Summary
Many common and rare variants associated with hematologic traits have been discovered through imputation on large-scale reference panels. However, the majority of genome-wide association studies (GWASs) have been conducted in Europeans, and determining causal variants has proved challenging. We performed a GWAS of total leukocyte, neutrophil, lymphocyte, monocyte, eosinophil, and basophil counts generated from 109,563,748 variants in the autosomes and the X chromosome in the Trans-Omics for Precision Medicine (TOPMed) program, which included data from 61,802 individuals of diverse ancestry. We discovered and replicated 7 leukocyte trait associations, including (1) the association between a chromosome X, pseudo-autosomal region (PAR), noncoding variant located between cytokine receptor genes (CSF2RA and CLRF2) and lower eosinophil count; and (2) associations between single variants found predominantly among African Americans at the S1PR3 (9q22.1) and HBB (11p15.4) loci and monocyte and lymphocyte counts, respectively. We further provide evidence indicating that the newly discovered eosinophil-lowering chromosome X PAR variant might be associated with reduced susceptibility to common allergic diseases such as atopic dermatitis and asthma. Additionally, we found a burden of very rare FLT3 (13q12.2) variants associated with monocyte counts. Together, these results emphasize the utility of whole-genome sequencing in diverse samples in identifying associations missed by European-ancestry-driven GWASs.
Keywords: blood-cell counts, whole-genome sequencing
Introduction
Counts of circulating white blood cells (WBCs) are important clinical parameters that are used for monitoring general disease activity and tolerance to therapies for oncological and rheumatologic diseases. WBCs are derived from hematopoietic stem cells and during differentiation are committed into two distinct lineages: myeloid (neutrophils, basophils, eosinophils, and monocytes) and lymphoid (lymphocytes). By studying the genetic determinants of WBC counts, we have been able to gain a more complete understanding of hematopoiesis and the complex roles of WBCs in both acute and chronic inflammation.1,2
Total and differential WBC counts are complex, polygenic, quantitative traits, and the genetic contribution to variance in WBC counts (heritability) is estimated at 50%–60%.3 Numerous recent studies have characterized both common (minor-allele frequency [MAF] greater than 5%) and infrequent (MAF between 0.5% and 5%) variation contributing to WBC counts in European, African, East Asian, and Hispanic populations.4, 5, 6, 7, 8 To date, most studies of the genetics of WBC counts have used a combination of study designs, including standard genome-wide genotyping arrays,3 exome sequencing,9 exome-chip genotyping,4,10 and application of genome-wide imputation using reference panels.5,6,8 An obvious gap in these study designs is a comprehensive, genome-wide interrogation of common and rare variation that could be missed by imputation-based approaches.
Whole-genome sequencing (WGS)-based analysis largely addresses these gaps, particularly in individuals of non-European origin. Importantly, WGS can assess population-specific variants,11 including variants that are often poorly imputed with standard reference panels and genotyping arrays.12 Here we utilized deep (∼30×) WGS data from 61,802 individuals, including African American (AA), East Asian (EAS), European American (EA), and Hispanic/Latino (HA) subjects. Data were generated as part of the National Heart, Lung and Blood Institute (NHLBI)-Trans-Omics for Precision Medicine (TOPMed) program investigating the genetics of WBC counts.
Material and methods
TOPMed samples
NHLBI’s TOPMed program comprises several parent studies. The parent studies that contributed to our analyses included Atherosclerosis Risk in Communities (ARIC),13 the Amish Complex Disease Research Program (Amish),14 BioMe Biobank (BioMe),15 Cardiovascular Artery Risk Development in Young Adults (CARDIA),16 the Cardiovascular Health Study (CHS),17 Genetic Epidemiology of COPD (COPDGene),18 the Framingham Heart Study (FHS),19 Genetic Study of Atherosclerosis Risk (GeneSTAR),20 Hispanic Community Health Study/Study of Latinos (HCHS/SOL),21 the Jackson Heart Study (JHS),22,23 Multi-Ethnic Study of Atherosclerosis (MESA),24 the San Antonio Family Heart Study (SAFS),25 and the Women’s Health Initiative (WHI).26 Additional information about the design of each study and the sampling of individuals within each cohort for WGS is available in the supplemental information. Participants included in these analyses (unique n = 61,865) are shown in Table S1, stratified by study, ancestry group (see supplemental methods), and WBC trait. For these analyses, 1% of participants are Asian, 23% are Black, 22% are Hispanic/Latino, and 54% are white. All studies were approved by the appropriate institutional review boards (IRBs), and informed consent was obtained from all participants.
TOPMed WGS and quality control
WGS was performed at an average depth of 38× by six sequencing centers (Broad Genomics, Northwest Genome Center, Illumina, New York Genome Center, Baylor, and McDonnell Genome Institute) with Illumina X10 technology and DNA from blood. Here we report analyses from the “Freeze 8” dataset, where reads were aligned to human-genome build GRCh38 through the use of a common pipeline across all sequencing centers. To perform variant quality control (QC) within the Freeze 8 dataset, we trained a support vector machine (SVM) classifier on known variant sites (positive labels) and Mendelian inconsistent variants (negative labels). Further variant filtering was done for variants with excess heterozygosity and Mendelian discordance. Sample QC measures included concordance between annotated and inferred genetic sex; concordance between prior array genotype data and TOPMed WGS data; and pedigree checks. Details regarding the genotype “freezes,” laboratory methods, data processing, and QC are described on the TOPMed website and in a common document accompanying each study’s dbGaP accession number.
WBC phenotype measurements and exclusion criteria
White blood cells, basophils, eosinophils, neutrophils, lymphocytes, and monocytes were counted in a subset of the TOPMed freeze 8 samples (Table S1) via automated clinical hematology analyzers. Each of the phenotypes is defined as the concentration of cell type in the blood and is measured in billions/liter. Trait-specific QC excluded participants with WBC count values > 100 × 109 cells/L (n = 5), neutrophil values > 75 × 109 cells/L (n = 1), monocyte values > 15 × 109 cells/L (n = 1), lymphocyte values > 150 × 109 cells/L (n = 1), eosinophil values > 20 × 109 cells/L (n = 1), and basophil values equal to 0.9 × 109 cells/L (n = 1). Additionally, in instances where multiple measurements were available, we kept only one measurement for each individual and each trait.
Single-variant association tests for quantitative traits
We performed genome-wide single-variant association tests by using a two-step linear mixed model (LMM). In the first step, we fit the ”null model” under the null hypothesis of no genetic association and did not include genetic variants in the model. We included sex, age, combined study by phase variable (e.g., WHI_2 refers to phase 2 of WHI study), and the first 11 PC-Air27 principal components (PCs) of genetic ancestry as fixed effects. To account for genetic relatedness, we included a 4th-degree sparse empirical kinship matrix (KM) computed with PC-Relate.28 In order to better control genomic inflation,29 we allowed for heteroscedasticity in the error variances by modeling separate residual variance components, one for each study, by ancestry group (e.g., WHI_White). Details on estimating the ancestry group are in the supplemental methods.
In order to improve power and appropriately control type I error in settings with non-normal phenotype distribution, we used a fully adjusted two-stage approach for fitting the null model.30 In stage 1, we fit an LMM with the observed phenotype values as the outcome, the fixed effects as covariates, a sparse KM, and heterogeneous residual variances. We applied a rank-based inverse-normal transformation to the residuals from the results of stage 1 and then rescaled them by the original variance. In stage 2, we fit another LMM by using the rescaled residuals obtained in stage 1 as the outcome and by using the same covariates, the same KM, and the same heterogeneous residual variance model as in stage 1. Finally, we used the output from stage 2 as the trait of interest to perform a score test of genetic association. In the association analyses, we included variants that had a minor-allele count (MAC) of at least 5, passed the TOPMed Informatics Research Center (IRC) quality filters, and had less than 10% of samples with a sequencing read depth of less than 10. A threshold level of 5 × 10−8 was used to determine statistical significance.
Single-variant association tests for basophil count as a binary trait
Instead of testing basophils as a continuous trait, we performed genome-wide single-variant association tests of basophils as a binary trait dichotomized at 0.05 × 109 cells/L (basophil3 ≥ 0.05 versus basophil < 0.05). We fit a generalized linear mixed model (GLMM) with binomial family and logit link via the penalized quasi-likelihood31 approach of GMMAT32 because our outcome was no longer quantitative. The same fixed-effect covariates and sparse KM as for quantitative-trait analysis were included. Because the variance model for a GLMM is specified by the binomial family and link function, we did not use heterogeneous residual variance groups or the two-stage rank-normalization procedure. We performed genome-wide association tests based on score statistics and saddlepoint approximation (SPA) of the p values.33,34 The SPA method has been shown to better control type I error even when the ratio of affected to control individuals is unbalanced, e.g., during the testing of low-frequency and rare variants, when the number of carriers is much lower than the sample size.
Conditional analyses
We performed conditional single-variant association tests where, in addition to adjusting for the fixed-effect covariates and sparse KM that were used in single-variant analyses, we adjusted for variants previously known to be associated with the outcomes (Table S3). First, we matched the known variants to TOPMed variants on the basis of position and alleles and selected the variants that passed the TOPMed IRC quality filters. We then used linkage disequilibrium (LD) (with a threshold of R2 > 0.8) to prune the set of matched variants for each trait separately and checked for collinearity of the pruned variants with the covariates. The final set of variants was included in the first round of conditional analyses. After the first round, we checked whether the remaining significant variants were near (within 1 Mb window) the known variants that failed the TOPMed IRC filters. These variants, in addition to the set of variants from the first round, were included in the second round of conditional analyses.
Gene-based aggregate rare-variant tests
To improve the power to detect rare-variant associations, we implemented several strategies of aggregating variants and testing for cumulative associations of gene-based groupings with the traits. We implemented a total of five strategies of variant groupings: three strategies included coding variants only, and two strategies included coding and noncoding variants from enhancer and promoter regions, but only those with “deleterious” consequences to the corresponding gene, “deleterious” being defined by various annotation-based filters; the details are provided in the supplemental methods. We performed aggregate tests by using the efficient variant-set mixed-model association test (SMMAT),35 which is more computationally efficient than SKAT-O (optimized SKAT [sequence kernel association test]) and more powerful than burden tests or SKAT alone. The SMMAT test used the same null model that was fit for the single-variant analyses, and the p value was constructed from a combination of the mixed-model burden p value with an asymptotically independent adjusted SKAT-like p value via Fisher’s method. In our analyses, we included non-monomorphic variants that had an MAF of less than 1% and that passed the same quality filters that were used for single-variant analyses. To upweight rarer variants, we used weights that are based on MAF and given by a beta distribution with parameters 1 and 25. We determined statistical significance by using a Bonferroni correction for the number of aggregate groups tested in each aggregation strategy.
Analyses of WBC-subtype proportions
In addition to analyzing counts of WBC subtypes, we also analyzed WBC-subtype proportions for all of the replicated, statistically significant WBC-subtype count signals. To do so, we identified samples whose WBC count and corresponding WBC-subtype count were collected at the same visit and divided the WBC-subtype count by the total measured WBC count. We excluded samples where the proportion of WBC-subtype count to WBC count was greater than 1. This proportion was treated as the phenotype and modeled similarly to the other phenotypes, i.e., with the two-step LMM described above.
Fine-mapping analyses
After conditional analyses, we carried out statistical fine mapping by using the following approach: because our conditional analyses implicated a single independent variant at each locus, we assumed a single causal variant at each locus. We then adapted the method proposed by Maller et al.36 to assign posterior inclusion probabilities (PIPs) to each variant and construct 95% credible sets. In brief, we considered all variants within 250 kb upstream and 250 kb downstream of the sentinel SNP and converted summary statistics into approximate Bayes factors (aBFs) as follows:
where and SE are the variant’s effect size and standard error, respectively, and represents the prior variance in allelic effects. As in Maller et al.,36 we set . We then calculated the PIP of each variant by dividing the variant’s aBF by the sum of the aBFs for all variants at the locus. We generated the 95% credible sets by ordering all variants (at a particular locus) from largest to smallest PIP and including variants until cumulative PIPs
Haplotype analyses
On the basis of the results of conditional single variant analyses, we performed haplotype analyses for the hemoglobin beta (HBB) region (rs334 and rs33930165) in total WBC counts and lymphocytes and the NRIP1 region (rs28574812 and rs2823002) in monocytes and total WBC counts. We constructed 2-SNP haplotypes from phased genotype data and identified haplotypes with non-zero frequencies. We counted the number of copies of each haplotype in each subject and included the number of copies of each non-reference haplotype as covariates in the model. The haplotype with the highest frequency was considered the reference haplotype. Using the null model from the single variant analyses, we performed association tests and report haplotype-specific results.
PheWAS analysis
We extracted phenome-wide association scanning (pheWAS) results for the seven new replicated signals from the UKBiobank (UKBB) and the BioVU biobank. The UKBB results were obtained from the UKBB ICD PheWeb hosted at the University of Michigan on the basis of 408,961 samples from white British participants. We considered the 1,261 phecodes with at least 100 affected individuals and an associated Bonferroni corrected threshold for significance of 0.05/1,261 = 3.96 × 10−5. BioVU is the Vanderbilt University Medical Center (VUMC) biobank that houses de-identified DNA samples linked to phenotypic data derived from the electronic health record (EHR) system of VUMC. The pheWAS lookups in BioVU were restricted to African Americans (n » 5,000). For the rs334 lookups, we had access to samples from ∼14,000 African Americans that were either heterozygous at rs334 or had two copies of the reference allele. Phenotypes were derived from billing codes of EHRs. Association between each binary phecode and a SNP was assessed using logistic regression, while adjusting for covariates of age, sex, genotyping array batch, and 10 principal components of ancestry. We considered the 726 phecodes with at least 100 cases with an associated Bonferroni corrected threshold for significance of 0.05/726 = 6.89×10−5.
UKBiobank analysis of asthma, COPD, and atopic dermatitis with rs28532112
We constructed phenotypes for chronic obstructive pulmonary disease (COPD), asthma, and atopic dermatitis (AD), as defined in Wu et al.,37 by using ICD10 codes to select a case group and a control group. The initial selected set of affected individuals and controls was purged of relatedness by the removal of one member of each related pair in an iterative fashion until no related subjects remained. Using the remaining group of affected individuals and remaining pool of controls, we selected a fixed number of control individuals for each affected individual, matched by sex, age, and ancestry. The fixed number used for the ratio of control individuals to affected individuals was adjusted to yield a total n in the range of 40,000 to 80,000 subjects. Association analyses were conducted with the OASIS pipeline.
TOPMed analysis of rs28532112 with asthma and asthma severity
TOPMed generated WGS on n = 869 subjects with asthma status (410 asthmatics and 459 non-asthmatic individuals) for the Barbados Asthma Genetics Study (BAGS)38,39 and n = 611 asthmatic subjects from the Severe Asthma Research Program (SARP)40 study. We used GENESIS to perform association tests for rs28532112 with asthma (in BAGS), and included age, batch, and sex as covariates. We also performed tests for association with asthma severity (in SARP), as measured by pre-forced expiratory volume1 (preFEV1), for rs28532112; we controlled for age, gender, and body mass index and stratified by ancestry (EA [n = 218] and AA [n = 393]).
ADRN analysis of rs28532112
To define genetic risk factors of AD, we performed WGS on 777 subjects from the Atopic Dermatitis Research Network (ADRN) of the National Institute of Allergy and Infectious Diseases as previously described.41 This includes 237 unaffected individuals, 491 individuals affected with AD without eczema herpeticum, and 49 individuals affected with AD with eczema herpeticum. To perform tests for association between rs28532112 and AD, we compared 491 AD-affected individuals to 237 unaffected individuals by using generalized logistic regression (GLM) and PLINK/Seq and adjusting for the first five PCs as covariates.
TOPMed analysis of rs28532112 with COPD and lung function
Details of the TOPMed analyses of COPD and lung function can be found in Zhao et al.42 In brief, the analysis involved 19,996 multi-ethnic individuals, including 12,314 EAs, 6,450 AAs, and 1,232 samples classified as “other,” from TOPMed. Phenotype harmonization of pulmonary-function-test measures, including pre-bronchodilator FEV1, forced vital capacity (FVC), and FEV1:FVC ratios, was conducted according to standard protocols. We incorporated covariate adjustment for age2, sex, height2, weight (FVC only), study, current smoking, former smoking, pack-years of smoking, first 10 PCs of ancestry, and sequencing center in an LMM framework to account for heterogenous variance across studies by using GENESIS. Case-control analyses incorporated covariate adjustment for age, sex, study, pack-years, whether an individual ever versus never smoked, first 10 PCs of ancestry, and sequencing center.
SOMAScan proteomic profiling and rs334 pQTL analysis
In JHS and MESA TOPMed participants, EDTA plasma samples collected at the respective baseline exams and stored in −70°C freezers were subjected to proteomic measurements with SOMAscan, a single-stranded DNA aptamer-based proteomics platform containing 1,305 aptamers. In JHS, samples (n = 2,054 AA) were run in three separate batches. Proteins were quantified in relative fluorescent units, the concentration of which is proportional to protein concentration in the plasma sample. Proteomic measurements were standardized to a set of control samples (pooled plasma) contained within each 96-well plate, and the resulting values were log transformed and scaled to a mean of 0 and standard deviation of 1. Association between rs334 genotype and protein values were assessed via linear mixed-effects models. In JHS, proteins were standardized within each batch and then inverse normalized across batches and adjusted for age, sex, and batch. MESA samples (n = 189 AA and 301 HA) were adjusted for age, sex, ethnicity (Hispanic yes or no), plate, and site. The cohort-specific results were meta-analyzed by inverse-variance weighting. A Bonferroni-adjusted significance threshold of 3.8 × 10−5 (0.05/1,301) was used.
rs28532112 plasma-protein association analysis
We further performed a targeted genotype plasma-protein quantification analysis to determine the association of rs28532112 with plasma concentrations of interleukin-3 (IL-3), soluble IL3R-alpha, granulocyte-macrophage colony-stimulating factor (GM-CSF), and thymic stromal lymphopoietin (TSLP) receptor and ligand by utilizing available SOMAscan data from 2,544 multi-ethnic JHS and MESA TOPMed samples. For soluble GM-CSF receptor alpha, we utilized a separate Olink Neurology proximity extension assay (PEA) panel measured in 1,328 multi-ethnic TOPMed WHI cohort samples, adjusted for age and ancestry.
rs334 and estimated lymphocyte subset analysis in JHS
Illumina MethylationEPIC array data (containing over 850,000 CpG methylation sites) from n = 1,756 JHS participants were generated from blood samples collected during the JHS baseline exam. Methylation levels were quantified in terms of the β value, for which the ratio of intensities between the methylated and un-methylated allele was used as the ratio of fluorescent signals. Methylation values were normalized with respect to background color intensity via the normal-exponential out-of-band (NOOB) method.43 Cell counts (granulocytes, monocytes, natural killer [NK], CD4+ T lymphocytes, naive CD8+ T lymphocytes, exhausted cytotoxic CD8+ T cells [defined as CD8 positive, CD28 negative, and CD45R negative], and plasmablasts) were estimated according to the method of Houseman et al.44 and Horvath et al.45 The association between estimated cell counts and rs334 carriers (excluding rs334 homozygotes), adjusted for age, sex, and 10 PCs of genetic ancestry, was assessed with generalized estimating equations in SAS 9.3 so that familial correlation would be accounted for.
Results
Single-nucleotide-variant-association results
In up to 61,802 multi-ethnic individuals (33,285 EA, 14,246 AA, 13,585 HA, and 686 EAS; Table S1), we performed genome-wide association tests for each single-nucleotide variant (SNV) or small insertion-deletion (indel) with a MAC > 5 for ∼109,563,748 association tests (Table S2) with 6 different phenotypes (total WBC, neutrophil, monocyte, lymphocyte , basophil, and eosinophil counts).
Across these traits, we observed 6,993 statistically significant associations (p < 5 × 10−8). Inspection of QQ plots and genomic inflation factors indicated well-calibrated p value distributions (Figures S3–S14). To determine whether any of these significant loci represent new associations for WBC-count traits, we performed genome-wide analyses conditioning on all known WBC-count-associated loci (see material and methods; Table S3). After conditional analysis, we observed 165 statistically significant associations, at a genome-wide threshold of 5 × 10−8 (Table S4), implicating 18 independent loci (that were at least 500 kb apart from each other) that have not been previously reported (Table S5). To replicate the findings from the conditional analysis, we performed lookups in independent samples from five different sources, representing self-identified European and European American, Hispanic American, and African American groups in up to 199,126 individuals (see material and methods). Of the 21 independent associations (at the 18 loci) that were submitted for replication, seven signals were robustly replicated (i.e., they met the following criteria: (1) consistent direction of effect between the discovery effect estimate and the meta-analyzed replication effect estimate; (2) p value < 0.05 in at least one replication cohort; and (3) p value < 0.05 across the meta-analyzed replication cohorts) in or near HBB, NRIP1, CSF2RA, S1PR3, NPHP3, and OPCML (Table 1). Several of the replicated lead variants showed large allele-frequency differences between populations (Table S5). In particular, the lead HBB and S1PR3 variants are found almost exclusively among individuals of African ancestry.
Table 1.
Replicated associations with white blood cell traits in TOPMed
| Trait | Chr | Position | Ref | Alt | rsID | Annotation | Betaa | SE | p value | Overall AFb | EA AFc | EA β | EA p | AA AFd | AA β | AA p | HA AFe | HA β | HA p | Replication β | Replication p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EOS | X | 1256294 | A | G | rs28532112 | intergenic CSF2RA | −0.009 | 0.001 | 1.08 × 10−11 | 0.2578 | 0.1579 | −0.0097 | 2.49 × 10−5 | 0.4276 | −0.0055 | 0.0079 | 0.2571 | −0.0139 | 0.0003 | −0.0901 | 4.82 × 10−6 |
| LYM | 11 | 5227002 | T | A | rs334 | missense HBB | −0.237 | 0.026 | 2.76 × 10−20 | 0.015 | 0.0003 | 0.0396 | 0.8985 | 0.0423 | −0.19 | 3.30 × 10−14 | 0.012 | −0.2797 | 1.41 × 10−6 | −0.1737 | 2.88 × 10−13 |
| MON | 21 | 15012619 | A | G | rs28574812 | intronic NRIP1 | −0.011 | 0.002 | 1.54 × 10−10 | 0.287 | 0.1592 | −0.0109 | 0.0006 | 0.5318 | −0.0102 | 0.0002 | 0.245 | −0.0101 | 0.0194 | −0.0231 | 2.19 × 10−7 |
| MON | 9 | 88921159 | C | A | rs28450540 | intergenic S1PR3 | −0.035 | 0.004 | 3.65 × 10−17 | 0.04 | 0.0009 | −0.0399 | 0.3284 | 0.117 | −0.033 | 3.44 × 10−12 | 0.0245 | −0.0532 | 5.19 × 10−5 | −0.2161 | 2.64 × 10−25 |
| WBC | 11 | 132819661 | GAC | G | rs79353195 | intronic OPCML | −0.135 | 0.025 | 4.00 × 10−8 | 0.0629 | 0.0465 | −0.1285 | 0.0006 | 0.0658 | −0.1072 | 0.0283 | 0.0629 | −0.1353 | 4.00 × 10−8 | −0.0207 | 0.0159 |
| WBC | 21 | 15001515 | A | T | rs2823002 | intronic NRIP1 | −0.079 | 0.014 | 3.75 × 10−8 | 0.3198 | 0.1625 | −0.0823 | 0.0001 | 0.6943 | −0.0828 | 0.0021 | 0.3198 | −0.0794 | 3.75 × 10−8 | −0.0186 | 0.0002 |
| WBC | 3 | 132710603 | C | T | rs62292471 | intronic NPHP3 | 0.1139 | 0.02 | 1.07 × 10−8 | 0.0965 | 0.1218 | 0.1104 | 5.64 × 10−6 | 0.025 | −0.0009 | 0.9906 | 0.0965 | 0.1139 | 1.07 × 10−8 | 0.0223 | 7.40 × 10−5 |
Effect sizes are represented in transformed trait values.
AF = allele frequency
EA = European American
AA = African American
HA = Hispanic American
HBB
At HBB on chromosome 11 (Figure 1A), we observed an association between lower lymphocyte counts and the missense mutation (rs334 [p.Glu6Val]) causing African sickle-cell disease (β = −0.237, p = 2.76 × 10−20). There was a nominal association of the rs334 sickle variant with increased neutrophil counts (β = 0.225, p = 4.83 × 10−6) and no association with total WBC counts (Table S6). The association with lymphocyte percentage (i.e., lymphocyte counts/total WBC counts) was also strong (β = −0.030, p = 2.12 × 10−25), as was the association with neutrophil percentage (β = 0.032, p = 1.59 × 10−21) (Table S7). The 95% fine-mapped credible set for lymphocyte count contained only the index variant, rs334, and PIP = 0.999 (Table S8). Exclusion of the very small number of rs334 homozygote individuals (n = 9) did not alter the association with lymphocyte count or with neutrophil percentage, suggesting that these associations are not driven by altered immune responsiveness or inflammation among individuals with sickle-cell disease. Through investigating a subset of the ∼3,400 African American individuals from JHS, we further demonstrated that additional covariate adjustment for other known sickle-cell-related or inflammation-related traits (red-cell indices, kidney function, D-dimer) did not substantively alter the rs334-lymphocyte count association (Table S9), suggesting that the lymphocyte association is not mediated through these other phenotypes. In a smaller subset of 1,458 JHS TOPMed individuals with lymphocyte and immune-cell subtype proportions estimated from genome-wide methylation data, heterozygosity of the rs334 sickle cell mutation was specifically associated with lower estimated levels of CD8+ T lymphocytes and NK cells (Table S10).
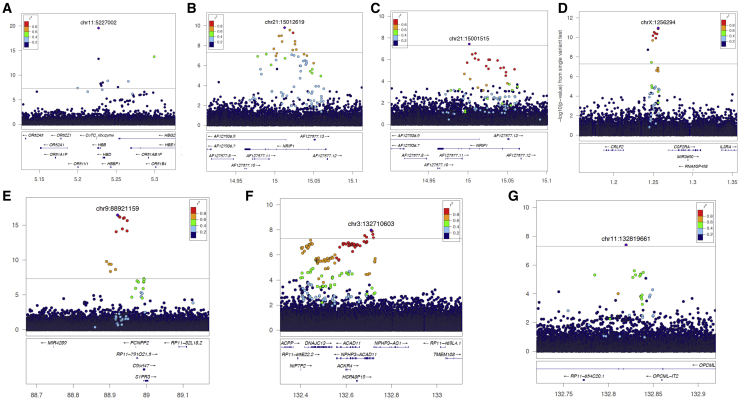
Figure 1.
Regional plots showing patterns of LD and evidence for association at the seven newly reported leukocyte-trait loci. Genomic coordinates are displayed on the horizontal axis, and −log10 p values are displayed on the vertical axis.
(A) rs334 (chr11: 5227002) with lymphocytes; (B) rs28574812 (chr21: 15012619) with monocytes; (C) rs2823002 (chr21: 15001515) with WBCs; (D) rs28532112 (chrX: 1256294) with eosinophils; (E) rs28450540 (chr9: 88921159) with monocytes; (F) rs62292471 (chr3: 132710603) with WBCs; and (G) rs79353195 (chr11: 132819661) with WBCs.
Interestingly, at the nucleotide position adjacent to rs334, we observed an association of the African HBB variant rs33930165 (encoding hemoglobin C, or p.Glu6Lys) with both higher total WBC count (β = 0.672,p = 8.81 × 10−12) and higher lymphocyte count (β = 0.351, p = 4.71 × 10−14). The associations of the HBB rs33930165 missense variant with higher total WBC and lymphocyte counts are consistent with a prior analysis conducted in n = 21,513 African Americans imputed to the TOPMed freeze 5b reference panel.12 The differing WBC-count phenotypic patterns of association between rs334 and rs33930165 in the current TOPMed analysis are summarized in Table S6. We also conducted a two-SNP haplotype analysis of rs334 and rs33930165 with both WBCs and lymphocytes (Table S12). The haplotype analysis was consistent with the single-variant association results in Table S4 (i.e., with the finding that the T allele at rs33930165 increases WBC count and that the A allele at rs334 decreases lymphocyte counts). Of note, both rs334 and rs33930165 are in very low LD (rsq < 0.001) and are present on distinct haplotypes in TOPMed (Table S13), consistent with prior evolutionary and population genetic data. Both variants are maintained at relatively high frequencies among populations such as those in sub-Saharan Africa, where malaria is endemic, but the geographic and population distributions and evolutionary histories of the rs334 hemoglobin S and rs33930165 hemoglobin C variant alleles are distinct.46,47
To further address the mechanism of the associations at HBB, we first assessed the relationship between rs334 and other potential immune-response-related genes in the genomic region. Although there are several type 1 interferon-inducible viral-related genes located about 400 kb centromeric to HBB on chromosome 11, we found no evidence of physical interaction between HBB and these neighboring genes when we used available promoter capture datasets in relevant blood cell types (with the HUGIN tool) nor any evidence of influence of rs334 on gene expression (eQTL) in whole blood when we used GTEx v8. It should be noted that the latter eQTL analysis might be limited by the small number of African American samples. Integration of additional functional-genomic information via the FATHMM, FANTOM5, and Roadmap annotation databases did not reveal evidence of a cis-regulatory role for rs334 (Table S11). Next, we performed proteome-wide analysis of rs334 genotype with ∼1,300 plasma proteins measured in African Americans and Hispanics from the MESA cohort and 2,045 African Americans from the JHS cohort (Table S12). Circulating levels of several red blood cell (RBC) (plasma hemoglobin, erythropoietin, and ephrin B248) and manganese proteins related to kidney function (cystatin C and β2-microglobulin) were significantly higher or lower (testican-2)49 among rs334 variant allele carriers (Bonferroni-corrected p value < 3.78 × 10−5). Moreover, several other proteins related to inflammatory response and lymphocyte activation or signal transduction (fractalkine/CX3CL1, sTNFsRII, and CD59) or response to viral infection (CD59 and testican-2)50 were significantly higher among rs334 variant carriers.
NRIP1
Two intronic variants within nuclear receptor interacting protein (NRIP1) (Figure 1B, Figure 1C) were associated with monocyte count (rs28574812, β = −0.011, p = 1.54 × 10−10) and total WBC count (rs2823002, β = −0.079, p = 3.75 × 10−8). A weaker association was observed between rs28574812 and monocyte percentage (β = −7.75 × 10−4, p = 2.36 × 10−4) (Table S7). The rs28574812 and rs2823002 variants are in strong LD in Europeans but are highly differentiated between African and non-African populations. In the 1000 Genomes Yoruba (YRI) West African population, rs28574812 has no other SNPs in strong LD (rsq > 0.8). Analysis of two-SNP haplotypes comprising rs28574812 and rs2823002 confirm the minor alleles of the two SNVs more commonly occur together in individuals of European ancestry. In addition, haplotype association analyses show that the two haplotypes containing the rs28574812 variant G allele are each associated with lower monocyte count, whereas the two haplotypes containing the rs2823002 variant T allele are each associated with lower WBC count (Table S15). This suggests allelic heterogeneity at this locus and that different alleles might be responsible for the monocyte and total WBC-count phenotypic associations. The haplotype association results are also consistent with our fine-mapping results, which pointed to different causal variants for each signal: the 95% credible set for the locus represented by rs28574812 contained 6 variants (Table S8), and the PIP for rs28574812 was 0.422; the 95% credible set for the locus represented by rs2823002 contained 55 variants (Table S8), and the PIP for rs2823002 was 0.611.
The mechanism of association of the NRIP1 intronic variant with hematologic traits is not immediately apparent. The protein (RIP140) encoded by NRIP1 interacts with hormone-dependent domains of nuclear receptors and is known to modulate transcriptional activity of the estrogen receptor (ESR1). It is a key regulator of gene expression via interaction with nuclear receptors, transcription factors, and other coregulators, each of which acts as either a coactivator or corepressor. GTEx version 8 did not report either the rs28574812 or rs2823002 variants as a statistically significant eQTL. Data from the Roadmap Epigenomics Consortium indicate elevated histone chromatin immunoprecipitation sequencing (ChIP-seq) experiments acetylated for H3K4me3 and H3K9ac across several types of cell line (Table S11). It has been reported that RIP-140 plays a role in the macrophage switching between classical M1 and alternative M2 subtypes51 and in the epigenetic responsiveness of monocytes to vitamin D,52 both of which are important for innate immunity and inflammatory diseases. Investigation of the NRIP1 region via Phenoscanner and the genome-wide association study (GWAS) catalog reveals that a distinct NRIP1 missense variant rs2229742 (common only in EUR) has been associated with hemoglobin, RBC count, systolic blood pressure, vitamin D levels, birthweight, and myopia. Additionally, NRIP1 gene expression signatures can predict survival in chronic lymphocytic leukemia.53
CLRF2-CSF2RA-IL3RA
Analyses of chromosome X found an association between rs28532112 and lower eosinophil count (β = −0.009, p = 1.08 × 10−11) within the intergenic region between cytokine-receptor-like factor 2 (CLRF2) and the region ∼20 kb upstream of colony-stimulating factor 2 receptor subunit alpha (CSF2RA), located in the pseudo-autosomal region (PAR1) of the X and Y chromosomes (Figure 1D). The association between rs28532112 and eosinophil percentage was similarly strong (β = −0.0013, p = 5.56 × 10−12) (Table S7). The 95% credible set for this locus contained 7 variants (Table S8), and the PIP for rs28532112 was 0.324. This region contains the genes encoding three related cytokine receptors, CRLF2 (the receptor for TSLP), CSF2RA (the receptor for colony-stimulating factor 2 (CSF-2) or GM-CSF), and IL3RA (the receptor for IL-3), all of which are involved in the regulation of hematopoiesis and type 2 inflammatory responses, including eosinophil production and function.54,55 Together with the interleukin 7 receptor-alpha, CRLF2 and TSLP activate STAT3 and STAT5, which polarize dendritic cells to induce type 2 inflammatory cytokines (IL-4, IL-5, and IL-13) and directly expand and/or activate Th2 cells, group 2 innate lymphoid cells, eosinophils, and basophils.56 CSF2RA encodes the alpha subunit of the heterodimeric receptor for GM-CSF, a cytokine that controls the production, differentiation, and function of granulocytes and macrophages. IL3RA encodes the IL-3-specific alpha subunit of the heterodimeric IL-3 receptor.
In GTEx version 8, rs28532112 was an eQTL associated with the nearby IL3RA (p = 4.8 × 10−9) in thyroid tissue and with CSF2RA in whole blood (p = 1.8 × 10−6), suggesting either IL3RA or CSF2RA as the potential causal gene(s) at this locus. Additional functional genomic annotation with FATHMM, FANTOM5, and Roadmap databases did not reveal additional regulatory features for rs28532112 (Table S11). We further performed a targeted genotype-plasma protein quantification analysis of IL-3, GM-CSF, and TSLP receptors and ligands by using available SOMAscan data from 2,544 African Americans and Hispanic/Latinos from the JHS and MESA TOPMed samples. Of the 5 proteins measured within the SOMAscan panel, there was no significant association between rs28532112 and circulating concentrations of IL-3, soluble IL3R-alpha, TSLP, TSLP receptor, or GM-CSF (all Bonferroni-corrected p > 0.05). GM-CSF receptor alpha is not one of the proteins included in the SOMAscan platform, but it was measured through a separate Olink PEA panel in 1,328 multi-ethnic TOPMed WHI cohort samples. In the WHI TOPMed samples, there was no evidence of association between rs28532112 genotype and plasma GM-CSF receptor alpha levels (β = −0.024, p = 0.47).
S1PR3
On chromosome 9 (Figure 1E), we identified an African-specific (MAF = 0.129) variant, rs28450540, associated with lower monocyte count (β = −0.035, p = 5.18×10−17). The association between rs28450540 and monocyte percentage was similarly strong (β = −0.004, p = 5.81 × 10−12) (Table S7). The 95% credible set for this locus contained 6 variants (Table S8), and the PIP for rs285450540 was 0.330. The rs28450540 variant is located 100 kb upstream of S1PR3, where a nearby SNP (rs567880204) was recently associated with monocyte count8 (LD between these two SNPs was low in AAs; rsq = 0.03). The broader region near S1PR3 was reported on extensively5 and was associated with a number of different blood-cell traits in individuals of European ancestry; such traits included monocyte counts, total WBC counts, lymphocyte counts, and platelet counts. The previously reported signals near S1PR3 include an S1PR3 missense variant associated with higher monocyte count. However, on the basis of our conditional analyses, the African-specific rs28450540 association with monocyte counts appears to be independent of these known signals, adding to the already complex allelic heterogeneity at this locus. Although GTEx contains very few African American samples, rs28450540 appears to be an eQTL for S1PR3 (p = 2.9 × 10−5) in whole blood. Functional annotation suggests that this variant is located in a putative enhancer (Table S11) in a number of different primary human cells and cell lines, including those of hematopoietic origin.
S1PR3 encodes one of five type G-protein-coupled receptors that mediate the biologic effects of sphingosine-1-phosphate (S1P), a chemoattractant for various blood and immune cells.57 An S1P gradient maintained between blood and other hematopoietic tissues (e.g., bone marrow and thymus lymph nodes) is an important mechanism for blood and immune-cell trafficking. S1pr3-deficient mice have defects in leukocyte recruitment and P-selectin-dependent leukocyte rolling, suggesting that S1PR3 mediates the chemotactic effect of S1P in bone-marrow-derived monocyte and macrophage recruitment during inflammation,58, 59, 60, 61 which might affect circulating monocyte count. Additionally, S1PR3 is highly expressed on hematopoietic stem and progenitor cells (HSPCs) and might affect egress of HSPCs from the bone marrow.62
NPHP3
An intronic variant (rs62292471) on chromosome 3 (Figure 1F) in NPHP3 was associated with increased total WBC count (β = 0.114, p = 1.07 × 10−8). The 95% credible set for this locus contained 33 variants (Table S8), and the PIP for rs62292471 was 0.150. GTEx version 8 reports rs62292471 as an eQTL for NPHP3 in cultured fibroblasts and in heart, adipose, lung, esophagus, and muscle tissue and as an eQTL for DNAJC13 in adipose, esophagus, and muscle tissue. The variant is located within an ENCODE distal enhancer region (Table S11). Another NPHP3 intronic variant, rs572076167 (not in 1000 Genomes), was associated with red-cell mean corpuscular hemoglobin (MCH) concentration,8 whereas rs17348614 was associated with lower MCH and mean corpuscular volume (MCV).5 Loss-of-function mutations in NPHP3 cause the congenital cystic kidney disorder nephronophthisis (NPH). Of the other genes located within a 1 Mb window surrounding NPHP3, the only one with a biologic connection to blood cells is ACKR4, which encodes a chemokine receptor that binds to dendritic-cell- and T-cell-activated chemokines, including CCL19, CCL21, and CCL25. ACKR4 belongs to the family of atypical chemokine receptors that includes DARC, which contains a well-characterized loss-of-function promoter variant (rs2814778) that is a major genetic determinant of WBC and neutrophil count in populations with African ancestry. By scavenging chemokines, ACKR4 regulates dendritic-cell trafficking to lymph nodes during inflammation.63
OPCML
We observed a 3-base indel (rs79353195) in the intron of OPCML (Figure 1G) associated with decreased total WBC count (β = −0.135, p = 4.00×10−9) with modest evidence for replication (p = 0.0159). The 95% credible set for this locus contained 48 variants (Table S8), and the PIP for rs79353195 was 0.868. This variant appears to be highly stratified across populations with MAF = 0.53 in EAS, MAF = 0.26 in SAS, MAF = 0.06 in AA, and MAF = 0.05 in EA populations. OPCML is a large gene that encodes an opioid-binding cell-adhesion molecule-like preprotein, a member of the IgLON immunoglobulin protein family highly expressed in the brain. Defects in OPCML are a cause of susceptibility to ovarian cancer (MIM: 167000). There is no apparent connection of this locus with blood cells or inflammation.
Results of association tests for aggregate rare variants
To improve the power to detect rare-variant associations, we implemented several strategies of aggregating variants and testing for cumulative associations of gene-based groupings with the traits (see materials and methods). In so doing, we detected statistically significant (p < 2.76 × 10−6) associations at four different genes (Table 2): MARCKSL1 (p = 2.98 × 10−7), TET2 (p = 6.21 × 10−9), FLT3 (p = 8.96 × 10−7), and CNKSR2 (p = 2.46 × 10−6). Replication analyses in independent samples from the UKBiobank (p = 2.42 × 10−18) and INTERVAL (p = 0.0003) studies confirmed the associations at FLT3. Analyses of the corresponding WBC-subtype proportions were consistent with these results (Table S16).
Table 2.
Gene-based rare variant associations with white blood cell counts in TOPMed
| Trait | Gene | Number of variants | Discovery p value | INTERVAL p value | UKBiobank p value |
|---|---|---|---|---|---|
| NEU | MARCKSL1 | 11 | 2.98 × 10−7 | 0.5576 | 0.1008 |
| LYM | CNKSR2 | 13 | 2.46 × 10−6 | 0.8067 | 0.8079 |
| MON | TET2 | 72 | 6.21 × 10−9 | 0.6984 | 0.6091 |
| MON | FLT3 | 65 | 8.96 × 10−7 | 0.0003 | 2.42 × 10−18 |
TET2 encodes a demethylation enzyme and epigenetic regulator64 that plays an important role in HSPC renewal, lineage commitment, and monocyte differentiation.65 The TET2 association with monocyte counts was spread across 72 rare coding variants (Figure S1). Because TET2 is a known driver gene for clonal hematopoiesis of indeterminate potential (CHIP), myeloid, and lymphoid malignancies, we compared our rare TET2 variants with those that are reported as somatic mutations66 or that appear in the COSMIC database. Indeed, we found that the majority of variants (53 of 72 variants) included in our aggregate-rare-variant test for TET2 were also reported as somatic in TOPMed, and these mutations largely drove the association with higher monocyte counts (Figure S1). These findings are further supported by the recent observation that somatic mutations of TET2 are commonly found among individuals referred to a hematology service for evaluation of monocytosis.67 By contrast, reports of germline TET2 loss-of-function variants associated with hematologic disease are quite rare.68, 69, 70
FLT3 encodes a receptor tyrosine kinase that regulates early hematopoiesis as well as the development of monocytes and dendritic cells. Somatic variants of FLT3 are found commonly in individuals with acute myeloid leukemia (AML) and generally consist of gain-of-function mutations involving either internal tandem duplications of the FLT3 juxtamembrane domain or point mutations located within the tyrosine kinase domain, both of which lead to constitutive activation of the FLT3 receptor.71 Because some TET2 somatic mutations had clearly passed quality control for germline variants, we also checked whether the rare variants contributing to the FLT3 signal were somatic. The COSMIC database identified some evidence for overlap between the TOPMed rare variants in FLT3, yet the presumably somatic mutations (12 of 65 variants) that contributed to the FLT3 association did not appear to drive the result (Figure S2). This observation is consistent with the recent discovery that common and low-frequency germline genetic variants of FLT3 are associated with monocyte count as well as risk of autoimmune disease.7,72 Interestingly, several of the rare FLT3 missense variants driving the association with monocyte count in TOPMed are located within the juxtamembrane or tyrosine kinase domains, which are also the most common location of somatic FLT3 mutations found in human cancers.
Results of phenome-wide association tests
Loci associated with WBC-count traits are often pleiotropically involved in autoimmune, allergic, infectious, and other blood-related diseases.4,5 Therefore, we additionally assessed whether any of the other newly identified WBC-count trait-associated variants are associated with clinical disease outcomes by using a combination of existing GWAS andPheWAS databases as well as evaluation of the X chromosome PAR locus and FLT3 aggregated rare variants in WGS-based datasets.
Most of our newly discovered autosomal loci associated with WBC-count traits, S1PR3, NRIP1, NPHP3, HBB, and OPCML, disproportionately impact individuals of African ancestry. Therefore, in addition to the large but Euro-centric UKBiobank (UKBB), we utilized PheWAS genotype and phenotype data from the more diverse BioVU EHR-based biobank at Vanderbilt University Medical Center. The latter includes African Americans genotyped on the Illumina MEGA array imputed to Haplotype Reference Consortium (HRC) reference genomes. For evaluation of HBB rs334 in BioVU, we excluded homozygous individuals because of the known relationship of sickle-cell disease (SCD) to clinical outcomes. For the S1PR3, NRIP1, and OPCML WBC-count trait-associated index variants, we found no evidence of significant association with disease outcomes in either UKBB or BioVU, whereas the NPHP3 index variant showed borderline association with myocardial infarction (Table S17). In the BioVU African American PheWAS, heterozygosity for rs334 was associated with several hemolytic anemia-related diagnosis codes, but not with any immune-related or infectious diseases (Table S17).
Eosinophils are classically associated with type 2 inflammation and are one of the hallmarks of allergic diseases such as asthma and AD. A subset of subjects with COPD might also be enriched for circulating eosinophils. Because the UKBiobank PheWAS lookup tool was restricted to the autosomes, we tested specifically for the X chromosome locus upstream of CSF2RA and its association with these three diseases in the UKBB imputed GWAS dataset (Table S18). We found a significant association with asthma in the UKBB (odds ratio [OR] = 0.94, p = 3.52 × 10−6); notably the association is as expected, i.e., the allele that decreases eosinophil count in our discovery is also associated with decreased risk for asthma. No associations were observed with COPD or AD in the UKBB.
Given the availability of several TOPMed lung-disease cohorts with data on asthma, COPD, and pulmonary function, we expanded our lookup of this variant in the following studies from TOPMed: BAGS for asthma, SARP for asthma severity, and ARIC, CHS, FHS, JHS, MESA, COPDGene, and EOCOPD for lung function and COPD (Table S18). In ∼20,000 multi-ethnic individuals,42 rs28532112 showed no association with spirometric measures of pulmonary function (FEV1, FVC, or FEV1:FVC ratio). There was a nominal association with COPD (p = 0.025) and severe COPD (p = 0.036), but paradoxically the minor allele associated with lower eosinophil count was associated with greater risk of COPD (OR = 1.09). In the BAGS asthma study there was no association of rs28532112 with asthma (p = 0.91), and no associations were noted for asthma severity in the SARP TOPMed study (p = 0.944 in 393 EAs, p = 0.144 in 218 AAs); however, we should note that these two asthma cohorts were underpowered and most likely unable to recapitulate the associations noted for asthma in the UKBB. Given the small sample size with a phecode definition of AD in the UKBB, we also took advantage of WGS data available from the ADRN, where in-depth phenotyping could help overcome the heterogeneity possible in the UKBB. In the ADRN samples, we found rs28532112 was associated with AD (OR 0.63; p = 0.008), and once again the association was as expected, i.e., the allele that decreases eosinophil count in our discovery was also associated with decreased risk for AD.
Because a low-frequency monocyte-count-associated variant of FLT3 (rs76428106) was recently associated with autoimmune thyroid disease (AITD),72 we used whole-exome sequencing to examine whether aggregated rare variants of FLT3 were associated with AITD among 200,000 UKBB individuals. Among 6,686 AITD-affected individuals and 179,346 individuals without AITD, there was no evidence of association by either burden (p = 0.81) or sequence kernel association test (p = 0.38). However, because of the relatively small number of cases in the UKBB, the lack of association might be due to low statistical power.
Discussion
By expanding coverage of the genome through deep WGS performed in a large sample of diverse individuals, we have identified several loci that are associated with WBC-count traits but that are distinct from variants previously identified through large GWASs. In each instance, the identified single variants (HBB, S1PR3, NPHP3, NRIP1, OPCML, and CSF2RA) are highly differentiated in allele frequency across ancestral populations; HBB and S1PR3 are essentially monomorphic in Europeans. The eosinophil-lowering CSF2RA variant located on the X chromosome may be associated with reduced susceptibility to asthma and AD. We also identified a burden of rare coding variants in FLT3 associated with monocyte count. These results demonstrate the utility of WGS in diverse cohorts of apparently healthy individuals for our further understanding of the genetic architecture of WBC traits and their relationship to immune-related disorders. Multi-omic data available from diverse TOPMed samples and other sources contributed to defining the likely causal genes or molecular mechanisms underlying these associations.
Despite its importance for hematologic traits and Mendelian disorders (e.g., Diamond-Blackfan anemia, hemophilia, and G6PD deficiency), the X chromosome has been under-studied in complex-trait genetics through GWASs, largely because of analytical issues and challenges related to imputation and sex-related differences in gene dosage. In particular, PAR1 on the X and Y chromosomes recombines in a sex-biased manner and thus has traditionally been ignored in linkage and association studies.73 The application of WGS allowed us to circumvent the need for genotype imputation and directly identify a common-variant association that prior GWASs had missed within PAR1. We have identified and replicated a common variant associated with lower eosinophil count within the X chromosome PAR1 region between CSF2RA (GM-CSF) and CRLF2 (cytokine receptor-like factor 2). The sentinel variant rs28532112 is about three times as common in African as in European populations, which might also have contributed to our ability to discover its association with eosinophil count. The recently completed gapless, end-to-end assembly of the human X chromosome reference sequence, including PARs,74 should additionally facilitate identification of X-linked or pseudo-autosomal variants newly associated with complex traits.
Several of the autosomal WBC-count trait-associated loci that we report (S1PR3, NPHP3, HBB, and NRIP1) contain other nearby variants that have been previously associated with hematologic traits, suggesting broader hematopoietic lineage regulation of these genomic regions. For example, previous studies in predominantly European ancestral populations have identified several genetic variants that occur within a ∼500 kb region upstream of S1PR3 and that are associated with WBC, RBC, and platelet traits.5,75 Although the rs334 variant encoding hemoglobin S is well known to affect RBC physiology, the mechanism of association for distinct variants within the NPHP3 and NRIP1 regions with RBC traits remains unclear.
Our results also extend the clinical importance of variants underlying WBC count and immune-related quantitative traits, particularly to non-European populations. The association of rs334 with a lower lymphocyte count, a higher proportion of neutrophils, and higher plasma levels of several immune- and kidney-related proteins adds to growing evidence that the carrier state of sickle-cell disease (p.Glu7Val [c.20A>T])-encoding hemoglobin S is associated with various medical phenotypes (RBC traits, higher D-dimer levels, and lower eGFR and hemoglobin A1c levels) and disease susceptibility (increased risk of chronic kidney disease and venous thromboembolic disease) in African Americans.76 There is evidence that, in addition to having a role in leukocyte recruitment and trafficking, S1PR3 is required for myeloid cell oxidative killing of microbial pathogens,77 is expressed on alveolar epithelial cells, and regulates epithelial integrity in lung disease.78 In this regard, the S1PR3 locus shows evidence of being under recent positive selection in African populations and might contribute to pulmonary edema and pathogenesis of severe malaria.79 Although our PheWAS did not show evidence that the African-specific variant S1PR3 rs28450540 was associated with any additional chronic-disease outcomes, the importance of rs28450540 or other variants at the S1PR3 locus for complications from other chronic lung and infectious disease or sickle-cell disease might require larger sample sizes.
The putative association of the X chromosome eosinophil-lowering variant with reduced risk of asthma and AD is consistent with prior studies demonstrating that genetically determined eosinophil count is associated with risk of allergic diseases in UKBB.5,8 These findings have potential implications for risk stratification or drug development for AD, asthma, or other allergic and lung diseases that disproportionately affect African Americans.80 Eosinophils and related type 2 inflammatory responses are particularly important at the barrier surfaces of skin and the respiratory and gastrointestinal tracts. Anti-TSLP antibodies such as Tezepelumab (AMG-157/MEDI9929) reduce levels of biomarkers of type 2 inflammation; such biomarkers include, for example, blood and sputum eosinophil counts.81 Moreover, mutations in CSF2RA are a cause of pulmonary surfactant metabolism dysfunction type 4 (SMDP4) (pulmonary alveolar proteinosis) (MIM: 300770), a rare lung disorder. Additional studies might be required for adequate characterization of the role of newly identified genetic variants at this important X-linked cytokine-receptor-gene-family locus for allergic, autoimmune, and pulmonary diseases, especially in the context of disease severity.
Additionally, our findings highlight one of the caveats inherent in WGS studies. The identification of a burden of rare variants in two genes (TET2 and FLT3) linked to leukemogenesis and clonal myeloid expansion and associated with monocyte count among a sample of largely unscreened individuals raises the question of distinguishing between germline, somatic, or clonal hematopoietic variants in blood-based next-generation sequencing studies involving complex traits or diseases. Because clonal hematopoiesis is an age-related condition, case-control WGS germline variant studies of aging-related conditions could be particularly prone to confounding by somatic variants.82 To avoid such potential confounding in blood-based genome-sequencing association studies, researchers might need to provide additional evidence of the germline or somatic origin by using clinical information, variant characteristics, and/or serial next-generation sequencing assays.83
In summary, using a WGS approach in diverse samples, we have identified and replicated 7 single-variant leukocyte-trait associations previously missed by GWASs; one of these is the association between a chromosome X PAR and both lower eosinophil count and reduced risk of allergic diseases. We extend the phenotypic profile of sickle-cell trait, which has previously been associated with RBC, kidney, and thrombosis-related biomarkers to include lymphocyte count and inflammation-related biomarkers. The identification of monocyte-specific associations, including an African-ancestry variant at the S1PR3 locus and a burden of very rare variants in FLT3, might warrant additional study in the context of infectious or autoimmune diseases, respectively.
Acknowledgments
Molecular data for the TOPMed program was supported by the National Heart, Lung and Blood Institute (NHLBI). Study-specific omics support information can be found in the supplement. Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination, were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). The project described was also supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant KL2TR002490 (L.M.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. L.M.R. was additionally funded by the NIH grants R01HL129132 and T32 HL129982. P.L.A. was funded by the NIH grant R01HL130733. P.S.d.V. was supported by American Heart Association grant number 18CDA34110116. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Declaration of interests
The authors declare no competing interests.
Published: September 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.08.007.
Contributor Information
Alexander P. Reiner, Email: apreiner@uw.edu.
Paul L. Auer, Email: pauer@mcw.edu.
Data and code availability
Data for each participating study can be accessed through dbGaP with the corresponding accession number (Amish, phs000956; ARIC, phs001211; BioMe, phs001644; BAGS, phs001143; CARDIA, phs001612; CHS, phs001368; COPDGene, phs000951; FHS, phs000974; GeneSTAR, phs001218; HCHS/SOL, phs001395; JHS, phs000964; MESA, phs001416; SAFS, phs001215; SARP, phs001446; and WHI, phs001237). Analysis results for the conditional single-variant analyses and the aggregate conditional analyses can be accessed through dbGaP: phs001974.
Web resources
OASIS pipeline, https://omicsoasis.github.io/
TOPMed website, www.nhlbiwgs.org
TOPMed whole-genome sequencing methods for freeze 8, https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-methods-freeze-8
University of Michigan pheweb server, https://pheweb.org/UKB-SAIGE/
Supplemental information
References
- 1.Morrell C.N., Aggrey A.A., Chapman L.M., Modjeski K.L. Emerging roles for platelets as immune and inflammatory cells. Blood. 2014;123:2759–2767. doi: 10.1182/blood-2013-11-462432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinod K., Wagner D.D. Thrombosis: tangled up in NETs. Blood. 2014;123:2768–2776. doi: 10.1182/blood-2013-10-463646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tajuddin S.M., Schick U.M., Eicher J.D., Chami N., Giri A., Brody J.A., Hill W.D., Kacprowski T., Li J., Lyytikäinen L.P. Large-scale exome-wide association identifies loci for white blood cell traits and pleiotropy with immune-mediated diseases. Am. J. Hum. Genet. 2016;99:22–39. doi: 10.1016/j.ajhg.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iotchkova V., Huang J., Morris J.A., Jain D., Barbieri C., Walter K., Min J.L., Chen L., Astle W., Cocca M., UK10K Consortium Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 2016;48:1303–1312. doi: 10.1038/ng.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain D., Hodonsky C.J., Schick U.M., Morrison J.V., Minnerath S., Brown L., Schurmann C., Liu Y., Auer P.L., Laurie C.A. Genome-wide association of white blood cell counts in Hispanic/Latino Americans: the Hispanic Community Health Study/Study of Latinos. Hum. Mol. Genet. 2017;26:1193–1204. doi: 10.1093/hmg/ddx024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M.-H., Raffield L.M., Mousas A., Sakaue S., Huffman J.E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., VA Million Veteran Program Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auer P.L., Johnsen J.M., Johnson A.D., Logsdon B.A., Lange L.A., Nalls M.A., Zhang G., Franceschini N., Fox K., Lange E.M. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auer P.L., Teumer A., Schick U., O’Shaughnessy A., Lo K.S., Chami N., Carlson C., de Denus S., Dubé M.P., Haessler J. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat. Genet. 2014;46:629–634. doi: 10.1038/ng.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffield L.M., Iyengar A.K., Wang B., Gaynor S.M., Spracklen C.N., Zhong X., Kowalski M.H., Salimi S., Polfus L.M., Benjamin E.J., TOPMed Inflammation Working Group. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Allelic Heterogeneity at the CRP Locus Identified by Whole-Genome Sequencing in Multi-ancestry Cohorts. Am. J. Hum. Genet. 2020;106:112–120. doi: 10.1016/j.ajhg.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalski M.H., Qian H., Hou Z., Rosen J.D., Tapia A.L., Shan Y., Jain D., Argos M., Arnett D.K., Avery C., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. TOPMed Hematology & Hemostasis Working Group Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. doi: 10.1371/journal.pgen.1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The ARIC investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 14.Mitchell B.D., McArdle P.F., Shen H., Rampersaud E., Pollin T.I., Bielak L.F., Jaquish C., Douglas J.A., Roy-Gagnon M.H., Sack P. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am. Heart J. 2008;155:823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman O., Kuivaniemi H., Tromp G., Faucett W.A., Li R., Manolio T.A., Sanderson S.C., Kannry J., Zinberg R., Basford M.A., eMERGE Network The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes G.H., Cutter G., Donahue R., Friedman G.D., Hulley S., Hunkeler E., Jacobs D.R., Jr., Liu K., Orden S., Pirie P. Recruitment in the coronary artery disease risk development in young adults (Cardia) study. Control. Clin. Trials. 1987;8(4, Suppl):68S–73S. doi: 10.1016/0197-2456(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 17.Fried L.P., Borhani N.O., Enright P., Furberg C.D., Gardin J.M., Kronmal R.A., Kuller L.H., Manolio T.A., Mittelmark M.B., Newman A. The cardiovascular health study: design and rationale. Ann. Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 18.Regan E.A., Hokanson J.E., Murphy J.R., Make B., Lynch D.A., Beaty T.H., Curran-Everett D., Silverman E.K., Crapo J.D. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J., D’Agostino R.B., Sr., Fox C.S., Larson M.G., Murabito J.M. The third generation cohort of the national heart, lung, and blood institute’s framingham heart study: design, recruitment, and initial examination. Am. J. Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 20.Becker D.M., Segal J., Vaidya D., Yanek L.R., Herrera-Galeano J.E., Bray P.F., Moy T.F., Becker L.C., Faraday N. Sex differences in platelet reactivity and response to low-dose aspirin therapy. JAMA. 2006;295:1420–1427. doi: 10.1001/jama.295.12.1420. [DOI] [PubMed] [Google Scholar]
- 21.Sorlie P.D., Avilés-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M. Design and implementation of the hispanic community health study/study of latinos. Ann. Epidemiol. 2010;20:629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor H.A., Jr., Wilson J.G., Jones D.W., Sarpong D.F., Srinivasan A., Garrison R.J., Nelson C., Wyatt S.B. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn. Dis. 2005;15(4, Suppl 6):S6–S4, 17. [PubMed] [Google Scholar]
- 23.Wilson J.G., Rotimi C.N., Ekunwe L., Royal C.D., Crump M.E., Wyatt S.B., Steffes M.W., Adeyemo A., Zhou J., Taylor H.A., Jr., Jaquish C. Study design for genetic analysis in the Jackson Heart Study. Ethn. Dis. 2005;15(4, Suppl 6):S6–S30, 37. [PubMed] [Google Scholar]
- 24.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R., Jr., Kronmal R., Liu K. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell B.D., Kammerer C.M., Blangero J., Mahaney M.C., Rainwater D.L., Dyke B., Hixson J.E., Henkel R.D., Sharp R.M., Comuzzie A.G. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 26.The Women’s Health Initiative Study Group Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 27.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernández-Rhodes L., Justice A.E., Graff M. Genetic diversity and association studies in us hispanic/latino populations: applications in the hispanic community health study/study of latinos. Am. J. Hum. Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sofer T., Zheng X., Gogarten S.M., Laurie C.A., Grinde K., Shaffer J.R., Shungin D., O’Connell J.R., Durazo-Arvizo R.A., Raffield L., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genet. Epidemiol. 2019;43:263–275. doi: 10.1002/gepi.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breslow N.E., Clayton D.G. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993;88:9–25. [Google Scholar]
- 32.Chen H., Wang C., Conomos M.P., Stilp A.M., Li Z., Sofer T., Szpiro A.A., Chen W., Brehm J.M., Celedón J.C. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dey R., Schmidt E.M., Abecasis G.R., Lee S. A fast and accurate algorithm to test for binary phenotypes and its application to phewas. Am. J. Hum. Genet. 2017;101:37–49. doi: 10.1016/j.ajhg.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Huffman J.E., Brody J.A., Wang C., Lee S., Li Z., Gogarten S.M., Sofer T., Bielak L.F., Bis J.C., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. TOPMed Hematology and Hemostasis Working Group Efficient variant set mixed model association tests for continuous and binary traits in large-scale whole-genome sequencing studies. Am. J. Hum. Genet. 2019;104:260–274. doi: 10.1016/j.ajhg.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., Morris A., Wellcome Trust Case Control Consortium Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu P., Gifford A., Meng X., Li X., Campbell H., Varley T., Zhao J., Carroll R., Bastarache L., Denny J.C. Mapping icd-10 and icd-10-cm codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform. 2019;7:e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathias R.A., Grant A.V., Rafaels N., Hand T., Gao L., Vergara C., Tsai Y.J., Yang M., Campbell M., Foster C. A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin. Immunol. 2010;125:336–346.e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes K.C., Neely J.D., Duffy D.L., Freidhoff L.R., Breazeale D.R., Schou C., Naidu R.P., Levett P.N., Renault B., Kucherlapati R. Linkage of asthma and total serum IgE concentration to markers on chromosome 12q: evidence from Afro-Caribbean and Caucasian populations. Genomics. 1996;37:41–50. doi: 10.1006/geno.1996.0518. [DOI] [PubMed] [Google Scholar]
- 40.Jarjour N.N., Erzurum S.C., Bleecker E.R., Calhoun W.J., Castro M., Comhair S.A., Chung K.F., Curran-Everett D., Dweik R.A., Fain S.B., NHLBI Severe Asthma Research Program (SARP) Severe asthma: lessons learned from the national heart, lung, and blood institute severe asthma research program. Am. J. Respir. Crit. Care Med. 2012;185:356–362. doi: 10.1164/rccm.201107-1317PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bin L., Malley C., Taylor P., Preethi Boorgula M., Chavan S., Daya M., Mathias M., Shankar G., Rafaels N., Vergara C. Whole genome sequencing identifies novel genetic mutations in patients with eczema herpeticum. Allergy. 2021;76:2510–2523. doi: 10.1111/all.14762. Published online February 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X., Qiao D., Yang C., Kasela S., Kim W., Ma Y., Shrine N., Batini C., Sofer T., Taliun S.A.G., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. TOPMed Lung Working Group Whole genome sequence analysis of pulmonary function and COPD in 19,996 multi-ethnic participants. Nat. Commun. 2020;11:5182. doi: 10.1038/s41467-020-18334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortin J.-P., Triche T.J., Jr., Hansen K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvath S., Levine A.J. Hiv-1 infection accelerates age according to the epigenetic clock. J. Infect. Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghansah A., Rockett K.A., Clark T.G., Wilson M.D., Koram K.A., Oduro A.R., Amenga-Etego L., Anyorigiya T., Hodgson A., Milligan P. Haplotype analyses of haemoglobin C and haemoglobin S and the dynamics of the evolutionary response to malaria in Kassena-Nankana District of Ghana. PLoS ONE. 2012;7:e34565. doi: 10.1371/journal.pone.0034565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esoh K., Wonkam A. Evolutionary history of sickle-cell mutation: implications for global genetic medicine. Hum. Mol. Genet. 2021;30(R1):R119–R128. doi: 10.1093/hmg/ddab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suenobu S., Takakura N., Inada T., Yamada Y., Yuasa H., Zhang X.Q., Sakano S., Oike Y., Suda T. A role of EphB4 receptor and its ligand, ephrin-B2, in erythropoiesis. Biochem. Biophys. Res. Commun. 2002;293:1124–1131. doi: 10.1016/S0006-291X(02)00330-3. [DOI] [PubMed] [Google Scholar]
- 49.Ngo D., Wen D., Gao Y., Keyes M.J., Drury E.R., Katz D.H., Benson M.D., Sinha S., Shen D., Farrell L.A. Circulating testican-2 is a podocyte-derived marker of kidney health. Proc. Natl. Acad. Sci. USA. 2020;117:25026–25035. doi: 10.1073/pnas.2009606117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn N., Kim W.-J., Kim N., Park H.W., Lee S.-W., Yoo J.-Y. The interferon-inducible proteoglycan testican-2/spock2 functions as a protective barrier against virus infection of lung epithelial cells. J. Virol. 2019;93:e00662-19. doi: 10.1128/JVI.00662-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y.-W., Lee B., Liu P.-S., Wei L.-N. Receptor-interacting protein 140 orchestrates the dynamics of macrophage m1/m2 polarization. J. Innate Immun. 2016;8:97–107. doi: 10.1159/000433539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilfinger J., Seuter S., Tuomainen T.-P., Virtanen J.K., Voutilainen S., Nurmi T., de Mello V.D., Uusitupa M., Carlberg C. Primary vitamin D receptor target genes as biomarkers for the vitamin D3 status in the hematopoietic system. J. Nutr. Biochem. 2014;25:875–884. doi: 10.1016/j.jnutbio.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Lapierre M., Castet-Nicolas A., Gitenay D., Jalaguier S., Teyssier C., Bret C., Cartron G., Moreaux J., Cavaillès V. Expression and role of RIP140/NRIP1 in chronic lymphocytic leukemia. J. Hematol. Oncol. 2015;8:20. doi: 10.1186/s13045-015-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dougan M., Dranoff G., Dougan S.K. Gm-csf, il-3, and il-5 family of cytokines: regulators of inflammation. Immunity. 2019;50:796–811. doi: 10.1016/j.immuni.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esnault S., Kelly E.A. Essential mechanisms of differential activation of eosinophils by il-3 compared to gm-csf and il-5. Crit. Rev. Immunol. 2016;36:429–444. doi: 10.1615/CritRevImmunol.2017020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reche P.A., Soumelis V., Gorman D.M., Clifford T., Liu Mr, Travis M., Zurawski S.M., Johnston J., Liu Y.J., Spits H. Human thymic stromal lymphopoietin preferentially stimulates myeloid cells. J. Immunol. 2001;167:336–343. doi: 10.4049/jimmunol.167.1.336. [DOI] [PubMed] [Google Scholar]
- 57.Rosen H., Goetzl E.J. Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat. Rev. Immunol. 2005;5:560–570. doi: 10.1038/nri1650. [DOI] [PubMed] [Google Scholar]
- 58.Nussbaum C., Bannenberg S., Keul P., Gräler M.H., Gonçalves-de-Albuquerque C.F., Korhonen H., von Wnuck Lipinski K., Heusch G., de Castro Faria Neto H.C., Rohwedder I. Sphingosine-1-phosphate receptor 3 promotes leukocyte rolling by mobilizing endothelial P-selectin. Nat. Commun. 2015;6:6416. doi: 10.1038/ncomms7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keul P., Lucke S., von Wnuck Lipinski K., Bode C., Gräler M., Heusch G., Levkau B. Sphingosine-1-phosphate receptor 3 promotes recruitment of monocyte/macrophages in inflammation and atherosclerosis. Circ. Res. 2011;108:314–323. doi: 10.1161/CIRCRESAHA.110.235028. [DOI] [PubMed] [Google Scholar]
- 60.Murakami K., Kohno M., Kadoya M., Nagahara H., Fujii W., Seno T., Yamamoto A., Oda R., Fujiwara H., Kubo T. Knock out of S1P3 receptor signaling attenuates inflammation and fibrosis in bleomycin-induced lung injury mice model. PLoS ONE. 2014;9:e106792. doi: 10.1371/journal.pone.0106792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L., Han Z., Tian L., Mai P., Zhang Y., Wang L., Li L. Sphingosine 1-phosphate receptor 2 and 3 mediate bone marrow-derived monocyte/macrophage motility in cholestatic liver injury in mice. Sci. Rep. 2015;5:13423. doi: 10.1038/srep13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogle M.E., Olingy C.E., Awojoodu A.O., Das A., Ortiz R.A., Cheung H.Y., Botchwey E.A. Sphingosine-1-phosphate receptor-3 supports hematopoietic stem and progenitor cell residence within the bone marrow niche. Stem Cells. 2017;35:1040–1052. doi: 10.1002/stem.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bryce S.A., Wilson R.A.M., Tiplady E.M., Asquith D.L., Bromley S.K., Luster A.D., Graham G.J., Nibbs R.J. Ackr4 on stromal cells scavenges ccl19 to enable ccr7-dependent trafficking of apcs from inflamed skin to lymph nodes. J. Immunol. 2016;196:3341–3353. doi: 10.4049/jimmunol.1501542. [DOI] [PubMed] [Google Scholar]
- 64.Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klug M., Schmidhofer S., Gebhard C., Andreesen R., Rehli M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013;14:R46. doi: 10.1186/gb-2013-14-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bick A.G., Weinstock J.S., Nandakumar S.K., Fulco C.P., Bao E.L., Zekavat S.M., Szeto M.D., Liao X., Leventhal M.J., Nasser J., NHLBI Trans-Omics for Precision Medicine Consortium Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–768. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cargo C., Cullen M., Taylor J., Short M., Glover P., Van Hoppe S., Smith A., Evans P., Crouch S. The use of targeted sequencing and flow cytometry to identify patients with a clinically significant monocytosis. Blood. 2019;133:1325–1334. doi: 10.1182/blood-2018-08-867333. [DOI] [PubMed] [Google Scholar]
- 68.Stremenova Spegarova J., Lawless D., Mohamad S.M.B., Engelhardt K.R., Doody G., Shrimpton J., Rensing-Ehl A., Ehl S., Rieux-Laucat F., Cargo C. Germline TET2 loss of function causes childhood immunodeficiency and lymphoma. Blood. 2020;136:1055–1066. doi: 10.1182/blood.2020005844. [DOI] [PubMed] [Google Scholar]
- 69.Kaasinen E., Kuismin O., Rajamäki K., Ristolainen H., Aavikko M., Kondelin J., Saarinen S., Berta D.G., Katainen R., Hirvonen E.A.M. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 2019;10:1252. doi: 10.1038/s41467-019-09198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duployez N., Goursaud L., Fenwarth L., Bories C., Marceau-Renaut A., Boyer T., Fournier E., Nibourel O., Roche-Lestienne C., Huet G. Familial myeloid malignancies with germline TET2 mutation. Leukemia. 2020;34:1450–1453. doi: 10.1038/s41375-019-0675-6. [DOI] [PubMed] [Google Scholar]
- 71.Kazi J.U., Rönnstrand L. Fms-like tyrosine kinase 3/flt3: from basic science to clinical implications. Physiol. Rev. 2019;99:1433–1466. doi: 10.1152/physrev.00029.2018. [DOI] [PubMed] [Google Scholar]
- 72.Saevarsdottir S., Olafsdottir T.A., Ivarsdottir E.V., Halldorsson G.H., Gunnarsdottir K., Sigurdsson A., Johannesson A., Sigurdsson J.K., Juliusdottir T., Lund S.H. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature. 2020;584:619–623. doi: 10.1038/s41586-020-2436-0. [DOI] [PubMed] [Google Scholar]
- 73.Flaquer A., Rappold G.A., Wienker T.F., Fischer C. The human pseudoautosomal regions: a review for genetic epidemiologists. Eur. J. Hum. Genet. 2008;16:771–779. doi: 10.1038/ejhg.2008.63. [DOI] [PubMed] [Google Scholar]
- 74.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vuckovic D., Bao E.L., Akbari P., Lareau C.A., Mousas A., Jiang T., Chen M.H., Raffield L.M., Tardaguila M., Huffman J.E., VA Million Veteran Program The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182:1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Naik R.P., Smith-Whitley K., Hassell K.L., Umeh N.I., de Montalembert M., Sahota P., Haywood C., Jr., Jenkins J., Lloyd-Puryear M.A., Joiner C.H. Clinical outcomes associated with sickle cell trait: a systematic review. Ann. Intern. Med. 2018;169:619–627. doi: 10.7326/M18-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hou J., Chen Q., Wu X., Zhao D., Reuveni H., Licht T., Xu M., Hu H., Hoeft A., Ben-Sasson S.A. S1pr3 signaling drives bacterial killing and is required for survival in bacterial sepsis. Am. J. Respir. Crit. Care Med. 2017;196:1559–1570. doi: 10.1164/rccm.201701-0241OC. [DOI] [PubMed] [Google Scholar]
- 78.Gon Y., Wood M.R., Kiosses W.B., Jo E., Sanna M.G., Chun J., Rosen H. Retraction for “S1P3 receptor-induced reorganization of epithelial tight junctions compromises lung barrier integrity and is potentiated by TNF”. Proc. Natl. Acad. Sci. USA. 2009;106:12561. doi: 10.1073/pnas.0906977106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Punsawad C., Viriyavejakul P. Expression of sphingosine kinase 1 and sphingosine 1-phosphate receptor 3 in malaria-associated acute lung injury/acute respiratory distress syndrome in a mouse model. PLoS ONE. 2019;14:e0222098. doi: 10.1371/journal.pone.0222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daya M., Barnes K.C. African American ancestry contribution to asthma and atopic dermatitis. Ann. Allergy Asthma Immunol. 2019;122:456–462. doi: 10.1016/j.anai.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakajima S., Kabata H., Kabashima K., Asano K. Anti-TSLP antibodies: Targeting a master regulator of type 2 immune responses. Allergol. Int. 2020;69:197–203. doi: 10.1016/j.alit.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 82.Holstege H., Hulsman M., van der Lee S.J., van den Akker E.B. The role of age-related clonal hematopoiesis in genetic sequencing studies. Am. J. Hum. Genet. 2020;107:575–576. doi: 10.1016/j.ajhg.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kraft I.L., Godley L.A. Identifying potential germline variants from sequencing hematopoietic malignancies. Blood. 2020;136:2498–2506. doi: 10.1182/blood.2020006910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for each participating study can be accessed through dbGaP with the corresponding accession number (Amish, phs000956; ARIC, phs001211; BioMe, phs001644; BAGS, phs001143; CARDIA, phs001612; CHS, phs001368; COPDGene, phs000951; FHS, phs000974; GeneSTAR, phs001218; HCHS/SOL, phs001395; JHS, phs000964; MESA, phs001416; SAFS, phs001215; SARP, phs001446; and WHI, phs001237). Analysis results for the conditional single-variant analyses and the aggregate conditional analyses can be accessed through dbGaP: phs001974.