Summary
Acute lymphoblastic leukemia (ALL) is the most common childhood cancer. Despite overlap between genetic risk loci for ALL and hematologic traits, the etiological relevance of dysregulated blood-cell homeostasis remains unclear. We investigated this question in a genome-wide association study (GWAS) of childhood ALL (2,666 affected individuals, 60,272 control individuals) and a multi-trait GWAS of nine blood-cell indices in the UK Biobank. We identified 3,000 blood-cell-trait-associated (p < 5.0 × 10−8) variants, explaining 4.0% to 23.9% of trait variation and including 115 loci associated with blood-cell ratios (LMR, lymphocyte-to-monocyte ratio; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio). ALL susceptibility was genetically correlated with lymphocyte counts (rg = 0.088, p = 4.0 × 10−4) and PLR (rg = −0.072, p = 0.0017). In Mendelian randomization analyses, genetically predicted increase in lymphocyte counts was associated with increased ALL risk (odds ratio [OR] = 1.16, p = 0.031) and strengthened after accounting for other cell types (OR = 1.43, p = 8.8 × 10−4). We observed positive associations with increasing LMR (OR = 1.22, p = 0.0017) and inverse effects for NLR (OR = 0.67, p = 3.1 × 10−4) and PLR (OR = 0.80, p = 0.002). Our study shows that a genetically induced shift toward higher lymphocyte counts, overall and in relation to monocytes, neutrophils, and platelets, confers an increased susceptibility to childhood ALL.
Keywords: acute lymphoblastic leukemia, blood-cell traits, GWAS, Mendelian randomization, lymphocytes
Introduction
The hematopoietic system is remarkably orchestrated and responsible for some of the most important physiological functions, such as the production of adaptive and innate immunity, nutrient transport, clearance of toxins, and wound healing. Genetic factors contribute significantly to inter-individual variation in blood-cell phenotypes, and heritability estimates for most blood-cell traits range from 50%–90% in twin studies to 30%–40% in population-based studies of array-based heritability.1, 2, 3, 4 Genome-wide association studies (GWASs) conducted in large population-based studies have revealed the highly polygenic nature of blood-cell traits, and over 5,000 independently associated genetic loci have been identified to date.4, 5, 6 Results from these studies have also provided insights into the genetic regulation of hematopoiesis and how dysregulation in blood-cell development can lead to disease.7 Genetic variants associated with blood-cell variation have been implicated in the risk of immune-related conditions, such as asthma, rheumatoid arthritis, and type 1 diabetes, and in rare blood disorders.4, 5, 6 Positive genetic correlation was found between counts of varying blood-cell types and the risk of myeloproliferative neoplasms, a group of diseases primarily of older age and characterized by the overproduction of mature myeloid cells.8 However, the contribution of heritable variation in blood-cell traits to the risk of other hematologic cancers has not been examined.
Acute lymphoblastic leukemia (ALL [MIM: 613065]) is a malignancy of white blood cells, developing from immature B cells or T cells, and is the most common cancer diagnosed in children under 15 years of age.9 Despite significant advances in treatment in recent decades and the corresponding improvements in survival rates,10 ALL remains one of the leading causes of pediatric cancer mortality in the United States.11 In addition, childhood ALL patients may endure severe toxicities during treatment, and survivors face long-term treatment-related morbidities and mortality.12,13 Thus, understanding the etiology of ALL remains important for identification of avenues for disease prevention as well as potential novel treatment targets.
In most cases, the development of ALL is thought to follow a two-hit model of leukemogenesis; in utero formation of a preleukemic clone and subsequent postnatal acquisition of secondary somatic mutations that drive progression to overt leukemia.14 Epidemiological studies have identified several genetic and non-genetic risk factors for ALL (reviewed in Williams et al.15 and Greaves14), but the biological mechanisms through which they promote leukemogenesis are largely unknown. GWASs of childhood ALL have revealed at least 12 common genetic risk loci to date, including at genes involved in hematopoiesis and early lymphoid development,16 such as ARID5B (MIM: 608538), IKZF1 (MIM: 603023), CEBPE (MIM: 600749), GATA3 (MIM: 131320), BMI1 (MIM: 164831), IKZF3 (MIM: 606221), and ERG (MIM: 165080).17, 18, 19, 20, 21, 22, 23, 24 Intriguingly, several childhood-ALL-risk regions have also been associated with variation in blood-cell traits4,6,22,23,25 and a recent phenome-wide association study (PheWAS) of childhood ALL identified platelet count as the most enriched trait among known ALL-risk loci.26 A comprehensive study of the role of blood-cell-trait variation in the etiology of childhood ALL is, therefore, warranted.
In this study, we utilize genome-wide data available from the UK Biobank (UKB) resource27 to perform a GWAS of blood-cell traits and apply the discovered loci to a GWAS of childhood ALL in 2,666 affected individuals and 60,272 control individuals of European ancestry. We assess the shared genetic architecture between blood-cell phenotypes and childhood ALL and conduct Mendelian randomization (MR) and mediation analyses to disentangle putative causal effects of variation in blood-cell homeostasis on ALL susceptibility.
Subjects and methods
Development of genetic instruments for blood-cell traits
The UKB is a population-based prospective cohort of over 500,000 individuals aged 40–69 years at enrollment in 2006–2010 who completed extensive questionnaires on health-related factors, underwent physical assessments, and provided blood samples.27 Blood samples collected in 4 mL EDTA vacutainers were analyzed with four Beckman Coulter LH750 instruments. The LH750 instrument is a quantitative, automated hematology analyzer and leukocyte differential counter for in vitro diagnostic use in clinical laboratories. Samples were analyzed at the UKB central laboratory within 24 h of blood draw.
Quality control (QC) steps for this dataset have been previously described.28 Briefly, genetic association analyses were restricted to individuals of predominantly European ancestry identified on the basis of self-report and refined by excluding samples with any of the first two genetic ancestry principal components (PCs) outside of 5 SD of the population mean. We removed samples with discordant self-reported and genetic sex, as well as one sample from each pair of first-degree relatives identified by using KING.29 Using a subset of genotyped autosomal variants with minor allele frequency (MAF) ≥ 0.01 and call rate ≥ 97%, we filtered samples with call rates < 97% or heterozygosity > 5 SD from the mean, leaving 413,810 individuals available for analysis.
We applied additional exclusions to optimize our dataset for developing genetic instruments for studies of cancer etiology by removing subjects with medical conditions that would alter blood-cell proportions by pathophysiological conditions (n = 13,597), such as pre-malignant myelodysplastic syndromes (MDS [MIM: 614286]), autoimmune diseases (MIM: 109100), and immunodeficiencies, including HIV (Figure S1). Blood counts (109 cells/L) outside of the LH750 reportable range and extreme outliers (>99th percentile) were excluded. Remaining values were converted to normalized Z scores with mean = 0 and SD = 1. In addition to overall blood-cell counts, we also examined relative concentrations: lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio (NLR), and platelet-to-lymphocyte ratio (PLR).
UKB participants were genotyped on the UK Biobank Affymetrix Axiom array (89%) or the UK BiLEVE array (11%) with imputation performed with the Haplotype Reference Consortium (HRC) and the merged UK10K and 1000 Genomes (1000G) phase 3 reference panels.27 We excluded variants that were out of Hardy-Weinberg equilibrium in cancer-free individuals (pHWE < 1 × 10−5) or had low imputation quality (INFO < 0.30). Analyses were restricted to 10,369,434 variants with MAF ≥ 0.005.
Genome-wide association analyses were conducted with linear regression in PLINK 2.0 (October 2017 version). Blood-cell traits were analyzed via a two-stage GWAS with a randomly sampled 70% of the cohort used for discovery and the remaining 30% reserved for replication followed by multi-trait analysis of GWAS (MTAG).30 Models for each trait were adjusted for age, age2, sex, genotyping array, the first 15 PCs, cigarette pack-years, blood-count device ID, and assay date. The resulting summary statistics were analyzed via MTAG, which has been shown to increase power to detect associations for correlated phenotypes by distinguishing between genetic correlation and correlations due to sample overlap or biases in GWAS effect sizes due to population stratification or cryptic relatedness.30 Genetic instruments were selected from MTAG results and defined as independent variants (linkage disequilibrium [LD] r2<0.05 in a clumping window of 10,000 kb) with p < 5 × 10−8 in the discovery stage and p < 0.05 and consistent direction of effect in the replication stage.
The functional relevance of the genetic instruments for blood-cell traits was assessed with in-silico functional annotations: combined annotation-dependent depletion (CADD) scores31 and RegulomeDB.32 We also explored associations with gene expression in whole blood in eQTLGen,33 a meta-analysis of 31,684 subjects, and immune-cell specific effects in DICE (Database of Immune Cell Expression),34 a dataset of 91 healthy blood donors; BLUEPRINT35 (n = 197 healthy blood donors); and CEDAR (Correlated Expression and Disease Association Research)36 (n = 322 healthy individuals from a cancer screening cohort). Gene expression datasets were accessed from the FUMA platform.37
Childhood acute lymphoblastic leukemia datasets
Genetic associations with childhood ALL were obtained from a meta-analysis of 2,666 affected individuals and 60,272 control individuals from two separate genome-wide scans38 (details in supplemental subjects and methods). The first GWAS consisted of a pooled dataset of 1,162 affected individuals and 1,229 control individuals from the California Cancer Records Linkage Project (CCRLP)21 with 56,112 additional control individuals from the Kaiser Permanente Genetic Epidemiology Research on Aging (GERA) cohort. Details of the CCRLP study and combined CCRLP/GERA GWAS have been previously described;21 the present analysis includes additional GERA control individuals and imputation with the HRC reference panel (version r1.1 2016).38 All CCRLP and GERA participants were genotyped on the Affymetrix Axiom World Array. The second ALL GWAS included 1,504 ALL-affected individuals from the Children’s Oncology Group (COG) and 2,931 cancer-free control individuals from the Wellcome Trust Case-Control Consortium (WTCCC), genotyped on either the Affymetrix Human SNP Array 6.0 (WTCCC, COG trials AALL0232 and P9904/9905)25 or the Affymetrix GeneChip Human Mapping 500K Array (COG P9906 and St. Jude Total Therapy XIIIB/XV).39 GWAS meta-analysis was restricted to individuals of predominantly European ancestry.
Standard QC steps were implemented,38 removing variants with pHWE < 1 × 10−5 in control individuals and imputation INFO < 0.30. We applied additional filters to minimize potential for bias due to the inclusion of external control individuals (Figure S1). Variants associated with control group (CCRLP versus GERA) at p < 1 × 10−5 were removed (n = 443). We also excluded variants if their MAF differed by >50% or ≥0.10 from the average MAF across CCRLP, GERA, and WTCCC control individuals (MAF ≥ 0.05, n = 3,029; MAF < 0.05, n = 198,632). Lastly, allele frequencies in CCRLP/GERA and WTCCC control individuals were compared to the gnomAD non-Finnish European reference dataset and variants with absolute MAF differences ≥ 0.10 were filtered out (n = 21,863).
Heritability and genetic correlation
We used LD score regression40 to estimate heritability (hg) for each blood-cell phenotype and for ALL, as well as the genetic correlation (rg) between each blood-cell phenotype and ALL. We used a reference panel of LD scores generated from all variants that passed QC with MAF > 0.0001 via a random sample of 10,000 European ancestry UKB participants. We used UKB LD scores to estimate hg for each blood-cell-trait phenotype and rg with ALL.
Mendelian randomization
We carried out Mendelian randomization (MR) analyses to investigate the potential causal relationship between blood-cell-trait variation and ALL. Genetic instruments excluded multi-allelic and non-inferable palindromic variants with intermediate allele frequencies (MAF > 0.42). To minimize potential for bias due to differences in allele frequencies between exposure (UKB) and outcome (ALL) populations, we restricted analyses to variants with MAF ≥ 0.01 and MAF difference < 0.10. For instruments that were unavailable in the ALL dataset (n = 294), LD proxies (r2 > 0.95) were obtained. MR analyses estimated odds ratios (ORs) and corresponding 95% confidence intervals (CIs) for a genetically predicted 1-SD increase in the normalized Z score for lymphocytes, monocytes, neutrophils, basophils, and eosinophils. For LMR, NLR, and PLR, effects were estimated per 1-unit increase in the ratio.
We used multiple MR estimators to strengthen inference by evaluating consistency in the observed effects. Maximum likelihood (ML) provides unbiased estimates in the absence of any horizontal pleiotropy, while inverse-variance weighted multiplicative random-effects (IVW-mre) accounts for non-directional pleiotropy.41,42 Weighted median (WM)43 provides unbiased estimates when up to 50% of the weights are from invalid instruments. Shrinkage-based MR RAPS (robust adjusted profile score)44,45 incorporates a robust loss function to limit the influence of invalid instruments. MR pleiotropy residual sum and outlier (PRESSO)46 regresses variant effects on the outcome on their exposure effects and compares the observed distance of all instruments to this regression line with the expected distance under the null hypothesis of no horizontal pleiotropy.46
To assess potential violations of MR assumptions, we examined the following diagnostic tests: (1) deviation of the MR Egger intercept from 0 (p < 0.05), indicative of directional horizontal pleiotropy; (2) I2GX < 0.90, indicative of regression dilution bias due to violation of the no measurement error (NOME) assumption;47 and (3) Cochran’s Q-statistic pQ < 0.05 or MR PRESSO pGlobal < 0.05, indicative of heterogeneity due to balanced horizontal pleiotropy. We also report MR PRESSO distortion p values, which test for significant differences between the original and pleiotropy-corrected effect estimates.
Next, we conducted multivariable MR (MVMR) analyses to estimate direct effects of specific blood-cell traits on ALL after accounting for related phenotypes. MVMR regresses SNP effects for all instruments across all exposures against the outcome together, weighting for the inverse variance of the outcome (MVMR-IVW). We also applied a modified analysis where the instruments are selected for each exposure on the basis of p < 5 × 10−8 and then all exposures for those SNPs are regressed together (MVMR-IVWmod). Feature selection was also performed via MV LASSO.
For ratios, we conducted summary-based mediation analysis to decompose the observed total MR effects into direct and indirect effects mediated by each of the component traits.48 For instance, for LMR, we quantified indirect effects on ALL risk that were mediated through regulation of lymphocyte and monocyte counts, as well as direct LMR effects on ALL.
Lastly, we applied MR-Clust,49 a heterogeneity-based clustering method for detecting distinct values of the causal effect that are evidenced by multiple genetic variants. MR-Clust assigns variants to K substantive clusters where all variants indicate the same causal effect, a null cluster, and a “junk” cluster, which includes non-null variants that do not fit into any of the substantive clusters. This approach may reveal different causal or pleiotropic pathways and identify previously undetected ALL-risk variants because of a reduced burden of multiple testing compared with GWASs. Variants were assigned to a cluster if their conditional probability of cluster membership was greater than 0.50. Clusters were formed with a minimum of four variants.
All statistical analyses were conducted with R (version 4.0.2). MR analyses were conducted with the TwoSampleMR R package (version 0.5.5).
Results
Genetic determinants of blood-cell traits
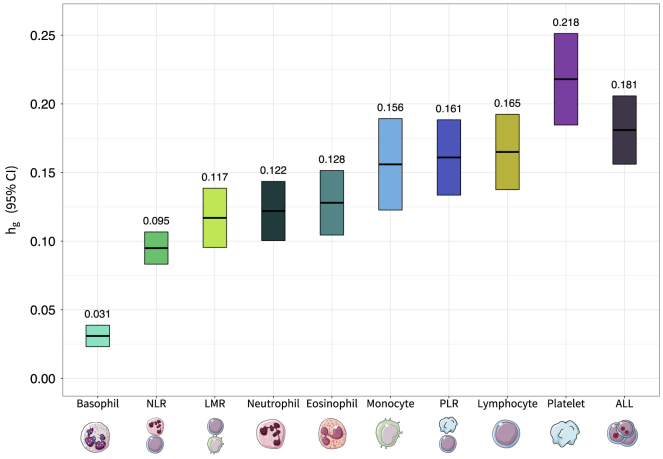
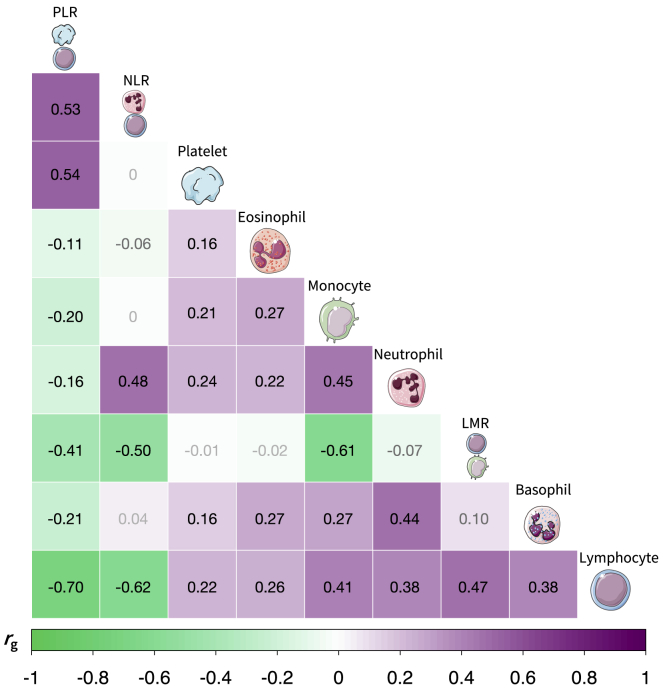
Genome-wide analyses revealed a substantial genetic contribution to blood-cell-trait variation. Heritability (hg) estimated from GWAS summary statistics on the full analytic cohort (median n = 335,030) ranged from 3.1% for basophils to 21.8% for platelets (Figure 1, Table S1). There was significant genetic correlation between all blood-cell populations, which supports our rationale for using MTAG to leverage this shared genetic basis (Figure 2, Table S2). Among non-composite traits, the largest correlations were observed between pairs of white blood cells: monocytes and neutrophils (rg = 0.45, SE = 0.023, p = 1.8 × 10−83), basophils and neutrophils (rg = 0.44, SE = 0.037, p = 4.0 × 10−33), and lymphocytes and monocytes (rg = 0.41, SE = 0.023, p = 1.3 × 10−68). Platelet counts were also significantly correlated with neutrophils (rg = 0.24, SE = 0.022, p = 1.7 × 10−26), lymphocytes (rg = 0.22, SE = 0.018, p = 3.8 × 10−35), and monocytes (rg = 0.21, SE = 0.020, p = 6.8 × 10−26). Cell-type ratios LMR and NLR were primarily correlated with their component traits and with each other. PLR was significantly correlated with all phenotypes, including monocytes (rg = −0.20, SE = 0.021, p = 2.5 × 10−20), neutrophils (rg = −0.16, SE = 0.029, p = 2.7 × 10−8), basophils (rg = −0.21, SE = 0.032, p = 1.1 × 10−10), and eosinophils (rg = −0.11, SE = 0.023, p = 6.3 × 10−7).
Figure 1.
Heritability for acute lymphoblastic leukemia and blood-cell subtypes
Array-based heritability (hg) for lymphocytes, monocytes, neutrophils, eosinophils, basophils, platelets, lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio (PLR), and acute lymphoblastic leukemia (ALL) estimated via LD score regression.
Figure 2.
Genetic correlation between blood-cell traits
Genetic correlation (rg) heat plot for lymphocytes, monocytes, neutrophils, eosinophils, basophils, platelets, lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio (NLR), and platelet-to-lymphocyte ratio (PLR) estimated via LD score regression. Associations with p < 1.4 × 10−3 were considered statistically significant after Bonferroni correction for 36 pairs tested, and corresponding rg estimates are labeled in black font.
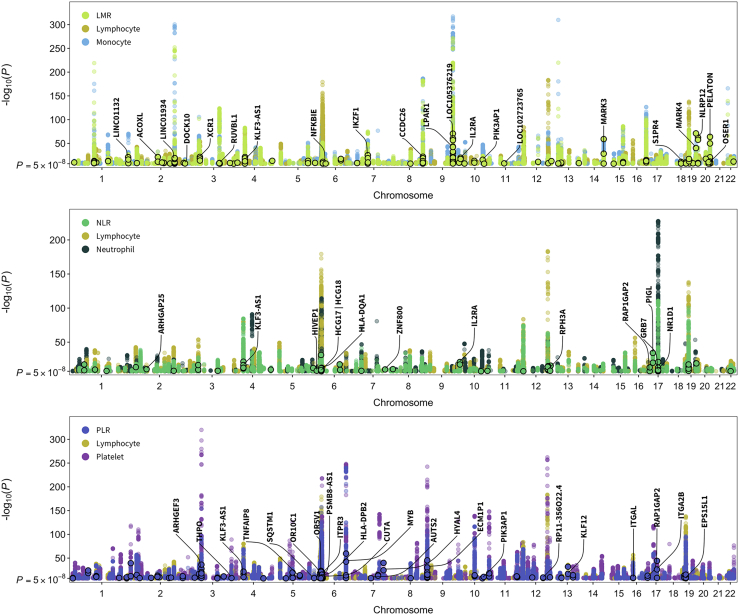
After applying our instrument selection criteria (discovery pMTAG < 5 × 10−8, replication pMTAG < 0.05; LD r2 < 0.05 within 10 Mb), we identified 3,000 variants that were independent within, but not across, hematological phenotypes (Table S3). Of these, 2,500 were associated with a single phenotype, 378 were associated with two, and 122 were instruments for three or more blood-cell traits. The number of available instruments ranged from 157 for basophils to 692 for platelets (Table S4). The proportion of trait variation accounted for by each set of instruments was estimated in the replication sample (100,284 to 100,764 individuals) and ranged between 5.1% for basophils to 24.4% for platelets (Table S4). Previous GWASs have not examined cell-type ratios, while we identified 770 instruments specifically for ratio phenotypes: LMR, NLR, and PLR. To assess whether these signals are captured by existing associations with cell counts or proportions, we performed clumping (LD r2 < 0.05 within 10 Mb) with loci reported in Vuckovic et al.,6 a meta-analysis of UK Biobank and Blood Cell Consortium cohorts. This yielded 225 independent, ratio-specific variants in 115 cytoband loci, including six missense mutations (Figure 3, Table S5).
Figure 3.
Manhattan plots for cell-type ratios and their component traits
Truncated Manhattan plots showing genome-wide significant (p < 5 × 10−8) associations for lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio (NLR), and platelet-to-lymphocyte ratio (PLR) and their component cell types. Points with black borders denote variants that were selected only as instruments for the given ratio trait and are not in linkage disequilibrium (r2 < 0.05 within 10 Mb) with previously reported loci for its component cell-type counts or proportions. Labeled genes contain variants with specific functional features (CADD score > 10, RegulomeDB rank 1–3a, missense mutations) and/or p < 1 × 10−20.
In-silico functional annotations identified overlap with multiple regulatory elements among all genetic instruments. A total of 324 variants were predicted to be in the top 10% of deleterious substitutions genome wide (CADD scores > 10),31 and 138 had significant (p < 0.05) evidence of overlap with open chromatin (FAIRE, DNase, Pol-II, CCCTC-binding factor (CTCF) [MIM: 604167], and MYC [MIM: 190080]) on the basis of ENCODE data from up to 14 cell types. Over 80% of all instruments (n = 2,405) were expression quantitative trait loci (eQTLs) in whole blood (false discovery rate [FDR] < 0.05) on the basis of results from eQTLGen33 (Table S6). Fewer immune cell eQTLs were identified, although these reference datasets were much smaller. The highest proportion of eQTLs was observed in monocytes (27.0%), T cells (23.6%), and neutrophils (21.1%), followed by B cells (11.4%) (Table S6). The proportion of immune-cell eQTLs was broadly similar across categories of instruments, ranging from 26% for neutrophils to 16% for platelets (Figure S2). For every instrument class, T cell eQTLs were the most common. Lymphocytes were the most prevalent instrument class among cell-specific immune eQTLs.
Among instruments specific to one phenotype with eQTL effects in >2 cell types (Figure S3), the largest number of target genes was observed for platelet-specific and monocyte-specific instruments. Instruments for >4 blood-cell traits with eQTL effects in >3 cell types (Figure S3) had a predominance of immune function genes in the human leukocyte antigen (HLA) region and a previously identified ALL-risk gene, BAK1 (MIM: 600516). Among instruments for >5 blood-cell traits with eQTL effects in a single cell/tissue type (whole blood) (Figure S4), notable findings included multiple ALL-risk genes (IKZF3, CDKN2A [MIM: 600160], CDKN2B [MIM: 600431], IRF1 and [MIM: 147575]) and FLT3 (MIM 136351), a receptor tyrosine kinase that serves as a key regulator of hematopoiesis and is frequently mutated in ALL and acute myeloid leukemia (AML [MIM: 601626]).
Impact of blood-cell variation on ALL risk
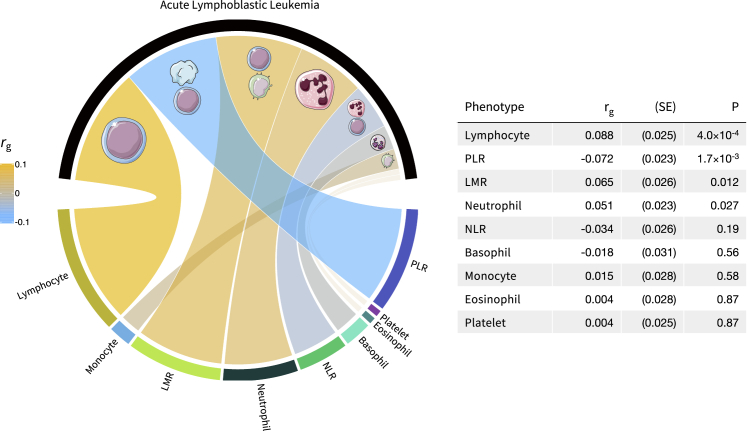
Associations between genetic determinants of blood-cell-trait variation and ALL susceptibility were investigated via a GWAS meta-analysis comprised of 2,666 affected individuals and 60,272 control individuals. Heritability of ALL was 18.1% (hg = 0.181, SE = 0.013), converted to the liability scale via Surveillance, Epidemiology, and End Results (SEER) Program estimates of ALL lifetime risk in non-Hispanic whites (0.15%) (Figure 1). At the genome-wide level, we observed positive correlations with ALL risk for increasing lymphocyte counts (rg = 0.088, SE = 0.025, p = 4.0 × 10−4), LMR (rg = 0.065, SE = 0.026, p = 0.012), and neutrophils (rg = 0.051, SE = 0.023, p = 0.027). Increasing PLR, corresponding to higher levels of platelets compared to lymphocytes, was inversely correlated (rg = −0.072, SE = 0.023, p = 1.7 × 10−3) with ALL risk (Figure 4).
Figure 4.
Genetic correlation between blood-cell subtypes and acute lymphoblastic leukemia
Circos plot depicting genome-wide genetic correlation (rg) estimates. The colors correspond to the direction of genetic correlation; warm shades depict positive correlations between increasing blood-cell counts or ratios and acute lymphoblastic leukemia risk, cool tones correspond to inverse associations, and faded gray shades correspond to null correlations. The width of each band in the Circos plot is proportional to the magnitude of the absolute value of the rg estimate.
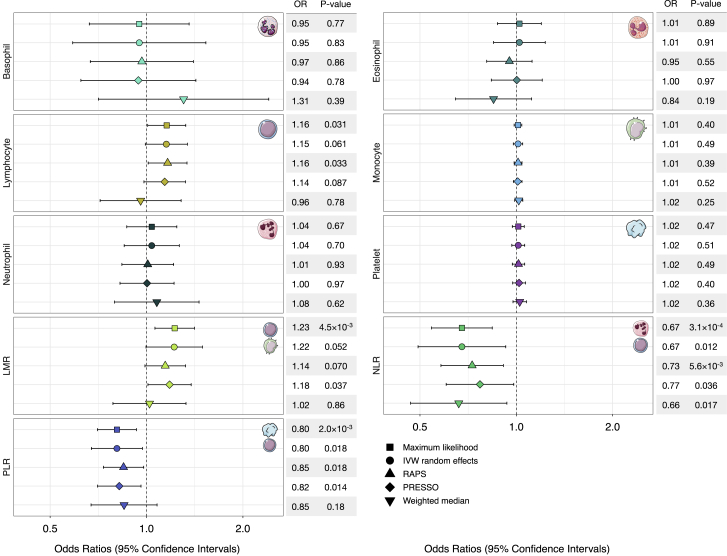
Next, we conducted MR analyses by using genetic instruments developed in the UKB to assess the putative causal relevance of blood-cell-trait variation in childhood ALL etiology (Figure 5, Table S7). We did not detect evidence of directional horizontal pleiotropy for any blood-cell traits (Table S8). However, there was indication of balanced horizontal pleiotropy for all phenotypes on the basis of Cochran’s Q (pQ < 0.05) and the PRESSO global test (pGlobal < 0.05). Among white blood cells, a 1-SD increase in lymphocyte counts was associated with a modest increase in ALL risk (ORML = 1.16, 95% CI 1.01–1.33, p = 0.035; ORIVW-mre = 1.15, 0.99–1.34, p = 0.061). This effect was slightly attenuated in pleiotropy-corrected analyses (ORPRESSO = 1.14, 0.98–1.32, p = 0.087; ORRAPS = 1.16, 1.01–1.34, p = 0.033), but the effect size distortion was not significant (pDist = 0.88). There was no significant association between counts of other white-blood-cell types (monocytes, neutrophils, basophils, eosinophils) or platelets and ALL risk (Figure 4, Table S7).
Figure 5.
Forest plots depicting Mendelian randomization results
Visualization of odds ratios (ORs) and 95% confidence intervals (CIs) for the effect of increasing blood-cell counts or blood-cell ratios on the risk of acute lymphoblastic leukemia (ALL). For each phenotype, association results based on five different Mendelian randomization estimators are shown.
Considering ratios, which indicate a genetic predisposition to a shift in the counts of one cell type relative to another, revealed several associations. An increase in LMR was associated with an approximately 22% increase in ALL risk (per 1-unit increase: ORML = 1.23, 1.07–1.41, p = 4.5 × 10−3; ORIVW-mre = 1.22, 1.00–1.50, p = 0.052). Accounting for the influence of potentially pleiotropic outliers slightly attenuated this effect (ORRAPS = 1.14, 0.99–1.32; ORPRESSO = 1.18, 1.01–1.38). An inverse association with ALL risk was observed for increasing NLR (ORML = 0.67, 0.54–0.83, p = 3.1 × 10−4; ORIVW-mre = 0.67, 0.49–0.92, p = 0.012), denoting a shift to higher levels of neutrophils compared to lymphocytes. Increased PLR was also associated with a lower risk of ALL (ORML = 0.80, 0.70–0.92, p = 2.0 × 10−3; ORIVW-mre = 0.80, 0.67–0.96, p = 0.012). Associations with ALL for both phenotypes remained stable in sensitivity analyses correcting for pleiotropy (NLR: ORPRESSO = 0.77, 0.66–0.98, p = 0.036; PLR: ORPRESSO = 0.82, 0.70–0.96, p = 0.014) and outliers (NLR: ORRAPS = 0.73, 0.58–0.91, p = 5.6 × 10−3; PLR: ORRAPS = 0.85, 0.73–0.98, p = 0.025).
In addition to analytically correcting for pleiotropy, we also conducted analyses by using a filtered set of genetic instruments excluding variants that showed evidence of heterogeneity on the basis of Cochran’s Q (Table S9). These sensitivity analyses confirmed our previous findings showing that an increase in lymphocyte counts (ORIVW-mre = 1.18, p = 7.4 × 10−3) and LMR (ORIVW-mre = 1.19, p = 0.016) conferred a modest increase in ALL risk, while increased NLR (ORIVW-mre = 0.67, p = 7.4 × 10−3) and PLR (ORIVW-mre = 0.82, p = 5.8 × 10−3) were associated with lower risk.
Assessment of additional diagnostic tests indicated that our analysis was robust against main threats to validity, including weak instrument bias (mean F-statistic > 60) and NOME violation (I2GX > 0.98) (Table S8). We used the MR Steiger directionality test50 to orient the causal effects and confirmed that instruments for blood-cell traits were affecting ALL susceptibility, not the reverse for all traits, including lymphocytes (p = 5.0 × 10−135), LMR (p = 5.4 × 10−98), NLR (p = 2.1 × 10−10), and PLR (p = 1.2 × 10−109). Our analyses were powered to at least 80% to detect a minimum OR of 1.17 (equivalent to 0.85) for LMR and PLR, OR of 1.20 for lymphocytes, and OR of 1.28 (equivalent to 0.78) for NLR (Figure S5).
Next, we conducted multivariable MR analyses to estimate independent direct effects on ALL for each blood-cell subtype (Table S10). Among lymphocytes, monocytes, neutrophils, and platelets, only lymphocytes were independently associated with ALL (ORMVMR = 1.18, 1.06–1.31, p = 3.3 × 10−3) on the basis of all variants and when restricting to instruments associated with each exposure (ORMVMR-mod = 1.43, 1.16–1.76, p = 8.8 × 10−4). This was confirmed via MV LASSO, which only retained lymphocytes. Among cell-type ratios, PLR was associated with ALL when considering all variants for all traits (ORMVMR = 0.90, 0.82–0.99, p = 0.033) but not in the instrument-specific analysis. PLR was the only trait selected by MV LASSO. Lastly, we explored the degree to which causal effects observed for ratio phenotypes were mediated by any of their component traits. We did not observe any statistically significant indirect effects, which suggests that the impact on ALL susceptibility observed for LMR, NLR, and PLR could not be attributed to effects on the counts of lymphocytes, monocytes, neutrophils, or platelets (Table S11).
Exploring mechanisms of ALL susceptibility
We applied MR-Clust49 to blood-cell traits associated with ALL to identify subgroups of variants with homogeneous causal effects and novel ALL-risk variants (Figure S6; Table S12). Clustering instruments for lymphocytes identified ten variants indicating a large effect of increasing lymphocyte counts on ALL (OR = 7.63). LMR instruments contained a cluster of nine variants (OR = 3.64). The largest cluster was identified for PLR, which had 18 variants (OR of 0.27 per 1-unit increase in the ratio). A cluster comprised of ten variants implied an extremely large inverse effect of NLR on ALL (OR = 0.039). The substantive clusters were largely distinct, but one variant was shared by all four traits (rs28447467: pALL = 0.026). Across all clusters, two variants were statistically significantly associated with ALL after correcting for the number of independent variants across all phenotypes tested (pALL < 5 × 10−5): rs6430608-C (OR = 1.28, 1.15–1.41, pALL = 2.5 × 10−6) near CXCR4 (MIM: 162643) on 2q22.1 and rs76428106-C (OR = 1.79, 1.36–2.35, PALL = 3.2 × 10−5) in FLT3 on 13q12.2. The former is an intergenic variant specific to NLR with cis effects on whole-blood gene expression of MCM6 (MIM: 601806) (peQTL = 4.0 × 10−28) and DARS1 (MIM: 603084) (peQTL = 3.7 × 10−37), based on data from eQTLGen.33 On the other hand, rs76428106, an intronic variant in FLT3 and an eQTL for FLT3 in whole blood (peQTL = 1.0 × 10−11)33 was included in substantive clusters for lymphocytes and PLR but assigned to the “junk” cluster for LMR. Annotation of variants in substantive clusters via PhenoScanner51 revealed a predominance of previously reported associations with blood-cell-trait variation, as well as autoimmune and allergic conditions, such as type 1 diabetes (MIM: 222100), Crohn disease (MIM: 266600), asthma (MIM: 600807), and IgA deficiency (MIM: 137100) (Table S12).
Notable instruments assigned to “junk” clusters for LMR, PLR, and NLR included established ALL-risk variants rs4948492 and rs4245597 (ARID5B, 10q21.2), rs2239630 (CEBPE, 14q11.2), rs78697948 (IKZF1, 7p12.2), and rs74756667 (8q24.2). These variants were also classified as outliers on the basis of Cochran’s Q, suggesting that their effects on ALL susceptibility are predominantly mediated through pathways other than regulation of blood-cell profiles. We formally tested this hypothesis via mediation analysis52 by decomposing the total SNP effect on ALL into direct and indirect (mediated) effects. For variants that were instruments for more than one blood-cell phenotype, mediation was only explored for phenotypes significantly associated with ALL. Mediator-outcome effects were obtained from MR results excluding outliers (Table S9). For rs4245597 (ARID5B), only a small proportion of its effect on ALL risk was mediated by blood-cell traits; 1.65% (0.79–2.51) was attributed to NLR and 0.84% (0.23–1.44) to PLR (Table S13). Mediated effects attributed to LMR were not statistically significant for rs4245597 (ARID5B; 0.75%), rs2239630 (2.16%; CEBPE), and rs78697948 (1.72%; IKZF1).
Modest, but statistically significant, mediated effects were observed for ALL-risk variants rs6430608 (NLR: 4.72%, 2.26–7.20) and rs76428106 (PLR: 2.43%, 0.68–4.19; lymphocytes: 2.51%, 0.67–4.35). The LMR-mediated effect of rs76428106 was larger (11.39%) but in the opposite direction from the effect of LMR on ALL, indicating pleiotropic effects consistent with the assignment of rs76428106 to the “junk” cluster for LMR. Of the six traits linked to this variant, its effect on monocytes (rs76428106-C: β = 0.484, p = 1.3 × 10−310) was by far the strongest. In MR analyses, monocyte counts were not implicated in ALL susceptibility, suggesting that rs76428106 may be influencing ALL via other pathways or broad effects on hematopoiesis.
Discussion
Hematopoiesis is a tightly regulated hierarchical process designed to maintain optimal physiological ranges. Abnormalities in blood counts may be indicative of systemic inflammation or the presence of infections and serve as indicators for a wide range of potentially adverse health conditions, including inborn defects in hematopoiesis. While the responsive and sensitive nature of blood-cell counts makes them useful clinical biomarkers, this poses a challenge for etiological studies. Elevated white-blood-cell counts are an established diagnostic feature of childhood ALL, reflecting the overproduction of immature lymphocytes, or lymphoblasts, in the bone marrow. However, blood counts at a single time point, particularly in cancer-affected individuals, may not be representative of the individual’s stable, pre-diagnostic blood-count profile, making it difficult to disentangle disease correlates from risk factors.
In this study, we leveraged the highly heritable nature of blood-cell variation to evaluate its role in ALL pathogenesis without the limitations inherent in observational blood-count measures. Our overarching finding is the convergence of genetic mechanisms resulting in increased lymphocyte counts and increased ALL susceptibility. Using genetic correlation and Mendelian randomization, we observed a significant positive relationship between a genetically predicted increase in lymphocyte counts and ratio of lymphocytes to monocytes (LMR) and risk of ALL. Conversely, genetic predisposition to an increased ratio of platelets to lymphocytes (PLR) and neutrophils to lymphocytes (NLR) was inversely associated with ALL risk. These effects were largely robust to analytic corrections for horizontal pleiotropy, and in some cases, the removal of instruments contributing to heterogeneity strengthened the observed associations. Taken together, these results reveal insights into ALL etiology and point to a specific shift in blood-cell homeostasis that confers an increased susceptibility.
However, the ways in which a genetic predisposition to over-production of lymphocytes may confer ALL risk are most likely multifactorial. Stable and consistent causal effect estimates for lymphocytes, PLR, and NLR do not imply a single biological mechanism, even if they are estimated with valid instruments that primarily regulate the target blood-cell trait. Acknowledging this, we propose two distinct, though not necessarily mutually exclusive, biological mechanisms related to the two-hit model of childhood ALL development that warrant further investigation. First, the initiating genetic lesions in childhood ALL, such as ETV6-RUNX1 (MIM: 600618 and 151385) gene fusions, arise prenatally in most cases and require additional somatic mutations to progress to overt leukemia.14,53 The presence of common alleles across the spectrum of variants that subtly tune lymphocyte production may lead to an elevated risk of ALL by increasing the reservoir of preleukemic cell clones, which in turn, may increase the chances of acquiring “second-hit” oncogenic events and progression to ALL.
Second, the “delayed infection” hypothesis posits that children who lack early microbial exposures may have an unmodulated immune network that results in dysregulated immune responses to infectious stimuli later in childhood and an increased risk of ALL.14 This is supported by epidemiological evidence, such as that proxies for early-life infectious exposure, including daycare attendance and higher birth order, are associated with a reduced risk of ALL,54,55 and by experimental models that demonstrated higher ALL incidence in mice with delayed exposure to pathogens.56,57 Further, children who develop ALL have been found to have different cytokine profiles at birth.58,59 Genetic variants that influence the blood-cell phenotypes associated with ALL risk in our study may confer their effects via modulation of neonatal immune development and of immune responses to infections in childhood that may trigger ALL development. A shift toward increased lymphocytes to neutrophils is suggestive of increased adaptive immunity and lymphocyte activation in response to infections. This is consistent with our findings for NLR, a marker of increased inflammation, which was associated with reduced ALL risk. Similarly, reduced immune-inflammatory responses and increased activation of lymphocytes would be denoted by a higher ratio of lymphocytes to monocytes and lymphocytes to platelets,60 both of which were associated with increased ALL risk in our study.
Previous studies have noted an overlap between ALL-risk loci and genomic regions associated with blood-cell phenotypes,21, 22, 23, 24 however in this study, we have systematically analyzed the contribution and causal effects of genetic variation across blood-cell traits in ALL etiology. In a recent PheWAS, ALL-risk variants were found to be enriched for regulation of platelet levels, but the overall association between platelet counts and ALL was null in Mendelian randomization and genetic score analyses with 223 platelet-associated variants.26 This is consistent with our findings via over 600 genetic instruments for platelets, which indicate that variation in platelet counts alone does not influence ALL susceptibility, whereas PLR, which captures dysregulation of platelets in relation to lymphocytes, has a significant impact.
Indeed, we identified the cell-type ratios LMR, NLR, and PLR as independent risk factors for ALL and found evidence that these ratios have distinct genetic mechanisms that are not captured by their component traits. In multivariable MR analyses that concurrently modeled the effects of lymphocyte, monocyte, neutrophil, and platelet counts on ALL, lymphocytes remained as the only independent risk factor and this association with ALL strengthened compared to univariate analyses. However, there was no evidence that the total MR effects for LMR, NLR, and PLR were mediated either by lymphocytes or by the other cell populations. This implies that while dysregulation of lymphocyte homeostasis seems to be a key factor, it should be considered in the broader context of other blood-cell subtypes.
In addition to identifying novel susceptibility pathways, our study also provides insights into the underlying mechanisms of several established ALL-risk variants in ARID5B, CEBPE, and IKZF1 and at chromosome 8q24. Despite significant associations with LMR and, in the case of ARID5B, with NLR and PLR, these variants were flagged as pleiotropic outliers in MR analyses, which mediation analyses subsequently confirmed. This supports that the overall effects of these loci on ALL risk are largely mediated by pathways other than regulation of blood-cell-trait variation, although we cannot rule out potential effects of these variants on early stages of hematopoiesis that may influence ALL development. Our MR-clustering analysis also identified two putative novel ALL-risk variants among genetic instruments for various blood-cell traits: rs6430608 on 2q22.1 and rs76428106 in FLT3 on 13q12.2.
Although additional studies are needed for confirmation of their association with ALL, the locus at FLT3 is of particular interest because this same variant was recently associated with an increased risk of autoimmune thyroid disease (MIM: 608173) and AML.61 The AML/ALL-risk-increasing allele, rs76428106-C, has a frequency of approximately 1% in the general population (1.3% in UKB, 1.4% in ALL GWAS) and is reported as a splicing QTL in GTEx (psQTL = 1.3 × 10−8). Indeed, rs76428106-C was shown to generate a cryptic splice site resulting in truncation of FLT3 but an increase in FLT3 ligand levels.61 Gain-of-function somatic mutations in FLT3 are relatively frequent in childhood ALL,62 and although rs76428106 has greater effects on the production of myeloid cells than lymphocytes in our analyses, its putative effects on ALL risk may largely occur via activation of the RAS/MAPK pathway. This activation is likely to be restricted to key developmental decisions of hematopoietic cells given the delimited expression of FLT3 to hematopoietic stem and progenitor cells.63 Less is known about the 2q22.1 variant, rs6430608, which is an eQTL for MCM6 in blood and CXCR4 in multiple tissues. MCM6 is upregulated in multiple cancers and is believed to regulate DNA replication and activate MAPK/ERK signaling.64 CXCR4 is a chemokine receptor that facilitates HIV-cell entry and regulates immune-cell migration, including retention of B cell precursors in the bone-marrow, and is being pursued as a therapeutic target in ALL and AML.65,66
Several limitations of this study should be acknowledged. First, genetic instruments were developed for blood-cell phenotypes measured in adult participants in the UK Biobank because of a paucity of adequately powered GWASs of blood-cell traits in newborns or children. Environmental exposures throughout the life course influence blood-cell dynamics, which has implications for the accuracy of genetic association estimates at different time points across the lifespan. Although studies of blood-cell development in pediatric populations should be pursued, the true underlying genetic architecture is not affected by age. This is also supported by studies of other complex traits, such as BMI, which showed that genetic risk scores developed in adults accurately predict weight gain in early childhood.67 Therefore, we would expect any error in our genetic instruments developed in adults to bias MR results toward the null.
Second, our analysis was limited to broad classes of cell types, such as lymphocytes, and in future studies, it will be important to distinguish between subpopulations of B cell and T cell lymphocytes. B cell precursor ALL, the most common subtype, most likely has distinct etiologic mechanisms from T cell ALL.15 Of relevance to our findings, the epidemiological evidence for the two-hit model of leukemogenesis is more compelling for B cell ALL than for T cell ALL14 and GWASs have revealed that hematopoietic transcription factor genes confer stronger effects on B cell ALL risk.24 We were also unable to characterize the effect of blood-cell traits on B cell ALL versus T cell ALL or on specific molecular subtypes or to explore the potential for germline-somatic interactions with specific ALL mutational signatures.
Finally, MTAG assumes that the variance-covariance matrix of effect sizes is homogeneous across all variants, which may be violated for SNPs that are null for one trait but non null for other traits.30 Replication is the best way to assess the credibility of observed associations; therefore our two-stage discovery and replication approach should minimize false positives. Furthermore, in a two-sample setting, false-positive instruments would bias MR estimates toward the null, not induce a spurious signal.
Despite some limitations, this study has important strengths that support the robustness of our findings. Our instrument development approach was optimized for Mendelian randomization studies of cancer etiology. The large sample size of the UK Biobank cohort allowed us to apply appropriate exclusions while retaining a sufficient number of participants for a two-stage discovery and replication analysis. Furthermore, applying the MTAG framework increased statistical power for identifying genetic determinants of specific blood-cell traits while taking into account the correlation between these phenotypes. This resulted in a set of strong genetic instruments explaining between 5% and 24% of variation in the target blood-cell trait. These variants were enriched for multiple regulatory features, and over 80% had significant effects on gene expression in whole blood and up to 27% of instruments were classified as eQTLs in immune-cell subtypes, albeit with a limited degree of cell-type specificity in the eQTL effects across instrument classes. In addition, we characterized the genetic determinants of blood-cell ratios, specifically LMR, NLR, and PLR, which have received considerably less attention in genetic association studies. A GWAS of PLR and NLR was conducted in 5,901 healthy Dutch individuals, which identified one significant locus for PLR.68 Examining these ratio phenotypes revealed additional ALL susceptibility pathways and helped contextualize the observed results for lymphocytes and platelets. Finally, the causal interpretation of our results depends on the credibility of fundamental MR assumptions, and to this end, we employed a range of MR-estimation methods and conducted multiple diagnostic tests to interrogate the robustness of our results with respect to confounding, horizontal pleiotropy, and weak instrument bias.
In conclusion, we demonstrate that a genetic propensity for overproduction of lymphocytes, particularly in relation to other blood-cell types, is associated with an increased risk of childhood ALL in individuals of predominantly European ancestry. It will be important to elucidate the underlying biological mechanisms of our findings and to assess their transferability to admixed and non-European ancestry populations.
Acknowledgments
This work was supported by research grants from the National Institutes of Health (NIH) National Cancer Institute (NCI): R03CA245998 (A.J.D. and L.K.), K99CA246076 (L.K.), R01CA155461 (J.L.W. and X.M.) and R01CA175737 (J.L.W. and X.M.). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP003862-04/DP003862; the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors. A subset of the CCRLP data used in this study was obtained from the California Biobank Program at the California Department of Public Health (CDPH), SIS request number 26, in accordance with Section 6555(b), 17CCR. The CDPH is not responsible for the results or conclusions drawn by the authors of this publication.
Declaration of interests
The authors declare no competing interests.
Published: August 31, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.08.004.
Data and code availability
This research was conducted with approved access to UK Biobank data under application number 14105 (PI: Witte) and in accordance with the UK Biobank Ethics and Governance Framework. UK Biobank data are publicly available by request from https://www.ukbiobank.ac.uk. Ethics approval for establishing the UK Biobank resource was obtained from the North West Centre for Research Ethics Committee (11/NW/0382). This study included the analysis of data derived from biospecimens from the California Biobank Program (CCRLP study). Any uploading of genomic data and/or sharing of these biospecimens or individual data derived from these biospecimens has been determined to violate the statutory scheme of the California Health and Safety Code Sections 124980(j); 124991(b), (g), and (h); and 103850 (a) and (d), which protect the confidential nature of biospecimens and individual data derived from biospecimens. This study was approved by institutional review boards at the California Health and Human Services Agency; University of Southern California; Yale University; and the University of California, San Francisco. The de-identified newborn dried blood spots for the CCRLP (California Biobank Program SIS request # 26) were obtained with a waiver of consent from the Committee for the Protection of Human Subjects of the State of California. This study makes use of data from the Kaiser Permanente (KP) Research Program on Genes, Environment, and Health (RPGEH) Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, available from dbGaP (dbGaP: phs000788.v1.p2). This study also makes use of data generated by the Wellcome Trust Case-Control Consortium available by request from the European Genotype Archive: https://ega-archive.org/ega (EGA: EGAD00000000021). Genotype data for COG ALL affected individuals are available for download from dbGaP (dbGaP: phs000638.v1.p1).
Web resources
FUMA platform, https://fuma.ctglab.nl
PhenoScanner database, http://www.phenoscanner.medschl.cam.ac.uk
R package for Mendelian randomization analyses, https://mrcieu.github.io/TwoSampleMR/index.html
RegulomeDB platform, https://regulomedb.org/regulome-search/
Supplemental information
References
- 1.Evans D.M., Frazer I.H., Martin N.G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 2.Garner C., Tatu T., Reittie J.E., Littlewood T., Darley J., Cervino S., Farrall M., Kelly P., Spector T.D., Thein S.L. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood. 2000;95:342–346. [PubMed] [Google Scholar]
- 3.Pilia G., Chen W.M., Scuteri A., Orrú M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429. doi: 10.1016/j.cell.2016.10.042. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CHARGE Consortium Hematology Working Group Meta-analysis of rare and common exome chip variants identifies S1PR4 and other loci influencing blood cell traits. Nat. Genet. 2016;48:867–876. doi: 10.1038/ng.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuckovic D., Bao E.L., Akbari P., Lareau C.A., Mousas A., Jiang T., Chen M.H., Raffield L.M., Tardaguila M., Huffman J.E. The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell. 2020;182:1214–1231. doi: 10.1016/j.cell.2020.08.008. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liggett L.A., Sankaran V.G. Unraveling Hematopoiesis through the Lens of Genomics. Cell. 2020;182:1384–1400. doi: 10.1016/j.cell.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao E.L., Nandakumar S.K., Liao X., Bick A.G., Karjalainen J., Tabaka M., Gan O.I., Havulinna A.S., Kiiskinen T.T.J., Lareau C.A. Inherited myeloproliferative neoplasm risk affects haematopoietic stem cells. Nature. 2020;586:769–775. doi: 10.1038/s41586-020-2786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linet M.S., Ries L.A., Smith M.A., Tarone R.E., Devesa S.S. Cancer surveillance series: recent trends in childhood cancer incidence and mortality in the United States. J. Natl. Cancer Inst. 1999;91:1051–1058. doi: 10.1093/jnci/91.12.1051. [DOI] [PubMed] [Google Scholar]
- 10.Hunger S.P., Lu X., Devidas M., Camitta B.M., Gaynon P.S., Winick N.J., Reaman G.H., Carroll W.L. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J. Clin. Oncol. 2012;30:1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtin S.C., Minino A.M., Anderson R.N. Declines in Cancer Death Rates Among Children and Adolescents in the United States, 1999-2014. NCHS Data Brief. 2016;257:1–8. [PubMed] [Google Scholar]
- 12.Turcotte L.M., Liu Q., Yasui Y., Arnold M.A., Hammond S., Howell R.M., Smith S.A., Weathers R.E., Henderson T.O., Gibson T.M. Temporal Trends in Treatment and Subsequent Neoplasm Risk Among 5-Year Survivors of Childhood Cancer, 1970-2015. JAMA. 2017;317:814–824. doi: 10.1001/jama.2017.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulrooney D.A., Hyun G., Ness K.K., Bhakta N., Pui C.H., Ehrhardt M.J., Krull K.R., Crom D.B., Chemaitilly W., Srivastava D.K. The changing burden of long-term health outcomes in survivors of childhood acute lymphoblastic leukaemia: a retrospective analysis of the St Jude Lifetime Cohort Study. Lancet Haematol. 2019;6:e306–e316. doi: 10.1016/S2352-3026(19)30050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2018;18:471–484. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams L.A., Yang J.J., Hirsch B.A., Marcotte E.L., Spector L.G. Is There Etiologic Heterogeneity between Subtypes of Childhood Acute Lymphoblastic Leukemia? A Review of Variation in Risk by Subtype. Cancer Epidemiol. Biomarkers Prev. 2019;28:846–856. doi: 10.1158/1055-9965.EPI-18-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gocho Y., Yang J.J. Genetic defects in hematopoietic transcription factors and predisposition to acute lymphoblastic leukemia. Blood. 2019;134:793–797. doi: 10.1182/blood.2018852400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaemmanuil E., Hosking F.J., Vijayakrishnan J., Price A., Olver B., Sheridan E., Kinsey S.E., Lightfoot T., Roman E., Irving J.A. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treviño L.R., Yang W., French D., Hunger S.P., Carroll W.L., Devidas M., Willman C., Neale G., Downing J., Raimondi S.C. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migliorini G., Fiege B., Hosking F.J., Ma Y., Kumar R., Sherborne A.L., da Silva Filho M.I., Vijayakrishnan J., Koehler R., Thomsen H. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122:3298–3307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 20.Xu H., Yang W., Perez-Andreu V., Devidas M., Fan Y., Cheng C., Pei D., Scheet P., Burchard E.G., Eng C. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J. Natl. Cancer Inst. 2013;105:733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiemels J.L., Walsh K.M., de Smith A.J., Metayer C., Gonseth S., Hansen H.M., Francis S.S., Ojha J., Smirnov I., Barcellos L. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat. Commun. 2018;9:286. doi: 10.1038/s41467-017-02596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Smith A.J., Walsh K.M., Francis S.S., Zhang C., Hansen H.M., Smirnov I., Morimoto L., Whitehead T.P., Kang A., Shao X. BMI1 enhancer polymorphism underlies chromosome 10p12.31 association with childhood acute lymphoblastic leukemia. Int. J. Cancer. 2018;143:2647–2658. doi: 10.1002/ijc.31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Smith A.J., Walsh K.M., Morimoto L.M., Francis S.S., Hansen H.M., Jeon S., Gonseth S., Chen M., Sun H., Luna-Fineman S. Heritable variation at the chromosome 21 gene ERG is associated with acute lymphoblastic leukemia risk in children with and without Down syndrome. Leukemia. 2019;33:2746–2751. doi: 10.1038/s41375-019-0514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian M., Xu H., Perez-Andreu V., Roberts K.G., Zhang H., Yang W., Zhang S., Zhao X., Smith C., Devidas M. Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in Hispanics. Blood. 2019;133:724–729. doi: 10.1182/blood-2018-07-862946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijayakrishnan J., Qian M., Studd J.B., Yang W., Kinnersley B., Law P.J., Broderick P., Raetz E.A., Allan J., Pui C.H. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat. Commun. 2019;10:5348. doi: 10.1038/s41467-019-13069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semmes E.C., Vijayakrishnan J., Zhang C., Hurst J.H., Houlston R.S., Walsh K.M. Leveraging Genome and Phenome-Wide Association Studies to Investigate Genetic Risk of Acute Lymphoblastic Leukemia. Cancer Epidemiol. Biomarkers Prev. 2020;29:1606–1614. doi: 10.1158/1055-9965.EPI-20-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachuri L., Johansson M., Rashkin S.R., Graff R.E., Bossé Y., Manem V., Caporaso N.E., Landi M.T., Christiani D.C., Vineis P. Immune-mediated genetic pathways resulting in pulmonary function impairment increase lung cancer susceptibility. Nat. Commun. 2020;11:27. doi: 10.1038/s41467-019-13855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong S., Boyle A.P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 2019;40:1292–1298. doi: 10.1002/humu.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Võsa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Kasela S. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. bioRxiv. 2018 doi: 10.1101/447367. [DOI] [Google Scholar]
- 34.Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175:1701–1715. doi: 10.1016/j.cell.2018.10.022. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martin D., Watt S., Yan Y., Kundu K., Ecker S. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414. doi: 10.1016/j.cell.2016.10.026. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momozawa Y., Dmitrieva J., Théâtre E., Deffontaine V., Rahmouni S., Charloteaux B., Crins F., Docampo E., Elansary M., Gori A.S. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun. 2018;9:2427. doi: 10.1038/s41467-018-04365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeon S., de Smith A.J., Li S., Chen M., Chan T.F., Muskens I.S., Morimoto L.M., DeWan A.T., Mancuso N., Metayer C. Genome-wide trans-ethnic meta-analysis identifies novel susceptibility loci for childhood acute lymphoblastic leukemia. medRxiv. 2021 doi: 10.1101/2021.05.07.21256849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowden J., Del Greco M F., Minelli C., Davey Smith G., Sheehan N., Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017;36:1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Q., Wang J., Hemani G., Bowden J., Small D.S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 2020;48:1742–1769. [Google Scholar]
- 45.Zhao Q., Chen Y., Wang J., Small D.S. Powerful three-sample genome-wide design and robust statistical inference in summary-data Mendelian randomization. Int. J. Epidemiol. 2019;48:1478–1492. doi: 10.1093/ije/dyz142. [DOI] [PubMed] [Google Scholar]
- 46.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowden J., Del Greco M F., Minelli C., Davey Smith G., Sheehan N.A., Thompson J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess S., Thompson D.J., Rees J.M.B., Day F.R., Perry J.R., Ong K.K. Dissecting Causal Pathways Using Mendelian Randomization with Summarized Genetic Data: Application to Age at Menarche and Risk of Breast Cancer. Genetics. 2017;207:481–487. doi: 10.1534/genetics.117.300191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foley C.N., Mason A.M., Kirk P.D.W., Burgess S. MR-Clust: Clustering of genetic variants in Mendelian randomization with similar causal estimates. Bioinformatics. 2020;37:531–541. doi: 10.1093/bioinformatics/btaa778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J., Butterworth A.S., Staley J.R. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kachuri L., Saarela O., Bojesen S.E., Davey Smith G., Liu G., Landi M.T., Caporaso N.E., Christiani D.C., Johansson M., Panico S. Mendelian Randomization and mediation analysis of leukocyte telomere length and risk of lung and head and neck cancers. Int. J. Epidemiol. 2019;48:751–766. doi: 10.1093/ije/dyy140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiemels J.L., Cazzaniga G., Daniotti M., Eden O.B., Addison G.M., Masera G., Saha V., Biondi A., Greaves M.F. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 54.Rudant J., Lightfoot T., Urayama K.Y., Petridou E., Dockerty J.D., Magnani C., Milne E., Spector L.G., Ashton L.J., Dessypris N. Childhood acute lymphoblastic leukemia and indicators of early immune stimulation: a Childhood Leukemia International Consortium study. Am. J. Epidemiol. 2015;181:549–562. doi: 10.1093/aje/kwu298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urayama K.Y., Ma X., Selvin S., Metayer C., Chokkalingam A.P., Wiemels J.L., Does M., Chang J., Wong A., Trachtenberg E., Buffler P.A. Early life exposure to infections and risk of childhood acute lymphoblastic leukemia. Int. J. Cancer. 2011;128:1632–1643. doi: 10.1002/ijc.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobaleda C., Vicente-Dueñas C., Sanchez-Garcia I. Infectious triggers and novel therapeutic opportunities in childhood B cell leukaemia. Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-021-00505-2. Published online February 8, 2021. [DOI] [PubMed] [Google Scholar]
- 57.Martín-Lorenzo A., Hauer J., Vicente-Dueñas C., Auer F., González-Herrero I., García-Ramírez I., Ginzel S., Thiele R., Constantinescu S.N., Bartenhagen C. Infection Exposure is a Causal Factor in B-cell Precursor Acute Lymphoblastic Leukemia as a Result of Pax5-Inherited Susceptibility. Cancer Discov. 2015;5:1328–1343. doi: 10.1158/2159-8290.CD-15-0892. [DOI] [PubMed] [Google Scholar]
- 58.Chang J.S., Zhou M., Buffler P.A., Chokkalingam A.P., Metayer C., Wiemels J.L. Profound deficit of IL10 at birth in children who develop childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomarkers Prev. 2011;20:1736–1740. doi: 10.1158/1055-9965.EPI-11-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Søegaard S.H., Rostgaard K., Skogstrand K., Wiemels J.L., Schmiegelow K., Hjalgrim H. Neonatal Inflammatory Markers Are Associated with Childhood B-cell Precursor Acute Lymphoblastic Leukemia. Cancer Res. 2018;78:5458–5463. doi: 10.1158/0008-5472.CAN-18-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gasparyan A.Y., Ayvazyan L., Mukanova U., Yessirkepov M., Kitas G.D. The Platelet-to-Lymphocyte Ratio as an Inflammatory Marker in Rheumatic Diseases. Ann. Lab. Med. 2019;39:345–357. doi: 10.3343/alm.2019.39.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saevarsdottir S., Olafsdottir T.A., Ivarsdottir E.V., Halldorsson G.H., Gunnarsdottir K., Sigurdsson A., Johannesson A., Sigurdsson J.K., Juliusdottir T., Lund S.H. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature. 2020;584:619–623. doi: 10.1038/s41586-020-2436-0. [DOI] [PubMed] [Google Scholar]
- 62.Annesley C.E., Brown P. The Biology and Targeting of FLT3 in Pediatric Leukemia. Front. Oncol. 2014;4:263. doi: 10.3389/fonc.2014.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazi J.U., Rönnstrand L. FMS-like Tyrosine Kinase 3/FLT3: From Basic Science to Clinical Implications. Physiol. Rev. 2019;99:1433–1466. doi: 10.1152/physrev.00029.2018. [DOI] [PubMed] [Google Scholar]
- 64.Liu M., Hu Q., Tu M., Wang X., Yang Z., Yang G., Luo R. MCM6 promotes metastasis of hepatocellular carcinoma via MEK/ERK pathway and serves as a novel serum biomarker for early recurrence. J. Exp. Clin. Cancer Res. 2018;37:10. doi: 10.1186/s13046-017-0669-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katsura M., Shoji F., Okamoto T., Shimamatsu S., Hirai F., Toyokawa G., Morodomi Y., Tagawa T., Oda Y., Maehara Y. Correlation between CXCR4/CXCR7/CXCL12 chemokine axis expression and prognosis in lymph-node-positive lung cancer patients. Cancer Sci. 2018;109:154–165. doi: 10.1111/cas.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cancilla D., Rettig M.P., DiPersio J.F. Targeting CXCR4 in AML and ALL. Front. Oncol. 2020;10:1672. doi: 10.3389/fonc.2020.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khera A.V., Chaffin M., Wade K.H., Zahid S., Brancale J., Xia R., Distefano M., Senol-Cosar O., Haas M.E., Bick A. Polygenic Prediction of Weight and Obesity Trajectories from Birth to Adulthood. Cell. 2019;177:587–596. doi: 10.1016/j.cell.2019.03.028. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin B.D., Carnero-Montoro E., Bell J.T., Boomsma D.I., de Geus E.J., Jansen R., Kluft C., Mangino M., Penninx B., Spector T.D. 2SNP heritability and effects of genetic variants for neutrophil-to-lymphocyte and platelet-to-lymphocyte ratio. J. Hum. Genet. 2017;62:979–988. doi: 10.1038/jhg.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This research was conducted with approved access to UK Biobank data under application number 14105 (PI: Witte) and in accordance with the UK Biobank Ethics and Governance Framework. UK Biobank data are publicly available by request from https://www.ukbiobank.ac.uk. Ethics approval for establishing the UK Biobank resource was obtained from the North West Centre for Research Ethics Committee (11/NW/0382). This study included the analysis of data derived from biospecimens from the California Biobank Program (CCRLP study). Any uploading of genomic data and/or sharing of these biospecimens or individual data derived from these biospecimens has been determined to violate the statutory scheme of the California Health and Safety Code Sections 124980(j); 124991(b), (g), and (h); and 103850 (a) and (d), which protect the confidential nature of biospecimens and individual data derived from biospecimens. This study was approved by institutional review boards at the California Health and Human Services Agency; University of Southern California; Yale University; and the University of California, San Francisco. The de-identified newborn dried blood spots for the CCRLP (California Biobank Program SIS request # 26) were obtained with a waiver of consent from the Committee for the Protection of Human Subjects of the State of California. This study makes use of data from the Kaiser Permanente (KP) Research Program on Genes, Environment, and Health (RPGEH) Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, available from dbGaP (dbGaP: phs000788.v1.p2). This study also makes use of data generated by the Wellcome Trust Case-Control Consortium available by request from the European Genotype Archive: https://ega-archive.org/ega (EGA: EGAD00000000021). Genotype data for COG ALL affected individuals are available for download from dbGaP (dbGaP: phs000638.v1.p1).