promotes kidney microvascular EC maturation and quiescence, supporting the need for carefully considered matrices in engineered systems [142]. The heterogeneous matrix microenvironment of site-specific ECMs, however, remains challenging to fully replicate. Future development in ECM protein design and spatial patterning would provide new tools to better model vascular and tissue functions.
Another important element is the original source of cells. Cell lines or isolated primary cells have so far been the predominant cell source in OOCs, but advances in stem cell technology have made a greater variety of cell types available to researchers. As a result, the prospect of patient-specific OOC systems populated with cells differentiated from patient-derived stem cells for applications in precision medicine has become more tenable [58,143]. Protocols for the differentiation of the many different endothelial subtypes are not yet available [144], but recent advances in vascular organoids [62] suggest renewable sources of these cells may be on the horizon. Incorporating these patient-derived cells into disease modelling OOC may provide new insight into the mechanisms of disease and the safety and efficacy of new therapeutic approaches.
In addition, blood proteins and cellular components would be crucial to incorporate in OOCs to optimize the culture system. We have recently shown that platelet rich plasma significantly enhanced vessel barrier and improve endothelial cell metabolism towards maturation [145]. These blood-derived components could be an alternative to replace current growth media and further improve OOCs. The incorporation of these components also enables the study of blood-endothelial interactions in healthy and diseased conditions. It appears that engineered microvessels have minimal interactions with normal human blood, suggesting normal vascular function. These perfusable vascular networks could be appropriate platforms for stepwise investigations of the interactions of various blood, cell, and protein components with the vessel wall during vascular activation, blood disorders, and even infectious disease progression.
In summary, existing vascular engineering technologies allow us to build vasculatures over the full range of diameters, curvatures, and potentially densities of blood vessels found in living organisms. Incorporating those technologies into a single engineered system encompassing the entire range of structures and cell types, however, remains an immense outstanding challenge. It is then important to define the goal of research projects, and wisely choose the appropriate OOC for approaching biological questions as well as drug and disease modeling. This approach has proven fruitful, but future advances are poised to further add to OOC device complexity and their ability to accurately replicate human physiology to gain a deeper understanding of vascular biology and to serve as powerful testbeds for modeling diseases and developing novel therapeutics.
Highlights.
Modeling vasculature requires heterogeneous structure and cell population.
Flow is a key driver for vascular formation and organ-on-a-chip models.
Patterning perfusable microvascular networks is an important achievement.
Fit for purpose OOC systems are needed to study vascular biology.
Acknowledgements:
We are thankful for support from the National Institute of Health R01 HL141570, UH2/UH3 DK107343, 1R01AI148802 and R01 AI141602 (to YZ), F32 HL143949 (to SR), support from the United Therapeutics Jenesis Innovative Research Award (to SR), support from the National Science Foundation Graduate Student Research Fellowship DGE 1762114 (to CH), and support from Institute of Stem Cell and Regenerative Medicine at the University of Washington (to YJS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Burggren WW, Cardiovascular Development and Angiogenesis in the Early Vertebrate Embryo, Cardiovasc. Eng. Technol 4 (2013) 234–245. 10.1007/s13239-013-0118-x. [DOI] [PubMed] [Google Scholar]
- [2].Patel-Hett S, D’Amore PA, Signal Transduction in Vasculogenesis and Developmental Angiogenesis, Int. J. Dev. Biol. 55 (2011) 353–363. 10.1387/ijdb.103213sp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heinke J, Patterson C, Moser M, Life is a pattern: vascular assembly within the embryo, Front. Biosci. Elite Ed. 4 (2012) 2269–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Culver JC, Dickinson ME, The Effects of Hemodynamic Force on Embryonic Development, Microcirculation. 17 (2010) 164–178. 10.1111/j.1549-8719.2010.00025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Han Y, Huang K, Yao Q-P, Jiang Z-L, Mechanobiology in vascular remodeling, Natl. Sci. Rev. 5 (2018) 933–946. 10.1093/nsr/nwx153. [DOI] [Google Scholar]
- [6].Carmeliet P, Jain RK, Molecular mechanisms and clinical applications of angiogenesis., Nature. 473 (2011) 298–307. 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rafii S, Butler JM, Ding B-S, Angiocrine functions of organ-specific endothelial cells, Nature. 529 (2016) 316–325. 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Betz C, Lenard A, Belting H-G, Affolter M, Cell behaviors and dynamics during angiogenesis, Development. 143 (2016) 2249–2260. 10.1242/dev.135616. [DOI] [PubMed] [Google Scholar]
- [9].Cleaver O, Melton DA, Endothelial signaling during development, Nat. Med. 9 (2003) 661–668. 10.1038/nm0603-661. [DOI] [PubMed] [Google Scholar]
- [10].Zhang B, Korolj A, Lai BFL, Radisic M, Advances in organ-on-a-chip engineering, Nat. Rev. Mater. 3 (2018) 257–278. 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- [11].Haddrick M, Simpson PB, Organ-on-a-chip technology: turning its potential for clinical benefit into reality, Drug Discov. Today. 24 (2019) 1217–1223. 10.1016/j.drudis.2019.03.011. [DOI] [PubMed] [Google Scholar]
- [12].Pollet AMAO, den Toonder JMJ, Recapitulating the Vasculature Using Organ-On-Chip Technology, Bioengineering. 7 (2020) 17. 10.3390/bioengineering7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lin DSY, Guo F, Zhang B, Modeling organ-specific vasculature with organ-on-a-chip devices, Nanotechnology. 30 (2019) 024002. 10.1088/1361-6528/aae7de. [DOI] [PubMed] [Google Scholar]
- [14].Lorthois S, Lauwers F, Cassot F, Tortuosity and other vessel attributes for arterioles and venules of the human cerebral cortex, Microvasc. Res 91 (2014) 99–109. 10.1016/j.mvr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- [15].Jambusaria A, Hong Z, Zhang L, Srivastava S, Jana A, Toth PT, Dai Y, Malik AB, Rehman J, Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation, ELife. 9 (2020) e51413. 10.7554/eLife.51413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marcu R, Choi YJ, Xue J, Fortin CL, Wang Y, Nagao RJ, Xu J, MacDonald JW, Bammler TK, Murry CE, Muczynski K, Stevens KR, Himmelfarb J, Schwartz SM, Zheng Y, Human Organ-Specific Endothelial Cell Heterogeneity, IScience. 4 (2018) 20–35. 10.1016/j.isci.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cleuren ACA, van der Ent MA, Jiang H, Hunker KL, Yee A, Siemieniak DR, Molema G, Aird WC, Ganesh SK, Ginsburg D, The in vivo endothelial cell translatome is highly heterogeneous across vascular beds, Proc. Natl. Acad. Sci. 116 (2019) 23618–23624. 10.1073/pnas.1912409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aird WC, Endothelial Cell Heterogeneity, Cold Spring Harb. Perspect. Med 2 (2012). 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aird William C, Phenotypic Heterogeneity of the Endothelium, Circ. Res. 100 (2007) 158–173. 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- [20].Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, Rautou P-E, Liver sinusoidal endothelial cells: Physiology and role in liver diseases, J. Hepatol. 66 (2017) 212–227. 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- [21].Kopp H-G, Avecilla ST, Hooper AT, Rafii S, The Bone Marrow Vascular Niche: Home of HSC Differentiation and Mobilization, Physiology. 20 (2005) 349–356. 10.1152/physiol.00025.2005. [DOI] [PubMed] [Google Scholar]
- [22].Inra CN, Zhou BO, Acar M, Murphy MM, Richardson J, Zhao Z, Morrison SJ, A perisinusoidal niche for extramedullary haematopoiesis in the spleen, Nature. 527 (2015) 466–471. 10.1038/nature15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Alabi RO, Farber G, Blobel CP, Intriguing Roles for Endothelial ADAM10/Notch Signaling in the Development of Organ-Specific Vascular Beds, Physiol. Rev. 98 (2018) 2025–2061. 10.1152/physrev.00029.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chiu JJ, Chien S, Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives, Physiol. Rev. 91 (2011) 327–387. 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chien S, Mechanotransduction and endothelial cell homeostasis: the wisdom of the cell, Am. J. Physiol.-Heart Circ. Physiol 292 (2007) H1209–H1224. 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- [26].Chen Z, Tzima E, PECAM-1 is necessary for flow-induced vascular remodeling, Arterioscler. Thromb. Vase. Biol. 29 (2009) 1067–1073. 10.1161/ATVBAHA.109.186692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chiu JJ, Wang DL, Usami S, Chien S, Skalak R, Effects of disturbed flow on endothelial cells, J. Biomech. Eng. 120 (1998) 2–8. 10.1115/1.2834303. [DOI] [PubMed] [Google Scholar]
- [28].Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA, A mechanosensory complex that mediates the endothelial cell response to fluid shear stress, Nature. 437 (2005) 426–431. 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- [29].Coon BG, Baeyens N, Han J, Budatha M, Ross TD, Fang JS, Yun S, Thomas JL, Schwartz MA, Intramembrane binding of VE-cadherin to VEGFR2 and VEGFR3 assembles the endothelial mechanosensory complex, J. Cell Biol. 208 (2015) 975–986. 10.1083/jcb.201408103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Conway DE, Coon BG, Budatha M, Arsenovic PT, Orsenigo F, Wessel F, Zhang J, Zhuang Z, Dejana E, Vestweber D, Schwartz MA, VE-Cadherin Phosphorylation Regulates Endothelial Fluid Shear Stress Responses through the Polarity Protein LGN, Curr. Biol. 27 (2017) 2219–2225.e5. 10.1016/j.cub.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hahn C, Schwartz MA, Mechanotransduction in vascular physiology and atherogenesis, Nat. Rev. Mol. Cell Biol. 10 (2009) 53–62. 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Baeyens N, Schwartz MA, Biomechanics of vascular mechanosensation and remodeling, Mol. Biol. Cell. 27 (2016) 7–11. 10.1091/mbc.E14-11-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pries AR, Neuhaus D, Gaehtgens P, Blood viscosity in tube flow: dependence on diameter and hematocrit., Am. J. Physiol. 263 (1992) H1770–8. 10.1152/ajpheart.1992.263.6.H1770. [DOI] [PubMed] [Google Scholar]
- [34].Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A, Piezol and Piezo2 are essential components of distinct mechanically activated cation channels, Science. 330 (2010) 55–60. 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dumont O, Loufrani L, Henrion D, Key role of the NO-pathway and matrix metalloprotease-9 in high blood flow-induced remodeling of rat resistance arteries, Arterioscler. Thromb. Vase. Biol. 27 (2007) 317–324. 10.1161/01.ATV.0000254684.80662.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Min E, Schwartz MA, Translocating transcription factors in fluid shear stress-mediated vascular remodeling and disease, Exp. Cell Res. (2019). 10.1016/J.YEXCR.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chien S, Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell, in: Am. J. Physiol. - Heart Circ. Physiol., Am J Physiol Heart Circ Physiol, 2007. 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- [38].Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM, Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation, Nature. 399 (1999) 601–605. 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- [39].Tarbell JM, Shear stress and the endothelial transport barrier, Cardiovasc. Res. 87 (2010) 320–330. 10.1093/cvr/cvq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jin RC, Loscalzo J, Vascular nitric oxide: formation and function, J. Blood Med. 1 (2010) 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Davis GE, Stratman AN, Sacharidou A, Koh W, Molecular Basis for Endothelial Lumen Formation and Tubulogenesis During Vasculogenesis and Angiogenic Sprouting, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Davis GE, Kim DJ, Meng C-X, Norden PR, Speichinger KR, Davis MT, Smith AO, Bowers SLK, Stratman AN, Control of vascular tube morphogenesis and maturation in 3D extracellular matrices by endothelial cells and pericytes, Methods Mol. Biol. Clifton NJ. 1066 (2013) 17–28. 10.1007/978-1-62703-604-7_2. [DOI] [PubMed] [Google Scholar]
- [43].Kamei M, Saunders WB, Bayless KJ, Dye L, Davis GE, Weinstein BM, Endothelial tubes assemble from intracellular vacuoles in vivo., Nature. 442 (2006) 453–456. 10.1038/nature04923. [DOI] [PubMed] [Google Scholar]
- [44].Xu K, Cleaver O, Tubulogenesis during blood vessel formation, Semin. Cell Dev. Biol. 22 (2011) 993–1004. 10.1016/j.semcdb.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sacharidou A, Koh W, Stratman AN, Mayo AM, Fisher KE, Davis GE, Endothelial lumen signaling complexes control 3D matrix-specific tubulogenesis through interdependent Cdc42- and MT1-MMP-mediated events, Blood. 115 (2010) 5259–5269. 10.1182/blood-2009-11-252692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stratman AN, Davis MJ, Davis GE, VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines, Blood. 117 (2011) 3709–3719. 10.1182/blood-2010-11-316752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stratman AN, Malotte KM, Mahan RD, Davis MJ, Davis GE, Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation, Blood. 114(2009) 5091–5101. 10.1182/blood-2009-05-222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Whisler JA, Chen MB, Kamm RD, Control of perfusable microvascular network morphology using a multiculture microfluidic system, Tissue Eng. Part C Methods. 20 (2014) 543–552. 10.1089/ten.TEC.2013.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jeon JS, Bersini S, Whisler JA, Chen MB, Dubini G, Charest JL, Moretti M, Kamm RD, Generation of 3D functional microvascular networks with mural cell-differentiated human mesenchymal stem cells in microfluidic vasculogenesis systems, Integr. Biol. Quant. Biosci. Nano Macro 6 (2014) 555–563. 10.1039/c3ib40267c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hsu Y-H, Moya ML, Abiri P, Hughes CCW, George SC, Lee AP, Full range physiological mass transport control in 3D tissue cultures., Lab. Chip. 13 (2013) 81–9. 10.1039/c2lc40787f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LYN, George SC, Lee AP, Hughes CCW, A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications, Lab. Chip. 17 (2017) 511–520. 10.1039/C6LC01422D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Phan DT, Bender RHF, Andrejecsk JW, Sobrino A, Hachey SJ, George SC, Hughes CC, Blood-brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood-central nervous system interface, Exp. Biol. Med. 242 (2017) 1669–1678. 10.1177/1535370217694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hachey SJ, Movsesyan S, Nguyen QH, Burton-Sojo G, Tankanzyan A, Wu J, Hoang T, Hatch MM, Zhao D, Celaya E, Gomez S, Chen GT, Davis RT, Nee K, Pervolarakis N, Lawson DA, Kessenbrock K, Lee AP, Waterman ML, Hughes CCW, An In Vitro Vascularized Micro-Tumor Model of Human Colorectal Cancer Recapitulates In Vivo Drug Responses, BioRxiv. (2020) 2020.03.03.973891. 10.1101/2020.03.03.973891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu Y, Sakolish C, Chen Z, Phan DTT, Bender RHF, Hughes CCW, Rusyn I, Human in vitro vascularized micro-organ and micro-tumor models are reproducible organ-on-a-chip platforms for studies of anticancer drugs, Toxicology. 445 (2020) 152601. 10.1016/j.tox.2020.152601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Palikuqi B, Nguyen DHT, Li G, Schreiner R, Pellegata AF, Liu Y, Redmond D, Geng F, Lin Y, Gomez-Salinero JM, Yokoyama M, Zumbo P, Zhang T, Kunar B, Witherspoon M, Han T, Tedeschi AM, Scottoni F, Lipkin SM, Dow L, Elemento O, Xiang JZ, Shido K, Spence JR, Zhou QJ, Schwartz RE, De Coppi P, Rabbany SY, Rafii S, Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis, Nature. 585 (2020) 426–432. 10.1038/s41586-020-2712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD, Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation., Proc. Natl. Acad. Sci. U. S. A. 112 (2015) 214–9. 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Campisi M, Shin Y, Osaki T, Hajal C, Chiono V, Kamm RD, 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes, Biomaterials. 180 (2018) 117–129. 10.1016/j.biomaterials.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cochrane A, Albers HJ, Passier R, Mummery CL, van den Berg A, Orlova VV, van der Meer AD, Advanced in vitro models of vascular biology: Human induced pluripotent stem cells and organ-on-chip technology, Adv. Drug Deliv. Rev. 140 (2019) 68–77. 10.1016/j.addr.2018.06.007. [DOI] [PubMed] [Google Scholar]
- [59].Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, Xia Y, Florido MHC, He W, Pan W, Prummer M, Warren CR, Jakob-Roetne R, Certa U, Jagasia R, Freskgard PO, Adatto I, Kling D, Huang P, L.l. Zon, E.L. Chaikof, R.E. Gerszten, M. Graf, R. lacone, C.A. Cowan, Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells, Nat. Cell Biol. 17 (2015) 994–1003. 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Xie C, Ritchie RP, Huang H, Zhang J, Chen YE, Underlying Molecular Mechanisms, 31 (2012) 1485–1494. 10.1161/ATVBAHA.110.221101.Smooth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wimmer RA, Leopoldi A, Aichinger M, Kerjaschki D, Penninger JM, Generation of blood vessel organoids from human pluripotent stem cells, Nat. Protoc 14 (2019) 3082–SI00. 10.1038/S41596-019-0213-z. [DOI] [PubMed] [Google Scholar]
- [62].Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hammerle M, Esk C, Bagley JA, Lindenhofer D, Chen G, Boehm M, Agu CA, Yang F, Fu B, Zuber J, Knoblich JA, Kerjaschki D, Penninger JM, Human blood vessel organoids as a model of diabetic vasculopathy, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM, Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2, Cell. 181 (2020) 905–913.e7. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim J, Koo B-K, Knoblich JA, Human organoids: model systems for human biology and medicine, Nat. Rev. Mol. Cell Biol. 21 (2020) 571–584. 10.1038/S41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Low JH, Li P, Chew EGY, Zhou B, Suzuki K, Zhang T, Lian MM, Liu M, Aizawa E, Rodriguez Esteban C, Yong KSM, Chen Q, Campistol JM, Fang M, Khor CC, Foo JN, Izpisua Belmonte JC, Xia Y, Generation of Human PSC-Derived Kidney Organoids with Patterned Nephron Segments and a De Novo Vascular Network, Cell Stem Cell. 25 (2019) 373–387.e9. 10.1016/j.stem.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Holloway EM, Wu JH, Czerwinski M, Sweet CW, Wu A, Tsai YH, Huang S, Stoddard AE, Capeling MM, Glass I, Spence JR, Differentiation of Human Intestinal Organoids with Endogenous Vascular Endothelial Cells, Dev. Cell. 54 (2020) 516–528.e7. 10.1016/j.devcel.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, Chapeton K, Patterson B, Yuan Y, He CS, Raredon MSB, Dengelegi J, Kim KY, Sun P, Zhong M, Lee S, Patra P, Hyder F, Niklason LE, Lee SH, Yoon YS, Park IH, Engineering of human brain organoids with a functional vascular-like system, Nat. Methods 16 (2019) 1169–1175. 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R, Flow-enhanced vascularization and maturation of kidney organoids in vitro, Nat. Methods. 16 (2019) 255–262. 10.1038/S41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA, Endothelial fluid shear stress sensing in vascular health and disease, J. Clin. Invest. 126 (2016) 821–828. 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA, Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1, Curr. Biol. 23 (2013) 1024–1030. 10.1016/j.cub.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kadohama T, Nishimura K, Hoshino Y, Sasajima T, Sumpio BE, Effects of different types of fluid shear stress on endothelial cell proliferation and survival, J. Cell. Physiol. 212 (2007) 244–251. 10.1002/jcp.21024. [DOI] [PubMed] [Google Scholar]
- [72].Moretti M, Prina-Mello A, Reid AJ, Barron V, Prendergast PJ, Endothelial cell alignment on cyclically-stretched silicone surfaces, J. Mater. Sci. Mater. Med. 15 (2004) 1159–1164. 10.1023/B:JMSM.0000046400.18607.72. [DOI] [PubMed] [Google Scholar]
- [73].Frangos JA, Eskin SG, Mclntire LV, Ives CL, Flow effects on prostacyclin production by cultured human endothelial cells, Science. 227 (1985) 1477–1479. 10.1126/science.3883488. [DOI] [PubMed] [Google Scholar]
- [74].Wang C, Baker BM, Chen CS, Schwartz MA, Endothelial cell sensing of flow direction, Arterioscler. Thromb. Vase. Biol. 33 (2013) 2130–2136. 10.1161/ATVBAHA.113.301826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kalucka J, De Rooij LPMH, Goveia J, Li X, Luo Y, Single-Cell Transcriptome Atlas of Murine Endothelial Cells Characterization of inter-and intra-tissue EC heterogeneity d Discovery tool for characterization of ECs in other datasets, (n.d.). 10.1016/j.cell.2020.01.015. [DOI] [Google Scholar]
- [76].Chavkin NW, Hirschi KK, Single Cell Analysis in Vascular Biology, Front. Cardiovasc. Med 7 (2020). 10.3389/fcvm.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Qiu J, Hirschi KK, Endothelial cell development and its application to regenerative medicine, Circ. Res. 125 (2019) 489–501. 10.1161/CIRCRESAHA.119.311405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mandrycky C, Hadland B, Zheng Y, 3D curvature-instructed endothelial flow response and tissue vascularization, Sci. Adv 6 (2020) 3629–3645. 10.1126/sciadv.abb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gerhardt H, VEGF and endothelial guidance in angiogenic sprouting, Organogenesis. 4 (2008) 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ribatti D, Crivellato E, “Sprouting angiogenesis”, a reappraisal, Dev. Biol. 372 (2012) 157–165. 10.1016/j.ydbio.2012.09.018. [DOI] [PubMed] [Google Scholar]
- [81].Rouwkema J, Khademhosseini A, Vascularization and Angiogenesis in Tissue Engineering: Beyond Creating Static Networks, Trends Biotechnol. 34 (2016) 733–745. 10.1016/j.tibtech.2016.03.002. [DOI] [PubMed] [Google Scholar]
- [82].Saberianpour S, Heidarzadeh M, Geranmayeh MH, Hosseinkhani H, Rahbarghazi R, Nouri M, Tissue engineering strategies for the induction of angiogenesis using biomaterials, J. Biol. Eng 12 (2018). 10.1186/s13036-018-0133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tien J, Microfluidic approaches for engineering vasculature, Curr. Opin. Chem. Eng. 3 (2014) 36—41. 10.1016/j.coche.2013.10.006. [DOI] [Google Scholar]
- [84].Kim S, Kim W, Lim S, Jeon JS, Vasculature-On-A-Chip for In Vitro Disease Models, Bioengineering. 4 (2017). 10.3390/bioengineering4010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chung S, Sudo R, Vickerman V, Zervantonakis IK, Kamm RD, Microfluidic Platforms for Studies of Angiogenesis, Cell Migration, and Cell–Cell Interactions, Ann. Biomed. Eng. 38 (2010) 1164–1177. 10.1007/s10439-010-9899-3. [DOI] [PubMed] [Google Scholar]
- [86].Nguyen D-HT, Stapleton SC, Yang MT, Cha SS, Choi CK, a Galie P, Chen CS, Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro., Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 6712–7. 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wong KHK, Chan JM, Kamm RD, Tien J, Microfluidic Models of Vascular Functions, Annu. Rev. Biomed. Eng 14 (2012) 205–230. 10.1146/annurev-bioeng-071811-150052. [DOI] [PubMed] [Google Scholar]
- [88].Song JW, Munn LL, Fluid forces control endothelial sprouting., Proc. Natl. Acad. Sci. U. S. A. 108 (2011) 15342–7. 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kim S, Chung M, Ahn J, Lee S, Jeon NL, Interstitial flow regulates the angiogenic response and phenotype of endothelial cells in a 3D culture model, Lab. Chip 16 (2016) 4189–4199. 10.1039/C6LC00910G. [DOI] [PubMed] [Google Scholar]
- [90].Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, Hirschi KK, Chen CS, A non-canonical Notch complex regulates adherens junctions and vascular barrier function, Nature. 552 (2017) 258. 10.1038/nature24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Fang JS, Coon BG, Gillis N, Chen Z, Giu J, Chittenden TW, Burt JM, Schwartz MA, Hirschi KK, Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification, Nat. Commun 8 (2017) 1–14. 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mack JJ, Mosqueiro TS, Archer BJ, Jones WM, Sunshine H, Faas GC, Briot A, Aragon RL, Su T, Romay MC, McDonald AI, Kuo C-H, Lizama CO, Lane TF, Zovein AC, Fang Y, Tarling EJ, de Aguiar Vallim TG, Navab M, Fogelman AM, Bouchard LS, Iruela-Arispe ML, NOTCH1 is a mechanosensor in adult arteries, Nat. Commun 8 (2017) 1620. 10.1038/s41467-017-01741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rayner SG, Zheng Y, Engineered Microvessels for the Study of Human Disease, J. Biomech. Eng. 138 (2016) 110801. 10.1115/1.4034428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Griffith LG, Naughton G, Tissue engineering - Current challenges and expanding opportunities, Science. 295 (2002). 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- [95].Pober JS, Sessa WC, Evolving functions of endothelial cells in inflammation, Nat. Rev. Immunol. 7 (2007) 803–815. 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- [96].Yau JW, Teoh H, Verma S, Endothelial cell control of thrombosis, BMC Cardiovasc. Disord. 15(2015) 130. 10.1186/s12872-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rodrigues M, Kosaric N, Bonham CA, Gurtner GC, Wound Healing: A Cellular Perspective, Physiol. Rev. 99 (2019) 665–706. 10.1152/physrev.00067.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Song H-HG, Rumma RT, Ozaki CK, Edelman ER, Chen CS, Vascular Tissue Engineering: Progress, Challenges, and Clinical Promise, Cell Stem Cell. 22 (2018) 340–354. 10.1016/j.stem.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fleischer S, Tavakol DN, Vunjak - Novakovic G, From Arteries to Capillaries: Approaches to Engineering Human Vasculature, Adv. Funct. Mater. 30 (2020) 1910811. 10.1002/adfm.201910811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Reconstituting organ-level lung functions on a chip., Science. 328 (2010) 1662–8. 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, Hamilton GA, Ingber DE, Microfabrication of human organs-on-chips, Nat. Protoc 8 (2013) 2135–2157. 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- [102].Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, Levy O, Gregory KE, Breault DT, Cabral JMS, Kasper DL, Novak R, Ingber DE, A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip, Nat. Biomed. Eng 3 (2019) 520–531. 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, Roberts K, Chung S, Novak R, Ingram M, Fatanat-Didar T, Koshy S, Weaver JC, Church GM, Ingber DE, Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip., Nat. Biomed. Eng 1 (2017). 10.1038/s41551-017-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A, Maoz BM, Jeanty SSF, Somayaji MR, Burt M, Calamari E, Chalkiadaki A, Cho A, Choe Y, Chou DB, Cronce M, Dauth S, Divio T, Fernandez-Alcon J, Ferrante T, Ferrier J, FitzGerald EA, Fleming R, Jalili-Firoozinezhad S, Grevesse T, Goss JA, Hamkins-lndik T, Henry O, Hinojosa C, Huffstater T, Jang KJ, Kujala V, Leng L, Mannix R, Milton Y, Nawroth J, Nestor BA, Ng CF, O’Connor B, Park TE, Sanchez H, Sliz J, Sontheimer-Phelps A, Swenor B, Thompson G, Touloumes GJ, Tranchemontagne Z, Wen N, Yadid M, Bahinski A, Hamilton GA, Levner D, Levy O, Przekwas A, Prantil-Baun R, Parker KK, Ingber DE, Robotic fluidic coupling and interrogation of multiple vascularized organ chips, Nat. Biomed. Eng 4 (2020) 407–420. 10.1038/s41551-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nguyen DHT, Lee E, Alimperti S, Norgard RJ, Wong A, Lee JJK, Eyckmans J, Stanger BZ, Chen CS, A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling, Sci. Adv 5 (2019). 10.1126/sciadv.aav6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD, In vitro microvessels for the study of angiogenesis and thrombosis., Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 9342–7. 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kotha SS, Hayes BJ, Phong KT, Redd MA, Bomsztyk K, Ramakrishnan A, Torok-Storb B, Zheng Y, Engineering a multicellular vascular niche to model hematopoietic cell trafficking., Stem Cell Res. Ther. 9 (2018) 77. 10.1186/s13287-018-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Redd MA, Zeinstra N, Qin W, Wei W, Martinson A, Wang Y, Wang RK, Murry CE, Zheng Y, Patterned human microvascular grafts enable rapid vascularization and increase perfusion in infarcted rat hearts., Nat. Commun. 10 (2019) 584. 10.1038/s41467-019-08388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rayner SG, Phong KT, Xue J, Lih D, Shankland SJ, Kelly EJ, Himmelfarb J, Zheng Y, Reconstructing the Human Renal Vascular-Tubular Unit In Vitro., Adv. Healthc. Mater 7 (2018) e1801120. 10.1002/adhm.201801120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Masse S, Kim J, Reis L, Momen A, Nunes SS, Wheeler AR, Nanthakumar K, Keller G, Sefton MV, Radisic M, Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis, Nat. Mater. 15 (2016) 669–678. 10.1038/nmat4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen A. a., Galie P. a., Yu X, Chaturvedi R, Bhatia SN, Chen CS, Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues, Nat. Mater 11 (2012) 768–774. 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lin NYC, Homan KA, Robinson SS, Kolesky DB, Duarte N, Moisan A, Lewis JA, Renal reabsorption in 3D vascularized proximal tubule models, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 5399–5404. 10.1073/pnas.1815208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yemeni S, Bliley JM, Campbell PG, Feinberg AW, 3D bioprinting of collagen to rebuild components of the human heart., Science. 365 (2019) 482–487. 10.1126/science.aav9051. [DOI] [PubMed] [Google Scholar]
- [114].Heintz KA, Bregenzer ME, Mantle JL, Lee KH, West JL, Slater JH, Fabrication of 3D Biomimetic Microfluidic Networks in Hydrogels., Adv. Healthc. Mater 5 (2016) 2153–60. 10.1002/adhm.201600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Arakawa C, Gunnarsson C, Howard C, Bernabeu M, Phong K, Yang E, DeForest CA, Smith JD, Zheng Y, Biophysical and biomolecular interactions of malaria-infected erythrocytes in engineered human capillaries, Sci. Adv 6 (2020). 10.1126/sciadv.aay7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF, Lam WA, Microvasculature-on-a-chip for the long-term study of endothelial barrier dysfunction and microvascular obstruction in disease, Nat. Biomed. Eng. 2 (2018) 453–463. 10.1038/s41551-018-0224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hesh CA, Qiu Y, Lam WA, Vascularized Microfluidics and the Blood-Endothelium Interface, Micromachines. 11 (2019). 10.3390/mi11010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Moses SR, Adorno JJ, Palmer AF, Song JW, Vessel-on-a-chip models for studying microvascular physiology, transport, and function in vitro, Am. J. Physiol. Cell Physiol. 320 (2021) C92–C105. 10.1152/ajpcell.00355.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zheng Y, Chen J, Lopez JA, Flow-driven assembly of VWF fibres and webs in in vitro microvessels., Nat. Commun. 6 (2015) 7858. 10.1038/ncomms8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Bernabeu M, Gunnarsson C, Vishnyakova M, Howard CC, Nagao RJ, Avril M, Taylor TE, Seydel KB, Zheng Y, Smith JD, Binding Heterogeneity of Plasmodium falciparum to Engineered 3D Brain Microvessels Is Mediated by EPCR and ICAM-1, MBio. 10(2019). 10.1128/mBio.00420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Koff RS, Schimmel EM, Energy metabolism: Tissue determinants and cellular corollaries. Edited by J.M. Kinney and H.N. Tucker. 562 pp. New York: Raven Press, 1992. $56, Hepatology. 17 (1993) 347–347. 10.1002/hep.1840170231. [DOI] [Google Scholar]
- [122].Palpant NJ, Pabon L, Friedman CE, Roberts M, Hadland B, Zaunbrecher RJ, Bernstein I, Zheng Y, Murry CE, Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells, Nat. Protoc 12 (2017) 15–31. 10.1038/nprot.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Roberts MA, Tran D, Coulombe KLK, Razumova M, Regnier M, Murry CE, Zheng Y, Stromal Cells in Dense Collagen Promote Cardiomyocyte and Microvascular Patterning in Engineered Human Heart Tissue, Tissue Eng. Part A. 22 (2016) 633–644. 10.1089/ten.tea.2015.0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Lee CS, Leong KW, Advances in microphysiological blood-brain barrier (BBB) models towards drug delivery, Curr. Opin. Biotechnol. 66 (2020) 78–87. 10.1016/j.copbio.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Petrosyan A, Cravedi P, Villani V, Angeletti A, Manrique J, Renieri A, De Filippo RE, Perin L, Da Sacco S, A glomerulus-on-a-chip to recapitulate the human glomerular filtration barrier, Nat. Commun. 10 (2019) 3656. 10.1038/s41467-019-11577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R, Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening, Trends Biotechnol. 34 (2016) 156–170. 10.1016/j.tibtech.2015.11.001. [DOI] [PubMed] [Google Scholar]
- [127].Zamani M, Karaca E, Huang NF, Multicellular Interactions in 3D Engineered Myocardial Tissue, Front. Cardiovasc. Med 5 (2018). 10.3389/fcvm.2018.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chen H, Zhang A, Wu JC, Harnessing cell pluripotency for cardiovascular regenerative medicine, Nat. Biomed. Eng 2 (2018) 392–398. 10.1038/s41551-018-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Blanchard JW, Bula M, Davila-Velderrain J, Akay LA, Zhu L, Frank A, Victor MB, Bonner JM, Mathys H, Lin Y-T, Ko T, Bennett DA, Cam HP, Kellis M, Tsai L-H, Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes, Nat. Med. 26 (2020) 952–963. 10.1038/s41591-020-0886-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Qian T, Maguire SE, Canfield SG, Bao X, Olson WR, Shusta EV, Palecek SP, Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells, Sci. Adv 3 (2017) e1701679. 10.1126/sciadv.1701679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lu TM, Barcia Duran JG, Houghton S, Rafii S, Redmond D, Lis R, Human Induced Pluripotent Stem Cell-Derived Brain Endothelial Cells: Current Controversies, Front. Physiol 12 (2021). 10.3389/fphys.2021.642812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ligresti G, Nagao RJ, Xue J, Choi YJ, Xu J, Ren S, Aburatani T, Anderson SK, MacDonald JW, Bammler TK, Schwartz SM, Muczynski KA, Duffield JS, Himmelfarb J, Zheng Y, A Novel Three-Dimensional Human Peritubular Microvascular System, J. Am. Soc. Nephrol. 27 (2016) 2370–2381. 10.1681/ASN.2015070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bovard D, Sandoz A, Luettich K, Frentzel S, Iskandar A, Marescotti D, Trivedi K, Guedj E, Dutertre Q, Peitsch MC, Hoeng J, A lung/liver-on-a-chip platform for acute and chronic toxicity studies, Lab. Chip 18 (2018) 3814–3829. 10.1039/C8LC01029C. [DOI] [PubMed] [Google Scholar]
- [134].Norona LM, Nguyen DG, Gerber DA, Presnell SC, LeCluyse EL, Editor’s Highlight: Modeling Compound-Induced Fibrogenesis In Vitro Using Three-Dimensional Bioprinted Human Liver Tissues, Toxicol. Sci. 154 (2016) 354–367. 10.1093/toxsci/kfw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Norona LM, Nguyen DG, Gerber DA, Presnell SC, Mosedale M, Watkins PB, Bioprinted liver provides early insight into the role of Kupffer cells in TGF-β1 and methotrexate-induced fibrogenesis, PLOS ONE. 14 (2019) e0208958. 10.1371/journal.pone.0208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Freag MS, Namgung B, Fernandez MER, Gherardi E, Sengupta S, Jang HL, Human Nonalcoholic Steatohepatitis on a Chip, Hepatol. Commun 5 (2021) 217–233. 10.1002/hep4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].March S, Hui EE, Underhill GH, Khetani S, Bhatia SN, Microenvironmental regulation of the sinusoidal endothelial cell phenotype in vitro., Hepatol. Baltim. Md 50 (2009) 920–8. 10.1002/hep.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C, A molecular atlas of cell types and zonation in the brain vasculature, Nature. 554 (2018) 475–480. 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- [139].Meng H, Tutino VM, Xiang J, Siddiqui A, High WSS or Low WSS? Complex Interactions of Hemodynamics with Intracranial Aneurysm Initiation, Growth, and Rupture: Toward a Unifying Hypothesis, Am. J. Neuroradiol 35 (2014) 1254–1262. 10.3174/ajnr.A3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Davis George E, Senger Donald R, Endothelial Extracellular Matrix, Circ. Res. 97 (2005) 1093–1107. 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- [141].Saldin LT, Cramer MC, Velankar SS, White LJ, Badylak SF, Extracellular matrix hydrogels from decellularized tissues: Structure and function, Acta Biomater. 49 (2017) 1–15. 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Nagao RJ, Xu J, Luo P, Xue J, Wang Y, Kotha S, Zeng W, Fu X, Himmelfarb J, Zheng Y, Decellularized Human Kidney Cortex Hydrogels Enhance Kidney Microvascular Endothelial Cell Maturation and Quiescence, Tissue Eng. Part A. 22 (2016) 1140–1150. 10.1089/ten.tea.2016.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Williamson A, Singh S, Fernekorn U, Schober A, The future of the patient-specific Body-on-a-chip, Lab. Chip. 13 (2013) 3471–3480. 10.1039/c3lc50237f. [DOI] [PubMed] [Google Scholar]
- [144].Jang S, Collin de I’Hortet A, Soto-Gutierrez A, Induced Pluripotent Stem Cell-Derived Endothelial Cells, Am. J. Pathol. 189 (2019) 502–512. 10.1016/j.ajpath.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Nagao RJ, Marcu R, Wang Y, Wang L, Arakawa C, DeForest C, Chen J, Lopez JA, Zheng Y, Transforming Endothelium with Platelet-Rich Plasma in Engineered Microvessels, Adv. Sci 6 (2019) 1901725. 10.1002/advs.201901725. [DOI] [PMC free article] [PubMed] [Google Scholar]


