Abstract
Keratins K5 and K14 are the hallmarks of mitotically active keratinocytes of stratified epithelia. They are transcribed at a high level and in a tissue-specific manner, enabling us to use the K14 gene to elucidate the regulatory mechanism underlying epidermis-specific transcription. We have identified four DNase I-hypersensitive sites (HSs) present in the 5′ regulatory sequences of the K14 gene under specific conditions where the gene is actively expressed. Two of these sites (HSsII and -III) are conserved in position and sequence within the human and mouse K14 genes. Using an in vivo transgenic approach and an in vitro keratinocyte culture approach, we have discovered that most of K14's transcriptional activity is restricted to a novel 700-bp regulatory domain encompassing these HSs. This enhancer is sufficient to confer epidermis-specific activity to a heterologous promoter in transfection assays in culture and in transgenic mice in vivo. A 125-bp DNA fragment encompassing HSsII harbors the majority of the transactivation activity in vitro, and electrophoretic mobility shift and mutational assays reveal a role for AP-1, ets, and AP-2 family members in orchestrating the keratinocyte-preferred expression of HSsII. The HSsII element also confers epidermal expressivity to a heterologous promoter in transgenic mice, although it is not sufficient on its own to fully restrict activity to keratinocytes. Within the HSsII element, the ets and AP-2 sites appear to be most critical in collaborating to regulate epidermal specificity in vivo.
The epidermis is a stratified squamous epithelium that resides at the skin surface. Its proliferative compartment is located in the innermost basal layer, where stem cells and transiently amplifying cells attach to an underlying basement membrane of extracellular matrix (16, 18). In a homeostatic and self-renewing tissue, transiently amplifying basal cells periodically withdraw from the cell cycle, lose their contact with basement membrane, and activate a program of terminal differentiation (16–18). As the cells move upward toward the surface, they progress through three distinct stages: spinous, granular, and stratum corneum (15).
Throughout all but the stratum corneum, unique sets of genes are turned on and off in a growth- and differentiation-specific manner. These include genes encoding structural cytoskeletal proteins, transcription factors, and products that are necessary for the barrier function of skin (16–18). Basal cells transcribe the genes encoding keratins 5 and 14 (K5 and K14), while cells in the spinous layer switch off these genes and induce K1 and K10, required for the marked mechanical resistance of these cells (15). In the granular layer, cells express loricrin, filaggrin, and transglutaminase, involved in assembly of the cornified envelope. This structure forms a sac to contain the keratin filaments and to provide a scaffold upon which specialized lipids are extruded and organized (15, 16, 18). Once the barrier is in place, keratinocytes cease their metabolic activity, becoming dead squames that are eventually sloughed from the skin surface, continually being replaced by inner cells differentiating and transiting outward (18).
Temporally and spatially compartmentalized, the epidermis provides a unique system to study the molecular switches that govern tissue-specific and differentiation-specific gene expression (4, 11). In some studies, the promoters and enhancers of epidermis-specific genes have been examined to elucidate the cis and trans elements that govern gene regulation. In other studies, transcription factors which are preferentially expressed in epidermal keratinocytes have been identified. Most epidermal promoters contain functional binding sites for the AP-2 family of transcription factors expressed not only in epidermis but also in a number of other cell types (5, 6, 26, 30, 32, 41, 48). Although addition of exogenous AP-2 to certain nonkeratinocyte cell types can enable them to activate otherwise epidermally restricted promoters, the AP-2 site in the promoter of at least one epidermal gene, K5, is largely dispensable for tissue-specific gene expression in transgenic mice (5, 6). Together with the broad expression patterns of AP-2 genes (32), these findings suggest that AP-2 alone is unlikely to confer epidermal specificity in vivo.
Functional Sp1 sites are also found in the promoters of epidermally expressed genes, although they do not appear to be involved in cell-type-specific gene expression (4, 5, 28). Additional factors include POU domain proteins, NF-κB, AP-1, and ets family members (10, 11, 13, 36, 38, 40, 49). The POU domain proteins Skn-1a/I and Tst-1/Oct-6 seem to suppress suprabasal expression of K5 and K14 genes (1, 12), while NF-κB may play a role in the ability of basal keratinocytes to respond to external cues (40). In contrast, AP-1 and ets proteins have been implicated predominantly in regulating suprabasal genes, although some evidence supports the notion that these proteins may also influence expression of basal epidermal genes (7, 24, 28, 34, 38, 44). Whether these findings are physiologically relevant remains unexplored.
Despite extensive research, no transcription factor that is strictly expressed in a keratinocyte-specific fashion has been found, and no transcription factor regulatory motif which on its own can confer keratinocyte-restricted activity to a heterologous promoter in vitro or in vivo has been identified. It could be that such regulatory proteins exist but have yet to be identified. Alternatively, keratinocyte-specific activity could be controlled by combinatorial action of more broadly expressed transcription factors, which in vivo are expressed simultaneously only in keratinocytes.
Since basal epidermal keratinocytes can be easily propagated in culture, our studies have focused on the K5 and K14 genes, which are abundantly transcribed in these cells in vitro (43). Furthermore, K14 and K5 induction in vivo occurs concomitantly with commitment of an embryonic basal cell to an epidermal fate, making them valuable markers of this cell type (6). Previously, we characterized the human K14 and K5 promoters, both of which possess a classical TATA box and contain functional AP-2 and Sp1 sites (5, 27). However, in the course of these analyses, we found that sequences residing within 2 kb 5′ upstream of the K14 promoter were also required for appreciable reporter gene activity in SCC-13 squamous cell carcinoma keratinocytes in vitro and in transgenic mice in vivo (27).
We now define the nature of these critical upstream sequences and explore more deeply their role in keratinocyte-specific gene expression. Specifically, we have (i) identified the regions of the K14 gene that display altered chromatin structure under conditions where the gene is active but not when it is silent; (ii) elucidated which of these regions are responsible for the bulk of keratinocyte-specific gene activity in vitro and in transgenic mice in vivo; (iii) uncovered a correlation between conservation of K14 gene sequences and keratinocyte-specific alterations in chromatin structure; (iv) demonstrated that a 700-bp regulatory domain encompassing these sequences confers keratinocyte specificity when coupled to a heterologous promoter in vitro and in vivo; (v) showed that this region harbors functional binding sites for the AP-1, AP-2, and ets families of transcription factors, of which the AP-2 and ets sites are key; and (vi) showed that a segment containing functional AP-2 and ets sites can give rise to keratinocyte-preferred gene expression in vivo, whereas an analogous segment in which these sites are mutated cannot. Our findings suggest that basal keratinocyte-specific and differentiation-specific genes are governed in a combinatorial fashion by similar sets of transcription factor families, whose individual members are differentially expressed during terminal differentiation, thereby leading to switches in the expression of structural epidermal genes.
MATERIALS AND METHODS
Isolation of nuclei and DNase I HSs analysis.
DNase I-hypersensitive sites (HSs) were identified as described by Bellard et al. (3). Cells were scraped from 10 100-cm2 plates and subjected to centrifugation at 3,000 × g for 10 min. The pellet was resuspended in 10 ml of buffer A (15 mM Tris HCl, pH 7.5, 60 mM KCl, 15 mM NaCl, 1 mM EDTA [pH 8.0], 1.9 M sucrose, 0.1% Triton X-100, 0.5 mM spermidine), and cells were lysed in a Dounce homogenizer (10 to 15 strokes). After lysis, 10 ml of buffer B (buffer A without Triton X-100) was added, and the suspension was subjected to centrifugation at 12,000 × g for 10 min at 4°C. Nuclei were resuspended in 1.5 ml of buffer C (15 mM Tris HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 0.34 M sucrose, 0.5 mM spermidine) and then pelleted again in a tabletop centrifuge at 12,000 × g for 5 min. Final pellets were resuspended in a small volume of DNase I assay buffer (40 mM Tris HCl [pH 7.5], 6 mM MgCl2). (All buffers also contained 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of leupeptin per ml, and 10 μg of pepstatin per ml, which were added fresh for each step.)
DNase I (7,500 U/ml; Pharmacia) was diluted in buffer containing 10 mM Tris HCl (pH 7.5), 50 mM NaCl, 10 mM CaCl2, and 62.5 mM MgCl2 to a final concentration range of 0 to 6 U/μl. Ten-microliter aliquots of these DNase I solutions (0, 1, 2, 4, and 6 U/μl) were added to 90 μl of the resuspended nuclei in 100-μl reaction volume. After incubation at 37°C for 10 min, reactions were quenched by adding 200 μl of stop buffer (50 mM Tris HCl [pH 7.5], 100 mM NaCl, 1% sodium dodecyl sulfate, 20 mM EDTA [pH 8.0]). Forty microliters of proteinase K (20 mg/ml) was added to each reaction, and nuclei were incubated overnight at 55°C to digest proteins. Following extraction with phenol-chloroform, DNA was precipitated with 3 volumes of ethanol. After the DNA pellet was washed with 70% ethanol, genomic DNA was resuspended in TE (10 mM Tris HCl [pH 8.0], 1 mM EDTA). Ten micrograms of genomic DNA was digested with the appropriate restriction endonucleases followed by electrophoresis through a 1% agarose gel. Digested DNA was then transferred to Zeta-Probe GT membrane (Bio-Rad), and the blot was hybridized with the random primer-labeled probe according to the manufacturer's protocol except that 10% dextran sulfate was added to the hybridization buffer to improve the signal.
Cell culture and transient transfection assays.
Human primary keratinocytes were grown in serum-free keratinocyte growth medium (Clonetics) and were split at passage 2 and/or 3 for all experimental purposes. mK, a spontaneously arising immortalized mouse keratinocyte cell line, was grown in low-Ca2+ medium (20). All other cells, including HepG2 (American Type Culture Collection, Manassas, Va.), NIH 3T3, and SCC-13 cells, primary human fibroblasts, and HeLa cells, were grown under standard conditions (5, 27). Transient transfections of all DNA constructs were carried out using FuGENE6 transfection reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. Cells were plated in six-well plates and transfected at 30 to 40% confluence. For each well, 2 μg of the luciferase reporter plasmid and 1 μg of a CMV-lacZ plasmid (internal control) were cotransfected to adjust for variations in the transfection efficiency. Cells were harvested 36 h after transfection, washed with phosphate-buffered saline (PBS), and lysed with luciferase reporter lysis buffer (Promega). Luciferase assays were performed with a luciferase assay system (Promega) using a luminometer. β-Galactosidase values were obtained using a chemiluminescence reporter gene assay system (Galactolight kit; Tropix) according to the manufacturer's protocol. Luciferase activity was then normalized against β-galactosidase values.
Generation of transgenic mice and β-galactosidase staining.
Transgenic mice were generated as before (27, 46). Mice were genotyped by PCR using two specific primers for the β-galactosidase gene, and genomic DNA was isolated from tails or toes. Embryos from transgenic females were isolated at embryonic day 15.5 (E15.5) or E16.5, washed in PBS, and fixed in tissue fixation buffer (Specialty Media) for 30 min. Following several washes with PBS, embryos were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Duration of the staining varied from a few hours to overnight, depending on the strength of the signal. No staining was seen in wild-type control embryos. Following staining, the embryos were washed in PBS, postfixed in 4% paraformaldehyde, and embedded in paraffin. Tissues from adult mice were embedded in OCT (optimal cutting temperature) compound and sectioned (10 μm). Frozen sections were fixed in 0.5% glutaraldehyde for 2 min and assayed for β-galactosidase activity. Sections were counterstained with eosin.
DNA constructs for transfection and transgenics.
Recombinant plasmids were constructed using DNA fragments generated by restriction enzyme digestion or by PCR. Mutants were created by using a two-step PCR approach (45). All junctions or PCR-generated fragments were verified by sequencing. The thymidine kinase (TK) minimal promoter (5) was cloned into the NheI and XhoI sites of the luciferase reporter pGL3-basic vector (Promega) to create the LTK construct. The various fragments of the K14 promoter/enhancer regions were cloned into the KpnI-SacI or KpnI-NheI site of the LTK construct. A 700-bp DNA fragment of the K14 5′ region (−2000 to −1300) was cloned upstream of the simian virus 40 (SV40) minimal promoter in the pGL3-promoter vector (Promega) and upstream of the K14 minimal promoter (−210 to +40) to create 700SV40Luc and 700K14mpLuc, respectively. To facilitate cloning, the vector pNASSβ (Clontech) was modified to contain several additional restriction enzymes sites. All constructs were then transferred from the LTK constructs using suitable restriction endonucleases to the modified pNASSβ to create the LTKLacZ plasmids.
Nuclear extract and EMSA.
Nuclear extracts were prepared according to Schreiber et al. (39), with slight modifications. Protease inhibitors PMSF (1 mM), leupeptin (10 μg/ml), and pepstatin (10 μg/ml) were added freshly at the beginning of each step of extraction. Nuclear extracts for HeLa cells were purchased from Santa Cruz and Promega. Electrophoretic mobility shift assays (EMSAs) were performed with 4 to 6 μg of nuclear extracts and 3 × 104 cpm of end-labeled double-stranded oligonucleotides. Typically, binding reactions were performed at room temperature for 20 min. All binding reactions were performed in 20 μl of DNA binding buffer (25 mM HEPES [pH 7.9], 75 mM KCl, 1 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.5 mM dithiothreitol, 0.5 mM PMSF); 1 μg of poly(dI-dC) was added to each reaction as nonspecific DNA. For supershift assays, 1 μl of the antibody was added to each reaction. All antibodies were purchased from Santa Cruz Biotechnology. For competition assays, 50- or 100-fold-excess cold oligonucleotides were incubated with each nuclear extract for 10 min before addition of radiolabeled probe. The protein-DNA complexes were resolved by gel electrophoresis on 5% polyacrylamide gels. After electrophoresis, gels were dried and visualized by autoradiography.
RESULTS
Identification of keratinocyte-specific DNase I HSs within the 5′ upstream sequence of the human K14 gene.
Previously we showed that 2.1 to 2.2 kb of 5′ upstream sequence of the human K14 gene was sufficient to target expression of foreign genes to the basal layer of transgenic mouse epidermis (reference 46 and 47 and references therein). To uncover clues as to the possible cis elements needed for K14 gene expression, we began by analyzing its chromatin conformation in two primary diploid cell types: epidermal keratinocytes, which express copious amounts of K14, and fibroblasts, which do not express the K14 gene. Nuclei isolated from cultured human keratinocytes and fibroblasts were treated with increasing amounts of DNase I, and after extraction of proteins, genomic DNAs were further digested into defined fragments by restriction endonuclease treatment.
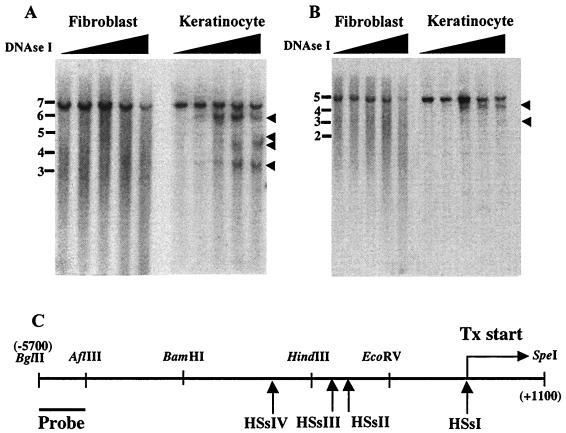
Southern analysis revealed that the chromatin encompassing the K14 gene was selectively hypersensitive to DNase I in keratinocytes in a fashion that was not seen in fibroblasts (Fig. 1). Four keratinocyte-specific HSs were found within a 6-kb stretch of sequence 5′ from and including the transcription initiation site of the K14 gene (29). A 600-bp radiolabeled probe corresponding to a BglII/AflIII fragment was used to localize these sites within the larger BglII/SpeI segment (Fig. 1C). The four HSs mapped approximately to the transcription initiation site (HSsI) and to sites at 1,300 bp (HSsII, faint), 1,700 (HSsIII) bp and 2,800 bp (HSsIV) 5′ upstream from this site (Fig. 1A). These sites were confirmed by repeating the mapping this time using BglII and EcoRV (Fig. 1B). Similar results were obtained, although it was somewhat more difficult to distinguish HSsII and -III, because of their close proximity to the EcoRV site.
FIG. 1.
Keratinocyte-specific DNase I HSs in the K14 chromosomal locus. Nuclei from fibroblasts and keratinocytes were treated with increasing amounts of DNase I (ascending triangle; see Materials and Methods for details). Following treatment, DNAs were digested with BglII-SpeI (A) or BglII-EcoRV (B) and subjected to Southern blotting with the radiolabeled probe indicated in panel C. Arrowheads in panels A and B denote the relative positions of HSs. Molecular mass markers (in kilobases) are indicated at the left. Arrows in panel C denote approximate locations of HSs. The transcription initiation site (Tx start) is indicated. Also shown is a partial restriction map of the K14 gene and its 5′ upstream regions: EcoRV (−1000), HindIII (−2200), BamHI (−4000), and AflIII (−5100). Note that HSsII was faint in panel A and visible clearly only upon longer exposures. HSsII and HSsIII are not well resolved in BglII-EcoRV digests (B).
The K14 promoter encompassing HSsI is dispensable for tissue-specific gene expression in transgenic mice.
We next focused on ascertaining the relative contribution of the four DNase HSs to K14 gene activity. Since HSsIV resides at −2800, beyond the −2200 limit sequence necessary for keratinocyte-specific gene expression in transgenic mice (46), we limited the present study to HSsI, -II, and -III. Previously, Leask et al. found that the K14 minimal promoter (−450 to +30), which encompasses HSsI, was not sufficient to provide epidermal-specific expression in keratinocytes or in mice (27). To determine if it was dispensable, we replaced it with the heterologous TK promoter, which on its own exhibits no expression in mice (reference 21 and references therein). The stick diagram in Fig. 2 illustrates 1850TKLacZ, the Lacz expression vector engineered for this test.
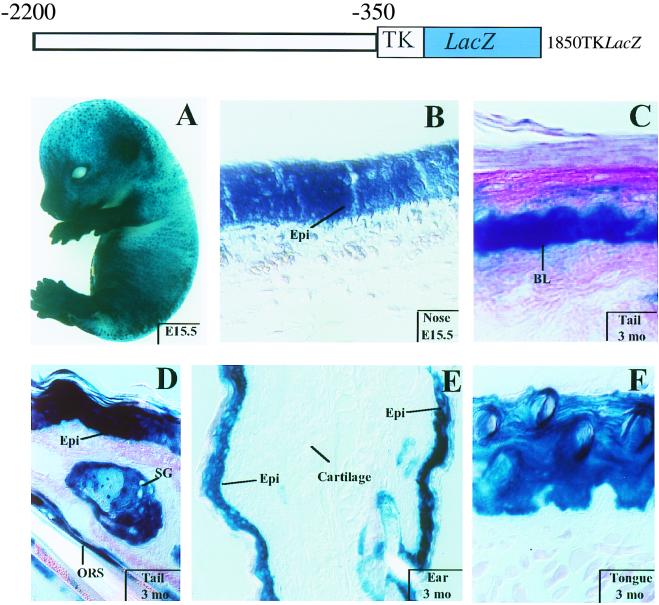
FIG. 2.
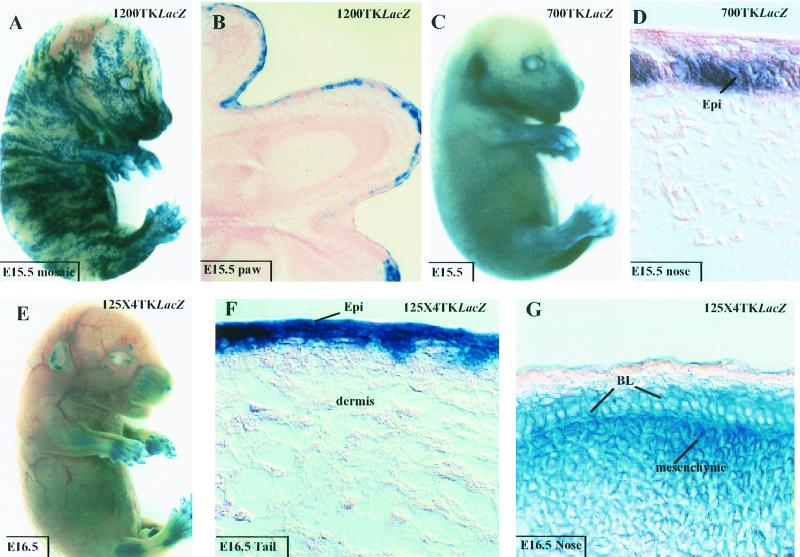
Expression of 1850TKLacZ in embryos and adult tissues. The 1850TKLacZ transgene contains the K14 5′ upstream sequences (−2200 to −350) linked to the heterologous TK minimal promoter. Embryos and skin sections from mice harboring this transgene were processed for β-galactosidase activity using X-Gal cleavage as an assay. (A) E15.5 transgenic embryo displaying intense blue staining in skin; (B to F) skin or tongue sections from transgenic animals at ages indicated at the lower right. Tissue area examined is noted on each frame. BL, basal layer; Epi, epidermis; SG, sebaceous gland; ORS, outer root sheath.
Three different transgenic animals expressed the transgene, as judged by β-galactosidase activity assays. Figure 2 provides representative data from these mice, all of which displayed similar and marked epidermis-specific transgene expression. Intense blue staining was detected within a few hours of incubation with X-Gal; in E15.5 embryos, it was most prominent over the snout, limbs, and ears, with appreciable but less staining over the dorsal region (Fig. 2A).
Sectioning of transgenic embryos and adult mice revealed epidermal and outer root sheath hair follicle specificity of transgene activity. In embryos, staining was detected in multiple epidermal layers (Fig. 2B), a feature also characteristic of endogenous K5 and K14 (6). In postnatal skin, staining was most prominent in the basal layer, sebaceous glands, and outer root sheath (Fig. 2C to E). Appreciable expression was also detected in other stratified squamous epithelia, e.g., tongue (Fig. 2F), but not in any tissues lacking endogenous K14.
While 1850TKLacZ retained the tissue-specific activity of its parent promoter, some staining was observed in the suprabasal cells of epidermis, a feature not seen in 2100K14LacZ mice or with endogenous K14 (S. Zinkel and E. Fuchs, unpublished data). Since the suprabasal cells are derived from differentiation of basal cells, the 1850TKLacZ mice seemed to lack some of the regulatory elements that control promoter downregulation in differentiating epidermis. The inability to downregulate differentiation-specific K14 promoter activity was also observed in other stratified squamous epithelia. Overall, however, our results demonstrate that the K14 minimal promoter is dispensable for tissue-specific gene expression, even though some features of differentiation-specific promoter regulation are altered when a heterologous promoter is used.
A 700-bp enhancer encompassing HSsII and HSsIII is selectively active in keratinocytes.
We next examined whether sequences encompassing HSsII and/or HSsIII might harbor clues to keratinocyte-specific gene expression. We began by performing transient transfection studies using K14 reporter gene constructs in cultured keratinocytes. We used three major changes over our past studies. First, recent improvements in gene transfection efficiencies enabled us to use primary keratinocytes, which express five to eight times the level of K14 and K5 proteins over the SCC-13 cell line previously used (25, 43). Second, we used the luciferase reporter gene, more sensitive than the chloramphenicol acetyltransferase gene previously employed. Finally, we used the TK promoter, enabling us to avoid consideration of elements in the K14 minimal promoter that we had already studied and which now appeared dispensable for keratinocyte specificity.
For our initial studies, five constructs were generated, each of which contained various segments of K14 upstream sequence encompassing HSsII and HSsIII, placed 5′ from the TK promoter and luciferase reporter gene (Fig. 3A). At 36 h posttransfection, keratinocytes were harvested for luciferase (test) and β-galactosidase (control) assays. All of these transgenes displayed robust (25- to 40-fold) activation compared with the TK promoter alone (Fig. 3B). Paring down the 5′ sequences from −2200 to −2000 and the 3′ sequences from −350 to −1300 did not result in appreciable loss of activity. This functional evidence defines an enhancer in the segment harboring HSsII and HSsIII (−2000 to −1300).
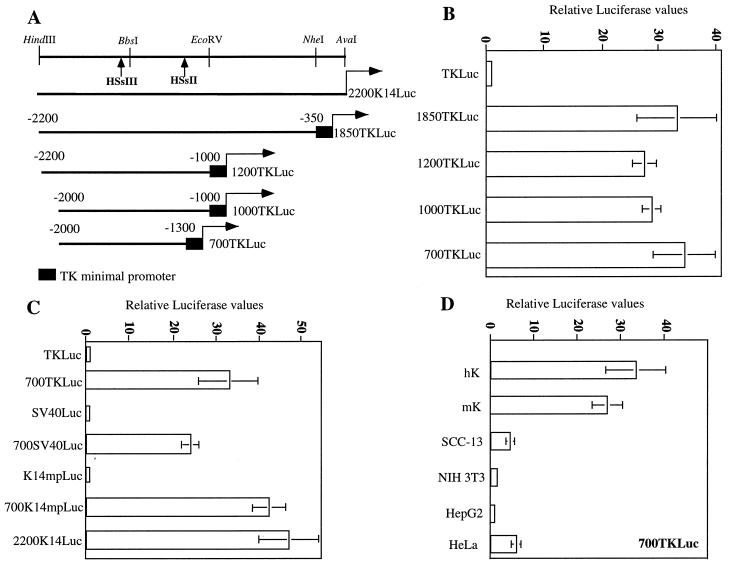
FIG. 3.
700TKLuc retains strong transcriptional activation in a keratinocyte-specific fashion in vitro (A) Diagram of the five initial luciferase constructs used for experiments shown in panels B to D. The partial restriction map of the K14 5′ upstream region shown includes the AvaI (+1), NheI (−350), EcoRV (−1000), BbsI (−1570), and HindIII (−2200) sites and the approximate positions of HSsII and HSsIII. (B) Luciferase activity data from primary human keratinocytes transfected with constructs shown in panel A. The value for the TKLuc construct was low and was arbitrarily set at 1. (C) Activity of the 700-bp enhancer (−2000 to −1300) in conjunction with different heterologous promoters. Activities of all the minimal promoters were low and set at 1. SV40, major early viral promoter; K14mp, K14 minimal promoter; TK, thymidine kinase minimal promoter. (D) Activity of 700TKLuc in different cell types and/or lines. hK, primary human keratinocytes; mK, immortalized mouse keratinocytes. In all transfection experiments, luciferase activities were corrected for β-galactosidase values from an internal control reporter, CMV-lacZ. Note that β-galactosidase activities were comparable in all lines tested. Activities represent the mean ± standard deviation of a minimum of three different experiments performed in duplicate.
The enhancer functioned robustly (20- to 45-fold) with two additional promoters, that of the SV40 T antigen and the K14 minimal promoter (K14 mp; −210 to +40) (Fig. 3C). Notably, the 700K14mpLuc displayed luciferase values comparable to those provided by the entire 2,200 bp of 5′ upstream K14 gene sequence. Thus, at least in cultured primary keratinocytes, the sequences encompassing −1300 to −350 and −2200 to −2000 did not possess essential activating or repressing potential.
To explore the cell type specificity of enhancer activity, we transfected 700TKLuc into primary human keratinocytes and a variety of cell lines, including a spontaneously immortalized line of mouse keratinocytes, squamous cell carcinoma cells (SCC-13), fibroblasts (NIH 3T3), hepatocytes (HepG2), and simple epithelial cells (HeLa). In all of these lines, overall transfection efficiencies varied somewhat, but the activity of the CMV-lacZ gene was strong and comparable, enabling us to use cotransfections of this control gene to correct for these variations. In contrast to CMV-lacZ, the activity of 700TKLuc was robust only in human and mouse keratinocytes (Fig. 3D). Notably, activity was distinctly modest in SSC-13 cells, the line previously used (5, 27) for all of our K14 and K5 promoter studies. Expression was also modest in simple epithelial (HeLa) cells and was not detected in hepatocytes (HepG2) or fibroblasts (NIH 3T3). These findings indicated that the 700-bp K14 enhancer fragment possessed marked cell-type specificity in vitro.
Delineating the key elements within the 700-bp keratinocyte enhancer.
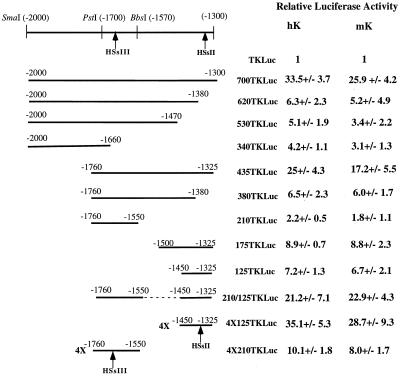
To assess the relative importance of sequences within the 700-bp K14 enhancer element, 5′ and 3′ deletions were made within the keratinocyte enhancer of 700TKLuc (−2000 to −1300), and the smaller segments were tested in human and mouse keratinocytes (Fig. 4). Even small (<100-bp) 3′ deletions severely impaired its activity. These small deletions likely removed the sequences contributing to HSsII, which mapped to the 3′ end of the enhancer. Larger deletions at this end had little or no additional effect, confirming the presence of key regulatory sequences localized to this end of the enhancer. In contrast, 240 bp could be removed from the 5′ end without appreciable loss of K14 enhancer activity. Internal deletions revealed an additional regulatory element, corresponding to the −1760 to −1550 region mapping to HSsIII. The −1760 to −1325 segment had near full activity, whereas the −1500 to −1325 enhancer had less than 25% the activity of the full enhancer.
FIG. 4.
Delineating the active elements within the 700-bp K14 enhancer. 700TKLuc was the parental construct used to create all deletion constructs. The partial restriction map of the enhancer indicates approximate locations of HSsII and HSsIII. Each offspring construct is named based on its relative proportion of the 700-bp enhancer. 4×125 indicates four copies of −1450 to −1325 multimerized in a head-to-tail fashion; 4×210 indicates four copies of −1760 to −1550 multimerized in a head-to-tail fashion. Each plasmid was cotransfected with CMV-lacZ plasmid in order to normalize all values against β-galactosidase control activity. Transfections were conducted with both primary human keratinocytes (hK) and mouse keratinocytes (mK). Cell extracts were subsequently assayed for luciferase and β-galactosidase. Numbers represent the relative luciferase values ± standard deviations of at least three independent experiments. All values are given relative to TKLuc, containing the minimal TK promoter, assigned a value of 1.
Neither the HSsIII nor the HSsII elements on their own were sufficient to confer high levels of reporter activity (Fig. 4). This was particularly true for HSsIII, which exhibited only 10 to 15% of full enhancer activity. However, when coupled to the HSsII element, activity was enhanced by nearly fourfold (Fig. 4). Together, HSsII and HSsIII elements accounted for most of the activity of the full 700-bp enhancer in vitro.
Multimerization of the −1760 to −1550 HSsIII segment only modestly enhanced the activity of the reporter gene over a single copy of the element, revealing relatively poor ability of this segment to operate alone as a keratinocyte enhancer. In contrast, multimerization of the −1450 to −1325 HSsII segment resulted in an enhancer with robust activity in both human and mouse keratinocytes (Fig. 4). This construct showed strong cell type-specific activity, as it was not active in either HepG2 cells or NIH 3T3 fibroblasts (data not shown). Thus, the HSsIII element was not strong on its own, while HSsII could act independently in eliciting keratinocyte-specific gene expression in vitro. Given the ability of HSsII to function autonomously, we proceeded to conduct an in-depth study of this 125-bp enhancer fragment.
The mouse K14 gene contains a DNase I HSs similar to HSsII, which displays significantly greater conservation than most other 5′ upstream sequences.
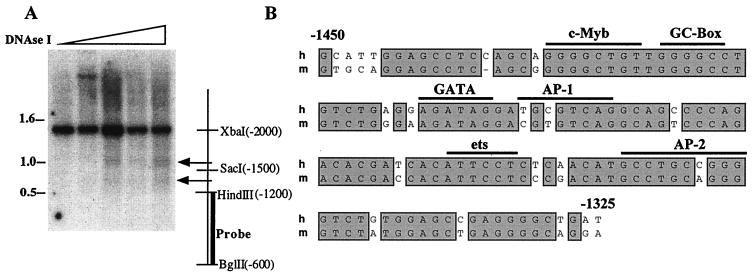
Cross-species comparison of cis elements provides a quick and powerful strategy to identify and confirm potential regulatory sequences. We therefore assessed whether the 5′ upstream region of the mK14 gene (E. Fogarty and E. Fuchs, unpublished data) possesses keratinocyte-specific, DNase I HSs positioned similarly to those in the human gene. Nuclei from mouse keratinocytes were treated with DNase I, and isolated genomic DNAs were then digested with XbaI (−2000) and BglII (−600). Southern analysis with a radiolabeled BglII-HindIII (−1200) probe identified two HSs located at approximately −1300 and −1700 (Fig. 5A). This corresponded well to the relative positions of HSsII and HSsIII of the human K14 gene. Similar to the human study, an HSs was also detected in the minimal promoter region (data not shown).
FIG. 5.
DNase I HSs mapping of the mouse K14 chromosomal locus and sequence comparisons of evolutionarily conserved HSs. (A) Following DNase I treatment of mouse keratinocyte nuclei, genomic DNAs were isolated, digested with XbaI-BglII, and subjected to Southern blot analysis with the radiolabeled probe indicated. Also shown are the relative positions of the two HSs (arrows) corresponding to HSsII and HSsIII of the human K14 gene and the restriction enzymes used to map the region. Molecular markers (in kilobases) are shown at left; the ascending triangle denotes increasing DNase I concentrations used in the assay. (B) Sequence alignment of the human (h) and mouse (m) HSsII regions. The sequence similarity is 80% over this stretch of DNA and 63% in the entire 700-bp enhancer element. Horizontal bars denote possible binding motifs for transcription factor family members.
The mouse DNase I HSs were similar in sequence to the corresponding HSs of the human K14 gene. In particular, mouse HSsII (at −1300) was 80% identical to the human sequence (Fig. 5B). Similarities dropped significantly in the regions flanking this site, although there were small stretches, including approximately 100 bp at −1700 (HSsIII), with relatively high identity between human and mouse (data not shown). The similarities in sequence and DNase I hypersensitivity within these regulatory elements of the K14 enhancer is likely to explain why the human K14 enhancer behaves faithfully in transgenic mice.
Analysis of the elements within the 125-bp HSsII segment of the K14 enhancer: binding of keratinocyte nuclear proteins and their preliminary characterization.
A computer analysis (35) of the −1325 to −1450 HSsII enhancer element revealed a number of putative sequence motifs for known classes of DNA binding proteins, including c-Myb, GATA, AP-1, AP-2, and ets family members (Fig. 5B). To explore whether these sites bind nuclear proteins, we conducted EMSAs using radiolabeled oligonucleotides corresponding to each of these sites in the human sequence and nuclear extracts from different cell types. Three oligonucleotides bound specific DNA-protein complexes and were chosen for further study.
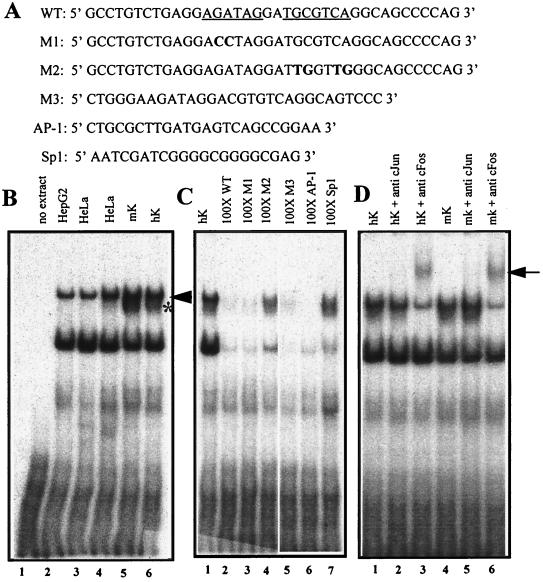
One of the oligonucleotides that bound nuclear proteins contained the putative AP-1 and GATA motifs (Fig. 6A). Two protein-DNA complexes were present in EMSAs performed with nuclear extracts from HepG2 cells, HeLa cells, and human and mouse keratinocytes (Fig. 6B, lanes 2 to 6). An additional band was generated only with nuclear extracts from keratinocytes (Fig. 6B, lanes 5 and 6). As expected, a 100-fold excess of unlabeled wild-type oligonucleotide competed effectively for the binding of all of keratinocyte proteins that had bound to its radiolabeled counterpart (Fig. 6C, lanes 1 and 2). This was also true for the M1 oligonucleotide containing mutations in the GATA element, ruling out GATA proteins as the source of these complexes (lane 3).
FIG. 6.
EMSAs of nuclear extracts showing binding of AP-1 to a motif within the 125-bp HSsII K14 enhancer. (A) The wild-type oligonucleotide (WT) corresponds to the human sequence and is shown at the top, with the GATA and AP-1 sites underlined. Mutant oligonucleotides are shown below, with mutations in bold. M1, GATA mutant; M2, AP-1 mutant; M3, corresponding WT mouse sequence (Fig. 5B); AP-1 and Sp1 are consensus oligonucleotides for these two transcription factor family members and were purchased from Promega. (B) EMSAs were performed with radiolabeled WT oligonucleotide and nuclear extracts (4 to 6 mg) from the cell type indicated above each lane. Arrowhead and asterisk denote broadly expressed and keratinocyte-specific AP-1 complexes, respectively; note that all complexes within this mobility range were supershifted with AP-1 antibodies (D). (C) EMSAs were performed as for panel B, this time using radiolabeled WT oligonucleotide and a 100-fold excess of the unlabeled double-stranded oligonucleotide indicated above each lane. Note that the AP-1 complexes were specifically competed only by oligonucleotides encompassing an AP-1 binding site, whereas most other complexes were competed nonspecifically with all oligonucleotides. (D) EMSAs were performed as for panel B, but anti-c-Jun and anti-c-Fos were added to the complexes prior to gel electrophoresis. The arrow indicates supershifts, reflective of complexes between AP-1, DNA, and antibody.
In contrast, a 100-fold excess of the M2 oligonucleotide, containing mutations in the AP-1 consensus site failed to compete, suggesting that a member(s) of the AP-1 family binds specifically to this human K14 oligonucleotide (Fig. 6C, lane 4). Even though the mouse and human K14 AP-1 sequences are not completely conserved, the M3 oligonucleotide corresponding to the wild-type mouse sequence appeared to be equally effective in competing for proteins binding to the corresponding human sequence (lane 5). A consensus AP-1 oligonucleotide was also effective, but a consensus Sp1 oligonucleotide, used as another control, was not (lanes 6 and 7, respectively). In all of these assays, the lower band always displayed some competition with all the oligonucleotides, suggesting that it was probably a non-sequence-specific DNA-protein complex. In contrast, the upper band(s) appeared to be specific for those oligonucleotides that contained an AP-1 sequence motif.
To confirm the identity of the upper DNA-protein complex, we incubated the EMSAs with antibodies against c-Jun and c-Fos. The c-Jun antibody is broadly reactive against c-Jun, JunB, and JunD proteins and the c-Fos antibody is broadly reactive against c-Fos, FosB, Fra1, and Fra2. Anti-cFos produced a marked supershift of the slowest-migrating DNA-protein complex(es) (Fig. 6D, arrow). Anti-c-Jun also produced a supershift, but it was not as effective as anti-c-Fos (not shown). Both antibodies appeared selective for the large DNA-protein complexes, which included the keratinocyte-specific complex (Fig. 6D). Whether the keratinocyte complex represents a novel isoform of AP-1 awaits further analysis.
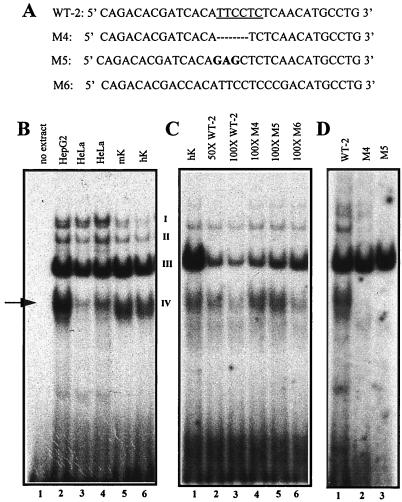
Another oligonucleotide exhibiting keratinocyte nuclear protein binding activity corresponded to sequences just 3′ of the AP-1 motif. This sequence encompassed a putative binding site for the ets family of DNA binding proteins (Fig. 5B). EMSAs with labeled oligonucleotide WT-2 produced four major complexes (I to IV), which were present in nuclear extracts from HepG2, HeLa cells, human and mouse keratinocytes (Fig. 7A and B). Band IV migrated slightly faster with keratinocyte nuclear extracts compared with extracts from HepG2 or HeLa (Fig. 7B). The proteins binding to this oligonucleotide were competed by unlabeled wild-type oligonucleotide but not by either of two oligonucleotides (M4 and M5) mutated within the putative ets sequence motif (Fig. 7C). Of the four major bands in these EMSAs, only band IV showed a marked difference in its ability to be competed for by wild-type versus ets mutant oligonucleotides. This complex was also competed for by M6, an oligonucleotide corresponding to the sequences from the equivalent mouse region (Fig. 7C, lane 6). Neither the M4 nor the M5 ets mutant oligonucleotides were able to generate a complex IV when they were subjected to an EMSA (Fig. 7D). Taken together, these data verified the specificity of the DNA-protein complex and indicated that an ets or ets-related protein was likely to bind to an ets sequence motif within the HSsII region.
FIG. 7.
EMSAs of nuclear extracts showing binding of ets family members to a motif within the K14 enhancer. (A) The wild-type oligonucleotide (WT-2) corresponding to the relevant sequence of the human enhancer is shown at the top, with the ets site underlined. The mutant oligonucleotides are shown below, indicating residues that were either deleted (TTCC in M4; dashed lines) or mutated (TTC>GAG; shown in bold in M5). M6, the WT oligonucleotide to the corresponding mouse sequence (Fig. 5B). (B) EMSAs were performed by incubating radiolabeled WT-2 oligonucleotide with nuclear extracts (4 to 6 μg) from the cell line indicated above each lane. The four DNA-protein complexes are denoted I, II, II, and IV (complex I was often faint). (C) EMSAs were performed as for panel B, this time adding a 50- or 100-fold excess of unlabeled competitor oligonucleotide as indicated above each lane. The arrow denotes complex IV, competed by human and mouse WT oligonucleotides. (D) EMSAs performed with radiolabeled WT-2, M4, and M5 oligonucleotides. Note that complex IV (arrow) is absent from the EMSAs conducted with mutants M4 and M5 (compare lane 1 with lanes 2 and 3).
We incubated the reactions with available antibodies against known ets family members, including ets-1, ets-2, fli-1, and elf-1, all of which are expressed in the epidermis (2). While none of the antibodies affected the formation of the complex (data not shown), it is possible that a novel family member of the ets family or a protein which binds to the ets binding motif may be responsible for the complex we have uncovered in this study. The keratinocyte-specific differences in complex IV migration patterns lend additional support for this notion.
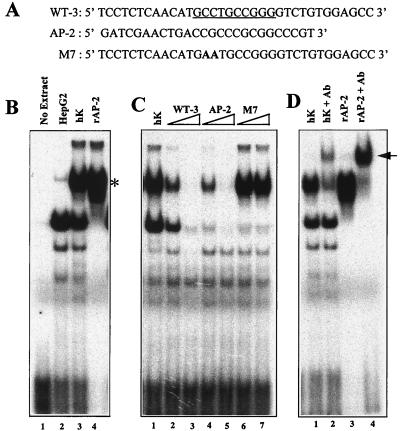
The final radiolabeled oligonucleotide that formed a stable complex with nuclear proteins from keratinocytes encompassed a putative AP-2 core binding site (Fig. 5B and 8A). Two major EMSA bands were produced; one was present in both HepG2 and keratinocyte nuclear extracts, and the other was unique to the keratinocyte extract (Fig. 8B, lanes 2 and 3, respectively). When the keratinocyte extract was replaced by purified recombinant AP-2 protein, a complex migrating at similar electrophoretic mobility was generated (lane 4), indicating the likelihood that the keratinocyte-specific complex contained an AP-2 protein.
FIG. 8.
EMSAs of nuclear extracts showing binding of AP-2 family members to a motif within the K14 enhancer. (A) The wild-type oligonucleotide (WT-3) corresponding to the relevant sequence of the human enhancer is shown at the top, with the AP-2 site underlined. An equivalent oligonucleotide (M7) mutated for the AP-2 site is shown below, with mutant residues in bold. AP-2, a consensus AP-2 oligonucleotide, purchased from Promega. (B) EMSAs were performed by incubating radiolabeled WT-3 oligonucleotide with either nuclear extracts (4 to 6 μg) from human hepatocytes (HepG2) or keratinocytes (hK) or recombinant AP-2 protein (rAP-2; Promega). The asterisk denotes an AP-2 complex, known to be absent in HepG2 cells (6). (C) EMSAs were performed as for panel B, this time adding a 50- or 100-fold excess of unlabeled competitor oligonucleotide as indicated by the triangle over each lane. (D) EMSAs performed with radiolabeled WT-3 as in panel B, this time adding AP-2 antibody to the complexes prior to gel electrophoresis. The arrow denotes supershifts.
To further establish the identity of the complexes generated with this oligonucleotide, we showed that M7, containing a mutated AP-2 core consensus sequence, failed to compete for the binding of the keratinocyte protein, in contrast to either the wild-type or a synthetic AP-2 oligonucleotide (Fig. 8C). Finally, antibodies against AP-2 proteins supershifted the complex, confirming the nature of the keratinocyte band. The faster-migrating complex, detected with both HepG2 and keratinocyte extracts, was competed for by all DNAs used in the test, including both the AP-2 synthetic and mutant oligonucleotides, revealing its nonspecific nature (Fig. 8B to D).
Within the 125-bp K14 enhancer element, several additional radiolabeled oligonucleotides were tested, including those spanning the putative c-Myb site as well as regions lacking putative binding sites for any known transcription factor families (Fig. 5B). None of these additional oligonucleotides produced specific EMSAs with keratinocyte proteins (data not shown). While we cannot rule out the possibility that less stable protein-DNA complexes might exist in vivo that we cannot detect by our in vitro assays, our EMSA data suggested that HSsII is generated upon binding of keratinocyte proteins to the AP-1, ets, and AP-2 consensus motifs that reside within this element.
Functional analysis of the sequences within the 125-bp K14 enhancer reveals a key role for the ets site.
We next mutagenized sequences across the 125-bp K14 enhancer and assessed the consequences of these mutations to keratinocyte-specific enhancer function. The underlying rationale for our mutagenesis was based on (i) whether a specific sequence had been shown by EMSA to bind a protein from keratinocyte nuclear extracts and (ii) whether a specific sequence was highly conserved between mouse and human (Fig. 9). The mutants spanned the entire 125-bp segment, enabling us to broadly assess the importance of elements within the enhancer. All mutations were tested in the context of the 125-bp enhancer, which with the TK promoter was used to drive luciferase reporter gene expression. Wherever mutations were also tested in the context of the 700-bp enhancer, the effects of the mutations were similar with the two constructs (data not shown).
FIG. 9.
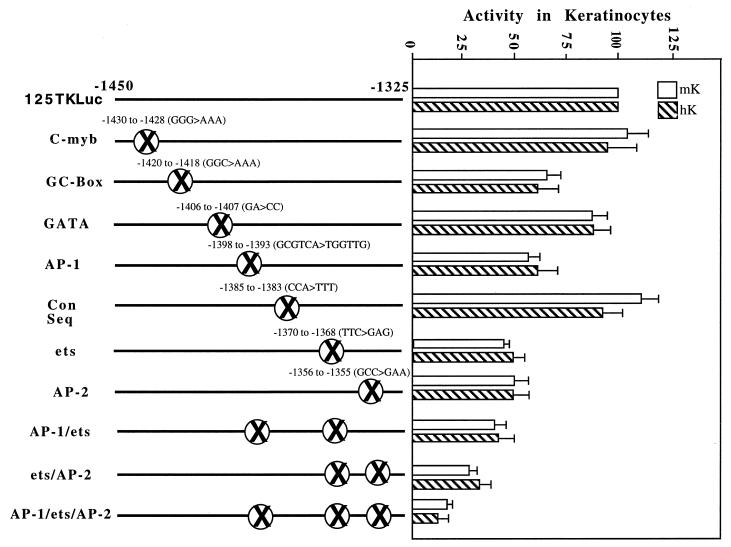
Functional analysis of the HSsII region. The 125-bp HSsII enhancer was scanned for (i) sequences conserved between mouse and human and (ii) putative binding motifs for any of the known transcription factor families. Based on this information, we engineered mutant 125-bp K14 enhancer constructs harboring mutations in one of the conserved sequences (Con Seq) within this region. Areas mutated are denoted by the circumscribed X's, with the mutated residues indicated above. Wild-type and mutant plasmids were transfected into human and mouse keratinocytes (hK and mK) along with a CMV-lacZ reporter construct, and luciferase activities were determined and normalized against the β-galactosidase values. Each experiment was repeated at least three times, and data represent averages with standard deviations. The activities of the 700TKLuc (not shown) and 125TKLuc were set at 100%.
Some mutations had no effect on enhancer activity, including the mutations abolishing the putative c-Myb and GATA binding motifs and a mutation in the middle of a highly conserved sequence between the AP-1 and ets binding motifs (Fig. 9). Mutation of a GC-rich sequence adjacent to the c-Myb sequence had a modest effect (60% ± 5% of the wild-type level) on enhancer activity. Based on sequence, we predicted that this motif might bind Sp1 or Krüppel-like family members, but EMSAs with a radiolabeled oligonucleotide and an anti-Sp1 antibody did not provide evidence that Sp1 binds to this site (data not shown).
Mutations that had the greatest effect on keratinocyte enhancer activity were those within the core binding site for transcription factors, which gave strong and specific signals in our EMSAs. Thus, mutation of the AP-1 and AP-2 sequence motifs each gave an approximately twofold reduction in enhancer activity, and mutation of the ets sequence motif gave a threefold reduction (Fig. 9). While double mutations of these binding sites did not eliminate enhancer activity, mutations of the ets/AP-2 sites reduced activity to ∼25% of wild-type levels, and mutations of the ets/AP-1 sites also had a marked effect. Mutations of all three sites reduced the activity of the 125-bp enhancer to less than 15% of its wild-type level. Since this level was comparable to the activity of 530TKLuc (−2000 to −1470), i.e., complete deletion of −1470 to −1325, the data indicated that at least in vitro, these three sites accounted for the bulk of the activity of the 125-bp enhancer element (−1450 to −1325), a segment sufficient on its own to confer strong keratinocyte-specific activity in culture.
Analysis of the upstream enhancer in transgenic mice.
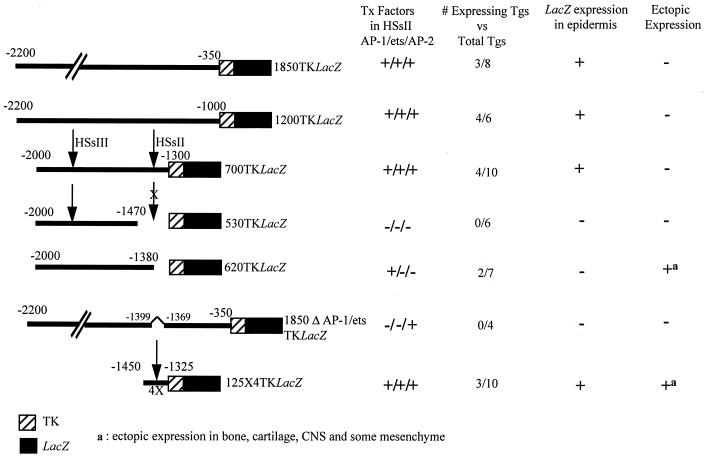
To assess whether our fine mapping of the K14 enhancer element in vitro was relevant to keratinocyte-specific gene expression in vivo, we engineered a series of additional constructs, depicted in Fig. 10, and used these to generate transgenic mice. Both 1200TKLacZ and 700TKLacZ showed marked LacZ gene expression in the skin of mouse embryos (Fig. 11A to D). In both cases, expression was largely if not solely restricted to keratinocytes. Due to variability in transgene site integration, determining or comparing levels of expression was not possible. However, the percentages of mice expressing transgenes were similar for the 1850TKLacZ, 1200TKLacZ, and the 700TKLacZ animals (Fig. 10).
FIG. 10.
Summary of β-galactosidase expression in mice and/or embryos harboring various TKLacZ transgenes. The nomenclature of the transgenes is described in the text. The HSs are denoted by arrows, and the presence of AP-1, ets and AP-2, sites is indicated to the right of each stick diagram. Transgenic embryos (Tgs) were scored as positive or negative after overnight incubation with X-Gal and visual inspection for blue staining. Embryos displaying transgene expression were sectioned to evaluate expression patterns in more detail. Wherever observed, ectopic expression was always in the CNS, cartilage, bone, and mesenchyme underlying the epidermis. Shown are the number of embryos and/or mice that expressed LacZ versus the total number of transgenic mice generated for each construct.
FIG. 11.
Expression of 1200TKLacZ, 700TKLacZ, and 125×4TKLacZ in embryos. Transgene expressed is indicated in the upper right of each frame, and animal age and tissue are indicated in the box on each frame. Animals were either processed intact for β-galactosidase activity or sectioned prior to processing (for methods, see reference 6). Note that animal panel A was a mosaic, as denoted by the patchy striped pattern of X-Gal staining. The embryo in panel C shows stronger expression over the nose, limbs, and ears than other body sites; however, sectioning revealed skin-specific expression with no CNS or bone or cartilage staining. Note that embryo in panel E shows signs of ectopic expression in the brain and bone or cartilage. This was confirmed in embryo sections (not shown). Abbreviations: BL, basal layer; Epi, epidermis.
Deletion of the 125-bp HSsII enhancer element, containing the AP-2, AP-1, and ets sites, appeared to abolish transgene expression altogether. Of six transgenic animals, none displayed detectable expression (530TKLacZ in Fig. 10). Furthermore, of seven transgenic mice harboring 620TKLacZ, containing the AP-1 site but lacking ets and AP-2 sites, and four harboring 1850ΔAP1/etsTKLacZ, mutated in the ets and AP-1 sites, none gave transgene expression in the epidermis (Fig. 10). This was in striking contrast to 700TKLacZ, where four of 10 animals displayed epidermal expression.
While the correlation between lack of AP-2 and/or ets sites and lack of epidermal enhancer activity is a negative one, it is consistent with our in vitro studies delineating the importance of these motifs to the 700-bp K14 enhancer activity. Moreover, two of the 620TKLacZ animals exhibited similar patterns of ectopic expression in underlying mesenchyme central nervous system (CNS), cartilage, and bone (data not shown). This argues against the notion that the transgenes simply integrated into chromosomally silent loci.
As a final test of our analysis of the K14 enhancer, we engineered transgenic mice expressing 125×4TKLacZ, containing four copies of the element encompassing the AP-1, AP-2, and ets binding sites. Expression was observed in 3 of 10 embryos generated, and in all cases, the epidermis was positive for transgene expression (Fig. 11E to G). This said, while some skin areas such as the tail exhibited only epidermal expression (Fig. 11F), other regions such as the nose displayed enhancer activity not only in the epidermis but also in underlying mesenchyme (Fig. 11G). In addition, CNS, cartilage, and bone were strongly positive for 125×4TKLacZ transgene expression (Fig. 11E and data not shown). This broadened expression pattern was seen in three independently generated 125×4TKLacZ embryos and resembled the ectopic expression of 620TKLacZ animals. When it is considered that 700TKLacZ and 125×TKLacZ animals faithfully express in epidermis but 620TKLacZ and 1850ΔAP1/etsTKLacZ animals do not, it seems likely that the epidermal specificity is due to the presence of the AP-2 and ets sites.
DISCUSSION
An epidermal enhancer with keratinocyte-specific chromatin structure.
We examined for the first time the relation between keratinocyte-specific changes in chromatin structure, specific functional regulatory sequences, and transcription factor family members that bind to these sequences. DNase I HS mapping has long provided a means of detecting perturbations in nucleosome arrangement that typically arise when nonhistone proteins bind to DNA upon transcriptional activation (9, 14). Our DNase I HSs mapping led to identification of a novel 700-bp segment encompassing two HSs, HSsII and HSsIII, which together are sufficient to confer epidermis-specific gene expression in transgenic mice in vivo and in primary keratinocytes in vitro. The importance of this segment had not been revealed from earlier mutagenesis and transfection studies, presumably due to the lack of sensitivity in measuring K14 transcriptional activity in SCC-13 cells and the absence of a systematic procedure to identify potentially key regulatory domains.
We also unveiled a strong correlation between the locations of HSsI, HSsII, and HSsIII and the functional relevance of these sequences to keratinocyte-specific gene expression in vivo and in vitro. Since the K14 gene resides within a compact locus of type I keratin genes, often separated by only a few kilobases (8, 29), and the limit segment necessary and sufficient for strong, epidermis-, developmental stage-, and differentiation-specific gene expression encompasses a mere 2,100 bp of 5′ upstream sequence, which excludes even HSsIV (27, 46, 47), it seems likely that these three HS regulatory elements account for the keratinocyte specificity of the K14 gene.
Specific roles of HSsI, HSsII, and HSsIII in epidermal specificity and in differentiation-specific gene expression in vitro and in vivo.
By replacing the K14 minimal promoter encompassing HSsI with one of several generic promoters, we have discovered that it plays a role in differentiation-specific regulation of gene expression in vivo and in vitro. Since the heterologous TK promoter otherwise functioned equivalently, the K14 minimal promoter is not required for epidermis-specific gene expression.
HSsII and HSsIII define a key 700-bp enhancer which is sufficient for keratinocyte-specific gene expression. HSsII appears to be indispensable, as judged by four major criteria: (i) little or no luciferase reporter activity was observed in cultured keratinocytes harboring promoter/enhancer constructs lacking this gene segment; (ii) no epidermis-specific β-galactosidase reporter gene expression was detected in 17 different independently derived transgenic animals that lacked part or all of these regulatory sequences; (iii) when a 125-bp segment encompassing HSsII was multimerized, it provided keratinocyte-specific enhancer activity in vitro that was comparable to the full-length 2,200-bp promoter/enhancer activity; and (iv) in transgenic mice, the multimerized 125-bp HSsII segment conferred enhancer activity in tissues that included, but was not limited to, the epidermis. In contrast, transgenes that contained HSsIII as well as HSsII faithfully targeted reporter gene expression, suggesting that a combination of HSsII and HSsIII is necessary to fully restrict gene expression to the epidermis in vivo.
Functional elements of the HSsII enhancer.
A number of other studies have implicated AP-1, AP-2, and ets family members in regulating the expression of viral and endogenous genes in various cultured keratinocyte cell lines (4, 10, 11, 24, 25, 26, 28, 41, 42, 49). Curiously, the genes studied in those reports include both basal and differentiation-specific epidermal genes. Our findings here establish a role for these factors in epidermis-specific gene expression in vivo. Our most persuasive evidence was the near quantitative loss of keratinocyte enhancer activity when all three sites were mutated and, conversely, near full enhancer activity when a 125-bp fragment encompassing these three motifs was multimerized. Additionally, our studies reveal an interesting correlation between keratinocyte-specific changes in chromatin structure and a DNA segment containing the functional AP-1, AP-2, and ets sites. Finally, our data strengthen the view that epidermis-specific gene expression is governed by multiple transcriptional regulatory sequences and factors.
A role for AP-2 in K14 gene expression was previously recognized due to the existence of an additional functional AP-2 site within the K14 promoter (26, 27). However, the importance of AP-1 and ets sites in K14 promoter/enhancer regulation was not appreciated from prior studies with SCC-13 keratinocytes grown in the presence of serum (27). Given that AP-1 and ets genes can be transcriptionally sensitive to serum-responsive factors, a feature which can also change upon immortalization, this could broadly account for discrepancies in studies that employ cultured cells to identify AP-1 and ets factors as regulators of gene expression. While primary keratinocytes and serum-free medium can eliminate some of these caveats, they still may not be sufficient to ensure accurate delineation of epidermis-specific regulatory elements. An illustration of this point is the ectopic expression in some mesenchyme achieved by the multimerized 125-bp HSsII enhancer, despite the fact that in cell culture, this transgene was expressed in keratinocytes and not in fibroblasts.
The AP-1 and AP-2 sites in the K14 enhancer displayed comparable activities in vitro, and when mutated, these sites behaved similarly in reducing enhancer activity. The finding of a putative AP-1 regulatory site in the K14 HSsII enhancer was intriguing, because in vitro studies with other epidermally expressed genes had also uncovered AP-1 as a potential component of keratinocyte-specific gene expression (38, 44, 49). However promising the in vitro studies, in vivo, the presence of the AP-1 site correlated with ectopic enhancer activity in bone, cartilage, CNS, and mesenchyme. Furthermore, while a number of AP-1 transcription factors, including c-Fos, JunB, and Fra-2, are expressed in the epidermis, gene knockout studies have not revealed overt defects in epidermal differentiation and/or gene expression (19, 23, 37). This said, all of our promoter/enhancer constructs that conferred epidermis-specific gene expression also contained the functional AP-1 site within the HSsII enhancer, and thus it is still formally possible that AP-1 plays a key role in epidermis-specific gene expression in vivo as it seems to do in vitro.
The ets binding site was the most critical both for the keratinocyte-specific and epidermis-specific activity of the K14 enhancer. Ablation of the ets site had the most deleterious effect on enhancer activity in vitro, and in vivo, a perfect correlation existed between the presence of the functional ets site and epidermal expressivity in mice. Fourteen of 34 transgenic mice that contained a functional ets site expressed the K14 enhancer-driven lacZ gene, and 17 of 17 different lines that lacked this site displayed no epidermal gene expression. It seems unlikely that the ets site alone can account for epidermal expressivity, as the ets family of transcription factors is a large one and includes many broadly expressed members (22). This said, many of these ets factors, including ESE-1, are expressed broadly in epithelia (2, 33, 34), and one of these, ESE-2, has an even more restricted pattern that includes the epidermis (33). However, none of these epithelial ets proteins has yet been detected in the basal epidermal layer. Overall, these issues raise the possibility that as yet unidentified ets family members, or other proteins that interact directly or indirectly with ets binding sites, may play a critical role in regulating the expression of K14 and other basally expressed epidermal genes in vivo.
Our integrated studies illuminate a combinatorial role for AP-2, ets, and possibly AP-1 family members in controlling preferential epidermal expression in vivo. However, elucidating the mechanism by which specific members of these families cooperate in governing transcriptional regulation in the epidermis may not be trivial. As is the case for the ets protein family, AP-1 and AP-2 factors also display remarkable regional and temporal variation in their expression patterns, which are broad and extend to many different cell types within the body, including the epidermis, brain, bone, cartilage, and mesenchyme, all relevant to the present study (2, 4, 13, 30, 31, 34, 36, 38, 50). Even within a particular tissue such as the epidermis, regional and differentiation-specific differences are evident in the expression patterns of AP-1, AP-2, and ets proteins (2, 4, 6, 28, 38, 50).
Summary.
We have identified structural differences in chromatin that exist within the locus of an epidermally expressed gene in primary keratinocytes but not in other cell types in vitro. Such differences provided a powerful method for identifying an epidermis-specific enhancer that is both necessary and sufficient for keratinocyte-specific gene expression. The enhancer contains AP-2, AP-1, and ets sites that all contribute to epidermis-specific and/or differentiation-specific transcriptional activity in keratinocytes in vitro and in vivo. No one factor or element is on its own sufficient for the faithful restriction of transcription to the epidermis. Rather, these regulatory sequences act combinatorially to confer keratinocyte-specific expression. In turn, it is the specific factors that bind to these sequences, or the specific context of the sequence motifs themselves, that influence the differential activity of these genes during the terminal differentiation process. Of the factors involved, our studies illuminate the importance of ets, AP-2, and AP-1 in conferring preferential epidermis expression both in vitro and in vivo. This study now paves the way for future studies aimed at elucidating the specific family members and the intricacies of the underlying mechanisms involved in this process.
ACKNOWLEDGMENTS
We thank Wenyu Bai for transgenic mouse generation, Jai Uttam for technical support, and Grazina Traska for assistance with cell culture. S.S. is the recipient of postdoctoral fellowship from the National Institutes of Health. E.F. is an Investigator of the Howard Hughes Medical Institute. This work was supported by funds from the Howard Hughes Medical Institute and from NIH grant RO1-AR31737.
REFERENCES
- 1.Andersen B, Weinberg W C, Rennekampff O, McEvilly R J, Bermingham J R, Jr, Hooshmand F, Vasilyev V, Hansbrough J F, Pittelkow M R, Yuspa S H, Rosenfeld M G. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 2.Andreoli J M, Jang S I, Chung E, Coticchia C M, Steinert P M, Markova N G. The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res. 1997;25:4287–4295. doi: 10.1093/nar/25.21.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellard M, Dretzen G, Giangrande A, Ramain P. Nuclease digestion of transcriptionally active chromatin. Methods Enzymol. 1989;170:317–436. doi: 10.1016/0076-6879(89)70054-9. [DOI] [PubMed] [Google Scholar]
- 4.Byrne C. Regulation of gene expression in developing epidermal epithelia. Bioessays. 1997;19:691–698. doi: 10.1002/bies.950190809. [DOI] [PubMed] [Google Scholar]
- 5.Byrne C, Fuchs E. Probing keratinocyte and differentiation specificity of the K5 promoter in vitro and in transgenic mice. Mol Cell Biol. 1993;13:3176–3190. doi: 10.1128/mcb.13.6.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 7.Casatorres J, Navarro J M, Blessing M, Jorcano J L. Analysis of the control of expression and tissue specificity of the keratin 5 gene, characteristic of basal keratinocytes. Fundamental role of an AP-1 element. J Biol Chem. 1994;269:20489–204996. [PubMed] [Google Scholar]
- 8.Ceratto N, Dobkin C, Carter M, Jenkins E, Yao X L, Cassiman J J, Aly M S, Bosco P, Leube R, Langbein L, Feo S, Romano V. Human type I cytokeratin genes are a compact cluster. Cytogenet Cell Genet. 1997;77:169–174. doi: 10.1159/000134566. [DOI] [PubMed] [Google Scholar]
- 9.Crossley M, Orkin S H. Regulation of the beta-globin locus. Curr Opin Genet Dev. 1993;3:232–237. doi: 10.1016/0959-437x(93)90028-n. [DOI] [PubMed] [Google Scholar]
- 10.DiSepio D, Jones A, Longley M A, Bundman D, Rothnagel J A, Roop D R. The proximal promoter of the mouse Ioricrin gene contains a functional AP-1 element and directs keratinocyte-specific but not differentiation-specific expression. J Biol Chem. 1995;270:10792–10799. doi: 10.1074/jbc.270.18.10792. [DOI] [PubMed] [Google Scholar]
- 11.Eckert R L, Crish J F, Banks E B, Welter J F. The epidermis: genes on-genes off. J Investig Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- 12.Faus I, Hsu H J, Fuchs E. Oct 6: a regulator of keratinocyte gene expression in stratified squamous epithelia. Mol Cell Biol. 1994;14:3263–3275. doi: 10.1128/mcb.14.5.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer D F, Gibbs S, van De Putte P, Backendorf C. Interdependent transcription control elements regulate the expression of the SPRR2A gene during keratinocyte terminal differentiation. Mol Cell Biol. 1996;16:5365–5374. doi: 10.1128/mcb.16.10.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs E. Epidermal differentiation and keratin gene expression. J Cell Sci. 1993;17:197–208. doi: 10.1242/jcs.1993.supplement_17.28. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs E, Byrne C. The epidermis: rising to the surface. Curr Opin Genet Dev. 1994;4:725–736. doi: 10.1016/0959-437x(94)90140-x. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs E V. Of mice and men: genetic disorders of the cytoskeleton. Keith R. Porter Lecture. Mol Biol Cell. 1997;8:189–203. doi: 10.1091/mbc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandarillas A, Watt F M. Changes in expression of members of the fos and jun families and myc network during terminal differentiation of human keratinocytes. Oncogene. 1995;11:1403–1407. [PubMed] [Google Scholar]
- 20.Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 21.Goldhamer D J, Brunk B P, Faerman A, King A, Shani M, Emerson C P J. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 22.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 23.Gruda M C, van Amsterdam J, Rizzo C A, Durham S K, Lira S, Bravo R. Expression of FosB during mouse development: normal development of FosB knockout mice. Oncogene. 1996;12:2177–285. [PubMed] [Google Scholar]
- 24.Jang S I, Steinert P M, Markova N G. Activator protein 1 activity is involved in the regulation of the cell type-specific expression from the proximal promoter of the human profilaggrin gene. J Biol Chem. 1996;271:24105–24114. doi: 10.1074/jbc.271.39.24105. [DOI] [PubMed] [Google Scholar]
- 25.Kim K H, Schwartz F, Fuchs E. Differences in keratin synthesis between normal epithelial cells and squamous cell carcinomas are mediated by vitamin A. Proc Natl Acad Sci USA. 1984;81:4280–4284. doi: 10.1073/pnas.81.14.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leask A, Byrne C, Fuchs E. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc Natl Acad Sci USA. 1991;88:7948–7952. doi: 10.1073/pnas.88.18.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leask A, Rosenberg M, Vassar R, Fuchs E. Regulation of a human epidermal keratin gene: sequences and nuclear factors involved in keratinocyte-specific transcription. Genes Dev. 1990;4:1985–1998. doi: 10.1101/gad.4.11.1985. [DOI] [PubMed] [Google Scholar]
- 28.Lee J H, Jang S I, Yang J M, Markova N G, Steinert P M. The proximal promoter of the human transglutaminase 3 gene. Stratified squamous epithelial-specific expression in cultured cells is mediated by binding of Sp1 and ets transcription factors to a proximal promoter element. J Biol Chem. 1996;271:4561–4568. [PubMed] [Google Scholar]
- 29.Marchuk D, McCrohon S, Fuchs E. Complete sequence of a gene encoding a human type I keratin: sequences homologous to enhancer elements in the regulatory region of the gene. Proc Natl Acad Sci USA. 1985;82:1609–1613. doi: 10.1073/pnas.82.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPherson L A, Weigel R J. AP2alpha and AP2gamma: a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell P J, Timmons P M, Hebert J M, Rigby P W J, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Cloning and characterization of a second AP-2 transcription factor: AP-2 beta. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 33.Oettgen P, Kas K, Dube A, Gu X, Grall F, Tharnrongsak U, Akbarali Y, Finger E, Boltax J, Endress G, Munger K, Kunsch C, Libermann T A. Characterization of ESE-2, a novel ESE-1-related ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J Biol Chem. 1999;274:29439–29452. doi: 10.1074/jbc.274.41.29439. [DOI] [PubMed] [Google Scholar]
- 34.Oettgen P, Alani R M, Barcinski M A, Brown L, Akbarali Y, Boltax J, Kunsch C, Munger K, Libermann T A. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol Cell Biol. 1997;17:4419–4433. doi: 10.1128/mcb.17.8.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi A, Jang S I, Ceci R, Steinert P M, Markova N G. Effect of AP-1 transcription factors on the regulation of transcription in normal human epidermal keratinocytes. J Investig Dermatol. 1998;110:34–40. doi: 10.1046/j.1523-1747.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- 37.Rutberg S E, Saez E, Glick A, Dlugosz A A, Spiegelman B M, Yuspa S H. Differentiation of mouse keratinocytes is accompanied by PKC-dependent changes in AP-1 proteins. Oncogene. 1996;13:167–176. [PubMed] [Google Scholar]
- 38.Sark M W, Fischer D F, de Meijer E, van de Putte P, Backendorf C. AP-1 and ets transcription factors regulate the expression of the human SPRR1A keratinocyte terminal differentiation marker. J Biol Chem. 1998;273:24683–24692. doi: 10.1074/jbc.273.38.24683. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seitz C S, Lin Q, Deng H, Khavari P A. Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc Natl Acad Sci USA. 1998;95:2307–2312. doi: 10.1073/pnas.95.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snape A M, Winning R S, Sargent T D. Transcription factor AP-2 is tissue-specific in Xenopus and closely related or identical to keratin transcription factor 1 (KTF-1) Development. 1991;113:283–293. doi: 10.1242/dev.113.1.283. [DOI] [PubMed] [Google Scholar]
- 42.Snape A M, Jonas E A, Sargent T C. KTF-1, a transcriptional activator of Xenopus embryonic keratin expression. Development. 1990;109:157–165. doi: 10.1242/dev.109.1.157. [DOI] [PubMed] [Google Scholar]
- 43.Stellmach V, Leask A, Fuchs E. Retinoid-mediated transcriptional regulation of keratin genes in human epidermal and squamous cell carcinoma cells. Proc Natl Acad Sci USA. 1991;88:4582–4586. doi: 10.1073/pnas.88.11.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takaoka A S, Yamada T, Gotoh M, Kanai Y, Imai K, Hirohashi S. Cloning and characterization of the human beta 4-integrin gene promoter and enhancers. J Biol Chem. 1998;273:33848–33855. doi: 10.1074/jbc.273.50.33848. [DOI] [PubMed] [Google Scholar]
- 45.Vallette F, Mege E, Reiss A, Adesnik M. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 1989;17:723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci USA. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Kinkel S, Polonsky K, Fuchs E. Transgenic studies with a keratin-promoter driven growth hormone transgene: prospects for gene therapy. Proc Natl Acad Sci USA. 1997;94:219–226. doi: 10.1073/pnas.94.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warshawsky D, Miller L. Tissue-specific in vivo protein-DNA interactions at the promoter region of the Xenopus 63 kDa keratin gene during metamorphosis. Nucleic Acids Res. 1995;23:4502–4509. doi: 10.1093/nar/23.21.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welter J F, Crish J F, Agarwal C, Eckert R L. Fos-related antigen (Fra-1), junB, and junD activate human involucrin promoter transcription by binding to proximal and distal AP-1 sites to mediate phorbol ester effects on promoter activity. J Biol Chem. 1995;270:12614–12622. doi: 10.1074/jbc.270.21.12614. [DOI] [PubMed] [Google Scholar]
- 50.Welter J F, Eckert R L. Differential expression of the fos and jun family members c-fos, fosB, Fra-1, Fra-2, c-jun, junB and junD during human epidermal keratinocyte differentiation. Oncogene. 1995;11:2681–2687. [PubMed] [Google Scholar]