Abstract
Quantifying gene expression in individual cells can substantially improve our understanding about complex genetically engineered cell products such as chimeric antigen receptor (CAR) T cells. Here we designed a single-cell RNA sequencing (scRNA-seq) approach to monitor the delivery of a CD19-CAR gene via lentiviral vectors (LVs), i.e., the conventional vesicular stomatitis virus (VSV)-LV and the CD8-targeted CD8-LV. LV-exposed human donor peripheral blood mononuclear cells (PBMCs) were evaluated for a panel of 400 immune response-related genes including LV-specific probes. The resulting data revealed a trimodal expression for the CAR and CD8A, demanding a careful distribution-based identification of CAR T cells and CD8+ lymphocytes in scRNA-seq analysis. The fraction of T cells expressing high CAR levels was in concordance with flow cytometry results. More than 97% of the cells hit by CD8-LV expressed the CD8A gene. Remarkably, the majority of the potential off-target cells were in fact on-target cells, resulting in a target cell selectivity of more than 99%. Beyond that, differential gene expression analysis revealed the upregulation of restriction factors in CAR-negative cells, thus explaining their protection from CAR gene transfer. In summary, we provide a workflow and subsetting approach for scRNA-seq enabling reliable distinction between transduced and untransduced cells during CAR T cell generation.
Keywords: CAR T cells, CAR, chimeric antigen receptor, CD19, lentiviral vector, scRNA sequencing, CD8-LV, VSV-LV, transduction
Graphical abstract
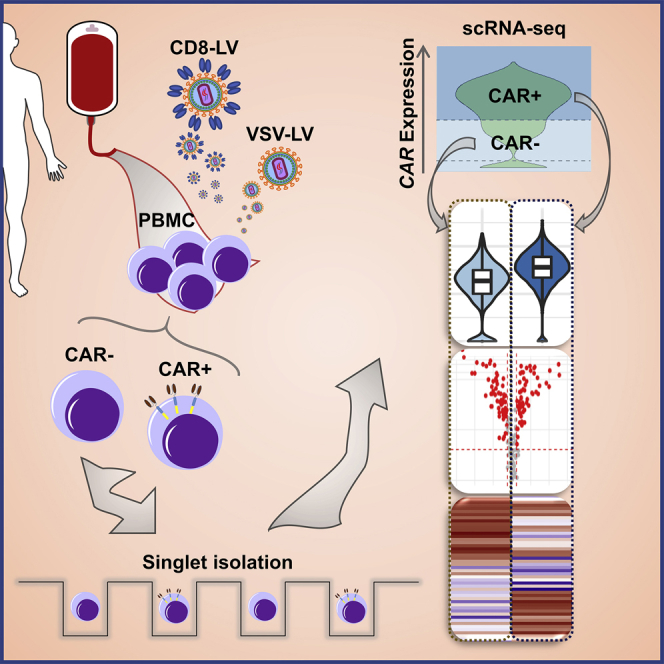
Buchholz and coworkers describe a process for a nanowell-based scRNA-seq pipeline, thereby making use of a CD8-specific lentiviral vector (LV) for identification and analysis of CAR T cells. Exposure to CD8-LV or VSV-LV and CAR expression altered the transcriptomes of individual lymphocytes. Upregulation of restriction factors correlated with protection from transduction.
Introduction
In the time since Gross et al. generated genetically modified T cells to express a chimeric antigen receptor (CAR), many advances have been made including the development of second-generation CARs, which has led to impressive clinical benefit, particularly for patients suffering from B cell lymphoma and acute lymphoblastic leukemia (ALL), through targeting the CD19 antigen.1, 2, 3, 4, 5 Further improvements, aiming at on one hand the expansion of CAR T cells to the treatment of other cancer entities including solid tumors and on the other hand the simplification of the manufacturing process, are the focus of ongoing research. Especially the complex manufacturing process, which results in a patient-specific, highly complex mixture of various T cell types, requires novel single-cell-based analysis tools to allow a better understanding and improvements.
A typical manufacturing process comprises the ex vivo delivery of the CD19-CAR gene into autologous T cells, subsequent CAR T cell expansion, and finally the adoptive transfer of the personalized medicinal product. Among the most frequently used gene delivery tools is the lentiviral vector (LV).6 Carrying two positive single-stranded RNA molecules, the therapeutic gene spans between two extreme long terminal repeats (LTRs). The 5'-LTR serves as a promoter during the production of LV, transcribing the viral genomic RNA (gRNA), whereas the 3'-LTR, also known as self-inactivating LTR (SIN), contains a common polyadenylation site of both viral gRNA and the transgene’s mRNA.7 Upon cellular entry, its gRNA is reverse transcribed and the transfer cassette integrates into the host genome. Consequently, only the therapeutic gene of interest is expressed via an internal promotor.6,8 Downstream of the therapeutic gene, the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) serves as an enhancing factor for increased gene expression.9
Viral envelope glycoproteins are responsible for the recognition and the engagement of particular host receptors, leading to particle entry. Conventional LVs are pseudotyped with vesicular stomatitis virus (VSV) glycoprotein G and accordingly enter cells through the low-density lipoprotein receptor (LDLR), which is expressed on T cells activated with anti-CD3, anti-CD28 antibodies and cultivated in the presence of cytokines.10,11 More advanced LVs are pseudotyped with engineered glycoproteins mediating selective entry into cell types expressing a target cell surface receptor of choice.12 For selective gene delivery into T lymphocyte subtypes, LVs with very high selectivity for CD4+ or CD8+ T cells have been described.13 The CD8 receptor-targeted LV (CD8-LV) enables specific transduction of CD8 cytotoxic T cells, not only ex vivo but also in vivo including CAR T cell generation in humanized mice.14,15 CAR T cell generation with LVs is a process covering ∼12 days. The first half includes T cell activation, incubation with vector particles, followed by vector entry and genomic integration, and finally the expression and cell surface transport of the CAR. The second half is mainly expansion of the CAR T cells to reach sufficient numbers for transplantation. The transduction process contains many unknowns on the cellular and molecular levels, such as why certain T cells become CAR-positive and others do not or what consequences the vector particle exposure has for T cells. Questions like this can now be addressed by single-cell RNA sequencing (scRNA-seq). However, as an emerging tool in the field, only a few studies have analyzed CAR T cells via scRNA-seq so far. These studies focused mainly on the diversity of CAR T cell phenotypes in pre-infusion products and correlated these to activities in patients16, 17, 18 or investigated the consequences of different CAR signaling domains and antigenic stimulation.19,20
None of these studies has looked closely into the process of LV-mediated transduction. Our scope was to establish a methodology that thoroughly investigates differences between transduced and non-transduced cells during product generation and profiles the consequences of different vector types applied. We have set up a nanowell-based scRNA-seq approach for LV-mediated CAR delivery making use of the high selectivity of CD8-LV for CD8+ T cells. In particular, we have performed a targeted gene amplification analysis of untransduced CD8-LV- and VSV-LV-generated CAR T cell products expanded for a short period of time. The CAR T cells were detected via 3'-end-targeted amplification of the transgene with customized primers annealing within the WPRE region. This approach enabled us to accurately associate changes in cellular gene expression caused by the CAR and/or exposure to the LV particles and demonstrate that CD8-LV has near-perfect selectivity for its target cells.
Results
Setting up the system
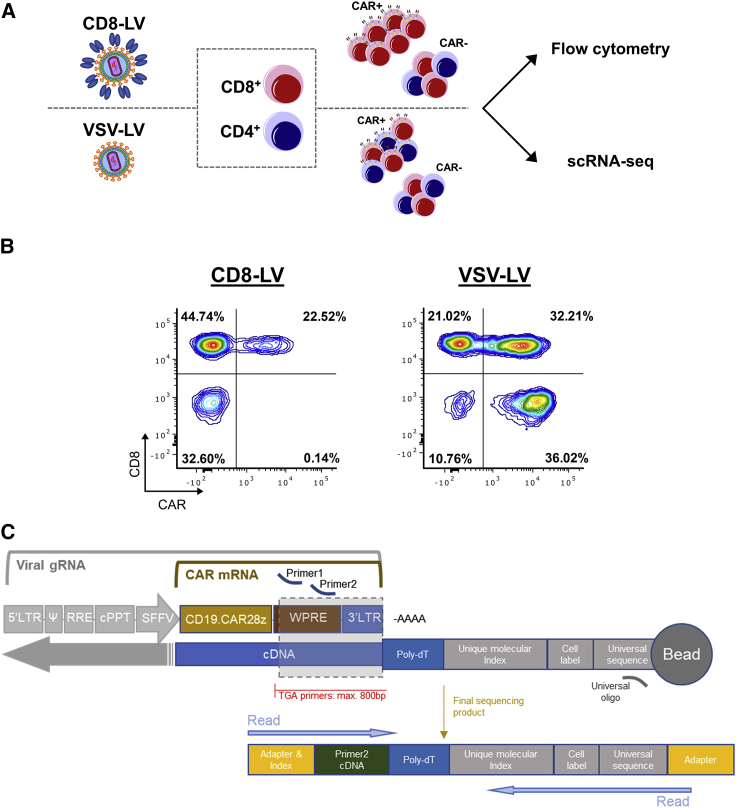
Human donor peripheral blood mononuclear cells (PBMCs) were used as the cell source for CAR T cell generation. The cells were activated and then incubated with CD8-LV or VSV-LV or left untransduced as control. After 6 days of cultivation in the presence of interleukin (IL)-7/IL-15, the cells were analyzed by flow cytometry and processed for scRNA-seq (Figure 1A). Vector doses were optimized such that a similar fraction of CD8+ CAR T cells was obtained with both vector types, while also leaving a significant fraction of cells CAR-negative (Table 1). According to flow cytometry, CD8-LV had generated 22.5% CAR T cells, all of which were CD8-positive. Thus, about one-third of the CD8 cells had been converted to CAR T cells (Figure 1B). With VSV-LV, similar fractions of CD8+ and CD4+ CAR T cells were obtained (Figure 1B). Thus, CAR T cell generation had worked out as expected. For scRNA-seq, we chose a microwell-based system for single-cell isolation and processing. Rather than utilizing a whole transcriptome approach, we opted for a targeted gene panel covering 399 human genes relevant for immune response to achieve high resolution of the sequencing results, thus allowing highly sensitive detection of differentially expressed genes. To detect CAR-positive T cells by scRNA-seq, the WPRE element adjacent to the 3'-LTR was identified as being at an ideal distance from the poly(A) tail to allow sensitive detection of mRNA transcribed from the integrated vector genome (Figure 1C).
Figure 1.
Methodological approach
(A) Experimental workflow. (B) Flow cytometry analysis of CAR T cells generated by CD8-LV or VSV-LV. (C) Location of customized primers (primer 1, primer 2) for amplification and subsequent detection of CAR mRNA and/or viral gRNA. Barcode elements required for cell and mRNA molecule identification are coupled with magnetic beads. After the cDNA synthesis, barcodes attach to the 3'-end of the targeted gene, enabling the matching of the paired-end reads performed by an Illumina sequencer.
Table 1.
Characteristics of the applied vectors and generated CAR T cells
| Sample | MOIa | Particles/cell | VCNb | Viabilityc |
|---|---|---|---|---|
| Untransduced | – | – | n.d. | 62.7% |
| CD8-LV | 0.045 | 12 × 103 | 0.91 ± 0.71d | 69.3% |
| VSV-LV | 33.3 | 5.8 × 103 | 3.94 ± 1.76 | 74.6% |
n.d., non-detectable.
As determined on MOLT cells.
VCN was measured in replicates from different samples generated using different batches of LVs on three donors, including the batch and donor used in the scRNA-seq experiment (untransduced n = 3, CD8-LV n = 7, VSV-LV n = 14) (mean ± standard deviation).
Determined by BD Rhapsody scanner upon staining with calcein AM and DRAQ7, before single-cell seeding.
VCN was quantified on DNA extracted from whole samples and extrapolated on CD8 cells based on their frequency determined by flow cytometry.
The high selectivity of CD8-LV is confirmed by scRNA sequencing
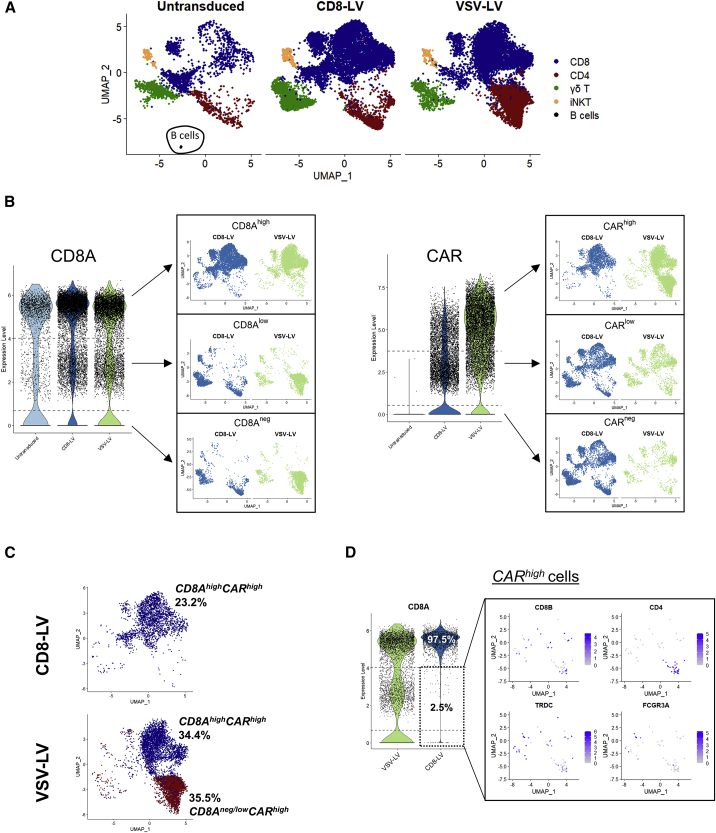
Single-cell mRNA was isolated, reverse transcribed, and amplified with the pool of primers from the Immune Response Panel (BD Biosciences) and the customized WPRE primers. Quality analysis of the generated cDNA libraries was within the expected range as determined by fragment analysis (Figure S1A). Low-quality cells (185 in total), which passed the Seven Bridges pre-processing filtering steps but showed too low or too high content of targeted genes and RNA molecule numbers, were eliminated from the analysis (Figure S1B). For the initial computational analysis, post-processed samples were merged in one Seurat object and principal component analysis was conducted, choosing the first 40 components for further analysis and uniform manifold approximation and projection (UMAP) plotting (Figures S1C and S2A). Unsupervised clustering identified 14 clusters (Figure S2B). The expression of the major T cell-associated markers was analyzed across the clusters, and, subsequently, clusters were merged depending on their identity (Figure S2C). The expected cell types such as CD4, CD8, γδ T cells, and some mixed population of natural killer T (NKT) cells were present, while transduced and untransduced samples showed similar cellular compositions (Figure 2A; Figure S2D). The only exception was a small cluster of B cells in the untransduced control sample (Figure 2A; Figure S2D). Presence of residual B cells during the first days of PBMC cultivation was expected, and their absence was previously demonstrated to be due to CAR T cell-mediated killing.14 The absence of B cells in the transduced samples thus confirmed the killing activity of the generated CAR T cells.
Figure 2.
CAR gene expression and CAR T cell subpopulations
(A) UMAP plots of the three samples colored for the major immune cell subtypes identified by the expression of marker genes. (B) Subsetting strategy based on the distribution of highly, low, and negatively expressing cells for CD8A and CAR genes and their projection into UMAP plots for cell population and purity evaluation. (C) Frequency of CAR T cells identified in scRNA sequencing based on the multimodal subsetting strategy. (D) CD8A expression of CARhigh cells across LV-exposed samples (left). Analysis of CD8Aneg/low/CARhigh cells identified in the CD8-LV sample for the expression of CD8, CD4, and TCRγδ.
When we plotted normalized gene expression data for CD8A-expressing cells, a trimodal distribution of CD8Ahigh, CD8Alow, and cells with undetectable or absent CD8A mRNA (CD8Aneg cells) became obvious in all samples (Figure 2B). Performing multimodal analysis, we identified the peaks of the modes as well as the antimodes, which we used as thresholds for gating of the populations (Figure S3). For the CD8Ahigh cells, this resulted in the most confined population of CD8 cells in UMAP plots (Figure 2B). On the other hand, CD8Alow and CD8Aneg cells comprised the rest of immune cell populations, including CD4 cells. While less pronounced, a similar tendency for a trimodal distribution was also observed for CAR T cells as determined by the CAR transgene expression (Figure 2B; Figure S3).
Next, we compared these data to the flow cytometry results generated from identical cell samples. Interestingly, we observed concordant CAR T cell frequencies between flow cytometry and scRNA data when we subset for CARhigh and CD8Ahigh in the scRNA plots. Then, 23.2% CAR T cells were identified by scRNA with CD8-LV (Figure 2C). Notably, our subsetting approach showed that 97.5% of CARhigh cells were CD8Ahigh cells in the CD8-LV sample (Figure 2D) and 2.5% of CARhigh cells (in total 59 cells) were CD8Aneg/low. These potential off-target cells consisted of 54.2% γδ T cells, 6.8% CD4/CD8 double-positive cells, 3.4% NKT cells, and 25.4% CD4 cells (Table 2; Figure S4). Of all these, only 28.8% were CD8Aneg and CD8Bneg double-negative cells (Table 2). Importantly, a close match in the frequencies of CD8 and CD4 CAR T cells between flow cytometry and scRNA analysis was also observed for CAR T cells generated with VSV-LV (Figure 2C), which nicely validated our gating strategy.
Table 2.
Identification of CARhighCD8Aneg/low cells after CD8-LV mediated transduction
| Cell type | Marker genes | Cell number |
|---|---|---|
| CD4 T cells | CD4posCD8Bneg | 15 |
| CD8Alow | 7 | |
| CD8Aneg | 8 | |
| CD8 T cells | CD8Bpos | 5 |
| CD8Alow | 5 | |
| CD4/CD8 T cells | CD4posCD8Bpos | 4 |
| CD8Alow | 2 | |
| CD8Aneg | 2 | |
| NKT | FCGR3Apos | 2 |
| CD8BposCD8Alow | 1 | |
| CD8BnegCD8Alow | 1 | |
| γδ T | TRDCpos | 32 |
| CD8BposCD8Alow | 7 | |
| CD8BposCD8Aneg | 7 | |
| CD8BnegCD8Alow | 9 | |
| CD8BnegCD8Aneg | 9 | |
| Remaining cells | CD8BnegCD4negTRDCnegFCGR3Aneg | 1 |
| CD8Alow | 1 |
Numbers in bold indicate total counts of the subfractions listed below, respectively.
The CARlow-expressing cells consisted of all identified immune cells, including CD8, CD4, γδ T, and NKT cells (Figures 2A and 2B). Comparison of their expression profile with that of CARneg cells revealed no significant differences between these two cell populations, whereas significant differences were obvious between CARlow and CARhigh cells (Figure S5). We therefore combined them with the CARneg cells in one group (CARneg/low) for further analysis. Notably, CARlow cells were substantially different from untransduced cells, which had not been exposed to vector particles (Figures S5A and S5B).
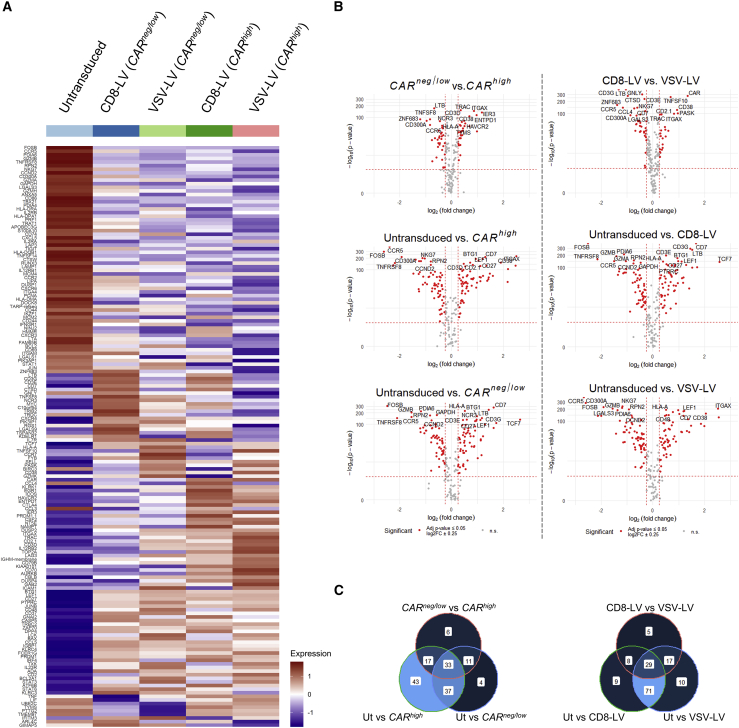
Differentially expressed genes in the CD8+ populations
By analyzing the isolated cell subsets of CD8 T cells, as described above, we observed alterations of gene expression profiles across the populations. The differentially expressed genes for each group identified by expression analysis are shown in the heatmap plot in Figure 3A. Intriguingly, we observed differences between the CARneg/low cells from the two vector types (Figure 3A). Moreover, we also observed significant differences in 58 genes between the CARhigh populations transduced by the two LVs (Figure 3A; Figure S6). Disregarding the LV type used for transduction, biggest differences were observed between untransduced (cells that were never exposed to LVs) and CARneg/low cells (LV-exposed cells) and also between untransduced and CARhigh cells, whereas the fold-change gene expression difference between CARneg/low and CARhigh cells was clearly less pronounced (Figure 3B). When comparing the particular vector types among each other or against untransduced, the differences from untransduced were in each case more pronounced, in terms of both the number of genes and the fold change, than the difference between CD8-LV- and VSV-LV-exposed cells (Figure 3B).
Figure 3.
Differentially expressed genes in the subpopulations
(A) Heatmap plots of 161 significantly differentially regulated genes identified for all groups compared (untransduced n = 1,444; CD8-LV [CARneg/low] n = 4,216; VSV-LV [CARneg/low] n = 1,739; CD8-LV [CARhigh] n = 2,257; VSV-LV [CARhigh] n = 3,084) by the FindAllMarkers function in Seurat (log2(fold change) ± 0.25, adjusted p value < 0.05). (B) Volcano plots comparing CARhigh, CARneg/low, and untransduced (Ut) cells, disregarding the LV used (left) and comparisons of the CD8+ populations in the CD8-LV, VSV-LV, and Ut samples (right). The fold change of CAR is not shown in every plot due to log2FC > 3. (C) Overlaps of differentially expressed genes identified based on CAR expression levels (left) and vector types (right).
The majority of the differentially expressed genes, 130 out of 151 either up- or downregulated, were identified when comparing untransduced with CARhigh cells (Figure 3C, left). From these, 70 genes were shared between the comparisons of untransduced with either CARhigh or CARneg/low cells, 33 genes were shared in all of the comparisons, including CARneg/low with CARhigh, whereas 43 were unique, comprising the biggest difference in that group of comparison. On the other hand, by comparing the whole CD8 cell groups between control and CD8-LV or VSV-LV, we found that most of the differentially expressed genes were shared between the vectors, when comparing control with transduced samples (100 out of 149), whereas 29 genes were commonly identified in all of the three comparisons (Figure 3C, right). Gene set enrichment analysis with the Gene Ontology (GO) Biological Process database revealed enrichment of genes especially related with cytokine-mediated signaling, T cell activation and immune response, and regulation of cell proliferation or apoptosis (Figure S7).
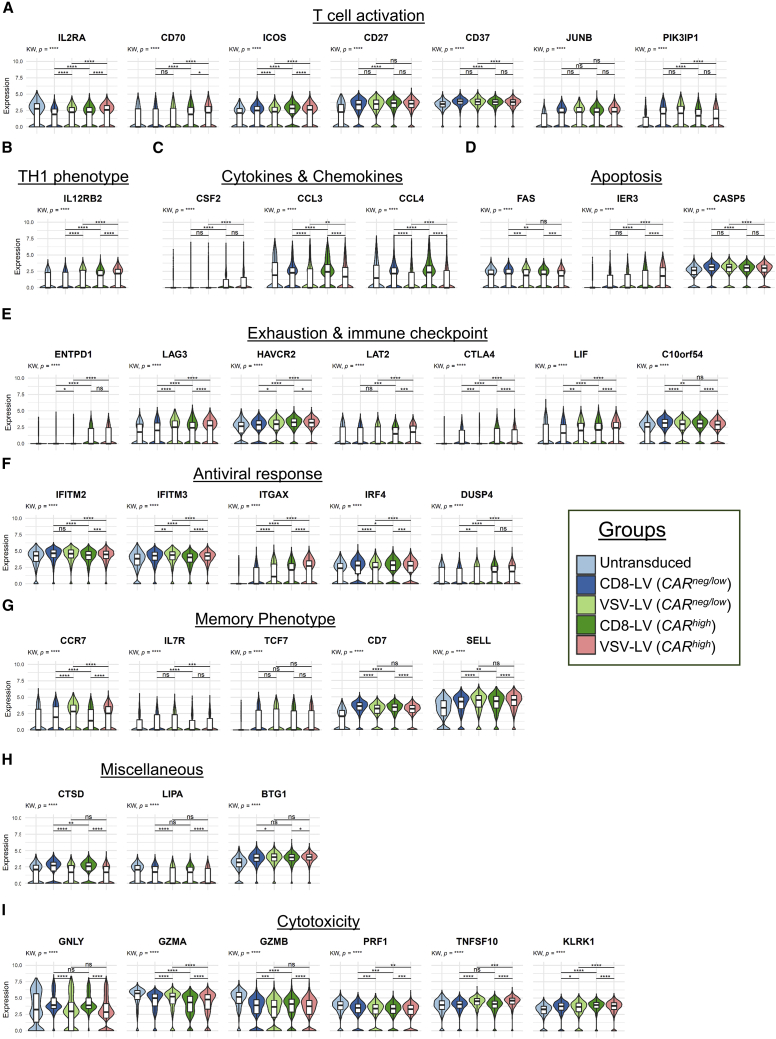
Looking closer into particular genes of CD8+ cells that show a significant level of up- or downmodulation between the different settings, we grouped them into topics related to CAR activities and vector-host interactions (Figure 4). With respect to T cell activation, upregulation of CD70, ICOS, and JUNB (part of the AP-1 transcription factor) selectively in the CARhigh cells is most likely a result of the CAR-mediated activities (Figure 4A). CARhigh CD8 cells favored a TH1-phenotype as exemplified by upregulated IL-12 receptor (IL12RB2) (Figure 4B) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (CSF2) (Figure 4C) as well as an unaltered expression of interferon-γ (IFNG) and STAT4 (Figure S8). Unchanged expression of TNF (Figure S8) and reduced levels of FAS (Figure 4D), which can trigger both T cell apoptosis as well as increased levels of IER3, are well in line with an anti-apoptotic profile in CARhigh cells (Figure 4D).21 The expression of several exhaustion and immune checkpoint markers (ENTPD1, LAG3, HAVCR2, LAT2, CTLA4) (Figure 4E) accompanied by the increase in co-stimulatory markers (ICOS, CD70, CD27) (Figure 4A) indicated early exhaustion of the CARhigh CD8 cells, potentially as a result of CAR tonic signaling.22 Finally, we found the mitogen-activated protein (MAP) kinase phosphatase-2 (DUSP4), which is known to inactivate MAP and extracellular signal-regulated kinase (ERK) kinases as well to promote TH1 response, to be significantly upregulated in CARhigh cells (Figure 4F).23,24
Figure 4.
Violin plots for genes of interest
Representative differentially expressed genes in CD8+ subpopulations clustered into topics related to CAR activities and vector-host interactions. (A) T cell activation. (B) TH1 phenotype. (C) Cytokines and chemokines. (D) Apoptosis. (E) Exhaustion and immune checkpoint. (F) Antiviral response. (G) Memory phenotype. (H) Miscellaneous. (I) Cytotoxicity. KW, Kruskal-Wallis multiple comparison test. Wilcoxon rank-sum test performed for pairwise comparisons. p values adjusted based on Bonferroni; ns, non-significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The bars in box plots indicate the 25th to 75th percentile range and the median value is shown with a horizontal line.
Besides being caused by CAR activities, differences between CARneg/low and CARhigh cells may have been caused by intrinsic factors present in particular cells preventing proper transduction by the LVs. Among these is the IL-2 receptor alpha subunit (IL2RA), which we found to be downmodulated in the CARneg/low cells not only compared to CARhigh cells but also compared to untransduced cells (Figure 4A), suggesting that their low activation level contributed to being protected from gene transfer. In line with this, PIK3IPI implicated in inhibition of T cell activation was upregulated in the CARneg/low population (Figure 4A), further confirming the low activation status of these cells.25 Although only slightly upregulated, both interferon-induced transmembrane proteins covered by the panel, IFITM2 and IFITM3, were significantly higher in CARneg/low cells than in untransduced or CARhigh cells (Figure 4F).
Besides changes in gene expression in the CARhigh cells only, we identified several genes that were up- or downregulated in both CARhigh and CARneg/low cells compared with untransduced cells. Thus, these changes were most likely due to the exposure to LV vectors. This referred to the negative regulators of proliferation and inhibitors of T cell activation CD37 and PIK3IPI, respectively (Figure 4A),25,26 as well as co-stimulatory and phenotype markers such as CD27, CD7, CD62L (SELL), and TCF7 (Figures 4A and 4G). Typical markers for apoptosis induction (CASP5) (Figure 4D) and exhaustion (LIF, C10orf54) (Figure 4E) were also induced upon exposure to LV particles. Of particular interest in this context are antiviral response factors. Indeed, BTG1, a cell cycle regulator,27 the CD11c gene (ITGAX), IFITM3, and in part also IRF4 were all upregulated upon exposure to LVs (Figures 4F and 4H).
Finally, gene profile variations were observed between the samples exposed to the two different LVs. In particular, ITGAX (Figure 4F) as well as the cytotoxic cytokine TRAIL gene (TNFSF10) (Figure 4I), CCR7, and CD62L (SELL) (Figure 4G) exhibited a tendency to be more upregulated in cells that had been exposed to VSV-LV than in those exposed to CD8-LV. On the other hand, the lysosomal enzyme lipase A (LIPA) was found to be more downmodulated in the VSV-LV sample (Figure 4H). Further examples for differences between these two groups referred to cathepsin D (CTSD) (Figure 4H), the chemokines CCL3 and CCL4 (Figure 4C), and granulysin (GNLY) (Figure 4I), which were selectively upregulated in CD8-LV-exposed cells.
Discussion
Deploying single-cell analysis tools in CAR T cell therapy can potentially not only improve the development and the optimization of the upstream processes but also enhance the clinical efficacy and reduce post-transfusion side effects by neutralizing product variations. In particular, failures in the subsetting of the exact CAR T cell population due to improper distinction between transduced and non-transduced cells can have immediate consequences for any biological conclusions drawn from the data. In this study, we have therefore started to follow early time points in CAR T cell generation, especially focusing on the transduction process with the final goal to improve the identification of the various subpopulations and to better understand why particular cells are becoming CAR T cells and others not.
Key for our analysis was the use of CD8-LV, which is known to be highly selective for the CD8+ T cells in human PBMCs.15 This facilitated identification of the transduced CD8+ T cells and their comparison to CAR T cells generated with the conventional VSV-LV. We decided to perform a targeted gene amplification and sequencing analysis covering ∼400 genes to establish our methodological approach. In particular, comparing to a typical whole transcriptome experiment requiring 50,000 reads/cell, we managed to efficiently sequence with ∼4,000 reads/cell without compromising the sequencing saturation and depth.28 This allowed us to increase the processed cell number by up to 10-fold compared to a typical whole transcriptome analysis (WTA) run and yet have a high resolution of the expressed genes.
Our approach identified a trimodal distribution not only in the expression of the CAR gene but also for CD8A. Notably, previous studies have not described this.16, 17, 18, 19, 20 Since the expression profiles of CARlow and CARneg cells were basically identical, they were merged for the downstream analyses (Figure S2A). The situation appears more complicated with the CD8Alow population. Indeed, the PBMCs used in this study include besides T lymphocytes also natural killer (NK) cells and dendritic cells (DCs). However, almost all CD8Alow cells were CD3+, thus rather excluding the presence of NK cells and DCs as explanation (Figure S2D). Instead, it is likely that transient fluctuations in mRNA levels that do not immediately convert in loss of the encoded proteins have accounted for this.29
In the current study, we have evaluated the selectivity of a receptor-targeted vector by both flow cytometry and scRNA-seq, thus on protein and mRNA levels. The obtained data on one hand were in perfect agreement with flow cytometry-based selectivity analysis and on the other hand confirmed the high, near-absolute selectivity of CD8-LV for its target cells (Figures 1B and 2D). Of the 59 (2.5%) potential off-target cells, 9 were in fact CD8+ or double-positive cells based on the expression of CD8B. Of the remaining 50 cells, 32 were γδ T cells, of which only 9 express neither CD8A nor CD8B; 2 cells were NKT cells; 15 were CD4 cells, of which 8 were negative for CD8 markers whereas 7 had CD8Alow expression (Table 2). This leaves just 17 cells that could be true off-target cells and thus an on-target rate of 99.28%. However, even these few cells could be target cells when considering the above-mentioned mRNA fluctuation. Combining barcoded antibody staining with expression profiling could be a straightforward next step to clarify this issue.
Our study revealed interesting differences in expression of particular genes between the groups. The data revealed transcriptomic alterations upon exposure to vector particles and expression of the CAR as the result of proper gene delivery and transduction. Thus, the exposure of cells to any of the two LVs resulted in alterations of their gene expression profiles, regardless of whether the cells were eventually properly transduced by the vector or not. These findings suggest that under the given experimental conditions exposure of T cells to LV particles results in stronger gene expression profile alterations than presence of the CAR. While CD8+ CARhigh cells exhibited an activated TH1 phenotype and an overall profile well in accordance with that observed in previous studies (Figure 4),16,17,20 CD8 CARneg/low cells expressed genes that potentially restricted cell viability, phenotype, and viral entry. These cells had upregulated genes involved in inhibition of T cell activation (PIK3IP1) and proliferation (CD37, BTG1) as well as promoting pyroptosis (CASP5), a type of programmed cell death resulting in inflammatory cytokine release (Figures 4A, 4D, and 4H).25,26,30,31 Particularly remarkable was the observation that two restriction factors (IFITM2, IFITM3) directly implicated in preventing viral entry were significantly increased in those cells that remained CARneg despite having been exposed to LVs (Figure 4F). The two interferon-induced transmembrane proteins expressed from IFITM2 and IFITM3 reside in endosomal compartments and inhibit the fusion of the viral envelope with the endosomal membrane, a process characteristic for the pH-dependent entry pathway of VSV-LV.32 IFITM3 has been investigated most and was recently identified as the main target to improve gene transfer into hematopoietic stem cells through the use of resveratrol as transduction enhancer.33, 34, 35 Although our data certainly support this strategy also for T lymphocytes, they also suggest that not only intracellular localization of IFITMs but also subtle changes in their overall expression levels may determine whether a particular T cell becomes properly transduced.35
Differences in gene profiles between the cells exposed to CD8-LV versus VSV-LV were also observed, although we previously demonstrated that CAR T cells generated with each of the two vector types were equally active in killing tumor target cells.15 These might be a consequence of the viral entry pathway used, toxicity of VSV-LV or the transduction of CD4 cells by VSV-LV thus affecting CD8 cells in a paracrine cytokine secretion-driven manner. CCL4 (MIP-1β), a major HIV suppressive factor, was highly upregulated in CD8 cells of the CD8-LV sample, indicating a possible antiviral response against the Nipah virus-derived envelope of CD8-LV (Figure 4C).36 In addition, CCR7 and CD62L were upregulated upon VSV-LV exposure, indicating a more pronounced central memory phenotype in these CAR T cells than in those generated with CD8-LV (Figure 4G). This is in line with granulysin (GNLY) increase in the CD8-LV sample, which is indicative for a more cytotoxic activity of these CAR T cells (Figure 4I).37 Although a central memory phenotype is beneficial for CAR T cell persistence,38 previous phenotype comparisons between CAR T cells generated with VSV-LV versus CD8-LV did not reveal significant differences.15 In the same direction, cathepsin-D peptidase (CTSD) was significantly increased in the CD8-LV sample (Figure 4H). It prevents oxidative stress-induced cell death, a feature relevant for CD3/CD28-activated T cells, in which the glycolysis pathway is elevated because of a skew to effector phenotype leading to production of reactive oxygen species (ROS).39,40 In the VSV-LV sample, a lysosomal lipase (LIPA) was downregulated, which correlates with the endosomal-related pH-dependent entry pathway of VSV-LV (Figure 4H).
Although further investigation will be required before final conclusions on up- or downregulation of particular genes can be drawn, we have here successfully established a scRNA-seq workflow for CAR T cells, generated with conventional or receptor-targeted LV, and managed to distinguish transduced from untransduced cells by implementing a customized pair of primers. Based on the distribution of gene expression, we propose a subsetting method for distinguishing CAR T cells in scRNA-seq analysis, wherever applicable.
Materials and methods
Cell culture
HEK293T (ATCC CRL-11268) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich, Munich, Germany) supplemented with 2 mM glutamine (Sigma-Aldrich) and 10% fetal calf serum (FCS) (Biochrom, Berlin, Germany). MOLT 4.8 cells were cultured in RPMI 1640 (Biowest, Nuaillé, France) supplemented with 10% FCS and 2 mM glutamine. All cell cultures were incubated at 37°C with 5% CO2 and 90% humidity.
Generation of lentiviral vectors
A detailed protocol describing the generation of T cell-targeted LVs was recently published.41 In brief, second-generation LVs were produced as previously described by co-transfecting 2 × 107 HEK293T cells per T125 flask with a plasmid cassette, consisting of packaging plasmid (pCMVΔR8.9) and transfer plasmid (pSEW-mycCD19-CAR-28z), in the presence of linear polyethylenimine (PEI). For the generation of VSV-pseudotyped LV we co-transfected the plasmid encoding the envelope G-glycoprotein (pMD2.G) and for the CD8-LV the plasmids encoding the mutated G-glycoprotein of the Nipah virus coupled with single-chain anti-CD8α (pCAGGS-NiV-Gd34-CD8) and the truncated fusion glycoprotein (pCAGGS-NiV-Fd22). LVs were harvested from culture supernatants, concentrated by a 24-h centrifugation through a 20% sucrose cushion at 4,500 × g (4°C), and re-suspended in 60 μL per flask Dulbecco’s phosphate-buffered saline (DPBS, Mg2+ and Ca2+ free), followed by aliquoting and freezing at −80°C. LVs were thawed only once for each experimental use.
Transduction units per mL (TU/mL) were calculated based on the titration of the vector stocks on MOLT 4.8 cells. The multiplicity of infection (MOI) applied in each experimental condition was calculated by correlating the transducing units added to the number of cells present in the PBMC culture. Particle numbers in the vector stocks were measured by nanoparticle tracking analysis using the NanoSight device as previously described.12
PBMC isolation, culture, and transduction
PBMCs were purified from a buffy coat derived from an anonymous donation collected at the German Red Cross blood donation center (DRK-Blutspendedienst Baden-Württemberg-Hessen, Frankfurt) by Pancoll (PAN-Biotech, Aidenbach, Germany) density gradient centrifugation and cryopreserved in 90% FCS and 10% dimethyl sulfoxide (DMSO). Cells were thawed at 37°C, washed, and activated for 72 h in a 6-well plate with pre-coated human recombinant anti-CD3 (1 μg/mL, clone OKT3, Miltenyi Biotec, Bergisch Gladbach, Germany), soluble anti-CD28 (3 μg/mL, clone 15E8, Miltenyi Biotec), supplemented with the cytokines IL-7 (25 IU/mL) and IL-15 (50 IU/mL), in RPMI medium with 10% FCS, 2 mM glutamine, 0.5% penicillin-streptomycin, and 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (Sigma-Aldrich). After activation, 8 × 104 cells per well were seeded in a 96-well plate and spinoculated with 1 μL of each LV stock at 850 × g and 32°C for 90 min. Medium was refreshed once after 72 h of inoculation. Final samples were collected and processed 6 days post-inoculation.
For quantification of the vector copy number (VCN), genomic DNA was isolated from at least 2.5 × 105 cells with the DNeasy blood and tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions, after elution with DNA-free water (Sigma-Aldrich). Real-time PCR was performed as previously described,42 with primer pairs specific for WPRE and human albumin (ALB). VCN was calculated by the formula . The efficiency of the standard curves was within a range of 90%–110%.
Flow cytometry
Cells were re-suspended, washed with buffer (PBS, 5% FCS, 2 mM NaN3), and stained for 30 min at 4°C with the antibodies anti-CD3 [fluorescein] (clone BW264/56, Miltenyi Biotec), anti-CD8 [VioBlue] (clone BW135/80, Miltenyi Biotec), and anti-c-myc [phycoerythrin] (9B11, Cell Signaling Technology, Danvers, MA, USA) and with a fixable viability dye (eFluor 780, Thermo Fisher Scientific, Waltham, MA, USA). After staining, cells were washed twice, fixed with 0.5% paraformaldehyde (PFA), and run in MACSQuant X (Miltenyi Biotec). Results were analyzed in FCS Express v.6 (De Novo Software, Pasadena, CA, USA).
Single-cell RNA isolation, library preparation, and sequencing
Single-cell isolation and mRNA processing were conducted with the BD Rhapsody platform (BD Biosciences) according to the manufacturer’s instructions (Doc IDs: 214062 and 210968, BD Biosciences). For viability evaluation, cell suspensions were incubated for 5 min at 37°C with 10 μM calcein AM (Thermo Fisher Scientific) and 1.5 μM DRAQ7 (BD Biosciences). Single cells were then captured in nanowell-containing cartridges; cellular mRNA was released after lysis and captured by poly(dT)-coated magnetic beads. Beads were subsampled to yield ∼31,000 total cells.
Reverse-transcribed cDNA was amplified with the Immune Response Panel (cat. 633750, BD Biosciences) of primers covering 399 genes. For detection of mRNA derived from the CAR gene in CAR T cells, we in silico predicted and confirmed by sequencing the poly(A) signal site within the 3'-SIN-LTR) of the transfer vector plasmid (Poly(A) Signal Miner).43 Primers binding within 800 bp away from the poly(A) signal, thus within the WPRE, were then customized by BD Biosciences (Franklin Lakes, NJ, USA) and added to the Immune Response Panel.
Libraries were indexed uniquely, and their quality and final length were assessed with the high-sensitivity NGS kit with the Fragment Analyzer (Agilent, Santa Clara, CA, USA). Data were analyzed in ProSize 2.0 (Advanced Analytical Technologies, Heidelberg, Germany) (Figure S1A). Libraries were then quantified by Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), pooled in a ratio defined by the cell number of each sample and the required sequencing depth, and loaded into a NextSeq High Output flowcell at 1 pM, spiked with 20% PhiX, and sequenced in NextSeq 550 (Illumina, San Diego, CA, USA).
Data analysis
The raw FASTQ files were processed with the bioinformatics pipeline of Seven Bridges Genomics (Charlestown, MA, USA). Overall, 21,527 cells passed the pre-processing step with a mean sequencing depth of 4,304 reads per cell. Recursive substitution error correction (RSEC)-adjusted molecule count matrices were generated and processed in R (v.4.0.3) with Seurat.44 Thresholds for filtering the low quality of cells and the number of principal components chosen for the UMAP analysis are shown in Figures S1B–S1D. The filtering metrics as well as the final number of putative cells per sample are listed in Table S1. Wilcoxon rank-sum test for differential gene expression analysis was performed with the FindAllMarkers and FindMarkers functions, applying the thresholds of log2(fold change) ± 0.25 and minimum fraction of expressing cells 25%. Violin plots were constructed with ggplot2 (v.3.3.3), and Kruskal-Wallis non-parametric test and Wilcoxon rank-sum pairwise test were performed with rstatix (v.0.7.0).45 GO Biological Process enrichment analysis on differentially expressed genes was performed with the Independent Enrichment Analysis tool of Appyters collection by defining the genes of the Immune Response Panel.46 For every multiple comparison, including the differential gene expression analysis and volcano plotting, p values were corrected based on Bonferroni. Multimodal analysis was conducted with the package multimode.47 Abbreviations used represent the following values: ns = non-significant, ∗p< 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Data availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE184895 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184895).48
Acknowledgments
The authors are grateful to Csaba Miskey (Paul-Ehrlich-Institut) for his advice on library generation and sequencing, to Manuela Gallet and Vanessa Riechert (both Paul-Ehrlich-Institut) for the qPCR, as well as to Vadir Lopez-Salmeron, Edyta Kowalczyk, and Siobhan Cashman (all BD Biosciences) for their advice on the Rhapsody system. This project has received funding to C.C. and C.J.B. from the European Union’s Horizon 2020 research and innovation program STACCATO under the Marie Skłodowska-Curie grant agreement No. 813453.
Author contributions
Conceptualization: C.J.B., F.T.C., and F.B.T. Data curation: F.T.C. Formal analysis: F.T.C. and E.A. Funding acquisition: C.J.B. and C.C. Investigation: F.T.C. and E.A. Methodology: C.J.B., F.T.C., and F.B.T. Project administration: C.J.B. Resources: C.J.B. and C.C. Supervision: C.J.B. Visualization: F.T.C. and E.A. Writing – original draft: F.T.C., E.A., and C.J.B. Writing – review & editing: C.J.B. and C.C.
Declaration of interests
C.J.B. is listed as co-inventor of the patent for the CD8-targeted lentiviral vector. All the other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.09.019.
Supplemental information
References
- 1.Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher J., Brentjens R.J., Gunset G., Rivière I., Sadelain M. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRzeta /CD28 receptor. Nat. Biotechnol. 2002;20:70–75. doi: 10.1038/nbt0102-70. [DOI] [PubMed] [Google Scholar]
- 3.Gross G., Gorochov G., Waks T., Eshhar Z. Generation of effector T cells expressing chimeric T cell receptor with antibody type-specificity. Transplant. Proc. 1989;21:127–130. [PubMed] [Google Scholar]
- 4.Schuster S.J., Svoboda J., Chong E.A., Nasta S.D., Mato A.R., Anak Ö., Brogdon J.L., Pruteanu-Malinici I., Bhoj V., Landsburg D. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med. 2017;377:2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milone M.C., O’Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32:1529–1541. doi: 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaiss A.-K., Son S., Chang L.-J. RNA 3’ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J. Virol. 2002;76:7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zufferey R., Donello J.E., Trono D., Hope T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amirache F., Lévy C., Costa C., Mangeot P.E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 12.Friedel T., Hanisch L.J., Muth A., Honegger A., Abken H., Plückthun A., Buchholz C.J., Schneider I.C. Receptor-targeted lentiviral vectors are exceptionally sensitive toward the biophysical properties of the displayed single-chain Fv. Protein Eng. Des. Sel. 2015;28:93–106. doi: 10.1093/protein/gzv005. [DOI] [PubMed] [Google Scholar]
- 13.Frank A.M., Buchholz C.J. Surface-Engineered Lentiviral Vectors for Selective Gene Transfer into Subtypes of Lymphocytes. Mol. Ther. Methods Clin. Dev. 2018;12:19–31. doi: 10.1016/j.omtm.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeiffer A., Thalheimer F.B., Hartmann S., Frank A.M., Bender R.R., Danisch S., Costa C., Wels W.S., Modlich U., Stripecke R. In vivo generation of human CD19-CAR T cells results in B-cell depletion and signs of cytokine release syndrome. EMBO Mol. Med. 2018;10:e9158. doi: 10.15252/emmm.201809158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamali A., Kapitza L., Schaser T., Johnston I.C.D., Buchholz C.J., Hartmann J. Highly Efficient and Selective CAR-Gene Transfer Using CD4- and CD8-Targeted Lentiviral Vectors. Mol. Ther. Methods Clin. Dev. 2019;13:371–379. doi: 10.1016/j.omtm.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Q., Han G., Puebla-Osorio N., Ma M.C.J., Strati P., Chasen B., Dai E., Dang M., Jain N., Yang H. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 2020;26:1878–1887. doi: 10.1038/s41591-020-1061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheih A., Voillet V., Hanafi L.A., DeBerg H.A., Yajima M., Hawkins R., Gersuk V., Riddell S.R., Maloney D.G., Wohlfahrt M.E. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 2020;11:219. doi: 10.1038/s41467-019-13880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai Z., Lundh S., Kim D., Woodhouse S., Barrett D.M., Myers R.M., Grupp S.A., Maus M.V., June C.H., Camara P.G. Single-cell multiomics dissection of basal and antigen-specific activation states of CD19-targeted CAR T cells. J. Immunother. Cancer. 2021;9:e002328. doi: 10.1136/jitc-2020-002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boroughs A.C., Larson R.C., Marjanovic N.D., Gosik K., Castano A.P., Porter C.B.M., Lorrey S.J., Ashenberg O., Jerby L., Hofree M. A Distinct Transcriptional Program in Human CAR T Cells Bearing the 4-1BB Signaling Domain Revealed by scRNA-Seq. Mol. Ther. 2020;28:2577–2592. doi: 10.1016/j.ymthe.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Peticone C., Kotsopoulou E., Göttgens B., Calero-Nieto F.J. Single-cell transcriptome analysis of CAR T-cell products reveals subpopulations, stimulation, and exhaustion signatures. OncoImmunology. 2021;10:1866287. doi: 10.1080/2162402X.2020.1866287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arlt A., Schäfer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur. J. Cell Biol. 2011;90:545–552. doi: 10.1016/j.ejcb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Long A.H., Haso W.M., Shern J.F., Wanhainen K.M., Murgai M., Ingaramo M., Smith J.P., Walker A.J., Kohler M.E., Venkateshwara V.R. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 2015;21:581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Mutairi M.S., Cadalbert L.C., McGachy H.A., Shweash M., Schroeder J., Kurnik M., Sloss C.M., Bryant C.E., Alexander J., Plevin R. MAP kinase phosphatase-2 plays a critical role in response to infection by Leishmania mexicana. PLoS Pathog. 2010;6:e1001192. doi: 10.1371/journal.ppat.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haddock A.N., Labuzan S.A., Haynes A.E., Hayes C.S., Kakareka K.M., Waddell D.S. Dual-specificity phosphatase 4 is upregulated during skeletal muscle atrophy and modulates extracellular signal-regulated kinase activity. Am. J. Physiol. Cell Physiol. 2019;316:C567–C581. doi: 10.1152/ajpcell.00234.2018. [DOI] [PubMed] [Google Scholar]
- 25.Uche U.U., Piccirillo A.R., Kataoka S., Grebinoski S.J., D’Cruz L.M., Kane L.P. PIK3IP1/TrIP restricts activation of T cells through inhibition of PI3K/Akt. J. Exp. Med. 2018;215:3165–3179. doi: 10.1084/jem.20172018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Spriel A.B., Puls K.L., Sofi M., Pouniotis D., Hochrein H., Orinska Z., Knobeloch K.-P., Plebanski M., Wright M.D. A regulatory role for CD37 in T cell proliferation. J. Immunol. 2004;172:2953–2961. doi: 10.4049/jimmunol.172.5.2953. [DOI] [PubMed] [Google Scholar]
- 27.Hwang S.S., Lim J., Yu Z., Kong P., Sefik E., Xu H., Harman C.C.D., Kim L.K., Lee G.R., Li H.-B., Flavell R.A. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020;367:1255–1260. doi: 10.1126/science.aax0194. [DOI] [PubMed] [Google Scholar]
- 28.Mair F., Erickson J.R., Voillet V., Simoni Y., Bi T., Tyznik A.J., Martin J., Gottardo R., Newell E.W., Prlic M. A Targeted Multi-omic Analysis Approach Measures Protein Expression and Low-Abundance Transcripts on the Single-Cell Level. Cell Rep. 2020;31:107499. doi: 10.1016/j.celrep.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erhard F., Baptista M.A.P., Krammer T., Hennig T., Lange M., Arampatzi P., Jürges C.S., Theis F.J., Saliba A.-E., Dölken L. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature. 2019;571:419–423. doi: 10.1038/s41586-019-1369-y. [DOI] [PubMed] [Google Scholar]
- 30.Guéhenneux F., Duret L., Callanan M.B., Bouhas R., Hayette S., Berthet C., Samarut C., Rimokh R., Birot A.M., Wang Q. Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia. 1997;11:370–375. doi: 10.1038/sj.leu.2400599. [DOI] [PubMed] [Google Scholar]
- 31.Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 32.Hornick A.L., Li N., Oakland M., McCray P.B., Jr., Sinn P.L. Human, Pig, and Mouse Interferon-Induced Transmembrane Proteins Partially Restrict Pseudotyped Lentiviral Vectors. Hum. Gene Ther. 2016;27:354–362. doi: 10.1089/hum.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao X., Li J., Winkler C.A., An P., Guo J.T. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front. Microbiol. 2019;9:3228. doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozog S., Timberlake N.D., Hermann K., Garijo O., Haworth K.G., Shi G., Glinkerman C.M., Schefter L.E., D’Souza S., Simpson E. Resveratrol trimer enhances gene delivery to hematopoietic stem cells by reducing antiviral restriction at endosomes. Blood. 2019;134:1298–1311. doi: 10.1182/blood.2019000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cocchi F., DeVico A.L., Garzino-Demo A., Arya S.K., Gallo R.C., Lusso P. Identification of RANTES, MIP-1 α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 37.Nicolet B.P., Guislain A., van Alphen F.P.J., Gomez-Eerland R., Schumacher T.N.M., van den Biggelaar M., Wolkers M.C. CD29 identifies IFN-γ-producing human CD8+ T cells with an increased cytotoxic potential. Proc. Natl. Acad. Sci. USA. 2020;117:6686–6696. doi: 10.1073/pnas.1913940117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klebanoff C.A., Gattinoni L., Torabi-Parizi P., Kerstann K., Cardones A.R., Finkelstein S.E., Palmer D.C., Antony P.A., Hwang S.T., Rosenberg S.A. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc. Natl. Acad. Sci. USA. 2005;102:9571–9576. doi: 10.1073/pnas.0503726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F., Zhang H. Lysosomal Acid Lipase in Lipid Metabolism and Beyond. Arterioscler. Thromb. Vasc. Biol. 2019;39:850–856. doi: 10.1161/ATVBAHA.119.312136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hah Y.S., Noh H.S., Ha J.H., Ahn J.S., Hahm J.R., Cho H.Y., Kim D.R. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012;323:208–214. doi: 10.1016/j.canlet.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Weidner T., Agarwal S., Perian S., Fusil F., Braun G., Hartmann J., Verhoeyen E., Buchholz C.J. Genetic in vivo engineering of human T lymphocytes in mouse models. Nat. Protoc. 2021;16:3210–3240. doi: 10.1038/s41596-021-00510-8. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal S., Hanauer J.D.S., Frank A.M., Riechert V., Thalheimer F.B., Buchholz C.J. In Vivo Generation of CAR T Cells Selectively in Human CD4+ Lymphocytes. Mol. Ther. 2020;28:1783–1794. doi: 10.1016/j.ymthe.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Han H., Li J., Wong L. An in-silico method for prediction of polyadenylation signals in human sequences. Genome informatics. International Conference on Genome Informatics. 2003;14:84–93. [PubMed] [Google Scholar]
- 44.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag) [Google Scholar]
- 46.Clarke D.J.B., Jeon M., Stein D.J., Moiseyev N., Kropiwnicki E., Dai C., Xie Z., Wojciechowicz M.L., Litz S., Hom J. Appyters: Turning Jupyter Notebooks into data-driven web apps. Patterns (NY) 2021;2:100213. doi: 10.1016/j.patter.2021.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ameijeiras-Alonso J., Crujeiras R.M., Rodriguez-Casal A. multimode: An R Package for Mode Assessment. J. Stat. Softw. 2021;97:1–32. [Google Scholar]
- 48.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE184895 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184895).48