Abstract
The ARF tumor suppressor protein stabilizes p53 by antagonizing its negative regulator, Mdm2 (Hdm2 in humans). Both mouse p19ARF and human p14ARF bind to the central region of Mdm2 (residues 210 to 304), a segment that does not overlap with its N-terminal p53-binding domain, nuclear import or export signals, or C-terminal RING domain required for Mdm2 E3 ubiquitin ligase activity. The N-terminal 37 amino acids of mouse p19ARF are necessary and sufficient for binding to Mdm2, localization of Mdm2 to nucleoli, and p53-dependent cell cycle arrest. Although a nucleolar localization signal (NrLS) maps within a different segment (residues 82 to 101) of the human p14ARF protein, binding to Mdm2 and nucleolar import of ARF-Mdm2 complexes are both required for cell cycle arrest induced by either the mouse or human ARF proteins. Because many codons of mouse ARF mRNA are not recognized by the most abundant bacterial tRNAs, we synthesized ARF minigenes containing preferred bacterial codons. Using bacterially produced ARF polypeptides and chemically synthesized peptides conjugated to Sepharose, residues 1 to 14 and 26 to 37 of mouse p19ARF were found to interact independently and cooperatively with Mdm2, while residues 15 to 25 were dispensable for binding. Paradoxically, residues 26 to 37 of mouse p19ARF are also essential for ARF nucleolar localization in the absence of Mdm2. However, the mobilization of the p19ARF-Mdm2 complex into nucleoli also requires a cryptic NrLS within the Mdm2 C-terminal RING domain. The Mdm2 NrLS is unmasked upon ARF binding, and its deletion prevents import of the ARF-Mdm2 complex into nucleoli. Collectively, the results suggest that ARF binding to Mdm2 induces a conformational change that facilitates nucleolar import of the ARF-Mdm2 complex and p53-dependent cell cycle arrest. Hence, the ARF-Mdm2 interaction can be viewed as bidirectional, with each protein being capable of regulating the subnuclear localization of the other.
Activation of the p53 transcription factor in response to oncogenic stress signals results in cell cycle arrest or apoptosis, thereby enabling cells to repair genotoxic damage or to be eliminated from the organism (reviewed in references 21 and 24). Loss of p53 function cancels these surveillance functions and strongly predisposes the cells to cancer development. Although p53 is a highly unstable protein, it accumulates in response to DNA damage or oncogenic signaling, largely through protein stabilization following disruption of its interaction with its negative regulator, Mdm2 (Hdm2 in humans) (12, 22). Mdm2 opposes p53 function at several levels. It can bind to the N-terminal transcriptional activation domain of p53 to block expression of p53-responsive genes (28, 30). Mdm2 has an intrinsic E3 ligase activity that conjugates ubiquitin to p53 (14, 15), and it may play a role in shuttling p53 from the nucleus to the cytoplasm, where p53 is degraded in cytoplasmic proteasomes (9, 37, 45). Although Mdm2-dependent degradation of p53 was reported to depend on the nuclear export signal (NES) of Mdm2 (37, 45), the interpretation is complicated by observations that unassembled p53 subunits can exit the nucleus independently (43). One possibility is that Mdm2-directed ubiquitination of p53 might disrupt the p53 tetramer, thereby facilitating Mdm2-independent export of p53 dimers or monomers to the cytoplasm. Regardless of the exact mechanism, stabilization of p53 occurs when the Mdm2-p53 interaction is blocked, either by posttranslational modifications of p53 (reviewed in references 10 and 33) or by the direct interaction of Mdm2 with the ARF tumor suppressor protein (18, 32, 44, 51).
ARF is encoded by the INK4a-ARF locus, which also specifies the cyclin D-dependent kinase inhibitor, p16INK4a (35). The N-terminal 62 amino acids of the 169-residue mouse p19ARF polypeptide (132-amino-acid p14ARF in humans) are encoded by a unique first exon, with the remaining residues being specified by exon 2, which also encodes the bulk of p16INK4a from an alternative reading frame. Mutations of the INK4a-ARF locus occur often in cancer cells, regardless of tumor type and patient age, at a frequency that approaches that of p53 (reviewed in references 40 and 41). ARF is activated by inappropriate proliferative signals induced by oncoproteins, such as Myc (52), E1A (6), E2F-1 (1), Ras (31), or v-Abl (36), and it in turn activates a p53-dependent stress response (19; reviewed in reference 41). The encoded ARF protein is nucleolar, and its binding sequesters Mdm2 in nucleoli, inhibits Mdm2 nuclear export, and thereby stabilizes p53 in the nucleoplasm (46, 47). As for p53 (7, 16, 20), loss of ARF alone (17, 19) or INK4a-ARF (39) in mice strongly predisposes to tumor development. Indeed, ARF inactivation or Mdm2 overexpression occurs more commonly in tumor cells that retain wild-type p53, in accord with the hypothesis that disruption of the ARF-Mdm2-p53 pathway is important in the life histories of most cancer cells.
The interaction of ARF with Mdm2 does not obligatorily affect the ability of Mdm2 to bind to p53, so formation of ternary complexes can also occur (18, 32, 44, 51). Hence, it remains unclear whether binary ARF-Mdm2 or ternary ARF-Mdm2-p53 complexes are physiologically relevant to ARF function. Based on studies in which Mdm2, but not p53, was seen to be mobilized into nucleoli following mouse p19ARF induction, we suggested that nucleolar sequestration of the binary ARF-Mdm2 complex was central to ARF action (47). In accord with this concept, a mouse ARF mutant lacking residues 26 to 37 that bound to Mdm2 but was defective in nucleolar localization neither mobilized Mdm2 to this compartment nor induced p53-dependent cell cycle arrest. An alternative interpretation is that the Mdm2-p53 complex normally exits the nucleus via the nucleolus, where ARF might act to negatively regulate transport (46). Previous findings that Mdm2 binds to rRNA and the ribosomal protein L5 (25) and that its exit from the nucleus can be blocked by competitive inhibitors of lentiviral Rev proteins (37), which affect a pathway used to export 5S rRNA, make this an attractive model. However, p53 has not been directly visualized in nucleoli after ARF induction, so transnucleolar export of Mdm2-p53 complexes, if it occurs, would have to be an extremely efficient and rapid process. The inability to trap p53 in the nucleolus following treatment of cells with the CRM1 inhibitor leptomycin (J. D. Weber and C. J. Sherr, unpublished observations) argues against this interpretation. Yet a third model stems from observations that overexpressed ARF, Mdm2, and p53 proteins could accumulate together in “nuclear bodies” within the nucleoplasm (50). A surfeit of Mdm2 can prevent ARF from localizing to the nucleolus (47), but whether this occurs under physiologic circumstances is unknown. For the latter model to be valid, it is not only necessary to propose that ternary ARF-Mdm2-p53 complexes retain transcriptional activity but also to discount observations that delocalized nucleoplasmic ARF mutants that can still bind Mdm2 are functionally handicapped. A further complication is that a nucleolar localization signal (NrLS) in human ARF is not confined to a region topologically analogous to that in the mouse protein but instead maps to an entirely different segment of p14ARF encompassed by residues 82 to 101 (50). Together, these findings pointed to the possibility that mouse p19ARF and human p14ARF might function in manners that were different from one another.
To address these issues, we evaluated a series of mouse and human ARF mutants for the ability to bind to Mdm2 or Hdm2, to import Mdm2 into the nucleolus, and to induce cell cycle arrest. Our data indicate that, despite differences in the positioning of mouse and human ARF NrLSs, nucleolar compartmentalization of the ARF-Mdm2 complex is central to the ability of both mouse and human ARFs to inhibit cell cycle progression. In agreement with recent data of others (24a), we find that mobilization of the ARF-Mdm2 complex depends not only on the ARF NrLS but also on a similar sequence within the C-terminal RING domain of Mdm2 that appears to be unmasked upon ARF binding. We propose that cooperative ARF binding to Mdm2 through two independent sites induces a conformational change that enables the cryptic Mdm2 NrLS to direct the nucleolar import of the ARF-Mdm2 complex.
MATERIALS AND METHODS
Cell culture and introduction of expression plasmids.
NIH 3T3 cells (ARF null; p53 wild type) maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum, 2 mM glutamine, and 100 U of penicillin and streptomycin (GIBCO/BRL, Gaithersburg, Md.) per ml were transfected with expression plasmids as previously described (52). Virus production and infection of cells were performed using retroviral helper and vector plasmids (29, 52) provided by Charles Sawyers (University of California, Los Angeles). Spodoptera frugiperda (Sf9) cells were maintained in Grace's medium supplemented with 5% fetal bovine serum and infected for 48 h with various baculoviruses before lysis (18).
Generation of ARF mutants.
A stop codon was inserted downstream of codon 37 in mouse ARF cDNA by PCR (see below). The sense (5′-GAATTCGATGGGTCGCAGGTTCTTGGT) and antisense (5′-GGATCCTTAGCTCGCTGTCCTGGGTCT) primers included the initiation and termination codons (underlined) flanked at their 5′ ends by EcoRI and BamHI consensus sequences, respectively. The purified PCR product was cloned into the EcoRI-BamHI sites of the pEGFP-C1 vector (Clontech, Palo Alto, Calif.) in frame with the C terminus of green fluorescent protein (GFP) to produce the plasmid GFP-ARF N37. Mouse ARF deletion mutants were constructed by using a pBluescript plasmid (Stratagene, La Jolla, Calif.) containing a hemagglutinin (HA)-tagged ARF cDNA template (35). Mutated sense and antisense oligonucleotides complementary to noncontiguous sequences flanking sites to be deleted were used. Two PCRs were performed with template ARF cDNA (200 ng) as follows: sense Δ1–14 (5′-GACTACGCTACCGGCCGCCCACTC) or Δ15–25 (5′-ATTCAGCGCGCGAAGTTCGTGCGA) mixed with T3 primer and antisense Δ1–14 (5′-GAGTGGGCGGCCGGTAGCGTAGTC) or Δ15–25 (5′-TCGCACGAACTTCGCGCGCTGAAT) mixed with T7 primer. The reaction buffer included 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM MgCl2, 0.1% gelatin, 80 μM (each) deoxynucleoside triphosphate, 500 ng of each primer, and 0.5 U of Taq DNA polymerase (Stratagene). Each of 25 cycles consisted of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The PCR products were isolated on 1% agarose gels and purified (with a gel extraction kit from Qiagen, Valencia, Calif.). Purified products from Δ1–14 and Δ15–25 reactions were mixed separately in reaction buffer along with T3 and T7 primers in the following two-step PCR: first, denaturation at 95°C for 1 min, annealing at 37°C for 1 min, and extension at 72°C for 2 min for 10 cycles, followed by denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min for 25 cycles. The final PCR products were ligated into pGEM-T cloning vectors (Promega, Madison, Wis.) for sequencing. Mutant ARF cDNAs were excised with EcoRI and subcloned into the EcoRI site of the pSRαMSV-tkneo retroviral vector (52) for expression in mammalian cells or into the EcoRI site of the pVL1393 baculovirus vector (Pharmingen, San Diego, Calif.) for expression in insect Sf9 cells. An HA-tagged ARF Δ1–14–Δ26–37 double-deletion mutant was constructed by incorporating template HA-ARF Δ26–37 cDNA, constructed as previously described (47), into the above-mentioned reactions with Δ1–14 primers.
Human ARF Δ82–101 was generated by Quickchange site-directed mutagenesis (Stratagene) as recommended by the manufacturer using sense (5′-GCTGCTCCACGGGGAGGGCTTCCT) and antisense (5′-AGGAAGCCCTCCCCGTGGAGCAGC) primers. The remaining human ARF mutants were constructed with mutated sense and antisense oligonucleotides complementary to wild-type p14ARF cDNA sequences as primers, analogous to the strategy employed above for construction of mouse p19ARF mutants. Two PCRs were performed with template human ARF cDNA (100 ng) as follows: sense Δ2–14 (5′-GGCGAGAACATGTGCGGCCCGCCG) or Δ26–37 (5′-GTTTTCGTGGTTGGGGCGCCCGCC) mixed with T7 primer and antisense Δ2–14 (5′-CGGCGGGCCGCACATGTTCTCGCC) or Δ26–37 (5′-GGCGGGCGCCCCAACCACGAAAAC) mixed with T3 primer. The reaction buffer included 20 mM Tris-HCl (pH 8.0), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg of bovine serum albumin, 200 μM (each) deoxynucleoside triphosphate, 1 μg of each primer, and 2.5 U of Pfu DNA polymerase (Stratagene). PCRs with T3 primer (25 cycles) or T7 primer (35 cycles) were run under the conditions described above for mouse ARF mutants. The p14ARF Δ2–14–Δ82–101 double-deletion mutant was constructed using template p14ARF Δ82–101 cDNA in the above-mentioned reaction with Δ2–14 primers. The PCR products were purified from 1% agarose gels, digested with BamHI and XhoI, and subcloned into pBluescript SK cloning vectors (Stratagene) for sequencing. Wild-type and mutant human ARF cDNAs were excised with BamHI and XhoI and subcloned into the BamHI-XhoI sites of pcDNA3.1 vector (Invitrogen, Carlsbad, Calif.) and into the ClaI site of the pSRαMSVtkCD8 (29) vector by blunt-end ligation (both for expression in mammalian cells).
Hdm2 mutant plasmid construction.
A nonomeric primer (5′-GGCCATATG) including an NdeI consensus site (underlined) was annealed to different regions of Hdm2 cDNA to provide ATG initiation codons. Translation initiation sites of this type were incorporated into sense primers that included Hdm2 residue 2 (5′-GGCCATATGTGCAATACCAACATG), residue 141 (5′-GGCCATATGCAAGAGCTTCAGGAA), residue 211 (5′-CATATGAGCAGTAGCAGTGAATCTACAGGG), or residue 278 (5′-GGCCATATGCAAGTTACTGTGTAT). Conversely, stop codons (underlined in antisense orientation) were inserted into BamHI-containing antisense primers at position 277 (5′-GGCGGATCCCTAATATACCTCATC), residue 305 (5′-GGATCCCTATTTCCAATAGTCAGCTAAGGA), or residue 351 (5′-GGCGGATCCCTATGAGTTTTCCAG). Appropriate sense and antisense primers were mixed with reaction buffer (see above) and template Hdm2 cDNA. A 25-cycle PCR consisting of denaturing at 95°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 2.5 min was utilized to construct the various Hdm2 mutants. The purified PCR products were ligated into pGEM-T cloning vectors for sequencing. Hdm2 1–276, Hdm2 140–350, Hdm2 277–491, and Hdm2 210–304 inserts were excised with NdeI and BamHI and subcloned into the NdeI-BamHI sites of the pET28a (Novagen, Madison, Wis.) bacterial expression vector in frame with the C terminus of a polyhistidine tag. Conversely, Hdm2 1–350 and 140–350 were excised with EcoRI and BamHI and subcloned into the EcoRI-BamHI site of the pcDNA3.1 mammalian expression vector. Hdm2 1–440 was generously provided by Karen Vousden (Frederick Cancer Research Center, Frederick, Md.). Hdm2 Δ466–473 was generated using mutated sense and antisense oligonucleotides as primers; these contained novel FspI restriction sites (underlined below) flanking codons 466 and 473. Two PCRs were performed with template Hdm2 cDNA (100 ng) as follows: sense Δ466–473 (5′-TCCCCCGGGTGCGCACCCTGCCCAGTATGTAGACAACCA) mixed with T7 primer and antisense Δ466–473 (5′-TCCCCCGGGTGCGCATGTAAAGCAGGCCATAAGATG) mixed with a primer containing the initiation codon (underlined) of Hdm2 (5′-ATGTGCAATACCAACATGTCTGTGTCTACC). Each of 35 cycles involved denaturation at 95°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min. The PCR products were digested with FspI and ligated to one another. Newly ligated Hdm2 Δ466–473 was excised with BamHI and XhoI and subcloned into the BamHI-XhoI sites of pcDNA3.1.
Synthetic ARF minigenes.
A synthetic (syn) minigene encoding the N-terminal 64 amino acids (N64) of mouse p19ARF was generated de novo by first annealing two long sense and antisense oligonucleotides that overlapped in an 18-bp region (underlined). The sense syn-ARF oligonucleotide (5′-GGCCGCATGGCATATGGG TCGCCG T TTCCTGG T TACTG TGCGCAT TCAGCG TGCGGGCCGCCCACTGCAAGAGCGTGTTTTCCTGGTGAAGTTCGTTCGCTCCCGTCGCCCGCGTACCGCTAGCTGCGCTCTGG) was mixed with an antisense syn-ARF oligonucleotide (5′-CGGTACCGGCGCGGATCCTTATTAACCTGGGCCCGGG T TACGG TGCGGACCGCGACGCAGGATGCGCTCCAGACGCAGCAGCATGTTAACGAAAGCCAGAGCGCAGCTAGCGG), and PCR was used to copy the single-stranded nonoverlapping ends. Each cycle (10 cycles total) consisted of denaturation at 95°C for 1 min, annealing at 37°C for 1 min, and extension at 72°C for 1 min. Following this initial reaction, sense and antisense oligonucleotides complementary to the N terminus (5′-GGGCCGCATGGCATATG) or the C terminus (5′-CGGTACCGGCGCGG) of the first PCR products were added to the reaction mixture, and 30 additional cycles were performed with denaturation at 95°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min. The final PCR product (designated syn-ARF N64) was isolated on a 1% agarose gel and purified. Following ligation into the pCRII cloning vector (Invitrogen), inserts were sequenced, excised with NdeI and BamHI, and subcloned into the NdeI-BamHI sites of pET28a bacterial expression vector in frame with the C terminus of the polyhistidine tag. Syn-ARF N37 was constructed by PCR using oligonucleotide primers complementary to the 5′ moiety of the syn-ARF N64 template (100 ng). A single reaction was performed with sense (5′-CATGGCATATGGGTCGCCGTTTC) and antisense (5′-CGGGATCCTTAGCTAGCGGTACG) primers. The cycles included denaturation at 95°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 30 s. The gel-purified PCR product was subcloned into the pCRII cloning vector for sequencing and into the NdeI-BamHI sites of pET28a in frame with the C terminus of the polyhistidine tag.
Bacterial gene expression.
For bacterial expression of syn-ARF N37, BL21 (DE3) cells (Stratagene) were transformed with pET28a-polyHIS-syn-ARF N37, cultured in Luria-Bertani medium containing 30 mg of kanamycin/liter, and induced with isopropylthiogalactoside (1 mM). The cells were harvested, resuspended in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 5 mM imidazole, and lysed by sonication (Branson Sonifier 450, Danbury, Ct.). The lysates were centrifuged at 20,000 × g for 15 min, and 6 M urea was added to the soluble fraction. The urea-containing crude lysate was filtered (0.45 μm pore size; Millipore, Bedford, Mass.), and loaded onto a 5-ml chelating Sepharose column freshly charged with 50 mM NiSO4 and equilibrated with 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5 mM imidazole, and 6 M urea. The column was washed with 10 volumes of equilibration buffer followed by 5 volumes of equilibration buffer containing 60 mM imidazole. Synthetic ARF N37 was eluted in equilibration buffer containing 350 mM imidazole, and fractions containing the polypeptide were dialyzed against 20 mM Tris-HCl (pH 8.0) containing 500 mM NaCl.
Polyhistidine-tagged Hdm2 proteins were expressed and purified in a similar way. However, after being loaded on nickel affinity columns, the bound proteins were eluted with a linear gradient of imidazole (5 to 500 mM). The purified Hdm2 proteins were refolded by dilution in 20 mM Tris-HCl (pH 8.0) containing 50 mM NaCl, followed by ultrafiltration (Biomax 5; 5-kDa exclusion; Millipore). The purity of all recombinant proteins was judged to be greater than 90% following their electrophoretic separation in denaturing gels containing sodium dodecyl sulfate (SDS) and staining with Coomassie brilliant blue.
FPLC.
Cyanogen bromide-activated Sepharose (Pharmacia, Piscataway, N.J.) was swollen in 1 mM HCl for 15 min, washed repeatedly with coupling buffer (100 mM NaHCO3 and 500 mM NaCl), and incubated with coupling buffer containing 2 to 5 mg of various ARF synthetic peptides or syn-ARF N37 at 4°C for 1 h. The Sepharose was blocked in 500 mM glycine (pH 8.0) at 4°C for 2 h and washed alternately with 100 mM sodium acetate and 500 mM NaCl (pH 4.0) and then with coupling buffer. Conjugated beads (5 ml) were poured into XK16 columns (Pharmacia Biotech, Uppsala, Sweden) and equilibrated with 25 mM Tris-HCl (pH 8.0). Purified Hdm2 140–350 and Hdm2 210–304 (25 μg of protein) were injected at a flow rate of 0.5 ml/min, washed with 20 ml of 25 mM Tris-HCl (pH 8.0) at 1.0 ml/min, and eluted with a 25-ml NaCl gradient (0 to 1.5 M) at 1.0 ml/min, followed by 20 ml of 100 mM glycine (pH 3.0) at 1.0 ml/min using BioLogic fast protein liquid affinity chromatography (FPLC) and BioLogicHR software (Bio-Rad, Hercules, Calif.). The collected protein fractions (1 ml) were precipitated with trichloroacetic acid, resuspended in 1 M Tris-HCl (pH 8.0), electrophoretically separated on denaturing polyacrylamide gels containing SDS, and visualized by Coomassie blue staining.
Immunofluorescence.
NIH 3T3 cells (3 × 104) seeded onto glass coverslips were cotransfected with plasmids encoding mouse p19ARF or the indicated ARF mutants together with T7 epitope-tagged Hdm2 (pCGT-T7Hdm2) (47). Cotransfections were also performed with pcDNA3 or pSRαMSV-tkCD8 plasmids (29) containing wild-type human p14ARF or p14ARF mutants in combination with pSRαMSV-Hdm2-tkneo (52). The cells were fixed 48 h after transfection with methanol-acetone (1:1 [vol/vol]) and stained for 1 h with either affinity-purified rabbit anti-p19ARF antibody (10 μg/ml) or anti-p14ARF antibody (3.2 μg/ml) (both directed to ARF C-terminal epitopes) (35) followed by a 30-min exposure to biotinylated anti-rabbit immunoglobulin and streptavidin-conjugated Texas red (both from Amersham, Arlington Heights, Ill.). T7 epitope-tagged Hdm2 was detected with monoclonal T7 antibody (Novagen) followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin (Amersham) or biotinylated anti-mouse immunoglobulin and streptavidin-conjugated Texas red. Untagged wild-type Hdm2 (in combination with human p14ARF) or Hdm2 mutants (in combination with GFP-p19ARF) were detected with monoclonal 2A10 antibody (Santa Cruz Inc., Santa Cruz, Calif.) followed by FITC-conjugated anti-mouse immunoglobulin or with biotinylated anti-mouse immunoglobulin and streptavidin-conjugated Texas red.
For measurement of DNA replication, 5-bromodeoxyuridine (BrdU) (10 μM) was added to the culture medium 24 h after transfection or infection. Cells were fixed in methanol-acetone (1:1 [vol/vol]) 24 h after the addition of BrdU, treated for 10 min with 1.5 N HCl, and stained for 1 h with mouse monoclonal anti-BrdU antibody (Amersham) followed by FITC-conjugated anti-mouse immunoglobulin. DNA was visualized with Hoechst dye. At least 100 cells were counted on each of three coverslips enumerated for each experimental condition. Fluorescence signals were detected using a BX50 microscope (Olympus, Lake Success, N.Y.) fitted with a Sensys 1400 charge-coupled device camera (Photometrics, Tucson, Ariz.).
ARF binding to Mdm2 (Hdm2).
Purified Hdm2 proteins were mixed for 1 h at 4°C with recombinant p19ARF produced in Sf9 cells in 0.1 ml of binding buffer containing 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, 0.4 U of aprotinin/ml, and 10 μg of leupeptin/ml. Antibody to the p19ARF C terminus, one of two antibodies (2A10 or SMP14) to Mdm2 (Santa Cruz Biotechnology), or nonimmune rabbit serum (NRS) was added to the binding reactions. In parallel, Sf9 cells coinfected for 48 h with baculoviruses encoding wild-type Mdm2 together with vectors encoding the indicated p19ARF mutants were lysed in binding buffer and incubated with antibodies as described above for 1 h at 4°C. Immune complexes were precipitated with protein A-Sepharose (Amersham) and washed under stringent conditions (18). The precipitated proteins were separated on denaturing polyacrylamide gels containing SDS and were transferred to Immobilon polyvinylidine difluoride membranes (Millipore) preactivated in methanol. Hdm2 or Mdm2 and ARF proteins were visualized by direct immunoblotting using monoclonal antibody 2A10 (which detects Mdm2, Hdm2 140–350, and Hdm2 277–491), rabbit polyclonal antibody SMP14 (for Hdm2 1–276), or antibodies to the ARF C terminus.
RESULTS
Mouse ARF residues 1 to 37 are necessary for cell cycle arrest.
The first exon (1β) of mouse ARF encodes residues 1 to 62 (N62) of the full-length 169-amino-acid protein. This segment of mouse p19ARF is both necessary and sufficient for the known functions of ARF, including nucleolar localization, binding to and nucleolar sequestration of Mdm2, p53 activation, and p53-dependent cell cycle arrest (18, 34, 47). To further pinpoint regions within the N62 domain necessary for ARF nucleolar localization and Mdm2 binding, we transfected ARF-null mouse NIH-3T3 fibroblasts with plasmids encoding different regions of p19ARF tagged at their N termini by GFP. When GFP was fused to full-length mouse ARF (not shown) or to the truncated N62 polypeptide, the chimeric protein localized to nucleoli (Fig. 1F), whereas unfused GFP remained predominantly cytoplasmic (Fig. 1B). As in previous studies (47), nucleoli were demarcated using antibodies to fibrillarin (data not shown). The nucleolar localization of full-length mouse ARF or ARF-N62 does not depend on the GFP tag and occurs in primary mouse embryo fibroblast (MEF) strains of various genetic backgrounds, including those lacking ARF, p53, or both p53 and Mdm2 (47). Importantly, these observations underscore the ability of p19ARF to localize to the nucleolus in the absence of Mdm2.
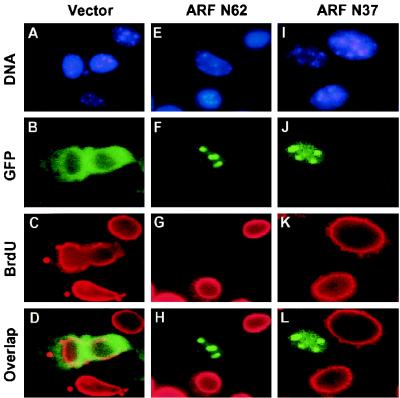
FIG. 1.
The N-terminal 37 amino acids of mouse p19ARF are sufficient for nucleolar localization and cell cycle arrest. NIH 3T3 cells were transfected with expression vectors encoding GFP (A to D), GFP-ARF N62 (E to H), or GFP-ARF N37 (I to L). Cells labeled for 24 h with BrdU 1 day posttransfection were fixed and analyzed by indirect immunofluorescence using a mouse monoclonal antibody to BrdU followed by biotinylated anti-mouse immunoglobulin and streptavidin Texas red (C, G, and K) and for GFP expression using an FITC filter (B, F, and J). Overlap staining is shown in panels D, H, and L. Nuclei were visualized by Hoechst dye (A, E, and I). Nucleolar staining was confirmed in parallel using antibodies to fibrillarin (data not shown) (47).
A GFP-ARF fusion protein containing only mouse ARF amino acids 1 to 37 (GFP ARF-N37) also localized to nucleoli (Fig. 1J), indicating that amino acids C-terminal to residue 37 were not required for this function. Moreover, when BrdU was introduced into the culture medium 1 day after transfection and scored for incorporation into replicated DNA 24 h later (Fig. 1), GFP alone did not affect S-phase entry (Fig. 1C and D). However, GFP-tagged ARF N62 (Fig. 1G and H) and ARF N37 (Fig. 1K and L) both induced cell cycle arrest. More than 90% of cells expressing these nucleolar ARF fusion proteins failed to incorporate BrdU, demonstrating that ARF N37 was biologically active.
Because they can be efficiently transfected by expression plasmids or readily infected by retroviruses, we used p53 wild-type ARF-null NIH 3T3 cells to document the effects of additional ARF mutants on cell cycle progression. We emphasize that similar data have been obtained using primary wild-type MEF strains infected with ARF retroviruses. In all cases, cell cycle arrest by active ARF mutants depended upon the presence of functional p53, which accumulated in the nucleoplasm of ARF-transfected cells. Given the strict correlations between these parameters as documented previously (47) and to achieve a reasonable economy of presentation, the results with p53 are not fully reiterated here, and only a complete data set using NIH 3T3 cells is presented below.
Two domains within ARF N37 bind to an internal segment of Hdm2.
The interaction between Mdm2 and ARF has been documented both in vitro and in vivo (18, 32, 44, 46, 47, 51), but defining the minimal interaction domains for both ARF and Mdm2 has thus far proven problematic. A diagram illustrating the known structural motifs within Mdm2 (or human Hdm2) together with additional molecular landmarks defined in this report is shown at the top of Fig. 2. The maps beneath similarly schematize domains within both mouse p19ARF and human p14ARF. A yeast two-hybrid interaction screen performed with human p14ARF as bait previously revealed interactions with the C-terminal moiety of Mdm2 (residues 208 to 491) lacking both the N-terminal p53 binding domain and additional sequences required both for nuclear localization (nuclear localization signal [NLS]) and nuclear export (NES). Conversely, others observed that deletion of Hdm2 residues 222 to 437 abolished p14ARF binding (44). However, a more complex interaction profile observed in cell lines engineered to express mouse ARF and various Mdm2 deletion mutants suggested that p19ARF engages multiple sites C-terminal to residue 155 in Mdm2 (32).
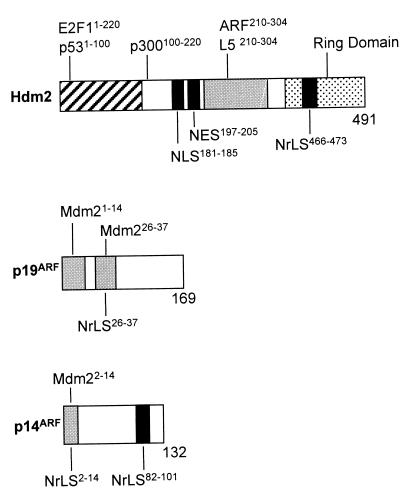
FIG. 2.
Schematic representations of Hdm2 or Mdm2 and ARF proteins. (Top) Hdm2 binding sites for p53 and E2F-1 (hatched), ARF and L5 (shaded), and p300 (open) are indicated. Amino acid domains in Hdm2 and Mdm2 required for these associations are indicated by superscripts. The NLS and NES sequences are similarly defined by solid bars. The RING domain (stippled) contains the NrLS (solid bar). (Middle) Shaded areas define the Mdm2 contact sites in mouse ARF; the segment from residues 26 to 37 also contains sequences required for nucleolar localization. (Bottom) The shaded bar defines the mapped Mdm2 binding site in human ARF, which is also required for nucleolar localization. A second NrLS is indicated by the black bar. An additional Mdm2 binding site within human p14ARF (see text) has not been mapped.
We expressed and purified three polyhistidine (His)-tagged Hdm2 truncation mutants from bacteria comprising amino acids 1 to 276, 140 to 350, and 277 to 491 and assayed their abilities to bind in vitro to full-length HA-tagged mouse p19ARF synthesized in insect Sf9 cells (Fig. 3A). Hdm2 1–276 failed to bind ARF (Fig. 3A, left), whereas Hdm2 277–491 bound relatively poorly (Fig. 3A, right). Consistent with the idea that the ARF binding domain(s) bridges these fragments (32), Hdm2 140–350 bound all of the available p19ARF under the same assay conditions (Fig. 3A, center). In the last reaction, the upper band corresponds to the input Hdm2 140–350 polypeptide, whereas the lower band represents a degradation product that also contains the His-tagged N terminus. Therefore, while amino acid sequences C-terminal to residue 350 are not required for ARF binding, the minimal Hdm2 interaction domain is smaller and appears not to require the distal C-terminal segment of the Hdm2 140–350 polypeptide (see below).
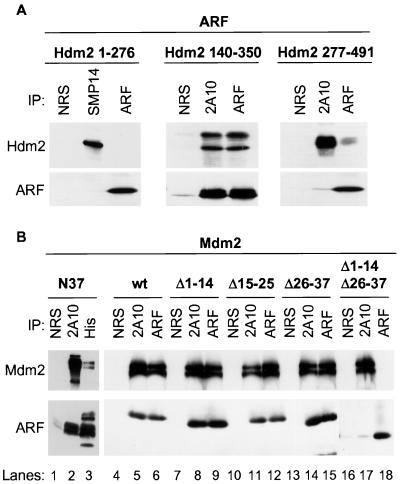
FIG. 3.
Two domains within the N terminus of mouse ARF and a central region of Mdm2 or Hdm2 are required to form the ARF-Mdm2 complex. (A) Polyhistidine-tagged Hdm2 proteins isolated from bacteria by nickel affinity chromatography were mixed for 1 h at 4°C with recombinant ARF protein prepared in baculovirus vector-infected insect Sf9 cells. Hdm2 and ARF proteins were immunoprecipitated (IP) with monoclonal antibodies to Hdm2 (SMP14 or 2A10 as indicated) or with antibody directed to the p19ARF C terminus (ARF) compared with precipitation with NRS. Precipitated proteins electrophoretically separated on denaturing gels were transferred to filters and immunoblotted with the same antibodies. (B) Polyhistidine-tagged syn-ARF N37 was mixed for 1 h at 4°C with recombinant Mdm2 produced in insect Sf9 cells (left). Sf9 cells were coinfected with baculoviruses encoding Mdm2 and the indicated ARF mutants. Mdm2 and ARF proteins were precipitated with 2A10 antibody to Mdm2 or with antibody to the ARF C terminus, whereas syn-ARF N37 was recovered using antibody to polyhistidine compared with NRS. The separated proteins were immunoblotted with the same antibodies.
ARF mRNA contains many codons poorly recognized by the most abundant bacterial tRNAs. We therefore generated a synthetic ARF N64 minigene by substituting 31 bacterial codons for their mammalian counterparts. Mass spectrometric analysis and peptide sequencing confirmed the predicted identity of the bacterial produced minigene-encoded N64 polypeptide (syn-ARF N64), which interacted with Hdm2 in a manner indistinguishable from that of the ARF N62 protein produced in Sf9 cells (data not shown). Using the synthetic minigene as a template, we then constructed syn-ARF N37 by PCR. The resulting bacterially synthesized protein was soluble and bound specifically to full-length Mdm2 produced in insect Sf9 cells (Fig. 3B, lanes 2 and 3). Interestingly, deletion of contiguous stretches of amino acids (Δ1 –14, Δ15–25, and Δ26–37) in the context of the full-length p19ARF protein did not affect ARF binding to full-length Mdm2 when these proteins were coexpressed in Sf9 cells (Fig. 3B, lanes 4 to 15). Since amino acid sequences C-terminal to ARF N62 do not contribute to Mdm2 binding (18), p19ARF likely contacts Mdm2 through more than one site within the N37 segment. In agreement with this concept, deletion of ARF residues 1 to 14 and 26 to 37 from the full-length protein resulted in the loss of Mdm2 binding (Fig. 3B, lanes 16 to 18). Identical results were obtained using Hdm2 140–350 purified from bacteria in place of full-length Mdm2 (data not shown).
These results suggested that two regions within the p19ARF N terminus (residues 1 to 14 and 26 to 37) contribute to the interaction with Mdm2. By covalently coupling ARF synthetic peptides (1–14, 15–25, 26–37, and 156–169) and syn-ARF N37 to Sepharose columns, we could use FPLC affinity chromatography to analyze their abilities to interact with purified Hdm2 140–350. The net charges at neutral pH of ARF peptides 1–14, 26–37, and 156–169 (used as a control) are very similar, so that major differences in the observed associations between basic ARF peptides and the acidic Hdm2 domain would not be likely to simply reflect electrostatic interactions. Elution of Hdm2 140–350 from ARF-Sepharose columns involved a gradient of increasing salt concentration (from 0.25 to 1.5 M) followed by a decrease in pH to below the pI of Hdm2 140–350 (pH ∼4). The Hdm2 140–350 polypeptide flowed through both the ARF 15–25 and ARF 156–169 peptide columns, showing no affinity for either resin (Fig. 4A). In contrast, Hdm2 140–350 bound strongly to the syn-ARF N37 column and could only be eluted by acid. The most conserved region of the mouse and human ARF proteins lies within amino acids 1 to 14, where 11 residues are identical and 2 of the remaining 3 are similar. Hdm2 140–350 bound to ARF 1–14 peptide columns composed of either the mouse or human ARF N termini, but it was partially eluted with salt (between 0.4 and 0.75 M and between 0.85 and 1.2 M NaCl, respectively) before being recovered at decreased pH (Fig. 4A). Similarly, Hdm2 140–350 bound to mouse ARF peptide 26–37 but was primarily eluted with NaCl (0.6 to 0.9 M) (Fig. 4A). These results agreed with the previous binding studies performed with ARF deletion mutants (Fig. 3B), indicating that p19ARF contains two noncontiguous binding sites for Hdm2 within residues 1–14 and 26–37, respectively. Moreover, the different elution profiles for the ARF peptide columns also suggested that residues 1–14 of both mouse and human ARFs might include a somewhat higher affinity-binding site for Hdm2 140–350 than mouse p19ARF residues 26 to 37.
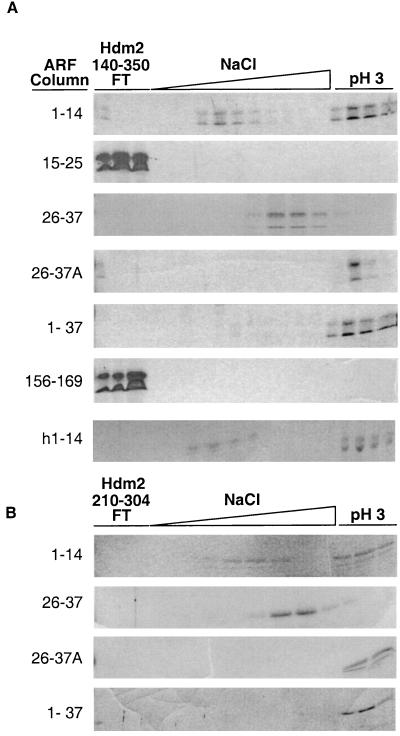
FIG. 4.
ARF-Mdm2 binding. (A) Mouse ARF peptides (1–14, 15–25, 26–37, and 156–169), recombinant syn-ARF N37, and human ARF 1–14 peptide (as indicated on the left) were coupled to Sepharose. Affinity-purified Hdm2 140–350 was injected onto the columns and eluted with an NaCl gradient and then with acid as indicated. Hdm2 140–350 preincubated with mouse soluble ARF 1–14 peptide was chromatographed on the ARF 26–37 peptide column (designated 26–37A on the left). Trichloroacetic acid-precipitated proteins were separated on denaturing polyacrylamide gels and stained with Coomassie brilliant blue G. (B) The same experiment was repeated using a smaller Hdm2 fragment including residues 210 to 304.
To determine whether Hdm2 binding to one ARF site might influence its association with another, Hdm2 140–350 was preincubated with the soluble ARF 1–14 peptide for 1 h at 4°C and then injected onto the ARF 26–37 peptide column (Fig. 4A, 26–37A). After elution of any unbound ARF peptide, a shift in the elution profile of bound Hdm2 140–350 was seen, with the majority now eluting only after the pH shift. This suggests that when Hdm2 140–350 binds to soluble ARF 1–14, its affinity for ARF 26–37 is increased, possibly through some conformational change.
ARF can inhibit the ability of Hdm2 to shuttle between the nucleus and cytoplasm by sequestering Hdm2 in the nucleolus (46, 47). It is also conceivable that ARF might block the intrinsic shuttling properties of Hdm2 by binding to its NES located between amino acids 197 and 205 (Fig. 2). To test this, we further truncated Hdm2 to include only residues 210 to 304, lacking both the NLS and NES. The elution profile of Hdm2 210–304 for ARF peptide and syn-ARF N37 affinity columns (Fig. 4B) was indistinguishable from that of Hdm2 140–350. Therefore, in agreement with the binding studies shown in Fig. 3A (middle), the ARF interaction domain of Hdm2 does not require residues 305 to 350 or the Hdm2 NLS and NES sequences.
Nucleolar localization of the ARF-Mdm2 complex is required for cell cycle arrest.
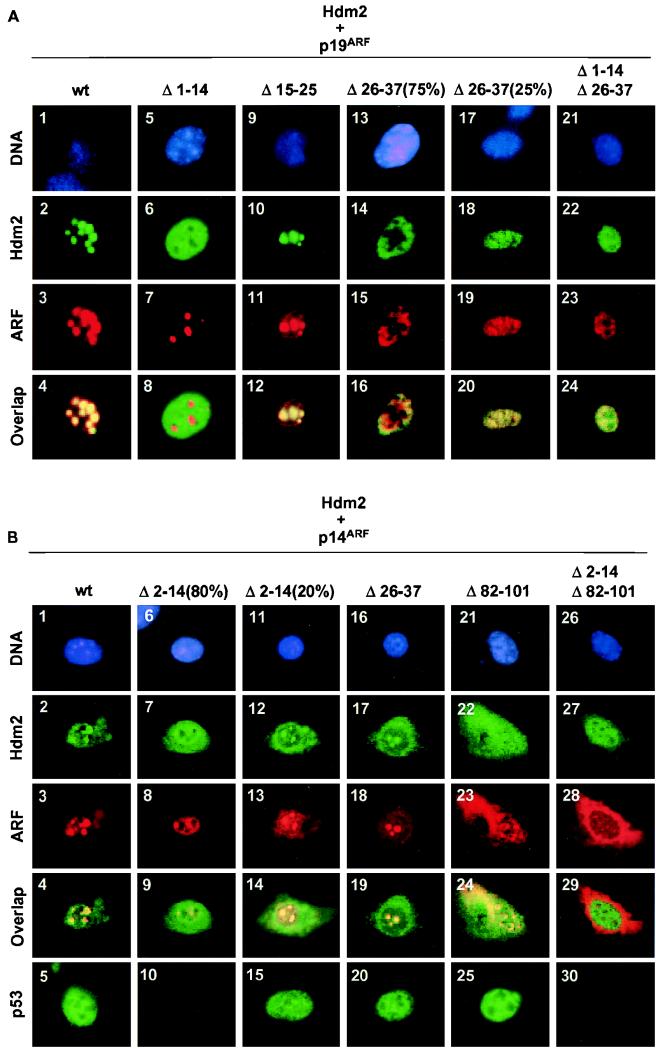
Because mouse ARF N37 retained all of the known biological properties of the full-length ARF protein, it not only contains binding sites for Mdm2 but should also have sequences that are important for p19ARF nucleolar compartmentalization. We previously found that an untruncated mouse ARF protein lacking amino acids 26 to 37 was impaired in its ability to localize to nucleoli and to induce cell cycle arrest, despite being able to bind Mdm2 (47). As shown in Fig. 5A, deletion of residues 1 to 14 (squares 5 to 8) or 15 to 25 (squares 9 to 12) from the full-length mouse p19ARF protein (squares 1 to 4) did not compromise its nucleolar localization (squares 3, 7, and 11). However, the ARF Δ26–37 mutant (squares 13 to 20) was excluded from the nucleoli of most of the transfected cells (75%) (square 15) while it localized to both the nucleoplasm and nucleoli of others (25%) (square 19). Therefore, although the p19ARF Δ26–37 mutant is impaired in its nucleolar localization relative to the wild-type protein, its enforced expression can sometimes bypass the block. Consistent with previous results, wild-type p19ARF and p19ARF Δ15–25 mobilized cotransfected Hdm2 to nucleoli (squares 2 and 10), but the p19ARF Δ26–37 mutant was largely defective (square 14). Hence, mouse ARF residues 26 to 37 not only contribute to Hdm2 binding (see above) but are also necessary in order for ARF to sequester Hdm2 in the nucleolus.
FIG. 5.
Nucleolar localization of the ARF-Mdm2 and -Hdm2 complexes. (A) NIH 3T3 cells were cotransfected with expression plasmids encoding T7-tagged Hdm2 and mouse ARF mutants as indicated. Hdm2 was detected with antibody to the T7 epitope, and p19ARF was detected with antibody to the C terminus. The numbered squares show nuclear DNA staining by Hoechst dye (top row; blue), Hdm2 fluorescence (second row; green), ARF fluorescence (third row; red), and Hdm2-ARF overlap (fourth row; yellow). In experiments performed with ARF Δ26–37, 75% of cells exhibited one staining pattern (squares 13 to 16), while the remaining 25% exhibited another (squares 17 to 20). (B) Cells were transfected with expression plasmids encoding Hdm2 and human ARF mutants as indicated. Hdm2 was detected with antibody 2A10, and p14ARF was detected with antibody to a C-terminal epitope. The organization of rows is similar to that in panel A except that representative staining of endogenous p53 (with antibody 421) expressed in transfected cells is also illustrated in the bottom row. Because the same cells could not be stained for all markers, cells stained for p53 are not the same as those illustrated in the columns directly above them. In data not shown, cells stained for p53 were costained for p14ARF to identify positive transfectants. In experiments performed with ARF Δ2–14, 80% of transfected cells exhibited one staining pattern (squares 6–10) and the remaining 20% exhibited another (squares 11 to 15).
The p19ARF mutant lacking residues 1 to 14 remained able to enter nucleoli (Fig. 5A, square 7), but it failed to import Hdm2 (square 6). Although p19ARF Δ1–14 coprecipitates with Mdm2 in Sf9 extracts (Fig. 3B) and binds Mdm2 in vitro (Fig. 4), its inability to relocalize Hdm2 to nucleoli points to a more complex interaction in vivo. The lower-affinity interaction between p19ARF Δ1–14 and Hdm2 and/or the inability of this mutant to induce a conformational change in Hdm2 (Fig. 4) (see the last section of Results below) may hamper the ability of ARF to sequester Hdm2 in the nucleolus. The key points here are that both Hdm2 binding and nucleolar localization are necessary for p19ARF-induced cell cycle arrest (Table 1). On the one hand, despite the fact that the p19ARF Δ1–14 mutant localized to nucleoli, its failure to mobilize Hdm2 to this compartment correlated with its inability to block DNA replication (Table 1). Moreover, on a cell-by-cell basis, that fraction of cells (75%) that expressed the ARF Δ26–37 mutant in the nucleoplasm incorporated BrdU, whereas those that exhibited both nucleolar and nucleoplasmic ARF staining did not. Therefore, the ARF Δ26–37 mutant behaves hypomorphically; although it is largely defective in localizing to nucleoli, importing Hdm2, and inducing cell cycle arrest, its gross overexpression can overcome the defects. As expected, the combined deletion of segments 1 to 14 and 26 to 37 from mouse ARF generated a mutant that was completely devoid of activity (Fig. 5A, squares 21 to 24, and Table 1).
TABLE 1.
Nucleolar localization of the ARF-Mdm2 complex correlates with growth arrest
| Retrovirus | ARF localizationa | Hdm2 localizationa | % BrdU-positive cellsb |
|---|---|---|---|
| Mouse | |||
| CD8 vector control | 96.4 ± 6.7 | ||
| ARF (wild type) | Nucleoli | Nucleoli | 7.6 ± 4.3 |
| ARF Δ1–14 | Nucleoli | Nucleoplasm | 81.2 ± 9.5 |
| ARF Δ15–25 | Nucleoli | Nucleoli, nucleoplasm | 11.6 ± 5.1 |
| ARF Δ26–37 (75%) | Nucleoplasm, cytoplasm | Nucleoplasm | 85.6 ± 8.5 |
| ARF Δ26–37 (25%) | Nucleoli, nucleoplasm, cytoplasm | Nucleoli, nucleoplasm | 16.1 ± 4.2 |
| ARF Δ1–14-Δ26–37 | Nucleoplasm, cytoplasm | Nucleoplasm, cytoplasm | 95.7 ± 6.0 |
| Human | |||
| CD8 vector control | 92.6 ± 4.1 | ||
| ARF (wild type) | Nucleoli | Nucleoli | 9.2 ± 4.0 |
| ARF Δ2–14 (80%) | Nucleoplasm, cytoplasm | Nucleoplasm, cytoplasm | 84.9 ± 7.1 |
| ARF Δ2–14 (20%) | Nucleoli, nucleoplasm, cytoplasm | Nucleoli, nucleoplasm, cytoplasm | 30.5 ± 5.8 |
| ARF Δ26–37 | Nucleoli, nucleoplasm | Nucleoli, nucleoplasm | 16.8 ± 8.3 |
| ARF Δ82–101 | Nucleoli, cytoplasm | Nucleoli, nucleoplasm, cytoplasm | 11.5 ± 5.6 |
| ARF Δ2–14-Δ82–101 | Cytoplasm | Nucleoplasm, cytoplasm | 92.6 ± 3.0 |
NIH 3T3 cells were cotransfected with expression vectors encoding T7-tagged Hdm2 and the indicated ARF mutants. Mouse ARF was detected using antibodies to the p19ARF C terminus, with simultaneous detection of Hdm2 using antibody to the T7 epitope. Human ARF was scored using antibodies to the p14ARF C terminus, with simultaneous detection of Hdm2 using monoclonal antibody 2A10. The relative distribution of staining is summarized, as documented in Fig. 5.
NIH 3T3 cells were infected in parallel with retroviruses encoding the same ARF mutants, but without Hdm2, and scored 24 h postinfection for BrdU incorporation into replicating DNA (24-h pulse, equivalent to one cell cycle). Three independent coverslips were scored, and over 100 cells were counted for each (± standard deviation).
Despite different localization signals, cell cycle arrest by human p14ARF also requires nucleolar import of Hdm2.
To assess functional differences between various domains within the mouse and human ARF proteins, we analyzed human p14ARF deletion mutants as well (Fig. 5B and Table 1). In this set of experiments, representative data illustrating p53 staining are included. Unlike its mouse ARF counterpart, the localization of human p14ARF Δ2–14 varied from cell to cell. A majority of transfected cells (80%) (Fig. 5B, squares 6 to 10) displayed primarily nucleoplasmic staining for p14ARF Δ2–14 (square 8). In this population, Hdm2 remained in the nucleoplasm and cytoplasm (square 7), p53 was not induced (square 10), and cells did not undergo proliferative arrest (Table 1). The remaining transfected cells exhibited some detectable nucleolar ARF staining (Fig. 5B, square 13), and in these, some Hdm2 was mobilized to the nucleolus (square 12), p53 staining was increased (square 15), and BrdU incorporation was significantly inhibited (Table 1). Thus, residues 2 to 14 in human p14ARF are not only necessary for Mdm2 binding but, unlike the cognate conserved region of mouse p19ARF, also contribute to p14ARF nucleolar compartmentalization. These data agree with recently obtained results of Lohrum et al. (personal communication), who found that incorporation of p14ARF residues 1 to 22 into the active site loop of thioredoxin mobilized it to nucleoli.
In addition, a second NrLS was previously mapped to residues 82 to 101 of human p14ARF (50). Deletion of amino acids 82 to 101 also resulted in significant delocalization of human ARF throughout the cells (Fig. 5B, square 23). Nonetheless, residual p14ARF Δ82–101 and Mdm2 costaining were seen in the nucleoli of transfected cells (square 24), p53 was induced (square 25), and most cells underwent arrest (Table 1). Together, these results and those described above are inconsistent with a previous suggestion that p14ARF nucleolar localization depends solely on the NrLS within residues 82 to 101 (50). Indeed, whereas both human p14ARF Δ2–14 and Δ82–101 behaved hypomorphically, deletion of both regions resulted in complete delocalization of ARF to the cytoplasm (Fig. 5B, square 28), no mobilization of Hdm2 to the nucleolus (Fig. 5B, squares 27 and 29), no induction of p53 (square 30), and no detectable cell cycle arrest (Table 1). In contrast to mouse ARF, deletion of amino acids 26 to 37 from human p14ARF had little effect on its ability to localize to the nucleolus or to induce cell cycle arrest (Fig. 5B, squares 16 to 20, and Table 1). Therefore, although the regions required for nucleolar localization of mouse and human ARF are different in their placement, it is clear that efficient nucleolar colocalization of either mouse p19ARF or human p14ARF with Mdm2 or Hdm2 is required for halting the cell cycle.
Mdm2 contributes to nucleolar localization of the ARF-Mdm2 complex.
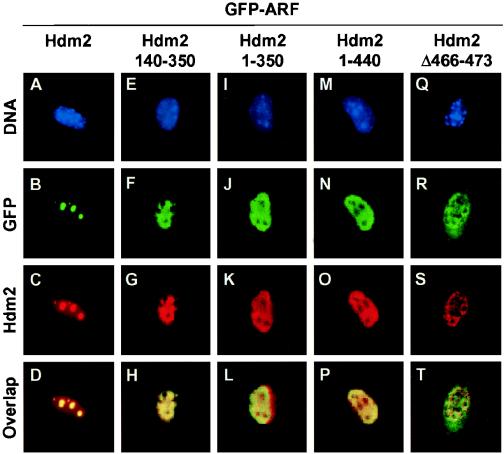
Mouse ARF N62 localizes to nucleoli in primary MEFs lacking both p53 and Mdm2, indicating that neither of the gene products is strictly essential for p19ARF nucleolar import (46, 47). However, our findings that particular sequences contributing to nucleolar localization of both mouse (residues 26 to 37) and human (residues 2 to 14) ARFs overlap segments that contact Mdm2 raised questions as to how the ARF NrLS can induce the nucleolar mobilization of the ARF-Mdm2 complex. Importantly, none of these data preclude the possibility that Mdm2 contributes to the nucleolar localization of the ARF-Mdm2 complex. Indeed, whereas GFP-p19ARF mobilized Hdm2 to nucleoli (Fig. 6A to D), its coexpression with the Hdm2 140–350 fragment containing the ARF binding site(s) resulted in retention of both ARF and this Hdm2 mutant in the nucleoplasm (Fig. 6E to H). At this point, we learned that Mdm2 contains a cryptic NrLS in its C-terminal RING domain (Fig. 2, residues 466 to 473) that appears to be unmasked upon ARF binding (24a). Specifically, an Hdm2 deletion mutant lacking residues 222 to 437 (which includes the ARF-binding domain) can relocalize to the nucleolus in the absence of p14ARF, whereas a C-terminal-truncation mutant (1 to 440) lacking the Hdm2 NrLS cannot be mobilized by human p14ARF to this compartment (e.g., Fig. 6M to P). Appending Hdm2 residues 466 to 473 to thioredoxin can reroute it to the nucleolus (24a). Conversely, deletion of these residues from full-length Hdm2 enables it to sequester mouse GFP-p19ARF in the nucleoplasm (Fig. 6Q to T). Therefore, whereas ARF localizes to nucleoli in the complete absence of Mdm2 (47), both ARF and Mdm2 contribute to nucleolar localization of the complex.
FIG. 6.
Hdm2 mutants lacking an NrLS in the RING domain remain in the nucleoplasm and sequester ARF within the same compartment. NIH 3T3 cells were transfected with expression plasmids encoding full-length Hdm2 and different Hdm2 mutants as indicated together with GFP-ARF. Hdm2 was detected with antibody 2A10. The numbered squares show nuclear DNA staining by Hoechst dye (top row; blue), GFP-ARF fluorescence (second row; green), Hdm2 fluorescence (third row; red), and Hdm2-ARF overlap (fourth row; yellow). The dark unstained regions within the nuclei correspond to nucleoli, as confirmed by using antibodies to fibrillarin (data not shown) (47).
DISCUSSION
The ARF-Mdm2 interaction is necessary for ARF to induce p53-dependent cell cycle arrest. Mdm2 binds to p53 (13, 28), ubiquitinates it (14, 15), and accelerates its shuttling from the nucleoplasm to the cytoplasm, where it is targeted for proteasomal degradation (12, 22, 37). ARF interferes with Mdm2-directed ubiquitination of p53 in vitro (15), and the ability of ARF to relocalize Mdm2 to the nucleolus allows transcriptionally active p53 to accumulate in the nucleoplasm in response to hyperproliferative signals (47). We have now attempted to pinpoint ARF-Mdm2 contact sites and NrLSs and to assay the functions of mutants lacking these segments in vivo. Given previous suggestions that the human and mouse ARF proteins might be functionally different from one another (50), we have also tried to compare the biological activities of mouse p19ARF mutants with those of their human p14ARF counterparts.
Mdm2 binding signals can overlap those required for ARF nucleolar localization.
The N-terminal 62 amino acids of mouse p19ARF retain all of the known functions of the full-length protein (18, 34, 47), and here we have further limited the active domain to amino acids 1 to 37. Given that GFP-p19ARF N37 localizes to nucleoli even in cells that lack Mdm2, and that ARF N37 binds to Mdm2 or Hdm2 and induces cell cycle arrest, this small mouse ARF polypeptide must contain both an NrLS and an Mdm2 binding site. Residues 26 to 37 of p19ARF are required for its efficient nucleolar localization, and their deletion results in a greatly reduced ability of ARF to mobilize Mdm2 to nucleoli and to arrest the cell cycle (47). We have noted that this region of the mouse ARF protein contains an R/K-R/K-X-R/K motif found in many other nucleolar proteins (Fig. 7). Interestingly, p19ARF Δ26–37 retained the ability to bind Mdm2 both in vitro and in vivo, leading us to reason previously that relocalization of Mdm2 to nucleoli by ARF was a necessary event and that binding to Mdm2 alone was not sufficient for ARF-induced cell cycle arrest (47).
FIG. 7.
Nucleolar localization signals. Regions necessary for nucleolar localization in various proteins are indicated and aligned around a conserved R/K-R/K-X-R/K amino acid motif. Duplicated signals in other proteins are indicated by shaded boxes. The importance of such sequences in nucleolar compartmentalization has been documented (2–5, 24a, 27, 38, 42, 47, 50).
Mouse p19ARF contains two noncontiguous binding sites for Hdm2 which are restricted to ARF residues 1 to 14 and 26 to 37. Immediately, this alerted us to the fact that residues 26 to 37 had a dual function. In turn, the region of Mdm2 that binds to p19ARF is limited to amino acids 210 to 304. This acidic domain lacks the N-terminal p53-binding portion, the C-terminal RING finger, and the NLS and NES required for Mdm2 nuclear import and exit (Fig. 2). An immobilized ARF 1–14 peptide bound to Hdm2 210–304 (or to longer Hdm2 or Mdm2 fragments containing this segment) with extremely high affinity, only being dissociated with acid. The immobilized ARF 26–37 peptide bound Hdm2 with lower affinity, undergoing dissociation in high salt at neutral pH. Preincubation of Hdm2 with the soluble ARF 1–14 peptide resulted in higher-affinity binding of this complex to the immobilized ARF 26–37 peptide than that observed with Hdm2 alone. This demonstrated an influence of ARF residues 1 to 14 on the Hdm2 binding properties of residues 26 to 37, presumably through an induced conformational change in Hdm2. Importantly, disruption of either of the two Mdm2 and Hdm2 contact sites within ARF produced a nonfunctional p19ARF protein. However, contrary to results obtained with p19ARF Δ26–37, which was handicapped in nucleolar compartmentalization, p19ARF Δ1–14 localized to nucleoli but was unable to mobilize Mdm2 or Hdm2 to the same compartment. Although the reduced affinity of p19ARF Δ1–14 for Hdm2 could potentially account for the latter result, our data rather suggest that the apparent conformational change in Mdm2 induced by cooperative ARF binding is required for relocalization of the ARF-Mdm2 complex to nucleoli (see below).
Signals required for nucleolar localization of human p14ARF are displayed somewhat differently from those in the mouse protein. An NrLS for p14ARF (Fig. 7) was previously mapped between amino acids 82 and 101 encoded by ARF exon 2 (50). Deletion of this region in the context of full-length p14ARF resulted in a significant redistribution of the human ARF protein to the cytoplasm. However, when overexpressed, some p14ARF Δ82–101 still gained access to nucleoli, thereby mobilizing Hdm2 to the same compartment and inducing cell cycle arrest. This implied that another NrLS is present elsewhere in p14ARF. Indeed, apart from binding Hdm2, residues 2 to 14 of human ARF also contribute to nucleolar localization. Deletion of the latter segment prevented p14ARF from entering nucleoli in most cells and resulted in its concordant inability to relocalize Hdm2. However, a significant fraction of cells (∼20%) that overexpressed p14ARF Δ2–14 displayed nucleolar localization of both ARF and Hdm2, and these underwent arrest. Therefore, human p14ARF contains two NrLSs located between residues 2 and 14 and 82 and 101, and like the NrLS contained in mouse p19ARF residues 26 to 37, the former overlaps with an Hdm2 binding site.
Given the strong conservation of amino acid sequences at the extreme N termini of the human and mouse ARF proteins, why is human p14ARF Δ2–14 partially handicapped in entering nucleoli while its mouse counterpart appears to be unaffected? There is no R/K-R/K-X-R/K motif within residues 2 to 14 of human p14ARF, but an R/K-X-R/K-R/K segment is present, similar to basic sequence blocks found in nucleolar fibrillarin and nucleolin. Within mouse p19ARF residues 1 to 14, this sequence is replaced by R-I-Q-R. It is conceivable that mouse ARF residues 1 to 14 still contribute weakly to nucleolar compartmentalization but that their effect is occluded by the nearby NrLS located between residues 26 and 37. This is consistent with observations that mouse p19ARF Δ26–37 could enter the nucleolus when overexpressed, whereas p19ARF Δ1–14-Δ26–37 was unable to do so. In these respects, human p14ARF Δ2–14 and p14ARF Δ82–101 and mouse p19ARF Δ26–37 behave hypomorphically; their gross overexpression can overcome their impaired ability to relocalize Hdm2 to the nucleolus.
We have not yet identified sequences outside the residue 2 to 14 domain of human p14ARF that contribute to Hdm2 binding, but the apparent ability of overexpressed p14ARF Δ2–14 to relocalize Hdm2 to nucleoli also indicates that another binding site exists elsewhere in the protein. Given that human p14ARF Δ2–14-Δ82–101 was functionally inert, the most parsimonious explanation would be that, like residues 26 to 37 in mouse ARF, the segment from 82 to 101 in the human protein also contributes to Hdm2 binding. Despite the differences in the disposition of signals within human p14ARF and mouse p19ARF, all the available data reinforce the view that nucleolar localization of Mdm2 and Hdm2 in a complex with either the mouse or human ARF protein is required to arrest the cell cycle.
A cryptic Mdm2 NrLS is required for nucleolar localization of the ARF-Mdm2 complex.
It seemed puzzling that both mouse p19ARF amino acids 26 to 37 and human p14ARF residues 2 to 14 could serve as both NrLSs and sites for Mdm2 binding. Indeed, we initially anticipated that the ARF NrLS would allow binding to a protein other than Mdm2 that in turn would facilitate the nucleolar transport of the ARF-Mdm2 complex or its tethering within that compartment. Surprisingly, however, Mdm2 itself contributes to nucleolar targeting. When coexpressed with p19ARF, Hdm2 mutants lacking residues C terminal to the ARF binding domain sequestered ARF in the nucleoplasm. Others mapped a cryptic NrLS (amino acids 466 to 473) in the C-terminal RING domain of Hdm2 that can function to relocalize Hdm2 to the nucleolus when its central domain (residues 222 to 437) is deleted (24a). Conversely, truncation of Hdm2 at residue 440 results in a protein that retains coexpressed ARF in the nucleoplasm. An Hdm2 mutant lacking only amino acids 466 to 473 behaves similarly. Therefore, mobilization of the ARF-Hdm2 complex to nucleoli depends, at least in part, on the Hdm2 NrLS. Tandem R/K-R/K-X-R/K motifs compose this segment (Fig. 7). Because Mdm2 or Hdm2 provides a crucial localization signal that determines the topological fate of ARF, the interaction between ARF and Mdm2 can be viewed as bidirectional, with each protein regulating transport of the other.
Hdm2 apparently undergoes a conformational change after contacting both ARF-binding sites, and this may unmask the Hdm2 NrLS, but how the NrLSs actually function in directing these proteins to the nucleolus remains unclear. One possibility is that the NrLS of Mdm2 or Hdm2 normally interacts with its central acidic domain and is revealed when ARF binds to the same region. Another idea is that ARF competes with a binding protein that retains Hdm2 in the nucleoplasm. However, gross overexpression of Mdm2 or Hdm2 does not appear to titrate a nucleoplasmic tethering protein and so allow Mdm2 or Hdm2 to enter nucleoli; instead, gross overexpression of Mdm2 or Hdm2 can generate “nuclear bodies” that trap coexpressed ARF in the nucleoplasm (L. Taylor, J. D. Weber, C. J. Sherr, and D. Bar-Sagi, unpublished observations). Our results and those of others (24a, 32, 46, 47) imply that nuclear-body formation (50) does not occur under physiologic circumstances and strongly argue against the interpretation that such structures are required for ARF-induced cell cycle arrest.
What is the role of the NrLS?
If in fact the NrLSs act as positive signals for nucleolar import, then these motifs may be necessary for binding to active transporters in a manner analogous to those of NLSs and NESs. There are similarities between the ARF and Hdm2 NrLSs that correspond to sequence motifs in other proteins that also gain access to the nucleolus (Fig. 7). Among the latter is 5S RNA-binding ribosomal protein L5, which can also interact with the central domain of Hdm2 (Fig. 2) (25) and could conceivably compete with ARF for Mdm2 binding. The fact that Mdm2 export to the cytoplasm can be blocked by ARF (46, 50) and by polypeptide inhibitors of lentiviral Rev transport (37) is also intriguing, given the presence of a related signature motif in the human immunodeficiency virus type 1 Rev protein itself (Fig. 7). ARF does not bind directly to the Mdm2 NES but associates in close proximity (Fig. 2). It may well prove that different Mdm2 binding proteins have differential effects on Mdm2 transport, with proteins like L5, for example, perhaps acting as positive coregulators of nuclear export and ARF functioning instead as an inhibitor. In turn, the possibility that ARF might affect the transport of proteins other than Mdm2 remains an open question.
Mice engineered to overexpress a myc transgene under the control of the immunoglobulin heavy chain enhancer (Eμ) develop pre-B and B-cell lymphomas, with a majority of the resulting tumors sustaining ARF deletion, p53 mutation, or Mdm2 overexpression (8). It was noted, however, that several tumors that overexpressed Mdm2 isoforms also sustained ARF deletion, pointing to a more complex biochemical interaction between ARF and Mdm2 than was previously thought. In addition to p53 and L5, Mdm2 can also bind to other p53 family members (49), E2F-1 (26), p300 (11), and the retinoblastoma protein (48), underscoring its potential for interaction with other targets. Human tumors can sustain amplification of Hdm2, resulting in the overexpression of various spliced forms. Interestingly, many of these Hdm2 proteins retain the ARF-binding acidic domain and truncate the NrLS-containing C terminus. Alterations of the RING domain might not only enhance Mdm2 and Hdm2 stability (23), presumably by canceling their E3 ubiquitin ligase activities (15), but might also act to antagonize ARF function. This may in effect provide dominant forms of Mdm2 and Hdm2 which bind to ARF and sequester it in the nucleoplasm, allowing remaining Mdm2 and Hdm2 proteins to target p53 or other Mdm2 and Hdm2 binding proteins. Identifying the sequences of potentially oncogenic spliced forms of Mdm2 and Hdm2 and determining their ability to circumvent ARF surveillance may help identify other targets in the ARF-Mdm2 pathway.
ACKNOWLEDGMENTS
We thank Karen Vousden for alerting us to the functional significance of the cryptic Mdm2 NrLS, for generously providing the Hdm2 1–440 mutant, and for exchanging manuscripts prior to submission for publication. We thank Esther Van de Kamp, Rose Mathew, and Ming Wang for excellent technical assistance; Jinjun Dang for his help in generating the Hdm2 mutant lacking residues 466 to 473; and John L. Cleveland for insightful criticism of the manuscript.
This work was supported in part by NIH grants P01 CA-71907 (M.F.R.), Cancer Center CORE grant CA-21765, the American Cancer Society (R.W.K.), and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. B.B. is supported by a Hal and Alma Reagan Fellowship. C.J.S. is an Investigator and J.D.W. is a Research Associate of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bates S, Phillips A C, Clarke P, Stott F, Peters G, Ludwig R L, Vousden K H. E2F-1 regulation of p14ARF links pRB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 2.Chan Y-L, Lin A, McNally J, Wool I G. The primary structure of rat ribosomal protein L5. J Biol Chem. 1987;262:12879–12886. [PubMed] [Google Scholar]
- 3.Chang J H, Dumbar T S, Olson M O. cDNA and deduced primary structure of rat protein B23, a nucleolar protein containing highly conserved sequences. J Biol Chem. 1988;263:12824–12827. [PubMed] [Google Scholar]
- 4.Cochrane A W, Perkins A, Rosen C A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang C V, Lee W M F. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 6.De Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 8.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-MDM2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by mdm2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giaccia A J, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 11.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 12.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 13.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 14.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 15.Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of MDM2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 17.Kamijo T, Bodner S, van de Kamp E, Randle D H, Sherr C J. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- 18.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 20.Kemp C J, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 21.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 22.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 23.Kubbutat M H G, Ludwig R L, Levine A J, Vousden K H. Analysis of the degradation function of Mdm2. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- 24.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 24a.Lohrum, M. A. E., M. Ashcroft, M. H. G. Kubbutat, and K. H. Vousden. Identification of a cryptic nucleolar localization signal in MDM2. Nat. Cell Biol., in press. [DOI] [PubMed]
- 25.Marechal V, Elenbaas B, Piette J, Nicolas J-C, Levine A J. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin K, Trouche D, Hagemeier C, Sorensen T S, LaThangue N B, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by mdm2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 27.Michael W M, Dreyfuss G. Distinct domains in ribosomal protein L5 mediate 5 S rRNA binding and nucleolar localization. J Biol Chem. 1996;271:11571–11574. doi: 10.1074/jbc.271.19.11571. [DOI] [PubMed] [Google Scholar]
- 28.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 29.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive leukemias. Mol Cell Biol. 1994;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 31.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 32.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R. The Ink4a tumor suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 33.Prives C. Signaling to p53:breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 34.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:3436–3440. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 36.Radfar A, Unnikrishnan I, Lee H-W, DePinho R A, Rosenberg N. p19Arf induces p53-dependent apoptosis during Abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth J, Dobbelstein M, Freedman D, Shenk T, Levine A J. Nucleocytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt C, Lipsius E, Kruppa J. Nuclear and nucleolar targeting of human ribosomal protein S6. Mol Biol Cell. 1995;6:1875–1885. doi: 10.1091/mbc.6.12.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 40.Sharpless N E, DePinho R A. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 41.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 42.Siomi H, Shida H, Nam S H, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type 1 pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 43.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao W, Levine A J. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao W, Levine A J. P19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 48.Xiao Z X, Chen J, Levine A, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interactions between the retinoblastoma protein and the oncoprotein mdm2. Nature. 1995;375:694–697. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 49.Zeng X, Chen L, Jost C A, Maya R, Keller D, Wang X, Kaelin W G, Jr, Oren M, Chen J, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppressor pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 52.Zindy F, Eischen C M, Randle D, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]