Abstract
Prostate cancer is still one of the most common malignancies in men all around the world. The mechanism of how prostate cancer initiates and develops is still not clear. Here in this study, we show that tumor suppressor ZBTB38 could suppress the migration and proliferation of prostate cancer cells. We find lower ZBTB38 expression in prostate cancer tissues, which also strongly predicts a poorer prognosis of prostate cancer. ZBTB38 binds DKK1 (Dickkopf WNT signaling pathway inhibitor 1) locus and promotes DKK1 expression in prostate cancer cell lines. Consistently, reduction of DKK1 expression significantly restores ZBTB38-mediated suppression of migration and proliferation of prostate cancer cell lines. Mechanistically, we find that ZBTB38 primarily binds the promoters of target genes, and differentially regulates the expression of 1818 genes. We also identify PRKDC (protein kinase, DNA-activated, catalytic subunit) as a ZBTB38-interacting protein that could repress the function of ZBTB38 in suppressing migration and proliferation of prostate cancer cells. Taken together, our results indicate that ZBTB38 could repress cell migration and proliferation in prostate cancer via promoting DKK1 expression, and also provide evidence supporting ZBTB38 as a potential prognosis marker for prostate cancer.
Subject terms: Tumour-suppressor proteins, Prostate cancer
Introduction
Prostate cancer is still one of the most common causes of cancer-related death in males, particularly in western countries [1]. The incidence of prostate cancer is increasing rapidly all around the world. Although comprehensively studied, the underlying mechanism of the initiation and development of prostate cancer is still not fully understood.
Zinc finger and BTB domain containing 38 (ZBTB38, also known as CIBZ, ZNF921m, or PPP1R171) is a new member of the ZBTB family located on chromosome 3q23. ZBTB (zinc finger and BTB domain protein family) family refers to a kind of proteins that contain multiple zinc finger domains at the C-terminus and a BTB domain at the N-terminus [2]. These proteins play important roles in growth, tumorigenesis, cytoskeleton organization, stem cell homeostasis, transcription regulation, hematopoiesis, chromatin remodeling, and protein degradation through synergistic effects [3]. ZBTB38 gene consists of 8 exons, and its transcription product contains BTB binding domain, CtBr binding domain, and C2H2 zinc finger domain, which has a high degree of homology among primates [4]. ZBTB38 is a new gene with unclear functions. Previous studies found that it can bind to methylated DNA to inhibit the transcription of insulin-like growth factor 2 (IGF-II) [4, 5]. In addition, ZBTB38 protein can regulate the cleavage and activation of caspase-3 (Caspase-3) through non-DNA methylation mechanisms [6]. Previous large-scale GWAS studies found that rs6763931, an SNP site located in ZBTB38 gene region, is a prostate cancer risk SNP (11, 12), which indicated the potential role of ZBTB38 in prostate cancer. Moreover, only one previous research last year showed the preliminary study on the role of ZBTB38 in the initiation and progression of prostate cancer [7]. However, the roles and mechanisms of ZBTB38 in prostate cancer are still unclear and need further research.
Dickkopf WNT Signaling Pathway Inhibitor 1 (DKK1) is a secreted protein with two cysteine-rich domains that could mediate protein–protein interactions, which is a member of the dickkopf family proteins. Previous studies showed that DKK1 could inhibit β-catenin-dependent Wnt signaling by inhibiting LRP5/6 interaction with Wnt, and play important roles in embryonic development and bone formation in adults [8–10]. It has been observed that DKK1 shows abnormal expression in various types of human cancers and may promote or inhibit proliferation and invasion in cancer cell lines [11–14]. The role and expression pattern of DKK1 in prostate cancer are complicated. Some results indicate that DKK1 expresses highly in prostate cancer tissues and promotes cancer cells proliferation and migration [15]. However, some researches later showed that DKK1 expression decreases in metastasis progression in prostate cancer, despite the high expression level in early development, indicating the potential tumor suppressor effect in advanced prostate cancer [16, 17]. Therefore, the role and expression regulating mechanism of DKK1 in prostate cancer remain to be further studied.
In this study, we studied the function and mechanism of ZBTB38 in the progression of prostate cancer systematically. We found that overexpression of ZBTB38 could repress the proliferation and migration of prostate cancer cells, and its expression is reversely correlated with prostate cancer disease progression. Mechanistically, our results indicated that PRKDC could interact with ZBTB38 and repress the function of ZBTB38. Knocking down PRKDC could promote DKK1 expression to suppress the malignant progression of prostate cancer cells.
Results
Lower ZBTB38 expression correlated with prostate cancer development
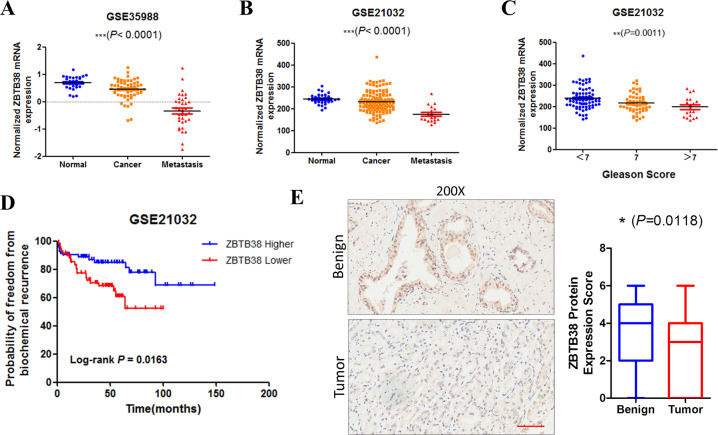
To explore the function of ZBTB38 in prostate cancer, we first determined the expression level of ZBTB38 in benign and cancer tissues. Microarray-based expression analyses using GSE35988 and GSE21032 data sets indicated significantly reduced ZBTB38 expression in primary and metastatic prostate cancer samples (Fig. 1A, B). Moreover, we found that ZBTB38 downregulation was also related to the progression and prognosis of prostate cancer. In Fig. 1C, we showed that prostate cancer samples with higher Gleason scores showed the lower level of ZBTB38 mRNA expression level. Patients with a lower ZBTB38 level (lower than the median value of ZBTB38 expression) showed a much poorer prognosis (Fig. 1D). To confirm the results from online data sets, we examined the expression of ZBTB38 by immunohistochemistry (IHC) in prostate cancer tissues. As shown in Fig. 1E, the expression of ZBTB38 is significantly lower in cancer tissues compared with benign prostatic epithelia. Our results above showed the lower expression level of ZBTB38 in prostate cancer and suggested the potentially important role of ZBTB38 in disease progression.
Fig. 1. ZBTB38 expression level correlates with prostate cancer progression and prognosis.
A, B ZBTB38 mRNA expression levels in prostate cancer samples from GSE35988 (A) and GSE21032 (B). One-way ANOVA analysis was used. C ZBTB38 mRNA expression levels in prostate cancer samples with different Gleason Score groups in GSE21032. One-way ANOVA analysis was used. D ZBTB38 mRNA level was associated with prostate cancer prognosis. A statistically significant increase in RFS was observed in patients with a higher ZBTB38 expression group (P = 0.0163). E Representative IHC images of ZBTB38 in prostate cancer tissues (N = 94 in Benign and cancer group, respectively; Scale bar: 100 μm).(**P < 0.01, ***P < 0.001).
Overexpression of ZBTB38 suppressed prostate cancer proliferation and migration
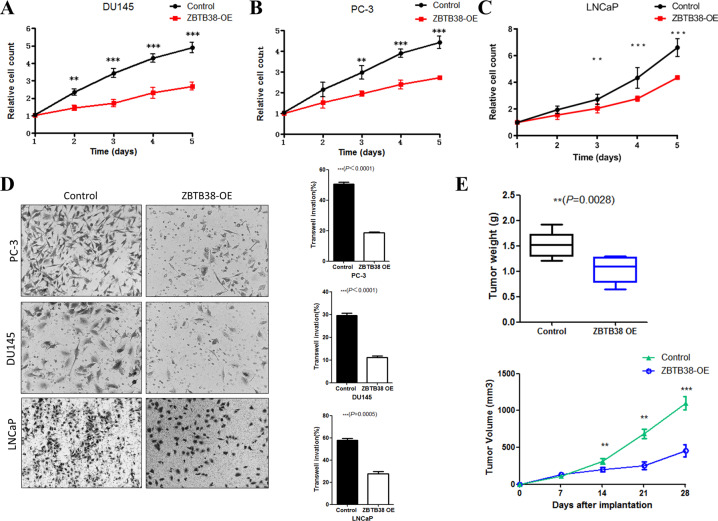
To further determine the role of ZBTB38 in prostate cancer progression, we overexpressed ZBTB38 in LNCaP, DU145, and PC-3 prostate cancer cell lines (Supplementary Fig. S1A). We first determined the effect of ZBTB38 on prostate cancer cell proliferation. Results showed that overexpression of ZBTB38 could significantly inhibit prostate cancer cell proliferation (Fig. 2A–C). Then we did transwell assays to explore the role of ZBTB38 in prostate cancer cell migration. As shown in Fig. 2D, overexpression of ZBTB38 in the three cell lines could greatly suppress the migration activities. Finally, results of the xenograft tumor model using DU145 cells also revealed that upregulation of ZBTB38 expression induced the growth inhibition of tumor in vivo (Fig. 2E).
Fig. 2. ZBTB38 suppresses prostate cancer cell proliferation and migration.
A Cell proliferation was measured at the indicated time points in DU145 cells (**P < 0.01, ***P < 0.001). B Cell proliferation was measured at the indicated time points in PC-3 cells (**P < 0.01, ***P < 0.001). C Cell proliferation was measured at the indicated time points in LNCaP cells (**P < 0.01, ***P < 0.001). D Transwell assay analyses of the indicated cell lines. E Xenograft analyses of DU145 derived tumors with control vector or ZBTB38 overexpression (**P < 0.01, ***P < 0.001).
ZBTB38 primarily binds the promoter regions of its target genes
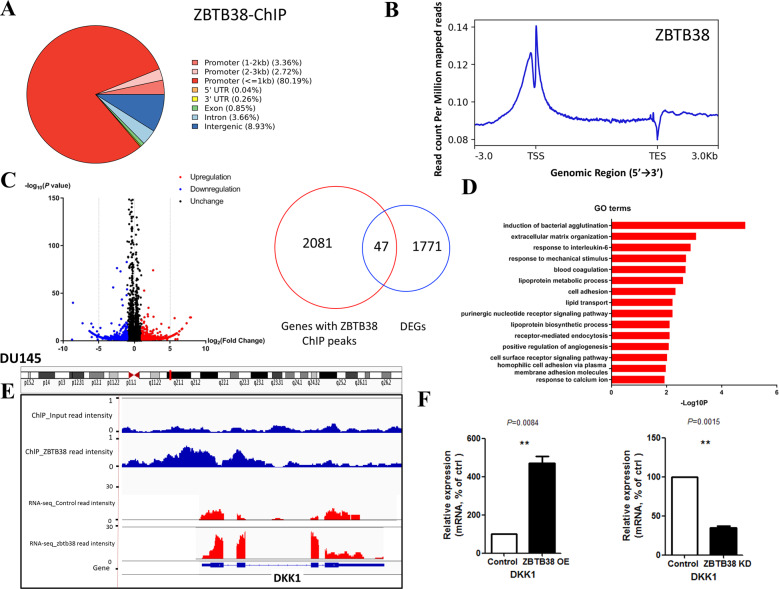
Previous studies have determined that ZBTB38 could serve as a transcription factor implicated in multiple developmental processes [5], we then carried out ChIP-seq assay in order to explore the binding sites of ZBTB38 in the genome. This experiment yielded a total of 2352 peaks covering 2128 genes, among which 86.27% of the binding sites were in the promoter regions (TSS ±3000 bp). Results of the signal plot analyses showed the binding of ZBTB38 in the TSS and 5′-end regions of its target genes primarily (Fig. 3A, B).
Fig. 3. ZBTB38 binds the promoters and regulates the expression of its target genes in DU145 cell.
A Genome distribution of HA- ZBTB38 ChIP-seq peaks in DU145 cells. B Signal plot of HA-ZBTB38 ChIP-seq signals in DU145 cells. C Volcano plot for DEGs in ZBTB38 overexpression DU145 RNA-seq. Right panel: the overlap of ChIP-seq peaks and DEGs. D GO analysis (biological process, BP) of downregulated genes after ZBTB38 overexpression in DU145 cells using the DAVID program. E A representative snapshot of ZBTB38 ChIP-seq and RNA-seq signals located in DKK1 gene locus. F RT-qPCR analyses of the mRNA levels of DKK1 gene in the DU145 cells with a control vector or ZBTB38 overexpression or ZBTB38 knockdown. (**P < 0.01, ***P < 0.001).
Results of functional enrichment analysis of ZBTB38-regulated genes
To further analyze the downstream genes and signaling pathways regulated by ZBTB38, we performed RNA-seq for the control and ZBTB38-overexpressed DU145 cell lines. After analyzing the results, we identified 1818 genes that were differentially expressed in each group (| log2 (fold change) | >1, P < 0.05, Fig. 3C). In total, 901 differentially expressed genes (DEGs) were upregulated on ZBTB38 overexpressing, whereas 917 were downregulated (Supplementary Table S1). Then, we did further analyses combining the results of RNA-seq and ChIP-seq, and found that 47 DEGs were bound by ZBTB38 at gene body regions or TSS (Fig. 3C), indicating a direct regulation by ZBTB38. To further analyze the gene function of DEGs, we did Gene ontology (GO) and KEGG analyses using the upregulated and downregulated DEGs, respectively. Results showed enrichment for gene sets in cell movement and adhesion (Fig. 3D, Supplementary Fig. 1B–D), consistent with our previous phenotypic analyses (Fig. 2).
ZBTB38 inhibits prostate cancer cell proliferation and migration via direct upregulation of DKK1 expression
After validating that ZBTB38 inhibits the proliferation and migration of prostate cancer cells, we then tried to address the underlining mechanisms. We explored the DEGs that were both directly bound by ZBTB38 and reported to be implicated in prostate cancer disease progression. Among all eligible candidate genes, DKK1 is a WNT signaling regulator that plays important role in multiple cancer types. Then we focused on DKK1, whose TSS was bound by ZBTB38 and whose expression was significantly upregulated upon ZBTB38 overexpression (Fig. 3E). The elevated expression of DKK1 was also confirmed by qRT-PCR (Fig. 3F).
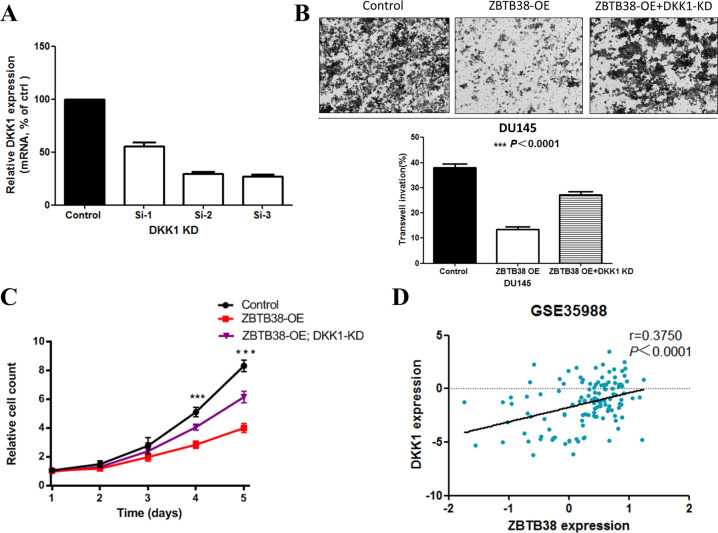
To further validate that DKK1 could mediate the tumor suppressor role of ZBTB38 in prostate cancer cells, we then knocked down DKK1 expression in DU145 cells (Fig. 4A). Results of transwell assays showed that the migration ability of ZBTB38-overexpressed DU145 was partially restored after DKK1 suppression (Fig. 4B). Similarly, knocking down DKK1 also partially mitigated the inhibition of cell proliferation caused by overexpression of ZBTB38 in DU145 (Fig. 4C). These results were also confirmed in PC-3 prostate cancer cell line (Supplementary Fig. 2A, B). Then we investigated the correlation between ZBTB38 and DDK1 expression in prostate cancer samples. We analyzed online publicly available data sets from GSE35988 and found that the mRNA expression level of ZBTB38 was positively correlated with DDK1 (Fig. 4D). All these data above indicate that upregulation of DKK1 is one of the potential mechanisms that mediated the tumor-suppressing function of ZBTB38.
Fig. 4. ZBTB38 suppresses prostate cancer cell proliferation and migration through promoting DKK1 expression.
A RT-qPCR showed the mRNA levels of DKK1 gene in the DU145 cells with a control vector or DKK1 knockdown SiRNA. B DKK1 knockdown rescued ZBTB38-mediated reduced cell migration. One-way ANOVA analysis was used. C DKK1 knockdown rescued ZBTB38-mediated reduced cell proliferation (**P < 0.01, ***P < 0.001). D Expression correlation analyses of ZBTB38 and DKK1 mRNA levels in prostate cancer samples from GSE35988.
PRKDC could interact with ZBTB38 and inhibit the upregulation of DKK1 expression
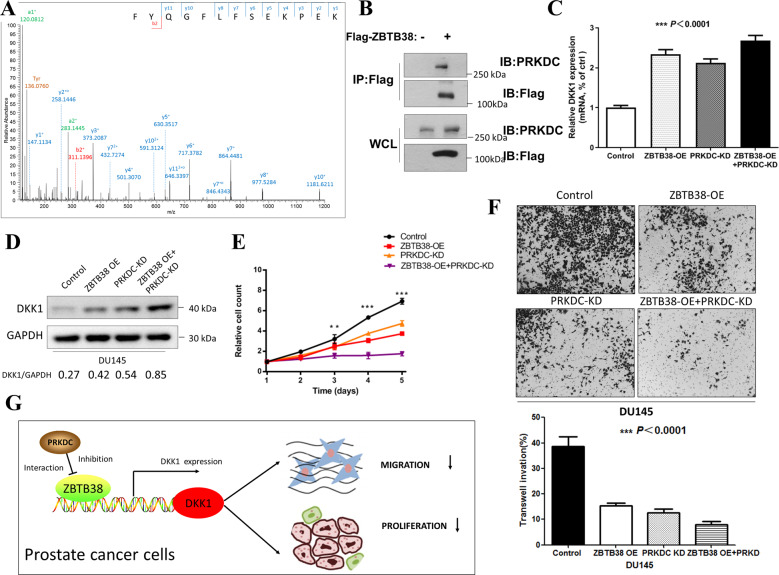
To explore deeply how ZBTB38 might regulate DKK1 expression, we performed Flag immunoprecipitation in DU145 cells with Flag-ZBTB38 expression followed by mass spectrometry analyses to identify interacting proteins of ZBTB38 (Supplementary Table S2). Among all possible proteins, we focused on PRKDC (Fig. 5A). PRKDC (Protein Kinase, DNA-Activated, Catalytic Polypeptide) is a nuclear protein serine/threonine kinase that is a molecular sensor of DNA damage, which has been shown to play important roles in the disease process of a variety of tumors, including breast cancer, colorectal cancer, and prostate cancer [18–20]. Particularly, a previous study confirmed that PRKDC is an independent prognostic factor in patients with prostate cancer and downregulation of PRKDC inhibited prostate cancer growth [19]. In the co-immunoprecipitation (co-IP) assay, we also confirmed the interaction between Flag-ZBTB38 and PRKDC (Fig. 5B). Importantly, we found that PRKDC could inhibit the tumor-suppressive function of ZBTB38, as PRKDC knockdown (Supplementary Fig. 2C) resulted in the increase of DKK1 expression and enhanced the tumor-suppressive function of ZBTB38 (Fig. 5C–F). Moreover, we assessed the expression levels of ZBTB38 in PRKDC knockdown DU145 cells to see whether PRKDC knockdown produced lower levels of ZBTB38 which could lead to the results above. As shown in Supplementary Fig. 2C, PRKDC knockdown did not influence ZBTB38 expression, thus excluding this possibility. Taken together, these findings uncovered the tumor-suppressive roles of ZBTB38 to suppress cell proliferation and migration in prostate cancer cells. We identified DKK1 as the direct downstream of ZBTB38, which could mediate the tumor-suppressing function. PRKDC could interact with ZBTB38 and potentially be involved in the regulation of DKK1 expression by ZBTB38 (Fig. 5G).
Fig. 5. PRKDC interacts with ZBTB38 and inhibits the function of ZBTB38.
A Mass spectrometry identification of a higher-energy collisional dissociation (HCD) MS/MS spectrum was recorded on the [M + 3H]3+ ion at m/z 540.94 of the human prKDC peptide FYQGFLFSEKPEK. Predicted a-, b-, and y-type ions (not including all) are listed above and below the peptide sequence, respectively. The cycle symbol means the neutral loss of water. B Co-IP analysis of the interaction between ZBTB38 and PRKDC. C PRKDC knockdown induced DKK1 upregulation in mRNA expression level. One-way ANOVA analysis was used. D PRKDC knockdown induced upregulation of DKK1 protein. E PRKDC knockdown reduced ZBTB38-mediated proliferation in DU145 cells (**P < 0.01, ***P < 0.001). F PRKDC knockdown reduced ZBTB38-mediated migration in DU145 cells. One-way ANOVA analysis was used. (ZBTB38 OE+PRKD means ZBTB38 oversexpression with PDKDC knockdown in DU145 cells) G Working model depicting the mechanism of ZBTB38/ PRKDC-mediated DKK1 promotion in prostate cancer.
Discussion
Prostate cancer is one of the most common malignant tumors in males and ranks sixth in cancer-related deaths [1]. Among all the cancer types, metastatic prostate cancer and the development of castration-resistant prostate cancer (CRPC) are the major catalysts of mortality in prostate cancer patients [21]. However, the molecular biological mechanisms of the progression of prostate cancer are still not clear, leading to poor prevention and treatment of such aggressive prostate cancer. Therefore, an in-depth study of the molecular mechanisms of prostate cancer disease progression will help find new drug targets and intervention strategies.
Here in this study, we focused on ZBTB38 and investigated its role in prostate cancer progression. We found that ZBTB38 expresses much lower in prostate cancer tissues, and its expression is reversely correlated with prostate cancer disease progression. Overexpression of ZBTB38 could repress the proliferation and migration of prostate cancer cells via direct promotion of DKK1 expression. Mechanistically, our results indicated that PRKDC is an important interacting protein of ZBTB38, which could repress the tumor-suppressive function of ZBTB38. Knocking down PRKDC could significantly promote DKK1 expression to suppress the malignant progression of prostate cancer cells. These findings are of importance since the molecular pathogenesis of prostate cancer is still poorly known, the identification of ZBTB38 as a novel tumor suppressor might provide novel possible targets for therapeutics.
ZBTB38 is a new member of the ZBTB family located on chromosome 3q23 with eight exons. Although previous research has revealed the tumor-suppressive role of ZBTB38 in prostate cancer [7], however, the mechanisms are still unclear. Besides confirming previous results, our results also showed the binding affinity of ZBTB38 using ChIP-seq analysis and identified DKK1 as the direct downstream of ZBTB38.expression in mediating its tumor-suppressive role. DKK1 could inhibit β-catenin-dependent WNT signaling by inhibiting LRP5/6 interaction with WNT, and play a role in embryonic development and bone formation in adults [8–10]. Our results suggest the novel role of DKK1 in prostate cancer, which could serve as the direct downstream of ZBTB38 in regulating prostate cancer cells proliferation and migration. However, the downstream genes and signaling pathways of DKK1 in prostate cancer are still unknown, which are also quite intriguing issues. It might be WNT pathway or some other novel mechanisms. In the future, we will focus on the role of DKK1 in prostate cancer, which could help us understand the disease better.
Our study for the first time determined the role of PRKDC as an interacting protein associated with the tumor suppressor ZBTB38 to promote DKK1 expression in prostate cancer. We used IP and co-IP assays to confirm the interaction between ZBTB38 and PRKDC. In vitro assay indicated that PRKDC knockdown resulted in an increase of DKK1 expression and enhanced the tumor-suppressive function of ZBTB38, which suggested that PRKDC could inhibit the tumor-suppressive function of ZBTB38. The interaction of PRKDC and ZBTB38 is quite interesting. PRKDC might phosphorylate some critical sites of ZBTB38, and we will focus on the concrete mechanisms in the future.
Nowadays, we still have no effective treatment strategies for patients with advanced prostate cancer (including CRPC and mCRPC). One of the critical reasons is that the mechanisms of how CRPC and mCRPC develop are still not quite clear. Our results indicated the tumor-suppressing role of ZBTB38 in both AR-positive prostate cancer and CRPC cells. It is possible that ZBTB38 also contributes to the development of advanced prostate cancer. In the next step, we will also focus on the potential function of ZBTB38 in this aspect.
Our study contained limitations. In our study, we make an investigation that ZBTB38 could interact with PRKDC to promote DKK1 expression to suppress prostate cancer cell proliferation and migration. The possible molecular mechanisms of how ZBTB38 interacts with PRKDC, and how the protein complex functions are still not clear and need further study. Moreover, the entire signaling pathways downstream of ZBTB38-DKK1 still need to uncover in the future.
Conclusions
Taken together, our findings in this study uncovered the tumor-suppressive roles of ZBTB38 to suppress cell proliferation and migration by upregulating DKK1 expression. We also identified that PRKDC could interact with ZBTB38 and regulate its tumor-suppressive roles in prostate cancer.
Materials and methods
Cell culture
We obtained LNCaP (AR positive), PC-3 (AR negative), and DU145 (AR negative) cells from ATCC (Bethesda, USA). All cells were tested for mycoplasma contamination and maintained in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum and antibiotics (0.1 mg/ml streptomycin and 100 units/ml penicillin) as previously [22]. All cell lines used in our study were authenticated by STR profiling and tested for mycoplasma contamination
Constructs and construction of stable cell lines
We used pPB-CAG-EBNXN vector (Sanger Institute) to deal with constructs and pPB-CAG-ires-Pac was generated as previously described [22, 23]. We ligated full-length ZBTB38, DKK1, and PRKDC into the multiple cloning sites of pPB-CAG-ires-Pac to generate pPB-CAG-ZBTB38-ires-Pac, pPB-CAG-DKK1-ires-Pac, and pPB-CAG-PRKDC-ires-Pac, respectively. Control, ZBTB38, PRKDC, or DKK1 overexpression stable cells were obtained as previously described and all stable cell lines were selected and identified by western blotting [22].
siRNA transfection
In this study, we knocked down gene expression using the individual set of three siRNAs (Shanghai Biotend Biotechnology Co., Ltd, China) against the target genes. The most effective single siRNAs were used for further experiments. Then, 3 × 105 cells per well were subjected to reverse transfection with 20 nM siRNA (Biotend, China) using Lipofectamine 3000 transfection reagent (Invitrogen), following the manufacturer’s instructions for siRNA transfection in six-well plates. The siRNA sequences are shown in Supplementary Table S3.
Antibodies and immunoblotting
In western blotting, cells were lysed in 1× SDS loading buffer (50 mM Tris-HCl pH6.8, 10% glycerol, 2% SDS, 0.05% bromophenol blue, and 1% 2-mercaptoethanol). Antibodies were listed as follows: anti-ZBTB38 antibody (21906-1-AP, Proteintech), anti-DKK1 (21112-1-AP, Proteintech), anti-PRKDC (SC-5282, Santa Cruz), anti-HA (51064-2-AP, Proteintech), anti-FLAG (20543-1-AP, Proteintech), anti-GAPDH (10494-1-AP, Proteintech), and anti-TUBULIN (ab134185, Abcam).
We did immunoblot as previously described [22]. In brief, all proteins were separated by SDS–PAGE and were transferred to polyvinylidene difluoride membranes (Millipore). Horseradish peroxidase-labeled secondary antibodies and an enhanced chemiluminescence system were used for signal detection. Protein was visualized using ChemiDoc XRS chemiluminescence detection and imaging system (Bio-Rad Laboratories) or KODAK film machine.
Tissue microarrays (TMAs) and IHC
TMAs were constructed using 94 benign prostate tissues and 94 prostate cancer tissues, and IHC was performed as described elsewhere [24]. In brief, slides were deparaffinized and heated in citrate buffer pH6 for antigenic retrieval. The primary antibody was ZBTB38 antibody (21906-1-AP, Proteintech, 1/600, 30 min). Immunohistochemistry was performed using the streptavidin-biotin-peroxidase method with diaminobenzidine as the chromogen (KitLSAB, Dakocytomotion, Glostrup, Denmark). Negative controls were obtained after omission of the primary antibody or incubation with an irrelevant antibody.
ZBTB38 staining was scored by two independent observers (including one pathologist) as described previously [24]. In brief, a positive reaction was scored in four grade categories depending on the intensity of the staining and the percentage of ZBTB38-positive cells. The sum of the intensity and percentage scores was used as the final score. The staining pattern was defined as follows: 0, negative; 1–2, weak; 3–4, moderate; and 5–6, strong.
Cell proliferation assay
Cell proliferation (MTS) assay was performed as described elsewhere [25]. In brief, we seeded all cells at 4000 cells/well (0.1 ml) in 96-well plates. All cells were incubated overnight at 37 °C for 5–6 days. At each indicated time point, cells were incubated with 20 μl of CellTiter 96 AQueous One solution reagent MTS (Promega) in 100 μl of RPMI-1640 for one hour. Cell number was then estimated using a microtiter plate reader (Bio-tek).
In vivo tumor growth assay
In tumor growth assay, we inoculated s.c. (subcutaneous) with 1 × 107 prostate cancer cells in six-week-old male athymic mice. Eight mice were in each group (Control group and ZBTB38-OE group) and all mice were sacrificed after 28 days. Tumor growth was monitored weekly, and tumor sizes were measured recorded in length × width2 (mm3). The Ethics Committee of Nanjing Medical University approved all animal use procedures.
Cell migration assays
We used Transwell (Corning) system in 24-well tissue culture plates to deal with cell migration assays as described previously [22].
Real-time RT-PCR assays
We extracted total RNAs from indicated cells in our study using TRIzol reagent (Invitrogen). The extracted RNA was then subjected to reverse transcription with reverse transcriptase (Fermentas). We used the Bio-Rad CFX96 system for quantitative real-time PCR, and normalized the relative gene expression to GAPDH as a control. Primer sequences used in the study were shown as follows:
DKK1-F: CTCGGTTCTCAATTCCAACG; DKK1-R: GCACTCCTCGTCCTCTG;
GAPDH-F: GCACCACCAACTGCTTA; GAPDH-R: AGTAGAGGCAGGGATGAT
ChIP-Seq assays
We did ChIP assays as previously described [26]. For HA-ZBTB38 ChIP, the chromatin was incubated with HA antibody (3724 S, Cell Signaling Technology) overnight at 4 °C, and additionally with pre-washed protein A/G agarose beads (SA032005, Smart-lifesciences) for 1 h. The DNA samples were sequenced by Illumina Novaseq PE150 (Rainbow-genome. Co., Ltd., Shanghai).
RNA-seq and analysis
We extracted total RNAs from cells in our study using TRIzol reagent (Invitrogen) and genomic DNA was removed using DNase I (TaKara). Then RNA was quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA sample (OD260/280 = 1.8~2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28 S:18 S ≥ 1.0, >10 μg) was used to construct sequencing library. The RNA-seq libraries were sequenced on the Illumina HiSeq 4000 sequencing platform at Shanghai Majorbio Bio-pharm Technology Co., Ltd (China).
EdgeR was used for examining the differential expression of RNA-Seq count data as previously described [27, 28]. DEGs were identified with the following criterion: fold change (FC) ≥ 2 or ≤0.5; P value <0.05. The Venn calculation result was analyzed using the online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
IP-mass spectrometry
We did all these experiments as described previously [22]. Cells were harvested with EBC lysis buffer supplemented with protease inhibitors (Selleck Chemicals) and phosphatase inhibitors (Selleck Chemicals). For IP, cell lysates were incubated with the FLAG antibody (1–2 µg). Then protein A-Sepharose beads (GE Healthcare) were added to a mixture of cell lysates and antibodies for 1 h. IP complexes were washed five times with NETN buffer. After washing, the IP samples were resolved by SDS–PAGE on a 4–20% polyacrylamide gel (Bio-Rad) and visualized using the Bio-Safe Coomassie Stain (Bio-Rad). The gel containing FLAG-ZBTB38 complex was excised and all peptide matches were filtered on the basis of mass accuracy, tryptic state (for trypsin), and XCorr, and confirmed by manual inspection. The results of mass spectrometry proteomics data have been provided in Supplementary Table S2 in the supplementary information.
Publicly available gene expression data sets and clinical data sets
Prostate adenocarcinoma samples from GSE21032 and GSE35988 were used in this study.
Statistical analyses
All experiments were done in triplicates at least and the results were presented as the average values ± standard error of mean. We did prognosis analysis after surgery using Kaplan–Meier method with log-rank test. Between-group variations in this study were evaluated by the use of the Student’s t test, data from more than two groups were analyzed by one-way analysis of variance followed by Dunnett’s test. Spearman’s correlation finished the exploration of the relationships. The variance between the groups that are being statistically compared is similar. A two-sided P < 0.05 was thought to have statistical significance. Graphpad 8.0 was used for the analyses.
Supplementary information
Acknowledgements
The authors thank Dr. Chao Huang and Dr. Jiabin Cai for the help in providing instructive suggestions.
Author contributions
J.W., G.D., W.L., and Q.Z.: conceptualization, methodology, software; G.D. and K.L.: data curation, writing–original draft preparation; H.Z., F. W., C.Z., C.F.: visualization, investigation; J.W.: supervision; Q.Z.: software, validation; J.W.: writing–reviewing and editing.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this study was supported by the National Natural Science Foundation of China (81802565 to J.W.), Natural Science Foundation of Jiangsu Province (BK20180216 to J.W.), Key Project of the Scientific Research Project of Nanjing Medical University Affiliated Suzhou Hospital (szslyy2017005 to J.W.), and Suzhou Gusu Key Health Talent of Suzhou, Youth Medical Talents program (GSWS2020072 to J.W.).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics statement
The authors state that they have been approved by the Ethics Committee of Nanjing Medical University and have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. Informed consent was obtained from all subjects.
Footnotes
Edited by Professor Stephen Tait
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guanxiong Ding, Wei Lu, Qing Zhang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04278-3.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lee SU, Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genom Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 4.Hong KW, Shin YB, Jin HS, Lim JE, Choi JY, Chang KT, et al. Alternative splicing of human height-related zinc finger and BTB domain-containing 38 gene through Alu exonization. Biochem Genet. 2011;49:283–291. doi: 10.1007/s10528-010-9406-4. [DOI] [PubMed] [Google Scholar]
- 5.Clayton P, Bonnemaire M, Dutailly P, Maisonobe P, Naudin L, Pham E, et al. Characterizing short stature by insulin-like growth factor axis status and genetic associations: results from the prospective, cross-sectional, epidemiogenetic EPIGROW study. J Clin Endocrinol Metab. 2013;98:E1122–1130. doi: 10.1210/jc.2012-4283. [DOI] [PubMed] [Google Scholar]
- 6.Oikawa Y, Matsuda E, Nishii T, Ishida Y, Kawaichi M. Down-regulation of CIBZ, a novel substrate of caspase-3, induces apoptosis. J Biol Chem. 2008;283:14242–14247. doi: 10.1074/jbc.M802257200. [DOI] [PubMed] [Google Scholar]
- 7.de Dieuleveult M, Marchal C, Jouinot A, Letessier A, Miotto B. Molecular and clinical relevance of ZBTB38 expression levels in prostate cancer. Cancers. 2020;12:1106. [DOI] [PMC free article] [PubMed]
- 8.Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, et al. DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Sarosi I, Cattley RC, Pretorius J, Asuncion F, Grisanti M, et al. Dkk1-mediated inhibition of Wnt signaling in bone results in osteopenia. Bone. 2006;39:754–766. doi: 10.1016/j.bone.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Ren S, Johnson BG, Kida Y, Ip C, Davidson KC, Lin S-L, et al. LRP-6 is a coreceptor for multiple fibrogenic signaling pathways in pericytes and myofibroblasts that are inhibited by DKK-1. Proc Natl Acad Sci. 2013;110:1440–1445. doi: 10.1073/pnas.1211179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang X, Zhang H, Li X, Li X, Cong M, Peng F, et al. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat Cell Biol. 2017;19:1274–1285. doi: 10.1038/ncb3613. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Zhang X, Zhao X, Jiang M, Gu M, Wang Z, et al. DKK1 promotes migration and invasion of non–small cell lung cancer via β-catenin signaling pathway. Tumor Biol. 2017;39:1010428317703820. doi: 10.1177/1010428317703820. [DOI] [PubMed] [Google Scholar]
- 13.Qiao L, Xu Z-l, Zhao T-j, Ye L-h, Zhang X-d. Dkk-1 secreted by mesenchymal stem cells inhibits growth of breast cancer cells via depression of Wnt signalling. Cancer Lett. 2008;269:67–77. doi: 10.1016/j.canlet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 14.Duan H, Yan Z, Chen W, Wu Y, Han J, Guo H, et al. TET1 inhibits EMT of ovarian cancer cells through activating Wnt/β-catenin signaling inhibitors DKK1 and SFRP2. Gynecol Oncol. 2017;147:408–417. doi: 10.1016/j.ygyno.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Thudi NK, Martin CK, Murahari S, Shu ST, Lanigan LG, Werbeck JL, et al. Dickkopf-1 (DKK-1) stimulated prostate cancer growth and metastasis and inhibited bone formation in osteoblastic bone metastases. Prostate. 2011;71:615–625. doi: 10.1002/pros.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall CL, Daignault SD, Shah RB, Pienta KJ, Keller ET. Dickkopf-1 expression increases early in prostate cancer development and decreases during progression from primary tumor to metastasis. Prostate. 2008;68:1396–1404. doi: 10.1002/pros.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Yu C, Dai J, Keller J, Hua A, Sottnik J, et al. Parathyroid hormone-related protein inhibits DKK1 expression through c-Jun-mediated inhibition of β-catenin activation of the DKK1 promoter in prostate cancer. Oncogene. 2014;33:2464–2477. doi: 10.1038/onc.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun S, Cheng S, Zhu Y, Zhang P, Liu N, Xu T, et al. Identification of PRKDC (protein kinase, dna-activated, catalytic polypeptide) as an essential gene for colorectal cancer (CRCs) cells. Gene. 2016;584:90–96. doi: 10.1016/j.gene.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Wang Y, Ning Y. Down-regulation of protein kinase, DNA-activated, catalytic polypeptide attenuates tumor progression and is an independent prognostic predictor of survival in prostate cancer. Urol Oncol. 2017;35:111 e115–111 e123. doi: 10.1016/j.urolonc.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Yang WK, Wen GM, Tang H, Wu CA, Wu YX, et al. High expression of PRKDC promotes breast cancer cell growth via p38 MAPK signaling and is associated with poor survival. Mol Genet Genom Med. 2019;7:e908. doi: 10.1002/mgg3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA. Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. J Natl Cancer Inst. 2003;95:661–668. doi: 10.1093/jnci/95.9.661. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, He C, Gao P, Wang S, Lv R, Zhou H, et al. HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene. 2020;39:1335–1346. doi: 10.1038/s41388-019-1065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong L, Tan L, Lv R, Shi Z, Xiong L, Wu F, et al. A primary role of TET proteins in establishment and maintenance of De Novo bivalency at CpG islands. Nucleic Acids Res. 2016;44:8682–8692. doi: 10.1093/nar/gkw529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Xiao Q, Chen X, Tong S, Sun J, Lv R, et al. LanCL1 protects prostate cancer cells from oxidative stress via suppression of JNK pathway. Cell Death Dis. 2018;9:197. doi: 10.1038/s41419-017-0207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Chen X, Tong S, Zhou H, Sun J, Gou Y, et al. Overexpression of WDFY2 inhibits prostate cancer cell growth and migration via inactivation of Akt pathway. Tumour Biol. 2017;39:1010428317704821. doi: 10.1177/1010428317704821. [DOI] [PubMed] [Google Scholar]
- 26.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.