Highlights
-
•
The cellular activity of USPs is functionally implicated in cancer progression.
-
•
USP8 negatively regulates the TRAF6-mediated signals for NF-κB and autophagy.
-
•
USP8KO SK-HEP-1 cells reveal increases of cancer progression in vivo and in vitro.
-
•
TCGA and transcriptome data support the functional role of USP8 in liver cancers.
Keywords: USP8, Autophagy, Toll-like receptor 4, TRAF6-mediated signal, Liver cancer progression
Abbreviations: USP, ubiquitin-specific protease; LIHC, liver hepatocellular carcinoma; TLR, toll-like receptor; TRAF6, TNF receptor associated factor 6; TAK1, TGF-beta activated kinase 1; TAB2, TAK1 binding protein 2; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; VEGF, vascular endothelial growth factor; MCP-1, monocyte chemotactic protein-1; ROS, reactive oxygen species; LPS, lipopolysaccharide; NSG, NOD scid gamma; HEK, human embryonic kidney; TCGA, the cancer genome atlas; GAPIA, gene expression profiling interactive analysis; IHC, immunohistochemistry; H&E, hematoxylin and eosin; DMSO, dimethyl sulfoxide; 3-MA, 3-methyladenine; CQ, chloroquine
Abstract
Herein, we aimed to elucidate the molecular and cellular mechanism in which ubiquitin-specific protease 8 (USP8) is implicated in liver cancer progression via TRAF6-mediated signal. USP8 induces the deubiquitination of TRAF6, TAB2, TAK1, p62, and BECN1, which are pivotal roles for NF-κB activation and autophagy induction. Notably, the LIHC patient with low USP8 mRNA expression showed markedly shorter survival time, whereas there was no significant difference in the other 18-human cancers. Importantly, the TCGA data analysis on LIHC and transcriptome analysis on the USP8 knockout (USP8KO) SK-HEP-1 cells revealed a significant correlation between USP8 and TRAF6, TAB2, TAK1, p62, and BECN1, and enhanced NF-κB-dependent and autophagy-related cancer progression/metastasis-related genes in response to LPS stimulation. Furthermore, USP8KO SK-HEP-1 cells showed an increase in cancer migration and invasion by TLR4 stimulation, and a marked increase of tumorigenicity and metastasis in xenografted NSG mice. The results demonstrate that USP8 is negatively implicated in the LIHC progression through the regulation of TRAF6-mediated signal for the activation of NF-κB activation and autophagy induction. Our findings provide useful insight into the LIHC pathogenesis of cancer progression.
Introduction
Ubiquitin-specific proteases (USPs) as a deubiquitinases, regulate multiple fundamental cellular processes including cell proliferation, cell cycle, and cancer progression by removing ubiquitin from the substrate proteins [1,2]. However, recent evidences have demonstrated that ubiquitin specific peptidase 8 (USP8) stabilizes oncogenes and proto-oncogenes, therefore promoting cancer progression and survival through the activation of multiple signaling pathways [1,3,4]. It is critically associated with the movement of the ubiquitin from oncogenes and proto-oncogenes, and thus protect the proteins from degradation [3,4]. Depending with the ubiquitination linkages, the ubiquitinated substrate proteins can either play part in the regulation of protein degradation pathway, like in the case of K48-linkage ubiquitination, or functional regulation of cellular signaling, like in the case of K63-linkage ubiquitination [5], [6], [7]. USP8 can cleave K6, K48, and K63 linkages in the ubiquitin chain of the substrate proteins [8,9]. However, while there are numerous studies on the regulation of K48-linkage ubiquitinated substrates by USP8 and the subsequent role in cancer progression [10,11], the implication of K63-linkage deubiquitination by USP8 in cancer progression is poorly understood.
Accumulating evidences have demonstrated that the autophagy and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activity induced by toll-like receptors (TLRs) functionally regulates cancer progression [12], [13], [14], [15], [16], [17], [18]. When TLRs are stimulated, TNF Receptor Associated Factor 6 (TRAF6) simultaneously induces the K63-linked ubiquitination of TAK1 Binding Protein 2 (TAB2) and TGF-Beta Activated Kinase 1 (TAK1) for the activation of NF-κB, and the K63-linked ubiquitination of BECN1 for autophagy induction, subsequently enhancing cancer progression [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. Interestingly, several deubiquitinating enzymes, such as A20 and USP14, have been reported to give raise to dampen cellular signals for the activation of NF-κB and autophagy induction through the deubiquitination of TRAF6-downstream molecules [23], [24], [25], [26]. A20 specifically targeting the K63-linked ubiquitin chain antagonizes the modifying function of TRAF6 on the ubiquitinated Beclin-1, thereby attenuating autophagy induction and cancer progression in response to TLR signaling [23,24]. Additionally, USP14 negatively regulates the autophagy induction by controlling K63 ubiquitination of Beclin 1 and the activation of NF-κB by controlling K63 ubiquitination of TAB2 [25,26]. The above findings strongly suggest that the functional identification of cellular USPs might help to understand their pathological mechanism and explore the relevance of USPs as therapeutic targets in cancer progression.
Liver hepatocellular carcinoma (LIHC) is a major cause of cancer-related mortality and cellular activation of NF-κB and autophagy has been reported to majorly affect its progression [27], [28], [29], [30], [31], [32]. Several cellular factors including vascular endothelial growth factor (VEGF) and monocyte chemotactic protein-1 (MCP-1) are regulated by reactive oxygen species (ROS) and NF-ĸB, and play an essential role in tumor angiogenesis and tumor aggressiveness [33], [34], [35]. On the other hand, accumulating evidence has demonstrated that autophagy is robustly activated in hepatocellular carcinoma (HCC tumor) under various cellular stressors and promotes tumor malignancy and progression [36,37]. Recent reports have also shown that TLRs stimulation enhance the production of cytokines, such as IL-6, CCL2/MCP-1, CCL20/MIP-3α, VEGFA, and MMP2, which are necessary for cancer cells migration and invasion, through the TRAF6-mediated ubiquitination of BECN1 for autophagy activation [15], [16], [17], [18]. However, while increasing evidence have suggested that NF-κB and autophagy functionally promotes tumorigenesis and cancer progression it is partly controversial whether the two are either positively or negatively implicated in the cancer progression [27], [28], [29], [30], [31], [32].
In the present study, we investigated the functional role of USP8 in the LIHC cancer progression in vitro and in vivo. The current work has addressed the molecular and cellular mechanism by which USP8 as a deubiquitinase is implicated in the TRAF6-mediated signaling for the activation of NF-κB and autophagy. Furthermore, we provide evidence using CRISPR/Cas9 knockout method, transcriptome analysis, TCGA data analysis, and xenografted NOD scid gamma (NSG) mice to support the molecular and cellular mechanism of USP8 in the regulation of liver cancer progression. Our data also provide useful insights in to the pathogenesis of cancer progression and might contribute to develop new therapeutic targets for LIHC.
Methods and materials
Animal experimentation
NOD/SCID/IL-2Rγnull (NSG) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained under specific pathogen-free conditions in accordance with the ethical guidelines for the care of these mice at the Laboratory Animal Research Center (LARC) of the Samsung Biomedical Research Institute (SBRI) and Samsung Medical Center (SMC), Seoul, South Korea. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the SMC (No. 20160617001). The NSG mice were 6–8 weeks old at the time of injection with cancer cell line. USP8KO (5 × 106 cells per mouse, n = 10) or control (Ctrl) SK-HEP-1 cells (5 × 106 cells per mouse, n = 10) were injected under the NSG mice skin (back area) within serum free DMEM. Final injection volume was 100 μl/mouse, cell suspension was 50 μl and 1:1 v/v mixture of ice-chilled Matrigel (BD Biosciences, La Jolla, CA, USA), and kept on ice until injection. Two weeks after cancer cell injection, tumor volume was measured by a caliper every 3, 4 days until 70 days after injection, and the volumes (mm3) were calculated by (length x width)2 × 0.5. Tumor growth curves are presented as average tumor volume ± SEM for each group in this study. All studies involving mice were approved by the Nemours IACUC.
Cells
Human embryonic kidney (HEK) 293T cells (ATCC, CRL-11268) were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, 11965092) with 10% fetal bovine serum (FBS). Human liver adenocarcinoma SK-HEP-1 cells (ATCC, HTB-52) were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM, HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, Cromwell, CT, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (HyClone) in a humidified atmosphere at 37 °C with and 5% CO2.
Generation of USP8-knockout (USP8KO) SK-HEP-1 cells by CRISPR/Cas9
The guide RNA sequences for CRISPR/Cas9 were designed as 5′-CACCG AATGGATGCTCGAAGAATGC -3′ and 3′- CTTACCTACGAGCTTCTTACG CAAA -5′ for human USP8. USP8KO SK-HEP-1 cells were generated as previously described.17,18 Complementary oligonucleotides to the guide RNAs (gRNAs) were annealed and cloned into a lentivirus CRISPR v2 vector (Addgene plasmid, 52961). Lenti CRISPR v2/gRNA was transfected into SK-HEP-1 cells using Lipofectamine 2000 according to the manufacturer's instructions. USP8KO SK-HEP-1 colonies were selected and confirmed by western blots, as previously described [17,18].
Antibodies and reagents
Anti-Myc (2276), anti-GAPDH (2118), and anti-LC3A/B (4108) were purchased from Cell Signaling Technology. Anti-HA (ab18181) was purchased from Abcam. Flag (SAB4200071), lipopolysaccharide (LPS; serotype 0128: B12), chloroquine (CQ; C6628), dimethyl sulfoxide (DMSO; 472301), puromycin (P8833), paraformaldehyde (P6148), Triton X-100 (T8787), 3-methyladenine (3-MA; M9281), gentamicin (G1272), deoxycholate (D6750), and Dulbecco's phosphate-buffered saline (DPBS; D8537) were purchased from Sigma-Aldrich. Lipofectamine 2000 (11668019) was purchased from Thermo Fisher Scientific.
Plasmids
Flag-TRAF6 (21624), Flag-HA-USP8 (22608), HA-p62 (28027), pRK6-HA-TAK1 (44160), Flag-BECN1 (24388) plasmids were purchased from Addgene. Using Flag-TRAF6 plasmid, a full-length Myc-TRAF6 construct was cloned into a pCMV-3Tag-7 vector (Agilent Technologies, 240202) and generated. Using Flag-HA-USP8 plasmid, a full-length Myc-USP8 or a full-length Flag-USP8 construct was cloned into a pCMV-3Tag-7 vector (Agilent Technologies, 240202) and generated. Using Flag-BECN1 plasmid, a full-length Myc-BECN1 construct was cloned into a pCMV-3Tag-7 vector (Agilent Technologies, 240202) and generated. Using HA-p62 plasmid, a full-length Myc-p62 construct was cloned into a pCMV-3Tag-7 vector (Agilent Technologies, 240202) and generated. Using pRK6-HA-TAK1 plasmid, a full-length Myc-TAK1 construct was cloned into a pCMV-3Tag-7 vector (Agilent Technologies, 240202) and generated. Flag-TAB1 and HA-Ub plasmids were obtained from Dr. J. H. Shim (University of Massachusetts Medical School, USA). Truncated mutants of Flag-TRAF6, Flag-TRAF6 110–522, Flag-TRAF6 260–522, and Flag-TRAF6 349-522, were generated as previously described [26]. Flag-USP8 C786S mutant was generated by site-directed mutagenesis as previously described [38].
Western Blotting (WB) and Immunoprecipitation (IP) assays
Western Blotting and IP assays were performed as previously described [[16], [17], [18],22,26]. Briefly, HEK-293T cells were seeded in 6-well plates, transfected, treated as described in the text and Figures. The cells were incubated for 38 to 48 h. Then, the cells were collected, and cell lysates were immunoprecipitated with anti-Myc, anti-Flag, or anti-HA antibody. The IP complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 6–10%) and immune-probed with anti-Myc, anti-Flag, or anti-HA antibody. For the ubiquitination assay, mock vector, Flag-TRAF6, Myc-USP8, Flag-TAB2, Myc-TRAF6, Myc-TAK1, Myc-p62, Flag-USP8, Myc-BECN1, and Flag-USP8 C786S mutant were transfected separately into HEK293T cells along with HA-tagged Ub, as described in the text and Figures. Cell lysates were immunoprecipitated with anti-Myc or anti-Flag antibody and probed with anti-Myc, anti-Flag, or anti-HA antibody (the different antibodies indicated in the text and Figures). Control (Ctrl) SK-HEP-1 and USP8KO SK-HEP-1 cells were treated with or without a vehicle or CQ (10 μM) or 3-MA (5 mM) in the presence or absence of LPS (10 μg/mL) for 6 h. The cell lysates were immunoblotted with anti-LC3A/B antibody and anti-GAPDH as a loading control.
Wound-healing migration assay
A wound-healing migration assay was performed following previous protocols [17,18,26]. Briefly, Ctrl SK-HEP-1 and USP8KO SK-HEP-1 cells were seeded in 12-well plates and cultured to reach confluence. The cell monolayers were gently scratched and washed with culture medium. After the floating cells and debris were removed, the cells were treated with a vehicle (DMSO), 3-MA (5 mM), and CQ (10 μM) in the presence or absence of LPS (10 μg/mL). Cell images were captured after culturing for different time periods as indicated in each experiment.
Transwell invasion assay
The Transwell Matrigel invasion assay was performed following previous protocols [17,18,26]. Briefly, Ctrl SK-HEP-1 and USP8KO SK-HEP-1 cells were suspended in a culture medium (200 μL) without FBS, and cells were added to the upper compartments of a 24-well Transwell® chamber containing polycarbonate filters with 8 mm pores and coated with 60 mL of Matrigel (Sigma Aldrich, E1270; 1:9 dilution). Culture medium with 10% FBS was added to the lower chambers and incubated for 24 h. Cells in the upper compartments were removed, washed with PBS, and fixed. The invaded cells were stained with 4,6-diamidino-2-phenylindole (Sigma-Aldrich, D9542) and quantified by counting the number of fluorescent cells.
The cancer genome atlas (TCGA) cancer patient survival and gene correlation analysis
The survival data of 20 different cancer patients was analyzed using OncoLnc (http://www.oncolnc.org/search_results/?q=USP8, http://www.oncolnc.org/search_results/?q=USP8). The gene correlation analysis between USP8 and TRAF6, TAK1 (MAP3K7), TAB2, BECN1, or p62 (SQSTM1) in LIHC was performed by using TCGA data (GAPIA, gene expression profiling interactive analysis; http://gepia.cancer-pku.cn/detail.php?gene=USP8).
RT-qPCR analysis of hCCL20 and hIL-6
Control (Ctrl) and USP8KO SK-HEP-1 cells were treated without or with 10 μg/mL LPS for 9 h. Total RNA was extracted, cDNA was obtained. Specific primers of hCCL20 and hIL-6 were purchased from Qiagen (Hilden, Germany). RT-qPCR analysis was performed, as previously described [16,17].
Transcriptome sequencing analysis
For RNA sequencing (RNA-seq), USP8KO and Ctrl SK-HEP-1 cells were treated with or without LPS (10 μg/ml) for different time periods (0 h, 6 h, and 9 h). Cells were dissolved in TRIzol reagent (Invitrogen) and RNA extraction was performed, as previously described [39]. RNA pellets were dried for 10 min at RT, and then dissolved in RNase-free water. The purified RNA quality was measured using an Agilent 4200 tape station. RNA-seq and cDNA libraries were constructed, as previously described [39]. RNA-seq was performed by Macrogen Inc., Korea, following previous protocols [40], [41], [42]. Differential gene expression analysis was performed using EdgeR [43]. The R package Heatmaps was used to generate the DEG heatmap. For gene ontology enrichment and functional annotation analysis, the GO database (http://www.geneontology.org/) was used. The biological pathways and functions were analyzed on the basis of q value through DAVID, and ClusterProfiler.
Histological analysis
Tumor and liver tissues were isolated from NSG mice xenografted with USP8KO or Ctrl SK-HEP-1 cells at the end of experiment and embedded in paraffin. The paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E). Immunohistochemistry (IHC) staining was performed on 4 μm tissue sections fixed on slide glass after deparaffinization with xylene and rehydration with ethanol. Antigen epitope retrieval was performed by heating in 0.01 M citrate buffer (pH 6.0) in a microwave for 15 min and cooling at room temperature. Endogenous peroxidase was inhibited with 3% H2O2 for 10 min at room temperature. The slides were washed with 1X Tris-buffered saline (TBS) containing 0.1% Tween-20 and incubated in 1% BSA in TBS buffer for 2 h. at room temperature. The slides were stained with primary antibodies, recombinant anti-CEACAM5 + CD66b + CEACAM1 + CEACAM6 (EPR20721; 1:4000, abcam), anti-mitochondria (MAB1273, clone 113-1; 1:100, Merckmillipore), and incubated at 40 °C for an overnight. The next day, the slides were washed and treated with Real Envision Detection System Peroxidase/DAB, Rabbit/Mouse kit (DAKO). Stained slides were observed using an Olympus CX41 light microscope (Olympus, Japan) with × 10/22 numeric aperture and × 40/ 0.75 numeric aperture objectives, and photographic images were collected with a Virtual Slide System (Aperio Technologies, Inc., CA, USA) and analyzed using AperioImageScope software (Aperio Technologies, Inc.).
Statistical analysis
The in vitro and in vivo data are expressed as the mean ± SEM of triplicate samples. Statistical significance was analyzed using ANOVA or Student's t-test in GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001 was considered statistically significant.
Results
USP8 induces the deubiquitination of TRAF6-associated proteins for the activation of NF-κB
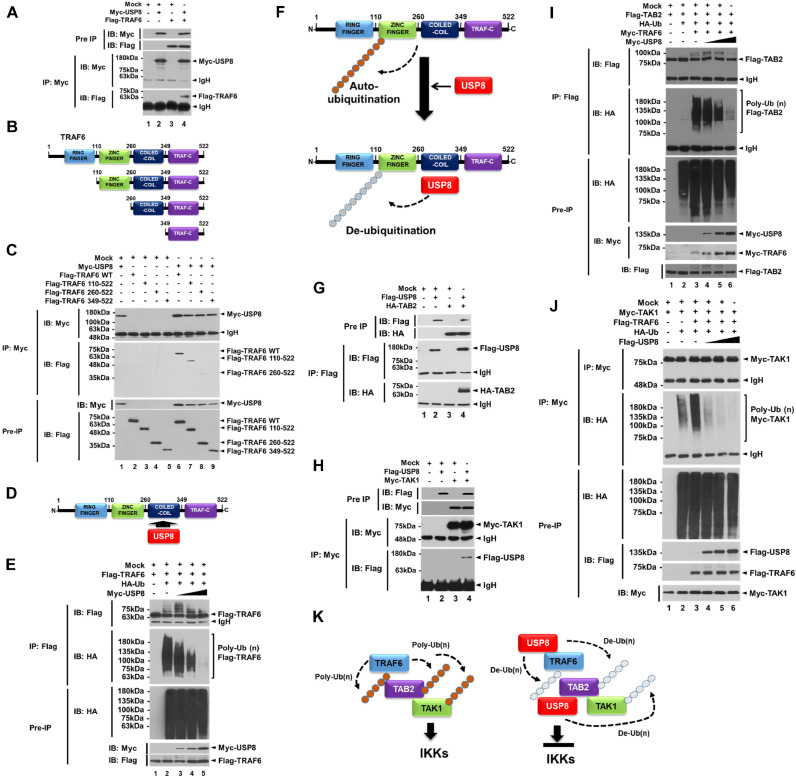
The processes of protein ubiquitination and deubiquitination are functionally implicated in TLR signaling [44,45], especially in TRAF6-mediated signaling for the activation of NF-κB and autophagy [[12], [13], [14], [15], [16], [17], [18],44,45]. To investigate whether USP8 is involved in the TRAF6-mediated signaling, we first examined the molecular association of USP8 with TRAF6. Our results showed that Myc-USP8 interacted with Flag-TRAF6 (Fig. 1A, lane 4). In addition, USP8 interacted with Flag-wild type (WT) TRAF6, Flag-TRAF6 110-522 truncated mutant, and Flag-TRAF6 260-522 truncated mutant (Fig. 1B, TRAF6 truncated mutants; Fig. 1C, lanes 6–8), whereas there was no significant interaction with Flag-TRAF6 349-522 truncated mutant (Fig. 1C, lane 9), indicating that USP8 interacts with the coiled-coil domain of TRAF6 (Fig. 1D). TRAF6 auto-ubiquitination serves as a key role for the molecular association with the TAK1-TAB1-TAB2 complex, thereby enhancing the activation of NF-κB [19,46]. Therefore, we examined whether USP8 induces the deubiquitination of TRAF6. The ubiquitination of TRAF6 could be seen in the absence of Myc-USP8 (Fig. 1E, lane 2), whereas the marked deubiquitination of TRAF6 was observed in the presence of Myc-USP8 in a dose-dependent manner (Fig. 1E, lane 3–5), indicating that USP8 induces the deubiquitination of TRAF6, as depicted in Fig. 1F. The ubiquitinated TRAF6 is associated with the TAK1-TAB1-TAB2 complex and induces the ubiquitination of TAB2 and TAK1, leading to the activation of NF-κB [19,46,47]. We further examined whether USP8 is involved in the deubiquitination of TAB2 and TAK1. Flag-USP8 interacted with HA-TAB2 (Fig. 1G, lane 4) or Myc-TAK1 (Fig. 1H, lane 4). The ubiquitination of TAB2 and TAK1 was markedly attenuated in the presence of Myc-USP8 or Flag-USP8 in a dose-dependent manner (Fig. 1I, lane 4, 6; Fig. 1J, lane 4, 6), as compared with that in the absence of Myc-USP8 or Flag-USP8 (Fig. 1I, lane 3; Fig. 1J, lane 3). Notably, the activation of NF-κB and production of pro-inflammatory cytokines, such as IL-6, TNF-α, and IL-1β, were also significantly attenuated in the USP8-overexpressed HEK293T (SFig. 1A, NF-κB; SFig. 1B, IL-6, SFig. 1C, TNF-α; SFig. 1D, IL-1β), whereas marked increase was observed in the USP8-knockdown (USP8KD) THP-1 cells (SFig. 2A) in response to LPS stimulation (SFig. 2B, NF-κB; SFig. 2C, IL-6). These results suggest that USP8 is negatively implicated in the TLR4-mediated signaling for the activation of NF-κB through the de-ubiquitination of TRAF6, TAB2, and TAK1, as depicted in Fig. 1K.
Fig. 1.
USP8 induces the deubiquitination of TRAF6, TAB2, and TAK1. (A) Immunoprecipitation (IP) assay was performed with anti-Myc antibody on HEK 293T cells transfected with mock, Myc-USP8, and Flag-TRAF6, as indicated. (B) Truncated mutants of TRAF6, TRAF6 110-522, TRAF6 260-522, and TRAF6 349-522. (C) IP assay was performed with anti-Myc antibody on HEK 293T cells transfected with mock, Myc-USP8, Flag-TRAF6 wild type (WT), and Flag-TRAF6 truncated mutants, Flag-TRAF6 110-522, Flag-TRAF6 260-522, and Flag-TRAF6 349-522, as indicated. (D) A schematic model for the interaction between TRAF6 and USP8. (E) Deubiquitination assay was performed on HEK 293T cells transfected with mock, Flag-TRAF6, HA-Ub, and different concentrations of Myc-USP8, as indicated. (F) A schematic model for the deubiquitination of TRAF6 by USP8. (G) IP assay was performed with anti-Flag antibody on HEK 293T cells transfected with mock, Flag-USP8, and HA-TAB2, as indicated. (H) IP assay was performed with anti-Myc antibody in HEK 293T cells transfected with mock, Flag-USP8, and Myc-TAK1, as indicated. (I) Deubiquitination assay was performed on HEK 293T cells transfected with mock, Flag-TAB2, HA-Ub, Myc-TRAF6, and different concentrations of Myc-USP8, as indicated. (J) Deubiquitination assay was performed on HEK 293T cells transfected with mock, Myc-TAK1, Flag-TRAF6, HA-Ub, and different concentrations of Flag-USP8, as indicated. (K) A schematic model of how USP8 induces the deubiquitination of TRAF6, TAB2, and TAK1 for the inhibition of the activation of IKKs.
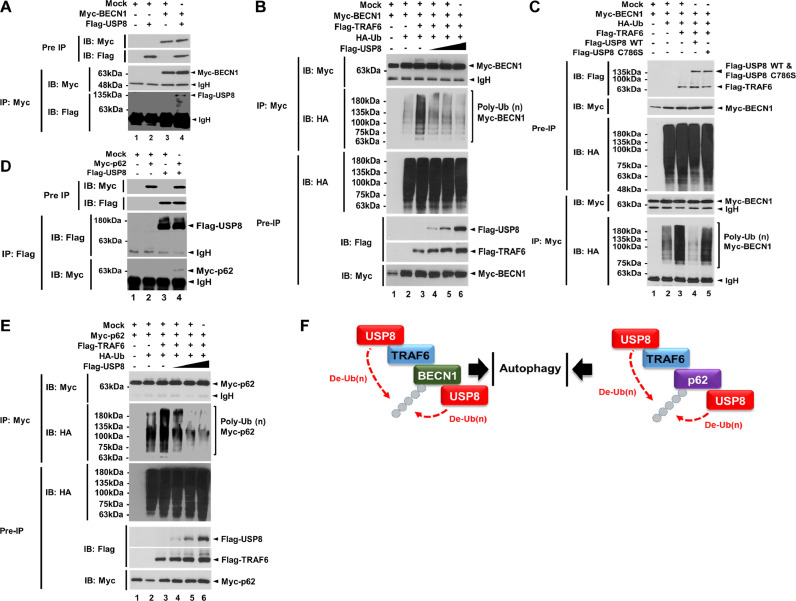
Fig. 2.
USP8 induces the deubiquitination of BECN1 and p62. (A) Immunoprecipitation (IP) assay was performed with anti-Myc antibody on HEK 293T cells transfected with mock, Myc-BECN1, and Flag-USP8, as indicated. (B) Deubiquitination assay was performed on HEK 293T cells transfected with mock, Myc-BECN1, Flag-TRAF6, HA-Ub, and different concentrations of Flag-USP8, as indicated. (C) Deubiquitination assay was performed on HEK 293T cells transfected with mock, Myc-BECN1, HA-Ub, Flag-TRAF6, Flag-USP8 wild type (WT), and Flag-USP8 C786S mutant, as indicated. (D) IP assay was performed with anti-Flag antibody on HEK 293T cells transfected with mock, Myc-p62, and Flag-USP8, as indicated. (E) Deubiquitination assay was performed in HEK 293T cells transfected with mock, Myc-p62, Flag-TRAF6, HA-Ub, and different concentrations of Flag-USP8, as indicated. (F) A schematic model for the deubiquitination of BECN1 (left) and p62 (right) by USP8.
USP8 induces deubiquitination of BECN1 and p62 protein related to autophagy induction
TRAF6 induces the ubiquitination of BECN1 and p62, therefore regulating autophagy induction [23,24,48,49]. Having shown that USP8 induces the deubiquitination of TRAF6, TAB2, and TAK1, and resulted in the attenuation of NF-κB activation induced by TLR4 stimulation, we further investigated whether USP8 induces the deubiquitination of BECN1 and p62. Flag-USP8 interacted with Myc-BECN1 (Fig. 2A, lane 4). Moreover, Myc-BECN1 was ubiquitinated in the absence of Flag-USP8, whereas significant deubiquitination of Myc-BECN1 could observed in the presence of Flag-USP8 in a dose-dependent manner (Fig. 2B, lane 4–6 vs. lane 3), indicating that USP8 induces the de-ubiquitination of BECN1. To evaluate whether the deubiquitination of BECN1 was dependent on the catalytic activity of USP8, we generated a catalytic mutant of USP8, USP8 C786S mutant [38], and performed an ubiquitination assay. Consistently, the wild-type USP8 induced the deubiquitination of BECN1 (Fig. 2C, lane 3 vs 4), whereas the catalytic mutant of USP8 did not (Fig. 2C, lane 5), indicating that the BECN1 deubiquitination by USP8 is dependent on the catalytic activity of USP8. The ubiquitination of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. TRAF6 interacts with p62, and the ubiquitination of p62 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress [48,49]. Consistently, we observed that Flag-TRAF6 interacted with Myc-p62 (SFig. 3, lane 4). In addition, Flag-USP8 interacted with Myc-p62 (Fig. 2D, lane 4). Myc-p62 was ubiquitinated in the absence of Flag-USP8 (Fig. 2E, lane 3), whereas marked deubiquitination could be seen in the presence of Flag-USP8 in a dose-dependent manner (Fig. 2E, lane 4–6 vs. lane 3), indicating that USP8 induces the deubiquitination of p62. Taken together, these results suggest that USP8 interacts with TRAF6, BECN1, and p62, induces the deubiquitination of these ubiquitinated proteins, and subsequently may be negatively implicated in the autophagy induction, as depicted in Fig. 2F.
USP8 is negatively implicated in the liver cancer progression induced by TLR4
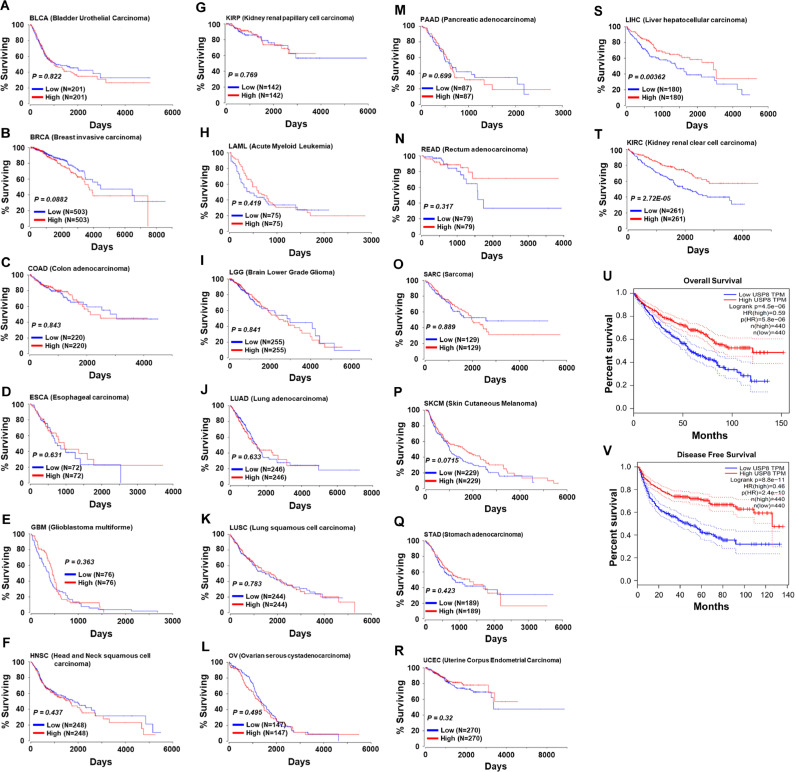
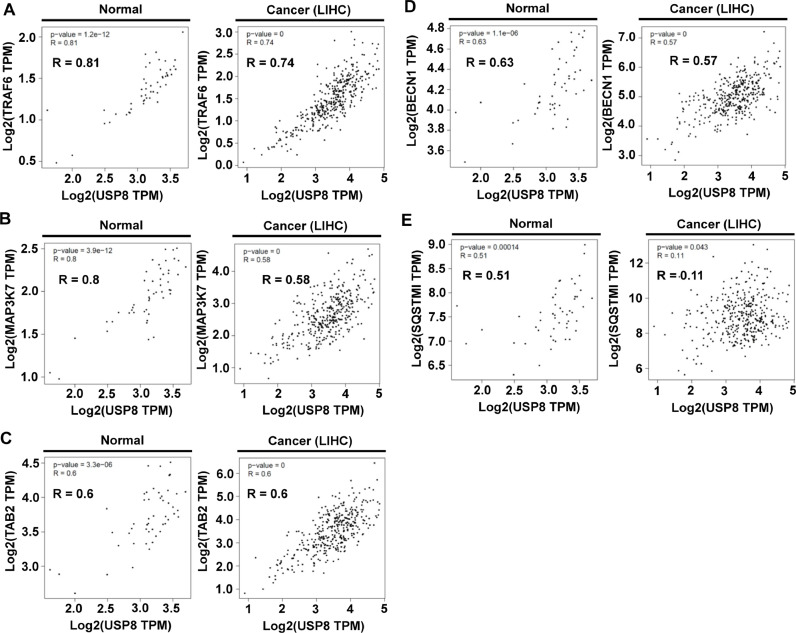
Autophagy promotes tumor progression at advanced stages of tumor development, and USP8 negatively regulates the autophagy by deubiquitinating p62 (SQSTM1) at K420 [36,[50], [51], [52]], supposing that USP8 may be negatively implicated in the tumor progression via the regulation of autophagy. In order to get an insight into the mechanism through which USP8 enhance tumor progression, we analyzed the relationship between USP8 expression and patient survival in 20-different human cancers provided by OncoRank (http://www.oncolnc.org). Kaplan-Meier analysis revealed that no significant associations in 18-different cancers patients (Fig. 3A–R), whereas patients with liver hepatocellular carcinoma (LIHC) and kidney renal clear cell carcinoma (KIRC) with low USP8 mRNA expression had significantly shorter survival time (Fig. 3S, LIHC; Fig. 3T, KIRC). The combined patient survival of LIHC and KIRC showed a markedly decrease in patients with USP8 mRNA expression (Fig. 3U, overall survival; Fig. 3V, disease free survival), indicating that USP8 might have an important role in LIHC and KIRC patient survival. Previous reports have suggested that autophagy activation promotes the progression of hepatocellular carcinoma and contributes to the tolerance of oxaliplatin via reactive oxygen species (ROS) modulation [53,54]. Moreover, it has been reported that TLR4 plays a pivotal role in HCC cancer tumorigenesis by promoting the malignant transformation of epithelial cells and tumor growth [54,55]. Based on the findings that USP8 induced the deubiquitination of TRAF6-associated proteins, such as TRAF6, TAK1 (MAP3K7), TAB2, BECN1, and p62 (SQSTM1), for the activation of NF-κB and autophagy in response to TLR4, the GEPIA database was used to analyze the correlation between USP8 and these genes in the normal and LIHC cells. Our results revealed that USP8 was positively correlated with TRAF6 (Fig. 4A, R = 0.81 in normal; R = 0.74 in LIHC), TAK1 (MAP3K7) (Fig. 4B, R = 0.8 in normal; R = 0.58 in LIHC) and TAB2 (Fig. 4C, R = 0.6 in normal; R = 0.6 in LIHC), and with BECN1 (Fig. 4D, R = 0.63 in normal; R = 0.57 in LIHC). There was a significant correlation between USP8 and p62 in the normal (Fig. 4E, R = 0.51 in normal), but not in the LIHC cancer (Fig. 4E, R = 0.11 in LIHC). Similar correlations between USP8 and TRAF6 (SFig. 4A, R = 0.81), TAB2 (SFig. 4B, R = 0.79) TAK1 (SFig. 4C, R = 0.75), or BECN1 (SFig. 4D, R = 0.79), but not p62 (SFig. 4E, R = 0.082) could be observed in the normal and KIRC cancer.
Fig. 3.
Kaplan-Meier survival analysis and logrank test show a significant association between the USP8 expression and the survival of LIHC patient. For Kaplan-Meier survival analysis on 20 different cancers (provided by OncoRank (http://www.oncolnc.org), (A) BLCA, Bladder Urothelial Carcinoma; (B) BRCA, Breast Invasive Carcinoma; (C) COAD, Colon Adenocarcinoma; (D) ESCA, Esophageal Carcinoma; (E) GBM, Glioblastoma Multiforme; (F) HNSC, Head and Neck Squamous Cell Carcinoma; (G) KIRP, Kidney Renal Papillary Cell Carcinoma; (H) LAML Acute Myeloid Leukemia; (I) LGG, Brain Lower Grade Glioma; (J) LUAD, Lung Adenocarcinoma; (K) LUSC, Lung Squamous Cell Carcinoma; (L) OV, Ovary Serous Cystadenocarcinoma; (M) PAAD, Pancreatic Adenocarcinoma; (N) READ, Rectum Adenocarcinoma; (O) SARC, Sarcoma; (P) SKCM, Skin Cutaneous Melanoma; (Q) STAD, Stomach Adenocarcinoma; (R) UCEC, Uterine Corpus Endometrial Carcinoma; (S) LIHC, Liver Hepatocellular Carcinoma; (T) KIRC, Kidney Renal Clear Cell Carcinoma, the patients were categorized into high-expression group (upper 50 percentile, red curve) and low-expression group (lower 50 percentile, blue curve) based on USP8 gene expression levels. (U and V) The combined Kaplan-Meier overall (U) or disease free (V) survival analysis was performed using GAPIA TCGA data. The number of patients in each group and the p-value were represented.
Fig. 4.
A significant correlation between USP8 and TRAF6, TAK1 (MAP3K7), TAB2, BECN1, or p62 (SQSTM1) is found in the normal and LIHC cells. The GEPIA TCGA database was used to analyze the correlation between USP8 and TRAF6, TAK1 (MAP3K7), TAB2, BECN1, and p62 (SQSTM1) in normal and LIHC cells. (A) USP8 and TRAF6 were positively correlated in normal (R = 0.81) and LIHC (R = 0.74). (B) USP8 and TAK1 were positively correlated in normal (R = 0.8) and LIHC (R = 0.58). (C) USP8 and TAB2 were positively correlated in normal (R = 0.6) and LIHC (R = 0.6). (D) USP8 and BECN1 were positively correlated in normal (R = 0.63) and LIHC (R = 0.57). (E) USP8 and p62 were positively correlated in normal (R = 0.51), but not in LIHC (R = 0.11). The p-value was represented in the inner panel of each figure.
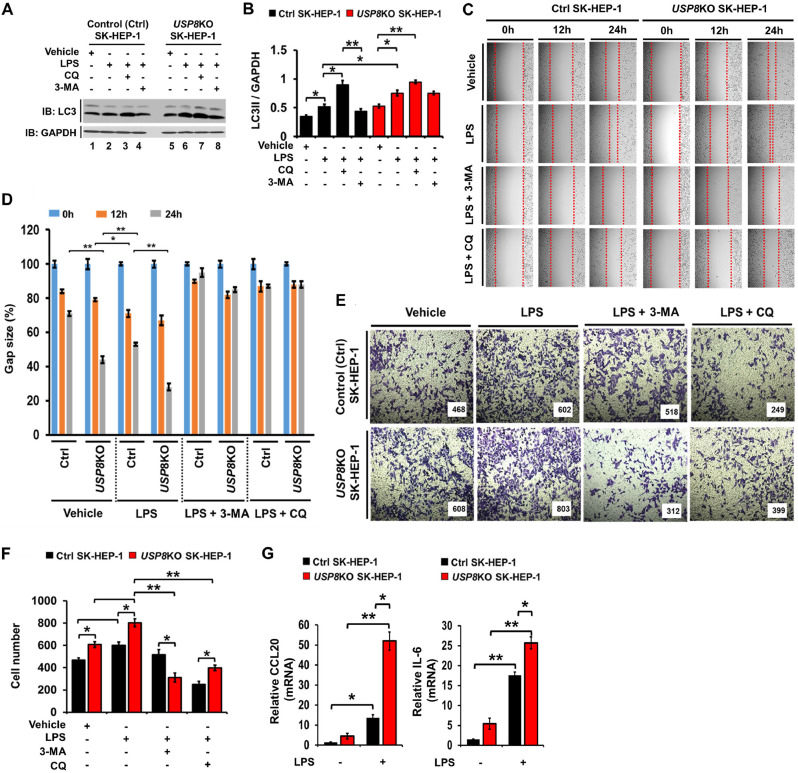
To verify the functional role of USP8 in liver cancer progression, USP8 knockout (KO) SK-HEP-1 human hepatic adenocarcinoma cell line was generated using CRISPR/Cas9 gene editing method (SFig. 5, lane 2), and the autophagy induction by TLR4 stimulation was assessed. The level of LC3-II was significantly increased in USP8KO SK-HEP-1 stimulated with LPS, as compared with that of Ctrl SK-HEP-1 (Fig. 5A, lane 2 vs. lane 6; Fig. 5B). As expected, autophagy inhibitors, such as chloroquine (CQ) and 3-Methyladenine (3-MA), induced the accumulation or decrease of LC3-II levels, respectively, in response to LPS stimulation (Fig. 5A and B in CQ or 3-MA treatments). In addition, upon LPS stimulation the migration in USP8KO SK-HEP-1 cells was significantly enhanced, as compared with that of Ctrl SK-HEP-1 cells (Fig. 5C and D, USP8KO SK-HEP-1 cells treated with LPS vs. Ctrl SK-HEP-1 cells treated with LPS). Consistently, the invasive ability induced by LPS stimulation in USP8KO SK-HEP-1 cells was markedly higher than that of Ctrl SK-HEP-1 cells (Fig. 5E and F, USP8KO SK-HEP-1 cells treated with LPS vs. Ctrl SK-HEP-1 cells treated with LPS). The production of CCL20, IL-6, MMP2, and CCL2, which are critical for enhanced invasion of cancer cells triggered by TLR4,3-induced autophagy [15], were also markedly increased in USP8KO SK-HEP-1 cells treated with LPS (Fig. 5G, CCL20, left and IL-6, right; SFig. 6A, MMP2; SFig. 6B, CCL2). These results suggest that USP8 is negatively involved in the liver cancer progression induced by TLR4.
Fig. 5.
USP8KO SK-HEP-1 cells exhibit increases of autophagy, migration, and invasion in response to TLR4 stimulation. (A) Control (Ctrl) and USP8KO SK-HEP-1 cells were treated with LPS, CQ, and 3-MA, as indicated. Cell lysates were immunoblotted with antibodies specific for LC3-I/-II or GAPDH. (B) LC3-II levels were analyzed with Image J quantification tool (*p < 0.05, **p < 0.01). Quantification performed from 3-independent experiments. (C and D) Ctrl and USP8KO SK-HEP-1 cells were seeded into 12-well cell culture plates. Confluent monolayers were scraped with a sterile yellow Gilson-pipette tip. The wound was then treated with vehicle (DMSO, <0.2% in culture medium), LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL) or CQ (10 μM) plus LPS (10 μg/mL) for different time periods as indicated. A representative experiment is shown (C). The residual gap between migrating cells from the opposing wound edge was expressed as a percentage of the initial scraped area (± SEM, n = 3) (D). *p < 0.05; **p < 0.01. (E and F) Ctrl and USP8KO SK-HEP-1 cells were suspended in DMEM medium including vehicle, LPS (10 μg/mL), 3-MA (5 mM) plus LPS (10 μg/mL) or CQ (10 μM) plus LPS (10 μg/mL) and placed ontop chambers of 24-transwell plates. After an overnight incubation, cells were fixed and stained with crystal violet (E). The migrating cells were counted. Results are presented as mean ± SEM of three independent experiments (F). *p < 0.05, **p < 0.01. (G) Ctrl and USP8KO SK-HEP-1 cells were treated without or with 10 μg/mL LPS, as indicated. Total RNA was extracted, cDNA was obtained, and RT-qPCR analysis performed with specific primers, such as, hCCL20 (left) and hIL-6 (right) (± SEM, n = 3; *p < 0.05, **p < 0.01).
Fig. 6.
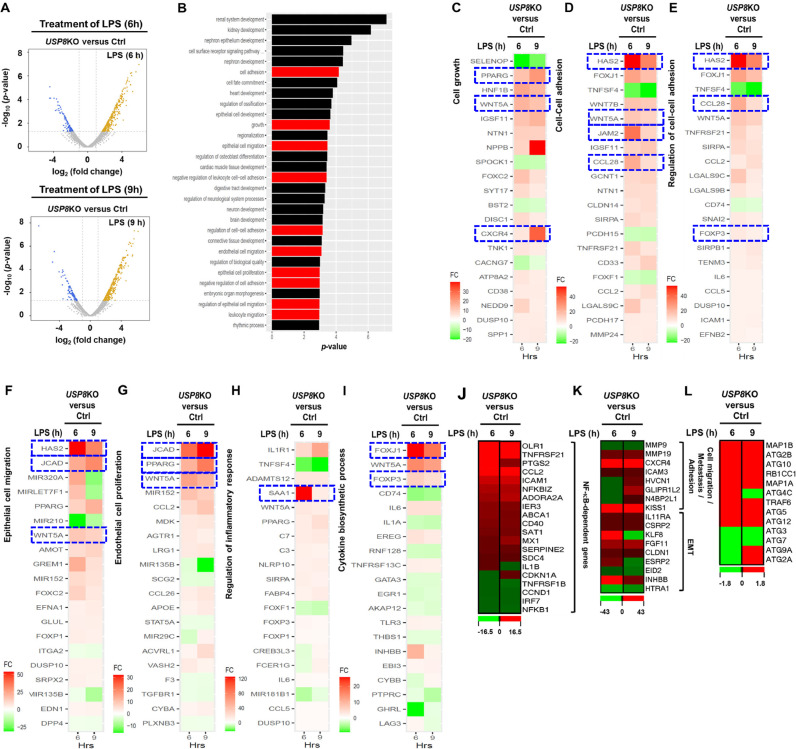
Differential expression analysis shows USP8 is critical in cancer progression. (A) The volcano plot shows that the genes were significantly up- and down-regulated in USP8KO SK-HEP-1 vs Ctrl SK-HEP-1 cells under 6 h (top) and 9 h (bottom) LPS stimulation. (B) Top 30 pathways enriched with the genes that are significantly differentially expressed both under 6 h and 9 h LPS stimulation. (C-L) The top 20 genes that are most significantly differentially expressed in USP8KO SK-HEP-1 vs Ctrl SK-HEP-1 cells that are related to cell growth (C), cell-cell adhesion (D and E), epithelial cell migration (F), endothelial cell proliferation (G), inflammatory response (H), cytokine biosynthetic process (I), NF-κB-dependent genes (J), cell migration and metastasis and adhesion and EMT(K), and autophagy (L).
USP8 negatively regulates liver cancer-related genes induced by TLR4
Based on the above results, we examined whether USP8 is affected on the expression of cancer-related genes in response to TLR4 stimulation. Ctrl SK-HEP-1 and USP8KO SK-HEP-1 cells were treated with or without LPS for different time periods (6- and 9 h), and transcriptome analysis was performed using RNA-Seq. Differentially Expressed Genes (DEG) analysis was performed using edgeR for 6 comparison combinations, as indicated in SFig. 7. 742 Genes satisfying the condition (p-value < 0.05) in at least one comparison combination were extracted, and represented as hierarchical clustering heat map (SFig. 7). To characterize the genes and pathways functionally associated with USP8, we performed a DEG analysis. Both for the cases of 6- and 9 h LPS stimulation, the number of upregulated genes in USP8KO SK-HEP-1 cells was higher (>74%, Fig. 6A), where the overlap of the upregulated/downregulated genes in the two conditions was significant (hypergeometric P < 10E-300). The genes (n = 285) that were significant in both conditions showed enrichment in the pathways strongly associated with cancer (Fig. 6A). Among the top 30 significantly enriched pathways, 10 pathways are cancer-related pathways, such as cell proliferation, adhesion and migration, angiogenesis, and immune response (Fig. 6B, red bars). The top 20 genes that are differentially significant in 7 pathways were represented (Fig. 6C–I). Notably, ten representative genes; HAS2, CXCR4, JAM2, CCL28, FOXJ1, JCAD, WNT5A, FOXP3, PPARG and SAA1 genes, which have been reported to promote cancer progression (HAS2 [56], CCL28 [57], JCAD [58], SAA1 [59], and PPARG [60]), carcinogenesis (CXCR4 [61]), adhesion (JAM2 [62]), cancer proliferation (FOXJ1 [63]), cancer malignancy (WNT5A [64]), or metastasis (FOXP3 [65] and PPARG [60]), were significant up-regulated in the USP8KO SK-HEP-1 cells treated with LPS (Fig. 6C–I, indicated as blue-dashed squares). Furthermore, NF-κB-dependent-, cell migration, metastasis and adhesion, and EMT-, and autophagy-related genes were markedly up-regulated in USP8KO SK-HEP-1 cells treated with LPS (Fig. 6J, NF-κB-dependent genes; Fig. 6K, cell migration, metastasis and adhesion, and EMT-related genes; Fig. 6L autophagy-related genes), suggesting that USP8 negatively regulates liver cancer-related genes induced by TLR4.
Fig. 7.
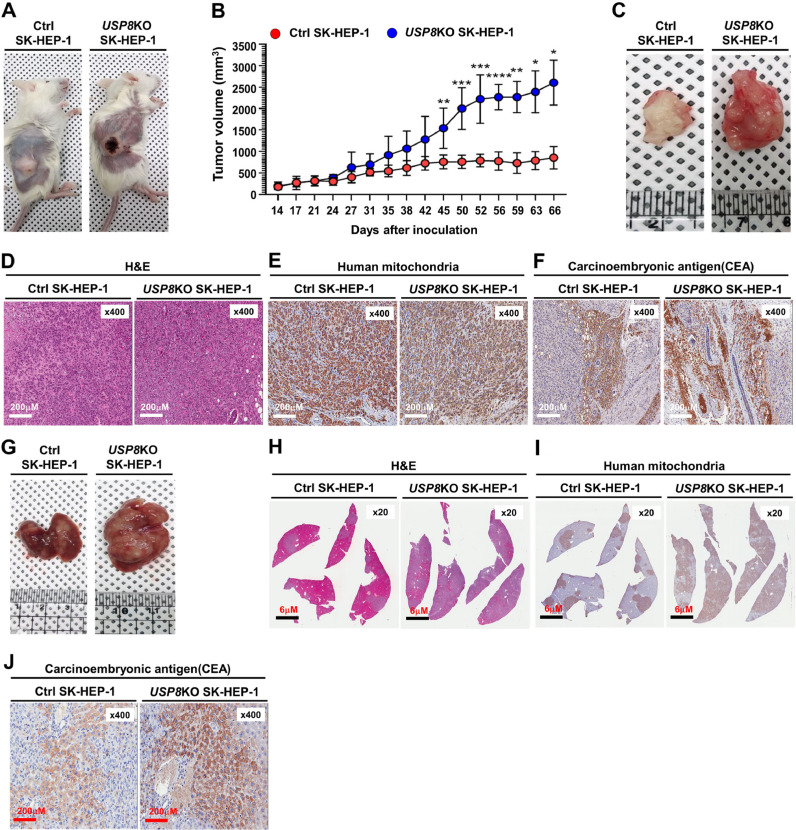
Tumorigenicity and metastasis of USP8KO SK-HEP-1 cells are increased in NSG mice. (A) The primary tumors (human xenografts) at day 66 post subcutaneous injection of Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells in NSG mice. (B) Xenograft tumor volumes of each group (Ctrl SK-HEP-1 injected group; USP8KO SK-HEP-1 cells injected group) were measured, at indicated times after post-injected day. Values are mean ± SEM. (n = 10 each, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 significant compared with Ctrl SK-HEP-1 and USP8KO SK-HEP-1, Student's t-test). (C) Macroscopic primary tumors formed after the subcutaneous injection of Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells at day 66 post injection in NSG mice. (D-F) The histological and pathological features of primary subcutaneous tumor cells derived from NSG mice xenografted with Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells were assessed by H&E staining (D), human mitochondria staining (E), and carcinoembryonic antigen (CEA) (F). (G) Macroscopic metastatic liver tumors formed by the subcutaneous injection of Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells day 66 post-injection in NSG mice. (H-J) The histological and pathological features of metastatic liver tumor cells derived from NSG mice xenografted with Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells were assessed by H&E staining (H), human mitochondria staining (I), and carcinoembryonic antigen (CEA) (J).
The tumorigenicity of USP8KO SK-HEP-1 cells is enhanced in xenografted NSG mice
To study USP8 function in an in vivo model, we subcutaneously injected Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells into NSG mice (n = 10, each). The percentage of visible tumor formation was ∼ 90% on day 66 post-injection in both Ctrl SK-HEP-1 cells injected group and USP8KO SK-HEP-1 cells injected group (Fig. 7A). A significant difference in the tumor mass of the USP8KO SK-HEP-1 cells injected group was observed on day 27 after injection of cells, as compared with that of Ctrl SK-HEP-1 cells injected group (Fig. 7B, Ctrl SK-HEP-1 vs. USP8KO SK-HEP-1). Consistently, the overall tumor growth rate and tumor masses at the end of the experiment on day 66 were significantly increased in USP8KO SK-HEP-1 cells injected group (Fig. 7C, Ctrl SK-HEP-1 vs. USP8KO SK-HEP-1). To characterize the morphology and tumor derived from human SK-HEP-1 cells, H&E staining and immunohistochemistry (IHC) assay with antibody to human mitochondria were performed in the tumor tissues derived from Ctrl SK-HEP-1 cells injected or USP8KO SK-HEP-1 injected NSG mice (Fig. 7D, H&E staining; Fig. 7E, human mitochondria). Neoplastic epithelial cells masses were observed in the USP8KO SK-HEP-1 injected tumor (Fig. 7D, Ctrl SK-HEP-1 vs. USP8KO SK-HEP-1). Histopathologic features of carcinoembryonic antigen (CEA) staining revealed marked elevation of CEA in the USP8KO SK-HEP-1 injected tumor, as compared with that of Ctrl SK-HEP-1 injected tumor (Fig. 7F, USP8KO SK-HEP-1 vs. Ctrl SK-HEP-1). To observe the liver metastasis of injected Ctrl SK-HEP-1 or USP8KO SK-HEP-1 cells into NSG mice, liver tissues were isolated from both groups, and IHC assay was performed on the liver tumor tissues. The tumor mass in the liver tissues was markedly increased in the USP8KO SK-HEP-1 injected NSG mice (Fig. 7G). Similar results with those of tumor tissue staining could be observed in H&E (Fig. 7H), human mitochondria (Fig. 7I), and CEA (Fig. 7J) staining. Taken together, these results suggest that USP8 negatively regulates the liver cancer formation and progression in vivo.
Discussion
Emerging evidence has recently suggested that ubiquitination and deubiquitination play a pivotal role in cancer development and progression through the regulation of cancer metabolism [1,10]. Ubiquitination is known as an enzymatic post-translational modification in which an ubiquitin is attached to a substrate protein, whereas deubiquitination is known as an opposing process where the ubiquitin is removed from the ubiquitinated substrate protein [1,[8], [9], [10],66]. The K63-linked ubiquitylation is known to regulate proteasome-independent events such as signal transduction, whereas K48-, K11-, or K29-linked ubiquitin chains are known to regulate proteasome-dependent events for the degradation [5], [6], [7]. Therefore, the de-ubiquitination by USPs is critically linked to cellular functions of ubiquitinated proteins [1,[8], [9], [10],66]. Herein, we report for the first time USP8 induced deubiquitination of TRAF6-assocaited proteins for the activation of NF-κB and induction of autophagy, and subsequent attenuation of liver cancer progression in vitro and in vivo.
Recently, TRAF6-mediated signaling have been reported to play pivotal roles in regulating cancer progression through the activation of NF-κB and the induction of autophagy in response to TLR4 stimulation [12], [13], [14], [15], [16], [17], [18]. Upon TLR4 stimulation, TRAF6 is auto-K63 ubiquitinated, the ubiquitinated TRAF6 interacts with TAB2 and induces the K63-linked ubiquitination of TAB2 [19,46,47]. The TRAF6-ubiqutinated TAB2 proteins complex further interact and induce the K63-linked ubiquitylation of TAK1, which is an ubiquitin-dependent kinase of MKK and IKK, for the activation of NF-κB [19,46,47], as also depicted in Fig. 8. Importantly, in this study, we found that USP8 interacts with TRAF6, TAB2, and TAK1 proteins, and induces their deubiquitination resulting in the inhibition of NF-κB activation in response to TLR4 stimulation (Fig. 8, left). Additionally, USP8 interacted with p62 and BECN1, which are key regulatory proteins for the induction of autophagy through the TRAF6-dependent ubiquitination (Fig. 8, right) [23,24,48,49], and induced the deubiquitination of p62 and BECN1. Based on these previous findings, we hypothesized that USP8 might be negatively implicated in the cancer progression induced by TLR4 stimulation through the inhibition of NF-κB activation and autophagy induction. Notably, the clinical pan-cancer TCGA data analysis revealed that the survival of LIHC and KIRC patients with the low USP8 mRNA levels was significantly decreased, whereas no significant difference was observed in 18 other cancers. Furthermore, there was a positive correlation between USP8 and TRAF6-associated proteins, such as TRAF6, TAB2, TAK1, and BECN1, with LIHC and KIRC. These results strongly suggested that USP8 might be functionally associated with the LIHC and KIRC cancer progression.
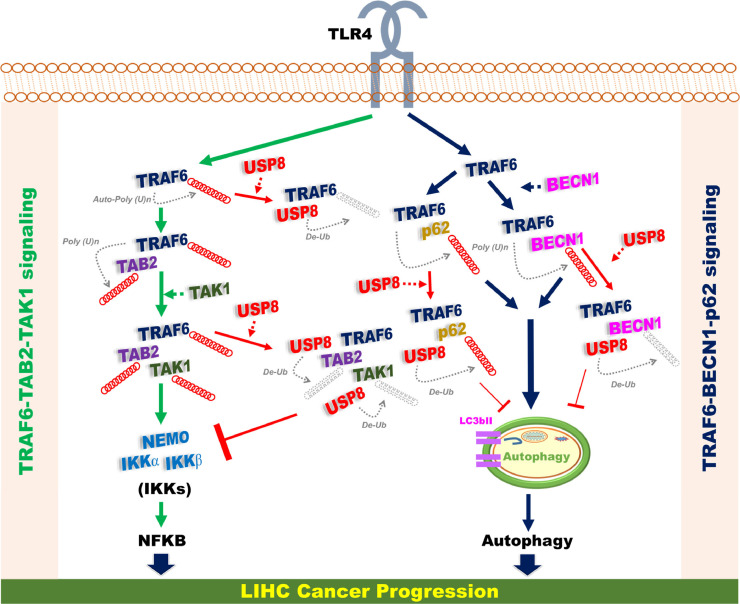
Fig. 8.
A schematic model of the functional role of USP8 in cancer progression via the regulation of TRAF6-mediated signaling for the activation of NF-κB and autophagy. Upon TLR4 stimulation, two signaling axis, TRAF6-TAB2-TAK1 signaling (left) for the activation of NF-κB and TRAF6-BECN1-p62 signaling (right) for the activation of autophagy, were activated via the TRAF6-mediated ubiquitination, as indicated. USP8 interacts with TRAF6 and TAB2 and TAK1, and induces the deubiquitination of these proteins, thereby inhibiting the activation of NF-κB. Simultaneously, USP8 interacts with BENC1 and p62, and induces the de-ubiquitination of BECN1 and p62, thereby inhibiting the induction of autophagy. Eventually, the USP8-mediated inhibition of NF-κB and autophagy negatively regulates LIHC cancer progression.
Recent studies have shown that the ubiquitination and de-ubiquitination have multiple roles in liver cancer progression [10,67,68]. Although it is still controversial whether cellular DUBs are either positively or negatively implicated in liver cancer progression and tumorigenicity, growing evidence suggest that DUBs may have pivotal roles altering liver cancer progression in response to various cellular contexts [67], [68], [69], [70], [71], [72]. Given that USP8 negatively regulates the TRAF6-associated signaling for the activation of NF-κB and autophagy, in the present study, we investigated the role of USP8 in liver cancer progression and tumorigenicity. Notably, USP8KO SK-HEP-1 human hepatic adenocarcinoma cell line significantly enhanced cancer migration and invasion in response to TLR4 stimulation, accompanying the autophagy induction. Furthermore, transcriptome analysis on USP8KO SK-HEP-1 cells revealed an increase in the number of genes related to growth, adhesion, migration, invasion, proliferation and metastasis, as well as NF-κB-dependent genes, in response to TLR4 stimulation. Importantly, the NSG mice xenografted with USP8KO SK-HEP-1 cells showed marked increases of the metastasis into liver and tumorigenicity, as compared to those of NSG mice xenografted with Ctrl SK-HEP-1 cells. These results strongly suggest that USP8 negatively regulates liver cancer progression and formation through the regulation of deubiquitination of cellular proteins related to TRAF6-mediated signaling for the activation of NF-κB activity and autophagy induction.
Taken together, we propose the molecular mechanism by which USP8 is negatively implicated in liver cancer progression regulated by TRAF6-mediated signaling, as depicted in Fig. 8. Upon TLR4 stimulation, two signaling axis, TRAF6-TAB2-TAK1 signaling (left) for the activation of NF-κB and TRAF6-BECN1-p62 signaling (right) for the activation of autophagy, are activated via the TRAF6-mediated ubiquitination, as indicated. USP8 interacts with TRAF6 and TAB2 and TAK1, and induces the de-ubiquitination of these proteins, thereby inhibits the activation of NF-κB. A previous report showed that USP8 protected against LPS-induced inflammatory response in vitro and in vivo via the reductions of TNF-α, IL-1β, PGE2, and NO [73]. In addition, USP8 interacted with TAK1 and deubiquitinated the K63‑linked ubiquitination of TAK1, resulted in the inhibition of intermittent hypoxia/reoxygenation (IHR)‑induced activation of NF-κB [74]. We suppose that these USP8 regulatory roles might be implicated in the TRAF6-TAB2-TAK1 signaling (Fig. 8, left). Simultaneously, USP8 interacts with BENC1 and p62, and induces the de-ubiquitination of BECN1 and p62, thereby inhibits the induction of autophagy (Fig. 8, right). Eventually, the USP8-mediated inhibition of NF-κB and autophagy negatively regulates LIHC cancer progression (Fig. 8). Although a huge progress has been made in exploring the roles of DUBs in cancer progression, including liver cancer progression, very little is known about the molecular and cellular mechanism by which USPs is implicated in the liver diseases including liver fibrosis, liver cirrhosis and final hepatocellular carcinoma. Our present study results provide insight into the pathological liver processes and future development of therapeutic agents based on DUB biology.
CRediT authorship contribution statement
Mi-Jeong Kim: Investigation, Writing – review & editing. Bongkum Choi: Investigation, Formal analysis, Writing – review & editing. Ji Young Kim: Investigation, Writing – review & editing. Yoon Min: Investigation, Writing – review & editing. Do Hee Kwon: Investigation, Writing – review & editing. Juhee Son: Investigation, Writing – review & editing. Ji Su Lee: Investigation, Writing – review & editing. Joo Sang Lee: Formal analysis, Writing – review & editing. Eunyoung Chun: Conceptualization, Writing – original draft, Supervision, Writing – review & editing. Ki-Young Lee: Conceptualization, Writing – original draft, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare no potential conflicts of interest
Acknowledgments
Funding
This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (NRF-2021R1F1A1049324 and NRF-2021R1A2C1094478).
Ethics approval for animal experiments
The research was approved by at the Laboratory Animal Research Center (LARC) of the Samsung Biomedical Research Institute (SBRI) and Samsung Medical Center (SMC), Seoul, South Korea. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the SMC (No. 20160617001).
Acknowledgment
We would like to thank Hyehwa Forum members for their helpful discussion
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101250.
Contributor Information
Eunyoung Chun, Email: echun@chamc.co.kr.
Ki-Young Lee, Email: thylee@skku.edu.
Appendix. Supplementary materials
References
- 1.Young M.J., Hsu K.C., Lin T.E., Chang W.C., Hung J.J. The role of ubiquitin-specific peptidases in cancer progression. J. Biomed. Sci. 2019;26:42. doi: 10.1186/s12929-019-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S., Liu Y., Zhou H. Advances in the development ubiquitin-specific peptidase (USP) inhibitors. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22094546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam M.T., Chen F., Chen H. The oncogenic role of ubiquitin specific peptidase (USP8) and its signaling pathways targeting for cancer therapeutics. Arch. Biochem. Biophys. 2021;701 doi: 10.1016/j.abb.2021.108811. [DOI] [PubMed] [Google Scholar]
- 4.Byun S., Lee S.Y., Lee J., Jeong C.H., Farrand L., Lim S., Reddy K., Kim J.Y., Lee M.H., Lee H.J. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin. Cancer Res. 2013;19:3894–3904. doi: 10.1158/1078-0432.CCR-12-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes-Turcu F.E., Wilkinson K.D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson K.D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 7.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yau R., Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 9.Lauwers E., Jacob C., André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 2009;185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun T., Liu Z., Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer. 2020;19:146. doi: 10.1186/s12943-020-01262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh Y.M., Lee S.B., Choi J., Suh H.Y., Shim S., Song Y.J., Kim B., Lee J.M., Oh S.J., Jeong Y., Cheong K.H., Song P.H., Kim K.A. USP8 modulates ubiquitination of LRIG1 for met degradation. Sci. Rep. 2014;4:4980. doi: 10.1038/srep04980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park M.H., Hong J.T. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15. doi: 10.3390/cells5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Y., Shen S., Verma I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhutia S.K., Mukhopadhyay S., Sinha N., Das D.N., Panda P.K., Patra S.K., Maiti T.K., Mandal M., Dent P., Wang X.Y., Das S.K., Sarkar D., Fisher P.B. Autophagy: cancer's friend or foe? Adv. Cancer. Res. 2013;118:61–95. doi: 10.1016/B978-0-12-407173-5.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan Z., Xie X., Cao H., Zhou X., Zhang X.D., Fan H., Liu Z. Autophagy facilitates TLR4-and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy. 2014;10:257–268. doi: 10.4161/auto.27162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min Y., Kim M.J., Lee S., Chun E., Lee K.Y. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy. 2018;14:1347–1358. doi: 10.1080/15548627.2018.1474995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M.J., Min Y., Im J.S., Son J., Lee J.S., Lee K.Y. p62 is negatively implicated in the TRAF6-BECN1 signaling axis for autophagy activation and cancer progression by Toll-Like receptor 4 (TLR4) Cells. 2020;9:1142. doi: 10.3390/cells9051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M.J., Min Y., Shim J.H., Chun E., Lee K.Y. CRBN is a negative regulator of bactericidal activity and autophagy activation through inhibiting the ubiquitination of ECSIT and BECN1. Front. Immunol. 2019;10:2203. doi: 10.3389/fimmu.2019.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishida S., Sanjo H., Akira S., Matsumoto K., Ninomiya-Tsuji J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells. 2005;10:447–454. doi: 10.1111/j.1365-2443.2005.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morlon A., Munnich A., Smahi A. TAB2, TRAF6 and TAK1 are involved in NF-kappaB activation induced by the TNF-receptor, Edar and its adaptator Edaradd. Hum. Mol. Genet. 2005;14:3751–3757. doi: 10.1093/hmg/ddi405. [DOI] [PubMed] [Google Scholar]
- 22.Moon G., Kim J., Min Y., Wi S.M., Shim J.H., Chun E., Lee K.Y. Phosphoinositide-dependent kinase-1 inhibits TRAF6 ubiquitination by interrupting the formation of TAK1-TAB2 complex in TLR4 signaling. Cell. Signal. 2015;27:2524–2533. doi: 10.1016/j.cellsig.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Shi C.S., Kehrl J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi C.S., Kehrl J.H. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010;6:986–987. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D., Shan B., Sun H., Xiao J., Zhu K., Xie X., Li X., Liang W., Lu X., Qian L., Yuan J. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016;30:1718–1730. doi: 10.1101/gad.285122.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min Y., Lee S., Kim M.J., Chun E., Lee K.Y. Ubiquitin-specific protease 14 negatively regulates toll-like receptor 4-mediated signaling and autophagy induction by inhibiting ubiquitination of TAK1-binding protein 2 and beclin 1. Front. Immunol. 2017;8:1827. doi: 10.3389/fimmu.2017.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur H., Bhalla S., Raghava G.P. Classification of early and late stage liver hepatocellular carcinoma patients from their genomics and epigenomics profiles. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0221476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luedde T., Schwabe R.F. NF-κB in the liver—Linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsharkawy A.M., Mann D.A. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 30.Dai T., Zhang D., Cai M., Wang C., Wu Z., Ying Z., Wu J., Li M., Xie D., Li J., Song L. Golgi phosphoprotein 3 (GOLPH3) promotes hepatocellular carcinoma cell aggressiveness by activating the NF-kappaB pathway. J. Pathol. 2015;235:490–501. doi: 10.1002/path.4479. [DOI] [PubMed] [Google Scholar]
- 31.Toshima T., Shirabe K., Matsumoto Y., Yoshiya S., Ikegami T., Yoshizumi T., Soejima Y., Ikeda T., Maehara Y. Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial beta-oxidation. J. Gastroenterol. 2014;49:907–916. doi: 10.1007/s00535-013-0835-9. [DOI] [PubMed] [Google Scholar]
- 32.Meng Y.C., Lou X.L., Yang L.Y., Li D., Hou Y.Q. Role of the autophagy-related marker LC3 expression in hepatocellular carcinoma: a meta-analysis. J. Cancer Res. Clin. Oncol. 2020;146:1103–1113. doi: 10.1007/s00432-020-03174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yancopoulos G.D., Davis S., Gale N.W., Rudge J.S., Wiegand S.J., Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 34.Moon D.O., Kim M.O., Kang S.H., Choi Y.H., Kim G.Y. Sulforaphane suppresses TNF-α-mediated activation of NF-κB and induces apoptosis through activation of reactive oxygen species-dependent caspase-3. Cancer Lett. 2009;274:132–142. doi: 10.1016/j.canlet.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu H., Bolati D., Higashiyama Y., Nishijima F., Shimizu K., Niwa T. Indoxyl sulfate upregulates renal expression of MCP-1 via production of ROS and activation of NF-κB, p53, ERK, and JNK in proximal tubular cells. Life Sci. 2012;90:525–530. doi: 10.1016/j.lfs.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19:1–16. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao X., Qian H., Wang S., Fulte S., Ding W.X. Autophagy and liver cancer. Clin. Mol. Hepatol. 2020;26:606–617. doi: 10.3350/cmh.2020.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell M.K., Sheng M. USP8 deubiquitinates SHANK3 to control synapse density and SHANK3 activity-dependent protein levels. J. Neurosci. 2018;38:5289–5301. doi: 10.1523/JNEUROSCI.3305-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leem G., Park J., Jeon M., Kim E.S., Kim S.W., Lee Y.J., Choi S.J., Choi B., Park S., Ju Y.S. 4-1BB co-stimulation further enhances anti-PD-1-mediated reinvigoration of exhausted CD39+ CD8 T cells from primary and metastatic sites of epithelial ovarian cancers. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe E.L., Doherty T.M., Karahashi H., Arditi M. Ubiquitination and de-ubiquitination: role in regulation of signaling by Toll-like receptors. J. Endotoxin Res. 2006;12:337–345. doi: 10.1179/096805106X118915. [DOI] [PubMed] [Google Scholar]
- 45.Kawai T., Akira S., signaling T.L.R. Elsevier; 2007. Seminars in Immunology; pp. 24–32. [DOI] [PubMed] [Google Scholar]
- 46.Walsh M.C., Kim G.K., Maurizio P.L., Molnar E.E., Choi Y. TRAF6 autoubiquitination-independent activation of the NFκB and MAPK pathways in response to IL-1 and RANKL. PLoS ONE. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ajibade A.A., Wang H.Y., Wang R.F. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013;34:307–316. doi: 10.1016/j.it.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Peng H., Yang J., Li G., You Q., Han W., Li T., Gao D., Xie X., Lee B.H., Du J. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27:657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Peng H., Xu Y., Xie X., Hu R. SQSTM1/p62 (sequestosome 1) senses cellular ubiquitin stress through E2-mediated ubiquitination. Autophagy. 2018;14:1072–1073. doi: 10.1080/15548627.2017.1332566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng H., Yang F., Hu Q., Sun J., Peng C., Zhao Y., Huang C. The ubiquitin-specific protease USP8 directly deubiquitinates SQSTM1/p62 to suppress its autophagic activity. Autophagy. 2020;16:698–708. doi: 10.1080/15548627.2019.1635381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo T., Fu J., Xu A., Su B., Ren Y., Li N., Zhu J., Zhao X., Dai R., Cao J. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12:1355–1371. doi: 10.1080/15548627.2015.1034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu M., Jiang L., Fu X., Wang W., Ma J., Tian T., Nan K., Liang X. Cytoplasmic liver kinase B1 promotes the growth of human lung adenocarcinoma by enhancing autophagy. Cancer Sci. 2018;109:3055–3067. doi: 10.1111/cas.13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding Z.B., Hui B., Shi Y.H., Zhou J., Peng Y.F., Gu C.Y., Yang H., Shi G.M., Ke A.W., Wang X.Y. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin. Cancer Res. 2011;17:6229–6238. doi: 10.1158/1078-0432.CCR-11-0816. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y.H., Ding Z.B., Zhou J., Hui B., Shi G.M., Ke A.W., Wang X.Y., Dai Z., Peng Y.F., Gu C.Y. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 55.Yang J., Li M., Zheng Q.C. Emerging role of Toll-like receptor 4 in hepatocellular carcinoma. J. Hepatocell. Carcinoma. 2015;2:11. doi: 10.2147/JHC.S44515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuda H., Kobayashi A., Xia B., Watabe M., Pai S.K., Hirota S., Xing F., Liu W., Pandey P.R., Fukuda K. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem–like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–547. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X.L., Liu K.Y., Lin F.J., Shi H.M., Ou Z.L. CCL28 promotes breast cancer growth and metastasis through MAPK-mediated cellular anti-apoptosis and pro-metastasis. Oncol. Rep. 2017;38:1393–1401. doi: 10.3892/or.2017.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye J., Li T.S., Xu G., Zhao Y.M., Zhang N.P., Fan J., Wu J. JCAD promotes progression of nonalcoholic steatohepatitis to liver cancer by inhibiting LATS2 kinase activity. Cancer Res. 2017;77:5287–5300. doi: 10.1158/0008-5472.CAN-17-0229. [DOI] [PubMed] [Google Scholar]
- 59.Ni X.C., Yi Y., Fu Y.P., He H.W., Cai X.Y., Wang J.X., Zhou J., Fan J., Qiu S.J. Serum amyloid A is a novel prognostic biomarker in hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2015;15:10713–10718. doi: 10.7314/apjcp.2014.15.24.10713. [DOI] [PubMed] [Google Scholar]
- 60.Li H., Sorenson A.L., Poczobutt J., Amin J., Joyal T., Sullivan T., Crossno Jr J.T., Weiser-Evans M.C., Nemenoff R.A. Activation of PPARγ in myeloid cells promotes lung cancer progression and metastasis. PLoS ONE. 2011;6:e28133. doi: 10.1371/journal.pone.0028133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghanem I., Riveiro M.E., Paradis V., Faivre S., de Parga P.M.V., Raymond E. Insights on the CXCL12-CXCR4 axis in hepatocellular carcinoma carcinogenesis. Am. J. Transl. Res. 2014;6:340. [PMC free article] [PubMed] [Google Scholar]
- 62.Lian S., Meng L., Xing X., Yang Y., Qu L., Shou C. PRL-3 promotes cell adhesion by interacting with JAM2 in colon cancer. Oncol. Lett. 2016;12:1661–1666. doi: 10.3892/ol.2016.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H.W., Huang X.D., Li H.C., He S., Ni R.Z., Chen C.H., Peng C., Wu G., Wang G.H., Wang Y.Y. Expression of FOXJ1 in hepatocellular carcinoma: correlation with patients' prognosis and tumor cell proliferation. Mol. Carcinog. 2013;52:647–659. doi: 10.1002/mc.21904. [DOI] [PubMed] [Google Scholar]
- 64.Famili F., Naber B., Vloemans S., De Haas E., Tiemessen M., Staal F. Discrete roles of canonical and non-canonical Wnt signaling in hematopoiesis and lymphopoiesis. Cell Death Dis. 2015;6:e1981. doi: 10.1038/cddis.2015.326. -e1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S., Liu Y., Li M.Y., Ng C.S., Yang S.l., Wang S., Zou C., Dong Y., Du J., Long X. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol. Cancer. 2017;16:1–12. doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilkinson K.D. Elsevier; 2000. Ubiquitination and Deubiquitination: Targeting of Proteins for Degradation by the proteasome, Seminars in Cell & Developmental Biology; pp. 141–148. [DOI] [PubMed] [Google Scholar]
- 67.Lv X.Y., Duan T., Li J. The multiple roles of deubiquitinases in liver cancer. Am. J. Cancer Res. 2020;10:1647. [PMC free article] [PubMed] [Google Scholar]
- 68.Dawson S.P. Hepatocellular carcinoma and the ubiquitin–proteasome system. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2008;1782:775–784. doi: 10.1016/j.bbadis.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Ni Q., Chen J., Li X., Xu X., Zhang N., Zhou A., Zhou B., Lu Q., Chen Z. Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim. Biophys. Sin. 2017;49:680–688. doi: 10.1093/abbs/gmx056. (Shanghai) [DOI] [PubMed] [Google Scholar]
- 70.Zhang S., Xie C., Li H., Zhang K., Li J., Wang X., Yin Z. Ubiquitin-specific protease 11 serves as a marker of poor prognosis and promotes metastasis in hepatocellular carcinoma. Lab. Investig. 2018;98:883–894. doi: 10.1038/s41374-018-0050-7. [DOI] [PubMed] [Google Scholar]
- 71.Wang J.H., Wei W., Guo Z.X., Shi M., Guo R.P. Decreased Cezanne expression is associated with the progression and poor prognosis in hepatocellular carcinoma. J. Transl. Med. 2015;13:1–10. doi: 10.1186/s12967-015-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang B., Liang X., Tang F., Zhang J., Zeng S., Jin S., Zhou L., Kudo Y., Qi G. Expression of USP22 and Survivin is an indicator of malignant behavior in hepatocellular carcinoma. Int. J. Oncol. 2015;47:2208–2216. doi: 10.3892/ijo.2015.3214. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J., Bi W., Zhang J., Xiao S., Zhou R., Tsang C.K., Lu D., Zhu L. USP8 protects against lipopolysaccharide-induced cognitive and motor deficits by modulating microglia phenotypes through TLR4/MyD88/NF-κB signaling pathway in mice. Brain Behav. Immun. 2020;88:582–596. doi: 10.1016/j.bbi.2020.04.052. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y., Luo Y., Wang Y., Liu H., Yang Y., Wang Q. Effect of deubiquitinase USP8 on hypoxia/reoxygenation‑induced inflammation by deubiquitination of TAK1 in renal tubular epithelial cells. Int. J. Mol. Med. 2018;42:3467–3476. doi: 10.3892/ijmm.2018.3881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.