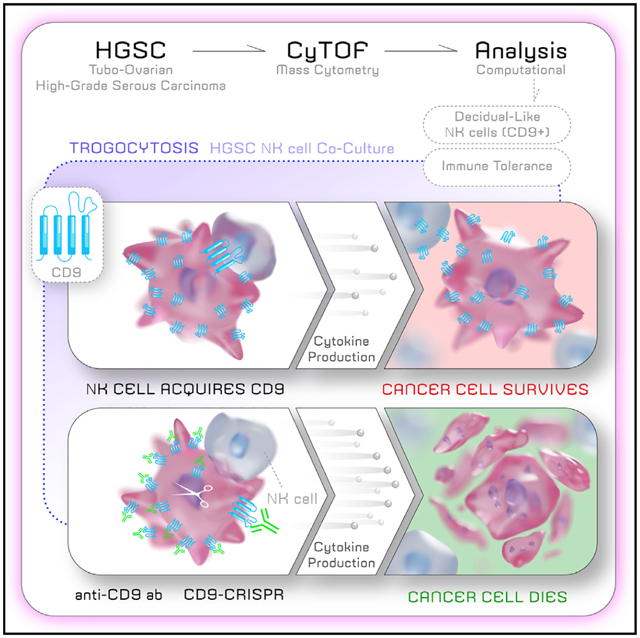
SUMMARY
Tubo-ovarian high-grade serous carcinoma (HGSC) is unresponsive to immune checkpoint blockade despite significant frequencies of exhausted T cells. Here we apply mass cytometry and uncover decidual-like natural killer (dl-NK) cell subpopulations (CD56+CD9+CXCR3+KIR+CD3−CD16−) in newly diagnosed HGSC samples that correlate with both tumor and transitioning epithelial-mesenchymal cell abundance. We show different combinatorial expression patterns of ligands for activating and inhibitory NK receptors within three HGSC tumor compartments: epithelial (E), transitioning epithelial-mesenchymal (EV), and mesenchymal (vimentin expressing [V]), with a more inhibitory ligand phenotype in V cells. In cocultures, NK-92 natural killer cells acquire CD9 from HGSC tumor cells by trogocytosis, resulting in reduced anti-tumor cytokine production and cytotoxicity. Cytotoxicity in these cocultures is restored with a CD9-blocking antibody or CD9 CRISPR knockout, thereby identifying mechanisms of immune suppression in HGSC. CD9 is widely expressed in HGSC tumors and so represents an important new therapeutic target with immediate relevance for NK immunotherapy.
Graphical Abstarct
In brief
In their mass cytometry study of newly diagnosed tubo-ovarian HGSC, Gonzalez et al. identify decidual-like NK cells that correlate with tumor cell abundance. CD9 distinguishes decidual NK cells from other NK cell phenotypes. In coculture, CD9 trogocytosis from ovarian tumor to NK cells confers NK cells with immune-suppressive properties.
INTRODUCTION
Tubo-ovarian high-grade serous carcinoma (HGSC) is the most lethal gynecologic malignancy mainly because of its advanced-stage diagnosis by which time it has metastasized to multiple sites, making curative treatment challenging. Standard of care is surgical debulking and platinum-based chemotherapy with a 70% to 80% likelihood of recurrence within 5 years (Singh et al., 2017; Bast et al., 2019; Bowtell et al., 2015; Matulonis et al., 2016). Recently, two new treatment modalities have been introduced into the clinic for women with HGSC. Poly (ADP-ribose) polymerase inhibitors (PARPi) exploit the paradigm of synthetic lethality through the administration of small molecules and are clinically approved as maintenance therapy (upfront) and treatment of recurrent plat inum-sensitive HGSC (Lord and Ashworth, 2017, Ashworth and Lord, 2018; Coleman et al., 2019; González-Martín et al., 2019; Mirza et al., 2020). Immunotherapy, the second treatment modality, is aimed at restoring the ability of the patient’s immune system to eradicate a tumor and largely focused on reactivation of T lymphocytes (Rodriguez et al., 2018). Although HGSC tumors show high frequencies of functionally exhausted T cells, with high levels of immune checkpoint proteins, responses to immunotherapy for HGSC have not shown clinical benefit (Rodriguez et al., 2018; Kandalaft et al., 2019; Zamarin et al., 2020; Pujade-Lauraine et al., 2021; Webb et al., 2015). A deeper understanding of the cell types within the HGSC immune microenvironment could assist in identifying mechanistic biomarkers to select patients likely to derive the most benefit from immunotherapy.
Natural killer (NK) cells are innate lymphocytes with potent cytotoxic activity against tumor- and virally infected cells. NK cell function results from the coordinated integration of intracellular signaling mediated by the combinatorial expression of multiple activating and inhibitory germline-encoded receptors (Orr and Lanier, 2010; Vivier et al., 2011; Wilk and Blish, 2018; Morvan and Lanier, 2016). NK cells produce an array of cytokines that regulate immune responses and are mechanistically distinct from T lymphocytes in that their cytotoxic activity occurs in an antigen-independent manner and without the need for prior sensitization. In tumors, these dual functions endow NK cells with roles in both immune surveillance to eradicate tumor cells and conversely with the creation of an immune-tolerant microenvironment facilitating tumor progression (Chiossone et al., 2018; Morvan and Lanier, 2016).
We previously applied mass cytometry (cytometry by time-of-flight [CyTOF]) to characterize primary HGSC surgical samples (patients who had not received chemotherapy treatment) and identified three tumor cell compartments: epithelial cells (E) expressing E-cadherin, mesenchymal cells (V) expressing vimentin, and EV cells with a phenotype transitioning between the epithelial and the mesenchymal compartments (Gonzalez et al., 2018). We now report CyTOF data acquired from the T and NK immune cell infiltrates from these specimens.
NK cells are at the center of immunotherapeutic approaches to exploit their tumor cell-killing activity (Daher and Rezvani, 2018; Li et al., 2018; Uppendahl et al., 2018; Lorenzo-Herrero et al., 2018; Rezvani, 2019; Rezvani et al., 2017). The single-cell data reported here identify mechanisms by which HGSC cells subvert the killing activity of NK cells. These findings merit urgent consideration for optimizing NK cell-based immunotherapy.
RESULTS
CyTOF analysis to identify T and NK cell phenotypes from HGSC tumor infiltrates
In our prior CyTOF study of primary HGSC surgical specimens, computational analysis focused on tumor cells (Tables S1 and S2) (Gonzalez et al., 2018). Here we report further analysis of these tumors with a CyTOF antibody panel designed to characterize T and NK cell subtypes (Table S3; STAR Methods). All steps for quality control and CyTOF processing were described previously (Gonzalez et al., 2018; V.D.G., Y.-W.H., and W.J.F., unpublished data) (STAR Methods). Viable immune cell populations were gated as cPARP−CD45+CD66− (STAR Methods). T and NK immune single-cell datasets were combined and subjected to unsupervised analysis with X-shift clustering (Samusik et al., 2016). Using 25 surface markers delineating T and NK cell phenotypes, 52 X-shift cell clusters (calculated for an optimal k of 30 nearest neighbors) were generated for subsequent analysis (Table S3).
Correlation analysis between tumor and immune cell clusters from HGSC tumors
To understand interactions between tumor and immune cells, we applied a network approach to compute correlations between the frequencies of these cell phenotypes (Gonzalez et al., 2018; Spitzer et al., 2017). Pairwise Spearman correlation coefficients (rs) were displayed on a hierarchically clustered heatmap (Figure 1A).
Figure 1. HGSC tumor and EV cell frequencies correlate with a dl-NK cell phenotype.
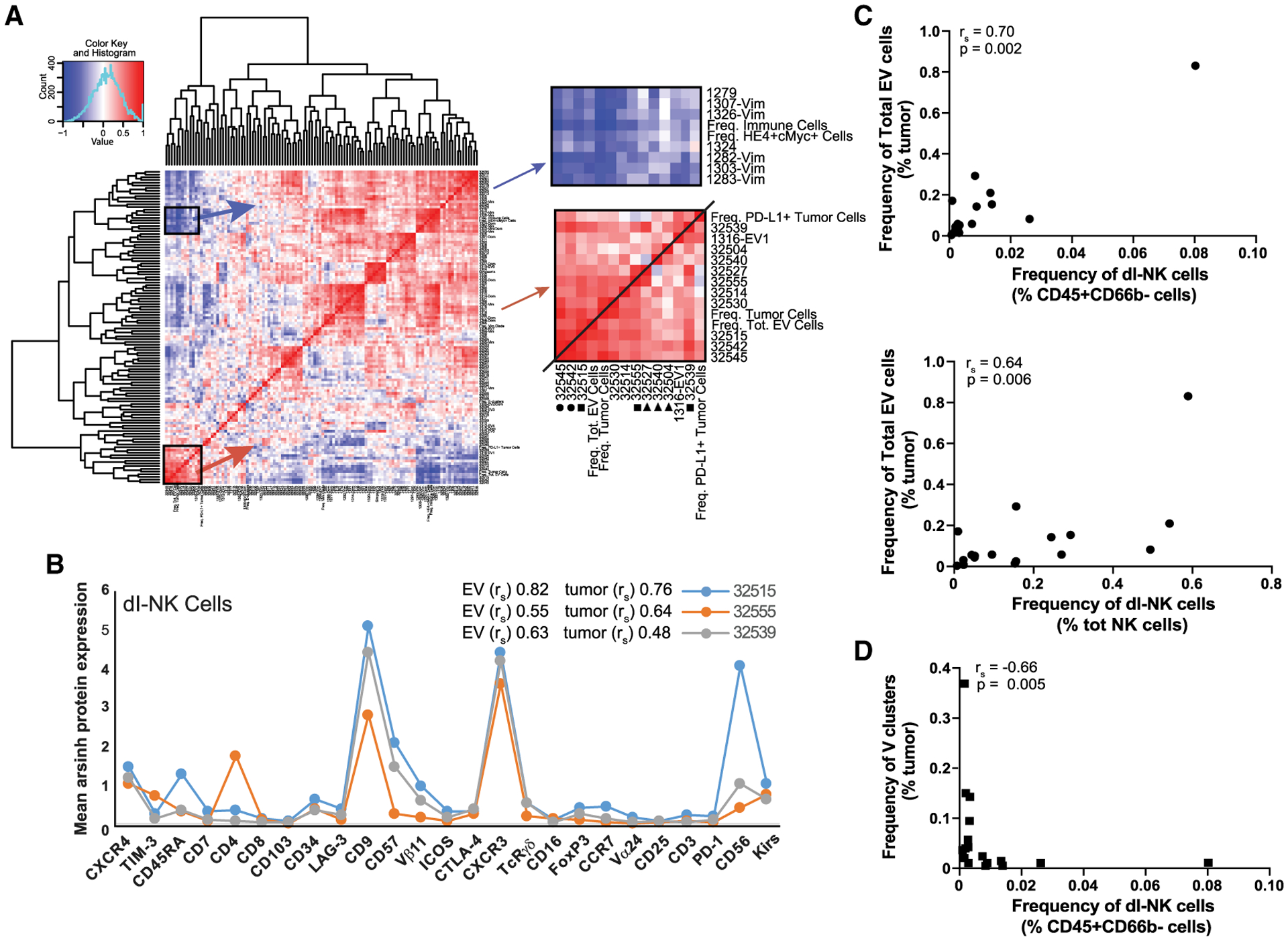
(A) Hierarchically organized heatmap showing pairwise Spearman correlations between tumor and immune cell clusters. Enlarged areas depict positive (red) and negative (blue) correlations of dl-NK cell clusters. Positive correlations between dl-NK cell clusters with total abundance of tumor and EV cells (square) and T cell clusters (circle). Three dl-NK cell clusters were found in all tumors (triangle).
(B) Protein expression patterns consistent with a dl-NK cell phenotype (asterisk) of the three positively correlated clusters (square).
(C) dl-NK cells manually gated from total immune cell infiltrate (top plot) and total NK cell population (bottom plot) correlate with EV cell abundance.
(D) dl-NK cells manually gated from total immune cell infiltrate correlate negatively with a subgroup of vimentin clusters.
Five immune cell clusters correlated positively with both total tumor and EV cell abundance, but not with E or V cell abundance (rs > 0.5) (Figure 1A). Three of these immune cell clusters (32515, 32539, and 32555) were CD3−CD16−CD56+CD9+CXCR3+ and killer immunoglobulin-like receptor (KIR)+, a phenotype resembling decidual NK (d-NK) cells (Koopman et al., 2003; Hanna et al., 2006) (Figure 1B). Notably, among the many NK cell phenotypes reported, CD9 expression is exclusive to decidual NK cells (Koopman et al., 2003; Horowitz et al., 2013). The combined cell frequency of these three decidual-like NK (dl-NK) clusters ranged from 1.3% to 28% of the immune cell population. Ratios of tumor to dl-NK cell ranged from 0.25 to 25 across the 17 tumors. Furthermore, correlations determined with manually gated dl-NK cells and EV tumor cells supported the unsupervised X-shift analysis (Figures 1C and S1A).
The remaining two immune cell clusters (32542 and 32545) exhibited a T cell phenotype but also displayed d-NK cell features (high levels of CD56, CXCR3, and CD9) (Figure S1B). Low levels of the invariant T cell receptor Vα24-Vβ11 implied that these cell clusters have natural killer T (NKT)-like functions (Kim et al., 2002; Bernstein et al., 2006). They were present in the immune cell infiltrates at a frequency of 0.1%–3.9%. Eight PD-1 expressing T cell clusters did not correlate with tumor or EV cell abundance (Table S8).
An additional three dl-NK cell clusters (32527, 32504, and d32540) correlated weakly with tumor or EV cell abundance (rs of 0.3–0.49) (Figures 1A and S1C). Cluster 32527 correlated with dl-NK cell clusters 32555 and 32539. Clusters 32504 and 32540 were found in all tumors with a combined frequency of 14%–86% of the immune cell infiltrate. To visualize the relationships between all T and all NK immune cell clusters, a minimum spanning tree was generated revealing that clusters 32504 and 32540 were phenotypically similar to both dl-NK and T cell clusters (Figure S1D).
Of the eight described immune cell clusters, five correlated negatively (rs > −0.6) with several vimentin clusters, consistent with published reports describing an inverse relationship between metastases and NK cell infiltrates (Figure 1D) (Lorenzo-Herrero et al., 2018; López-Soto et al., 2017).
d-NK cells are critical in the first trimester of pregnancy, conferring immune tolerance toward the hemi-allogeneic fetus (Koopman et al., 2003; Hanna et al., 2006; Jabrane-Ferrat, 2019). Here, identification of tumor infiltrating dl-NK cells, in some cases at very high frequencies, indicates that immune tolerance overrides potential NK cell-killing activity. This led us to hypothesize that the same features of d-NK cell-mediated immune tolerance could be coopted for HGSC tumor maintenance and progression.
NK receptor ligand expression across newly diagnosed HGSC tumors
To determine whether there were differences in how E, EV, and V intratumor cell compartments modulated NK cell function toward an immune-tolerant state, we measured frequencies of cells expressing NK receptor ligands and two ADAMs (a disinte-grin and metalloproteinase) across the three compartments (Table S4; STAR Methods). We modified our CyTOF panel to include antibodies against the following twelve NK receptor ligands, as well as ADAM10 and ADAM17 (Table S4): (1) ULBP1, ULBP2/5/6, ULPBP3, ULPBP4, and MICA/B that bind to the NKG2D-activating NK receptor (Dhar and Wu, 2018; Raulet et al., 2013); (2) ADAM10 and ADAM17 involved in NK ligand and NK receptor shedding (Boutet et al., 2009; Dhar and Wu, 2018; Ferrari de Andrade et al., 2018); (3) nectin-like ligands CD111, CD112, CD113, CD155, and nectin-4, which bind the activating NK receptor, CD226 (also known as DNAM1), and the inhibitory NK receptors, T cell immune receptor with immunoglobulin and ITIM domains (TIGIT) and CD96 (also known as TACTILE) (Martinet and Smyth, 2015; Fabre-Lafay et al., 2007; Sanchez-Correa et al., 2019; Reches et al., 2020); (4) human leukocyte antigen (HLA) class 1 molecules A, B, and C that bind to inhibitory KIRs (Wroblewski et al., 2019; Morvan and Lanier, 2016); and (5) HLA-E, a non-classical HLA class 1 molecule that binds to the NK inhibitory receptor heterodimer CD94/NKG2A with greater affinity than to the activating CD94/NKG2C heterodimer (Kamiya et al., 2019).
Tumor cells were manually gated from CyTOF datasets to exclude immune, angiogenic, and stromal cells (CD45−,CD31−, and FAP−) (Gonzalez et al., 2018). The resultant single-cell data files were combined and clustered using the X-shift algorithm. Spatial relationships between tumor cells expressing NK receptor ligands within the 56 X-shift tumor cell clusters were visualized by force directed layouts (FDLs) (Figures 2A–2C, STAR Methods). The resultant FDLs corroborated the presence of the E, V, and EV compartments (Figure 2A). Overall, FDLs revealed that NK-activating and NK-inhibitory receptor ligands were expressed at variable levels within pockets of tumor cells, rather than evenly interspersed, in the three compartments (Figures 2A–2C).
Figure 2. Expression patterns of NK receptor ligands.
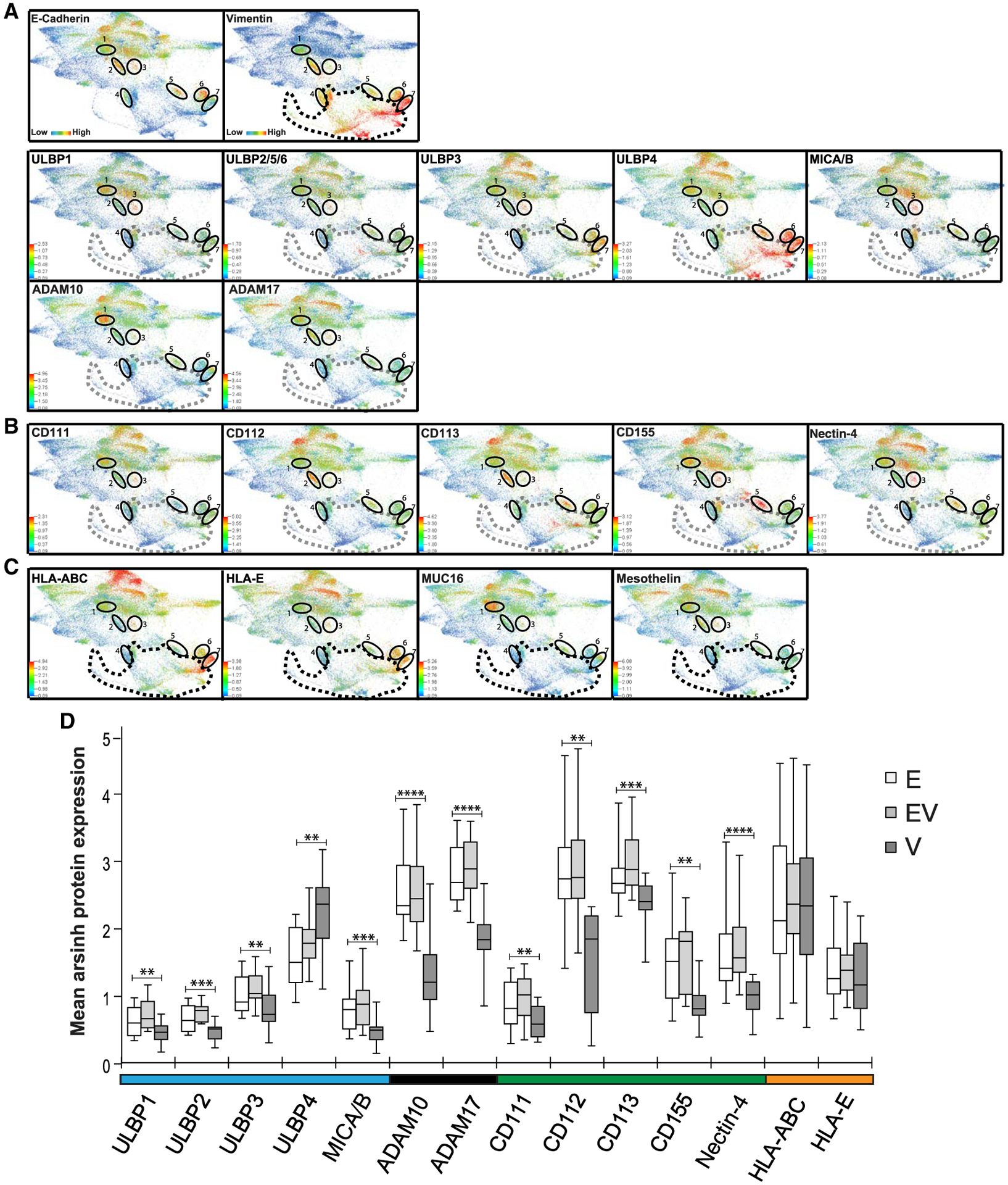
FDLs are composites of twelve newly diagnosed HGSC tumors colored for the proteins indicated.
(A) E-cadherin and vimentin (row 1), NKG2D-activating receptor ligands (row 2), and ADAMs (row 3), with EV clusters 1–7 encircled.
(B) Nectin-family ligands.
(C) Inhibitory ligands: HLA-ABC, HLA-E, and tumor-associated antigens CA125 and mesothelin.
(D) Expression levels of NK receptor ligands and ADAMs across E, EV, and V tumor compartments.
Medians and interquartile ranges. **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Previous studies demonstrated that upregulation of ligands for the NKG2D-activating NK receptor is a major mechanism by which NK cells are able to detect and eradicate tumor cells (Dhar and Wu, 2018; Raulet et al., 2013). Apart from ULPBP4, the highest levels of NKG2D receptor ligands were found in the E and EV tumor compartments, with minimal levels in the V compartment (Figure 2A). This is consistent with an immune-surveillance role for NK cells within the E and EV tumor cell compartments. The high levels of ULBP4 throughout the V cell compartment are anomalies but consistent with a recent study describing ULBP4 as a functional outlier within the ULBP family (Zöller et al., 2018). ADAM10 and ADAM17 were observed in the E compartment and in the EV1 and EV2 subsets. They were often coexpressed with NKG2D ligands, suggesting an attempt by tumor cells to nullify NK cell-killing activity by promoting NK ligand shedding (Ferrari de Andrade et al., 2018; Dhar and Wu, 2018; Raulet et al., 2013). The nectin family of ligands also exhibited variable expression patterns with pockets of E and EV tumor cells coexpressing different combinations (Figure 2B).
Similarly, HLA-A, HLA-B, HLA-C, and HLA-E inhibitory NK receptor ligands were coexpressed in all three tumor compartments, thus contributing to suppression of tumor cell destruction by NK cells (Figure 2C) (Morvan and Lanier, 2016). By contrast, major histocompatibility complex class I (MHC class I) molecules expressed by tumor cells targets them for destruction by cytotoxic CD8 T lymphocytes (CTLs) and occurs in an antigen-dependent mechanism, whereby MHC class I molecules present peptide fragments from tumor-associated antigens (TAAs) to CD8 CTLs (Brennick et al., 2017; Vyas et al., 2008). Well-established TAAs in HGSC are MUC16 and mesothelin (Bast et al., 2019). Expression of these TAAs and HLAs was mostly mutually exclusive, indicating minimal tumor cell destruction by CD8 CTLs (Figures 2C and S2). These data suggest that HGSC cells may have evolved dual mechanisms of escape from killing activity by NK and CD8 T cells.
Expression levels of NK receptor ligands and ADAMs across E, EV, and V tumor compartments
Median expression levels for the NK receptor ligands and ADAMs did not differ significantly between E and EV compartments (Figure 2D). However, within the V compartment, levels were all statistically lower (except for ULBP4), enabling V cells to escape immune surveillance more easily, consistent with previously reported experimental models (Table S5) (López-Soto et al., 2017; Lorenzo-Herrero et al., 2018). There were no statistical differences between tumor compartments for expression of HLA-ABC and HLA-E.
Quantifying combinatorial diversity of NK receptor ligand expression
The combinatorial expression patterns of activating and inhibitory receptors endow NK cells with a high degree of phenotypic and functional diversity (Wilk and Blish, 2018; Horowitz et al., 2013). In turn, a correspondingly complex repertoire of activating and/or inhibitory ligands present on tumor cells can affect NK receptor activity (Morvan and Lanier, 2016). We applied Boolean analysis to measure the frequency of E, EV, and V tumor cells with distinct combinatorial expression patterns for the twelve NK receptor ligands and ADAM proteases. We assessed 214 (16,384) NK receptor ligand combinations. Using a threshold cell frequency of >1% for cells in any compartment in any sample expressing an NK receptor ligand/ADAM combination, 163 NK receptor ligand combinations were expressed by tumor cells across the 12 HGSCs (Figure 3A; STAR Methods).
Figure 3. Combinatorial diversity of NK receptor ligands within HGSC tumor compartments.
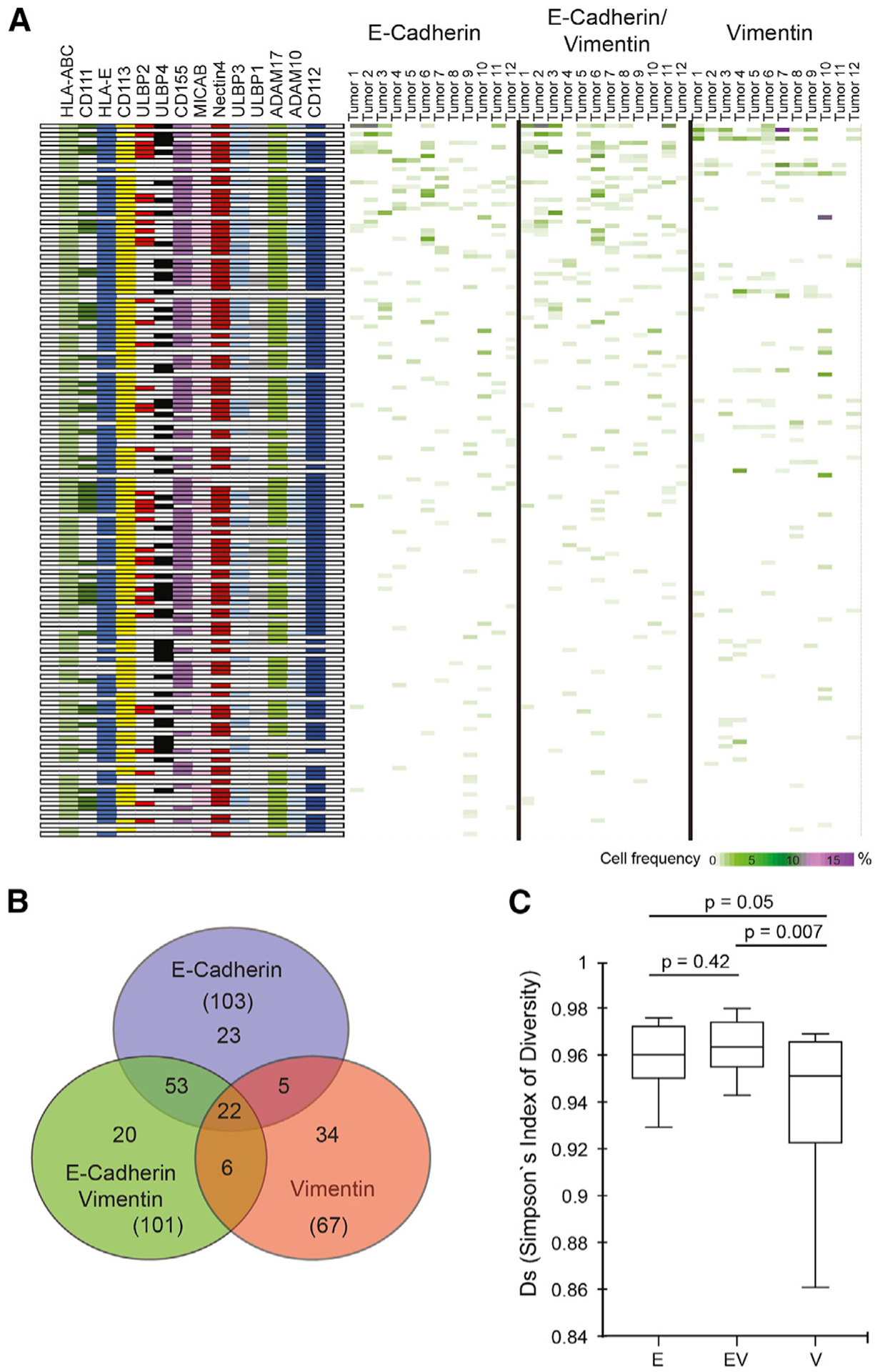
(A) Boolean computation for combinations of 12 NK receptor ligands and ADAMs (rows, left panel). Frequencies of E, EV, and V tumor cells expressing ligand combinations (rows) for each of 12 tumor samples (columns). Rows ranked highest (top) to lowest total cell frequency (heatmaps; right panels).
(B) Distinct and overlapping NK receptor ligand/ADAM combinations between tumor compartments.
(C) Simpson’s inverse index of diversity for NK receptor ligand/ADAM combinations with medians and interquartile ranges. E versus V (p = 0.05) and EV versus V (p = 0.007).
Boolean analysis demonstrated that cells in the E and EV tumor compartments had greater combinatorial diversity for NK receptor ligands/ADAMs (103 and 101 phenotypes, respectively) than the V tumor compartment (67 phenotypes) (Figure 3B). There were more shared phenotypes between E and EV (53) compartments than between E and V (5) and EV and V (6) compartments. These data suggest differential regulation of the immune microenvironment orchestrated by tumor cells in each of these compartments.
To quantify NK receptor ligand combinatorial diversity in the tumor cell compartments, we used Simpson’s index of diversity (Figure 3C). This index is often used in ecology to quantify biodiversity within a natural habitat and was recently applied to NK cells and ovarian tumor CyTOF datasets (Gonzalez et al., 2018; Horowitz et al., 2013). When applied here, Simpson’s index of diversity was significantly higher in the E and EV compartments compared with the V compartment, consistent with the Boolean analysis.
NK receptor ligand expression across HGSC cell lines
With the goal of providing more reliable in vitro models of HGSC, Domcke et al. (2013) presented a group of ovarian cell lines ranked by the concordance of their genetics with resected HGSC tumors. We analyzed thirteen of the top-ranked cell lines with our tumor/NK receptor ligand CyTOF antibody panel (Table S4). Data analysis performed with X-shift clustering and FDL visualization revealed HGSC cell lines that phenocopied E, EV, and V tumor compartments (Figure S3). Patterns of NK receptor ligand/ADAM expression levels, although not identical, were comparable to the E, V, and EV counterparts in tumors. We selected OVCAR4, Kuramochi, and TYK-nu, representing E, EV, and V HGSC tumor cells, respectively.
Carboplatin changes expression levels of NK receptor ligands
Activation of the DNA damage response with genotoxic agents increases the expression of ligands for NKG2D and DNAM1, thereby making a stressed cell more susceptible to NK cell killing (Cerboni et al., 2014; Gasser et al., 2005). Accordingly, the three preceding HGSC cell lines were exposed to carboplatin, a genotoxic agent that is part of the standard-of-care regimen for women with HGSC (Bast et al., 2019; Bowtell et al., 2015; Matulonis et al., 2016). After one week of exposure, HGSC cell lines were processed for CyTOF using the tumor/NK receptor ligand antibody panel (Table S4).
The DNA damage response to carboplatin treatment was confirmed by a recognized increase in pH2AX (Krenning et al., 2019) (Figures 4A–4C). Of the NK-activating receptor ligands measured, only ULBP2 increased in OVCAR4 cells after carboplatin exposure (24% to 50%), whereas of the inhibitory NK receptors measured, HLA-E increased in all three cell lines and HLA-ABC increased in Kuramochi cells.
Figure 4. Responses to carboplatin across E, EV, and V HGSC cell lines.
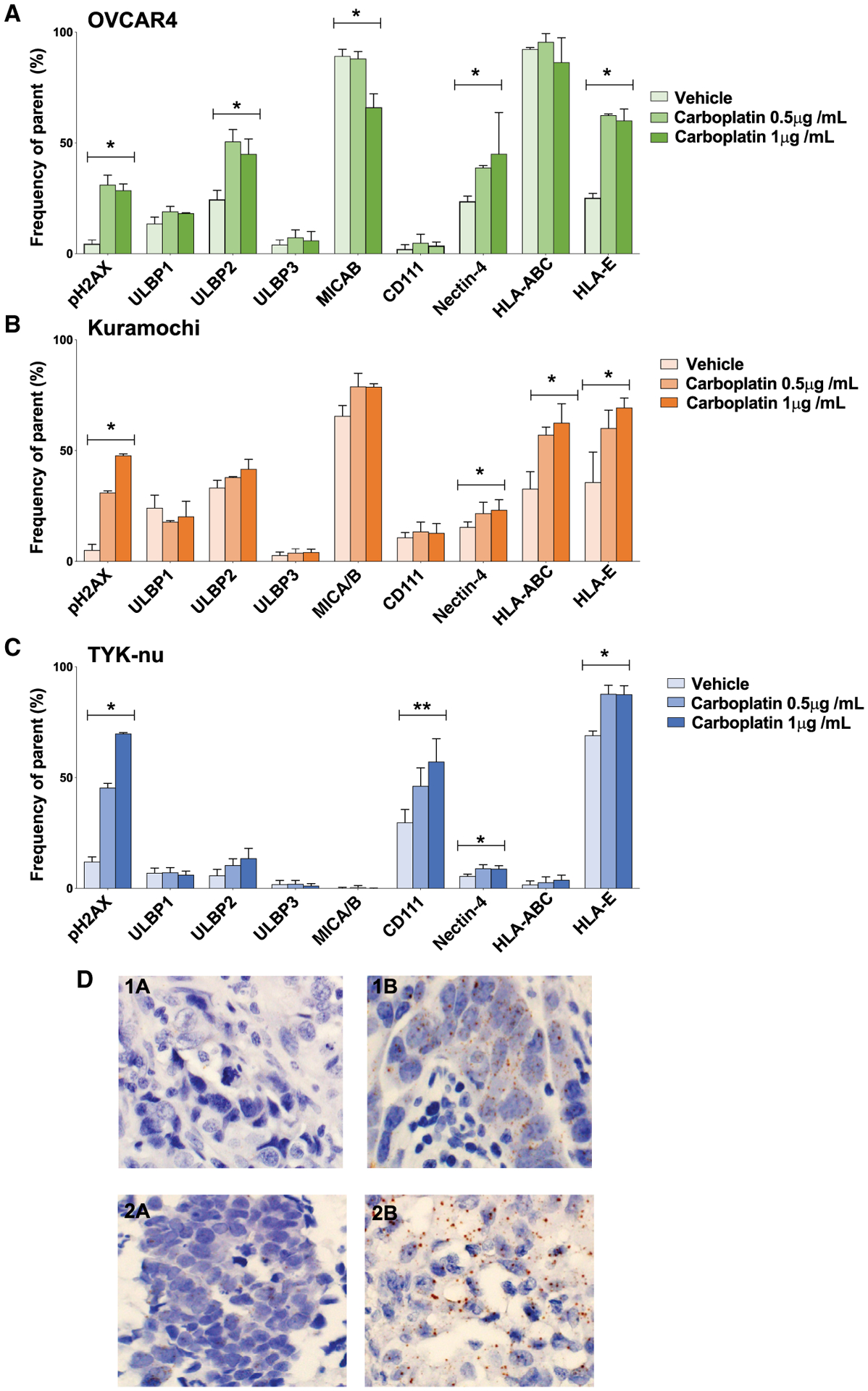
Cell lines were exposed to carboplatin or vehicle (H2O) as indicated. Frequencies of HGSC cells (y axis) expressing NK receptor ligands and pH2AX (x axis).
(A–C) OVCAR4 (A), Kuramochi (B), and TYK-nu cell lines (C). Triplicates with standard deviations. *p ≤ 0.05, **p ≤ 0.005 for overall ANOVA.
(D) Nectin-4 mRNA expression in tumor tissue after NACT (brown staining 1B and 2B). Absent before NACT in matched samples (1A and 2A).
Carboplatin also increased the frequency of HGSC cells expressing nectin-4, a ligand for the inhibitory TIGIT receptor (Reches et al., 2020). For two carboplatin concentrations, OVCAR4 frequencies increased from 24% to 39% and 45%, Kuramochi frequencies increased from 15% to 22% and 23%, and TYK-nu cells frequencies increased from 6% to 9% for both carboplatin concentrations (Figures 4B and 4C). In TYK-nu cells, carboplatin mediated a significant increase in CD111, a ligand for the inhibitory CD96 receptor. CD111 has an additional role enhancing signaling through TIGIT (Martinet and Smyth, 2015). Overall, these data support a role for carboplatin in tumor-mediated immune suppression.
Nectin-4 levels increase after carboplatin-based neoadjuvant chemotherapy (NACT)
To assess the clinical relevance of the preceding data, we measured nectin-4 RNA expression in ten archival patient-matched HGSC tumor sections pre- and post-NACT. Using RNAscope, four of the ten tumor pairs revealed large increases in the percentage of nectin-4 staining tumor cells post-NACT (Figure 4D; Table S6; STAR Methods). These results suggest that nectin-4, an inhibitory ligand for TIGIT, can be increased in response to carboplatin.
HGSC-NK-92 cell line cocultures to model the HGSC immune-tolerant microenvironment
The positive association between dl-NK cell subpopulations with overall tumor and EV cell abundance indicated that dl-NK cells have immune tolerance toward the tumor (Figures 1A–1C). To determine how the tumor cells might create this immune-tolerant environment, we set up cocultures of OVCAR4, Kuramochi, and TYK-nu with the human NK-92 cell line, the latter chosen for its clinical relevance (Suck et al., 2016; Rezvani et al., 2017). After coculture, we performed CyTOF analysis with an NK cell antibody panel (Table S7).
CD9 expression in NK-92 cells increases after coculture with HGSC cell lines
After coculture with HGSC cell lines, up to 60% of NK-92 cells expressed CD9 with the greatest induction in OVCAR4 cells (Figure 5A). With a membrane barrier (transwell, 3 μm pore size) between the two cell lines, CD9 expression on NK-92 cells was reduced to background monoculture levels (<4%). This demonstrated the requirement for physical contact between HGSC tumor cells and NK-92 cells (Figure 5A; STAR Methods). Treatment of HGSC tumor cells with carboplatin did not significantly alter CD9 uptake in coculture experiments (Figure S4A; STAR Methods).
Figure 5. CD9 trogocytosis.
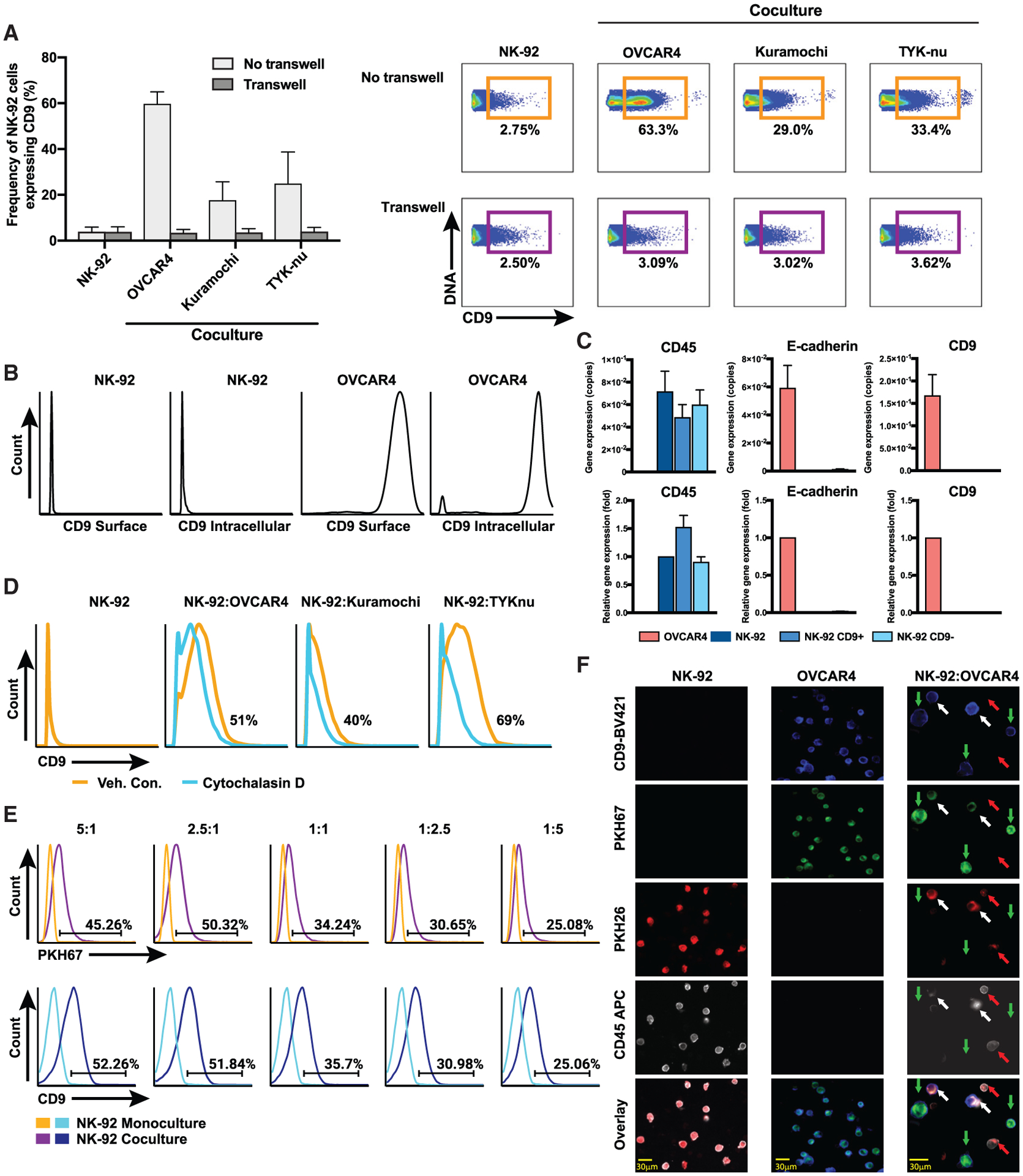
HGSC and NK-92 cell lines were cocultured at a ratio of effector (NK-92) to target (HGSC cell line) of 1:1 for 6 h unless otherwise indicated (STAR Methods).
(A) Frequency of CD9+ NK-92 cells after coculture with and without transwell (left panel). Mean and standard deviations for n = 4. Exemplary 2D flow plots for CD9+ NK-92 cells after coculture (right panel).
(B) Extra- and intracellular CD9 protein expression absent from the NK-92 cells and present at high levels in the OVCAR4 cells.
(C) Quantitative real-time PCR of FACS-purified CD9+ and CD9− NK-92 cells after coculture with OVCAR4 cells. Copy number (top plots), fold gene expression changes (bottom plots). CD9 were transcripts exclusive to OVCAR4 cells. Controls: CD45 (positive for NK-92, negative for OVCAR4) and E-cadherin (negative for NK-92, positive for OVCAR4).
(D) Flow cytometry shows cytochalasin D partially inhibits trogocytosis from HGSC cell lines.
(E) Transfer of CD9+ membrane fragments from OVCAR4 cells onto NK-92 cells. Cocultures between OVCAR4 cells pre-stained with fluorescent membrane dye PKH67 and NK-92 cells at different target:effector ratios. PKH67 frequencies (top histograms) and CD9 frequencies (bottom histograms).
(F) Microscopy shows trogocytosis. OVCAR4 cells and NK-92 cells stained with PKH67 (green) and PKH26 (red), respectively; cocultured for 3 h; and stained with CD45 and CD9 antibodies. Images (from a Keyence BZ-X800 microscope) for cells grown in monoculture 20× and coculture 60×. NK-92 cells that acquired CD9 from OVCAR4 cells are indicated with white arrows. Images were enhanced for brightness and contrast to optimize the printed image.
One potential explanation for the appearance of CD9 expression on the surface of NK-92 cells is that they retain intracellular CD9 pools that are induced to traffic to the cell surface during coculture with HGSC cells. To address this, we stained NK-92 and OVCAR4 cells grown in monoculture with a CD9 antibody (STAR Methods). Sequential cell staining for CD9 (surface and, following permeabilization, intracellular) showed that NK-92 cells were devoid of both surface and intracellular CD9. By contrast, OVCAR4 cells expressed robust levels of CD9 in both cellular locations (Figure 5B; STAR Methods).
To establish that CD9 displayed on NK-92 cells during coculture was not endogenously produced, CD9+ NK-92 cells and CD9− NK-92 cells underwent fluorescence-activated cell sorting (FACS) after coculture with the OVCAR4 cell line and were analyzed for CD9 transcripts (Figure 5C). Transcripts were not detected in CD9+ or control NK-92 cells. Robust levels of CD9 transcripts were seen in the OVCAR4 cell line (Figure 5C).
CD9 expression across HGSC primary tumors and cell lines
To determine the prevalence of CD9 expression in HGSC, we screened 17 primary tumors and 11 cell lines. For both sources of HGSC cells, there were high frequencies of CD9-expressing cells with high levels of CD9 protein expression (Figures 6A, 6B, and S5). CD9 protein expression levels did not change after exposing cell lines to carboplatin (Figure S4B; STAR Methods).
Figure 6. Comparisons of CD9 trogocytosis from non-HGSC cells and NK cells from other sources.
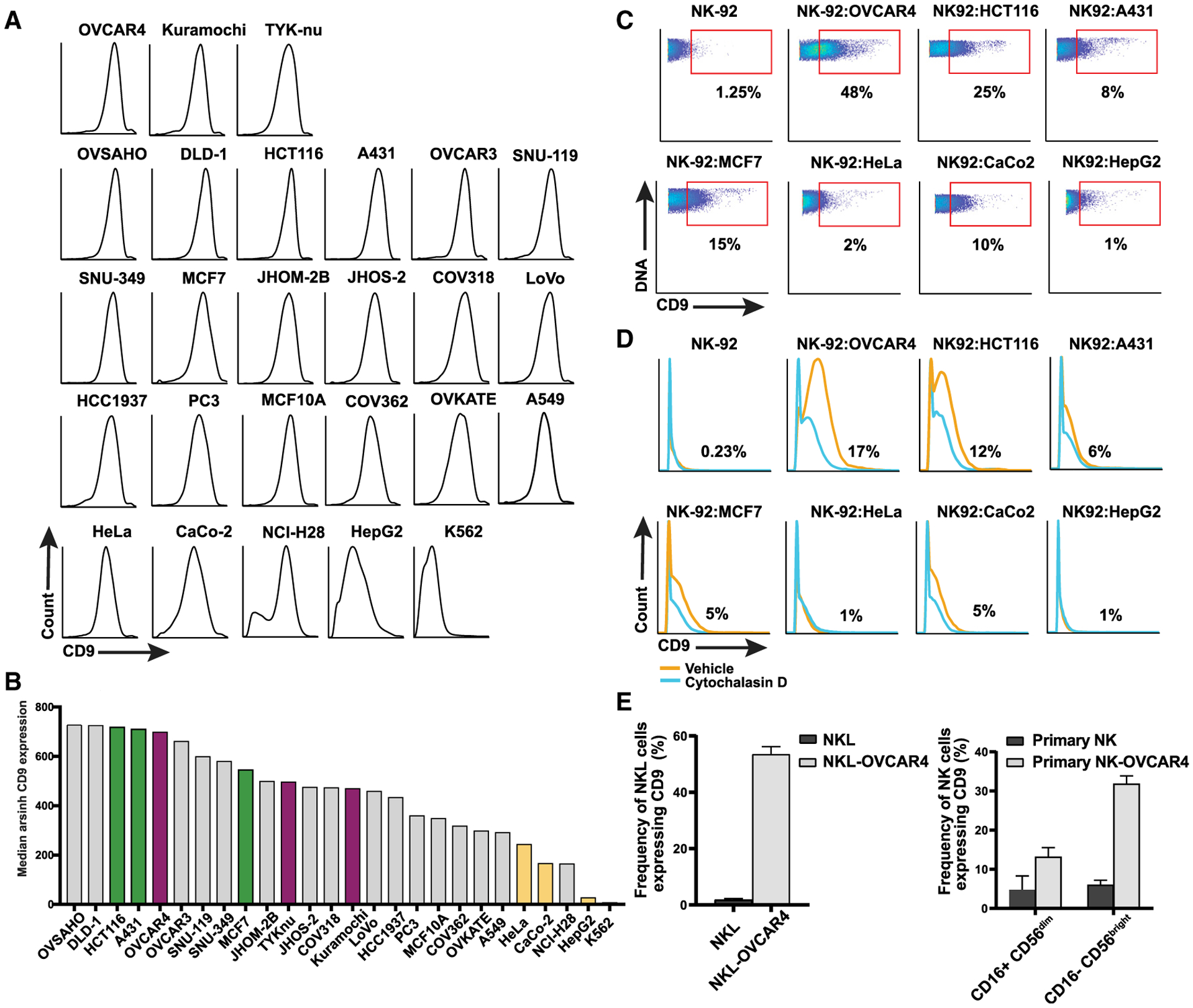
(A) CD9 expression levels in 11 HGSC and 15 non-HGSC tumor cell lines (STAR Methods).
(B) Cell lines ranked by level of CD9 expression. Cell lines selected for coculture with NK-92 cells: HGSC (magenta) and non-HGSC with high levels of CD9 (green) and with lower levels of CD9 (yellow).
(C) Representative flow plots showing frequency of CD9+ NK-92 cells after coculture with non-HGSC tumor cell lines.
(D) Cytochalasin D partially inhibits CD9 trogocytosis from non-HGSC tumor cell lines.
(E) CD9 trogocytosis by the NKL cell line and primary NK cells in peripheral blood mononuclear cells (PBMCs).
Trogocytosis as the mechanism by which NK-92 cells acquire CD9
Because there was no evidence of de novo synthesis of CD9 by NK-92 cells, we hypothesized that during coculture, CD9 must be transferred to NK-92 cells by trogocytosis (Joly and Hudrisier, 2003; Dance, 2019). This process takes place within minutes of cell-cell contact and involves the transfer of plasma membrane fragments from one cell to another, including anchored proteins therein. After coculture with OVCAR4 cells (15, 30, 60, 120, and 360 min), CD9 was detected as early as 15 min on NK-92 cells with a steady increase up to 360 min (Figure S6A). These data are consistent with trogocytosis. as previously reported in other systems.
Many studies have shown that inhibitors of actin polymerization block trogocytosis but that the effects of these inhibitors vary depending on cell type (Aucher et al., 2008; Gary et al., 2012). From pilot experiments, cytochalasin D was selected as the optimal inhibitor (STAR Methods). Its inclusion in coculture experiments with NK-92 and the HGSC cell lines reduced the frequencies of CD9+ NK-92 cells by 40%–69% (Figure 5D).
To establish that plasma membrane fragments containing CD9 were transferred from OVCAR4 cells, they were labeled with PKH67, a green-fluorescent lipophilic membrane dye, before coculture with NK-92 cells. Cells were stained with antibodies against CD45 (for gating NK-92 cells) and CD9 and processed for fluorescence-based flow cytometry (STAR Methods). At the highest ratios of OVCAR4 to NK-92 cells, PKH67 and CD9 were codetected in ~50% NK-92 cells. As cell ratios decreased, so did the capture of OVCAR4 membrane fragments by NK-92 cells (Figure 5E).
We performed microscopy to visualize CD9 contained within the membrane fragments that were transferred from OVCAR4 to NK-92 cells (Figure 5F). Each cell line was labeled with a lipophilic fluorescent dye: OVCAR4 with PKH67 (green) and NK-92 with PKH26 (red). After coculture for×h, cells were stained with antibodies against CD9 (blue) and CD45 (white). OVCAR4 cells were only visible in the green (PKH67) and blue (CD9) detection channels, whereas a proportion of NK-92 cells was visible in all four channels, confirming that NK-92 cells trogocytosed plasma membrane fragments containing anchored CD9 from OVCAR4 cells (Figure 5F).
Evaluation of NK-92 trogocytosis from non-HGSC cell lines
To determine whether NK-92 trogocytosis of CD9 could occur with non-HGSC tumor cells, we measured the frequency of CD9-expressing cells in 15 non-HGSC tumor cell lines (Figure 6A). Because it was previously shown that the expression level of a plasma membrane protein was not a determinant of the efficiency of its transfer to a recipient cell, we chose three tumor cell lines with high expression (HCT116, A431, and MCF7) and three with lower CD9 expression (HeLa, CaCo-2, and HepG2) for trogocytosis experiments (Figure 6B) (Daubeuf et al., 2010). NK-92-mediated trogocytosis from these cell lines was varied, confirming the observation of Daubeuf et al. (2010). Apart from the HCT116 colorectal cancer cell line, CD9 uptake was less pronounced than that seen with HGSC cell lines (Figures 5C and 6C). Cytochalasin D inhibition of CD9 uptake was most marked for HCT116, MCF7, and CaCo-2 cell lines (Figure 6D).
We also demonstrated CD9 trogocytosis by the NKL cell line and primary NK cells (CD16+CD56dim and CD16−CD56bright), providing additional evidence of a generalizable mechanism by which NK cells can exert immune tolerance (Figures 6E and S6B).
CD9+ NK-92 cells have a more immunosuppressive intracellular cytokine profile
d-NK cells exert immune tolerance through their poor cytotoxic responses (Crespo et al., 2017; Hanna et al., 2006; Jabrane-Ferrat, 2019). Thus, we hypothesized that upon acquiring CD9, NK-92 cells could exhibit similar immune tolerance. To this end, we assembled a CyTOF antibody panel to measure degranulation, production of cytolytic proteins, and intracellular cytokine production (ICP) (Bryceson et al., 2010; Siebert et al., 2008; Fauriat et al., 2010) (Table S7). NK-92 cells were analyzed in monoculture, after coculture with HGSC cell lines, and in the presence and absence of phorbol-12-myristate-13-acetate (PMA). Exposing cells to PMA is a convenient method for measuring NK cell function, because it bypasses upstream signaling through NK cell receptors (Shabrish et al., 2016) (STAR Methods). The K562 erythroleukemic cell line, which only expresses ligands for activating NK receptors, was used as a control for NK cell functional activation (Tremblay-McLean et al., 2019). A CD9 antibody in the CyTOF panel enabled us to gate out CD9+ from CD9− NK-92 cells. For intracellular proteins, two metrics were measured: (1) changes in frequency of NK-92 cells expressing a specific cytokine and (2) median ICP expression levels (Figures 7A and 7B).
Figure 7. Functional effects of CD9.
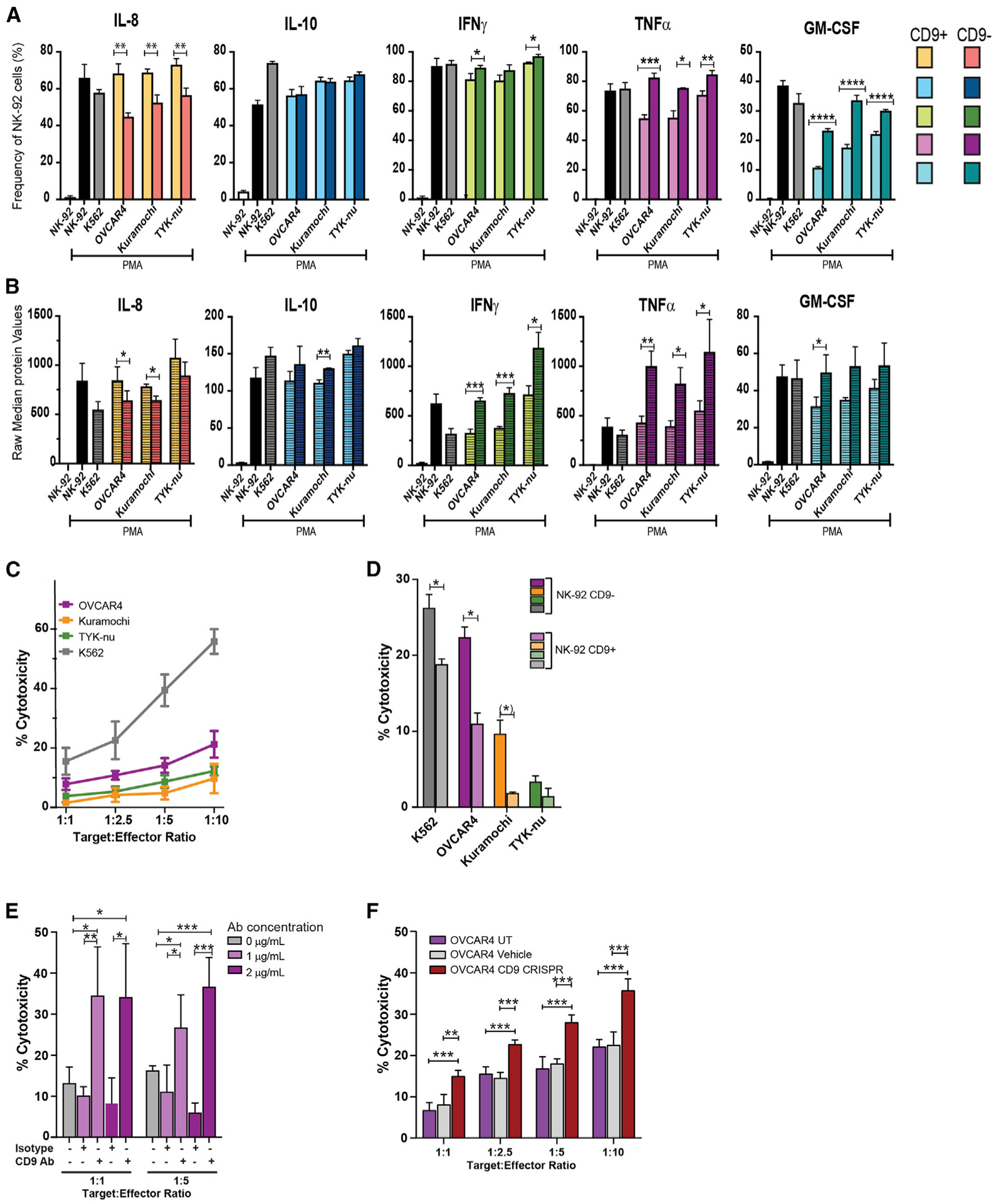
NK-92 cells were cocultured with HGSC cell lines (1:1) for 6 h, treated with brefeldin A/monensin and PMA/ionomycin or vehicle, and analyzed with the NK cell antibody panel (K562 cells as positive control) (Table S7; STAR Methods). CD9+ and CD9− NK-92 cells were manually gated with CD45+.
(A) Frequencies of CD9+ and CD9− NK-92 cells producing intracellular cytokines are indicated. Student’s two-tailed t test (also for B), showing statistically significant differences between CD9+ and CD9− NK-92 cells. *p ≤ 0.01, **p ≤ 0.001, ***p ≤ 0.0001, ****p ≤ 0.00001.
(B) Levels of intracellular cytokines produced by CD9+ and CD9− NK-92 cells. CD9+ NK-92 cells produced lower levels of anti-tumor cytokines (see text).
(C) Cytotoxicity, measured by the calcein release assay, was suppressed toward HGSC cell lines compared with K562 cells (positive control) (STAR Methods). Student’s two-tailed t test (also for D and E). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
(D) CD9+ NK-92 cells that underwent FACS after coculture have reduced cytotoxicity for HGSC cells compared with CD9− NK-92 cells. Statistical significance was determined with a two-tailed t test. *p ≤ 0.03, (*) p = 0.06.
(E) CD9-blocking antibody significantly increased NK-92 cytotoxicity toward OVCAR4 cells.
(F) CD9 CRISPR knockout in OVCAR4 cells significantly increased NK-92 cytotoxicity in coculture. UT, untreated; vehicle, nucleofector solution. Statistical significance was determined with a two-tailed t test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
(A–D) Mean of triplicates and (E and F) mean of quadruplicates with standard deviations.
For a subset of proteins, no differences were observed among NK-92 cells grown in mono- or coculture with or without PMA. Thus, high levels of granzyme B, perforin, and MIP1b were produced by >85% of NK-92 cells under all conditions with no differences between CD9+ and CD9− NK-92 cells. Cell frequencies and median expression levels of CD107a, a marker for degranulation, and production of MIP1α were both increased in response to PMA, but no differences were seen between CD9+ and CD9− NK-92 cells (Alter et al., 2004). Vascular endothelial growth factor (VEGF) levels were constitutively high in >90% of NK-92 cells grown in monoculture and coculture (Figure S6C).
After coculture with all three HGSC cell lines and PMA stimulation, both cell frequencies and amounts of interleukin (IL)-8 were statistically greater in CD9+ compared with CD9− NK-92 cells (Figures 7A and 7B). IL-8 is a proangiogenic factor produced by d-NK cells with a role in vascularizing the placenta (Jabrane-Ferrat, 2019) and, in the context of malignancy, a role in promoting the tumor angiogenic system (Yoneda et al., 1998). By contrast, after coculture and PMA stimulation, both cell frequencies and amounts of the anti-tumor cytokines tumor necrosis factor alpha (TNF-α), granulocyte macrophage colony-stimulating factor (GM-CSF), and interferon gamma (IFNγ) were statistically lower in CD9+ compared with CD9− NK-92 cells (Figures 7A and 7B). Similar frequencies (~60%) of CD9+ and CD9− NK-92 cells produced the immunosuppressive cytokine IL-10, but its expression was reduced in CD9+ NK-92 cells after coculture with the Kuramochi cell line (Figure 7A). These ICP data indicate that NK-92 cells that acquire CD9 are functionally more immune tolerant.
HGSC cell lines are poor targets for NK-92-mediated cytotoxicity
NK-92 cells were cocultured with OVCAR4, Kuramochi, and TYK-nu cell lines, and cytotoxicity was determined by the calcein release assay (Neri et al., 2001) (Figures 7C–7F; STAR Methods). Compared with the control K562 cell line, NK-92 cytotoxicity was significantly reduced toward all three HGSC cell lines (Figure 7C). Furthermore, the magnitude of attenuation trended with stage of tumor progression such that OVCAR4 cells were more and TYK-nu cells were less susceptible to NK-92 cell-mediated killing. This is consistent with the more immunosuppressive NK receptor ligand profile for V cells (Figures 2, 3, and 7C). Carboplatin did not mediate statistically significant changes in NK cell cytotoxicity (Figure S4C).
CD9+ NK-92 cells that had undergone FACS exhibit reduced cytotoxicity
To directly compare cytotoxicity of CD9+ with CD9− NK-92 cells, CD9+ NK-92 cells that had undergone FACS were cocultured with OVCAR4 (Figure 7D). CD9+ had significantly attenuated cytotoxicity compared with CD9− NK-92 cells. The TYK-nu cell line was resilient to both CD9+ and CD9− NK-92 cell-mediated killing, consistent with its mesenchymal phenotype.
CD9 antibody blockade increases NK-92 cell-mediated cytotoxicity
To determine the contribution of CD9 to the attenuated cytotoxicity of NK-92 cells toward HGSC cells, we performed the calcein release assay in the presence of a CD9-blocking antibody that significantly increased NK-92-mediated cytotoxicity for OVCAR4 cells (Figure 7E; STAR Methods).
CRISPR knockout of CD9 in OVCAR4 cells
To confirm the role of CD9 in NK cell-mediated immunosuppression, we generated a CD9 CRISPR knockout targeting exon 5 in OVCAR4 cells (STAR Methods). Sanger sequencing and flow cytometry confirmed 87% knockout of both the CD9 gene and the CD9 protein, respectively (Figures S6D and S6E). Cocultures between OVCAR4-CD9 CRISPR knockout cells with NK-92 cells resulted in 50% and 63% increases in NK-92 cytotoxicity (target:effector ratios of 1:5 and 1:10) (Figure 7F; STAR Methods). Overall, three independent approaches confirmed the role of CD9 in NK cell-mediated immune suppression (Figures 7D–7F).
DISCUSSION
This CyTOF study of newly diagnosed HGSC tumors identified dl-NK cell subpopulations that positively correlated with the overall abundance of tumor and EV cells (Figures 1A–1C). Previously, dl-NK cells were identified in colorectal and lung tumors, among other tumors (Bruno et al., 2018; Albini and Noonan, 2021).
Decidual NK cells express the tetraspanin CD9 and are functionally distinct from peripheral NK cells (Jabrane-Ferrat, 2019; Koopman et al., 2003). They comprise 70% of the total lymphocyte population during the first trimester of pregnancy. They produce a range of secretory proteins critical for placental decidualization, vascularization, and crucially, creation of a privileged immune-tolerant maternal-fetal compartment (Hanna et al., 2006; Jabrane-Ferrat, 2019). d-NK cells are poorly cytotoxic but contain cytotoxic granules that can be transiently activated to provide immunity to infection during pregnancy (Jabrane-Ferrat, 2019; Vento-Tormo et al., 2018; Tilburgs et al., 2015).
This study was based on the recognition that HGSC tumor cells promote immune tolerance to support tumor expansion. The identification and characterization of dl-NK cells described here supports the notion that HGSC tumor cells have a role in modulating NK cells toward a dl-NK phenotype. This included transfer of the tetraspanin CD9, a characteristic marker that classifies d-NK cells, by trogocytosis, as well as coexpression of NK receptor ligands that increase immunosuppression of NK cells.
To investigate how tumor and immune cells interact, we simultaneously measured the expression of twelve NK receptor ligands and two ADAM proteases on tumor cells, and E and EV tumor cells expressed more NKG2D-activating receptor ligands than V cells (Figures 2 and 3). This is consistent with previous reports that described upregulation of NKG2D ligands in tumor cells undergoing the epithelial-to-mesenchymal transition (EMT) rendering a majority susceptible to NK cell killing. Only tumor cells with low levels of NKG2D ligands escaped and progressed toward a metastatic state (López-Soto et al., 2017; Huergo-Zapico et al., 2018). Furthermore, the greater the number of infiltrating dl-NK cells, the greater the restraint on the EMT to V cells, consistent with the negative correlations we observed between dl-NK cells and V cells (Figure 1D).
In addition, the greater number of NK receptor ligand combinations observed in E and EV tumor cells could provide more opportunities for escape from immune surveillance, allowing transition into poor-prognosis V cells. V tumor cells may then switch to alternative, more stringent mechanisms of immune escape, including high expression levels of ULBP4, a functional outlier within the ULBP family (Figure 2) (Zöller et al., 2018; Dhar and Wu, 2018; Labani-Motlagh et al., 2016) (Figures 2B, 2D, and 3).
Numerous studies documented upregulation of NK-activating receptor ligands in cells exposed to genotoxic agents, epigenetic modifiers, and radiation (Gasser et al., 2005; Dhar and Wu, 2018). In contrast, carboplatin induced a more inhibitory NK receptor ligand phenotype (increased HLA-E, nectin-4, HLA-ABC, and CD111) in HGSC cell lines. Of these, nectin-4 was reported to have a role in HGSC metastasis and chemotherapeutic resistance (Derycke et al., 2010) (Figure 4). With independent roles for nectin-4 and CD111 in adhesion, cell movement, and stem cell biology, our data have uncovered a potentially far greater contribution for carboplatin in tumor-mediated immune suppression (Belaaloui et al., 2003; Martinet and Smyth, 2015; Derycke et al., 2010). The increase in nectin-4 expression post-NACT in a small subgroup of HGSC tumors warrants a follow-up study with a larger cohort (Figure 4D; Table S6). Characterizing NK receptor ligand expression, especially after carboplatin-based chemotherapy, may be a valuable companion diagnostic for determining patient suitability for NK immunotherapy.
To ascertain how HGSC cells might modulate NK cells toward an immunosuppressive role, we cocultured HGSC cell lines with the molecularly well-characterized NK-92 cell line that expresses CD56 and lacks inhibitory KIR receptors. Unmodified NK-92 cells or NK-92 cells genetically engineered to express chimeric antigen receptors or activating NK receptors such as NKG2D have satisfied safety criteria in several early-phase clinical trials (Rezvani, 2019; Daher and Rezvani, 2018; Rezvani et al., 2017; Suck et al., 2016).
Coculture data confirmed that CD9 protein was transferred from HGSC cells and, to a lesser extent, from non-HGSC cells to NK-92 cells by NK-92-mediated trogocytosis (Figures 5, 6C, and 6D). This indicates the potential ease with which therapeutically administered NK cells could be reprogrammed toward a dl-NK cell phenotype. Trogocytosis has been observed for T, B, and NK cells and basophils (Domaica et al., 2009; Gary et al., 2012; Joly and Hudrisier, 2003; McCann et al., 2007; Miyake et al., 2017; Tabiasco et al., 2002). Coculture experiments between d-NK cells and extravillous trophoblasts and between peripheral NK and melanoma cells resulted in transfer of HLA-G onto the NK cells, enhancing their immune tolerance (Caumartin et al., 2007; Tilburgs et al., 2015). A recent study using a mouse model of leukemia identified trogocytosis as a mechanism by which chimeric antigen receptor T (CAR-T) cells lost their efficacy by acquiring the CD19 target they were engineered to seek from the tumor cells they were programmed to kill (Hamieh et al., 2019). It was reported that trogocytosis-mediated therapeutic resistance could account for the observed tumor cell antigen loss in patients undergoing CAR-T cell therapy (Hamieh et al., 2019). In the same study, tetraspanin CD81, complexed with CD19, was jointly acquired by the CAR-T cells. This raises the question as to whether tetraspanins have a central role in trogocytosis. Altogether, these studies provide increasing evidence of trogocytosis playing a key role in immune tolerance and therapeutic resistance.
Although the exclusivity of CD9 protein and mRNA expression in the decidual NK cells is well established, its functional role in these cells is unclear (Koopman et al., 2003; Kopcow et al., 2005). In our study, uptake of CD9 in NK-92 cells by trogocytosis coincided with decreased anti-tumor cytokine production, increased production of pro-angiogenic IL-8, and suppressed cytotoxicity (Figures 7A and 7B). CD9 trogocytosis was greatest for OVCAR4 cells, yet OVCAR4 cells were subject to a small but consistently greater susceptibility to NK cell cytotoxicity (Figures 5A and 7C). This suggests that CD9-mediated immune suppression may be more important for E cells to counter immune surveillance mediated by higher expression levels of NK-activating receptor ligands (Figures 2, 3, and 5A). Critically, in cocultures with a CD9-blocking antibody, CD9 CRISPR knockout, and CD9− NK-92 cells that had undergone FACS, there were significant increases in NK-92 cytotoxicity, providing strong evidence that CD9 confers NK-92 cells with immunosuppressive properties (Figures 7D–7F).
We observed high levels of CD9 in HGSC tumor cells, suggesting that in vivo, these tumor cells could be a likely source of CD9 not only for intratumoral NK cell-mediated trogocytosis but also potentially for other tumor-infiltrating immune cell types, such as T cells (Figure S1B). The presence of high frequencies of two CD9+ immune cell clusters (CD3−CD56LoCD9+CXCR3+CD4+) in all tumors with only weak tumor cell correlations suggests they could be precursors for the strongly correlated dl-NK and T cells and signify plasticity between these CD9 NK and CD9 T cell phenotypes (Figures S1C and S1D).
CD9 shows ubiquitous distribution and is involved in multiple cellular functions, such as proliferation, motility, and adhesion, with major roles in immune cell synapse formation, which may be critical in regulating the HGSC tumor microenvironment (Reyes et al., 2018; Lorico et al., 2021). CD9 directly associates with ADAM17 and inhibits its cleavage activity toward surface protein ectodomains (Gutiérrez-López et al., 2011). Thus, transfer of CD9 from HGSC tumor cells onto intratumoral NK cells could reactivate ADAM17 cleavage activity and indirectly facilitate immune escape by NK ligand and/or receptor shedding (Lanier, 2015; Boutet et al., 2009; Raulet et al., 2013).
Future studies will be directed toward determining the mechanism by which CD9, and any other cotransferred proteins, suppresses NK cell function. To expand the coculture data, a transcriptomic, proteomic, and functional characterization of intratumoral NK cells in HGSC will be undertaken.
Our results emphasize the critical need to evaluate both CD9 and NK receptor ligand expression within HGSC tumors to stratify those patients most likely to benefit from NK immunotherapy. Furthermore, abundant expression of CD9 on HGSC tumor cells presents the distinct possibility that NK immunotherapy could be seriously compromised through CD9 trogocytosis. The data from this study identify mechanisms of immune suppression in HGSC and provide evidence that CD9 could serve as a drug target with immediate relevance for NK immunotherapy.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Wendy J. Fantl (wjfantl@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Two datasets; 17 CyTOF FCS files for NK/T cell tumor infiltrates and 12 CyTOF FCS files for tumor cells have been deposited at Mendeley Data and will be made publicly available as of the date of publication. The link to download the datasets is listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified antibodies conjugated to metal isotopes, see Tables S3, S4, and S7 | Various | Various |
| BV421 mouse anti-human CD9 (clone M-L13) | BD Biosciences | Cat#743047; RRID: AB_2741243 |
| PE mouse anti-human CD9 (clone M-L13) | BD Biosciences | Cat#555372; RRID: AB_395774 |
| APC anti-human CD45 (clone HI30) | BioLegend | Cat#304037; RRID: AB_2562049 |
| Anti-PE-165Ho (clone PE001) | Fluidigm | Cat#3165015B; RRID: AB_2714168 |
| Purified anti-human CD11c (3.9) | BioLegend | Cat#301601: RRID: AB_314171 |
| Anti-human CD14 (M5E2)-160Gd | Fluidigm | Cat#3160001B; RRID: AB_2687634 |
| Anti-human CD19 (HIB19)-169TM | Fluidigm | Cat#3169011B; RRID; AB_2893034 |
| Mouse IgG1 kappa (clone 15–6E10A7) | abcam | Cat#ab170190; RRID: AB_2736870 |
| Mouse anti-CD9 antibody (clone MEM-61) | abcam | Cat#ab2215; RRID: AB_302894 |
| Biological samples | ||
| Newly diagnosed chemo naive HGSC tumors prepared as single cell suspensions, see Table S1 | Indivumed | https://www.indivumed.com/ |
| Archival FFPE HGSC tumor samples | Stanford Pathology | https://med.stanford.edu/pathology.html |
| Peripheral blood mononuclear cells (PBMCs) | Stanford Blood Center | https://stanfordbloodcenter.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium heparin | Sigma-Aldrich | Cat#2106-15VL |
| Benzonase | Sigma-Aldrich | Car#E8263-25KU |
| Antibody stabilization solution | Candor Bioscience | Cat#131050 |
| Live/Dead Fixable Near-IR Dead Cell Stain Kit, for 633 or 635 nm excitation | Thermo Fisher Scientific | Cat#L10119 |
| Human TruStain FcX (FC Receptor blocking solution) | BioLegend | Cat#422302 |
| eBioscience Permeabilization Buffer (10X) | Thermo Fisher Scientific | Cat#00-8333-56 |
| Cell-ID intercalator-Ir | Fluidigm | Cat#201192B |
| Cell-ID intercalator-103Rh-2000 μM | Fluidigm | Cat#201103B |
| Palladium isotopes as nitrate salts | Trace Sciences International | N/A |
| Calibration Beads, EQ Four Element | Fluidigm | Cat#201078 |
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15711 |
| 32% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15714 |
| HistoGel | Thermo Fisher Scientific | Cat#HG-4000–012 |
| Cisplatin | Sigma-Aldrich | Cat#P4394 |
| Carboplatin | Sigma-Aldrich | Cat#2538 |
| eBioscience Cell Stimulation Cocktail (500x) | Thermo Fisher Scientific | Cat#00-4970-93 |
| eBioscience Brefeldin A solution (1000X) | Thermo Fisher Scientific | Cat#00-4506-51 |
| eBioscience Monensin solution (1000X) | Thermo Fisher Scientific | Cat#00-4505-51 |
| PKH67 Green Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling | Sigma Aldrich | Cat#MINI67–1KT |
| DAPI solution | BD Biosciences | Cat#564907 |
| PKH26 Red Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling | Sigma-Aldrich | Cat#MINI26–1KT |
| Cytochalasin D | Sigma-Aldrich | Cat#C2618 |
| Calcein-AM | Thermo Fisher Scientific | Cat#C3100MP |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | QIAGEN | Cat#51304 |
| GeneRead DNaseq Targeted Ovarian V2 panel | QIAGEN | Custom |
| MaxPar conjugation kit | Fluidigm | N/A |
| RNAscope 2.5 HD assay-brown kit with Hs-Nectin4 probe | ACD bio | N/A |
| Corning 96 well TC-treated microplates | Thermo Fisher Scientific | Cat#3799 |
| Corning 96 well black polystyrene microplate | Thermo Fisher Scientific | Cat#3603 |
| Corning HTS Transwell −96 Permeable Support System | Thermo Fisher Scientific | Cat#09-761-80 |
| miRNeasy isolation kit | QIAGEN | Cat#74004 |
| High-Capacity cDNA Reverse Transcription kit | Applied Biosystems, ThermoFisher Scientific | Cat#4368814 |
| TaqMan gene expression assay:
Hs00170423_m1 (CDH1-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4453320 |
| TaqMan gene expression assay:
Hs00894716_m1 (PTPRC-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4448892 |
| TaqMan gene expression assay:
Hs01124022_m1 (CD9-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4453320 |
| TaqMan gene expression assay:
Hs02758991_g1 (GADPH-VIC) |
Applied Biosystems, ThermoFisher Scientific | Cat#4448489 |
| TaqMan Gene Expression Master Mix | Applied Biosystems, Thermo Fisher Scientific | Cat#4370048 |
| Gene Knockout kit V2 targeting CD9 | Synthego | N/A |
| CAS9 2NLS Nuclease | Synthego | N/A |
| SE. Cell Line 4D-Nucleofector X Kit S | Lonza | Cat#V4XC-1032 |
| Taq PCR Master Mix kit | QIAGEN | Cat#201443 |
| QIAquick PCR purification kit | QIAGEN | Cat#28104 |
| Deposited data | ||
| CyTOF datasets of NK and T cell infiltrate and tumor cells for ovarian tumors | Mendeley Data | https://dx.doi.org/10.17632/mtbgnz7yk5.1 |
| Experimental models: Cell lines | ||
| OVCAR4 | Fox Chase Cancer Center | N/A |
| Kuramochi | JCRB Cell Bank | JCRB0098 |
| TYK-nu | JCRB Cell Bank | JCRB0234.0 |
| NK-92 | ATCC | CRL-2407 |
| PC3 | Brooks Lab, Stanford | N/A |
| SNU-349 | Fan Lab, Stanford | N/A |
| K562 | ATCC | CCL-243 |
| HeLa | ATCC | CCL-2 |
| A431 | ATCC | CRL-1555 |
| NCI-H28 | ATCC | CRL-5820 |
| HEPG2 | ATCC | HB-8065 |
| HCT116 | ATCC | CCL-247 |
| MCF7 | ATCC | HTB-22 |
| MCF10A | ATCC | CRL-10317 |
| CaCo-2 | ATCC | HTB-37 |
| OVCAR3 | ATCC | HTB-161 |
| HCC1937 | ATCC | CRL-2336 |
| LoVo | ATCC | CCL-229 |
| A549 | ATCC | CCL-185 |
| OVSAHO | JCRB Cell Bank | JCRB1046 |
| OVKATE | JCRB Cell Bank | JCRB1044 |
| SNU-119 | Seoul National University - Korea | N/A |
| JHOS-2 | RIKEN BRC Cell Bank | RBRC-RCB1521 |
| JHOM-2B | RIKEN BRC Cell Bank | RBRC-RCB1682 |
| COV362 | Public Health England | 7071910 |
| DLD-1 | Horizon Discovery | HD PAR-008 |
| COV318 | Sigma Aldrich | 7071903 |
| NKL | Chen Lab, Institute of Biomedical Science (Taiwan) | N/A |
| Oligonucleotides | ||
| CD9 multi-guide RNA
probe1: GCGACAUACCGCAUAGUGGA |
Synthego | N/A |
| CD9 multi-guide RNA
probe2: CUUGGUUUUCAGCUUGUUGU |
Synthego | N/A |
| CD9 multi-guide RNA
probe3: CUGCCCAUUGUAGGUGAUUA |
Synthego | N/A |
| Primer: CD9
Forward GAGCCAAGTTAGGAGCCAAGT |
Integrated DNA Technologies (IDT) | Custom |
| Primer: CD9
Reverse CGAGTACGTCCTTCTTGGGG |
Integrated DNA Technologies (IDT) | Custom |
| Primer: CD9 DNA
sequencing CCTGAGAGAAGGCAGTGCTA |
Integrated DNA Technologies (IDT) | Custom |
| Software and algorithms | ||
| CellEngine analysis software | CellCarta | https://cellengine.com/#/ |
| Vortex | Samusik et al., 2016 | https://github.com/nolanlab/vortex/ |
| Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/, Version 9 |
| Inference of CRIPSR Edits (ICE) | Synthego (Hsiau et al., 2019) | https://ice.synthego.com/ |
| MATLAB - Normalizer | (Finck et al., 2013) | https://github.com/nolanlab/bead-normalization/wiki/Normalizing-FCS-Files |
| MATLAB – Single Cell Debarcoder | (Zunder et al., 2015) | https://github.com/nolanlab/single-celldebarcoder |
| R environment | R-project | https://www.r-project.org |
| Premessa R package | (Zunder et al., 2015) | https://github.com/ParkerICI/premessa |
| Cytobank | (Kotecha et al., 2010) | https://cytobank.org/ |
| Microsoft excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| FlowJo | BD Biosciences | https://www.flowjo.com/ |
| Other | ||
| CyTOF2 mass cytometer | Fluidigm | N/A |
| Sony SH800 cell sorter | Sony Biotechnology | N/A |
| 7900HT Fast Real-Time PCR System | Stanford Genomics | N/A |
| Keyence BZ-X800 microscope | Keyence | N/A |
| BD LSRII flow cytometer | BD Biosciences | N/A |
| 4D-Nucleofector unit | Lonza | N/A |
| ABI 3130xl sequencer | Stanford Protein and Nucleic Acid Facility | N/A |
| Tecan Infinite M1000 microplate reader | Stanford High-Throughput Bioscience Center | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Newly diagnosed chemo naive HGSC tumors for CyTOF
Deidentified newly diagnosed chemo naive HGSC tumors prepared as single cell suspensions for CyTOF analysis collected over a two-year period were purchased from Indivumed (Hamburg, Germany). Information of patients are shown in Table S1. Tumor samples were collected in compliance with the Helsinki declaration and all patients provided written informed consent. The use of human tissue was approved and complied with data protection for patient confidentiality. Institutional review board approval was obtained at the Physicians Association in Hamburg, Germany.
Tumor slices for RNAScope
Ten patient-matched pairs of deidentified archival FFPE HGSC tumor blocks before and after NACT were obtained from the Pathology archives with Institutional Review Board (IRB) approval.
Peripheral blood mononuclear cells (PBMCs)
Viably frozen PBMCs from healthy donors were obtained via the Stanford Blood Center. For trogocytosis experiments, PBMCs were thawed in cell culture medium (RPMI-1640, 10% FBS, 1% L-glutamine, 1% pen/strep) supplemented with 20 U/mL sodium heparin and 0.025 U/mL benzonase.
Cell lines
Cell lines were authenticated by short tandem repeat (STR) profiling performed by the Stanford Functional Genomics Facility.
OVCAR4 cells were maintained in M199 medium supplemented with 5% FBS, 1% L-glutamine, and 1% pen/strep. Kuramochi, SNU-349, NCI-H28, HCC1937, OVSAHO, OVKATE, and SNU-119 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 1% L-glutamine, and 1% pen/strep. TYK-nu and HEPG2 cells were maintained in EMEM medium supplemented with 10% FBS, 1% L-glutamine, and 1% pen/strep. NK-92 and NKL cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 1% L-glutamine, 1% pen/strep, and 200 U/mL IL-2. PC3, LoVo, and A549 cells were maintained in F-12K medium supplemented with 10% FBS, 1% L-glutamine, and 1% pen/strep. K562 cells were maintained in IMDM medium supplemented with 10% FBS, and 1% pen/strep. HeLa, A431, COV362, and COV318 cells were maintained in DMEM medium supplemented with 10% FBS, 1% L-glutamine, and 1% pen/strep. HCT116 and OVCAR-3 cells were maintained in McCoy’s medium supplemented with 10% FBS, 1% L-glutamine, and 1% pen/strep. MCF7 cells were maintained in EMEM medium supplemented with 10% FBS, 1% L-gluta-mine, 1% pen/strep, and 0.01 mg/mL human recombinant insulin. MCF10A cells were maintained in DMEM/F12 medium supplemented with 5% horse serum, 10 μg/mL human insulin, 20 ng/mL hEGF, 100 ng/mL Cholera toxin, and 0.5 μg/mL hydrocortisone. CaCo-2 cells were maintained in DMEM medium supplemented with 20% FBS, 1% L-glutamine, and 1% pen/strep. JHOS-2 and JHOM-2B cells were maintained in DMEM/F12 medium supplemented with 10% FBS, 0.1 mM non-essential amino acids (NEAA), and 1% pen/strep. DLD-1 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 1% L-glutamine, 1% pen/strep, and 25 mM sodium bicarbonate.
METHOD DETAILS
Genomic sequencing and analysis for TP53 and BRCA1/2
DNA was extracted and enriched through multiplex PCR (QIAGEN QIAmp DNA Mini-Kit and QIAGEN GeneRead DNaseq Targeted Ovarian V2 Panel, respectively). The TrueSeq protocol was used to make an indexed Illumina sequencing library from the pooled sample amplicons. The subsequent protocols for sequencing were described previously (Gonzalez et al., 2018). The pathogenic variants were noted (Table S2).
Antibodies for CyTOF
Antibodies were either purchased pre-conjugated or conjugated in-house as previously reported (Gonzalez et al., 2018) In brief, for in-house conjugations, antibodies in carrier-free PBS were conjugated to metal-chelated polymers (MaxPAR antibody conjugation kit, Fluidigm) according to the manufacturer’s protocol or to bismuth with our protocol (Han et al., 2017). Metal-labeled antibodies were diluted to 0.2–0.4 mg/mL in antibody stabilization solution (CANDOR Biosciences) and stored at 4°C. Each antibody was titrated using cell lines and primary human samples as positive and negative controls. Antibody concentrations used in experiments were based on an optimal signal-to-noise ratio. Three CyTOF antibody panels were used in this study to characterize: i) tumor T and NK cells (Table S3) ii) tumor NK receptor ligand expression (Table S4) and iii) NK cell receptor and intracellular cytokine expression (Table S7).
Antibodies for fluorescence flow cytometry
Antibodies against CD9 BV421, CD9 PE, from BD and CD45 APC from BioLegend were used to detect CD9 and CD45. The same antibody clones were used for CyTOF (Tables S3 and S7). Near-IR fixable LIVE/DEAD stain from Thermo Fisher Scientific was used to distinguish dead cells.
Sample processing and antibody staining for CyTOF
Frozen, fixed single-cell suspensions of HGSC tumors or cell lines were thawed at room temperature. For each sample, 1 × 106 cells were aliquoted into cluster tubes in 96 well plates and subjected to pre-permeabilization palladium barcoding (Gonzalez et al., 2018). After barcoding, pooled cells were pelleted and incubated with TruStain FcX Fc receptor block (BioLegend) to prevent non-specific antibody binding, for 10min at room temperature. Cells were then incubated with antibodies against surface markers for 45min at room temperature. Cells were permeabilized at 4°C with methanol or 1x Permeabilization Buffer (eBioscience, Thermo Fisher Scientific), (only for NK cell CyTOF antibody panel Table S7) on ice for 10min. Cells were subsequently stained with antibodies against intracellular markers for 1h at room temperature, washed, and incubated with the 191/193Ir DNA intercalator (Fluidigm) at 4°C overnight. Cells were washed and resuspended in a solution of normalization beads before introduction into the CyTOF 2 (Bendall et al., 2011; Gonzalez et al., 2018).
Treatment of cell lines with carboplatin
Cells were seeded in 10 cm dishes (300K for OVCAR4, 350K for Kuramochi and 150K for TYK-nu) and grown in recommended media. On day 2, carboplatin (0.5 μg/mL or 1 μg/mL) or vehicle (H20) was added. On day 5, media was aspirated and replaced with fresh media containing the same concentrations of carboplatin or vehicle. On day 7, cells were harvested for measuring expression levels of NK receptor ligands. The inclusion of an antibody against pH2AX was used to verify carboplatin-mediated DNA damage.
For other assays, cells treated with carboplatin according to these conditions were used to measure CD9 expression levels, CD9 uptake, and NK-92 cell cytotoxicity (See below).
Measurements of nectin 4 RNA expression by RNAscope
Nectin-4 RNA probes were validated based on nectin-4 protein expression in HGSC cell lines determined by CyTOF (Figures 4A–4C and S3C). We used two positive cell lines (OVCAR3 and SNU119) and two negative HGSC cell lines (COV318 and TYK-nu) for this validation. We grew and pelleted 4 to 6 million cells per cell line. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 20min at room temperature, washed with PBS twice, embedded in HistoGel (TM) (Thermo Fisher Scientific), and transferred to 70% ethanol for paraffin-embedding and sectioning at 5-μm thickness.
Formalin-fixed paraffin-embedded (FFPE) HGSC tissue sections (4 μm thick) were subjected to RNA In Situ Hybridization. The sections underwent the manufacturer’s recommended FFPE sample preparation and standard pretreatment. Post-pretreatment, the RNAscope® 2.5 HD assay protocol was performed using the RNAscope® 2.5 HD Assay-Brown kit (ACD bio) with Hs-nectin-4 probe (nucleotides 798 – 1699 of the human gene for nectin-4, accession number NM_030916.2). UBC was used as a positive control RNA probe and DapB as a negative non-specific control probe. DapB recognizes a bacterial species that is not part of the human microbiome.
Determination of intracellular pools of CD9
NK-92 and OVCAR4 cells were stained with cisplatin (Sigma Aldrich), fixed with 1.6% paraformaldehyde (Electron Microscopy Sciences), washed, incubated with TruStain FcX Fc receptor block (BioLegend) (as above) and then incubated with CD9-PE (Becton Dickinson) for 45min at room temperature. Cells were washed and stained with anti-PE-165Ho (Fluidigm) for 30min at room temperature. Following secondary antibody staining, cells were permeabilized with 1x Permeabilization Buffer (eBioscience, Thermo Fisher Scientific), on ice for 10min. Cells were subsequently stained with CD9–156Gd (Fluidigm) (to detect intracellular CD9) for 1h at room temperature. Cells were processed and introduced into the CyTOF 2 as described above.
Cocultures
The HGSC cell lines, OVCAR4, Kuramochi and TYK-nu cells, were each cocultured with the NK-92 or NKL cell lines, at an effector:target ratio of 1:1 for 6h unless otherwise indicated, at 37°C in a humidified cell culture incubator. HGSC cells (100,000/well) were seeded in U-bottom 96-well plates (Corning, Costar) with NK-92 cells (100,000/well). When evaluating CD9 uptake by primary NK cells, after coculture with of OVCAR4 with PBMCs, we gated NK cells out of the population: CD45+ CD3−, CD19−, CD14−, CD11c− CD7+. We used antibodies against CD16 and CD56 to identify the two peripheral NK cell phenotypes (CD16+CD56dim and CD16−CD56bright). Assays were performed with biological and technical replicates.
In some cases, OVCAR4, Kuramochi and TYK-nu cells were cocultured with NK-92 in a 96-well dual-chamber transwell plate with 3 μm micropores (Corning, Costar). HGSC cells (100,000/well) were placed into the lower chamber, and NK-92 cells (100,000/well) were placed into the upper chamber. The cells were cultured at 37°C for 6h in a humidified cell culture incubator.
For intracellular cytokine production, PMA / Ionomycin cell stimulation cocktail (500x) (eBioscience, Thermo Fisher Scientific), was added during the last 4h. The protein transport inhibitors, Brefeldin A and Monensin (eBioscience, Thermo Fisher Scientific), were used at a final concentration of 3 μg/mL and 2 μM, respectively. There were two positive controls; i) NK-92 cells grown in monoculture −/+ PMA and ii) coculture of the K562 cell line (HLA null erythroleukemic) (Tremblay-McLean et al., 2019) with NK-92 cells. CD107a-151Eu antibody (Fluidigm) (1 μl) was added to each well as a marker for degranulation. All experiments were performed with biological and technical triplicates with details described for specific assays.
Plasma membrane uptake assay
OVCAR4 cells were labeled with PKH67 (Sigma Aldrich) prior to coculture with NK-92 cells. In brief, OVCAR4 cells were washed with serum free media and resuspended in diluent C. A 2x working solution of PKH67 was prepared immediately prior to use. Cells were mixed with PKH67 working solution for a final concentration of 5 × 106 cells/mL in 20 μM PKH67 and incubated for 5min at room temperature. The labeling was quenched with an equal volume of fetal bovine serum, incubated for 1min, and washed three times with 10mL of complete media. Cells were seeded in U-bottom 96-well plates (Corning, Costar), and cocultured with NK-92 cells at target:effector ratios 5:1, 2.5:1, 1:1, 1:2.5 and 1:5 for 24h at 37°C. Cells were then stained with CD45 and CD9 antibodies and processed for flow cytometry.
Cell sorting by FACS
OVCAR4 cells were cocultured with NK-92 or NKL cells at a 1:1 ratio in U-bottom 96 well plates for 24h at 37°C. Cocultured cells were harvested and washed with serum-free RPMI-1640 medium. Cells (2 × 106) were stained in 90 μL of serum-free RPMI-1640 medium with 5 μL of PE mouse anti-human CD9 (BD PharMingen, clone M-L13) and 5 μL of APC anti-human CD45 antibody (BioLegend, clone HI30) for 30min on ice. The cells were washed with serum-free RPMI-1640 medium and stained with DAPI (BD PharMingen, 0.1 μg/mL) in 1mL of serum-free RPMI-1640 medium for 10min at RT, quenched and washed with complete RPMI-1640 media (supplemented with 10% FBS, 2 mM L-glutamine and 1% pen strep). Cells were resuspended in 1mL complete RPMI-1640 media and filtered before sorting. CD9+ and CD9− NK cells were collected in FBS. Cells grown in monoculture were used as controls.
Cells were sorted on a Sony SH800 cell sorter, according to the following steps: i) FSC- versus FSC-A for singlets ii) DAPI for live cells iii) CD45 for NK cells and iv) CD9 versus CD45 to collect CD9+ and CD9− NK cells.
Quantitative real-time PCR
Total mRNA was isolated with a miRNeasy isolation kit (QIAGEN) from monocultures of OVCAR4 and NK-92 cells and FACS-sorted CD9+ and CD9− NK-92 cells after coculture with OVCAR4 cells. cDNA was generated using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR for E-cadherin (CDH1) Hs00170423_m1, CD45 (PTPRC) Hs00894716_m1, and CD9 Hs01124022 was performed with the TaqMan gene expression assay (Applied Biosystems) and the 7900HT fast real-Time PCR system (Applied Biosystems).
Imaging trogocytosis
OVCAR4 and NK-92 cells were labeled with the membrane dyes PKH67 and PKH26 (Sigma Aldrich), respectively, prior to coculture. Cells were washed with serum free media and resuspended in Diluent C. A 2x working solution of each membrane dyes was prepared immediately prior to use. Cells (5 × 106cells/mL) were mixed with their respective working solution of dye for a final concentration of 20 μM. After a 5min incubation at room temperature the labeling was quenched with an equal volume of fetal bovine serum, incubated for 1min, and washed three times with 10mL of complete media. Cells were seeded in U-bottom 96-well plates (Corning, Costar), and cocultured with NK-92 cells at an effector:target ratio 1:1 for 3h at 37°C. Cells were then fixed with a final concentration of 1.6% paraformaldehyde, stained with CD45 and CD9 antibodies and seeded on microscope slides for imaging on a Keyence BZ-X800 microscope.
Inhibition of trogocytosis
NK-92 cells were pre-treated with cytochalasin D (10 μM in complete media) for 2h (Aucher et al., 2008). They were then cocultured with cancer cell lines as indicated in a ratio of 1:1 in the continuous presence of cytochalasin D for a further 2h after which cells were stained with antibodies against CD9, CD45 and processed prior to CyTOF analysis as described above.
CRISPR knockout of CD9 in OVCAR4 cells
Knockout of CD9 in OVCAR4 cells was performed using the Gene Knockout kit v2 (Synthego) with three CD9 exon 5 multi-guide RNA probes: G*C*G*ACAUACCGCAUAGUGGA, C*U*U*GGUUUUCAGCUUGUUGU, C*U*G*CCCAUUGUAGGUGAUUA. The lyophilized pellet of sgRNAs was rehydrated in nuclease-free 1x TE buffer (100mM) and diluted with nuclease-free water to a working solution (30μM). To maximize transfection efficiency, while minimizing cell death, the following variables were investigated using a green fluorescent protein construct (Lonza) and flow cytometry as a readout: cell number, nucleofector solutions, programs on the 4D-Nucleofector Unit.
Cas9 ribonucleoprotein (RNP) complexes were assembled immediately before nucleofection by mixing multi-guide sgRNAs with Cas9 protein (has two nuclear localization sequences) (20μM) at a 6:1 molar ratio in Cell Line Nucleofector solution (Lonza) for 10 min at room temperature. OVCAR4 cells (3×105) were harvested and washed with Dulbecco’s phosphate buffered saline (DBPS) prior to resuspension in Nucleofector solution (5μl). Cell suspensions and pre-complexed RNPs (25μL) were mixed, transferred to Nucleocuvette strips (Lonza), and placed in the 4D Nucleofector Unit (Lonza) calibrated for program EH-100. Immediately after nucleofection, cells were mixed with 70μL of warm complete M199 medium (supplemented with 5% FBS, 2mM L-glutamine and 1% pen strep). 50μL of the cell suspension were transferred to each well of a 24-well plate. Cells were incubated at 37°C, media (complete M199) replaced at 24h followed by incubation for another 48h.
DNA sequence analysis to confirm CD9 knockout
OVCAR4 cells (5 × 104) were harvested after 72h and washed with DPBS. Genomic DNA was extracted using QIAamp DNA Mini Kit (QIAGEN). PCR amplification of the target sequence was performed with the Taq PCR Master Mix Kit (QIAGEN) and primer sequences GAGCCAAGTTAGGAGCCAAGT and CGAGTACGTCCTTCTTGGGG (5′ - 3′, forward and reverse, respectively). The PCR product generated from cells after nucleofection with sgRNAs or controls (untreated and nucleofector solution (vehicle)) was purified with QIAquick PCR Purification Kit (QIAGEN). Sanger Sequencing analysis was performed using a 5′ to 3′ forward primer CCTGAGAGAAGGCAGTGCTA on the ABI 3130xl instrument in the Stanford Protein and Nucleic Acid Facility. CD9 knockout was determined using Inference of CRISPR Edits (ICE) (Synthego). The results were Indel percentage, 87%, and a Knockout-Score, 82.
Calcein-AM release cytotoxicity assay
OVCAR4, Kuramochi, TYK-nu and K562 (control) target cells were washed in PBS and resuspended in calcein-acetoxymethyl (calcein-AM; Thermo Fisher Scientific) staining solution (2.5 μM in PBS) at a cell density of 1×106/mL and incubated for 30min at 37°C (Lorenzo-Herrero et al., 2020; Ramadoss et al., 2020). When performing the cytotoxicity assay with CD9 knockout OVCAR4 cells, controls were: i) untreated OVCAR4 and ii) OVCAR4 cells incubated with Nucleofector solution.
Target cells were seeded in U-bottom 96-well plates (Corning, Costar), and cocultured with NK-92 cells at increasing effector:target ratios 1:1, 2.5:1, 5:1 and 10:1 for 4h at 37°C. For the cytotoxicity assay with CD9 blocking antibody, after both cell lines were plated either control mouse IgG1 kappa (Abcam, clone: 15–6E10A7) or purified mouse monoclonal CD9 antibody (Abcam, clone: MEM-61) were added to the coculture at the concentrations indicated for the duration of the incubation. Cells were then spun down and 100 μL of supernatant were transferred to a black-walled 96 well plate (Corning, Costar). Calcein release was measured from the fluorescent signal using 485 nm excitation wavelength and 530 nm emission wavelength (Ex/Em Calcein: 494/517 nm) with a Tecan Infinite M1000 fluorescent plate reader. Control wells contained HGSC target cells alone (spontaneous lysis) or with 2% Triton X-100 (maximum lysis). Specific killing was calculated using the equation: specific killing = (lysis of coculture - spontaneous lysis) / (maximum lysis - spontaneous lysis) × 100%. The assay was performed with biological and technical quadruplicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data and statistical analysis were implemented with Microsoft Excel, MATLAB, R and GraphPad Prism 9. CyTOF datasets were evaluated with software available from Cytobank (Kotecha et al., 2010), CellEngine (cellengine.com) and FlowJo. Number of replicates (n), statistical tests used, and statistical significance per experiments are shown in figure legends.
Initial assessment of data quality
Initial data quality was assessed by determining dead and apoptotic cells which were excluded from further analysis. Viable cells, defined as cisplatin negative and cleaved PARP negative were used for experiments (Gonzalez et al., 2018). For cell lines treated with carboplatin, Cell-ID intercalator 103rhodium (Fluidigm) was used for live-dead viability staining. For experiments with newly diagnosed HGSC tumors, tumor cells were gated as CD45−/CD31−/FAP− and immune cells were gated as CD45+ CD66− as described previously (Gonzalez et al., 2018).
Clustering of tumor, T and NK cells
Manually gated CD45+CD66− (immune) and CD45−/CD31−/FAP− (tumor) cells from each HGSC tumor were pooled for clustering with X-shift (Samusik et al., 2016), a density-based clustering algorithm, using the Vortex clustering environment (Key Resources table). Markers for clustering the immune and tumor cells are shown in Tables S3 and S4, respectively.
Correlation network analysis
Spearman pairwise correlation coefficients (rs) were computed between: i) cell frequencies for 56 tumor cell clusters, ii) frequency of 52 T and NK cell clusters, iii) total tumor cell abundance, iv) total EV cell abundance, v) total E-cadherin cell abundance and vi) other features previously described (Gonzalez et al., 2018). A hierarchically ordered heat-map was generated in R.
Force directed layout visualization
Force directed layouts were generated from a composite of all 12 HGSC tumors. After merging all single cell data files, cells were clustered. 10,000 single cells were computationally sampled from each of the 56 tumor cell clusters. Each cell was connected on a 10-nearest-neighbor graph. This graph was subjected to a force-directed layout (FDL) that placed groups of phenotypically related cells adjacent to one another (Samusik et al., 2016). Repeat samplings generated comparable results. Layouts were colored for expression of E-cadherin, vimentin, NK receptor ligands or ADAM10 and ADAM17.
Combinatorial expression for NK receptor ligands in E, EV and V tumor compartments
The tumor population CD45−/FAP−/CD31− was the parent population for gating the E, EV and V tumor compartments (Gonzalez et al., 2018). Frequencies of tumor cell subpopulations defined by their combinatorial expression patterns of the twelve NK receptor ligands and two ADAM proteases were determined with MATLAB and FlowJo. For this analysis, the frequency of tumor cells expressing each of these proteins was determined in each compartment on a per sample basis. Combinations used in the analysis were based on a threshold frequency of > 1% for cells in any compartment in any sample.
Simpson’s index of diversity
The Simpson’s index of diversity, D was calculated in Excel with the formula D = 1 – (∑n(n – 1)/N(N – 1)) where N is the total number of tumor cell subpopulations with a specific NK receptor ligand combination (163) and n is the number of times a subpopulation is present in the E, EV and V compartments of each of the 12 tumors.
Supplementary Material
Highlights.
Decidual-like NK cells correlate with tumor cell abundance in tubo-ovarian HGSC
Combinatorial NK receptor ligand expression levels differ across tumor compartments
NK cells acquire CD9 from HGSC tumor cells via trogocytosis
CD9 suppresses anti-tumor cytokine production and cytotoxicity in NK-92 cells
ACKNOWLEDGMENTS
The authors thank Drs. Catherine Blish, James D. Brenton, and B. Matija Peterlin for critical reading of the manuscript. We thank Drs. Catherine Blish and Nancy Zhao for help with the calcein release assay, Rob Monroe for help designing nectin-4 and control probes for RNAscope, and Pauline Chu for sectioning cell pellets for RNAscope. We thank Shawn Hollahan at paradigm_shiftin@mac.com for artwork. This work was supported by funding from the Department of Defense (W81XWH-12-1-0591); NCI (1R01CA234553, R21CA231280); the 2019 Cancer Innovation Award, supported by the Stanford Cancer Institute, an NCI-designated Comprehensive Cancer Center; BRCA Foundation and the V Foundation for Cancer Research; a gift from the Gray Foundation; the Department of Urology at Stanford University; NHLBI (P01HL10879709); NIAID (U19AI057229); and a PICI Bedside to Bench grant. A.D.-G. thanks the Fundacion Alfonso Martin Escudero for his postdoctoral fellowship.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109632.
DECLARATION OF INTERESTS
The authors declare no competing interests. We have a published patent related to this study: WO2021/050200, PCT/US2020/046195.
REFERENCES
- Albini A, and Noonan DM (2021). Decidual-Like NK Cell Polarization: From Cancer Killing to Cancer Nurturing. Cancer Discov. 11, 28–33. [DOI] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, and Altfeld M (2004). CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294, 15–22. [DOI] [PubMed] [Google Scholar]
- Ashworth A, and Lord CJ (2018). Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol 15, 564–576. [DOI] [PubMed] [Google Scholar]
- Aucher A, Magdeleine E, Joly E, and Hudrisier D (2008). Capture of plasma membrane fragments from target cells by trogocytosis requires signaling in T cells but not in B cells. Blood 111, 5621–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC Jr., Matulonis UA, Sood AK, Ahmed AA, Amobi AE, Balkwill FR, Wielgos-Bonvallet M, Bowtell DDL, Brenton JD, Brugge JS, et al. (2019). Critical Questions in Ovarian Cancer Research and Treatment: Report of an American Association for Cancer Research Special Conference. Cancer 125, 1963–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaaloui G, Imbert AM, Bardin F, Tonnelle C, Dubreuil P, Lopez M, and Chabannon C (2003). Functional characterization of human CD34+ cells that express low or high levels of the membrane antigen CD111 (nectin 1). Leukemia 17, 1137–1145. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir E-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HB, Plasterer MC, Schiff SE, Kitchen CM, Kitchen S, and Zack JA (2006). CD4 expression on activated NK cells: ligation of CD4 induces cytokine expression and cell migration. J. Immunol 177, 3669–3676. [DOI] [PubMed] [Google Scholar]
- Boutet P, Agüera-González S, Atkinson S, Pennington CJ, Edwards DR, Murphy G, Reyburn HT, and Valés-Gómez M (2009). Cutting edge: the metalloproteinase ADAM17/TNF-alpha-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J. Immunol 182, 49–53. [DOI] [PubMed] [Google Scholar]
- Bowtell DD, Böhm S, Ahmed AA, Aspuria P-J, Bast RC Jr., Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et al. (2015). Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennick CA, George MM, Corwin WL, Srivastava PK, and Ebrahimi-Nik H (2017). Neoepitopes as cancer immunotherapy targets: key challenges and opportunities. Immunotherapy 9, 361–371. [DOI] [PubMed] [Google Scholar]
- Bruno A, Bassani B, D’Urso DG, Pitaku I, Cassinotti E, Pelosi G, Boni L, Dominioni L, Noonan DM, Mortara L, and Albini A (2018). Angiogenin and the MMP9-TIMP2 axis are up-regulated in proangiogenic, decidual NK-like cells from patients with colorectal cancer. FASEB J. 32, 5365–5377. [DOI] [PubMed] [Google Scholar]
- Bryceson YT, Fauriat C, Nunes JM, Wood SM, Björkström NK, Long EO, and Ljunggren HG (2010). Functional analysis of human NK cells by flow cytometry. Methods Mol. Biol 612, 335–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumartin J, Favier B, Daouya M, Guillard C, Moreau P, Carosella ED, and LeMaoult J (2007). Trogocytosis-based generation of suppressive NK cells. EMBO J. 26, 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni C, Fionda C, Soriani A, Zingoni A, Doria M, Cippitelli M, and Santoni A (2014). The DNA Damage Response: A Common Pathway in the Regulation of NKG2D and DNAM-1 Ligand Expression in Normal, Infected, and Cancer Cells. Front. Immunol 4, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiossone L, Dumas PY, Vienne M, and Vivier E (2018). Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol 18, 671–688. [DOI] [PubMed] [Google Scholar]
- Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, Okamoto A, Moore KN, Efrat Ben-Baruch N, Werner TL, et al. (2019). Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med 381, 2403–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo AC, van der Zwan A, Ramalho-Santos J, Strominger JL, and Tilburgs T (2017). Cytotoxic potential of decidual NK cells and CD8+ T cells awakened by infections. J. Reprod. Immunol 119, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher M, and Rezvani K (2018). Next generation natural killer cells for cancer immunotherapy: the promise of genetic engineering. Curr. Opin. Immunol 51, 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance A (2019). Core Concept: Cells nibble one another via the under-appreciated process of trogocytosis. Proc. Natl. Acad. Sci. USA 116, 17608–17610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubeuf S, Aucher A, Bordier C, Salles A, Serre L, Gaibelet G, Faye JC, Favre G, Joly E, and Hudrisier D (2010). Preferential transfer of certain plasma membrane proteins onto T and B cells by trogocytosis. PLoS ONE 5, e8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derycke MS, Pambuccian SE, Gilks CB, Kalloger SE, Ghidouche A, Lopez M, Bliss RL, Geller MA, Argenta PA, Harrington KM, and Skubitz AP (2010). Nectin 4 overexpression in ovarian cancer tissues and serum: potential role as a serum biomarker. Am. J. Clin. Pathol 134, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar P, and Wu JD (2018). NKG2D and its ligands in cancer. Curr. Opin. Immunol 51, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaica CI, Fuertes MB, Rossi LE, Girart MV, Avila DE, Rabinovich GA, and Zwirner NW (2009). Tumour-experienced T cells promote NK cell activity through trogocytosis of NKG2D and NKp46 ligands. EMBO Rep. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, and Schultz N (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre-Lafay S, Monville F, Garrido-Urbani S, Berruyer-Pouyet C, Ginestier C, Reymond N, Finetti P, Sauvan R, Adélaïde J, Geneix J, et al. (2007). Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 7, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C, Long EO, Ljunggren HG, and Bryceson YT (2010). Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 115, 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari de Andrade L, Tay RE, Pan D, Luoma AM, Ito Y, Badrinath S, Tsoucas D, Franz B, May KF Jr., Harvey CJ, et al. (2018). Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science 359, 1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, and Bendall SC (2013). Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Voelkl S, Palmisano R, Ullrich E, Bosch JJ, and Mackensen A (2012). Antigen-specific transfer of functional programmed death ligand 1 from human APCs onto CD8+ T cells via trogocytosis. J. Immunol 188, 744–752. [DOI] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, and Raulet DH (2005). The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436, 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez VD, Samusik N, Chen TJ, Savig ES, Aghaeepour N, Quigley DA, Huang YW, Giangarrà V, Borowsky AD, Hubbard NE, et al. (2018). Commonly Occurring Cell Subsets in High-Grade Serous Ovarian Tumors Identified by Single-Cell Mass Cytometry. Cell Rep. 22, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, McCormick C, Lorusso D, Hoskins P, Freyer G, et al. ; PRIMA/ENGOT-OV26/GOG-3012 Investigators (2019). Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med 381, 2391–2402. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-López MD, Gilsanz A, Yáñez-Mó M, Ovalle S, Lafuente EM, Domínguez C, Monk PN, González-Alvaro I, Sánchez-Madrid F, and Cabañas C (2011). The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell. Mol. Life Sci 68, 3275–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J, Eyquem J, Zhao Z, Whitlock BM, Miele MM, et al. (2019). CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Chen S-Y, Gonzalez VD, Zunder ER, Fantl WJ, and Nolan GP (2017). Atomic Mass Tag of Bismuth-209 for Increasing the Immunoassay Multiplexing Capacity of Mass Cytometry. Cytometry A 91, 1150–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, et al. (2006). Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med 12, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. (2013). Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med 5, 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, and Rich Stoner R (2019). Inference of CRISPR Edits from Sanger. Trace Data bioRxiv, 251082. [DOI] [PubMed] [Google Scholar]
- Huergo-Zapico L, Parodi M, Cantoni C, Lavarello C, Fernández-Martínez JL, Petretto A, DeAndrés-Galiana EJ, Balsamo M, López-Soto A, Pietra G, et al. (2018). NK-cell Editing Mediates Epithelial-to-Mesenchymal Transition via Phenotypic and Proteomic Changes in Melanoma Cell Lines. Cancer Res. 78, 3913–3925. [DOI] [PubMed] [Google Scholar]
- Jabrane-Ferrat N (2019). Features of Human Decidual NK Cells in Healthy Pregnancy and During Viral Infection. Front. Immunol 10, 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly E, and Hudrisier D (2003). What is trogocytosis and what is its purpose? Nat. Immunol 4, 815. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Seow SV, Wong D, Robinson M, and Campana D (2019). Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest 129, 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandalaft LE, Odunsi K, and Coukos G (2019). Immunotherapy in Ovarian Cancer: Are We There Yet? J. Clin. Oncol 37, 2460–2471. [DOI] [PubMed] [Google Scholar]
- Kim CH, Butcher EC, and Johnston B (2002). Distinct subsets of human Valpha24-invariant NKT cells: cytokine responses and chemokine receptor expression. Trends Immunol. 23, 516–519. [DOI] [PubMed] [Google Scholar]
- Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD, Park PJ, and Strominger JL (2003). Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med 198, 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopcow HD, Allan DS, Chen X, Rybalov B, Andzelm MM, Ge B, and Strominger JL (2005). Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc. Natl. Acad. Sci. USA 102, 15563–15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, and Irish JM (2010). Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom Chapter, 10:Unit 10.17. 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenning L, van den Berg J, and Medema RH (2019). Life or Death after a Break: What Determines the Choice? Mol. Cell 76, 346–358. [DOI] [PubMed] [Google Scholar]
- Labani-Motlagh A, Israelsson P, Ottander U, Lundin E, Nagaev I, Nagaeva O, Dehlin E, Baranov V, and Mincheva-Nilsson L (2016). Differential expression of ligands for NKG2D and DNAM-1 receptors by epithelial ovarian cancer-derived exosomes and its influence on NK cell cytotoxicity. Tumour Biol. 37, 5455–5466. [DOI] [PubMed] [Google Scholar]
- Lanier LL (2015). NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res 3, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hermanson DL, Moriarity BS, and Kaufman DS (2018). Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 23, 181–192.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Soto A, Gonzalez S, Smyth MJ, and Galluzzi L (2017). Control of Metastasis by NK Cells. Cancer Cell 32, 135–154. [DOI] [PubMed] [Google Scholar]
- Lord CJ, and Ashworth A (2017). PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Herrero S, López-Soto A, Sordo-Bahamonde C, Gonzalez-Rodriguez AP, Vitale M, and Gonzalez S (2018). NK Cell-Based Immunotherapy in Cancer Metastasis. Cancers (Basel) 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Herrero S, Sordo-Bahamonde C, González S, and López-Soto A (2020). Evaluation of NK cell cytotoxic activity against malignant cells by the calcein assay. Methods Enzymol 631, 483–495. [DOI] [PubMed] [Google Scholar]
- Lorico A, Lorico-Rappa M, Karbanová J, Corbeil D, and Pizzorno G (2021). CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. (May-wood) 246, 1121–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet L, and Smyth MJ (2015). Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol 15, 243–254. [DOI] [PubMed] [Google Scholar]
- Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, and Karlan BY (2016). Ovarian cancer. Nat. Rev. Dis. Primers 2, 16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann FE, Eissmann P, Onfelt B, Leung R, and Davis DM (2007). The activating NKG2D ligand MHC class I-related chain A transfers from target cells to NK cells in a manner that allows functional consequences. J. Immunol 178, 3418–3426. [DOI] [PubMed] [Google Scholar]
- Mirza MR, Coleman RL, González-Martín A, Moore KN, Colombo N, Ray-Coquard I, and Pignata S (2020). The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann. Oncol 31, 1148–1159. [DOI] [PubMed] [Google Scholar]
- Miyake K, Shiozawa N, Nagao T, Yoshikawa S, Yamanishi Y, and Karasuyama H (2017). Trogocytosis of peptide-MHC class II complexes from den dritic cells confers antigen-presenting ability on basophils. Proc. Natl. Acad. Sci. USA 114, 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan MG, and Lanier LL (2016). NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19. [DOI] [PubMed] [Google Scholar]
- Neri S, Mariani E, Meneghetti A, Cattini L, and Facchini A (2001). Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab. Immunol 8, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MT, and Lanier LL (2010). Natural killer cell education and tolerance. Cell 142, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujade-Lauraine E, Fujiwara K, Ledermann JA, Oza AM, Kristeleit R, Ray-Coquard IL, Richardson GE, Sessa C, Yonemori K, Banerjee S, et al. (2021). Avelumab alone or in combination with chemotherapy versus chemotherapy alone in platinum-resistant or platinum-refractory ovarian cancer (JAVELIN Ovarian 200): an open-label, three-arm, randomised, phase×study. Lancet Oncol. 22, 1034–1046. [DOI] [PubMed] [Google Scholar]
- Ramadoss NS, Zhao NQ, Richardson BA, Grant PM, Kim PS, and Blish CA (2020). Enhancing natural killer cell function with gp41-targeting bispecific antibodies to combat HIV infection. AIDS 34, 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH, Gasser S, Gowen BG, Deng W, and Jung H (2013). Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol 31, 413–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reches A, Ophir Y, Stein N, Kol I, Isaacson B, Charpak Amikam Y, Elnekave A, Tsukerman P, Kucan Brlic P, Lenac T, et al. (2020). Nectin4 is a novel TIGIT ligand which combines checkpoint inhibition and tumor specificity. J. Immunother. Cancer 8, e000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R, Cardeñes B, Machado-Pineda Y, and Cabañas C (2018). Tetraspanin CD9: A Key Regulator of Cell Adhesion in the Immune System. Front. Immunol 9, 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K (2019). Adoptive cell therapy using engineered natural killer cells. Bone Marrow Transplant. 54 (Suppl 2), 785–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K, Rouce R, Liu E, and Shpall E (2017). Engineering Natural Killer Cells for Cancer Immunotherapy. Mol. Ther 25, 1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM, Galpin KJC, McCloskey CW, and Vanderhyden BC (2018). The Tumor Microenvironment of Epithelial Ovarian Cancer and Its Influence on Response to Immunotherapy. Cancers (Basel) 10, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samusik N, Good Z, Spitzer MH, Davis KL, and Nolan GP (2016). Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Correa B, Valhondo I, Hassouneh F, Lopez-Sejas N, Pera A, Bergua JM, Arcos MJ, Bañas H, Casas-Avilés I, Durán E, et al. (2019). DNAM-1 and the TIGIT/PVRIG/TACTILE Axis: Novel Immune Checkpoints for Natural Killer Cell-Based Cancer Immunotherapy. Cancers (Basel) 11, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabrish S, Gupta M, and Madkaikar M (2016). A Modified NK Cell Degranulation Assay Applicable for Routine Evaluation of NK Cell Function. J. Immunol. Res 2016, 3769590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert JC, Inokuma M, Waid DM, Pennock ND, Vaitaitis GM, Disis ML, Dunne JF, Wagner DH Jr., and Maecker HT (2008). An analytical workflow for investigating cytokine profiles. Cytometry A 73, 289–298. [DOI] [PubMed] [Google Scholar]
- Singh N, McCluggage WG, and Gilks CB (2017). High-grade serous carcinoma of tubo-ovarian origin: recent developments. Histopathology 71, 339–356. [DOI] [PubMed] [Google Scholar]
- Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. (2017). Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck G, Linn YC, and Tonn T (2016). Natural Killer Cells for Therapy of Leukemia. Transfus. Med. Hemother 43, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabiasco J, Espinosa E, Hudrisier D, Joly E, Fournié JJ, and Vercellone A (2002). Active trans-synaptic capture of membrane fragments by natural killer cells. Eur. J. Immunol 32, 1502–1508. [DOI] [PubMed] [Google Scholar]
- Tilburgs T, Evans JH, Crespo AC, and Strominger JL (2015). The HLA-G cycle provides for both NK tolerance and immunity at the maternal-fetal interface. Proc. Natl. Acad. Sci. USA 112, 13312–13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay-McLean A, Coenraads S, Kiani Z, Dupuy FP, and Bernard NF (2019). Expression of ligands for activating natural killer cell receptors on cell lines commonly used to assess natural killer cell function. BMC Immunol. 20, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppendahl LD, Dahl CM, Miller JS, Felices M, and Geller MA (2018). Natural Killer Cell-Based Immunotherapy in Gynecologic Malignancy: A Review. Front. Immunol 8, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polański K, Goncalves A, et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, and Ugolini S (2011). Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas JM, Van der Veen AG, and Ploegh HL (2008). The known unknowns of antigen processing and presentation. Nat. Rev. Immunol 8, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JR, Milne K, and Nelson BH (2015). PD-1 and CD103 Are Widely Coexpressed on Prognostically Favorable Intraepithelial CD8 T Cells in Human Ovarian Cancer. Cancer Immunol. Res 3, 926–935. [DOI] [PubMed] [Google Scholar]
- Wilk AJ, and Blish CA (2018). Diversification of human NK cells: Lessons from deep profiling. J. Leukoc. Biol 103, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski EE, Parham P, and Guethlein LA (2019). Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC. Front. Immunol 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, and Fidler IJ (1998). Expression of angiogenesis-related genes and progression of human ovarian carcinomas in nude mice. J. Natl. Cancer Inst 90, 447–454. [DOI] [PubMed] [Google Scholar]
- Zamarin D, Burger RA, Sill MW Jr., Powell DJ Jr., Lankes HA, Feldman MD, Zivanovic O, Gunderson C, Ko E, Mathews C, et al. (2020). Randomized Phase II Trial of Nivolumab Versus Nivolumab and Ipilimumab for Recurrent or Persistent Ovarian Cancer: An NRG Oncology Study. J. Clin. Oncol 38, 1814–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller T, Wittenbrink M, Hoffmeister M, and Steinle A (2018). Cutting an NKG2D Ligand Short: Cellular Processing of the Peculiar Human NKG2D Ligand ULBP4. Front. Immunol 9, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir E-AD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. (2015). Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc 10, 316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Two datasets; 17 CyTOF FCS files for NK/T cell tumor infiltrates and 12 CyTOF FCS files for tumor cells have been deposited at Mendeley Data and will be made publicly available as of the date of publication. The link to download the datasets is listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified antibodies conjugated to metal isotopes, see Tables S3, S4, and S7 | Various | Various |
| BV421 mouse anti-human CD9 (clone M-L13) | BD Biosciences | Cat#743047; RRID: AB_2741243 |
| PE mouse anti-human CD9 (clone M-L13) | BD Biosciences | Cat#555372; RRID: AB_395774 |
| APC anti-human CD45 (clone HI30) | BioLegend | Cat#304037; RRID: AB_2562049 |
| Anti-PE-165Ho (clone PE001) | Fluidigm | Cat#3165015B; RRID: AB_2714168 |
| Purified anti-human CD11c (3.9) | BioLegend | Cat#301601: RRID: AB_314171 |
| Anti-human CD14 (M5E2)-160Gd | Fluidigm | Cat#3160001B; RRID: AB_2687634 |
| Anti-human CD19 (HIB19)-169TM | Fluidigm | Cat#3169011B; RRID; AB_2893034 |
| Mouse IgG1 kappa (clone 15–6E10A7) | abcam | Cat#ab170190; RRID: AB_2736870 |
| Mouse anti-CD9 antibody (clone MEM-61) | abcam | Cat#ab2215; RRID: AB_302894 |
| Biological samples | ||
| Newly diagnosed chemo naive HGSC tumors prepared as single cell suspensions, see Table S1 | Indivumed | https://www.indivumed.com/ |
| Archival FFPE HGSC tumor samples | Stanford Pathology | https://med.stanford.edu/pathology.html |
| Peripheral blood mononuclear cells (PBMCs) | Stanford Blood Center | https://stanfordbloodcenter.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium heparin | Sigma-Aldrich | Cat#2106-15VL |
| Benzonase | Sigma-Aldrich | Car#E8263-25KU |
| Antibody stabilization solution | Candor Bioscience | Cat#131050 |
| Live/Dead Fixable Near-IR Dead Cell Stain Kit, for 633 or 635 nm excitation | Thermo Fisher Scientific | Cat#L10119 |
| Human TruStain FcX (FC Receptor blocking solution) | BioLegend | Cat#422302 |
| eBioscience Permeabilization Buffer (10X) | Thermo Fisher Scientific | Cat#00-8333-56 |
| Cell-ID intercalator-Ir | Fluidigm | Cat#201192B |
| Cell-ID intercalator-103Rh-2000 μM | Fluidigm | Cat#201103B |
| Palladium isotopes as nitrate salts | Trace Sciences International | N/A |
| Calibration Beads, EQ Four Element | Fluidigm | Cat#201078 |
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15711 |
| 32% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15714 |
| HistoGel | Thermo Fisher Scientific | Cat#HG-4000–012 |
| Cisplatin | Sigma-Aldrich | Cat#P4394 |
| Carboplatin | Sigma-Aldrich | Cat#2538 |
| eBioscience Cell Stimulation Cocktail (500x) | Thermo Fisher Scientific | Cat#00-4970-93 |
| eBioscience Brefeldin A solution (1000X) | Thermo Fisher Scientific | Cat#00-4506-51 |
| eBioscience Monensin solution (1000X) | Thermo Fisher Scientific | Cat#00-4505-51 |
| PKH67 Green Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling | Sigma Aldrich | Cat#MINI67–1KT |
| DAPI solution | BD Biosciences | Cat#564907 |
| PKH26 Red Fluorescent Cell Linker Mini Kit for General Cell Membrane Labeling | Sigma-Aldrich | Cat#MINI26–1KT |
| Cytochalasin D | Sigma-Aldrich | Cat#C2618 |
| Calcein-AM | Thermo Fisher Scientific | Cat#C3100MP |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | QIAGEN | Cat#51304 |
| GeneRead DNaseq Targeted Ovarian V2 panel | QIAGEN | Custom |
| MaxPar conjugation kit | Fluidigm | N/A |
| RNAscope 2.5 HD assay-brown kit with Hs-Nectin4 probe | ACD bio | N/A |
| Corning 96 well TC-treated microplates | Thermo Fisher Scientific | Cat#3799 |
| Corning 96 well black polystyrene microplate | Thermo Fisher Scientific | Cat#3603 |
| Corning HTS Transwell −96 Permeable Support System | Thermo Fisher Scientific | Cat#09-761-80 |
| miRNeasy isolation kit | QIAGEN | Cat#74004 |
| High-Capacity cDNA Reverse Transcription kit | Applied Biosystems, ThermoFisher Scientific | Cat#4368814 |
| TaqMan gene expression assay:
Hs00170423_m1 (CDH1-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4453320 |
| TaqMan gene expression assay:
Hs00894716_m1 (PTPRC-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4448892 |
| TaqMan gene expression assay:
Hs01124022_m1 (CD9-FAM) |
Applied Biosystems, ThermoFisher Scientific | Cat#4453320 |
| TaqMan gene expression assay:
Hs02758991_g1 (GADPH-VIC) |
Applied Biosystems, ThermoFisher Scientific | Cat#4448489 |
| TaqMan Gene Expression Master Mix | Applied Biosystems, Thermo Fisher Scientific | Cat#4370048 |
| Gene Knockout kit V2 targeting CD9 | Synthego | N/A |
| CAS9 2NLS Nuclease | Synthego | N/A |
| SE. Cell Line 4D-Nucleofector X Kit S | Lonza | Cat#V4XC-1032 |
| Taq PCR Master Mix kit | QIAGEN | Cat#201443 |
| QIAquick PCR purification kit | QIAGEN | Cat#28104 |
| Deposited data | ||
| CyTOF datasets of NK and T cell infiltrate and tumor cells for ovarian tumors | Mendeley Data | https://dx.doi.org/10.17632/mtbgnz7yk5.1 |
| Experimental models: Cell lines | ||
| OVCAR4 | Fox Chase Cancer Center | N/A |
| Kuramochi | JCRB Cell Bank | JCRB0098 |
| TYK-nu | JCRB Cell Bank | JCRB0234.0 |
| NK-92 | ATCC | CRL-2407 |
| PC3 | Brooks Lab, Stanford | N/A |
| SNU-349 | Fan Lab, Stanford | N/A |
| K562 | ATCC | CCL-243 |
| HeLa | ATCC | CCL-2 |
| A431 | ATCC | CRL-1555 |
| NCI-H28 | ATCC | CRL-5820 |
| HEPG2 | ATCC | HB-8065 |
| HCT116 | ATCC | CCL-247 |
| MCF7 | ATCC | HTB-22 |
| MCF10A | ATCC | CRL-10317 |
| CaCo-2 | ATCC | HTB-37 |
| OVCAR3 | ATCC | HTB-161 |
| HCC1937 | ATCC | CRL-2336 |
| LoVo | ATCC | CCL-229 |
| A549 | ATCC | CCL-185 |
| OVSAHO | JCRB Cell Bank | JCRB1046 |
| OVKATE | JCRB Cell Bank | JCRB1044 |
| SNU-119 | Seoul National University - Korea | N/A |
| JHOS-2 | RIKEN BRC Cell Bank | RBRC-RCB1521 |
| JHOM-2B | RIKEN BRC Cell Bank | RBRC-RCB1682 |
| COV362 | Public Health England | 7071910 |
| DLD-1 | Horizon Discovery | HD PAR-008 |
| COV318 | Sigma Aldrich | 7071903 |
| NKL | Chen Lab, Institute of Biomedical Science (Taiwan) | N/A |
| Oligonucleotides | ||
| CD9 multi-guide RNA
probe1: GCGACAUACCGCAUAGUGGA |
Synthego | N/A |
| CD9 multi-guide RNA
probe2: CUUGGUUUUCAGCUUGUUGU |
Synthego | N/A |
| CD9 multi-guide RNA
probe3: CUGCCCAUUGUAGGUGAUUA |
Synthego | N/A |
| Primer: CD9
Forward GAGCCAAGTTAGGAGCCAAGT |
Integrated DNA Technologies (IDT) | Custom |
| Primer: CD9
Reverse CGAGTACGTCCTTCTTGGGG |
Integrated DNA Technologies (IDT) | Custom |
| Primer: CD9 DNA
sequencing CCTGAGAGAAGGCAGTGCTA |
Integrated DNA Technologies (IDT) | Custom |
| Software and algorithms | ||
| CellEngine analysis software | CellCarta | https://cellengine.com/#/ |
| Vortex | Samusik et al., 2016 | https://github.com/nolanlab/vortex/ |
| Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/, Version 9 |
| Inference of CRIPSR Edits (ICE) | Synthego (Hsiau et al., 2019) | https://ice.synthego.com/ |
| MATLAB - Normalizer | (Finck et al., 2013) | https://github.com/nolanlab/bead-normalization/wiki/Normalizing-FCS-Files |
| MATLAB – Single Cell Debarcoder | (Zunder et al., 2015) | https://github.com/nolanlab/single-celldebarcoder |
| R environment | R-project | https://www.r-project.org |
| Premessa R package | (Zunder et al., 2015) | https://github.com/ParkerICI/premessa |
| Cytobank | (Kotecha et al., 2010) | https://cytobank.org/ |
| Microsoft excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| FlowJo | BD Biosciences | https://www.flowjo.com/ |
| Other | ||
| CyTOF2 mass cytometer | Fluidigm | N/A |
| Sony SH800 cell sorter | Sony Biotechnology | N/A |
| 7900HT Fast Real-Time PCR System | Stanford Genomics | N/A |
| Keyence BZ-X800 microscope | Keyence | N/A |
| BD LSRII flow cytometer | BD Biosciences | N/A |
| 4D-Nucleofector unit | Lonza | N/A |
| ABI 3130xl sequencer | Stanford Protein and Nucleic Acid Facility | N/A |
| Tecan Infinite M1000 microplate reader | Stanford High-Throughput Bioscience Center | N/A |


