Summary
Congenital diaphragmatic hernia (CDH) is a severe congenital anomaly that is often accompanied by other anomalies. Although the role of genetics in the pathogenesis of CDH has been established, only a small number of disease-associated genes have been identified. To further investigate the genetics of CDH, we analyzed de novo coding variants in 827 proband-parent trios and confirmed an overall significant enrichment of damaging de novo variants, especially in constrained genes. We identified LONP1 (lon peptidase 1, mitochondrial) and ALYREF (Aly/REF export factor) as candidate CDH-associated genes on the basis of de novo variants at a false discovery rate below 0.05. We also performed ultra-rare variant association analyses in 748 affected individuals and 11,220 ancestry-matched population control individuals and identified LONP1 as a risk gene contributing to CDH through both de novo and ultra-rare inherited largely heterozygous variants clustered in the core of the domains and segregating with CDH in affected familial individuals. Approximately 3% of our CDH cohort who are heterozygous with ultra-rare predicted damaging variants in LONP1 have a range of clinical phenotypes, including other anomalies in some individuals and higher mortality and requirement for extracorporeal membrane oxygenation. Mice with lung epithelium-specific deletion of Lonp1 die immediately after birth, most likely because of the observed severe reduction of lung growth, a known contributor to the high mortality in humans. Our findings of both de novo and inherited rare variants in the same gene may have implications in the design and analysis for other genetic studies of congenital anomalies.
Keywords: congenital diaphragmatic hernia, de novo variants, ALYREF, LONP1
Introduction
Congenital diaphragmatic hernia (CDH [MIM: 142340]) affects approximately 3 per 10,000 neonates.1,2 Approximately 40% of CDH-affected individuals occur with additional congenital anomalies besides common secondary anomalies (dextrocardia and lung hypoplasia).3 The most common additional anomalies4,5 are structural heart defects (11%–15%) and musculoskeletal malformations (15%–20%), including limb deficiency, club foot, and omphalocele.6 However, anomalies of almost every organ have been described in association with CDH. Despite advances in care, including improved prenatal diagnosis, fetal interventions, extracorporeal membrane oxygenation (ECMO), and gentle ventilation, CDH continues to be associated with at least 20% mortality and significant long-term morbidity, including feeding difficulties, pulmonary hypertension and other respiratory complications, and neurocognitive deficits.3,7,8
The complexity of the phenotypes associated with CDH is mirrored by the complexity of the genetics, which are heterogeneous; approximately 30% of individuals with CDH have an identifiable major genetic contributor. Typically, each gene or copy-number variant (CNV) associated with CDH accounts for at most 1%–2% of affected individuals.9 The full spectrum of genomic variants has been associated with CDH, including chromosome aneuploidies (10%), copy-number variants (CNVs) (3%–10%), monogenic conditions (10%–22%), and emerging evidence for oligogenic causes1 (CNVs and individual genes10).
Although CDH-affected familial individuals have been described, CDH most commonly occurs in individuals without a family history of CDH, and sibling recurrence risk in CDH-affected isolated individuals is less than 1%.11 Most likely because of the historically high mortality and low reproductive fitness, CDH is often due to de novo CNVs and single gene variants. However, dominant inheritance has been described with transmission of an incompletely penetrant variant from an unaffected parent or parent with a subclinical diaphragm defect.12 CDH has also been described in individuals with bi-allelic variants such as Donnai-Barrow syndrome13 (MIM: 222448). The occurrence of discordant monogenic twins suggests a role for stochastic events after fertilization.11
A genetic diagnosis for probands with CDH can inform prognosis and guide medical management. Some genetic conditions associated with CDH are associated with an increased risk for additional anomalies, increased mortality, and increased morbidity, including neurocognitive disabilities that may benefit from early intervention.3 Over the past decade, advances in genomic sequencing technology have helped to define the genes associated with CDH. We and others have shown that de novo variants with large effect size contribute to 10%–22% of CDH-affected individuals with enrichment of de novo likely damaging variants in CDH-affected individuals with an additional anomaly (complex CDH).9,14,15 We also demonstrated a higher burden of de novo likely damaging (LD) variants in females compared to males, supporting a “female protective model.”9 Most recently, in a cohort with long-term developmental outcome data,3 we demonstrated that de novo LD variants are associated with poorer neurodevelopmental outcomes as well as a higher prevalence of pulmonary hypertension (PH).
To expand upon our knowledge of the diverse genetic etiologies of CDH, we performed whole-genome sequencing (WGS) or exome sequencing of 827 CDH proband-parent trios. We confirmed an overall enrichment of damaging de novo variants in constrained genes and identified LONP1 (lon peptidase 1, mitochondrial [MIM: 605490]) and ALYREF (Aly/REF export factor [MIM: 604171]) as candidate CDH-associated genes with recurrent ultra-rare and de novo variants.
Material and methods
Participant recruitment and control datasets
Study participants were enrolled as fetuses, neonates, children, and adults with a radiologically confirmed diaphragm defect by the DHREAMS study16 (Diaphragmatic Hernia Research and Exploration; Advancing Molecular Science) or Boston Children’s Hospital/Massachusetts General Hospital (BCH/MGH) as described previously.14 Clinical data were prospectively collected from medical records and entered into a central research electronic data capture (REDCap) database.17 Probands and both parents provided a blood, skin biopsy, or saliva specimen for trio genetic analysis. All studies were approved by the institutional review boards at each participating institution and the CUIMC Institutional Review Board (IRB), and signed informed consent was obtained.
A total of 827 children with CDH and their parents had WGS or exome sequencing in the current study. A subset of trios (n = 574) has been described in our previous study.3,9
Participants with only a diaphragm defect were classified as having isolated CDH, while participants with at least one additional major congenital anomaly (e.g., congenital heart defect, central nervous system anomaly, gastrointestinal anomaly, skeletal anomaly, genitourinary anomaly, cleft lip/palate), moderate to severe developmental delay, or other neuropsychiatric phenotypes at last contact were classified as having complex CDH. Pulmonary hypoplasia, cardiac displacement, and intestinal herniation were considered to be part of the diaphragm defect sequence and were not considered independent malformations. Data on the child’s current and past health, including family history of congenital anomalies, postoperative pulmonary hypertension, mortality or survival status prior to initial discharge, and extracorporeal membrane oxygenation (ECMO) intake, were gathered as described previously.3
The control group consisted of unaffected parents from the Simons Powering Autism Research for Knowledge (SPARK) study18 (exomes) and Latinx samples from Washington Heights-Hamilton Heights-Inwood Community Aging Project (WHICAP) study19 (exomes).
WGS and exome data analysis
There are 233 CDH trios processed with WGS that were not included in previous studies3,9 (Table S1). Of these 233 previously unpublished trios, one trio was processed at Baylor College of Medicine Human Genome Sequencing Center and 232 trios were processed at Broad Institute Genomic Services. The genomic libraries of 219 affected individuals were prepared by TruSeq DNA PCR-Free Library Prep Kit (Illumina), while 14 were prepared by TruSeq DNA PCR-Plus Library Prep Kit (Illumina). Both libraries had an average fragment length about 350 bp and sequenced as paired-end of 150 bp on Illumina HiSeq X platform. Exome sequencing was performed in 20 CDH trios that were not previously published.3,9 Among these, the coding exons of nine trios were captured with Agilent Sure Select Human All Exon Kit v.2 (Agilent Technologies), ten trios with NimbleGen SeqCap EZ Human Exome .v3 kit (Regeneron NimbleGen), and one trio with NimbleGen SeqCap EZ Human Exome v.2 kit (Roche NimbleGen). Exomes of SPARK cohort were captured with a slightly modified version of the IDT xGen Exome Research Panel v.1.0 identical to the previous study.20 Whole-exome sequencing of the WHICAP cohort was performed at Columbia University with the Roche SeqCap EZ Exome Probes v.3.0 Target Enrichment Probes.21
Exome and WGS data of CDH-affected and control individuals were processed with a pipeline implementing GATK Best Practice v.4.0 as previously described.9,22 Specifically, reads of affected individuals' exomes were mapped to human genome GRCh37 reference with Burrows-Wheeler Aligner Maximal Exact Match (BWA-MEM),23 while reads of WGS data for affected individuals and SPARK and WHICAP control individuals were mapped to GRCh38; duplicated reads were marked with Picard;24 variants were called with GATK25 (v.4.0) HaplotypeCaller for generation of gVCF files for joint genotyping. All samples within the same batch (Table S1) were jointly genotyped and variant quality score recalibration (VQSR) was performed with GATK. To combine all affected individuals for further analysis, we lifted over the GRCh37 variants to GRCh38 by using CrossMap26 (v.0.3.0). We used common SNP genotypes within exome regions to validate familial relationships via KING27 and ancestries via peddy28 (v.0.4.3) in CDH-affected individuals, SPARK control individuals, and WHICAP control individuals.
De novo variants were defined as a variant present in the offspring with homozygous reference genotypes in both parents. Here, we limited WGS to coding regions on the basis of coding sequences and canonical splice sites of all GENCODE v.27 coding genes. We took a series of stringent filters to identify de novo variants as described previously:9 VQSR tranche ≤ 99.8 for single-nucleotide variants (SNVs) and ≤ 99.0 for indels, GATK’s FisherStrand ≤ 25, and quality by depth ≥ 2. We required the candidate de novo variants in probands to have ≥5 reads supporting the alternative allele, ≥20% alternative allele fraction, Phred-scaled genotype likelihood ≥ 60 (GQ), and population allele frequency ≤ 0.01% in gnomAD v.2.1.1; we required both parents to have ≥10 reference reads, <5% alternative allele fraction, and GQ ≥ 30. We applied DeepVariant29 to all candidate de novo variants for in silico confirmation and only included the ones with PASS from DeepVariant for downstream analysis.
To reduce batch effects in combined datasets from different sources30 in analysis of rare variants, we targeted ultra-rare variants located in xGen-captured protein-coding regions for non-Latinx populations and in regions targeted by xGen and SeqCap EZ v.3.0 for Latinx population. We used the following criteria to minimize technical artifacts and select ultra-rare variants:22 cohort allele frequency (AF) < 0.5% and population cohort < 1 × 10−5 across all genomes in gnomAD v.3.0; mappability = 1; >90% target region with depth ≥ 10; overlapped with segmental duplication regions < 95%; genotype quality > 30, allele balance > 20%, and depth > 10 in affected individuals.
We used Ensembl Variant Effect Predictor31 (VEP, Ensemble 102) and ANNOVAR32 to annotate variant function, variant population frequencies, and in silico predictions of deleteriousness. All coding SNVs and indels were classified as synonymous, missense, inframe, or likely gene disrupting (LGD, which includes frameshift indels, canonical splice site, or nonsense variants). We defined predicted damaging missense (D-mis) on the basis of CADD33 score v.1.3. All de novo variants and inherited variants in candidate risk genes were manually inspected in the Integrative Genome Viewer (IGV). A total of 179 variants were selected for validation via Sanger sequencing; all of them were confirmed (Table S2). To compare the clinical outcomes between affected individuals with deleterious variants in candidate genes and with likely damaging (LD) variants, we defined LD variants as in our previous study:3 (1) de novo LGD or deleterious missense variants in genes that are constrained (ExAC pLI ≥ 0.9) and highly expressed in developing diaphragm,34 (2) de novo LGD or deleterious missense variants in known risk genes for CDH or commonly comorbid disorders (congenital heart disease [CHD, MIM: 600001] and neurodevelopmental delay [NDD, MIM: 618354]), (3) plausible deleterious missense variants in known risk genes for CDH or commonly comorbid disorders (CHD and NDD), (4) deletions in constrained (ExAC pLI ≥ 0.9) or haploinsufficient genes from ClinGen genome dosage map,35 or (5) CNVs implicated in known syndromes. We classified CDH-affected individuals into two genetic groups: (1) LD, if the affected individual carried at least one de novo LD variant, or (2) non-LD, if the affected individual carried no such variants.
De novo CNVs were identified via an inhouse pipeline of read depth-based algorithm based on CNVnator36 v.0.3.3 in WGS trios as described in our previous study.3 The de novo CNV segments were validated by the additional pair-end/split-read (PE/SR) evidence via Lumpy37 v.0.2.13 and SVtyper38 v.0.1.4. Only the CNVs supported by both read depth (RD) and PE/SR were included in downstream analysis. We mapped de novo CNVs on GENCODE v.29 protein-coding genes with at least 1 bp in the shared interval. The GENCODE genes were annotated with variant intolerance metric by ExAC pLI,39 haploinsufficiency metric by Episcore,40 haploinsufficiency and triplosensitivity of genes from ClinGen genome dosage map,35 and CNV syndromes from DECIPHER41 v.11.1.
Quantitative PCR
We performed experimental validation of putative de novo genic CNVs by using quantitative PCR (qPCR). All PCR primers were designed for the selected genes located within the de novo CNVs and synthesized by IdtDNA. All qPCR reactions were performed in a total of 10 mL volume, comprising 5 mL 2× SYBR Green I Master Mix (Promega), 1 mL 10 nM of each primer, and 2 mL of 1:20 diluted cDNA in 96-well plates with CFX Connect Real-Time PCR Detection System (Bio-Rad). All reactions were performed in triplicate, and the conditions were 5 min at 95°C and then 40 cycles of 95°C at 15 s and 60°C at 30 s. The relative copy numbers were calculated via the standard curve method relative to the b-actin housekeeping gene. We used five-serial 4-fold dilutions of DNA samples to construct the standard curves for each primer.
Statistical analysis
Burden of de novo variants
The baseline mutation rates for different classes of de novo variants were calculated in each GENCODE coding gene via the published trinucleotide sequence context,42 and we calculated the rate in protein-coding regions that are uniquely mappable as previously described mutation model.9,18 The observed number of variants of various types (e.g., synonymous, missense, LGD) in each gene set and affected group was compared with the baseline expectation via Poisson test. In all analyses, constrained genes were defined by ExAC pLI39 score of >0.5, and all remaining genes were treated as other genes. We used a less stringent pLI threshold than previously suggested39 for defining constrained genes because it captures more known haploinsufficient genes important for heart and diaphragm development. We compared the observed number of variants in affected females versus males and affected complex versus isolated individuals by using the binormal test.
extTADA analysis
To identify risk genes based on de novo variants, we used an empirical Bayesian method: extTADA43 (extended transmission and de novo association). The extTADA model was developed on the basis of a previous integrated empirical Bayesian model, TADA,44 and estimates mean effect sizes and risk-gene proportions from the genetic data via MCMC (Markov chain Monte Carlo) process (for details, see supplemental note). To inform the parameter estimation with prior knowledge of developmental disorders, we stratify the genes into constrained genes (ExAC pLI score > 0.5) and non-constrained genes (other genes) and then estimate the parameters by using the extTADA model to each group of genes. After estimating posterior probability of association (PPA) of individual genes in each group, we combined both groups to calculate a final false discovery rate (FDR) for each gene by using extTADA’s procedure.
Gene-based case-control association analysis of ultra-rare variants
To identify risk genes based both on de novo and rare inherited variants, we performed a gene-based association test comparing the frequency of ultra-rare deleterious variants in CDH-affected individuals with control individuals without considering de novo status. Samples with read depth coverage ≥ 10× for 80% in exomes and 90% in genomes of the targeted regions were included in the analysis (Figure S1). Relatedness was checked via KING,27 and only unrelated affected and control individuals were included in the association tests (Figure S2). To control for confounding from genetic ancestry, we selected ancestry-matched control individuals by using SPARK exomes and Latinx WHICAP exomes to reach a fixed affected/control individual ratio in each population ancestry inferred by peddy28 (Figure S3). Specifically, for a specific ancestry (), consider number of CDH-affected individuals, number of control individuals, and the fold control individuals to affected individuals (yi/xi). We chose the minimized among all ancestries. In each genetic-ancestry group control individuals (), we ranked the Euclidean distance between each affected individual and control individuals, which were calculated from the top three principal-component analysis (PCA) eigenvectors, and selected control individuals from to ensure the same proportions in affected and control individuals. After filtering to reduce the impact of false positive variants, we tested for similarity of the ultra-rare synonymous variant rate among affected and control individuals in specific genetic-ancestry groups, assuming that ultra-rare synonymous variants are mostly neutral with respect to disease status.
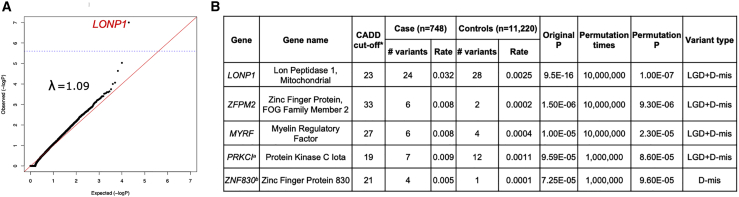
To identify CDH-risk genes, we tested the burden of ultra-rare deleterious variants (AF < 1 × 10−5 across all gnomAD v.3.0 genomes, LGD or D-mis) in each protein-coding gene in affected individuals compared to control individuals. To improve statistical power, we searched for a gene-specific CADD33 score threshold for defining D-mis that maximized the burden of ultra-rare deleterious variants in affected individuals compared to control individuals and used permutations to calculate statistical significance with the variable threshold test.22,45 For the binomial tests in each permutation, we used the binom.test function in R to calculate p values. We performed two association tests, one with LGD and D-mis variants combined and the other with D-mis variants alone, to account for different modes of action. We defined the threshold for genome-wide significance by Bonferroni correction for multiple testing (as two tests for each gene with 20,000 protein-coding genes, threshold p value = 1.25 × 10−6). We checked for inflation by using a quantile-quantile (Q-Q) plot and calculated the genomic control factor (lambda [λ]) by using QQperm in R. Lambda equal to 1 indicates no deviation from the expected distribution.
Protein modeling
We searched the LONP1 canonical sequence (UniProt: P36776-1) in UniProt and obtained the structural model of the human mitochondrial LONP1 monomer (encompassing only the residue range 413–951) by using SWISS-MODEL server46 with SWISS-MODEL Template Library (SMTL) ID 6u5z.1 as template. The 3D structure was visualized with PyMOL molecular viewer (The PyMOL Molecular Graphics System, v.1.2r3pre, Schrödinger).
Mice
All mice were housed in American Association for Accreditation of Laboratory Animal Care accredited facilities and laboratories at University of California, San Diego. All animal experiments were conducted under approved guidelines for the Care and Use of Laboratory Animals. Lonp1fl and Shhcre mice have both been described previously47 (International Mouse Strain Resource J:204812). All mice were maintained on a C57BL/6J background, and littermates were used as controls for minimization of potential genetic background effects.
Results
Cohort characteristics
Participants were recruited as part of the multi-site DHREAMS study (n = 748) and from BCH/MGH (n = 79). We performed WGS on 734 proband-parent trios and exome sequencing on 93 trios. In total, we analyzed 827 trios with WGS or exome sequencing.
In the cohort, there were 486 (59%) male probands (Table 1), consistent with a higher prevalence of CDH in males.9,48,49 The genetically determined ancestries (Figure S3A) were European (73.4%), admixed American (hereafter referred to as Latinx; 18.5%), African (3.7%), East Asian (1.8%), and South Asian (2.5%). Among the 277 (33.5%) CDH-affected complex individuals, the most frequent additional anomalies were CHD (n = 144), NDD (n = 54), skeletal anomalies (n = 46), genitourinary anomalies (n = 46), and gastrointestinal anomalies (n = 42). A total of 533 (64.4%) probands had isolated CDH without additional anomalies at the time of last follow up. The most common type of CDH was left-sided Bochdalek (Table 1).
Table 1.
Clinical summary of 827 CDH probands
| Number | Percent | ||
|---|---|---|---|
| Sex | male | 486 | 58.8% |
| female | 341 | 41.2% | |
| Genetic ancestry | African | 31 | 3.7% |
| Latinx | 153 | 18.5% | |
| European | 607 | 73.4% | |
| East Asian | 15 | 1.8% | |
| South Asian | 21 | 2.5% | |
| CDH classification | isolated | 533 | 64.4% |
| complex | 277 | 33.5% | |
| unknown | 17 | 2.1% | |
| CDH side | left | 645 | 78.0% |
| right | 119 | 14.4% | |
| bilateral/center/eventration/other | 38 | 4.6% | |
| unknown | 25 | 3.0% | |
| Timing of enrollment | fetal | 53 | 6.4% |
| neonatal | 464 | 56.1% | |
| child | 285 | 34.5% | |
| adult | 2 | 0.2% | |
| not specified | 23 | 2.8% | |
| Additional anomalies in CDH-affected complex individuals (n = 277) | cardiovascular | 144 | 52.0% |
| neurodevelopmentala | 54 | 19.5% | |
| skeletal | 46 | 16.6% | |
| genitourinary | 46 | 16.6% | |
| gastrointestinal | 42 | 15.2% | |
| pulmonary defectsb | 18 | 6.5% | |
| cleft lip or palate and/or micrognathia | 11 | 4.0% | |
Neurodevelopmental conditions include congenital abnormalities in the central nervous system and developmental delay or neuropsychiatric disorders on the basis of the follow-up developmental evaluations.
Does not include pulmonary hypoplasia or hypertension.
Burden of de novo coding variants
We identified 1,153 de novo protein-coding variants in 619 (74.8%) probands, including 1,058 SNVs and 95 indels (Table S2). The average number of de novo coding variants per proband is 1.39. The number of de novo coding variants across probands closely follows a Poisson distribution (Figure S4). Transition-to-transversion ratio of de novo SNVs was 2.75. We classified variants that were likely gene disrupting (LGD) or predicted damaging missense (“D-mis” with CADD score ≥ 25) as damaging variants. A total of 418 damaging variants (126 LGD and 292 D-mis) were identified in 318 (38.4%) affected individuals, including 83 (10%) probands harboring two or more such variants.
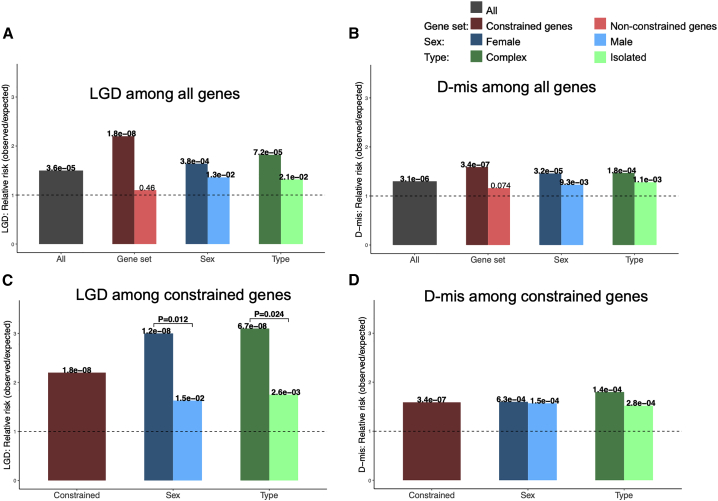
We analyzed the burden of de novo variants in CDH probands by comparing the observed number of variants to the expected number based on the background mutation rate. Consistent with previous studies on CDH9 and other developmental disorders,50, 51, 52 both de novo LGD (0.15 per affected individual) and D-mis variants (0.35 per affected individual) were significantly enriched in probands (relative risk [RR] = 1.5, p = 3.6 × 10−5 for LGD; RR = 1.3, p = 3.1 × 10−6; Figures 1A and 1B; Table S3), while the frequency of synonymous variants (0.30 per affected individual) closely matches the expectation (RR = 0.9, p = 0.12; Table S3). The burden of LGD variants is mostly located in constrained (ExAC39 pLI > 0.5) genes (RR = 2.2, p = 1.8 × 10−8). It is marginally higher in females than in male probands (RR = 3.0 versus 1.36, p = 0.012) and marginally higher in CDH-affected complex individuals than in isolated individuals (RR = 3.1 versus 1.75, p = 0.024; Figure 1C; Table S3).
Figure 1.
Burden of de novo coding variants in CDH compared to expectation
(A–D) LGD among all genes (A); D-mis among all genes (B); LGD among constrained genes (C); D-mis among constrained genes (D). p values between CDH-affected individuals and expectation by Poisson test are labeled for each bar. Significant p values between females and males and complex and isolated individuals by binormal test are labeled.
To identify CDH-risk genes by de novo variants, we applied extTADA43 to the data of 827 CDH trios. extTADA assumes a model of genetic architecture compatible with the observed burden and recurrence of de novo damaging variants and estimates an FDR for each gene via MCMC. From the burden analysis of de novo variants in CDH and previous studies,52 we reasoned that the constrained genes (ExAC pLI > 0.5) drive the higher burden of de novo damaging variants and are more likely to be plausible risk genes. We stratified the data into the constrained gene set and the non-constrained gene set (Table S4) and estimated extTADA priors (mean RR and prior probability of being a risk gene) in these two gene sets separately. Constrained genes had a higher prior of risk genes than non-constrained genes (0.037 versus 0.006). Meanwhile, both LGD and D-mis had higher relative risks in constrained genes than non-constrained genes (18.30 versus 5.24 for LGD; 10.01 versus 3.81 for D-mis). We estimated the Bayes factor of individual genes within each gene group and then combined the genes from two groups together to calculate FDR. We identified three genes with FDR < 0.05: MYRF (myelin regulatory factor [MIM: 608329]), LONP1, and ALYREF. Five of six MYRF de novo variants were described in our previous study.9 We identified three participants harboring de novo D-mis variants in LONP1 and two participants for de novo LGD variants in ALYREF. Of two participants with an ALYREF LGD variant, one had an isolated left-side CDH and the other had right-side CDH and ventricular septal defect. There were nine additional genes with ≥2 de novo predicted deleterious variants (HSD17B10 [MIM: 300256], GATA4 [MIM: 600576], SYMPK [MIM: 602388], PTPN11 [MIM: 176876], WT1 [MIM: 607102], FAM83H [MIM: 611927], CACNA1H [MIM: 607904], SEPSECS [MIM: 613009], and ZFYVE26 [MIM: 612012]) (Table 2). Of these, three are known CDH-associated genes (MYRF, GATA4, and WT1). All de novo variants in these genes are heterozygous.
Table 2.
Top CDH-associated genes predicted by pLI-stratified extTADA with ≥2 de novo predicted deleterious variants
| Gene | Gene name | #D-mis | #LGD | PPA | FDR | pLI |
|---|---|---|---|---|---|---|
| MYRFa | myelin regulatory factor | 3 | 3 | 1.00 | 3.97 × 10−6 | 1 |
| LONP1 | lon peptidase 1, mitochondrial | 3 | 0 | 0.97 | 0.014 | 1 |
| ALYREF | Aly/REF export factor | 0 | 2 | 0.93 | 0.033 | 0.83 |
| HSD17B10 | hydroxysteroid 17-beta dehydrogenase 10 | 1 | 1 | 0.87 | 0.056 | 0.89 |
| GATA4a | GATA-binding protein 4 | 1 | 1 | 0.86 | 0.072 | 0.8 |
| SYMPK | symplekin | 1 | 1 | 0.82 | 0.090 | 1 |
| PTPN11 | protein tyrosine phosphatase non-receptor type 11 | 2 | 0 | 0.79 | 0.11 | 1 |
| WT1a | WT1 transcription factor | 2 | 0 | 0.78 | 0.12 | 1 |
| FAM83H | family with sequence similarity 83 member H | 2 | 0 | 0.75 | 0.13 | 0.89 |
| CACNA1H | calcium voltage-gated channel subunit alpha1 H | 2 | 0 | 0.63 | 0.16 | 0 |
| SEPSECS | Sep (O-Phosphoserine) TRNA:Sec (Selenocysteine) TRNA synthase | 0 | 2 | 0.23 | 0.66 | 0 |
| ZFYVE26 | zinc finger FYVE-type containing 26 | 2 | 0 | 0.09 | 0.72 | 0 |
#D-mis, number of de novo D-mis; #LGD, number of de novo LGD; PPA, posterior probability of association; FDR, false discovery rate.
Known CDH-risk genes.
Recurrent genes in de novo CNVs
We applied CNVnator to call CNVs from WGS data and used customized filters to identify de novo CNVs. We performed experimental validation of 25 putative de novo genic CNVs, including all nine small CNVs (<5 kb), by using qPCR. 22 of 25 (88%) reported de novo CNVs in affected individuals were confirmed by qPCR. Removing the three false positive CNVs, there were 87 de novo CNVs identified in 734 CDH-affected individuals with WGS and an average of 0.12 per affected individual (Table S5). Among them, there were 54 (62%) deletions ranging from 2,096 bp to 33.7 Mb and 33 (38%) duplications ranging from 1,165 bp to 24.9 Mb. Seven samples carried known syndromic CNVs in DECIPHER41 dataset, one of which was heterozygous for a 16p13.11 microduplication, two of which were heterozygous for a 17q12 deletion associated with renal cysts and diabetes (RCAD), three of which were heterozygous for 21q22 duplication in the critical region for Down syndrome, and one of which was heterozygous for 22q11 deletion associated with DiGeorge syndrome. No recurrent genes were identified between de novo SNVs and CNVs. Four CNVs were recurrent (Table S6), two of which encompass single genes CSMD1 (CUB and sushi multiple domains 1 [MIM: 608397]) and GPHN (gephyrin [MIM: 603930]).
Candidate risk gene LONP1 contributes to CDH risk through both de novo and rare inherited variants
To identify additional risk genes that may contribute through rare inherited variants, we performed a gene-based, case-control association analysis of ultra-rare variants. Specifically, we used exome data from the SPARK (unaffected parents) and Latinx WHICAP samples as control individuals. Quality control procedures included at least 10× depth of sequence coverage across the target regions (Figure S1) and detection of cryptic relatedness among all CDH participants and control individuals (Figure S2). To prevent confounding by genetic ancestry, we performed PCA by peddy to infer genetic ancestry of all CDH-affected and control individuals and selected matching control individuals (15-fold of affected individuals numbers in each specific genetic-ancestry group) to reach a fixed affected/control individual ratio. With the same genetic-ancestry proportion in affected and control individuals (77% Europeans, 14.7% Latinx, 4.1% Africans, 2% East Asians, and 2.1% South Asians; Figure S3; Table S7), we selected 748 affected individuals and 11,220 control individuals for downstream analysis. We filtered the ultra-rare variant call sets of affected and control individuals in each genetic-ancestry group by empirical filters to reduce false positive calls and minimize technical batch effects across datasets. After filtering, the average numbers of ultra-rare (AF < 1 × 10−5 across all gnomAD v.3.0 genomes) synonymous variants per subject in affected and control individuals are nearly identical in everyone (enrichment rate = 1, p = 1) and specific ancestral groups (Table S8). Furthermore, a gene-level burden test confined to ultra-rare synonymous variants was consistent with a global null model in Q-Q plot (Figure S5), indicating that technical batch effects would most likely have minimal impact on genetic analyses. We then performed a variable threshold association test22,45 to identify risk genes on the basis of enrichment of ultra-rare damaging variants in individual genes. For each gene, we tested enrichment of LGD and D-mis variants together or just D-mis variants in order to account for potential different biological modes of action. In the variable threshold test, we determined a gene-specific optimal CADD score threshold to define D-mis in order to maximize the power of the association test and then estimated type I error rate by permutations. The overall result from the case-control association did not show inflation from the null model (l = 1.09; Figure 2A). The association of LONP1 (p = 1 × 10−7; Figure 2) exceeded the Bonferroni-corrected significance threshold (1.25 × 10−6, account for two tests in each gene). Three of the 24 ultra-rare deleterious variants in LONP1 were known de novo variants. Two known CDH-risk genes, ZFPM2 (zinc finger protein, FOG family member 2 [MIM: 603693]) and MYRF, fell just below the cutoff for genome-wide significance (Table S9).
Figure 2.
Gene-based association analysis with 748 CDH-affected individuals and 11,220 control individuals across all populations
(A) Results of a binomial test confined to ultra-rare LGD and D-mis variants or D-mis only variants in 18,939 protein-coding genes. Horizontal blue line indicates the Bonferroni-corrected threshold for significance.
(B) Complete list of top association genes with permutation p values < 1 × 10−4. ∗, a gene-specific CADD score threshold for defining D-mis that maximized the burden of ultra-rare deleterious variants in affected individuals compared to control individuals; #, numbers of deleterious variants; a, MIM: 600539; b, no MIM number.
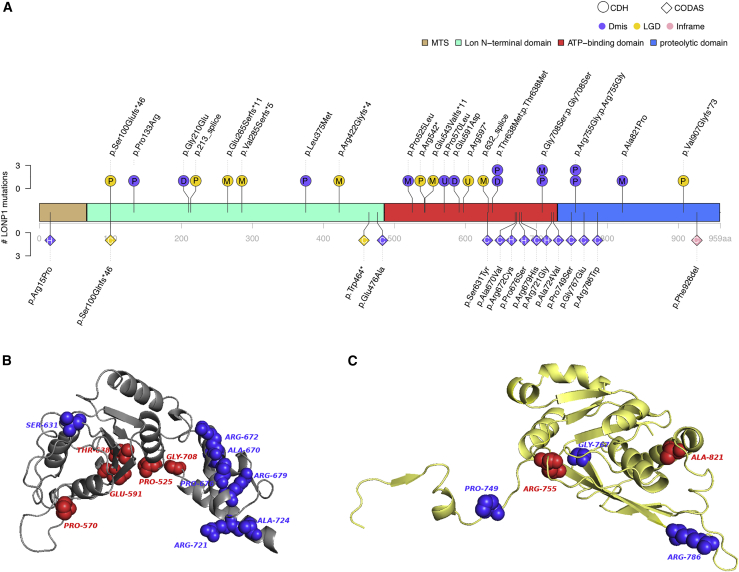
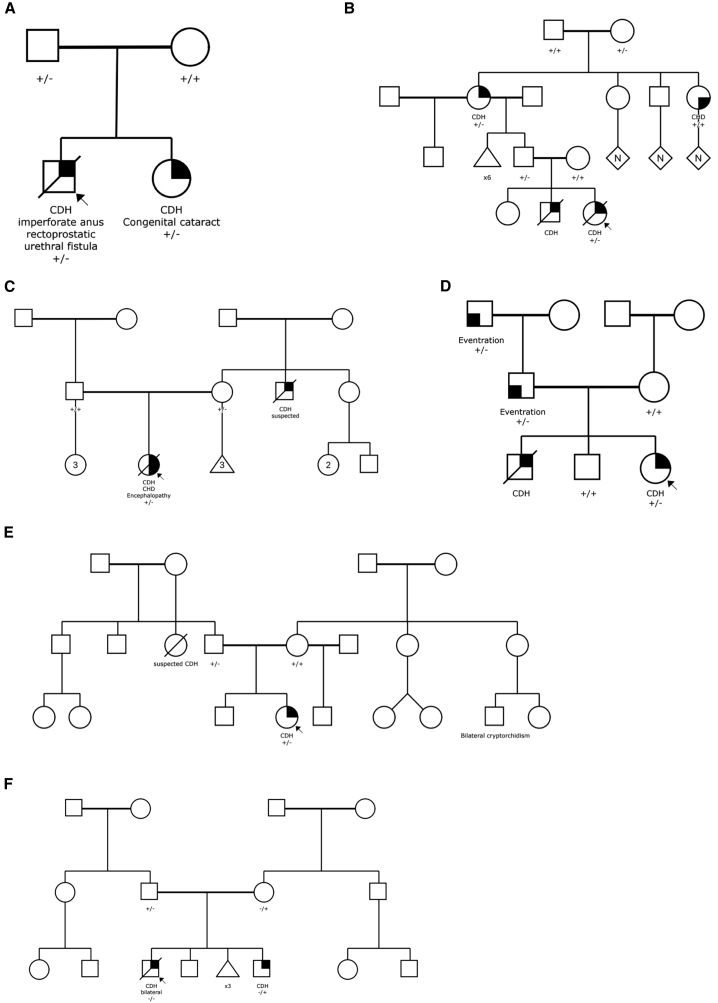
The association of LONP1 is due to both LGD and D-mis variants. We screened the whole cohort (Figure 3 and Table S10), including CDH relatives (n = 1) and exome sequencing singletons (n = 2), for ultra-rare damaging missense (CADD ≥ 25) and LGD in LONP1 (GenBank: NM_004793.3). A total of 23 CDH-affected individuals in 829 affected individuals (2.8%) carry 24 LONP1 variants, including ten LGD and 14 D-mis variants. Among 22 LONP1 variants excluding two of unknown inheritance variants in singletons, there are three (13.6%) de novo variants (all D-mis) and 19 (86.4%) inherited variants, 36.8% of which are from mothers (n = 7). Of 19 inherited variants, eight parents carrying LONP1 variants have a family history of CDH or diaphragm eventration (n = 4) or other congenital anomaly (n = 4; brain abnormality, cerebral palsy, cleft palate, skeletal abnormality) segregating with the LONP1 variant. Three inherited variants (c.1913C>T [p.Thr638Met], c.2122G>A [p.Gly708Ser], and c.2263C>G [p.Arg755Gly]) are each observed twice in the cohort on different probands. Familial segregation was established in six familial CDH-affected individuals for c.398C>G (p.Pro133Arg), c.639−1G>T (p.213_splice), c.1264del (p.Arg422Glyfs∗4), c.1574C>T (p.Pro525Leu), c.1913C>T (p.Thr638Met), and c.2720delinsGA (p.Val907Glyfs∗73) (Figure 4). One proband (01-1279) harbors bi-allelic heterozygous variants with c.1574C>T (p.Pro525Leu) inherited from the mother and c.2263C>G (p.Arg755Gly) inherited from the father (Figure 4). The participant with bi-allelic variants required ECMO and died at 8–9 h after birth with severe bilateral CDH with near complete diaphragm agenesis, bilateral lung hypoplasia, and no additional anomalies (Figure S6). All other detected variants were observed in the heterozygous state.
Figure 3.
Differential clustering of missense variants within LONP1 in CDH and CODAS syndrome
(A) Variant locations in LONP1 (GenBank: NM_004793.3) of CDH and CODAS syndrome. There are three main domains in LONP1: N-terminal Lon domain, ATP-binding domain, and proteolytic domain. Positions indicated at upper structure are variants in CDH. Deleterious heterozygous variants such as LGD and missense with CADD ≥ 25 and allele frequency < 1 × 10−5 across all gnomAD genomes in CDH are presented. Deleterious missense is presented in purple, LGD in yellow, and inframe in pink. Inheritance patterns were labeled in circles of variants (P, paternal; M, maternal; D, de novo; U, singleton unknown). Positions at lower structure are variants in published CODAS syndrome samples. CODAS syndrome is caused by bi-allelic variants in LONP1 (homozygous [H] or compound heterozygous [C] variants) in the diamonds.
(B and C) Predicted 3D structure of LONP1 protein with SWISS-Model. (B) CDH (red)- and CODAS (blue)-associated amino acids in ATPase domain (gray). CODAS-associated amino acids (Ala670–Ala724) are clustered at alpha-helix in ATPase domain. (C) CDH (red)- and CODAS (blue)-associated amino acids in protease domain (yellow). CDH-associated amino acid Ala821 is located at alpha-helix.
Figure 4.
Pedigree of CDH-affected familial individuals and carrying LONP1 deleterious variants
(A–F) Family 01-0670, p. Pro133Arg (A); family 04-0022, p.213_splice (B); family 1733, p.Arg422Glyfs∗4 (C); family 01-0513, p.Thr638Met (D); family 01-0732, p.Val907Glyfs∗73 (E); family 01-1279 carries bi-allelic heterozygous variants with c.1574C>T (p.Pro525Leu) inherited from the mother and c.2263C>G (p.Arg755Gly) inherited from the father (F).
Previous studies reported bi-allelic variants in LONP1 in cerebral, ocular, dental, auricular, and skeletal (CODAS) syndrome53,54 (MIM: 600373). We compared the locations of the predicted-damaging missense positions in CDH-affected and CODAS syndrome-affected individuals (Figure 3, Table S11). No variants overlap between CDH-affected and CODAS syndrome-affected individuals. LONP1 contains three functional domains. CDH-associated damaging variants are concentrated at the core of the domains. Bi-allelic variants observed in CODAS syndrome are located on the junction of ATP-binding and proteolytic domains (Figure 3, Table S11). The 23 CDH probands with LONP1 variants did not have features of CODAS syndrome.
Phenotype of CDH probands with LONP1 variants
We identified 24 ultra-rare heterozygous variants in 23 sporadic or familial CDH participants (Table S10). The majority (n = 17; 73.9%) are of European ancestry, and 13 (56.5%) are female (Table S10). Sixteen (70%) were enrolled as neonates. Fourteen of the 23 have a family history of congenital anomalies (Table S10), six of whom had a family history of CDH (Figure 4). Nine (39.1%) are CDH-affected complex individuals. Six of nine affected complex individuals have CHD in addition to CDH. We compared the clinical outcomes or phenotypes in CDH probands with LONP1 damaging variants and other CDH probands (Table 3). Compared to CDH probands without LONP1 ultra-rare damaging variants, those with a heterozygous LONP1 damaging variant experienced higher neonatal mortality rate prior to initial hospital discharge (69% versus 16%, p = 6.4 × 10−6) and greater need for ECMO (56% versus 28%, p = 2.3 × 10−2). Compared to CDH probands with other likely damaging variants defined in our previous study,3 those with a heterozygous LONP1 damaging variant had higher neonatal mortality rate prior to discharge (69% versus 24%, p = 1.8 × 10−3) and trended toward greater need for ECMO (56% versus 30%, p = 0.077).
Table 3.
CDH cases with heterozygous LONP1 deleterious rare variants are associated with higher mortality and need for ECMO
| CDH w/LONP1 deleterious variants (n = 23) |
CDH w/o LONP1 deleterious variants (n = 806) |
w/LONP1 versus w/o LONP1 deleterious variants |
CDH w/likely damaging variants (n = 98) |
w/LONP1 deleterious variants versus w/likely damaging variants |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total n | Categorized n | % | Total n | Categorized n | % | p value | Total n | Categorized n | % | p value | |
| Male | 23 | 10 | 43% | 806 | 477 | 59% | 0.14 | 98 | 47 | 48% | 0.82 |
| Complex | 23 | 9 | 39% | 789 | 269 | 34% | 0.66 | 96 | 50 | 52% | 0.35 |
| Familial CDH | 19 | 6 | 32% | 806 | 61 | 8% | 2.7 × 10−2∗ | 98 | 4 | 4% | 1.2 × 10−3∗ |
| Neonatal death prior to discharge | 16 | 11 | 69% | 450 | 72 | 16% | 6.4 × 10−6∗ | 55 | 13 | 24% | 1.8 × 10−3∗ |
| ECMO | 16 | 9 | 56% | 442 | 124 | 28% | 2.3 × 10−2∗ | 53 | 16 | 30% | 0.077 |
| PH at 1 month | 11 | 7 | 64% | 340 | 188 | 55% | 0.76 | 41 | 29 | 71% | 0.72 |
| PH at 3 months | 6 | 2 | 33% | 260 | 100 | 39% | 1 | 29 | 16 | 55% | 0.4 |
p values with asterisks highlight significance. ECMO, extracorporeal membrane oxygenation; PH, pulmonary hypertension.
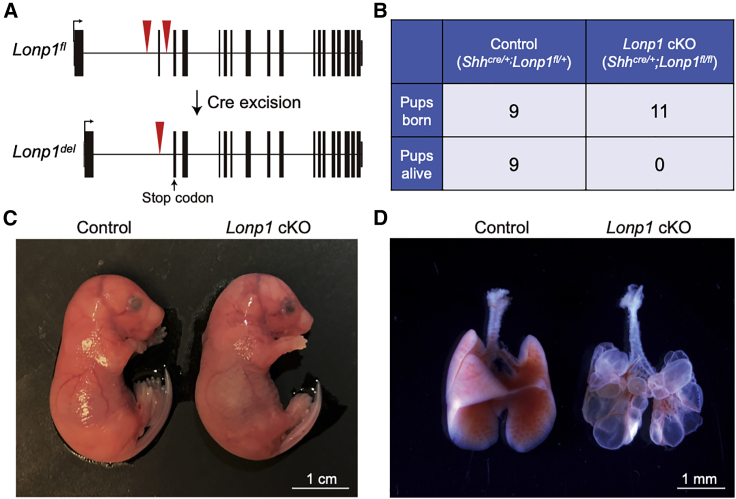
Inactivation of Lonp1 in mouse embryonic lung epithelium leads to disrupted lung development and full lethality at birth
The high rate of mortality and need for ECMO in CDH-affected individuals is predominantly due to abnormal lung function. Our hypothesis was that impaired or partial loss of LONP1 function in CDH-affected individuals might contribute directly to abnormal lung development, independent of its role in diaphragm formation. To test this hypothesis, we inactivated Lonp1 in the embryonic lung epithelium in mice. We achieved this by generating Shhcre/+;Lonp1fl/fl (hereafter Lonp1 cKO for conditional knockout) embryos by using existing alleles47 (International Mouse Strain Resource J:204812). In the mutant, the Cre recombinase expressed specifically in the epithelium drove deletion of key exons resulting in Lonp1 inactivation at the onset of lung initiation (Figure 5A). Although the mutants are externally normal in size, they exhibited 100% lethality at birth (Figures 5B and 5C). Upon dissection, the mutant lung was composed of large fluid-filled sacs, missing the normal airways and alveoli present in the control lung, indicating a severe reduction of lung growth (Figure 5D). This lung defect most likely contributed to embryonic lethality at birth in these mutant mice.
Figure 5.
Inactivation of Lonp1 in mice led to disrupted lung development and lethality at birth
(A) Gene structure of mouse Lonp1fl conditional allele before and after cre-mediated recombination of the loxP sites (red triangles). Recombination led to a premature stop codon (arrow) in the second exon.
(B) Number of embryos genotyped at perinatal stage, showing 100% lethality of the mutant embryos.
(C) Representative mutant and control embryos at embryonic day (E) 18.5, the day of birth.
(D) Representative mutant and control lungs at E18.5. Scale bars as indicated.
Discussion
In the current study of 827 CDH trios, we confirmed there is an overall enrichment of damaging de novo variants, particularly in constrained genes. We identified LONP1 and ALYREF as candidate risk genes on the basis of enrichment of de novo variants. By case-control association, we also confirmed LONP1 as a genome-wide significant candidate gene contributing to CDH risk through both de novo and inherited damaging variants. We demonstrated segregation of a LONP1 variant with diaphragm defect in five families. We found that CDH individuals with heterozygous ultra-rare damaging variants in LONP1 have clinical phenotypes frequently including CHD or skeletal anomalies, frequently requiring ECMO, and having a higher mortality than the rest of our CDH cohort. In addition, we confirmed MYRF and ZFPM2 as genes associated with CDH.9,14,55,56 In a mouse model with knockout of Lonp1 only in the embryonic lung epithelium with an intact diaphragm, we demonstrated reduced pulmonary growth and branching, resulting in perinatal lethality that suggests that the higher mortality rate and need for ECMO in human is due to a primary effect of LONP1 on pulmonary development in addition to diaphragm development.
The burden of damaging de novo variants in CDH is consistent with previous studies,9,14,15 and damaging de novo variants are more frequent in complex CDH compared to isolated CDH-affected individuals. Similar patterns have been observed in complex CHD with other congenital anomalies or neurodevelopmental disorders compared with isolated CHD50 and autism with/without intellectual disability.57 Deleterious de novo variants are more frequent in many severe early-onset diseases with reduced reproductive fitness compared to the general population.58 The higher frequency of de novo LGD variants in CDH-affected females relative to males supports the “female protective model” similar to autism,52,59,60 which means that risk variants have larger effects in males than in females so that females require a higher burden to reach the same diagnostic threshold as males.
Both de novo and rare inherited variant analyses highlight LONP1 as a CDH candidate risk gene. Approximately 3% of individuals in our CDH cohort are heterozygous for LONP1 rare variants. Three variants (p.Thr638Met, p.Gly708Ser, and p.Arg755Gly) are recurrently and independently found in unrelated families. Each of the recurrent variants is observed both in isolated and complex CDH-affected individuals (Table S10), suggesting that other genetic modifiers and/or environmental factors most likely determine the variable expressivity. CDH probands with LONP1 variants had higher mortality in the neonatal period compared with other children with CDH. Bi-allelic variants in LONP1 have been reported in CODAS, a multi-system developmental disorder characterized by cerebral, ocular, dental, auricular, and skeletal anomalies.61 The LONP1 holoenzyme is a homohexamer with six identical subunits. Each subunit consists of a mitochondrial-targeting sequence (MTS), a substrate recognition and binding (N) domain, an ATPase (AAA+) domain, and a proteolytic (P) domain. Bi-allelic missense variants reported in CODAS individuals are mostly located in the junction of ATP-binding and proteolytic domains of LONP1, while the heterozygous variants identified in CDH-affected individuals are located in the main domains of LONP1. Notably, there are no overlapping variants between CDH- and CODAS-affected individuals. Most of the variants in CODAS are located in the alpha-helix and may affect the interactions of subunits.61 Variants in CDH may interrupt the proteolytic and ATP-binding domains, resulting in the dysfunction of LONP1. Homozygous deletion of Lonp1 in mice is embryonic lethal because of progressive loss of mtDNA with subsequent failure to meet energy requirements for embryonic development.62 Heterozygous Lonp1+/− mice develop normally without obvious abnormalities.62 Analysis of Lonp1 expression in heterozygous mice indicated a 50% reduction at both RNA and protein levels in these animals. These data suggest different mechanisms of LONP1 in diseases with bi-allelic and monoallelic variants. Of note, one CDH-affected individual carried bi-allelic variants (p.Pro525Leu and p.Arg755Gly). No additional phenotypes were noted, perhaps because the baby died at 8–9 h after birth with severe bilateral CDH (Figure S6).
LONP1 is a nuclear-encoded mitochondrial protease. Besides binding of mtDNA,63 LONP1 was discovered as an ATP-dependent protease involved in the degradation of misfolded or damaged proteins.64, 65, 66 Accumulation of misfolded proteins has been observed in the impaired lungs of developing mice with deletion of other ATP-dependent proteins.67 The immature lung development and neonatal respiratory failure of our Lonp1 cKO mice could be due to accumulation of misfolded proteins and activation of the unfolded protein response (UPR) pathway.68 UPR activation during development could lead to reduced cell proliferation and cause other congenital anomalies, including CHD.69
LONP1 also acts as a chaperone that interacts with other mitochondrial proteins to regulate several cellular processes.70 Lon expression may stimulate cell proliferation71 and Lon downregulation may impair mitochondrial structure and function and cause apoptosis.72,73 Alterations in cell proliferation, differentiation, and migration can all lead to CDH. Myogenic cell differentiation and migration are essential during formation of the diaphragm.74 Myogenic differentiation requires increased expression of mitochondrial biogenesis-related genes, including Lon.75 The variants could cause an increased probability of failure of myogenesis during embryonic development, consequently resulting in the hernia.
The neonatal mortality of probands with LONP1 deleterious variants is much higher than CDH neonates without LONP1 deleterious variants or CDH neonates with likely damaging variants in genes other than LONP1. CDH neonates with LONP1 deleterious variants frequently required ECMO. In mice with Lonp1 knockout at the onset of lung development, 100% newborn pups died shortly after birth and had severe pulmonary defects. Thus, LONP1 could represent a class of CDH-risk genes associated with high mortality due to primary developmental effects on the lung, resulting in more severe pulmonary defects than would occur secondarily to lung compression by herniated abdominal viscera alone. This suggests that we should try to differentiate primary from secondary developmental effects on the lung as we phenotype newborns with CDH and as we investigate the mechanisms of action of CDH candidate genes.
The RNA-binding protein ALYREF plays a key role in nuclear export through binding to the 5′ and the 3′ regions of mRNA.76,77 It acts as an RNA 5-methylcytosine (m5C) adaptor to regulate the m5C modification.78,79 Disruption of ALYREF could affect the m5C modification, resulting in abnormal cell proliferation and migration.79 Previous studies50 identified several RNA-binding proteins (RBPs) playing essential roles in autism and congenital birth defects, including CHD. RBFOX2, an RBP that regulates alternative splicing, is critical for zebrafish heart development,80 and de novo variants in RBFOX2 are associated with congenital heart defects.50 Dozens of RBPs have established roles in autism spectrum disorder. RBFOX1,81,82 an RNA splicing factor, regulates expression of large genetic networks during early neuronal development including autism. The other RBPs, such as FMRP,83 CELF4, and CELF6,84 have also been implicated in autism. As an RBP, ALYREF may play a similar role in congenital anomalies and neurodevelopmental disorders. Two de novo LGDs in ALYREF were identified in our CDH cohort. One had an isolated CDH, and the other had CDH and a ventricular septal defect. Similarly, two CDH-affected individuals carried de novo variants in SYMPK (Table S9), another RBP identified with FDR < 0.1 in extTADA. One had a de novo predicted deleterious missense variant and isolated CDH, and the other had a de novo LGD with complex CDH with CHD, central nervous system anomaly, and genitourinary anomaly.
We found further support for the previously reported CDH-associated genes ZFPM2 and MYRF. We have identified six ultra-rare LGD variants in ZFPM2 in our CDH cohort, accounting for 0.7% of our CDH-affected individuals (Figure S7, Table S9). Three were CDH-affected complex individuals, all with minor cardiac malformations. Specifically, two females had atrial septal defects and one male had an enlarged aortic root. The other three heterozygotes had isolated CDH. ZFPM2 is expressed in the septum transversum of the diaphragm during early development, and Fog2−/− mice generated through chemical mutagenesis have been shown to have diaphragmatic eventration and pulmonary hypoplasia.55 ZFPM2 physically interacts with NR2F285 and GATA4,86 two other components of the retinoid signaling pathway implicated in diaphragm and lung development.87 Our results further support the pleiotropic role of ZFPM2 in the development of CDH.
MYRF was implicated in our previous de novo variant report9 as a gene for cardiac-urogenital syndrome (MIM: 618280), and we identified one more additional de novo variant in this cohort (Figure S8, Table S9). There are now more than ten variants implicated in CDH with additional anomalies (HGMD professional 2021.1). MYRF is highly expressed in mesothelial cells. Mesothelial cells are a key cellular component of the diaphragm. They are derived from the mesoderm of the pleuroperitoneal folds (PPFs) through cell proliferation, migration, and epithelial-to-mesenchymal transition.88,89 Single-cell analysis90 in fetal gonads suggests the cells that highly express MYRF also express WT1 and NR2F2, two genes associated with diaphragmatic hernia. Previously, we also demonstrated9 that individuals with pathogenic variants in MYRF have decreased expression of GATA4. WT1, NR2F2, and GATA4 are all important in retinoic acid (RA) signaling in the developing diaphragm.1 Therefore, the damaging variants in MYRF may affect the RA signaling pathway, leading to diaphragmatic hernia and other anomalies.
Among the 734 CDH trios with WGS data, we identified a total of 87 de novo CNVs and four of them are recurrent genes or CNVs. Given the rarity of de novo CNVs and small sample size, there were limited data for analysis of the differential burden between CDH-affected and control individuals in this study. Future studies with larger sample sizes will improve the power to analyze CNVs and structural variants in CDH. Our data suggest there is genetic heterogeneity underlying CDH pathogenesis including both de novo9 and ultra-rare inherited variants. In addition, the phenotypic heterogeneity associated with LONP1 variants is notable, encompassing a wide range of anomalies, including CDH, CHD, and skeletal, ophthalmologic, and other anomalies. Such genetic heterogeneity is similar to other structural congenital anomalies, such as CHD,50,57 and neurological conditions, including autism18 and NDD.91 The time at which the single variants occurred cannot be determined because we did not have access to multiple tissues in this study, leaving us unable to fully evaluate the possibility of mosaicism, and somatic mutations after fertilization could play a role. We hypothesize that a dosage effect along a spectrum of missense variants, LGD and number of variants, and a threshold of sensitivity of different organs could explain the distinction between CODAS and CDH. However, a dominant-negative mechanism for missense variants in CDH is an alternative explanation for variable phenotypes. Nevertheless, understanding the molecular mechanisms will require further functional studies.
In summary, our analysis of de novo and ultra-rare inherited variants identified two CDH candidate genes, LONP1 and ALYREF, and confirmed previous associations of MYRF and ZFPM2 with CDH. The identification of specific highly risk genes would enhance prenatal or early postnatal counseling and decision-making, especially with rapid turnaround of WGS or exome sequencing results. It is likely that transmitted rare variants also contribute to other CDH-affected individuals in our cohort, but we require a larger sample size to identify these genes confidently. Future studies will also leverage data from other developmental disorders and integrating genomic data during development.
Acknowledgments
We would like to thank the patients and their families for their generous contribution. Staff with technical assistance of Columbia University, Boston Children’s Hospital, Massachusetts General Hospital, and clinical coordinators across the DHREAMS centers are acknowledged in the supplemental note. The whole-genome sequencing data were generated through NIH Gabriella Miller Kids First Pediatric Research Program (X01HL132366, X01HL136998, X01HL155060). This work was supported by NIH grants R01HD057036 (L.Y., J.W., W.K.C.), R03HL138352 (A.K., W.K.C., Y.S.), R01GM120609 (H.Q., Y.S.), UL1 RR024156 (W.K.C.), P01HD068250 (P.K.D., F.A.H., J.M.W., W.K.C., Y.S., J.M.Z., D.J.M., X.S.) and NSFC81501295 (L.Y.). Additional funding support was provided by grants from CHERUBS, CDHUK, and the National Greek Orthodox Ladies Philoptochos Society and generous donations from the Williams family, Wheeler Foundation, Vanech Family Foundation, Larsen family, Wilke family, and many other families. Whole-genome sequencing data can be obtained from dbGaP through accession dbGaP: phs001110. The WHICAP study is supported by funding from NIA RF1AG054023 (B.N.V.). Biogen provided support for whole-exome sequencing for the WHICAP cohort.
Declaration of interests
The authors declare no competing interests.
Published: September 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.08.011.
Contributor Information
Yufeng Shen, Email: ys2411@cumc.columbia.edu.
Wendy K. Chung, Email: wkc15@cumc.columbia.edu.
Data and code availability
Data for damaging variants identified are listed in the supplementary tables. The whole-genome sequencing and exome sequencing CDH data used in this study are available at the database of Genotypes and Phenotypes (dbGaP: phs001110.v2.p1). The SPARK data are available under managed access from Simons Foundation Autism Research Initiative (SRARI). The WHICAP dataset analyzed for the manuscript is available from the author B.N.V. on request.
Web resources
ClinGen genome dosage map, https://dosage.clinicalgenome.org
DECIPHER, https://www.deciphergenomics.org
DHREAMS study, http://www.cdhgenetics.com/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
Integrative Genome Viewer (IGV), http://software.broadinstitute.org/software/igv
Mouse Genome Informatics (MGI), http://www.informatics.jax.org
OMIM, https://www.omim.org/
PyMOL molecular viewer, https://pymol.org/2/
The Human Protein Atlas, https://www.proteinatlas.org/
Supplemental information
References
- 1.Yu L., Hernan R.R., Wynn J., Chung W.K. The influence of genetics in congenital diaphragmatic hernia. Semin. Perinatol. 2020;44:151169. doi: 10.1053/j.semperi.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kardon G., Ackerman K.G., McCulley D.J., Shen Y., Wynn J., Shang L., Bogenschutz E., Sun X., Chung W.K. Congenital diaphragmatic hernias: from genes to mechanisms to therapies. Dis. Model. Mech. 2017;10:955–970. doi: 10.1242/dmm.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiao L., Wynn J., Yu L., Hernan R., Zhou X., Duron V., Aspelund G., Farkouh-Karoleski C., Zygumunt A., Krishnan U.S. Likely damaging de novo variants in congenital diaphragmatic hernia patients are associated with worse clinical outcomes. Genet. Med. 2020;22:2020–2028. doi: 10.1038/s41436-020-0908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montalva L., Lauriti G., Zani A. Congenital heart disease associated with congenital diaphragmatic hernia: A systematic review on incidence, prenatal diagnosis, management, and outcome. J. Pediatr. Surg. 2019;54:909–919. doi: 10.1016/j.jpedsurg.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Lin A.E., Pober B.R., Adatia I. Congenital diaphragmatic hernia and associated cardiovascular malformations: type, frequency, and impact on management. Am. J. Med. Genet. C. Semin. Med. Genet. 2007;145C:201–216. doi: 10.1002/ajmg.c.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosiński P., Wielgoś M. Congenital diaphragmatic hernia: pathogenesis, prenatal diagnosis and management - literature review. Ginekol. Pol. 2017;88:24–30. doi: 10.5603/GP.a2017.0005. [DOI] [PubMed] [Google Scholar]
- 7.Wynn J., Aspelund G., Zygmunt A., Stolar C.J., Mychaliska G., Butcher J., Lim F.Y., Gratton T., Potoka D., Brennan K. Developmental outcomes of children with congenital diaphragmatic hernia: a multicenter prospective study. J. Pediatr. Surg. 2013;48:1995–2004. doi: 10.1016/j.jpedsurg.2013.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wynn J., Krishnan U., Aspelund G., Zhang Y., Duong J., Stolar C.J., Hahn E., Pietsch J., Chung D., Moore D. Outcomes of congenital diaphragmatic hernia in the modern era of management. J. Pediatr. 2013;163:114–119.e1. doi: 10.1016/j.jpeds.2012.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi H., Yu L., Zhou X., Wynn J., Zhao H., Guo Y., Zhu N., Kitaygorodsky A., Hernan R., Aspelund G. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogenschutz E.L., Fox Z.D., Farrell A., Wynn J., Moore B., Yu L., Aspelund G., Marth G., Yandell M., Shen Y. Deep whole-genome sequencing of multiple proband tissues and parental blood reveals the complex genetic etiology of congenital diaphragmatic hernias. HGG Adv. 2020;1:100008. doi: 10.1016/j.xhgg.2020.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pober B.R., Lin A., Russell M., Ackerman K.G., Chakravorty S., Strauss B., Westgate M.N., Wilson J., Donahoe P.K., Holmes L.B. Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am. J. Med. Genet. A. 2005;138A:81–88. doi: 10.1002/ajmg.a.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L., Wynn J., Cheung Y.H., Shen Y., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum. Genet. 2013;132:285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantarci S., Al-Gazali L., Hill R.S., Donnai D., Black G.C., Bieth E., Chassaing N., Lacombe D., Devriendt K., Teebi A. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007;39:957–959. doi: 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longoni M., High F.A., Qi H., Joy M.P., Hila R., Coletti C.M., Wynn J., Loscertales M., Shan L., Bult C.J. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum. Genet. 2017;136:679–691. doi: 10.1007/s00439-017-1774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L., Sawle A.D., Wynn J., Aspelund G., Stolar C.J., Arkovitz M.S., Potoka D., Azarow K.S., Mychaliska G.B., Shen Y., Chung W.K. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum. Mol. Genet. 2015;24:4764–4773. doi: 10.1093/hmg/ddv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu L., Wynn J., Ma L., Guha S., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D. De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet. 2012;49:650–659. doi: 10.1136/jmedgenet-2012-101135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feliciano P., Zhou X., Astrovskaya I., Turner T.N., Wang T., Brueggeman L., Barnard R., Hsieh A., Snyder L.G., Muzny D.M. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom. Med. 2019;4:19. doi: 10.1038/s41525-019-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang M.X., Cross P., Andrews H., Jacobs D.M., Small S., Bell K., Merchant C., Lantigua R., Costa R., Stern Y., Mayeux R. Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology. 2001;56:49–56. doi: 10.1212/wnl.56.1.49. [DOI] [PubMed] [Google Scholar]
- 20.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.D., Liu D., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghavan N.S., Brickman A.M., Andrews H., Manly J.J., Schupf N., Lantigua R., Wolock C.J., Kamalakaran S., Petrovski S., Tosto G. Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2018;5:832–842. doi: 10.1002/acn3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N., Swietlik E.M., Welch C.L., Pauciulo M.W., Hagen J.J., Zhou X., Guo Y., Karten J., Pandya D., Tilly T. Rare variant analysis of 4241 pulmonary arterial hypertension cases from an international consortium implicates FBLN2, PDGFD, and rare de novo variants in PAH. Genome Med. 2021;13:80. doi: 10.1186/s13073-021-00891-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11.10.11–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H., Sun Z., Wang J., Huang H., Kocher J.P., Wang L. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen B.S., Quinlan A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poplin R., Chang P.C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 30.Tom J.A., Reeder J., Forrest W.F., Graham R.R., Hunkapiller J., Behrens T.W., Bhangale T.R. Identifying and mitigating batch effects in whole genome sequencing data. BMC Bioinformatics. 2017;18:351. doi: 10.1186/s12859-017-1756-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell M.K., Longoni M., Wells J., Maalouf F.I., Tracy A.A., Loscertales M., Ackerman K.G., Pober B.R., Lage K., Bult C.J., Donahoe P.K. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc. Natl. Acad. Sci. USA. 2012;109:2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L. ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., Marth G.T., Quinlan A.R., Hall I.M. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teschendorff A.E., Zhu T., Breeze C.E., Beck S. EPISCORE: cell type deconvolution of bulk tissue DNA methylomes from single-cell RNA-Seq data. Genome Biol. 2020;21:221. doi: 10.1186/s13059-020-02126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen H.T., Bryois J., Kim A., Dobbyn A., Huckins L.M., Munoz-Manchado A.B., Ruderfer D.M., Genovese G., Fromer M., Xu X. Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med. 2017;9:114. doi: 10.1186/s13073-017-0497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He X., Sanders S.J., Liu L., De Rubeis S., Lim E.T., Sutcliffe J.S., Schellenberg G.D., Gibbs R.A., Daly M.J., Buxbaum J.D. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris K.S., Zhang Z., McManus M.T., Harfe B.D., Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinton C.F., Siffel C., Correa A., Shapira S.K. Survival Disparities Associated with Congenital Diaphragmatic Hernia. Birth Defects Res. 2017;109:816–823. doi: 10.1002/bdr2.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leeuwen L., Mous D.S., van Rosmalen J., Olieman J.F., Andriessen L., Gischler S.J., Joosten K.F.M., Wijnen R.M.H., Tibboel D., IJsselstijn H., Spoel M. Congenital Diaphragmatic Hernia and Growth to 12 Years. Pediatrics. 2017;140:e20163659. doi: 10.1542/peds.2016-3659. [DOI] [PubMed] [Google Scholar]
- 50.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R., Zeng X., Qi H., Chang W., Sierant M.C. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauss K.A., Jinks R.N., Puffenberger E.G., Venkatesh S., Singh K., Cheng I., Mikita N., Thilagavathi J., Lee J., Sarafianos S. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Am. J. Hum. Genet. 2015;96:121–135. doi: 10.1016/j.ajhg.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shebib S.M., Reed M.H., Shuckett E.P., Cross H.G., Perry J.B., Chudley A.E. Newly recognized syndrome of cerebral, ocular, dental, auricular, skeletal anomalies: CODAS syndrome--a case report. Am. J. Med. Genet. 1991;40:88–93. doi: 10.1002/ajmg.1320400118. [DOI] [PubMed] [Google Scholar]
- 55.Ackerman K.G., Herron B.J., Vargas S.O., Huang H., Tevosian S.G., Kochilas L., Rao C., Pober B.R., Babiuk R.P., Epstein J.A. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bleyl S.B., Moshrefi A., Shaw G.M., Saijoh Y., Schoenwolf G.C., Pennacchio L.A., Slavotinek A.M. Candidate genes for congenital diaphragmatic hernia from animal models: sequencing of FOG2 and PDGFRalpha reveals rare variants in diaphragmatic hernia patients. Eur. J. Hum. Genet. 2007;15:950–958. doi: 10.1038/sj.ejhg.5201872. [DOI] [PubMed] [Google Scholar]
- 57.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacquemont S., Coe B.P., Hersch M., Duyzend M.H., Krumm N., Bergmann S., Beckmann J.S., Rosenfeld J.A., Eichler E.E. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B., Ji T., Zhou X., Wang J., Wang X., Wang J., Zhu D., Zhang X., Sham P.C., Zhang X. CNV analysis in Chinese children of mental retardation highlights a sex differentiation in parental contribution to de novo and inherited mutational burdens. Sci. Rep. 2016;6:25954. doi: 10.1038/srep25954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibellini L., De Gaetano A., Mandrioli M., Van Tongeren E., Bortolotti C.A., Cossarizza A., Pinti M. The biology of Lonp1: More than a mitochondrial protease. Int. Rev. Cell Mol. Biol. 2020;354:1–61. doi: 10.1016/bs.ircmb.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Quirós P.M., Español Y., Acín-Pérez R., Rodríguez F., Bárcena C., Watanabe K., Calvo E., Loureiro M., Fernández-García M.S., Fueyo A. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 63.Matsushima Y., Goto Y., Kaguni L.S. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc. Natl. Acad. Sci. USA. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gur E., Sauer R.T. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He L., Luo D., Yang F., Li C., Zhang X., Deng H., Zhang J.R. Multiple domains of bacterial and human Lon proteases define substrate selectivity. Emerg. Microbes Infect. 2018;7:149. doi: 10.1038/s41426-018-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mikita N., Cheng I., Fishovitz J., Huang J., Lee I. Processive degradation of unstructured protein by Escherichia coli Lon occurs via the slow, sequential delivery of multiple scissile sites followed by rapid and synchronized peptide bond cleavage events. Biochemistry. 2013;52:5629–5644. doi: 10.1021/bi4008319. [DOI] [PubMed] [Google Scholar]
- 67.Flodby P., Li C., Liu Y., Wang H., Marconett C.N., Laird-Offringa I.A., Minoo P., Lee A.S., Zhou B. The 78-kD Glucose-Regulated Protein Regulates Endoplasmic Reticulum Homeostasis and Distal Epithelial Cell Survival during Lung Development. Am. J. Respir. Cell Mol. Biol. 2016;55:135–149. doi: 10.1165/rcmb.2015-0327OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pareek G., Pallanck L.J. Inactivation of Lon protease reveals a link between mitochondrial unfolded protein stress and mitochondrial translation inhibition. Cell Death Dis. 2018;9:1168. doi: 10.1038/s41419-018-1213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi H., O’Reilly V.C., Moreau J.L., Bewes T.R., Yam M.X., Chapman B.E., Grieve S.M., Stocker R., Graham R.M., Chapman G. Gestational stress induces the unfolded protein response, resulting in heart defects. Development. 2016;143:2561–2572. doi: 10.1242/dev.136820. [DOI] [PubMed] [Google Scholar]
- 70.Kao T.Y., Chiu Y.C., Fang W.C., Cheng C.W., Kuo C.Y., Juan H.F., Wu S.H., Lee A.Y. Mitochondrial Lon regulates apoptosis through the association with Hsp60-mtHsp70 complex. Cell Death Dis. 2015;6:e1642. doi: 10.1038/cddis.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luciakova K., Sokolikova B., Chloupkova M., Nelson B.D. Enhanced mitochondrial biogenesis is associated with increased expression of the mitochondrial ATP-dependent Lon protease. FEBS Lett. 1999;444:186–188. doi: 10.1016/s0014-5793(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 72.Gibellini L., Pinti M., Boraldi F., Giorgio V., Bernardi P., Bartolomeo R., Nasi M., De Biasi S., Missiroli S., Carnevale G. Silencing of mitochondrial Lon protease deeply impairs mitochondrial proteome and function in colon cancer cells. FASEB J. 2014;28:5122–5135. doi: 10.1096/fj.14-255869. [DOI] [PubMed] [Google Scholar]
- 73.Bota D.A., Ngo J.K., Davies K.J. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic. Biol. Med. 2005;38:665–677. doi: 10.1016/j.freeradbiomed.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 74.Babiuk R.P., Zhang W., Clugston R., Allan D.W., Greer J.J. Embryological origins and development of the rat diaphragm. J. Comp. Neurol. 2003;455:477–487. doi: 10.1002/cne.10503. [DOI] [PubMed] [Google Scholar]
- 75.Bota D.A., Davies K.J. Mitochondrial Lon protease in human disease and aging: Including an etiologic classification of Lon-related diseases and disorders. Free Radic. Biol. Med. 2016;100:188–198. doi: 10.1016/j.freeradbiomed.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi M., Zhang H., Wu X., He Z., Wang L., Yin S., Tian B., Li G., Cheng H. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 2017;45:9640–9653. doi: 10.1093/nar/gkx597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan J., Wang K., Du X., Wang J., Chen S., Wang Y., Shi M., Zhang L., Wu X., Zheng D. ALYREF links 3′-end processing to nuclear export of non-polyadenylated mRNAs. EMBO J. 2019;38:e99910. doi: 10.15252/embj.201899910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y.S., Yang W.L., Zhao Y.L., Yang Y.G. Dynamic transcriptomic m5 C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA. 2021;12:e1639. doi: 10.1002/wrna.1639. [DOI] [PubMed] [Google Scholar]
- 80.Gallagher T.L., Arribere J.A., Geurts P.A., Exner C.R., McDonald K.L., Dill K.K., Marr H.L., Adkar S.S., Garnett A.T., Amacher S.L., Conboy J.G. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 2011;359:251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bill B.R., Lowe J.K., Dybuncio C.T., Fogel B.L. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int. Rev. Neurobiol. 2013;113:251–267. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee J.A., Damianov A., Lin C.H., Fontes M., Parikshak N.N., Anderson E.S., Geschwind D.H., Black D.L., Martin K.C. Cytoplasmic Rbfox1 Regulates the Expression of Synaptic and Autism-Related Genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernández E., Rajan N., Bagni C. The FMRP regulon: from targets to disease convergence. Front. Neurosci. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dougherty J.D., Maloney S.E., Wozniak D.F., Rieger M.A., Sonnenblick L., Coppola G., Mahieu N.G., Zhang J., Cai J., Patti G.J. The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors. J. Neurosci. 2013;33:2732–2753. doi: 10.1523/JNEUROSCI.4762-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huggins G.S., Bacani C.J., Boltax J., Aikawa R., Leiden J.M. Friend of GATA 2 physically interacts with chicken ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and represses COUP-TF2-dependent activation of the atrial natriuretic factor promoter. J. Biol. Chem. 2001;276:28029–28036. doi: 10.1074/jbc.M103577200. [DOI] [PubMed] [Google Scholar]
- 86.Svensson E.C., Tufts R.L., Polk C.E., Leiden J.M. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goumy C., Gouas L., Marceau G., Coste K., Veronese L., Gallot D., Sapin V., Vago P., Tchirkov A. Retinoid pathway and congenital diaphragmatic hernia: hypothesis from the analysis of chromosomal abnormalities. Fetal Diagn. Ther. 2010;28:129–139. doi: 10.1159/000313331. [DOI] [PubMed] [Google Scholar]
- 88.Carmona R., Cañete A., Cano E., Ariza L., Rojas A., Muñoz-Chápuli R. Conditional deletion of WT1 in the septum transversum mesenchyme causes congenital diaphragmatic hernia in mice. eLife. 2016;5:e16009. doi: 10.7554/eLife.16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sefton E.M., Gallardo M., Kardon G. Developmental origin and morphogenesis of the diaphragm, an essential mammalian muscle. Dev. Biol. 2018;440:64–73. doi: 10.1016/j.ydbio.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hamanaka K., Takata A., Uchiyama Y., Miyatake S., Miyake N., Mitsuhashi S., Iwama K., Fujita A., Imagawa E., Alkanaq A.N. MYRF haploinsufficiency causes 46,XY and 46,XX disorders of sex development: bioinformatics consideration. Hum. Mol. Genet. 2019;28:2319–2329. doi: 10.1093/hmg/ddz066. [DOI] [PubMed] [Google Scholar]
- 91.Wang T., Hoekzema K., Vecchio D., Wu H., Sulovari A., Coe B.P., Gillentine M.A., Wilfert A.B., Perez-Jurado L.A., Kvarnung M. Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders. Nat. Commun. 2020;11:4932. doi: 10.1038/s41467-020-18723-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for damaging variants identified are listed in the supplementary tables. The whole-genome sequencing and exome sequencing CDH data used in this study are available at the database of Genotypes and Phenotypes (dbGaP: phs001110.v2.p1). The SPARK data are available under managed access from Simons Foundation Autism Research Initiative (SRARI). The WHICAP dataset analyzed for the manuscript is available from the author B.N.V. on request.