Summary
Up to 80% of BRCA1 and BRCA2 genetic variants remain of uncertain clinical significance (VUSs). Only variants classified as pathogenic or likely pathogenic can guide breast and ovarian cancer prevention measures and treatment by PARP inhibitors. We report the first results of the ongoing French national COVAR (cosegregation variant) study, the aim of which is to classify BRCA1/2 VUSs. The classification method was a multifactorial model combining different associations between VUSs and cancer, including cosegregation data. At this time, among the 653 variants selected, 101 (15%) distinct variants shared by 1,624 families were classified as pathogenic/likely pathogenic or benign/likely benign by the COVAR study. Sixty-six of the 101 (65%) variants classified by COVAR would have remained VUSs without cosegregation data. Of note, among the 34 variants classified as pathogenic by COVAR, 16 remained VUSs or likely pathogenic when following the ACMG/AMP variant classification guidelines. Although the initiation and organization of cosegregation analyses require a considerable effort, the growing number of available genetic tests results in an increasing number of families sharing a particular variant, and thereby increases the power of such analyses. Here we demonstrate that variant cosegregation analyses are a powerful tool for the classification of variants in the BRCA1/2 breast-ovarian cancer predisposition genes.
Keywords: BRCA1, BRCA2, variant of uncertain significance, cosegregation data, classification, clinical, multifactorial model
Introduction
Identification of the BRCA1 (MIM: 113705) and BRCA2 (MIM: 600185) genes 25 years ago opened up a new field in cancer genetics and BRCA1/2 testing is now a paradigm in predictive medicine. Pathogenic germline BRCA1/BRCA2 monoallelic variants are associated with a high risk of breast and ovarian cancer.1 They are classified as pathogenic when they have a putative loss-of-function effect, usually when a stop codon is introduced by nonsense substitutions, frameshift insertions/deletions (indels), complete splice defects, or large gene rearrangements (LGRs).2, 3, 4, 5, 6 However, many numerous identified variants, such as missense, synonymous, or variants surrounding coding exons even at canonical splicing positions, are difficult to classify without complementary analyses and, in many cases, remain unclassified.7, 8, 9, 10, 11 An international effort to classify BRCA1/2 variants was initiated in 2009 through “The Evidence-based Network for the Interpretation of Germline Mutant Alleles” (ENIGMA) and has recently been extended to other breast cancer predisposition genes.12
In view of the increasing genetic testing capacity due to the use of next generation sequencing (NGS) and with the advent of PARP inhibitors (PARPi) for the treatment of individuals affected with a tumor driven by BRCA1/BRCA2 inactivation, the number of affected individuals tested and the number of variants detected are continuously increasing.13,14 In April 2021, about 40,000 and 25,000 BRCA1/BRCA2 different germline variants have been reported worldwide in the BRCAexchange and ClinVar databases, respectively, of which nearly 80% and 50%, respectively, are unclassified in term of their pathogenic effect15 (Table S1). However, due to the individual impacts of PARPi prescriptions and prophylactic decisions such as mastectomy and oophorectomy, a reliable classification of variants as either pathogenic or non-pathogenic is of utmost importance. In a survey of 3,672 women who underwent BRCA1/BRCA2 testing in 2014–2015, Kurian et al. reported that 51% of women carrying a variant of uncertain significance (VUS) underwent bilateral prophylactic mastectomy even in the absence of family history of cancer.16 In March 2016, the journalist Jeremy Lange reported in The New York Times the case of a woman whose doctors had conflicting information on her BRCA1/BRCA2 test result. “The situation is ripe for overinterpretation and misinterpretation,” said a geneticist interviewed.
The large size of the BRCA1 and BRCA2 proteins, the complexity of their functions, and the lack of reliable surrogate markers of their activity, such as pathological immunohistochemistry or a comprehensive functional assay, have led to the VUS classification approach based on a multifactorial model, which has been validated for BRCA1/2 variant classification in previous studies.11,17,18 For each VUS, this model comprises a prior probability (PriorP) of pathogenicity based on Align-GVGD prediction score and combined likelihoods of pathogenicity derived from measurements of associations between VUS and cancer: personal and family cancer history, breast tumor pathology, co-occurrence of VUS with a known pathogenic variant of the same gene, and cosegregation data.11,17,19, 20, 21 Plon et al. proposed a classification of cancer predisposition gene variants based on five levels of likelihood of pathogenicity, known as the IARC-5-tier classes: (1) benign (BV), (2) likely benign (LBV), (3) of uncertain significance, (4) likely pathogenic (LPV), and (5) pathogenic (PV).22
The COVAR (cosegregation variant) study has been set up within the French Unicancer Genetics Group (UGG) which brings together cancer genetics clinics and laboratories. Here, we report the first results of the COVAR study showing the value of cosegregation analyzes for classification of BRCA1/BRCA2 variants. In addition, by comparing the COVAR results to the variant scoring obtained by following the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines, we demonstrate the power of the COVAR approach.
Subjects and methods
UGG cancer genetics clinic and laboratory network, FrOG (French OncoGenetics) Database
The French UGG (see Web resources) was established in 1991 to contribute to the identification of cancer predisposing genes and estimation of cancer risks based on national (GENEPSO, GEMO, GENESIS) and international (IBCCS, CIMBA) genetic epidemiological studies and to define recommendations for genetic testing and management of individuals at risk.1,2,5,23, 24, 25, 26, 27, 28 In 2020, the UGG consists of a network of 148 public/private cancer genetics clinics throughout France and 26 academic molecular genetics laboratories, 17 of which routinely perform breast-ovarian cancer genetic testing.
Between 1994 and 2018, about 260,000 index cases (ICs) were sequenced for BRCA1/BRCA2 in the UGG cancer genetics clinic and laboratory network. The sequencing was performed by specific gene analyzes until 2017 and by multigene panel thereafter.26,29 ICs correspond to the first case to be tested in their families and are mostly women with a personal history of breast and/or ovarian cancer. Sequencing technologies evolved during this period, but genetic testing always included the search for substitutions and indels on all BRCA1/2 coding exons and exon/intron boundaries, as well as the search for LGR. All identified variants (except for common polymorphisms) were reported in a dedicated BRCA1/BRCA2 database initially called UMD-BRCA1/BRCA25 (BRCAShare30) and now called FrOG (French OncoGenetics) database. This database is a helpful everyday tool for molecular geneticists and an essential resource for the classification of variants. Affected individuals are pseudonymously registered in the database by means of a family ID assigned by the submitting laboratory.5 Patients can therefore be mapped back through their family ID for variant classification updates and research study invitations.
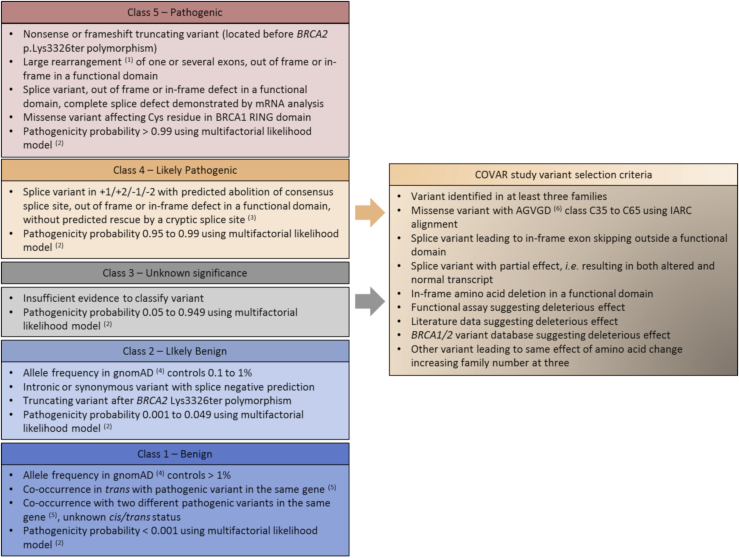
A working group composed of the UGG molecular geneticists is in charge of the classification of variants. The curation is under the responsibility of S.M.C. Systematic classification criteria have been introduced in the framework of the IARC-5-classes of pathogenicity.22 The principles of classification are reported in Figure 1 and in Béroud et al.30
Figure 1.
Variant classification criteria in the framework of the 5 classes of likelihood of pathogenicity and COVAR study variant selection criteria
The variant classification or selection for COVAR study relies on the presence of at least one of the listed criteria.
(1) For large duplications, tandem status disrupting the gene must be demonstrated by mRNA analysis or breakpoint sequencing, otherwise this variant should be considered to be a VUS.
(2) Multifactorial likelihood model proposed by Goldgar et al.17
(3) This classification criterion has been used since January 2019. Before 2019, these splice variants were classified in class 5 and were confirmed by mRNA analysis.
(4) Total gnomAD cohort or subpopulations except for Finnish, Ashkenazi Jewish, and “Other” groups due to the possibility of a founder effect and/or the small number of subjects. gnomAD started to be used as the control database for classification criteria in July 2017. Between December 2014 and June 2017, allele frequency was determined from the ExAC database, with the same thresholds and population groups for classification. Between 2012 and November 2014, allele frequency was determined from the 1000 Genomes Project and Exome Sequencing Project databases, with only one threshold, allele frequency > 1% for classification in class 1 variant, and only the total cohort of both projects.
(5) With no clinical or biological signs of Fanconi anemia.
(6) Align-GVGD prediction algorithm classifies variants from C0 to C65 classes; C0 corresponds to the least likely functional variants and C65 corresponds to the most likely functional variants.31 The last species of IARC alignment, purple sea urchin, is removed for Align-GVGD prediction when it is the only species with an amino acid change; in this case, the inclusion criterion corresponds to Align-GVGD class C35 to C65 AND deleterious prediction by SIFT algorithm.32
IARC, International Agency for Research on Cancer.
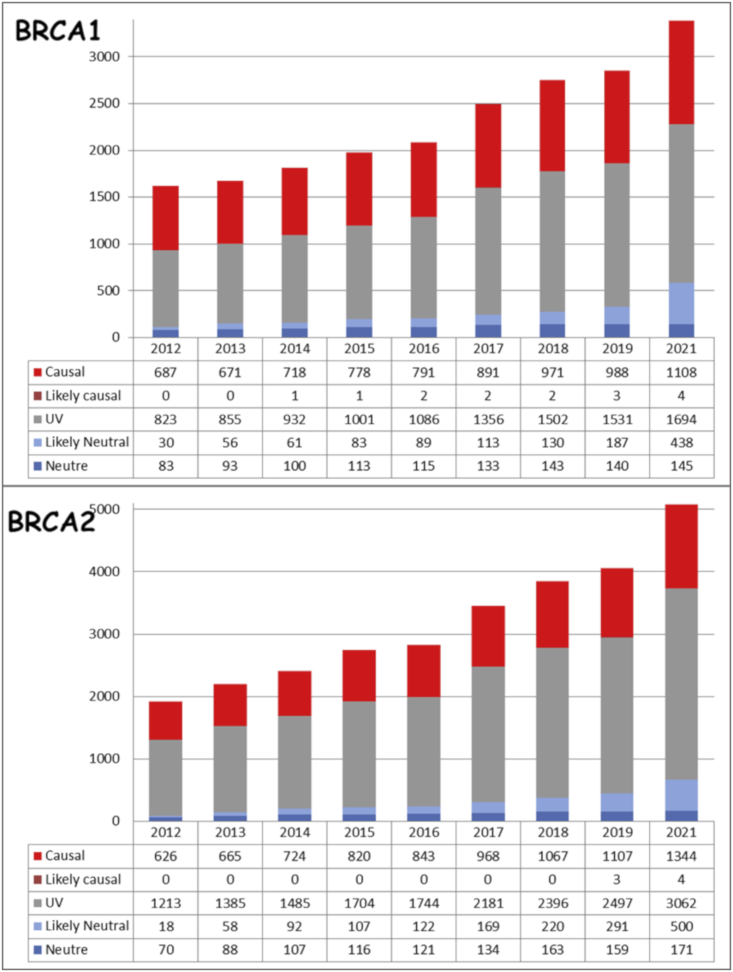
In April 2021, of the 3,389 distinct BRCA1 variants reported, 1,112 variants were classified as LPV/PV (class 4/5), 583 as BV/LBV (class 1/2), and 1,694 as class 3 (VUS), this last class representing 50% of all variants (Figure 2). A similar class distribution was observed for BRCA2: within 5,081 distinct BRCA2 variants reported, 3,062 were classified as class 3 (60%).
Figure 2.
Increase of the number of BRCA1 and BRCA2 variants in the FrOG database since 2012, not including the result of the COVAR study classification
The COVAR study and variant selection
The COVAR study was initiated in order to increase the proportion of classified variants by using cosegregation data within families. The study was designed to focus on sampling available relatives of the ICs and testing whether or not they carried the variant found in the IC with reference to their cancer phenotype. The model used here includes the absence/presence and age at diagnosis of BRCA-associated cancers (breast, ovarian, pancreas for BRCA1 and BRCA2, and prostate for BRCA2 only).1,33, 34, 35 The model compares the probability of observing the pedigree phenotypes and variant genotypes under the hypothesis that the variant is pathogenic with that under the hypothesis that the variant is benign with respect to risk.17,19 If this likelihood ratio (LR) is greater than 1, this indicates evidence in favor of a pathogenic effect of the variant and, conversely, if it is less than 1 this provides evidence that the variant is benign. For a given variant, combined cosegregation likelihood of pathogenicity is calculated by multiplying the LR obtained from families sharing the same variant (or different variants when the same impact at the mRNA and/or protein level has been demonstrated for each variant).
Variant selection is discussed by the variant classification working group according to the expected classification power. It is based on the number of families carrying the same variant, the number of potential participants among the relatives, and the level of PriorP, because a lower level of evidence is required to reach a final classification of PV or LPV when PriorP is high (Figure 1). A variant may be selected in the COVAR study when it has been identified in at least three families, whatever its PriorP. A minimum PriorP of 0.66 is sufficient to select a variant, even if it has been identified in only one family; the threshold of 0.66 corresponds to the Align-GVGD C35 to C55 prediction classes. LPVs are rare and systematically selected. Most selected variants are VUSs. Splice variants with equivocal effect are also selected, e.g., the deep intronic variant BRCA2 c.6937+594T>G.36,37 Variant selection is an ongoing process; thus, a variant not initially selected may be subsequently selected based on identification of new families carrying the same variant.
Ethics declaration
The COVAR study was authorized by Ethics Committee in 2011 (Comite de protection des personnes Ile de France III, Ref: Am5677-1-2940) and is registered in the international clinical trials platform (see Web resources). Informed consent for genetic testing was obtained from all individuals undergoing testing.
Affected individual inclusion and study participation
ICs carrying a variant selected in the COVAR study are invited to participate in the study by medical geneticists during the test result disclosure consultation, or later if the variant has been selected subsequently. Inclusion of relatives is prioritized as follows: individuals with a personal history of breast and/or ovarian cancer, older cancer-free women/males when they can be useful to infer genotypes. Potential participants receive invitation letters from the cancer genetics clinic that manage them. Relatives agreeing to participate in the study receive full information about the study by mail and can receive oral information from a genetic counselor upon request. After providing their written consent, they receive a salivary kit for DNA testing (Oragene, DNA Genotek) and a short clinical questionnaire that includes sex, cancer status (site and age at diagnosis), age at interview for unaffected relatives, and age at possible prophylactic mastectomy and oophorectomy. They provide their permission to access medical files in order to retrieve pathology reports. Breast cancer details are collected.38 For relatives not included in the study and whose cancer status is useful for cosegregation analysis, efforts are made to obtain cancer validation by pathology report, medical record, or bona fide clinical history. When a variant has been classified as BV or LBV, the ICs concerned are informed by mail and are invited to inform their relatives; when classified as LPV or PV, all study participants, regardless of individual test results, are invited to attend a cancer genetics clinic to perform diagnostic testing on a second sample.
Statistical model
Cosegregation LR is assessed using the statistical model developed by Thompson et al.19 and implemented in the COOL-webserver based on an improved penetrance model, developed by Belman et al.39 In addition to the cosegregation LRs derived in this way, the multifactorial LR for each family includes LR co-occurrence,17 personal and family cancer history,40,41 and breast pathology LRs38 as previously published.11 All these LR are multiplied to obtain the “combined LR.” As previously mentioned by Goldgar et al., since analysis of cosegregation is conditional on the phenotypes in the family, the data on cosegregation can be considered independent of the data on family history.17 Combined LRs of families sharing the same variant are multiplied. The PriorP of each variant is based on the Align-GVGD prediction class score determined by phylogenetic conservation of the modified amino acid, physicochemical change score for missense variants, and the MaxEntScan splicing site prediction tool for suspected splicing defects.21 PriorP and LRs are combined to calculate posterior probabilities using a Bayesian formula:11,17,18
For in-frame indels located in the same functional domain, the highest Align-GVGD PriorP is chosen.18 Combined LR (or LR causality) thresholds used are those defined by Goldgar et al.: >1,000:1 considered to be pathogenic (class 5) and <1:100 considered to be benign (class 1).17 It means that the Posterior Probability of pathogenicity is calculated only when the combined LR is greater than 1,000:1 or lower than 1:100. Variant classification is based on the Posterior Probability of pathogenicity, with values <0.001, between 0.001 and 0.049, between 0.95 and 0.99, or >0.99 corresponding to BV, LBV, LPV, or PV, respectively.22
Results
Variant selection and patient inclusion
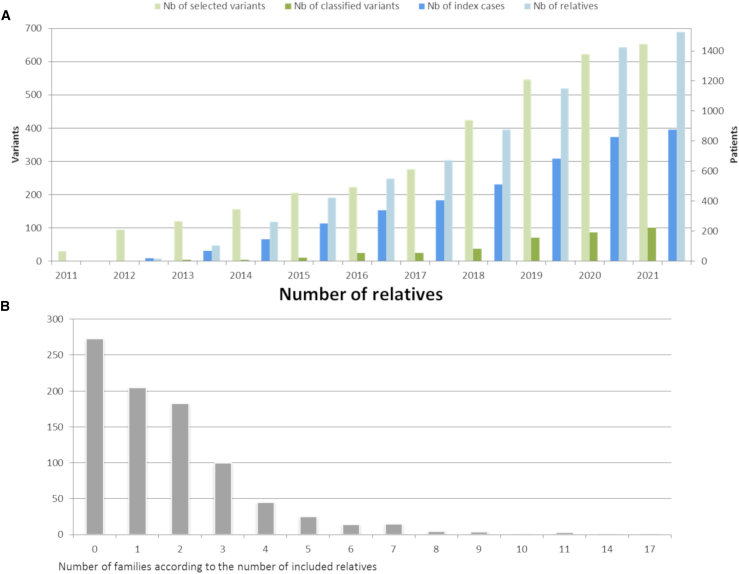
From December 2011 to April 2021, 645 VUSs (214 BRCA1 and 431 BRCA2) and 8 LPVs (4 BRCA1 and 4 BRCA2) were selected for the COVAR study (Figure 3A). These 653 variants included 518 missense, 53 intronic, 34 in-frame deletions/insertions, 22 synonymous, 14 nonsense or frameshift indels, and 12 in-frame LGR.
Figure 3.
Patients and variants in the COVAR study from December 2011 to November 2019
(A) Number of affected individuals and variants included and number of variants classified.
(B) Number of families participating in the COVAR study according to the number of relatives included.
By April 2021, 876 ICs carrying a selected variant (283 BRCA1 and 593 BRCA2) and 1,525 relatives had been included (Figure 3A). Among the 876 participating ICs, no relative was included for 273 ICs, one relative was included for 205 ICs, and more than one relative was included for 398 ICs (Figure 3B).
Variant classification
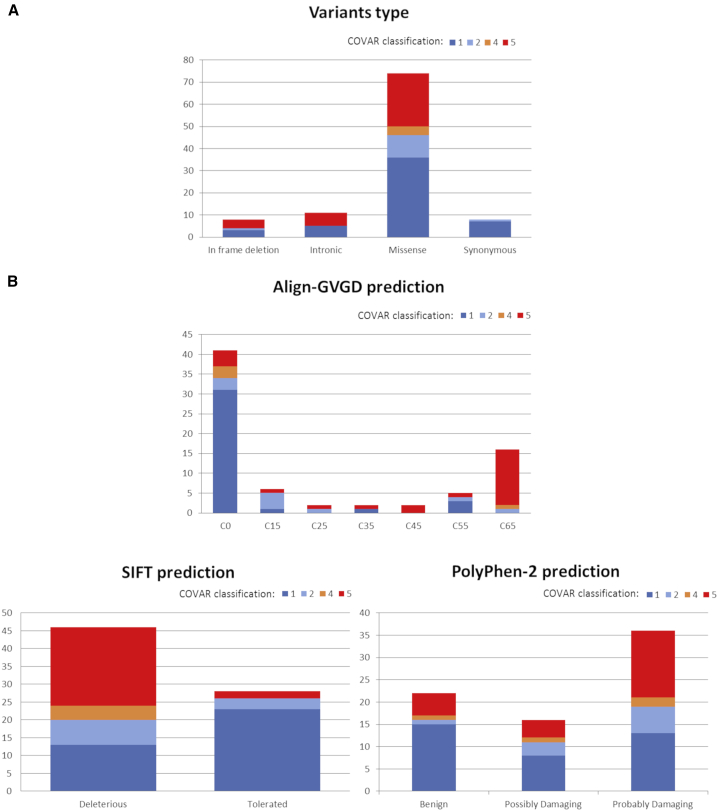
At the time of the study report, COVAR allowed the classification into clinically useful categories of 101 variants (15% of the selected variants) shared by 1,624 families registered in the UGG clinics network. Thirty-four variants (17 BRCA1, 17 BRCA2, 34%) were classified as PV; 4 BRCA1 variants (4%) as LPV, and 63 variants (25 BRCA1, 38 BRCA2, 62%) were classified as BV or LBV (Tables 1, S2, S3, S4, and S5). Table 1 reports LRs calculated for each component of each variant and its class with or without including cosegregation data. Of note, 29 of the 38 (76%) COVAR-classified PVs/LPVs and 37 of the 63 (59%) COVAR-classified BVs/LBVs, overall 66 of the 101 (65%) COVAR-classified variants would have remained VUSs without cosegregation data. Classified variant types were 74 missense, 11 intronic, and 8 synonymous variants, 6 in-frame indels, and 2 LGRs (Figure 4A). Each type contained variants classified either as pathogenic or benign, except for synonymous variants that were all classified as BV or LBV. Although 8 LPVs were selected for analysis, none of them were re-classified.
Table 1.
Variants classified by the COVAR study
| Gene | Exon/Intron | Nucleotide nomenclature | Protein nomenclature | Prior Probability of pathogenicity | Nb of families | Family history LR | Co-occurrence LR | Pathology LR | Combined LR without cosegregation | IARC 5-tier class without co-segregation dataa | Nb of families with co-segregation data | Varsome Classb | Cosegregation LR | Combined LR | Posterior Probability of pathogenicity | IARC 5-tier class |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PATHOGENIC | ||||||||||||||||
| BRCA1 | 3 | c.121C>T | p.His41Tyr | 0.81 | 4 | 4.88 | – | 73.42 | 358.04 | – | 3 | P | 128.396 | 45,970.26 | 0.9999949 | 5 |
| BRCA1 | I8 | c.547+1G>T | – | 0.97 | 4 | 1.87 | – | 703.797 | 1,313.41 | – | 4 | P | 204.08743 | 268,050.27 | 0.99999988 | 5c |
| BRCA1 | 16 | c.4963T>G | p.Ser1655Ala | 0.03 | 9 | 1.23 | – | 19.78 | 24.36 | 3 | 5 | LP | 93.2268842 | 2,271.10 | 0.98596299 | 4 |
| BRCA1 | 17 | c.4994T>A | p.Val1665Glu | 0.81 | 15 | 5.89 | – | 41.24 | 243.08 | – | 3 | LP | 53.95 | 13,114.14 | 0.99998211 | 5 |
| BRCA1 | 17 | c.5017_5019del | p.His1673del | 0.81 | 6 | 0.49 | – | 7.98 | 3.92 | 3 | 3 | LP | 488.41099 | 1,914.77 | 0.99987751 | 5 |
| BRCA1 | 17 | c.5062G>T | p.Val1688Phe | 0.03 | 1 | 11.69 | – | 11.79 | 137.84 | 3 | 1 | LP | 24.025 | 3,311.52 | 0.9903305 | 5 |
| BRCA1 | 17 | c.5072C>T | p.Thr1691Ile | 0.81 | 4 | 3.87 | – | 23.23 | 89.99 | – | 2 | P | 22.24 | 2,001.76 | 0.999882832 | 5 |
| BRCA1 | 18 | c.5116G>A | p.Gly1706Arg | 0.81 | 17 | 3.17 | – | 11.79 | 37.34 | – | 2 | P | 61.93 | 2,312.62 | 0.99989858 | 5 |
| BRCA1 | 18 | c.5117G>A | p.Gly1706Glu | 0.81 | 10 | 2.61 | – | 3.12 | 8.14 | 3 | 4 | LP | 7.37 | 59.97 | 0.99610374 | 4c |
| BRCA1 | 18 | c.5144G>A | p.Ser1715Asn | 0.66 | 4 | 6.42 | – | – | 6.42 | 3 | 2 | P | 167.97 | 1,078.99 | 0.99952279 | 5d |
| BRCA1 | 18 | c.5145C>G | p.Ser1715Arg | 0.81 | 3 | 6.78 | – | 13.91 | 94.28 | – | 2 | LP | 28.80 | 2,715.44 | 0.99991362 | 5d |
| BRCA1 | 20 | c.5213G>T | p.Gly1738Val | 0.81 | 2 | 6.05 | – | 6.21 | 37.55 | – | 1 | LP | 44.08 | 1,655.39 | 0.99985832 | 5 |
| BRCA1 | 20 | c.5216A>G | p.Asp1739Gly | 0.81 | 9 | 0.75 | – | 1009.26 | 754.45 | – | 2 | LP | 10.57 | 7,976.71 | 0.99997059 | 5 |
| BRCA1 | 20 | c.5216A>T | p.Asp1739Val | 0.81 | 5 | 5.39 | – | 71.42 | 384.68 | – | 3 | LP | 3.34 | 1,285.88 | 0.99981762 | 5 |
| BRCA1 | 20 | c.5254G>C | p.Ala1752Pro | 0.03 | 6 | 3.80 | – | 26.97 | 102.45 | 3 | 3 | LP | 5.88794143 | 603.24 | 0.94912744 | 4 |
| BRCA1 | 20 | c.5255C>A | p.Ala1752Glu | 0.03 | 2 | 10.98 | – | 10.61 | 116.44 | 3 | 1 | LP | 14.53517 | 1,692.47 | 0.98125391 | 4 |
| BRCA1 | 21 | c.5309G>Te | p.Gly1770Val | 0.03 | 33 | 2.48 | – | 5.599E+10 | 4.66E+12 | 5 | 11 | P | 3,668.96 | 1.71E+16 | 0.999999999 | 5d |
| BRCA1 | 21 | c.5309_5310delinsTTe | p.Gly1770Val | 0.03 | 2 | – | 5 | 1 | LP | 5 | ||||||
| BRCA1 | 23 | c.5426T>G | p.Val1809Gly | 0.66 | 16 | 2.36 | – | 129.20 | 304.56 | – | 8 | P | 60.2 | 18,334.43 | 0.9999719 | 5 |
| BRCA1 | 23 | c.5434C>G | p.Pro1812Ala | 0.03 | 12 | 5.94 | – | 34.64 | 205.76 | 3 | 5 | P | 79.1602119 | 16,288.35 | 0.99801887 | 5 |
| BRCA1 | 24 | c.5509T>C | p.Trp1837Arg | 0.81 | 3 | 6.81 | – | – | 6.81 | – | 2 | LP | 297.01 | 2,021.43 | 0.99988397 | 5d |
| BRCA2 | 3 | c.92G>C | p.Trp31Ser | 0.81 | 4 | 2.99 | – | 3.12 | 9.31 | – | 2 | P | 355.52 | 3,309.48 | 0.99992913 | 5 |
| BRCA2 | 3 | c.68-?_316+?delf | p.Asp23_Leu105del | 0.5 | 4 | 1,381.48 | – | 15.01 | 20,737.45 | 5 | 1 | P | 1.42701E+23 | 2.96E+27 | 1 | 5 |
| BRCA2 | 3 | c.156_157insAluf | – | 0.5 | 6 | 0 | P | 5 | ||||||||
| BRCA2 | I3 | c.316+1G>Tf | – | 0.5 | 1 | 0 | P | 5 | ||||||||
| BRCA2 | I3 | c.316+2T>Cf | – | 0.5 | 2 | 0 | P | 5d | ||||||||
| BRCA2 | I3 | c.316+4delf | – | 0.5 | 2 | 1 | LP | 5 | ||||||||
| BRCA2 | I3 | c.316+5G>Af | – | 0.5 | 2 | 1 | LP | 5 | ||||||||
| BRCA2 | I3 | c.316+5G>Cf | – | 0.5 | 28 | 10 | LP | 5 | ||||||||
| BRCA2 | 16 | c.7787G>T | p.Gly2596Val | 0.5 | 4 | 0.27 | – | 3.35 | 0.89 | 3 | 3 | VUS | 556.66 | 497.61 | 0.99799442 | 5 |
| BRCA2 | 16 | c.7795_7797delg | p.Glu2599del | 0.81 | 101 | 229.56 | – | 0.93 | 212.46 | – | 14 | P | 10,545,472.1 | 2,240,517,687 | 1 | 5 |
| BRCA2 | 17 | c.7975A>G | p.Arg2659Gly | 0.81 | 22 | 3.58 | – | 0.56 | 2.02 | 3 | 14 | P | 1,114.89 | 2,253.70 | 0.99989593 | 5 |
| BRCA2 | 18 | c.8009C>G | p.Ser2670Trp | 0.29 | 12 | 0.87 | – | 10.19 | 8.88 | 3 | 5 | LP | 819.97 | 7,283.30 | 0.99966396 | 5 |
| BRCA2 | 18 | c.8009C>T | p.Ser2670Leu | 0.29 | 15 | 8.47 | – | 0.87 | 7.33 | 3 | 5 | LP | 271.63 | 1,990.34 | 0.99877143 | 5 |
| BRCA2 | 18 | c.8057T>C | p.Leu2686Pro | 0.66 | 18 | 2.40 | – | 54.91 | 131.96 | – | 6 | VUS | 27.92 | 3,684.35 | 0.9998602 | 5 |
| BRCA2 | 23 | c.9004G>A | p.Glu3002Lys | 0.66 | 25 | 1.16 | – | 7.20 | 8.36 | 3 | 11 | P | 301.09 | 2,518.20 | 0.999795471 | 5 |
| BRCA2 | 24 | c.9154C>T | p.Arg3052Trp | 0.81 | 16 | 0.99 | – | 12.07 | 11.996 | – | 6 | P | 102.26 | 1,820.69 | 0.99987118 | 5c |
| BRCA2 | 25 | c.9371A>T | p.Asn3124Ile | 0.81 | 16 | 1.67 | – | 8.32 | 5.40 | – | 6 | P | 207.49 | 2,876.22 | 0.99991845 | 5c |
| BENIGN | ||||||||||||||||
| BRCA1 | 3 | c.92T>A | p.Ile31Asn | 0.66 | 8 | 0.23 | – | 0.004 | 0.0008 | 2 | 3 | VUS | 0.12 | 9.78E−5 | 0.000189741 | 1 |
| BRCA1 | 6 | c.243A>G | p.Gln81= | 0.02 | 18 | 0.03 | – | 0.3906892 | 0.0102 | – | 3 | B | 2.05213421 | 0.0209869 | 0.00042812 | 1c,d |
| BRCA1 | I6 | c.301+6T>C | – | 0.34 | 5 | 1.94 | 0.001776 | 0.30 | 0.01 | – | 2 | VUS | 1.6324848 | 0.00167665 | 0.00086298 | 1 |
| BRCA1 | 10 | c.670+8C>T | – | 0.5 | 11 | 0.32 | – | 0.0006 | 0.0002 | 1 | 4 | LB | 2.39 | 0.0005 | 0.000450112 | 1 |
| BRCA1 | 11 | c.693G>A | p.Thr231= | 0.02 | 20 | 0.45 | – | 0.35 | 0.16 | – | 3 | LB | 0.002 | 0.0003 | 5.8992E−06 | 1c |
| BRCA1 | 11 | c.1065G>A | p.Lys355= | 0.02 | 40 | 1.27 | 0.006087 | 0.0008 | 0.001 | 1 | 4 | LB | 0.09 | 5.31E−07 | 1.0831E−08 | 1c |
| BRCA1 | 11 | c.1242_1262del | p.Asp414_Asp420del | 0.02 | 3 | 1.82 | – | 0.08 | 0.15 | 3 | 2 | LP | 0.09 | 0.01 | 0.00027235 | 1 |
| BRCA1 | 11 | c.1384G>A | p.Gly462Arg | 0.02 | 9 | 0.22 | – | 0.002 | 0.0005 | 1 | 5 | VUS | 0.95 | 0.0005 | 9.5128E−06 | 1 |
| BRCA1 | 11 | c.1844C>T | p.Ser615Phe | 0.03 | 4 | 0.19 | – | 0.21 | 0.04 | – | 2 | VUS | 0.03 | 0.001 | 3.735E−05 | 1 |
| BRCA1 | 11 | c.2083G>T | p.Asp695Tyr | 0.03 | 45 | 0.31 | – | 0.0003 | 8.03E−05 | 1 | 13 | VUS | 9.36E−08 | 7.52E−12 | 2.325E−13 | 1 |
| BRCA1 | 11 | c.2662C>T | p.His888Tyr | 0.03 | 19 | 0.55 | – | 0.0004 | 0.0002 | 1 | 4 | VUS | 0.008 | 1.87E−06 | 5.7872E−08 | 1 |
| BRCA1 | 11 | c.2798G>A | p.Gly933Asp | 0.03 | 9 | 0.22 | – | 0.01 | 0.003 | 1 | 2 | VUS | 3.47 | 0.01 | 0.00034753 | 1 |
| BRCA1 | 11 | c.2884G>A | p.Glu962Lys | 0.03 | 9 | 0.07 | – | 0.05 | 0.003 | 1 | 2 | LB | 0.39 | 0.001 | 4.1619E−05 | 1c |
| BRCA1 | 11 | c.2935C>T | p.Arg979Cys | 0.02 | 16 | 0.31 | – | 0.0004 | 0.0001 | 1 | 3 | VUS | 3.22 | 0.0004 | 7.6066E−06 | 1 |
| BRCA1 | 11 | c.3327_3329del | p.Lys1110del | 0.02 | 5 | 0.06 | – | 0.21 | 0.012 | – | 2 | LP | 0.49 | 0.006 | 0.000122496 | 1 |
| BRCA1 | 11 | c.3708T>G | p.Asn1236Lys | 0.03 | 49 | 0.94 | 0.008 | 5.07E−05 | 4.78E−05 | 1 | 9 | VUS | 0.12 | 4.51E−08 | 1.3946E−09 | 1 |
| BRCA1 | 11 | c.3891_3893del | p.Ser1298del | 0.02 | 9 | 0.24 | – | 0.05 | 0.01 | 1 | 2 | LP | 1.28 | 0.01 | 0.00028881 | 1 |
| BRCA1 | 14 | c.4417T>C | p.Ser1473Pro | 0.03 | 20 | 0.14 | – | 0.003 | 0.000459893 | 1 | 2 | VUS | 1.195 | 0.00054971 | 1.7001E−05 | 1 |
| BRCA1 | I14 | c.4485-96A>G | – | 0.5 | 4 | 0.29 | – | 0.009 | 0.003 | 2 | 3 | VUS | 0.017 | 4.26E−05 | 4.2602E−05 | 1 |
| BRCA1 | 17 | c.4993G>A | p.Val1665Met | 0.29 | 20 | 0.42 | – | 0.16 | 0.07 | – | 5 | LB | 0.007 | 0.00050058 | 0.00020442 | 1 |
| BRCA1 | 17 | c.5005G>T | p.Ala1669Ser | 0.03 | 7 | 1.09 | – | 0.04 | 0.04 | – | 3 | VUS | 0.25 | 0.01 | 0.00033277 | 1 |
| BRCA1 | 17 | c.5071A>G | p.Thr1691Ala | 0.66 | 16 | 1.36 | – | 0.02 | 0.02 | – | 4 | LP | 0.13 | 0.003 | 0.006215 | 2 |
| BRCA1 | 18 | c.5117G>C | p.Gly1706Ala | 0.66 | 76 | 0.35 | – | 1.19E−05 | 4.14E−06 | 1 | 12 | VUS | 1.47E−06 | 6.09E−12 | 1.1816E−11 | 1c |
| BRCA1 | 24 | c.5531T>C | p.Leu1844Pro | 0.03 | 3 | 0.14 | – | 1.07 | 0.15 | – | 2 | VUS | 0.0001 | 1.91E−05 | 5.9194E−07 | 1 |
| BRCA1 | 24 | c.5531T>G | p.Leu1844Arg | 0.03 | 3 | 0.15 | – | 0.002 | 0.0003 | 1 | 2 | LB | 0.15 | 4.1067E−05 | 1.2701E−06 | 1c |
| BRCA2 | 3 | c.122C>T | p.Pro41Leu | 0.03 | 16 | 0.40 | – | 2.77 | 1.11 | 3 | 2 | VUS | 0.05 | 0.06 | 0.001881892 | 2 |
| BRCA2 | 3 | c.231T>G | p.Thr77= | 0.01 | 51 | 0.49 | 0.00002 | 0.79 | 0.38 | 1 | 5 | LB | 26.95 | 0.0002 | 1.7357E−06 | 1c |
| BRCA2 | 5 | c.433_435del | p.Val145del | 0.5 | 12 | 0.18 | – | 0.20 | 0.04 | – | 4 | VUS | 0.02835067 | 0.0010593 | 0.00105818 | 2 |
| BRCA2 | 10 | c.800G>A | p.Gly267Glu | 0.03 | 22 | 0.53 | 0.006 | 0.05 | 0.02 | 1 | 6 | VUS | 5.05 | 0.0007 | 2.1713E−05 | 1 |
| BRCA2 | 10 | c.831T>G | p.Asn277Lys | 0.02 | 21 | 0.42 | 0.006 | 0.81 | 0.34 | 1 | 4 | VUS | 0.03 | 5.83E−05 | 1.1898E−06 | 1 |
| BRCA2 | 10 | c.927A>Gh | p.Ser309= | 0.02 | 13 | 0.12 | – | 0.33 | 0.04 | – | 5 | LB | 0.18 | 0.007 | 0.000150187 | 1c |
| BRCA2 | 10 | c.1012G>A | p.Ala338Thr | 0.03 | 8 | 0.02 | – | 0.58 | 0.012 | – | 2 | LB | 0.33877369 | 0.00418389 | 0.00012938 | 1 |
| BRCA2 | 10 | c.1244A>G | p.His415Arg | 0.02 | 7 | 0.67 | – | 0.36 | 0.24 | – | 2 | VUS | 0.23 | 0.06 | 0.00115238 | 2 |
| BRCA2 | 10 | c.1564G>C | p.Gly522Arg | 0.03 | 9 | 0.32 | – | 0.44 | 0.14040988 | – | 3 | LB | 0.04183382 | 0.00587388 | 0.00018163 | 1 |
| BRCA2 | 10 | c.1798T>C | p.Tyr600His | 0.03 | 28 | 0.59 | – | 0.09 | 0.05 | – | 2 | LB | 0.13 | 0.007 | 0.00021097 | 1 |
| BRCA2 | 11 | c.2771A>T | p.Asn924Ile | 0.02 | 29 | 0.75 | – | 0.01 | 0.01 | 1 | 9 | VUS | 0.42 | 0.004 | 8.8819E−05 | 1 |
| BRCA2 | 11 | c.2944A>C | p.Ile982Leu | 0.03 | 18 | 0.50 | – | 0.099 | 0.05 | – | 3 | LB | 0.01205085 | 0.00059572 | 1.8424E−05 | 1 |
| BRCA2 | 11 | c.3262C>T | p.Pro1088Ser | 0.02 | 7 | 0.83 | – | – | 0.83 | – | 1 | VUS | 0.001 | 0.0008 | 1.6999E−05 | 1 |
| BRCA2 | 11 | c.5552T>G | p.Ile1851Ser | 0.03 | 13 | 1.24 | – | – | 1.24 | – | 7 | VUS | 0.02 | 0.02 | 0.00067659 | 1 |
| BRCA2 | 11 | c.5634C>G | p.Asn1878Lys | 0.03 | 22 | 0.62 | – | 0.31 | 0.19 | – | 4 | VUS | 0.03 | 0.006 | 0.00017755 | 1 |
| BRCA2 | 11 | c.5975C>T | p.Ser1992Leu | 0.29 | 21 | 0.28 | – | 0.53 | 0.15 | 3 | 3 | VUS | 0.16817314 | 0.02496567 | 0.01009431 | 2 |
| BRCA2 | 11 | c.6322C>T | p.Arg2108Cys | 0.02 | 41 | 0.97 | 0.00001 | 0.57 | 0.55 | – | 3 | LB | 5.96 | 0.03 | 0.00058336 | 1 |
| BRCA2 | I12 | c.6937+594T>G | – | 0.5 | 31 | 0.92 | 1.78E−8 | 0.06 | 1.06E−09 | 1 | 12 | LB | 115.71 | 1.23E−7 | 1.2263E−07 | 1c |
| BRCA2 | 14 | c.7057G>C | p.Gly2353Arg | 0.03 | 11 | 0.69 | – | 0.27 | 0.19 | – | 4 | VUS | 0.02 | 0.003 | 0.00010507 | 1 |
| BRCA2 | 14 | c.7219G>C | p.Val2407Leu | 0.03 | 5 | 0.16 | – | 1.44 | 0.23 | – | 3 | VUS | 0.20491405 | 0.04770388 | 0.0014732 | 2 |
| BRCA2 | 15 | c.7481G>A | p.Arg2494Gln | 0.66 | 7 | 0.19 | 0.0022 | 0.51 | 0.098 | – | 4 | VUS | 0.61 | 0.0001 | 0.00025681 | 1 |
| BRCA2 | 15 | c.7504C>T | p.Arg2502Cys | 0.03 | 22 | 0.61 | – | 0.19 | 0.12 | – | 3 | LB | 0.09 | 0.01 | 0.00031486 | 1 |
| BRCA2 | 16 | c.7759C>Th | p.Leu2587Phe | 0.29 | 13 | 0.12 | – | 0.33 | 0.04 | – | 5 | VUS | 0.18 | 0.007 | 0.0029973 | 2 |
| BRCA2 | 17 | c.7915C>G | p.Pro2639Ala | 0.29 | 6 | 0.15 | – | 0.61 | 0.09 | – | 3 | VUS | 0.03 | 0.003 | 0.00123378 | 2 |
| BRCA2 | 17 | c.7928C>G | p.Ala2643Gly | 0.64 | 33 | 0.58 | 0.0076 | 0.03 | 0.02 | – | 9 | VUS | 0.01 | 1.68E−06 | 2.9926E−06 | 1 |
| BRCA2 | 17 | c.7933A>G | p.Arg2645Gly | 0.29 | 16 | 0.24 | – | 0.05 | 0.01 | 2 | 7 | VUS | 2.93 | 0.03 | 0.01329559 | 2 |
| BRCA2 | 17 | c.7954G>A | p.Val2652Met | 0.29 | 27 | 0.52 | 0.005 | 0.76 | 0.002 | 1 | 4 | VUS | 1.29609452 | 0.00251463 | 0.00102605 | 2 |
| BRCA2 | 18 | c.7992T>A | p.Ile2664= | 0.02 | 8 | 0.82 | – | 0.35 | 0.28 | – | 3 | LB | 0.00753887 | 0.00214733 | 4.3821E−05 | 1c |
| BRCA2 | 18 | c.8010G>A | p.Ser2670= | 0.02 | 13 | 0.33 | – | 1.91 | 0.62 | – | 2 | LB | 0.097 | 0.06 | 0.00123121 | 2c |
| BRCA2 | 18 | c.8084C>T | p.Ser2695Leu | 0.03 | 14 | 0.43 | 0.003 | 0.10 | 0.0001 | – | 4 | VUS | 0.11852903 | 1.6682E−05 | 5.1595E−07 | 1 |
| BRCA2 | 18 | c.8111C>T | p.Ser2704Phe | 0.03 | 7 | 0.64 | – | 0.13 | 0.08 | – | 3 | VUS | 0.04636364 | 0.00387602 | 0.00011986 | 1 |
| BRCA2 | 23 | c.9104A>C | p.Tyr3035Ser | 0.66 | 31 | 0.64 | 0.007 | 0.32 | 0.20 | 2 | 9 | VUS | 0.33 | 0.0005 | 0.00092147 | 1 |
| BRCA2 | 23 | c.9116C>T | p.Pro3039Leu | 0.03 | 24 | 0.4 | – | 0.02 | 0.00754 | 1 | 7 | VUS | 0.55 | 0.004 | 0.00012765 | 1 |
| BRCA2 | 24 | c.9206G>T | p.Cys3069Phe | 0.03 | 3 | 0.07 | – | 0.59 | 0.04 | – | 3 | VUS | 0.03255207 | 0.00143349 | 4.4333E−05 | 1 |
| BRCA2 | 25 | c.9275A>G | p.Tyr3092Cys | 0.81 | 30 | 0.36 | 0.008 | 2.001 | 0.0056 | 2 | 9 | VUS | 1.44 | 0.008 | 0.03490992 | 2 |
| BRCA2 | 25 | c.9501+3A>T | – | 0.34 | 30 | 0.49 | – | 0.20 | 0.09920669 | – | 7 | VUS | 0.001 | 0.0001 | 7.3072E−05 | 1 |
| BRCA2 | 26 | c.9583A>G | p.Thr3195Ala | 0.03 | 26 | 0.42 | 0.005 | 0.16 | 0.0003 | 1 | 5 | LB | 0.26 | 7.6865E−05 | 2.3773E−06 | 1 |
| BRCA2 | 26 | c.9606G>C | p.Pro3202= | 0.02 | 21 | 0.40 | – | 0.04 | 0.015 | – | 3 | B | 0.26 | 0.0037923 | 7.7388E−05 | 1c |
Classifications without or with cosegregation data are in bold. See Table S3 for details of ClinVar, BRCAexchange and ACMG classification.
Dash (–) means that the posterior probability of pathogenicity was not calculated because the combined LR was not <0.01 or >1,000.
This variant was also classified by ACMG using Varsome.
This variant was also classified by BRCAexchange.
This variant was also classified by ClinVar.
BRCA1 variants leading to p.Gly1770Val were combined.
BRCA2 variants leading to complete loss of exon were combined.
This pathogenic variant probably has a French origin because we find it in many families, particularly in the North of France.
Both variants were always observed altogether. Their cosegregation in 4 meiosis supports that they are in cis.
Figure 4.
Classified variants type and in silico prediction
(A) Classified variants according to the type of variation.
(B) Classified variants according to the in silico predicted protein effect: Align-GVGD, SIFT, or PolyPhen-2.
Variant classification and pathogenicity prediction tools
A good concordance was observed between the Align-GVGD prediction tool for extreme classes and the COVAR study classification concerning amino acid changes, i.e., most C65 variants were classified as pathogenic and C0 variants as benign (Figure 4B). However, six variants with a predicted pathogenic effect ranging from Align-GVGD class C35 to C65 were demonstrated to be benign or likely benign: BRCA1 c.92T>A (p.Ile31Asn) located in the RING domain and c.5071A>G (p.Thr1691Ala) and c.5117G>C (p.Gly1706Ala) located in the BRCT1 domain; BRCA2 c.7481G>A (p.Arg2494Gln) located in the Helical domain and c.9104A>C (p.Tyr3035Ser) and c.9275A>G (p.Tyr3092Cys) located in the oligonucleotide binding 2 and 3 (OB2/3) domains, respectively. Conversely, nine variants with Align-GVGD class C0 to C25 were demonstrated to be pathogenic: BRCA1 c.4963T>G (p.Ser1655Ala) and c.5062G>T (p.Val1688Phe) located in the BRCT1 domain, c.5254G>C (p.Ala1752Pro) and c.5255C>A (p.Ala1752Glu) located in the linker between BRCT1 and BRCT2 domains, c.5309G>T (p.Gly1770Val), c.5309_5310delinsTT (p.Gly1770Val), and c.5434C>G (p.Pro1812Ala) located in the BRCT2 domain; BRCA2 c.8009C>T (p.Ser2670Leu) and c.8009C>G (p.Ser2670Trp) located in the OB1 domain. Poor concordance was observed between the COVAR study classification and SIFT and PolyPhen-2 prediction tools, especially for variants with a predicted pathogenic effect, almost half of which were classified by the COVAR study as BVs or LBVs (Figure 4B).
Specific variants
The BRCA2 c.6937+594T>G deep intronic variant previously reported to be associated with a cryptic exon inclusion between exons 12 and 13 was classified as a BV, like the BRCA2 c.9501+3A>T variant leading to partial exon 25 skipping.
Some variants were classified as pathogenic by combining pathogenicity likelihoods, as they have the same putative protein effect: BRCA1 c.5309G>T and c.5309_5310delinsTT that both result in Gly 1770 to Val substitution (a splice defect was excluded for both of these variants); BRCA2 c.68−?_316+?del (corresponding to exon 3 deletion), c.156_157insAlu, c.316+1G>T, c.316+2T>C, c.316+4del, c.316+5G>A, and c.316+5G>G that all lead to the complete loss of the in-frame exon 3.
Some variants located in the same codon but resulting in different amino acid changes were classified in different classes, e.g., three variants in BRCA1 1706 codon located in BRCT1 domain: BRCA1 c.5116G>A (p.Gly1706Arg), c.5117G>A (p.Gly1706Glu), and c.5117G>C (p.Gly1706Ala) were classified as PV, LPV, and BV, respectively.
Two BRCA2 variants were systematically identified in cis: BRCA2 c.927A>G (p.Ser309=) and c.7759C>T (p.Leu2587Phe), classified as BV and LBV, respectively.
Comparison of COVAR and ACMG/AMP classification
We compared the COVAR-classification results with ACMG/AMP scoring by using two bioinformatics tools, Varsome and InterVar (Tables 1 and S3).42,43 All variants could be compared with Varsome, but only 82 of the 101 COVAR classified variants could be assessed with InterVar, as InterVar does not process all variant types (indels, LGRs, intronic variants). Among the 34 variants classified as PV by the COVAR study, only 4 (11.8%) variants were classified in this category by Varsome and none by InterVar. As cosegregation data can be included in Varsome, we subsequently compared our results for the 34 COVAR-classified PVs with Varsome integrating cosegregation data. The number of Varsome-classified PVs increased from 4 (11.8%) to 18 (52.9% of the COVAR-classified PVs). The other COVAR-classified PVs were classified as LPV by Varsome or InterVar or remained VUSs. At the other extreme, among the 51 variants classified as benign by the COVAR study, two were classified similarly by Varsome, and only one was classified as benign by InterVar. The majority of the other COVAR-classified BVs remained VUSs or LBVs with both tools. In addition, it should be noted that nine COVAR-classified BVs were classified as LPVs by Varsome or Intervar.
Discussion
COVAR classification or the power of cosegregation analysis
Among the 653 BRCA1/2 variants selected for the COVAR study, 101 (15%) variants identified in 1,624 families had been classified at the time of this first report; 38% were classified as PVs or LPVs and 62% were classified as BVs or LBVs. Most of the selected and classified VUSs were missense variants, but other variant types were also classified: in-frame indels, LGRs, and synonymous and intronic variants. Among the 101 classified variants, 5 had never been previously reported, 13 were new for ClinVar and not reviewed in BRCAexchange, and 79 (78.2%) were present but not reviewed in the BRCAexchange database. Twenty-three (32.7%) and 59 (58.4%) variants were also reported in ClinVar as VUSs or “conflicting interpretation,” respectively (Table S3).
A very wide range of cosegregation LR values was observed (1.4E+23 to 9.4E−08), illustrating the power of cosegregation analyzes in variant classification provided that sufficient data have been collected. No other measurement of association between VUSs and cancer was more powerful in our dataset (Tables 1 and S3). Indeed, 66 of the 101 (65%) variants classified by COVAR would have remained VUSs without cosegregation data.
Specific variants
Combining data for different genetic variants with a common protein impact allows the classification of a greater number of variants by increasing the statistical power of cosegregation. However, mRNA analyses must be performed before combining variant data in order to ensure that all variants have a similar splicing profile. We confirmed that the two BRCA1 variants leading to the amino acid Gly1770 to Val substitution did not result in a splicing defect. Moreover, in order to classify the BRCA2 exon 3 deletion, we combined only BRCA2 variants leading to complete loss of exon 3.44
Conversely, variants located on the same codon but leading to different amino acid changes were classified as pathogenic or benign, e.g., BRCA1 variants located on codons 1691 or 1706. The class of each BRCA1 1691 or 1706 codon variant was concordant with previously published functional assays.45, 46, 47, 48, 49 Similarly, there was a good correlation between our classification of BRCA2 missense variants and functional assay results when available.50, 51, 52
Prior probability
Although cosegregation analysis is a powerful variant classification tool, the multifactorial model of pathogenicity that includes cosegregation data may have a number of limitations, especially the major influence of PriorP on posterior probability, as the better concordance observed between our classification and Align-GVGD prediction than SIFT or PolyPhen-2 prediction could be explained by the high weight of PriorP, because the Align-GVGD prediction score was used to determine PriorP. However, interestingly, some missense variants predicted as pathogenic by Align-GVGD were classified as benign and some variants predicted as benign were classified as pathogenic after cosegregation analysis. Lindor et al. pointed out that PriorP should not be used alone but combined with other arguments such as cosegregation, in a multifactorial model.18 Re-evaluation of these PriorP is ongoing.41
An example of the impact of PriorP on variant class is the observation for the two variants systematically identified in cis, BRCA2 c.927A>G (p.Ser309=) and c.7759C>T (p.Leu2587Phe), classified as BV and LBV, respectively. As all data and consequently all evidence of pathogenicity for these two variants are identical (since they always occur together), the only difference in posterior pathogenicity probability calculation for these variants was their PriorP (C0 versus C15).
Combined LR thresholds to estimate posterior probability
Combined LR thresholds used for classification of variants were those defined by Goldgar et al. in 2004,17 a combined LR greater than 1,000:1 for PV and lower than 1:100 for BV. The calculation of posterior probability was not performed when combined LR were between these thresholds, to ensure a sufficient level of evidence from observational data. Recent studies used the multifactorial model with prior probability truncated at 0.10 and 0.90 for minimum and maximum prior probability, respectively, which leads to combined LR lower than 0.5 or greater than 2 to reach class 2 or class 4, respectively; in these studies the estimation of posterior probability was not performed only for combined LR between 0.5 and 2.11,21 We could have classified 100 additional variants toward causality or neutrality if posterior probability had been calculated outside the range of 0.5–2 instead of the range of 1:100–1,000:1. However, it may lead to variant misclassification because of a low level of observational data and an increased weight of prior probability. This misclassification, especially in the PV or LPV classes, can have clinical consequences leading to inappropriate prophylactic surgery decisions for carriers and unsafe surveillance discontinuation for non-carriers.7,53
Multifactorial model parameters
A last limitation is that the model does not consider the possibility of de novo variants and may therefore classify a variant as benign because of the absence of a family history of cancers although it is pathogenic. However, this constitutes a minor limitation, as the BRCA1/2 de novo variant rate is estimated to be only 0.3% [0.1%; 0.7%] and de novo status should be detected by genetic testing of both parents when available.54
Note that the scoring for cosegregation data used in this study was designed for high penetrance variants. Some BRCA1/2 PVs with moderate penetrance could remain classified as VUS. This class uncertainty for moderate penetrance variants has been previously reported for the BRCA1 c.5096G>A (p.Arg1699Gln) PV, which was demonstrated to be associated with intermediate breast and ovarian cancer risks on the basis of the large volume of cosegregation data collected: using the standard multifactorial model for high penetrance variants, the BRCA1 c.5096G>A (p.Arg1699Gln) would have remained classified as a VUS; Spurdle et al. integrated in the model a lower level of BRCA1 variant penetrance so this variant could be classified as pathogenic.55,56 Adaptation of the cosegregation statistical model developed by Thompson et al., modifying the value of cancer risks and introducing polygenic risk scores for breast cancer risk and subsequent ovarian cancer risk, should improve the classification of variants with moderate penetrance.57 Adaptation of the statistical model will be no doubt required for VUSs of genes such as CHEK2 and ATM that are associated with moderate risk. The multifactorial likelihood model of pathogenicity could also be improved by incorporating a complementary LR to the LR-pathology based on tumor status of a germline variant, as somatic loss of the wild-type allele would suggest pathogenicity and conversely loss of variant non-pathogenicity (S.M.C., E.R., and D.S.-L., unpublished data).58 A parameter called somatic to germline ratio (SGR) has recently been used for TP53 variant classification.59 Increasing indications for BRCA1/2 gene analyses of tumors in order to guide PARPi therapy, thereby increasing the information regarding the wild-type allele, will allow the introduction of this information into the likelihood model.
ACMG/AMP classification
In 2015, the ACMG/AMP published general guidelines for gene variant classification based on various types of evidence, including population data, bioinformatics prediction tools, functional data and, when available, cosegregation data.60 We compared our results to the ACMG/AMP classification using Varsome and Intervar, the bioinformatics tools that were developed to standardize scoring. Many discrepancies were observed between our classification and the ACMG/AMP classification and between variant scoring by the two tools. This observation was expected as recent studies already showed discrepancies between classification based on the multifactorial model used in our study and ACMG/AMP classification.61,62 Most variants classified as pathogenic in our study remained classified as LP or VUSs while most BVs remained LB or VUSs. The greatest classification discrepancy concerned the 12 COVAR-classified BVs that were classified as LPVs by Varsome or InterVar. Three of these 12 variants were in-frame amino acid deletions classified as LPVs by Varsome. The weight applied to this variant type may be excessive in the ACMG/AMP scoring. Six other COVAR-classified BVs were also classified as LPVs by InterVar because it took into account reporting of the variant as pathogenic in public databases with no available evidence of pathogenicity as a supporting element for pathogenicity. Uncurated public database classification should not be used as a classification criterion to avoid misclassification based on insufficient data. Counting meiosis has been proposed for ranking of supporting evidence of pathogenicity based on cosegregation data in the ACMG/AMP classification.60,63 It allowed us to input cosegregation data in the Varsome-tool after counting meiosis, resulting in better concordance for our 34 classified PVs, increasing the number of Varsome-classified PVs from 4 (11.8%) to 18 (52.9%). We used the same levels of cosegregation as proposed by Jarvik and Browning in 201663 to consider cosegregation data as supporting, moderate, or strong evidence of pathogenicity for ACMG/AMP classification. We could not include cosegregation data for comparison of BV classification, as counting meiosis is unable to generate any evidence in favor of a benign effect.64 The ACMG/AMP scoring system is globally based on discrete variables with somewhat subjective ranking of evidence. Cosegregation data computed in a quantitative multifactorial model, as performed in this study, allow more robust classification.
Functional assays
A large-scale functional assay based on cell survival after BRCA1 disruption has been recently developed to classify nearly 4,000 known or possible BRCA1 variants located in RING and BRCT domains.47 All variants were generated by saturation genome editing with CRISPR-Cas9 system in human haploid cell lines, allowing an identical genomic background for all tested variants. This functional assay is very promising: it gives accurate results for a large number of variants. However, because the reporting test is cell survival, a surrogate marker of the BRCA1 role in DNA repair by homologous recombination, other BRCA1-altered functions could be missed. Moreover, in vitro data alone should not be used as the basis for medical advice until they have been clinically validated.65 Such functional assays can be used to prioritize variants selected for clinical and genetic studies such as the COVAR study. A “LR functional assay” would be helpful to implement functional data in the multifactorial model but there is no international consensus yet on which assays and which weights should be used.
Artificial intelligence
Finally, artificial intelligence (AI) is being developed for more and more clinical applications including genetic variant classification. At the present time, algorithms are being developed to integrate literature data, gene and protein structure, and species alignment (Corona-AI, REVEL,…). Using machine-learning on the ClinVar BRCA1/2 variant database, Favalli et al. developed an interesting variant classification algorithm. However, AI variant classification still requires critical examination by geneticists, especially with regard to the quality of the data on which AI tools are trained.66
Conclusion
In conclusion, the first series of results of the COVAR study show that cosegregation is a highly powerful tool for BRCA1/2 variant classification. Cosegregation studies, i.e., linkage studies, for VUS classification may be as useful as they were 25 years ago for the identification of a large of disease-related genes, including BRCA1/2. However, in order to be feasible, such studies must be organized by clinical and laboratory networks at a nationwide or even worldwide level such as ENIGMA. Although the initiation and organization of these studies requires considerable effort, the growing number of available genetic tests results in an increasing number of families sharing a particular variant, thereby increasing their power. Here we demonstrate that variant cosegregation studies, i.e., “linkage studies 2.0” for reference to the linkage studies which led to the localization of the BRCA1 and BRCA2 genes 30 years ago,67,68 are a powerful tool for the classification of variants in the BRCA1/2 breast-ovarian cancer predisposition genes.
Data availability
The dataset in support of the current study has not been deposited in a public repository because it is composed of patient data including pedigrees which are indirectly identifying. The data are available on request from the corresponding author. A large part of the variants have been deposited in BRCAShare (http://www.umd.be/), and all variants will be annotated in the upcoming FrOG (French OncoGenetics) database, as listed in Table S4 and all material and data supporting this study is included within the manuscript (or by request to the authors if this is the case).
Acknowledgments
This article is dedicated to the memory of Olga Sinilnikova.
S.M.C., L. Golmard, M. Léone, F.D., M. Guillaud-Bataille, F.R., E.R., N.D., A. Buisson, N. Basset, M.S., P.V., C. Garrec, M.P., M. Gay-Bellile, T.N.N.T., O. Caron, C. Lasset, A. Remenieras, N.B.-K., and L. Castéra are members of the variant classification working group. The authors thank the clinicians and biologists (Table S6) involved in the UGG. This work was funded by grants from the French National Cancer Institute (2013-1-BCB-01-531_ICH-1) and the French Ligue contre le cancer 92. The COVAR study is supported by the GENETICANCER and “Cercle Olympe” associations and AstraZeneca.
Declaration of interests
D.S.-L. and the Institut Curie have received honoraria for her participation in education meetings organized by AstraZeneca or Tesaro. The remaining authors declare no conflict of interest.
Published: September 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.09.003.
Web resources
BRCAShare, http://www.umd.be/
French Unicancer Genetics Group (UGG) of cancer genetics clinic and laboratory network, http://www.unicancer.fr/la-recherche-unicancer/les-groupes-transversaux/groupe-genetique-cancer-ggc
International clinical trials platform, https://clinicaltrials.gov/ct2/show/NCT01689584
OMIM, https://www.omim.org/
Supplemental Information
References
- 1.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., BRCA1 and BRCA2 Cohort Consortium Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 2.Lesueur F., Mebirouk N., Jiao Y., Barjhoux L., Belotti M., Laurent M., Léone M., Houdayer C., Bressac-de Paillerets B., Vaur D., GEMO Study Collaborators GEMO, a National Resource to Study Genetic Modifiers of Breast and Ovarian Cancer Risk in BRCA1 and BRCA2 Pathogenic Variant Carriers. Front. Oncol. 2018;8:490. doi: 10.3389/fonc.2018.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebbeck T.R., Friebel T.M., Friedman E., Hamann U., Huo D., Kwong A., Olah E., Olopade O.I., Solano A.R., Teo S.-H., EMBRACE. GEMO Study Collaborators. HEBON Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018;39:593–620. doi: 10.1002/humu.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouleau E., Jesson B., Briaux A., Nogues C., Chabaud V., Demange L., Sokolowska J., Coulet F., Barouk-Simonet E., Bignon Y.J. Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res. Treat. 2012;133:1179–1190. doi: 10.1007/s10549-012-2009-5. [DOI] [PubMed] [Google Scholar]
- 5.Caputo S., Benboudjema L., Sinilnikova O., Rouleau E., Béroud C., Lidereau R., French BRCA GGC Consortium Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40:D992–D1002. doi: 10.1093/nar/gkr1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputo S.M., Telly D., Briaux A., Sesen J., Ceppi M., Bonnet F., Bourdon V., Coulet F., Castera L., Delnatte C., French Covar Group Collaborators 5¢ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints. Cancers (Basel) 2021;13:3171. doi: 10.3390/cancers13133171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Hoya M., Soukarieh O., López-Perolio I., Vega A., Walker L.C., van Ierland Y., Baralle D., Santamariña M., Lattimore V., Wijnen J. Combined genetic and splicing analysis of BRCA1 c.[594-2A>C; 641A>G] highlights the relevance of naturally occurring in-frame transcripts for developing disease gene variant classification algorithms. Hum. Mol. Genet. 2016;25:2256–2268. doi: 10.1093/hmg/ddw094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meulemans L., Mesman R.L.S., Caputo S.M., Krieger S., Guillaud-Bataille M., Caux-Moncoutier V., Léone M., Boutry-Kryza N., Sokolowska J., Révillion F. Skipping nonsense to maintain function: the paradigm of BRCA2 exon 12. Cancer Res. 2020;80:1374–1386. doi: 10.1158/0008-5472.CAN-19-2491. [DOI] [PubMed] [Google Scholar]
- 9.Tubeuf H., Caputo S.M., Sullivan T., Rondeaux J., Krieger S., Caux-Moncoutier V., Hauchard J., Castelain G., Fiévet A., Meulemans L. Calibration of Pathogenicity Due to Variant-Induced Leaky Splicing Defects by Using BRCA2 Exon 3 as a Model System. Cancer Res. 2020;80:3593–3605. doi: 10.1158/0008-5472.CAN-20-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller D., Rouleau E., Schultz I., Caputo S., Lefol C., Bièche I., Caron O., Noguès C., Limacher J.M., Demange L. An entire exon 3 germ-line rearrangement in the BRCA2 gene: pathogenic relevance of exon 3 deletion in breast cancer predisposition. BMC Med. Genet. 2011;12:121. doi: 10.1186/1471-2350-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons M.T., Tudini E., Li H., Hahnen E., Wappenschmidt B., Feliubadaló L., Aalfs C.M., Agata S., Aittomäki K., Alducci E., KConFab Investigators Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019;40:1557–1578. doi: 10.1002/humu.23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spurdle A.B., Healey S., Devereau A., Hogervorst F.B., Monteiro A.N., Nathanson K.L., Radice P., Stoppa-Lyonnet D., Tavtigian S., Wappenschmidt B., ENIGMA ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat. 2012;33:2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couch F.J., Nathanson K.L., Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343:1466–1470. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colas C., Golmard L., de Pauw A., Caputo S.M., Stoppa-Lyonnet D. “Decoding hereditary breast cancer” benefits and questions from multigene panel testing. Breast. 2019;45:29–35. doi: 10.1016/j.breast.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Cline M.S., Liao R.G., Parsons M.T., Paten B., Alquaddoomi F., Antoniou A., Baxter S., Brody L., Cook-Deegan R., Coffin A., BRCA Challenge Authors BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752. doi: 10.1371/journal.pgen.1007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian A.W., Li Y., Hamilton A.S., Ward K.C., Hawley S.T., Morrow M., McLeod M.C., Jagsi R., Katz S.J. Gaps in Incorporating Germline Genetic Testing Into Treatment Decision-Making for Early-Stage Breast Cancer. J. Clin. Oncol. 2017;35:2232–2239. doi: 10.1200/JCO.2016.71.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldgar D.E., Easton D.F., Deffenbaugh A.M., Monteiro A.N.A., Tavtigian S.V., Couch F.J., Breast Cancer Information Core (BIC) Steering Committee Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindor N.M., Guidugli L., Wang X., Vallée M.P., Monteiro A.N.A., Tavtigian S., Goldgar D.E., Couch F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum. Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson D., Easton D.F., Goldgar D.E. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 2003;73:652–655. doi: 10.1086/378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavtigian S.V., Byrnes G.B., Goldgar D.E., Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008;29:1342–1354. doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallée M.P., Di Sera T.L., Nix D.A., Paquette A.M., Parsons M.T., Bell R., Hoffman A., Hogervorst F.B.L., Goldgar D.E., Spurdle A.B., Tavtigian S.V. Adding In Silico Assessment of Potential Splice Aberration to the Integrated Evaluation of BRCA Gene Unclassified Variants. Hum. Mutat. 2016;37:627–639. doi: 10.1002/humu.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B.L., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V., IARC Unclassified Genetic Variants Working Group Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazoyer S., Lalle P., Narod S.A., Bignon Y.J., Courjal F., Jamot B., Dutrillaux B., Stoppa-Lyonnett D., Sobol H. Linkage analysis of 19 French breast cancer families, with five chromosome 17q markers. Am. J. Hum. Genet. 1993;52:754–760. [PMC free article] [PubMed] [Google Scholar]
- 24.Eisinger F., Bressac B., Castaigne D., Cottu P.-H., Lansac J., Lefranc J.-P., Lesur A., Noguès C., Pierret J., Puy-Pernias S. [Identification and management of hereditary breast-ovarian cancers (2004 update)] Pathol. Biol. (Paris) 2006;54:230–250. doi: 10.1016/j.patbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Sinilnikova O.M., Dondon M.-G., Eon-Marchais S., Damiola F., Barjhoux L., Marcou M., Verny-Pierre C., Sornin V., Toulemonde L., Beauvallet J. GENESIS: a French national resource to study the missing heritability of breast cancer. BMC Cancer. 2016;16:13. doi: 10.1186/s12885-015-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretta J., Berthet P., Bonadona V., Caron O., Cohen-Haguenauer O., Colas C., Corsini C., Cusin V., De Pauw A., Delnatte C. [The French Genetic and Cancer Consortium guidelines for multigene panel analysis in hereditary breast and ovarian cancer predisposition] Bull Cancer. 2018;105:907–917. doi: 10.1016/j.bulcan.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Girard E., Eon-Marchais S., Olaso R., Renault A.-L., Damiola F., Dondon M.-G., Barjhoux L., Goidin D., Meyer V., Le Gal D. Familial breast cancer and DNA repair genes: Insights into known and novel susceptibility genes from the GENESIS study, and implications for multigene panel testing. Int. J. Cancer. 2019;144:1962–1974. doi: 10.1002/ijc.31921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao Y., Lesueur F., Azencott C.-A., Laurent M., Mebirouk N., Laborde L., Beauvallet J., Dondon M.-G., Eon-Marchais S., Laugé A., GEMO Study Collaborators. GENEPSO Study Collaborators A new hybrid record linkage process to make epidemiological databases interoperable: application to the GEMO and GENEPSO studies involving BRCA1 and BRCA2 mutation carriers. BMC Med. Res. Methodol. 2021;21:155. doi: 10.1186/s12874-021-01299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Institut National Du Cancer . 2020. Oncogénétique en 2018 - Consultations, laboratoires et suivi (2017 et 2018) [Google Scholar]
- 30.Béroud C., Letovsky S.I., Braastad C.D., Caputo S.M., Beaudoux O., Bignon Y.J., Bressac-De Paillerets B., Bronner M., Buell C.M., Collod-Béroud G., Laboratory Corporation of America Variant Classification Group. Quest Diagnostics Variant Classification Group. UNICANCER Genetic Group BRCA Laboratory Network BRCA Share: A Collection of Clinical BRCA Gene Variants. Hum. Mutat. 2016;37:1318–1328. doi: 10.1002/humu.23113. [DOI] [PubMed] [Google Scholar]
- 31.Tavtigian S.V., Byrnes G.B., Goldgar D.E., Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008;29:1342–1354. doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Easton D.F., Pharoah P.D.P., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocci E., Milne R.L., Méndez-Villamil E.Y., Hopper J.L., John E.M., Andrulis I.L., Chung W.K., Daly M., Buys S.S., Malats N., Goldgar D.E. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol. Biomarkers Prev. 2013;22:803–811. doi: 10.1158/1055-9965.EPI-12-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoniou A.C., Cunningham A.P., Peto J., Evans D.G., Lalloo F., Narod S.A., Risch H.A., Eyfjord J.E., Hopper J.L., Southey M.C. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br. J. Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaildrat P., Krieger S., Théry J.-C., Killian A., Rousselin A., Berthet P., Frébourg T., Hardouin A., Martins A., Tosi M. The BRCA1 c.5434C->G (p.Pro1812Ala) variant induces a deleterious exon 23 skipping by affecting exonic splicing regulatory elements. J. Med. Genet. 2010;47:398–403. doi: 10.1136/jmg.2009.074047. [DOI] [PubMed] [Google Scholar]
- 37.Anczuków O., Buisson M., Léoné M., Coutanson C., Lasset C., Calender A., Sinilnikova O.M., Mazoyer S. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin. Cancer Res. 2012;18:4903–4909. doi: 10.1158/1078-0432.CCR-12-1100. [DOI] [PubMed] [Google Scholar]
- 38.Spurdle A.B., Couch F.J., Parsons M.T., McGuffog L., Barrowdale D., Bolla M.K., Wang Q., Healey S., Schmutzler R., Wappenschmidt B., ABCTB Investigators. EMBRACE Group. GENICA Network. HEBON Group. kConFab Investigators Refined histopathological predictors of BRCA1 and BRCA2 mutation status: a large-scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res. 2014;16:3419. doi: 10.1186/s13058-014-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belman S., Parsons M.T., Spurdle A.B., Goldgar D.E., Feng B.-J. Considerations in assessing germline variant pathogenicity using cosegregation analysis. Genet. Med. 2020;22:2052–2059. doi: 10.1038/s41436-020-0920-4. [DOI] [PubMed] [Google Scholar]
- 40.Easton D.F., Deffenbaugh A.M., Pruss D., Frye C., Wenstrup R.J., Allen-Brady K., Tavtigian S.V., Monteiro A.N.A., Iversen E.S., Couch F.J., Goldgar D.E. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., LaDuca H., Pesaran T., Chao E.C., Dolinsky J.S., Parsons M., Spurdle A.B., Polley E.C., Shimelis H., Hart S.N. Classification of variants of uncertain significance in BRCA1 and BRCA2 using personal and family history of cancer from individuals in a large hereditary cancer multigene panel testing cohort. Genet. Med. 2019;22:701–708. doi: 10.1038/s41436-019-0729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R., Massouras A. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caputo S.M., Léone M., Damiola F., Ehlen A., Carreira A., Gaidrat P., Martins A., Brandão R.D., Peixoto A., Vega A., French COVAR group collaborators Full in-frame exon 3 skipping of BRCA2 confers high risk of breast and/or ovarian cancer. Oncotarget. 2018;9:17334–17348. doi: 10.18632/oncotarget.24671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millot G.A., Carvalho M.A., Caputo S.M., Vreeswijk M.P.G., Brown M.A., Webb M., Rouleau E., Neuhausen S.L., Hansen Tv., Galli A., ENIGMA Consortium Functional Assay Working Group A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 2012;33:1526–1537. doi: 10.1002/humu.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thouvenot P., Ben Yamin B., Fourrière L., Lescure A., Boudier T., Del Nery E., Chauchereau A., Goldgar D.E., Houdayer C., Stoppa-Lyonnet D. Functional Assessment of Genetic Variants with Outcomes Adapted to Clinical Decision-Making. PLoS Genet. 2016;12:e1006096. doi: 10.1371/journal.pgen.1006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandes V.C., Golubeva V.A., Di Pietro G., Shields C., Amankwah K., Nepomuceno T.C., de Gregoriis G., Abreu R.B.V., Harro C., Gomes T.T. Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation. J. Biol. Chem. 2019;294:5980–5992. doi: 10.1074/jbc.RA118.005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petitalot A., Dardillac E., Jacquet E., Nhiri N., Guirouilh-Barbat J., Julien P., Bouazzaoui I., Bonte D., Feunteun J., Schnell J.A., UNICANCER Genetic Group BRCA network Combining Homologous Recombination and Phosphopeptide-binding Data to Predict the Impact of BRCA1 BRCT Variants on Cancer Risk. Mol. Cancer Res. 2019;17:54–69. doi: 10.1158/1541-7786.MCR-17-0357. [DOI] [PubMed] [Google Scholar]
- 50.Biswas K., Das R., Eggington J.M., Qiao H., North S.L., Stauffer S., Burkett S.S., Martin B.K., Southon E., Sizemore S.C. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum. Mol. Genet. 2012;21:3993–4006. doi: 10.1093/hmg/dds222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mesman R.L.S., Calléja F.M.G.R., Hendriks G., Morolli B., Misovic B., Devilee P., van Asperen C.J., Vrieling H., Vreeswijk M.P.G. The functional impact of variants of uncertain significance in BRCA2. Genet. Med. 2019;21:293–302. doi: 10.1038/s41436-018-0052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson M.E., Hu C., Lee K.Y., LaDuca H., Fulk K., Durda K.M., Deckman A.M., Goldgar D.E., Monteiro A.N.A., Gnanaolivu R. Strong functional data for pathogenicity or neutrality classify BRCA2 DNA-binding-domain variants of uncertain significance. Am. J. Hum. Genet. 2021;108:458–468. doi: 10.1016/j.ajhg.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mighton C., Charames G.S., Wang M., Zakoor K.-R., Wong A., Shickh S., Watkins N., Lebo M.S., Bombard Y., Lerner-Ellis J. Variant classification changes over time in BRCA1 and BRCA2. Genet. Med. 2019;21:2248–2254. doi: 10.1038/s41436-019-0493-2. [DOI] [PubMed] [Google Scholar]
- 54.Golmard L., Delnatte C., Laugé A., Moncoutier V., Lefol C., Abidallah K., Tenreiro H., Copigny F., Giraudeau M., Guy C. Breast and ovarian cancer predisposition due to de novo BRCA1 and BRCA2 mutations. Oncogene. 2016;35:1324–1327. doi: 10.1038/onc.2015.181. [DOI] [PubMed] [Google Scholar]
- 55.Spurdle A.B., Whiley P.J., Thompson B., Feng B., Healey S., Brown M.A., Pettigrew C., Van Asperen C.J., Ausems M.G., Kattentidt-Mouravieva A.A., kConFab. Dutch Belgium UV Consortium. German Consortium of Hereditary Breast and Ovarian Cancer. French COVAR group collaborators. ENIGMA Consortium BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J. Med. Genet. 2012;49:525–532. doi: 10.1136/jmedgenet-2012-101037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moghadasi S., Meeks H.D., Vreeswijk M.P., Janssen L.A., Borg Å., Ehrencrona H., Paulsson-Karlsson Y., Wappenschmidt B., Engel C., Gehrig A. The BRCA1 c. 5096G>A p.Arg1699Gln (R1699Q) intermediate risk variant: breast and ovarian cancer risk estimation and recommendations for clinical management from the ENIGMA consortium. J. Med. Genet. 2018;55:15–20. doi: 10.1136/jmedgenet-2017-104560. [DOI] [PubMed] [Google Scholar]
- 57.Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.-H., Wang Q., Bolla M.K., ABCTB Investigators. kConFab/AOCS Investigators. NBCS Collaborators Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spearman A.D., Sweet K., Zhou X.-P., McLennan J., Couch F.J., Toland A.E. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J. Clin. Oncol. 2008;26:5393–5400. doi: 10.1200/JCO.2008.17.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fortuno C., Cipponi A., Ballinger M.L., Tavtigian S.V., Olivier M., Ruparel V., Haupt Y., Haupt S., Study I.S.K., Tucker K. A quantitative model to predict pathogenicity of missense variants in the TP53 gene. Hum. Mutat. 2019;40:788–800. doi: 10.1002/humu.23739. [DOI] [PubMed] [Google Scholar]
- 60.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J.-S., Oh S., Park S.K., Lee M.-H., Lee J.W., Kim S.-W., Son B.H., Noh D.-Y., Lee J.E., Park H.-L. Reclassification of BRCA1 and BRCA2 variants of uncertain significance: a multifactorial analysis of multicentre prospective cohort. J. Med. Genet. 2018;55:794–802. doi: 10.1136/jmedgenet-2018-105565. [DOI] [PubMed] [Google Scholar]
- 62.Dines J.N., Shirts B.H., Slavin T.P., Walsh T., King M.-C., Fowler D.M., Pritchard C.C. Systematic misclassification of missense variants in BRCA1 and BRCA2 “coldspots”. Genet. Med. 2020;22:825–830. doi: 10.1038/s41436-019-0740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jarvik G.P., Browning B.L. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am. J. Hum. Genet. 2016;98:1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rañola J.M.O., Liu Q., Rosenthal E.A., Shirts B.H. A comparison of cosegregation analysis methods for the clinical setting. Fam. Cancer. 2018;17:295–302. doi: 10.1007/s10689-017-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chanock S.J. The paradox of mutations and cancer. Science. 2018;362:893–894. doi: 10.1126/science.aav5697. [DOI] [PubMed] [Google Scholar]
- 66.Favalli V., Tini G., Bonetti E., Vozza G., Guida A., Gandini S., Pelicci P.G., Mazzarella L. Machine learning-based reclassification of germline variants of unknown significance: The RENOVO algorithm. Am. J. Hum. Genet. 2021;108:682–695. doi: 10.1016/j.ajhg.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall J.M., Lee M.K., Newman B., Morrow J.E., Anderson L.A., Huey B., King M.C. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 68.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N., Nguyen K., Seal S., Tran T., Averill D. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset in support of the current study has not been deposited in a public repository because it is composed of patient data including pedigrees which are indirectly identifying. The data are available on request from the corresponding author. A large part of the variants have been deposited in BRCAShare (http://www.umd.be/), and all variants will be annotated in the upcoming FrOG (French OncoGenetics) database, as listed in Table S4 and all material and data supporting this study is included within the manuscript (or by request to the authors if this is the case).