Abstract
Invariant natural killer T (iNKT) cells are highly conserved innate-like T lymphocytes that originate from CD4+CD8+ double-positive (DP) thymocytes. Here, we report that serine/arginine splicing factor 1 (SRSF1) intrinsically regulates iNKT cell development by directly targeting Myb and balancing the abundance of short and long isoforms. Conditional ablation of SRSF1 in DP cells led to a substantially diminished iNKT cell pool due to defects in proliferation, survival, and TCRα rearrangement. The transition from stage 0 to stage 1 of iNKT cells was substantially blocked, and the iNKT2 subset was notably diminished in SRSF1-deficient mice. SRSF1 deficiency resulted in aberrant expression of a series of regulators that are tightly correlated with iNKT cell development and iNKT2 differentiation, including Myb, PLZF, Gata3, ICOS, and CD5. In particular, we found that SRSF1 directly binds and regulates pre-mRNA alternative splicing of Myb and that the expression of the short isoform of Myb is substantially reduced in SRSF1-deficient DP and iNKT cells. Strikingly, ectopic expression of the Myb short isoform partially rectified the defects caused by ablation of SRSF1. Furthermore, we confirmed that the SRSF1-deficient mice exhibited resistance to acute liver injury upon α-GalCer and Con A induction. Our findings thus uncovered a previously unknown role of SRSF1 as an essential post-transcriptional regulator in iNKT cell development and functional differentiation, providing new clinical insights into iNKT-correlated disease.
Keywords: Invariant natural killer T cell, SRSF1, Development, Function, Alternative splicing
Subject terms: NKT cells, Immunology
Introduction
Invariant natural killer T (iNKT) cells, also named type I or classic NKT cells, are specialized T lymphocytes that paly crucial roles in both innate and adaptive immune responses [1]. These cells express the invariant T cell antigen receptor (TCR) α-chain (Vα14-Jα18 in mice, Vα24-Jα18 in humans) with a limited set of TCRβ-chains [2]. Progenitors from CD4+CD8+ double-positive (DP) thymocytes are positively selected by glycolipids in complex with CD1d, an MHC-class-I-like molecule, resulting in successful lineage commitment of iNKT cells [3–5]. Based on the expression of CD24, CD44, and NK1.1, iNKT cells can be further sequentially subcategorized into four developmental stages: stage 0, stage 1, stage 2, and stage 3 [6, 7]. Given the limitations of this linear stage developmental model, iNKT cells were defined as three functional subsets in a new transcription factor (TF)-based classification, termed iNKT1 (PLZFloRORγtloT-bethi), iNKT2 (PLZFhiRORγtloT-betlo), and iNKT17 (PLZFmedRORγthiT-betlo) [8]. Recently, single cell-based studies of iNKT cells have shown that both the iNKT1 and iNKT2 populations display high heterogeneity [9, 10], suggesting that the complicated regulatory mechanisms of iNKT cell development need to be further disclosed.
iNKT1 cells express the TF T-bet and predominantly secrete interferon-γ (IFNγ), which occurs in all stages of development and is mainly found at stage 3; iNKT2 cells express TF PLZF and Gata3 and produce high levels of interleukin 4 (IL-4) at stage 2; and iNKT17 cells express TF RORγt and primarily secrete interleukin 17 (IL-17) at stage 2 [8]. Upon stimulation, iNKT cells secrete cytokines instantly without the need for prior activation [8, 11]. These cytokines have protective or detrimental roles in autoimmune diseases, infection, and tumor surveillance [12, 13]. Accumulating studies have demonstrated that iNKT cells not only have destructive roles in promoting liver injury but also play a novel role in orchestrating the repair process during sterile injury [14–17].
Over the years, various TFs involved in the development and function of iNKT cells have been well characterized [7, 18–20]. Promyelocytic leukemia zinc finger (PLZF; encoded by Zbtb16) is a master regulator of iNKT cells that is maintained at elevated levels in stages 0–2 [21, 22]. Other TFs, including Myb [23], ICOS [24, 25], Gata3 [26], Egr2 [27], and T-bet [28], also promote iNKT cell development, whereas Klf2 [8, 29], Id2, and Id3 [30, 31] negatively control iNKT cell differentiation. These findings indicate that regulation at the transcriptional layer is responsible for iNKT cell formation. However, post-transcriptional events that drive the iNKT cell development program remain poorly understood.
RNA-binding protein (RBP)-mediated alternative splicing (AS) has been progressively considered a major determinant in controlling gene expression and cell fate by post-transcriptional regulation [32, 33]. Serine/arginine splicing factor 1 (SRSF1) is an archetypical RBP of the serine and arginine-rich (SR) protein family and a key splicing regulator that controls mRNA metabolism, splicing, decay, nuclear export, and translation [34]. Although SRSF1 has been extensively investigated, its function and target specificity in immune cells are still unclear.
In this study, we explored the critical requirement of SRSF1 in iNKT cells by a conditional knockout approach. Our findings reveal that SRSF1 serves as a post-transcriptional regulator of both iNKT cell development and function, indicating the therapeutic potential in iNKT-related immune disorders.
Materials and methods
Mice
Srsf1fl/fl mice were generously provided by Dr. Xiang-Dong Fu (University of California, San Diego). The targeting strategy has been previously described [35]. LCK-Cre (Stock No: 006889) [36] and B6SJL (CD45.1+) mice were obtained from The Jackson Laboratory. The Srsf1fl/fl mice and all strains used in this study had a C57BL/6 genetic background. For phenotypic and functional analysis, the mice were selected from 6- to 7-week-old littermates. For the bone marrow chimeric model, 6- to 10-week-old B6SJL recipients were used, and age matching was ensured for each individual experiment. Mice were kept under specific pathogen-free conditions with controlled temperature (22 ± 1 °C) and humidity (50 ± 10%) and exposed to a constant 12-h light-dark cycle in the animal facilities at China Agricultural University. All institutional and national guidelines for the care and use of laboratory animals were followed.
Cell isolation
Single-cell suspensions were prepared from the thymus and spleen as previously described [37]. For isolation of liver-derived lymphocytes, the liver was thoroughly dissected with RP10 (RPMI 1640 medium containing 10% FBS) and then filtered through a 70-μm cell strainer to obtain a single-cell suspension. Liver lymphocytes were centrifuged at 500 × g for 5 min, pellets were then suspended in 48% Percoll (17089101, Gentihold), and red blood cells were removed with ACK lysis buffer as previously described [38]. Mononuclear cells at the bottom of the solutions were collected for subsequent assays.
Flow cytometry
The following fluorochrome-labeled monoclonal antibodies were used: anti-TCRβ (H57-597), anti-CD24 (M1/69), anti-NK1.1 (PK136), anti-CD3e (145-2C11), anti-CD4 (RM4-5), anti-CD8a (53-6.7), anti-B220 (RA3-6B2), anti-Ly6G (RB6-BC5), anti-CD11b (M1/70), anti-CD11c (N418), anti-Ter119 (TER-119), anti-TCRγδ (GL-3), anti-CD45.2 (104), anti-CD1d (1B1), anti-PLZF (Mags.21F7), anti-RORγt (AFKJS-9), anti-T-bet (4B10), anti-Gata3 (TWAJ), anti-IFNγ (XMG1.2), and anti-IL-4 (11B11) were from eBioscience; anti-CD44 (IM7), anti-CD45.1 (A20), and anti-IL-17A (TC11-18H10) were from BD Biosciences; anti-ICOS (C398.4A) was from BioLegend; and anti-CD5 (53-7.3) was from Abcam. For surface staining, cells were stained with PBS57/CD1d tetramer, followed by staining with the corresponding antibodies in FACS buffer (PBS + 2% FBS). For intranuclear staining of the transcription factors PLZF, T-bet, Gata3, and RORγt, cells were fixed and permeabilized with Foxp3/Transcription Factor Staining Buffer Set (00-5523-00, Thermo Fisher Scientific) following the manufacturer’s instructions. For the analysis of cytokines, including IFNγ, IL-17A, and IL-4, cells were fixed and permeabilized with a Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (554714, BD Biosciences). Samples were acquired with FACSVerse or LSRFortessa (BD Biosciences) following the manufacturer’s instructions. Cell sorting was performed with a FACSAria II sorter (BD Biosciences) as previously described [39]. The flow cytometry data were analyzed with FlowJo software (Version 10.4.0, Tree Star, Inc.).
Detection of cytokines in vitro
For analysis of cytokine secretion of thymic iNKT cells, thymocytes were stimulated in vitro with PMA (100 ng/ml) and ionomycin (50 μg/ml) diluted in RPMI medium supplemented with 10% FBS, GolgiStop (1000×, 554724, BD Biosciences), and GolgiPlug (1000×, 555029, BD Biosciences) for 4 h. Cells were washed, stained with surface markers, and fixed with BD Cytofix/Cytoperm (554722, BD Biosciences), followed by intracellular staining with anti-IFNγ, anti-IL-4, and anti-IL-17A.
In vivo BrdU incorporation and apoptosis assays
For determination of the proliferation rate of iNKT cells, mice were injected i.p. with 1 mg BrdU (B5002, Sigma) in 200 μl of PBS at 0, 24, and 48 h. Then, the mice were euthanized at 72 h post-injection, and single-cell suspensions were collected from relevant tissues [40]. Thymocytes were surface stained with relative antibodies, fixed, and permeabilized as described above. BrdU incorporation was measured using a BrdU Flow Kit (557891, BD Biosciences) according to the manufacturer’s instructions.
For apoptosis detection, single-cell suspensions were stained with surface markers and resuspended in Annexin V binding buffer (BD Biosciences) containing fluorochrome-conjugated Annexin V (BMS306FI/300, eBioscience). Apoptotic cells were measured according to the manufacturer’s instructions.
Bone marrow chimeric mice
Bone marrow chimeras were generated as previously described [37]. Briefly, bone marrow (BM) cells from femurs and tibias were harvested and mixed at a 1:1 ratio with B6SJL (CD45.1+) bone marrow cells as donors for chimeric mice. A total of 2 × 106 donor cells were injected intravenously into lethally irradiated (8 Gray) B6SJL recipients. Chimeric mice were analyzed 9 weeks post-transplantation.
Retroviral transduction
The full-length CDS encoding the short isoform of Myb (Mybp75) or long isoform of Myb (Mybp89) was cloned into the pMigR1 vector for retroviral gene transduction. For the retrovirus package, HEK293T cells were transfected with the indicated plasmids by Lipofectamine 3000 (L3000015, Invitrogen) [41]. Briefly, retroviruses bearing Mybp75, Mybp89, or pMigR1 along with pCL-Eco were packaged with HEK293T cells and collected at 48 and 72 h after transfection. The retrovirus-containing medium was filtered through 0.45 µm filters and loaded onto 24-well nontissue culture plates (351147, Falcon) precoated with 10 μg/well RetroNectin (T100A, TaKaRa). Next, BM cells were isolated from ctrl or LCKCre/+Srsf1fl/fl mice (CD45.2+) and then stained with a biotinylated lineage-positive antibody cocktail (anti-CD45R, anti-Ly6G, anti-Ter119, anti-CD11b, anti-CD11c, anti-NK1.1, anti-CD3e, anti-CD4, anti-CD8, anti-TCRγδ) followed by depletion with DynabeadsTM M-280 Streptavidin (60210, Invitrogen). Lineage-negative BM cells were cultured overnight in IMDM containing 15% FBS, 100 μg/ml streptomycin and penicillin, 50 μM 2-mercaptoethanol, 20 ng/ml TPO (315-14, PeproTech), and 50 ng/ml SCF (250-03, PeproTech) on the plate described above. Then, the cells were centrifuged at 1000 × g for 90 min at 30 °C in the presence of 8 μg/ml polybrene (H9268, Sigma). After spinofection, the cells were cultured at 37 °C in a 5% CO2 incubator for another 2 h and then replaced with full IMDM containing the components and cytokines described above for 24–48 h. The infected cells containing 2000–5000 GFP+ cells were sorted and transplanted into irradiated (7.5 Gray) B6SJL mice. The recipients were analyzed at 8 to 10 weeks post-transplantation.
RNA isolation, reverse transcription, and quantitative RT-PCR
Cells were lysed by TRIzol (Invitrogen), and total RNA was extracted using a modified protocol with an RNeasy Mini Kit (74106, Qiagen) or RNeasy Micro Kit (74004, Qiagen). RNA was then reverse transcribed using a FastKing RT Kit (with gDNase) (KR106, Tiangen) following the manufacturer’s instructions. Relative gene expression levels were analyzed by quantitative RT-PCR (qPCR) with SYBR Green Master Mix (Tiangen) on a CFX96 ConnectTM Real-Time System (Bio-Rad). Hprt1 was used to normalize the expression of other genes, and the 2–ΔΔCT method was used to calculate the levels of target mRNAs. All primers used are listed in Table S2.
TCR rearrangement assay
The 7AAD−CD4+CD8+DP thymocytes were sorted from the thymi of LCKCre/+Srsf1fl/fl and ctrl mice. Total RNA was extracted and reverse transcribed as described above. The cDNA transcripts were assessed by qPCR with the forward primers Vα14, Vα8, and Vα3 paired with the reverse primers Jα56, Jα18, Jα9, or Cα (primer sequences are listed in Table S2). The relative expression of each PCR product (after normalization to Hprt1) in cells from the ctrl mice was arbitrarily set to one, and its relative expression in cells from LCKCre/+Srsf1fl/fl mice was normalized accordingly.
Semiquantitative PCR analysis
The cDNA was subjected to semiquantitative PCR with the defined parameters: the initial denaturation was set at 95 °C for 2 min, followed by the indicated cycles (iNKT, 35 cycles; DP, 33 cycles; cell line, 33 cycles), including denaturation (30 s at 94 °C), annealing (30 s at 59 °C), and extension (30 s at 72 °C). For analysis of the PCR product, 1.5% agarose gels were used to separate the two isoforms, and the levels of the two isoforms were measured by Image Lab Software (Version 5.1, Bio-Rad). Each sample was assayed at least three times to ensure reliability. The primer sequences were as follows: Myb, 5′-CAAGGTGCATGATCGTCCAC-3′ and 5′-GTCTGGTCTCGACATGGTGT-3′.
Splicing detection in vitro by minigene assays
For generation of the Myb minigene plasmid for alternative splicing analysis in vitro, a 3442bp DNA fragment from exon 9 to exon 10 (chr10:21,142,993–21,146,434, mm10) of the Myb gene was amplified directly by PCR from genomic DNA and then cloned into the pcDNA3.1 vector (V79020, Thermo Fisher Scientific) by using the BamHI and XhoI sites. The plasmid containing the mutant binding site of the Myb minigene described above was made by overlap-extension PCR. For generation of the expression plasmid of Srsf1, HA-tagged primer sets were used to amplify the coding region of Srsf1, and the PCR product was cloned into the pcDNA3.1 vector by using the EcoRI and XbaI sites. HEK293T cells were transfected with the Srsf1-expressing plasmid together with the indicated minigene plasmids using Lipofectamine 2000 following the manufacturer’s instructions. Cells were harvested for RNA or protein analysis at 48 h post-transfection. Semiquantitative PCR was performed with primers for amplifying the indicated transcripts, and the desired DNA products were isolated and sequenced for further confirmation.
Western blot
Western blot analyses were carried out as previously described [37]. Briefly, 7AAD−CD4+CD8+DP cells were isolated from the thymi of LCKCre/+Srsf1fl/fl and ctrl mice. Total protein was isolated from cells by lysis with ice-cold RIPA buffer. Equal amounts of protein were resolved by 10% SDS-PAGE (Invitrogen) and transferred onto PVDF membranes using an iBlot (Cat #IPVH00010, Merck Millipore). Membranes were blocked with 5% milk in TBS containing Tween-20 (Sigma-Aldrich) and immunoblotted with anti-Myb (05-175, Sigma) and anti-GAPDH (#5174, Cell Signaling Technology), followed by goat anti-rabbit IgG-HRP (ZB-2301, ZSGB-BIO) secondary antibodies. Protein bands were developed by incubation with Immobilon Western Chemiluminescent HRP Substrate (Cat #WBKLS0100, Merck Millipore).
Murine hepatitis model
For the induced liver injury model, mice were injected i.v. with PBS-dissolved Con A at a sublethal dose of 15 mg/kg or with 30 ng α-GalCer (KRN7000, Enzo Life Science) dissolved in PBS with 0.5% Tween-20 (vehicle). At the indicated time points, the mice were sacrificed, and serum was collected. The levels of IL-4, IFNγ, and TNFα in serum were measured by ELISAs. ALT and AST in the serum from the α-GalCer-induced or Con A-induced mice were measured by an automatic biochemical analyzer (Cobas6000, Roche) according to the manufacturer’s instructions. Liver tissues were fixed in formaldehyde solution and analyzed with H&E (hematoxylin and eosin) staining. For the survival rate, mice were injected i.v. with a lethal dose of Con A (30 mg/kg) and monitored every 30 min after administration.
ELISA
Analysis of the serum levels of cytokines was performed by enzyme-linked immunosorbent assay kits purchased from Biolegend according to the manufacturer’s protocol. Briefly, a standard sandwich ELISA, antibody-antigen-antibody, was used to assess TNFα, IL-4, and IFNγ levels in serum. The serum was coated in a 96-well plate pretreated with capture antibodies, followed by incubation with detection antibodies and avidin-HRP solution. Then, the samples were incubated with TMB Substrate Solution, and the reaction was finally blocked with Stop Solution. The absorbance at 450 nm was measured with a microplate reader (Tecan).
RNA sequencing and data analysis
Total RNA was extracted from the 7AAD−CD4+CD8+ DP cells of LCKCre/+Srsf1fl/fl and ctrl mice. Then, the RNA samples were subjected to paired-end RNA-Seq using an Illumina HiSeq Xten platform. The paired-end reads were assessed for quality using FastQC (version 0.11.5); adaptors and low-quality reads were trimmed by Trimmomatic 0.36. Next, clean reads were mapped to the mm10 mouse reference sequence using STAR (version 0.6.1). Differentially expressed genes (DEGs) were analyzed in R using DESeq2 (version 1.24.0). Significant DEGs were detected according to the following criteria: P < 0.001 and |log2FC| > 0.5. Gene Ontology term enrichment analysis was performed by DAVID 6.7 (https://david.ncifcrf.gov).
For the analysis of alternative splicing, rMATS (version 4.0.2) was used to calculate the five different categories of AS events: skipped exons (SE), retained introns (RI), mutually exclusive exons (MXE), alternative 5′ splice sites (A5SS), and alternative 3′ splice sites (A3SS). Significantly differentially expressed AS events were defined for those with an FDR < 0.0001 and |IncLevelDifference| > 0.05. For visualization purposes, Sashimi plots of the IGV genome browser were used to plot RNA-Seq densities along exons and junctions for multiple samples. RNA-Seq data were deposited in the Gene Expression Omnibus under accession number GSE156013.
scRNA‐Seq data and ImmGen gene expression analysis
Single-cell sequencing data of thymic iNKT cells were downloaded from published scRNA-seq data (GSE152786). Data analysis was performed using functions available in the Seurat (v3.1.4) package of R as described by Krovi et al. [9].
Relative expression of Srsf1 and selected genes in T and iNKT cell populations (Fig. 1A) were analyzed by publicly available data from the Immunological Genome Project (ImmGen) gene expression database (GSE15907).
Fig. 1.
SRSF1 is dynamically expressed in iNKT cell subsets and potentially binds multiple iNKT-related genes. A Analysis of Srsf1 expression in various T cell subsets by published microarray data (GSE15907). B The expression level of Srsf1 in thymic iNKT cells is illustrated in dot plots by published single-cell RNA-seq data (GSE152786). Left: Uniform manifold approximation and projection (UMAP) of iNKT cells was colored by inferred cluster identity; middle: The expression of Srsf1 is plotted along a colorimetric gradient, with red corresponding to high expression; right: Violin plots showing the aggregate expression level of Srsf1 from Cluster 0 to Cluster 10. C Overlapping SRSF1-binding genes in thymocytes (GSE141349) and iNKT-related genes. Sixty overlapping genes were identified, and iNKT-associated regulators in each category are shown
Statistical analysis
Statistical analysis was performed using Prism 7.0 (GraphPad Software), and the error bar is shown as the mean ± SD. For normally distributed data, statistical significance was determined by unpaired two-sided Student’s t test. For analysis of groups that showed differing variance, an unpaired two-sided Welch’s t test was used. For multiple comparisons, statistical analysis was performed by one-way ANOVA. Kaplan–Meier analysis was used, and the log-rank test was employed to determine any significant difference between the survival curves of the two groups. Statistical significance was considered as follows: *P < 0.05; **P < 0.01; ***P < 0.001. NS (nonsignificant) was defined as P ≥ 0.05.
Results
SRSF1 exhibits dynamic expression in iNKT cell subsets and binds iNKT-related genes
We recently reported that conditional ablation of SRSF1 from the DP stage resulted in impaired conventional T cell development. Given that iNKT cells also originate from DP thymocytes, we wondered whether SRSF1 plays a critical role in iNKT cell development and differentiation. We thus examined the expression of Srsf1 from DP and various T cell populations by analyzing published data (GSE15907) from the ImmGen database (Fig. 1A). Compared with other iNKT genes, Srsf1 exhibits dynamic expression in iNKT cells and reaches a peak in the CD44-NK1.1− and CD44+NK1.1− iNKT subsets. Moreover, we performed an Srsf1 expression assay in distinct clusters from recently published single-cell RNA-seq data (GSE152786) [9]. The results indicated that Srsf1 is dynamically expressed and exhibits relatively high abundance in Clusters 0 (stage 0), 1, 2, and 3 (belonging to iNKT2 subsets), which is in accordance with the result from the analysis of the ImmGen data above (Fig. 1B). We further found that 60 genes involved in iNKT cell development and function were directly bound by SRSF1 (Fig. 1C) through overlapping analysis of iNKT cell regulatory genes (collected from published literatures, Table S1) with our CLIP-seq data in thymocytes (GSE141349). These data collectively indicated the potential roles of SRSF1 in regulating iNKT cells.
SRSF1 is essential for iNKT cell development
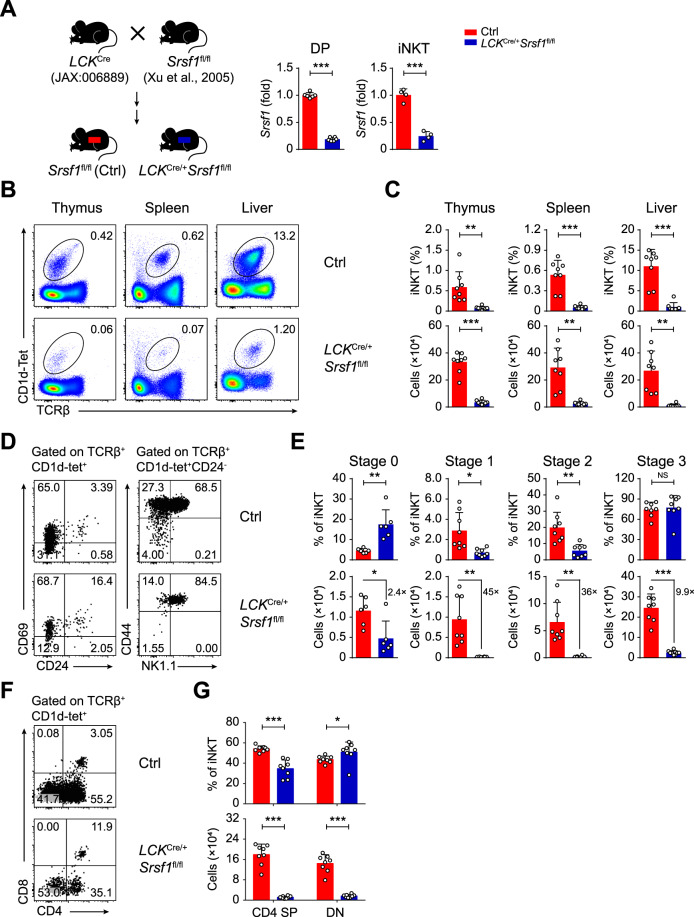
To extensively elucidate the function of SRSF1 in iNKT cells, we bred Srsf1fl/fl mice [35] with LCKCre/+ mice (JAX: 006889) [36] for conditional ablation of SRSF1 from DP thymocytes, which are the origin of iNKT cells. The deletion efficiency was further confirmed in both DP and iNKT cells (Fig. 2A). We next evaluated the iNKT cell populations in the thymus, spleen, and liver from the LCKCre/+Srsf1fl/fl mice and their littermate Srsf1fl/fl controls (henceforth called ctrl). The frequency and numbers of iNKT cells in all these tissues from the SRSF1-deficient mice exhibited a remarkable reduction (Fig. 2B, C). The absolute numbers of stage 0 iNKT cells were significantly decreased in the LCKCre/+Srsf1fl/fl thymi, although the frequency of cells in stage 0 was increased (Fig. 2D, E). However, the frequency and numbers of thymic iNKT cells from stage 1 to stage 2 were significantly decreased in the LCKCre/+Srsf1fl/fl mice. Accordingly, the numbers of stage 3 iNKT cells were also diminished even though their frequency was not changed (Fig. 2D, E). Moreover, the numbers of iNKT cells at stages 1–3 in the periphery, such as spleens and livers, from the LCKCre/+Srsf1fl/fl mice were substantially diminished (Supplementary Fig. 1A, B). Given that stage 2 cells are predominantly CD4+ and stage 3 cells are mostly CD4− [42], we detected CD4 expression accordingly. Reduced frequency and absolute numbers of CD4+ iNKT cells were observed in thymi (Fig. 2F, G), spleens, and livers (Supplementary Fig. 1C, D). Collectively, these data demonstrated that SRSF1 is crucial for the development of iNKT cells.
Fig. 2.
Ablation of SRSF1 impairs the development of iNKT cells. A Schematic graphs showing the strategy for generating the mouse model (left). The source of each strain was marked. The LCKCre/+Srsf1fl/fl mice with conditional ablation of SRSF1 and littermate Srsf1fl/fl mice (ctrl) were used throughout the study. Srsf1 mRNA expression in thymic DP and iNKT cells (right) was analyzed with qPCR. The relative expression of Srsf1 (after normalization to Hprt1) in cells from the ctrl mice was arbitrarily set to one, and its relative expression in cells from the LCKCre/+Srsf1fl/fl mice was normalized accordingly (n ≥ 4). B, C Representative pseudocolor plots (B) showing TCRβ+CD1d-Tet+ iNKT cells from thymi, spleens, and livers. The frequency and numbers of each population are shown in (C) (n = 8). D, E Developmental stage analysis. A representative dot plot (D) shows CD24 and CD69 expression in total thymic iNKT cells; TCRβ+CD1d-Tet+CD24− cells were further analyzed by CD44 and NK1.1. The frequency and numbers of each stage are shown in (E) (n ≥ 6). F, G Flow cytometric analysis of CD4 and CD8 expression among TCRβ+CD1d-Tet+ cells in the thymus of the ctrl and LCKCre/+Srsf1fl/fl mice (F). Frequency and cell numbers of each subset were shown in (G) (n = 8). Data were pooled from at least three independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when the variance between the groups was unequal. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD
SRSF1 modulates iNKT cell development in a cell-intrinsic manner
Next, we found that the expression level of CD1d in DP thymocytes was comparable between the LCKCre/+Srsf1fl/fl and ctrl mice (Supplementary Fig. 2A, B), reflecting that SRSF1 is dispensable for the expression of CD1d in DP thymocytes, which provides a microenvironment to support the positive selection of iNKT cells [7]. We further determined whether SRSF1 intrinsically controls iNKT cell development by using bone marrow chimeric mice as described in Supplementary Fig. 2C. Donor-contributed iNKT cells from recipients were analyzed at 9 weeks post-transplantation. We found that the SRSF1-deficient donor cells developed fewer iNKT cells in the thymus, spleen, and liver than the control cells (Supplementary Fig. 2D, E). In addition, the frequency of iNKT cells derived from the LCKCre/+Srsf1fl/fl donors (presenting as CD45.1−) at stages 1 to 3 in thymi (Supplementary Fig. 2F, G) was substantially reduced compared with those from the ctrl donors, as well as in spleens and livers (Supplementary Fig. 2H, I). Together, these results indicated a cell-intrinsic requirement of SRSF1 for iNKT cell development.
SRSF1 controls the differentiation of iNKT functional subsets
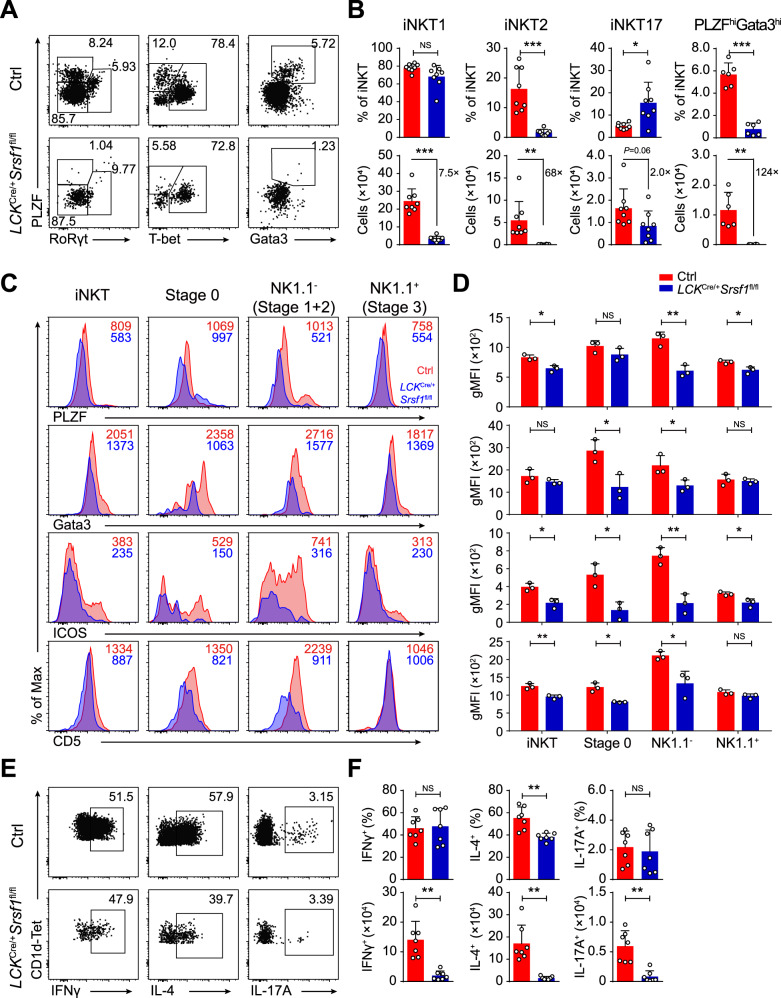
We next investigated whether iNKT functional subsets were affected in the SRSF1-deficient mice. The frequency and numbers of iNKT2 cells in the thymus were significantly impaired in the LCKCre/+Srsf1fl/fl mice (Fig. 3A, B). Although the frequency of iNKT1 cells was not altered and the frequency of iNKT17 cells was significantly increased, the absolute numbers of both subsets were notably reduced in thymi due to diminished total numbers of iNKT cells (Fig. 3A, B). The numbers of iNKT cells in all subsets of spleens from the LCKCre/+Srsf1fl/fl mice were notably diminished (Supplementary Fig. 3). These defects in functional differentiation may be partially caused by reduced expression of the essential regulators PLZF, Gata3, ICOS, and CD5 in relative developmental stages (Fig. 3C, D). Accordingly, the SRSF1-deficient iNKT cells showed severely reduced IL-4 production due to diminished iNKT2 cell proportions after 4 h of stimulation with PMA and ionomycin, but no significant difference in the percentages of IFNγ and IL-17A was observed (Fig. 3E, F). These data demonstrated that SRSF1 is required for the functional differentiation of iNKT cells.
Fig. 3.
SRSF1 is indispensable for iNKT cell functional differentiation. A, B Analysis of functional subsets in thymic iNKT cells. Representative dot plots (A) show iNKT1, iNKT2, iNKT17, and PLZFhiGata3hi cells. The frequency and numbers of the indicated subsets are shown in (B) (n ≥ 6). C, D Representative histograms (C) show the expression of PLZF, Gata3, ICOS, and CD5 in iNKT cells at distinct developmental stages by flow cytometry. The geometric mean fluorescence intensity (gMFI) of PLZF, Gata3, ICOS, and CD5 is shown in (D) (n = 3). E, F Representative dot plots (E) showing IFNγ+, IL-4+, and IL-17A+ populations in thymic iNKT cells stimulated in vitro with PMA and ionomycin for 4 h. Frequency and numbers are shown in (F) (n = 7). Data were pooled from at least two independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when variance between the groups was unequal. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD
SRSF1 is required for iNKT cell proliferation and survival
Given that blunted proliferation or increased apoptosis may account for rare iNKT cells, we then performed proliferation and apoptosis analyses on both DP and iNKT cells. BrdU incorporation in DP thymocytes and total iNKT cells exhibited no obvious differences between the genotypes of mice (Fig. 4A, B). However, when assessing the proportion of BrdU+ cells in distinct iNKT subsets, we found defective proliferation in thymic iNKT cells from stages 1 and 2 to stage 3 (CD24− iNKT cells) but not in stage 0 (CD24+ iNKT cells), suggesting that SRSF1 affected iNKT cell proliferation in stages 1, 2, and 3 (Fig. 4A, B). Based on the Annexin V staining results, the SRSF1-deficient mice not only showed increased apoptosis of DP cells and total iNKT cells but also exhibited elevated apoptosis after stage 0 during iNKT cell differentiation (Fig. 4C, D). Accordingly, the expression of the antiapoptotic genes Bcl2l1, Bcl2, and Mcl1 was lower, whereas the proapoptotic gene Bid was expressed at an aberrantly higher level in the SRSF1-deficient DP cells than in the control cells (Fig. 4E). In addition, we examined the expression of TCR signal response genes, which are also related to iNKT cell survival. The mRNA expression of Zap70, Cd5, and Cd40lg was dramatically decreased in the SRSF1-deficient iNKT cells, but Egr2 and Nr4a1 levels were not altered (Fig. 4F). Thus, our results indicated that SRSF1 maintains the survival of DP thymocytes and iNKT cells from stage 1 to stage 3.
Fig. 4.
SRSF1 deficiency impairs proliferation, survival, and TCRα rearrangement in iNKT and DP cells. A, B Proliferation assay. Representative dot plot (A) and frequency (B) of BrdU+ cells are shown in DP thymocytes and total; stage 0, NK1.1− (stage 1 and 2), and NK1.1+ (stage 3) thymic iNKT cells, respectively (n = 6). C, D Apoptosis assay. Representative histogram (C) and frequency (D) of Annexin V+ cells are shown in DP thymocytes and total; stage 0, NK1.1− (stage 1 and 2), and NK1.1+ (stage 3) thymic iNKT cells, respectively (n = 6). E Apoptosis-related gene expression in DP thymocytes. The relative expression of each transcript was normalized as described in Fig. 2A (n = 6). F Analysis of the expression of genes related to the TCR signaling response in thymic iNKT cells. The relative expression of each transcript was normalized accordingly as described in Fig. 2A (n = 6). G Schematic graph showing selected proximal, central, and distal Vα and Jα segments. H Analysis of transcripts of Vα to Jα rearrangements (constant α-region) segments and Vα14 and Vα3 rearrangements to Jα56, Jα18, or Jα9 by sorted DP thymocytes, presented relative to the expression of Hprt1 transcript (n = 6). I qPCR analysis of rearrangement-related gene expression in DP thymocytes. The relative expression of each transcript was normalized accordingly as described above (n = 6). Data were pooled from at least three independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when variance between the groups was unequal. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD
SRSF1 deficiency impairs TCRα rearrangement
As indicated by the schematic map of TCR Vα and Jα gene loci (Fig. 4G), the canonical iNKT TCR rearrangement joins Vα14 to the distal Jα18 region and requires secondary rearrangements [43]. Increased apoptosis of DP thymocytes may account for impaired distal TCRα rearrangements due to the limited time for the generation of multiple DNA excision circles [44], which would in turn lead to fewer iNKT cells and skewing of the TCR repertoire. We then examined whether TCRα rearrangements were affected in the absence of SRSF1. The results demonstrated that both proximal and distal TCRα rearrangements were impaired in the SRSF1-deficient DP cells (Fig. 4H). Correspondingly, the TCR rearrangement-related gene Dclre1c showed downregulated expression and Dntt showed upregulated expression in the SRSF1-deficient DP thymocytes, but other key regulators, Rag1, Rag2, and Lig4, were not significantly changed (Fig. 4I).
SRSF1 globally alters the transcription of genes in DP cells
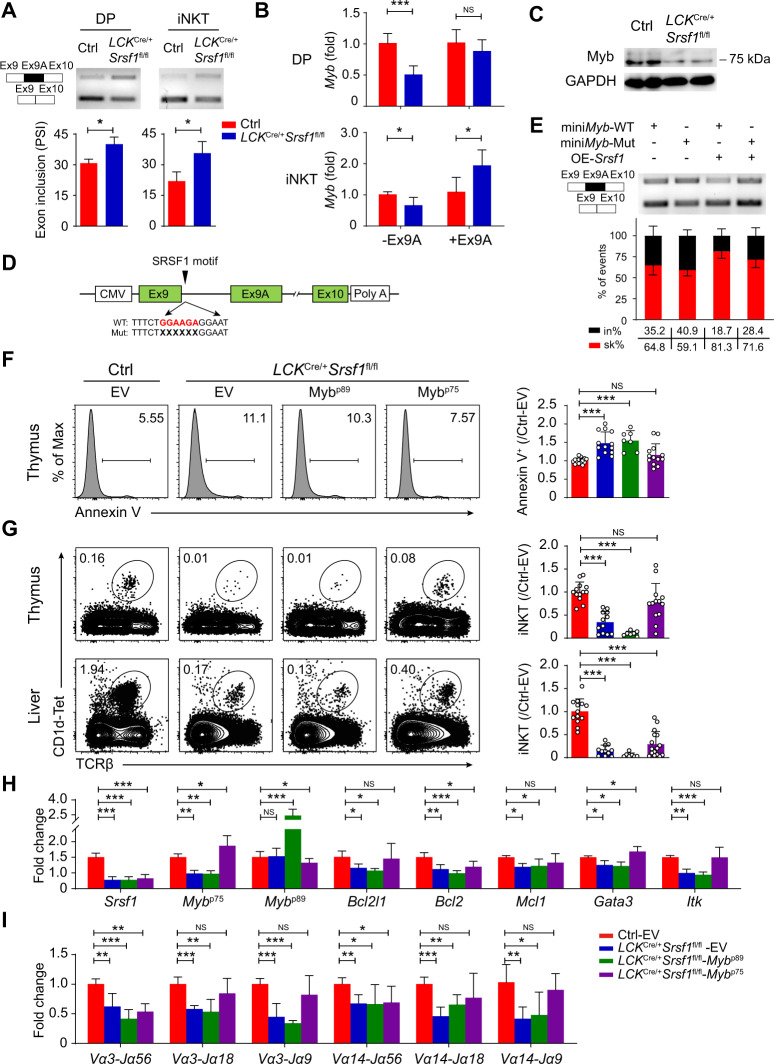
We next explored the underlying mechanism of SRSF1 regulation on iNKT cell development. DP thymocytes sorted from the LCKCre/+Srsf1fl/fl and ctrl mice were used for RNA-seq. A total of 330 genes with upregulated and 579 genes with downregulated expression were identified in the SRSF1-deficient DP thymocytes (Fig. 5A). Gene Ontology (GO) enrichment analysis revealed that these differentially expressed genes (DEGs) were significantly enriched in the regulation of T cell differentiation, apoptosis, T-helper 2 cell differentiation, and RNA splicing (Fig. 5B). Among these DEGs, the expression of Myb, Dclre1c, Itk, and Icos was downregulated in the SRSF1-deficient DP thymocytes (Fig. 5C). We further validated these DEGs by qPCR and observed a significant reduction in Myb expression, which is involved in promoting early stages of iNKT cell development [23], in the SRSF1-deficient DP cells (Fig. 5D). The expression of iNKT-related genes, including the Tec kinases Itk and Txk and the costimulator Icos, was remarkably decreased in the SRSF1-deficient DP cells (Fig. 5D). These data revealed that loss of SRSF1 leads to aberrant expression of a set of gene transcription programs for iNKT cells.
Fig. 5.
SRSF1 modulates the expression of a series of iNKT cell-related genes. A Volcano plot depicting SRSF1-regulated genes in DP thymocytes. Differentially expressed genes (DEGs) were identified from LCKCre/+Srsf1fl/fl versus ctrl samples (blue: downregulated; red: upregulated; gray: unchanged). B Representative Gene Ontology (GO) terms of the biological process categories enriched in DEGs. C Heatmap showing representative DEGs. D Quantification of selected DEGs in DP thymocytes. The relative expression of each transcript was normalized accordingly as described in Fig. 2A (n = 6). Data were pooled from three independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when variance between the groups was unequal. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD. E Bar chart showing SRSF1-regulated alternative splicing (AS) events in DP thymocytes, classified into five categories: skipped exon (SE), retained intron (RI), mutually exclusive exon (MXE), alternative 5’ splice site (A5SS), and alternative 3’ splice site (A3SS). F Venn diagram showing the overlapping genes of DEGs in (A) and AS genes in (E) and SRSF1-binding genes in Fig. 1C. G Analysis of Myb expression and exon-exon junctions. “Sashimi plots” show read coverage and exon-exon junctions (numbers on arches indicate junction reads), and the alternative exons are shaded with yellow columns. The lower panel indicates SRSF1-binding sites identified by CLIP-seq. The black inverted triangle denotes the SRSF1-binding site in introns 9–10 of Myb pre-mRNA. H Analysis of Myb-binding genes (ChIP-seq, GSE66122) correlated with iNKT cells. Red dots represent iNKT cell development-related genes, green dots represent iNKT cell function- and effector differentiation-related genes, and blue dots denote apoptosis-related genes
To elucidate the underlying molecular mechanisms, we then screened SRSF1-controlled alternative splicing (AS) events. A total of 3,231 events, which could be classified into five categories, were identified, and the majority of these AS events were skipped more in the DP thymocytes than the control thymocytes (Fig. 5E). To further strengthen the connection between SRSF1 and activation of the iNKT cell program, we analyzed DEGs from RNA-seq, AS events, and CLIP-seq of SRSF1 and found 111 DEGs with AS that were directly bound and regulated by SRSF1 (Fig. 5F). These findings thus suggested that SRSF1 controls the iNKT cell regulatory gene expression program through both direct and indirect regulatory mechanisms. In particular, Myb mRNA was differently spliced in the absence of SRSF1, reflecting the increased inclusion of exon 9 A (Fig. 5G). Given that Myb deficiency leads to a severe reduction in iNKT cells in the thymus [23], we analyzed the published ChIP-seq data of Myb (GSE66122) and observed that a series of DEGs fall into Myb-binding genes (Fig. 5H and Supplementary Fig. 4A–D), which are related to iNKT cell differentiation, function, and survival. Based on our findings above, we speculated that Myb may serve as an essential downstream target of SRSF1 linked to iNKT cell differentiation.
SRSF1 regulates AS of Myb during iNKT cell development
We next confirmed that SRSF1 deficiency led to increased inclusion of exon 9A in both DP and iNKT cells by semiquantitative PCR (Fig. 6A). Moreover, qPCR results showed that the expression of the Myb short isoform (−Ex9A; encodes the p75 isoform, which is dominantly expressed in hematopoietic cells) was significantly decreased in the SRSF1-deficient DP and iNKT cells, whereas the expression of the Myb long isoform (+Ex9A; encodes the p89 isoform, which is a less abundant transcript) was notably elevated in iNKT cells (Fig. 6B). As a result, the reduced protein level of the p75 isoform in the SRSF1-deficient DP thymocytes was further confirmed by western blots (Fig. 6C). To further examine whether SRSF1 directly regulates the splicing of Myb, we generated a splicing minigene reporter containing Myb pre-mRNA in intron 9 with a WT/mutated SRSF1-binding site (named WT or Mut) (Fig. 6D). Overexpression of SRSF1 led to more skipping of exon 9A of miniMyb-WT (PSI changed from 23.1 to 4.7%), while impaired skipping was detected in exon 9A of miniMyb-Mut (PSI changed from 4.7 to 17.9%) (Fig. 6E). These data strongly supported the notion that SRSF1 directly binds to the alternative region of Myb to control its splicing.
Fig. 6.
SRSF1-mediated alternative splicing of Myb mRNA controls iNKT cell formation. A Semiquantitative PCR validation of AS of Myb in DP and iNKT cells. The structure of PCR products is indicated schematically on the left. Alternative exon 9A is marked with black. Cumulative data are shown in the lower panel (n = 3). B qPCR analysis of the ratio of Myb transcripts with exon 9A (+Ex9A) and without exon 9A (−Ex9A) in DP and iNKT cells. The relative expression of each transcript was normalized accordingly as described in Fig. 2A (n ≥ 5). C Western blot analysis of the protein level of Myb in DP thymocytes. D Graphical representation of Myb minigenes. The black arrow denotes the SRSF1-binding site in intron 9 of Myb. The potential SRSF1-binding motif is marked in red characters, and the specific deletion mutations are indicated with multiple “X”. E Constructs with (WT) or without (Mut) SRSF1-binding site were applied for splicing assays with/without SRSF1 overexpression (OE) in HEK293T cells. The percentages of inclusion (in%, black) and skipping (sk%, red) within exon 9A of Myb transcripts are presented (n = 5). F Apoptosis of DP thymocytes in chimeras with forced expression of Mybp75 or Mybp89. Annexin V+ donor-derived DP cells in thymi from recipients were detected. Representative dot plot and the ratio (the frequency of ctrl-EV was set to one) of Annexin V+ cells in DP thymocytes are shown (n ≥ 7). G Analysis of iNKT cells in chimeras with forced expression of Mybp75 or Mybp89. Donor-derived iNKT cells in thymi and livers from recipients were detected. Representative dot plot and the ratio (the frequency of ctrl-EV was set to one) of iNKT cells are shown (n ≥ 7). H, I Analysis of the expression of genes impaired by ablation of SRSF1 (H) and TCRα rearrangement (I) in chimeric DP thymocytes. The relative expression of each transcript was normalized accordingly as described in Fig. 2A (n = 5). Data are pooled from at least three independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when variance between the groups was unequal. For multiple comparisons, data were analyzed by one-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD
Given that loss of the less abundant p89 isoform does not have any deleterious effects on hematopoiesis and development [45] and our results also indicated that SRSF1 tends to promote p75 isoform expression via AS, we thus questioned whether forced expression of Mybp75 could rectify the defects in iNKT cells. To achieve this goal, we employed bone marrow chimeric mice overexpressing Mybp75, Mybp89, or empty vector (EV). After 9 weeks of reconstitution, the LCKCre/+Srsf1fl/fl DP cells with forced expression of Mybp75 exhibited less apoptosis than the cells transduced with EV retrovirus but not Mybp89 (Fig. 6F). Moreover, ectopic expression of the p75 isoform in the LCKCre/+Srsf1fl/fl mice notably restored the proportion of iNKT cells (Fig. 6G). The expression of genes impaired by deactivated SRSF1, including Myb, Bcl2l1, Gata3, and Itk, was significantly rectified (Fig. 6H). Moreover, defective TCRα rearrangements were substantially rectified in the DP thymocytes transduced with the p75 retrovirus (Fig. 6I). However, overexpression of the p89 isoform did not result in phenotypic rescue in the SRSF1-deficient iNKT and DP cells (Fig. 6F–I). These findings strongly support that SRSF1-mediated splicing of Myb mRNA is a crucial regulatory event during iNKT cell development.
LCKCre/+Srsf1fl/fl mice were highly tolerant of acute hepatic injury
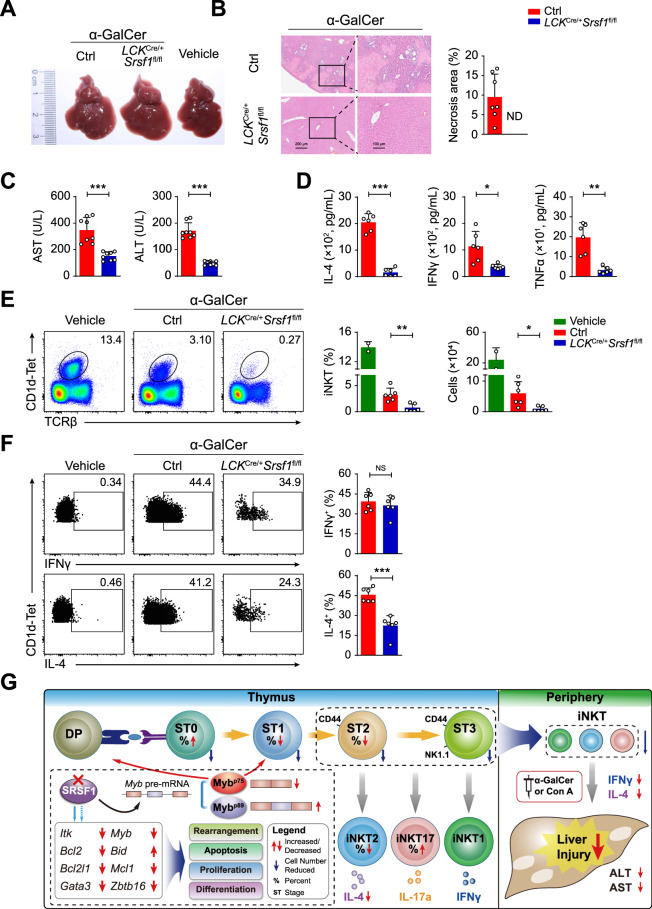
As a key immune regulator, iNKT cells in the liver are tightly associated with multiple liver dysfunctions [46–49]. To test the role of SRSF1 in the regulation of iNKT cell functions, we adopted α-GalCer-induced and Con A-induced hepatic injury models, which are well documented to experimentally resemble human autoimmune acute hepatitis [16, 49]. In the α-GalCer-induced groups, the ctrl mice exhibited aggravated liver injury compared with the LCKCre/+Srsf1fl/fl mice by liver morphology assays (Fig. 7A) and H&E staining (Fig. 7B). Two indices for liver damage, AST and ALT levels, were remarkably decreased in the serum of the LCKCre/+Srsf1fl/fl mice than the ctrl mice (Fig. 7C). The levels of IL-4, IFNγ, and TNFα in serum were higher in the ctrl mice than in the LCKCre/+Srsf1fl/fl mice (Fig. 7D). Correspondingly, a notable reduction in iNKT cells in the livers of the LCKCre/+Srsf1fl/fl mice was detected (Fig. 7E). Given that α-GalCer presented by CD1d provides specific iNKT-dependent activation, we further confirmed cytokine production at 2 h after α-GalCer stimulation in vivo. In accordance with the results in Fig. 3E, IL-4 expression was significantly decreased, but IFNγ was not, in iNKT cells from the SRSF1-deficient mice (Fig. 7F). Thus, we believe that both diminished iNKT numbers and defective IL-4 production in the SRSF1-deficient mice contributed to resistance to acute liver injury.
Fig. 7.
SRSF1-deficient mice exhibit resistance to α-GalCer-induced liver injury. A Representative images of livers at 3 d post-α-GalCer injection. The white spots are indicative of severe necrosis observed in the ctrl. B H&E staining of liver sections at 24 h post-α-GalCer injection. The percentages of necrotic areas are shown on the right (n = 7). C AST and ALT levels in serum were measured at 12 h post-α-GalCer injection (n = 8). D IL-4, IFNγ, and TNFα levels in serum were measured by ELISA at 2 h post i.v. injection with 30 ng α-GalCer (n = 6). E Representative pseudocolor plots showing iNKT cells in livers at 2 h post-injection with α-GalCer or vehicle. The frequency and numbers of iNKT cells are shown accordingly (vehicle: n = 2; Ctrl: n = 6; LCKCre/+Srsf1fl/fl: n = 6). F IL-4 and IFNγ in liver iNKT cells were measured by ICS. The frequency was shown accordingly (n = 6). G Proposed working model. A schematic illustration showed that SRSF1 promotes iNKT cell development via post-transcriptionally regulating genes involved in TCRα arrangement, survival, proliferation, and functional differentiation. In particular, SRSF1-mediated AS of Myb pre-mRNA is crucial for normal iNKT cell development. Due to a diminished iNKT cell pool in the periphery and reduced cytokine secretion, SRSF1-deficient mice exhibit slight clinical symptoms in acute liver injury upon α-GalCer or Con An induction. Data were pooled from at least two independent experiments. Statistical significance was determined by unpaired two-sided Student’s t test for normally distributed data, or an unpaired two-sided Welch’s t test was used when variance between the groups was unequal. *P < 0.05; **P < 0.01; ***P < 0.001; NS denotes not significant. Data are the mean ± SD
Similarly, after challenge with a low dose of Con A (15 mg/kg), the results were consistent with those in the α-GalCer-induced models (Supplementary Fig. 5A–E). Furthermore, when challenged with a lethal dose of Con A (30 mg/kg), all the ctrl mice died within 20 h, while approximately 40% of the LCKCre/+Srsf1fl/fl mice survived (Supplementary Fig. 5F). These results collectively demonstrated that conditional ablation of SRSF1 weakened pathologic symptoms in iNKT-related acute hepatic injury.
Discussion
The regulation of iNKT cell development is a complicated process involved in a well-established transcription factor program [20], whereas the molecular mechanism by which iNKT cell differentiation is regulated post-transcriptionally is still unclear. In this study, we validated that RBP SRSF1 is crucial for iNKT cell development and cytokine production and affects multifunctional factors to form competent iNKT cells.
Previous studies have indicated that SRSF1 is essential for controlling proliferation- and survival-related events [50–53]. In terms of lymphocytes, conditional deletion of SRSF1 in T cells resulted in increased apoptosis due to decreased expression of the antiapoptotic gene Bcl2l1 [52]. Similarly, SRSF1 deficiency accelerated apoptosis of both DP thymocytes and iNKT cells. Furthermore, SRSF1-deficient cells exhibited aberrantly increased expression of the proapoptotic gene Bid and decreased expression of the antiapoptotic genes Bcl2l1, Bcl2, and Mcl1, reflecting a shortened lifespan of these DP cells, which has effects on TCRα rearrangements of progenitors, ultimately resulting in a reduction in iNKT cell numbers [44]. Thus, the universal role of SRSF1 in cell survival is also essential for iNKT cell commitment.
Myb is essential for iNKT cell development and secretion of IL-4, a Th2-type cytokine [23, 54]. Similarly, we found that the SRSF1-deficient mice showed compromised iNKT cell generation and IL-4 production. By bioinformatics analysis, we found that 111 AS targets of SRSF1 were differentially expressed in DP thymocytes. Among them, we focused on the Myb gene, as its dominant mRNA product (short isoform MybP75) was remarkably switched to the exon-inclusion isoform (long isoform MybP89) in the absence of SRSF1, which was further validated by in vitro assays. Next, we selected the SRSF1-binding site (GA-enriched region) with the highest enrichment peak in introns 9–9A, which has been applied for mutagenesis in minigene analysis. Our results showed that fewer exon 9A skips were exhibited after this motif was mutated, indicating that this site located in intron 9–9A functions as an essential element to recruit SRSF1 for repression of exon 9 splicing with downstream exons. Given that specific effects on regulated splicing by one SR protein depend on a complex set of relationships with multiple other SR proteins or RBPs, the precise mechanism of Myb splicing still needs to be further elucidated. We also found that ectopic expression of the short isoform of MybP75 could partially rectify the defects of iNKT cells due to SRSF1 deficiency but not Mybp89. These observations demonstrated that SRSF1-dependent AS of Myb is crucial for iNKT cell differentiation. We also observed that SRSF1 modulates the expression of Dclre1c, a key regulator involved in remodeling and repair of break ends during V(D)J recombination [55]. Accordingly, the SRSF1-deficient DP cells exhibited defective TCRα rearrangements, which have also been observed in Myb-deficient DP cells [23]. In addition, we observed that SRSF1 deficiency impaired the expression of a series of iNKT transcription factors (TFs). For instance, SRSF1 indirectly controls the expression of PLZF, a master regulator of iNKT cell development [21, 22]. SRSF1 also maintains Gata3 expression, and the SRSF1-deficient mice phenocopied Gata3 deficiency, with a reduction in CD4+ iNKT cells and iNKT2 cell differentiation [8, 26]. Thus, SRSF1 directly or indirectly modulates the gene network involved in sustaining iNKT cell development. Furthermore, our recent study revealed that SRSF1 safeguards the late-stage development of conventional T cells by directly binding and regulating Irf7 and Il27ra expression, which is critical in the response to type I interferon signaling during T cell terminal mutation [56]. These findings collectively suggested that SRSF1 serves as a critical post-transcriptional regulator in both conventional T and iNKT cell intrathymic development by modulating distinct targets in a stage-specific manner.
SRSF1 exerted oncogenic roles via the control of AS of cancer-related genes overexpressed in human tumors. The liver harbors many iNKT cells, which are closely related to liver dysfunction [46]. Con A-induced liver injury in mice is regarded as a classic model to mimic pathogenic mechanisms and pathological changes in patients with liver inflammatory diseases [14, 16]. The α-GalCer-induced hepatitis mouse model with CD1d shows specific iNKT-dependent activation that resembles human autoimmune acute hepatitis [49]. When challenged with Con A or α-GalCer, the SRSF1-deficient mice showed more resistance than the ctrl mice. In addition, the SRSF1-deficient mice exhibited attenuated cytokine production, including that of IL-4, IFNγ, and TNFα, due to fewer iNKT cells and defects in IL-4 production by iNKT cells. Based on these findings, our study sheds light on SRSF1 as a potential clinical therapeutic target for diagnosis and therapy.
In summary, SRSF1 acts as a key post-transcriptional regulator to maintain TCRα arrangement, proliferation, survival, and functional differentiation of iNKT cells. By utilizing multipronged mechanisms, SRSF1 controls the expression of genes involved in iNKT differentiation and survival, including Myb, Itk, Gata3, Zbtb16, Bcl2, Bcl2l1, Mcl1, Bid, and TCR signaling. Furthermore, SRSF1-deficient mice were highly tolerant of induced liver injury, which is tightly associated with impaired iNKT cell numbers and function (Fig. 7G). The induced animal models verified the relevance between SRSF1 expression level and liver injury. Thus, our study not only provides new insight into SRSF1-dependent post-transcriptional regulation in iNKT cells but also sheds light on the clinical diagnosis and therapy of iNKT-related liver diseases.
Supplementary information
Acknowledgements
This work was supported in part by grants from the National Key Research and Development Program of China (2017YFA0104401), the National Natural Scientific Foundation of China (32130039, 31970831, and 31630038), and the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology from China Agricultural University (2021SKLAB6-3, 2021SKLAB6-4, 2019SKLAB6-6, and 2019SKLAB6-7).
Author contributions
Jingjing L., M.Y., and C.J. performed the major experiments and analyzed the data; M.Y., Z.W., and F.W. analyzed the high-throughput data; D.W., Z.Q., Y.Y., G.Y., Z.S., W.G., Juanjuan L., S.L., Y.J., and T.Z. assisted with the overall experiments; S.Y. designed and supervised the experiments with constructive suggestions from H.-H.X. and Y.X.; Y.Y., Jingjing L., and S.Y. wrote the manuscript with revisions from all authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jingjing Liu, Menghao You.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00766-w.
References
- 1.Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: how NKT cells, MAIT cells and gammadelta T cells emerge. Nat Rev Immunol. 2020;12:756–70. doi: 10.1038/s41577-020-0345-y. [DOI] [PubMed] [Google Scholar]
- 2.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adachi Y, Koseki H, Zijlstra M, Taniguchi M. Positive selection of invariant V alpha 14+ T cells by non-major histocompatibility complex-encoded class I-like molecules expressed on bone marrow-derived cells. Proc Natl Acad Sci USA. 1995;92:1200–4. doi: 10.1073/pnas.92.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 5.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–4. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 6.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 7.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–18. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 8.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–54. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harsha Krovi S, Zhang J, Michaels-Foster MJ, Brunetti T, Loh L, Scott-Browne J, et al. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat Commun. 2020;11:6238. doi: 10.1038/s41467-020-20073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranek T, Lebrigand K, de Amat Herbozo C, Gonzalez L, Bogard G, Dietrich C, et al. High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. 2020;32:108116. doi: 10.1016/j.celrep.2020.108116. [DOI] [PubMed] [Google Scholar]
- 11.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci USA. 2008;105:11287–92. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 13.Crosby CM, Kronenberg M. Tissue-specific functions of invariant natural killer T cells. Nat Rev Immunol. 2018;18:559–74. doi: 10.1038/s41577-018-0034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Z, Wei H, Sun R, Tian Z. The roles of innate immune cells in liver injury and regeneration. Cell Mol Immunol. 2007;4:241–52. [PubMed] [Google Scholar]
- 15.Wang H, Feng D, Park O, Yin S, Gao B. Invariant NKT cell activation induces neutrophil accumulation and hepatitis: opposite regulation by IL-4 and IFN-gamma. Hepatology. 2013;58:1474–85. doi: 10.1002/hep.26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heymann F, Hamesch K, Weiskirchen R, Tacke F. The concanavalin A model of acute hepatitis in mice. Lab Anim. 2015;49(1 Suppl):12–20. doi: 10.1177/0023677215572841. [DOI] [PubMed] [Google Scholar]
- 17.Liew PX, Lee WY, Kubes P. iNKT cells orchestrate a switch from inflammation to resolution of sterile liver injury. Immunity. 2017;47:752–65 e5. doi: 10.1016/j.immuni.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Shissler SC, Webb TJ. The ins and outs of type I iNKT cell development. Mol Immunol. 2019;105:116–30. doi: 10.1016/j.molimm.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 20.Gapin L. Development of invariant natural killer T cells. Curr Opin Immunol. 2016;39:68–74. doi: 10.1016/j.coi.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–64. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11:435–41. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikenheiser DJ, Stumhofer JS. ICOS co-stimulation: friend or foe? Front Immunol. 2016;7:304. doi: 10.3389/fimmu.2016.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akbari O, Stock P, Meyer EH, Freeman GJ, Sharpe AH, Umetsu DT, et al. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J Immunol. 2008;180:5448–56. doi: 10.4049/jimmunol.180.8.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–9. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 27.Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–71. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–94. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 29.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–16. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Wu D, Jiang N, Zhuang Y. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol. 2013;191:5052–64. doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Roy S, Kim YM, Li S, Zhang B, Love C, et al. Id2 collaborates with Id3 To suppress invariant NKT and innate-like tumors. J Immunol. 2017;198:3136–48. doi: 10.4049/jimmunol.1601935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–30. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong SSR. Proteins: binders, regulators, and connectors of RNA. Mol Cells. 2017;40:1–9. doi: 10.14348/molcells.2017.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res. 2014;12:1195–204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 36.Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci USA. 1995;92:12070–4. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Cui Z, Wang F, Yao Y, Yu G, Liu J, et al. Lrp5 and Lrp6 are required for maintaining self-renewal and differentiation of hematopoietic stem cells. FASEB J. 2019;33:5615–25. doi: 10.1096/fj.201802072R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TJ, Park G, Kim J, Lim SA, Kim J, Im K, et al. CD160 serves as a negative regulator of NKT cells in acute hepatic injury. Nat Commun. 2019;10:3258. doi: 10.1038/s41467-019-10320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Y, Guo W, Chen J, Guo P, Yu G, Liu J, et al. Long noncoding RNA Malat1 is not essential for T cell development and response to LCMV infection. RNA Biol. 2018;15:1477–86. doi: 10.1080/15476286.2018.1551705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedel R, Berry R, Mallevaey T, Matsuda JL, Zhang J, Godfrey DI, et al. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc Natl Acad Sci USA. 2014;111:E119–28. doi: 10.1073/pnas.1320777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–7. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 43.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–76. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 44.Das R, Sant’Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker SJ, Kumar A, Reddy EP. p89c-Myb is not required for fetal or adult hematopoiesis. Genesis. 2010;48:309–16. doi: 10.1002/dvg.20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu S, Zhang H, Bai L. NKT cells in liver diseases. Front Med. 2018;12:249–61. doi: 10.1007/s11684-018-0622-3. [DOI] [PubMed] [Google Scholar]
- 47.Bandyopadhyay K, Marrero I, Kumar V. NKT cell subsets as key participants in liver physiology and pathology. Cell Mol Immunol. 2016;13:337–46. doi: 10.1038/cmi.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, He W, Shi X, He X, Dou L, Gao Y. The role of CD1d and MR1 restricted T cells in the liver. Front Immunol. 2018;9:2424. doi: 10.3389/fimmu.2018.02424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dennert G, Aswad F. The role of NKT cells in animal models of autoimmune hepatitis. Crit Rev Immunol. 2006;26:453–73. doi: 10.1615/CritRevImmunol.v26.i5.50. [DOI] [PubMed] [Google Scholar]
- 50.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–8. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Wang R, Li X, Yu L, Hua D, Sun C, et al. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129:676–93. doi: 10.1172/JCI120279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsuyama T, Martin-Delgado IJ, Krishfield SM, Kyttaris VC, Moulton VR. Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus. Rheumatology. 2020;59:2146–55. doi: 10.1093/rheumatology/keaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–14. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakata Y, Brignier AC, Jin S, Shen Y, Rudnick SI, Sugita M, et al. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood. 2010;116:1280–90. doi: 10.1182/blood-2009-05-223255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–90. doi: 10.1016/S1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 56.Qi Z, Wang F, Yu G, Wang D, Yao Y, You M, et al. SRSF1 serves as a critical posttranscriptional regulator at the late stage of thymocyte development. Sci Adv. 2021;16:eabf0753. doi: 10.1126/sciadv.abf0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.