Abstract
The new generation of cell-free gene expression systems enables the prototyping and engineering of biological systems in vitro over a remarkable scope of applications and physical scales. As the utilization of DNA-directed in vitro protein synthesis expands in scope, developing more powerful cell-free transcription–translation (TXTL) platforms remains a major goal to either execute larger DNA programs or improve cell-free biomanufacturing capabilities. In this work, we report the capabilities of the all-E. coli TXTL toolbox 3.0, a multipurpose cell-free expression system specifically developed for synthetic biology. In non-fed batch-mode reactions, the synthesis of the fluorescent reporter protein eGFP (enhanced green fluorescent protein) reaches 4 mg/ml. In synthetic cells, consisting of liposomes loaded with a TXTL reaction, eGFP is produced at concentrations of >8 mg/ml when the chemical building blocks feeding the reaction diffuse through membrane channels to facilitate exchanges with the outer solution. The bacteriophage T7, encoded by a genome of 40 kb and ∼60 genes, is produced at a concentration of 1013 PFU/ml (plaque forming unit/ml). This TXTL system extends the current cell-free expression capabilities by offering unique strength and properties, for testing regulatory elements and circuits, biomanufacturing biologics or building synthetic cells.
Keywords: cell-free transcription–translation, gene circuits, bacteriophages, synthetic cell
1. Introduction
The emerging enthusiasm for cell-free gene expression as a multipurpose bioengineering technology arises from several major improvements made in the last two decades. These advances have been predominantly made for cell-free expression systems from Escherichia coli, the most common model organism (1–5). First, cell-free transcription–translation (TXTL: cell-free transcription–translation) systems have become powerful enough to enable executing genetic programs composed of several genes relevant to natural living systems (6, 7) and integrating new technologies like CRISPR (clustered regularly interspaced short palindromic repeats) (8–11). The current TXTL technology is characterized by a fast experimental turnover and high-throughput settings (12), which facilitates the rapid prototyping of regulatory elements and gene networks (13–22). Besides gene circuits, the strength of the current TXTL platforms is leveraged to develop new biomanufacturing methods that offer the unique speed of delivery and portability (23–27). Second, the preparation of TXTL systems, from E. coli especially, has been streamlined and reported in detail (28–30), which considerably improved the affordability and accessibility to these systems. Third, TXTL carries by nature a high degree of safety, making it ideal for applications outside laboratories, such as education (11, 31). Altogether, cell-free gene expression has been shaped into a versatile and user-friendly tool covering an ever-growing scope of applications (6, 7, 32–34). Although TXTL systems from a broad variety of organisms are being developed (35), the cell-free gene expression based on E. coli lysate remains the major test bed due to its strength and the knowledge of this system.
Among the different E. coli platforms developed, the all-E. coli TXTL system, now commercially available under the name myTXTL, was devised to incorporate a broad transcription repertoire that comprises the seven E. coli sigma factors in addition to the routinely used T7 and T3 bacteriophage ribonucleic acid (RNA) polymerases and promoters (36, 37). In its second version (38), a new adenosine triphosphate (ATP) regeneration was used to raise protein synthesis up to 2.3 mg/ml in batch mode (39). By means of its extensive transcription capabilities and its strength, this TXTL system has been employed in many different physical settings for synthetic biology purposes or to address fundamental questions (Figure 1). This system has proven effective for prototyping short deoxyribonucleic acid (DNA) (13–17) and gene circuits (18–20, 40, 41), biomanufacturing (38, 42–46) and building synthetic cells (38, 47–54). It is also a convenient platform to interrogate biological systems at a basic level, including reconstituting dynamical systems (19), emulating pattern formation (20, 40), showing decision-making based on a few molecules in regulatory networks (55) and revealing the importance of molecular crowding in two dimensions for the self-assembly of cytoskeletal proteins (49, 50).
Figure 1.
Overview of the all-E. coli TXTL system and its applications. References are given as examples of the toolbox usage.
In this article, we present the major capabilities of the new version of the all-E. coli TXTL toolbox, focusing on protein synthesis through the transcription repertoire, the synthesis of the phage T7 used as a reference for the processing of large DNA programs and protein synthesis in synthetic cell systems. Other characteristics of this toolbox have been reported in version 2.0 (38) such as messenger RNA and protein degradation and are not discussed in this work. When appropriate, we highlight the main differences between the three versions of the system, toolbox 1.0 published in 2012 (36), toolbox 2.0 published in 2016 (38) and toolbox 3.0 reported in this article.
2. Materials and methods
2.1. TXTL system and batch-mode reactions
The preparation of the cell-free expression system used in this work has been described thoroughly in several articles (28, 36, 38, 56). The toolbox 2.0 (38) is commercialized under the name myTXTL (Arbor Biosciences). Compared to the toolbox 2.0 (38), two major modifications were made. First, the cells (E. coli strain BL21 Rosetta2, Millipore Sigma) were grown at 40°C instead of 37°C during lysate preparation. Second, cell-free reactions were supplemented with 60 mM maltodextrin (Sigma Aldrich 419672) and 30 mM d-ribose (Sigma Aldrich R-7500) instead of just maltodextrin. Supplementary Table S1 summarizes the references that describe, in detail, the preparation of this system and the differences between the toolboxes versions. Cell-free reactions were carried out in a volume of 2–20 µl at 29–30°C. The reactions were either assembled by hand (10–20 µl) or dispensed on 96-well plates using a Beckman Labcyte Echo liquid dispenser (2–5 µl). Quantitative measurements were carried out with the reporter protein deGFP (d-enhanced green fluorescent protein) (25.4 kDa, 1 mg/ml = 39.37 µM). deGFP is a variant of the reporter eGFP (enhanced green fluorescent protein) that is more translatable in cell-free systems. The excitation and emission spectra, as well as fluorescence properties of deGFP and eGFP, are identical, as reported before (36). The fluorescence of deGFP produced in batch-mode reaction was measured on an H1m plate reader (Biotek Instruments, 96-well plate). Endpoint measurements were carried out after 15–20 h of incubation. Pure recombinant eGFP with a His tag (from either Cell Biolabs Inc. or Biovision) was used for quantification (linear calibration of the plate reader and microscope as described before (38)). Error bars are the standard deviations from at least three replicates.
2.2. DNA part lists and plasmid preparation
The DNA parts used in this work are available at Arbor Biosciences and are reported in Supplementary Table S2 (plasmids) and Supplementary Table S3 (linear). GenBank files of the plasmids used in this work are available as supplementary material. Unless specified, the plasmids contain the highly efficient untranslated region named UTR1 (36). The plasmids were amplified using standard mini or midi prep kits and further cleaned up with a polymerase chain reaction (PCR) purification kit and eluted in autoclaved water. The concentration of the DNA stock solutions was quantified on a NanoDrop spectrophotometer. Linear DNA templates were amplified by standard PCR from the respective plasmids, cleaned up using a PCR purification kit and eluted in autoclaved water. P70a-gamS was obtained from Twist Biosciences and amplified by PCR.
2.3. TXTL synthesis of phages
The T7 genomic DNA was purchased from Boca Scientific. The chi6 short DNA (Integrated DNA Technologies) was added at a concentration of 3 µM to prevent the degradation of the linear T7 DNA (57). dNTPs (deoxynucleotide triphosphates) (Invitrogen) were added to a concentration of 0.1 mM each to enable genome replication, as described before (44). The PEG8000 (Sigma Aldrich) concentration was increased from 2% (2.5 mM) to 3.5% (4.3 mM) to emulate molecular crowding (43). Bacteriophages were counted by the standard plaque-forming assay using the E. coli strain B for T7. Cells were grown in Luria–Bertani (LB) broth at 37°C. The plates were prepared as follows: each sample was added to a solution composed of 5 ml of 0.6% liquid LB-agar (45°C) and 50 µl of cell culture, poured on a 1.1% solid LB-agar plate. Plates were incubated at 37°C, and plaques were counted after 6 h.
2.4. TXTL-based synthetic cells
The cell-free reactions were encapsulated into large unilamellar phospholipid vesicles by the water-in-oil emulsion transfer method (48). Briefly, phospholipids (Avanti Polar Lipids, PC 840051, PE-PEG5000 880200) were dissolved in mineral oil (Sigma-Aldrich M5904) at a total concentration of 2 mg/ml (molar proportion: 99.33% PC and 0.66% PE-PEG5000). A few microliters of cell-free reaction was added to 0.5 ml of the phospholipid solution. This solution was vortexed for 5–10 s to create an emulsion. About 100–250 µl of the emulsion was placed on top of 20 µl of the feeding solution. The vesicles are formed by the centrifugation of the biphasic solution for 20 s at 4000 rpm. The phospholipid vesicles were observed with a CCD (charge-coupled device) camera mounted on an inverted microscope (Olympus IX-81) equipped with the proper set of fluorescence filters. The feeding solution contained the same components as the reaction except for the DNA and lysate that were replaced by water. Pure alpha-hemolysin (AH) was purchased from Sigma Aldrich.
2.5. Materials and resources availability statement
The list of plasmids used in this work (available at Arbor Biosciences) is summarized in Supplementary Table S2. The protocols for the Cell-Free Expression (CFE) system are available in the references provided and in Supplementary Table S1. Other materials are available on reasonable request.
3. Results and discussion
3.1. Overall picture of the all-E. coli TXTL system
To build a versatile toolbox that does not rely only on the T7 promoter and polymerase, our original goal was to develop an all-E. coli TXTL system that integrates a broad transcription repertoire so as to execute circuits composed of different regulatory elements. The primary transcription is achieved by the housekeeping sigma factor σ70 and the core RNA polymerase, both provided by the lysate, which also brings all the necessary components for translation. Transcription by the six other sigma factors and the two bacteriophage RNA polymerases T3 and T7 are performed via transcriptional activation cascades. The second goal was to achieve a protein synthesis level large enough to enable the expression of large genetic programs and biomanufacturing of biologics. To this end, the toolbox incorporates a chemical ATP regeneration based on a phosphate donor and a carbohydrate to exploit the glycolytic pathway of the lysate (38, 39). Several other functionalities were developed: (i) protein degradation via the ClpXP proteases (38), (ii) tunable messenger RNA degradation via the interferase MazF (58), (iii) protection of linear DNA templates such as PCR products via the GamS protein (28) and the chi6 short linear dsDNA (57). These functionalities, valid for the new version of the system reported here, have been already described thoroughly. Lysates are prepared on a regular basis and tested for leftovers of living E. coli cells by plating the equivalent of 100–200 µl of cell-free reaction on LB-agar petri dishes without antibiotics. As reported several times before, no colonies are observed when this control is done (Supplementary Figure S1), making it a true cell-free expression system.
Compared to the toolbox 2.0 (38), the toolbox 3.0 reported in this article incorporates two changes that enable protein synthesis >3 mg/ml. First, during the lysate preparation, the cells are grown at 40°C instead of 37°C. It was demonstrated previously that increasing the temperature of E. coli cultures can improve cell-free protein synthesis yields (59). The second modification applies to the reaction. Rather than just adding maltodextrin as a carbohydrate source to exploit glycolysis in the lysate (39), a mixture of maltodextrin (60 mM) and d-ribose (30 mM) is added to the reaction. We assume that maltodextrin and d-ribose improve ATP regeneration, but their role would need to be clarified by a study outside the scope of this work. It is the combination of these two changes (temperature and carbohydrate mixture) that enables cell-free protein synthesis to reach up to 4 mg/ml.
3.2. Protein synthesis yields and time course
We measured protein synthesis in batch mode for the seven E. coli transcription factors, the two bacteriophage RNA polymerases T3 and T7, and linear PCR products for both σ70 and T7, and we compared the results to the two previous versions of the toolbox (Table 1). Except for σ70 already present in the lysate, the synthesis of the reporter protein deGFP was achieved through a transcriptional activation cascade and specific promoters for each transcription factor or RNA polymerase. Each transcription factor or RNA polymerase was expressed through the strong E. coli promoter P70a and the untranslated region UTR1 (36), which originates from the bacteriophage T7 (60). The performance of all the sigma factors and bacteriophage RNA polymerases was largely greater than for the toolboxes 1.0 and 2.0 (Table 1). For toolbox 3.0, the maximum protein synthesis concentration was observed for the T7 cascade and topped 4 mg/ml. The effects of varying maltodextrin and d-ribose show the synergy produced by the two carbohydrates (Supplementary Figure S2). The effect of the temperature of cell growth cultures during the lysate preparation was measured in the case of the T7 cascade to show that a temperature of 40°C also contributes to the increased performance of the toolbox 3.0 (Supplementary Figure S3).
Table 1.
Endpoint measurements of the reporter protein deGFP concentration for each of the transcription (TX) factors and RNA polymerases of the all-E. coli TXTL toolboxes (TBs) 1.0 (36), 2.0 (38) and 3.0 (this work)
| TX factors plasmid | Reporter plasmid | TB 1.0 deGFP µM (mg/ml) |
TB 2.0 deGFP µM (mg/ml) |
TB 3.0 (this work) deGFP µM (mg/ml) |
|---|---|---|---|---|
| Plasmids | ||||
| σ70 | P70a-degfp | 25 (0.63) | 81 (2.05) | 130 (3.3) |
| P70a-σ19 | P19a-degfp | 7 (0.18) | 35 (0.89) | 54 (1.27) |
| P70a-σ24 | P24a-degfp | 11 (0.28) | 70 (1.78) | 95 (2.41) |
| P70a-σ28 | P28a-degfp | 21 (0.53) | 77 (1.95) | 122 (3.1) |
| P70a-σ32 | P32a-degfp | 19 (0.48) | 89 (2.26) | 100 (2.54) |
| P70a-σ38 | P38a-degfp | 13 (0.33) | 75 (1.90) | 96 (2.43) |
| P70a-σ54/ntrC | P54a-degfp | 5 (0.13) | 27 (0.68) | 58 (1.47) |
| P70a-T3rnap | T3p14-degfp | 27 (0.69) | 74 (1.88) | 129 (3.27) |
| P70a-T7rnap | T7p14-degfp | 29 (0.74) | 87 (2.20) | 160 (4.05) |
| PCR | ||||
| σ70 | P70a-degfp | NA | 50 (1.27) | 70 (1.77) |
| P70a-T7rnap | T7p14-degfp | NA | 36 (0.91) | 117 (2.97) |
The values shown in this table are the average maximum protein synthesis yields for each transcription. Variability in cell-free protein synthesis for each transcription is shown in the error bars of each histogram plot (Supplementary figures). The transcription factor σ70 and the core RNA polymerase are present in the lysate. The other TX factors and bacteriophages RNA polymerases (first column) are expressed from plasmids or PCR products, through the promoter P70a. The reporter gene degfp is expressed through two-stage transcriptional activation cascades via the respective promoters (second column; 1 mg/ml deGFP = 39.4 μM).
The concentration of plasmids was varied for each transcription factor to find the optimal settings (Supplementary Figures S4–S12). When linear PCR-amplified DNA templates were used, protein synthesis reached 1.77 and 2.97 mg/ml for σ70 and T7, respectively (Table 1). To achieve such a level of protein synthesis with linear DNA, both chi6 (3 µM) and P70a-gamS (linear, 1 nM) were added to the reaction. The protein GamS is dynamically synthesized to inhibit linear DNA degradation (61), concurrently with the expression of the reporter gene. These results were obtained using a liquid dispenser for reactions of volume 2 µl incubated on a 96-well plate, sterile disposable plastic wares and freshly prepared solutions. When prepared by hand, the protein synthesis yield of the reactions is usually lower and can reach, in the case of the T7 cascade, 3–3.5 mg/ml (Supplementary Figure S13). The variability of cell-free protein synthesis yields across different batches of TXTL systems was small (Supplementary Figure S13). The differences in protein synthesis via the T7 transcriptional activation cascade for the three toolboxes appear clearly on sodium dodecyl sulfate polyacrylamide gel electrophoresis (Supplementary Figure S14).
We measured similar increased productions of the reporter proteins deCFP and mCherry through the T7 transcriptional activation cascade (Supplementary Figure S15), thus showing that our observations are not specific and limited to deGFP. With a batch-mode protein synthesis of 3–4 mg/ml and a time course of almost 1 day, semi-continuous TXTL reactions, based on dialysis, become less relevant to implement for several reasons. Semi-continuous TXTL reactions are not cost-effective, not easy to handle and generally less reproducible than non-fed batch-mode reactions. Instead, semi-continuous TXTL was carried out in synthetic cells as discussed thereafter.
We measured the time course of protein synthesis through the T7 cascade for plasmids and linear DNA and compared it to the toolbox 2.0 settings (Figure 2). Two major differences appeared in the time course between the two versions of the system. For the toolbox 3.0, protein accumulation is characterized by a greater synthesis rate and a longer synthesis time, which explain the larger yield at the end of incubation. These results show that the toolbox 3.0 is a long-lived TXTL system that can express genes for periods of time up to 20 h instead of 12–15 h for the toolbox 2.0. This was observed for both plasmids and linear DNA templates (Figure 2).
Figure 2.
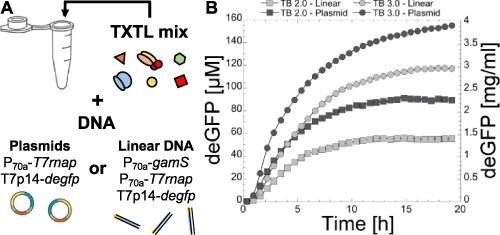
Time course of deGFP synthesis via the T7 transcriptional activation cascade. (A) Schematic of the experiment achieved in batch mode using either plasmids or linear DNA. (B) Graph showing the time course of deGFP synthesis in the conditions of the toolbox 2.0 (TB 2.0) (38) and toolbox 3.0 (TB 3.0, this work). The variability envelop is shown for each curve. Plasmids: 0.2 nM P70a-T7rnap, 4 nM T7p14-degfp. Linear DNA: 1 nM P70a-gamS, 0.2 nM P70a-T7rnap, 4 nM T7p14-degfp.
3.3. TXTL of the bacteriophage T7
We demonstrated previously that several E. coli bacteriophages can be synthesized from their genomes in both the toolboxes 1.0 and 2.0 (38, 44). To quantify phage synthesis in the toolbox 3.0 settings, we tested the synthesis of phage T7 as it also achieves genome replication, using the plaque assay as described before (44). We use the T7 genome as a benchmark to challenge the capabilities of the all-E. coli TXTL system for processing large gene sets and recapitulating self-assembly. The 40-kb genome of T7 encodes for ∼60 genes, including its own DNA replication genes. DNA replication is achieved by adding dNTPs to the cell-free reaction, as shown before (44). In the toolbox 3.0, we measured a concentration of 1013 PFU/ml (plaque forming unit/ml) (Figure 3A–C), which is on the order of 10 000 and 100 times larger than in the toolboxes 1.0 (≈109 PFU/ml) and 2.0 (≈1011 PFU/ml), respectively.
Figure 3.
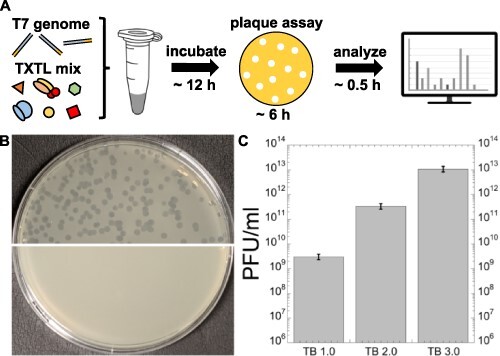
Cell-free expression and synthesis of the phage T7. (A) Schematic of the experiment. The cell-free reaction also contained 3 µM of chi6 short DNA to prevent degradation of the linear T7 genome and 0.1 mM of each of the dNTPs for genome replication. (B) Image of plaques. Top half: results from a TXTL reaction synthesizing the phage T7. Bottom half: negative control showing a lawn of E. coli cells. (C) Plot of the measured PFU/ml for the three versions of the toolbox.
3.4. TXTL synthetic cells
TXTL is the most common approach to build, from the ground up, genetically programmed synthetic cells, minimal cells in particular (62–74). Minimal cells consist of a TXTL reaction encapsulated in a cell-sized compartment, such as a liposome made of a phospholipid bilayer. Plasmids or linear DNA are added to the reaction so as to construct biological functions by expressing specific gene sets inside liposomes. (Figure 4A). This approach to synthetic cells has proven effective to emulate several natural mechanisms found in living cells (71, 75–80). To determine how large protein synthesis can be in synthetic cells in the toolbox 3.0 conditions, we expressed the gene degfp via the T7 cascade with and without the membrane channel AH (Figure 4B). AH forms channels of 1.4 nm diameter into the phospholipid bilayer, which corresponds to a molecular mass cutoff of ∼3–5 kDa (81). Thus, AH enables feeding the compartmentalized cell-free reaction with the necessary small nutrients, such as ATP and amino acids, by diffusion through the membrane. To quantify the fluorescence of deGFP, we made a calibration (Supplementary Figure S16) as described previously (38). Because it is difficult to get above 100 µM with pure eGFP, the calibration was carried out between 10 and 100 µM and found to be linear in this range. Given that the synthesis of the reporter in the liposomes was >100 µM, we used a neutral density filter to verify that above 100 µM the fluorescence intensity for eGFP concentrations >100 µM is still linearly proportional to the calibration (Supplementary Figure S17). This approach allowed us to keep the same illumination intensity. We measured the average concentration of deGFP for populations of 50–100 liposomes with diameters ranging from 1 to 20 µm. When AH was not added to the reaction and the external solution, we measured an average concentration of ∼180–190 µM (≈4.7 mg/ml), which is slightly larger than our measurements in batch-mode test-tube reactions (Figure 4C). When AH was added to the encapsulated cell-free reaction and the external feeding solution at a concentration of 0.1 µM, we measured an average concentration of 330 µM (≈8.4 mg/ml) (Figure 4C). The difference in protein synthesis with and without AH was more pronounced for small liposomes. The variability in the fluorescence intensity for any given population was also slightly smaller when AH was used. We found that a concentration of 0.1 µM AH was optimal. The time course of protein synthesis in the liposomes shows that AH enables a greater synthesis rate in the first hours of expression (Figure 4C).
Figure 4.
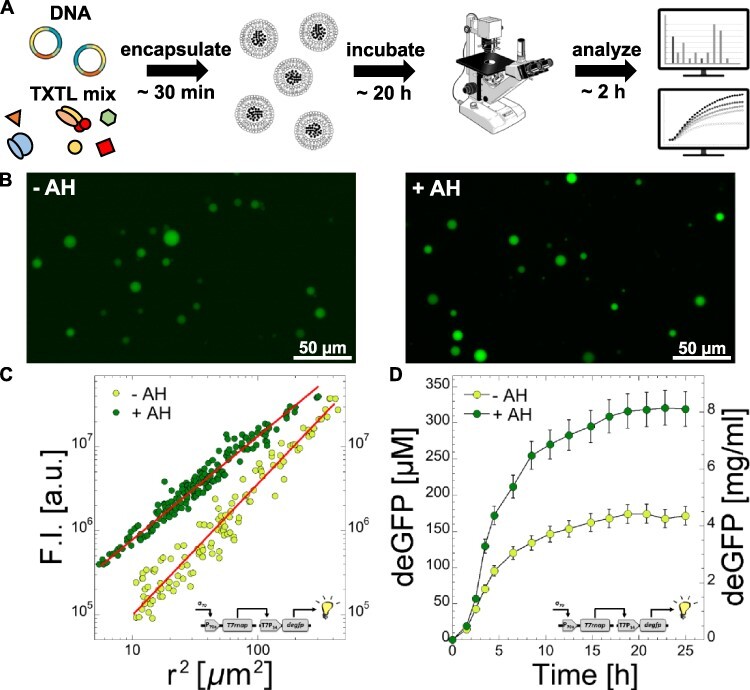
Cell-free expression and synthesis of deGFP in synthetic cells. (A) Schematic of the experiment (plasmids: P70a-T7rnap fixed at 0.2 nM, T7p14-degfp fixed at 4 nM). (B) Fluorescence images of the liposomes, with and without AH, taken after 20 h of incubation with a 40× objective. (C) Fluorescence intensity (F.I.) as a function of the square of the radius for two populations of liposomes, one without AH (−AH) and one with AH (+AH, 0.1 µM added to the inner and outer solutions). Linear fits: F.I. = −2.29 × 106 + 77 452 r2 (−AH, R = 0.96968) and F.I. = −6.42 × 105 + 1.379 × 105r2 (+AH, R = 0.97959). (D) Time course of deGFP synthesis in synthetic cells, without and with AH added to the solution.
4. Conclusion
Although major advances have been made in the optimization of cell-free gene expression systems, the exploration of their capabilities is far from complete and will be central to the development of new synthetic biology applications. In this work, we showed that an E. coli-based TXTL system is capable of producing a reporter protein at a concentration of 4 mg/ml in non-fed batch-mode reactions, a concentration not observed before in prokaryotic TXTL. In a semi-continuous synthetic cell setting, the synthesis of deGFP attains >8 mg/ml. No strain engineering was required to get such synthesis yields. The strength of the toolbox 3.0 should facilitate expressing large DNA programs encoding for biosynthesis pathways or for biological functions to build synthetic cells.
Supplementary Material
Contributor Information
David Garenne, School of Physics and Astronomy, University of Minnesota, Minneapolis, MN, USA.
Seth Thompson, School of Physics and Astronomy, University of Minnesota, Minneapolis, MN, USA.
Amaury Brisson, School of Physics and Astronomy, University of Minnesota, Minneapolis, MN, USA.
Aset Khakimzhan, School of Physics and Astronomy, University of Minnesota, Minneapolis, MN, USA.
Vincent Noireaux, School of Physics and Astronomy, University of Minnesota, Minneapolis, MN, USA.
Supplementary data
Supplementary data are available at SYNBIO online.
Data availability
Data are available on reasonable request.
Funding
National Science Foundation [NSF MCB-1844152, EF-1934496, CBET-1916030].
Author contributions
D.G.: experiments and manuscript editing, S.T.: experiments and manuscript editing, A.B.: experiments, A.K.: experiments and manuscript editing and V.N.: manuscript writing and editing.
Conflict of interest statement
The authors declare the following competing financial interest(s): Noireaux laboratory receives royalties from Arbor Biosciences, a distributor of myTXTL cell-free protein expression kit. Vincent Noireaux consults with Arbor on other cell-free expression topics.
References
- 1. Cole S.D., Miklos A.E., Chiao A.C., Sun Z.Z. and Lux M.W. (2020) Methodologies for preparation of prokaryotic extracts for cell-free expression systems. Synth. Syst. Biotechnol., 5, 252–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu D.V., Zawada J.F. and Swartz J.R. (2005) Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol. Prog., 21, 460–465.doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 3. Voloshin A.M. and Swartz J.R. (2005) Efficient and scalable method for scaling up cell free protein synthesis in batch mode. Biotechnol. Bioeng., 91, 516–521. [DOI] [PubMed] [Google Scholar]
- 4. Jewett M.C., Calhoun K.A., Voloshin A., Wuu J.J. and Swartz J.R. (2008) An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol., 4, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang W.C., Patel K.G., Wong H.E. and Swartz J.R. (2012) Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog., 28, 413–420. [DOI] [PubMed] [Google Scholar]
- 6. Silverman A.D., Karim A.S. and Jewett M.C. (2020) Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet., 28, 413–420.doi: 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 7. Garenne D. and Noireaux V. (2019) Cell-free transcription–translation: engineering biology from the nanometer to the millimeter scale. Curr. Opin. Biotechnol., 58, 19–27. [DOI] [PubMed] [Google Scholar]
- 8. Maxwell C.S., Jacobsen T., Marshall R., Noireaux V. and Beisel C.L. (2018) A detailed cell-free transcription-translation-based assay to decipher CRISPR protospacer-adjacent motifs. Methods, 143, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marshall R., Maxwell C.S., Collins S.P., Jacobsen T., Luo M.L., Begemann M.B., Gray B.N., January E., Singer A., He Y.. et al. (2018) Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell, 69, 146–157.e3.doi: 10.1016/j.molcel.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wandera K.G., Collins S.P., Wimmer F., Marshall R., Noireaux V. and Beisel C.L. (2020) An enhanced assay to characterize anti-CRISPR proteins using a cell-free transcription-translation system. Methods, 172, 42–50. [DOI] [PubMed] [Google Scholar]
- 11. Collias D., Marshall R., Collins S.P., Beisel C.L. and Noireaux V. (2019) An educational module to explore CRISPR technologies with a cell-free transcription-translation system. Synth. Biol., 4, ysz005.doi: 10.1093/synbio/ysz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borkowski O., Koch M., Zettor A., Pandi A., Batista A.C., Soudier P. and Faulon J.L. (2020) Large scale active-learning-guided exploration for in vitro protein production optimization. Nat. Commun., 11, 1872.doi: 10.1038/s41467-020-15798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agrawal D.K., Marshall R., Noireaux V. and Sontag E.D. (2019) In vitro implementation of robust gene regulation in a synthetic biomolecular integral controller. Nat. Commun., 10, 5760.doi: 10.1038/s41467-019-13626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Westbrook A., Tang X., Marshall R., Maxwell C.S., Chappell J., Agrawal D.K., Dunlop M.J., Noireaux V., Beisel C.L., Lucks J.. et al. (2019) Distinct timescales of RNA regulators enable the construction of a genetic pulse generator. Biotechnol. Bioeng., 116, 1139–1151.doi: 10.1002/bit.26918. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi M.K., Hayes C.A., Chappell J., Sun Z.Z., Murray R.M., Noireaux V. and Lucks J.B. (2015) Characterizing and prototyping genetic networks with cell-free transcription-translation reactions. Methods, 86, 60–72. [DOI] [PubMed] [Google Scholar]
- 16. Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V., Spring K.J., Al-Khabouri S., Fall C.P., Noireaux V.. et al. (2015) Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth. Biol., 4, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Z.Z., Yeung E., Hayes C.A., Noireaux V. and Murray R.M. (2014) Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol., 3, 387–397. [DOI] [PubMed] [Google Scholar]
- 18. Niederholtmeyer H., Sun Z.Z., Hori Y., Yeung E., Verpoorte A., Murray R.M. and Maerkl S.J. (2015) Rapid cell-free forward engineering of novel genetic ring oscillators. Elife., 4, e09771.doi: 10.7554/elife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karzbrun E., Tayar A.M., Noireaux V. and Bar-Ziv R.H. (2014) Programmable on-chip DNA compartments as artificial cells. Science (80-), 345, 829–832. [DOI] [PubMed] [Google Scholar]
- 20. Tayar A.M., Karzbrun E., Noireaux V. and Bar-Ziv R.H. (2017) Synchrony and pattern formation of coupled genetic oscillators on a chip of artificial cells. Proc. Natl. Acad. Sci. USA, 114, 11609–11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moore S.J., MacDonald J.T., Wienecke S., Ishwarbhai A., Tsipa A., Aw R., Kylilis N., Bell D.J., McClymont D.W., Jensen K.. et al. (2018) Rapid acquisition and model-based analysis of cell-free transcription–translation reactions from nonmodel bacteria. Proc. Natl. Acad. Sci., 115, E4340–E4349.doi: 10.1073/pnas.1715806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karim A.S., Dudley Q.M., Juminaga A., Yuan Y., Crowe S.A., Heggestad J.T., Garg S., Abdalla T., Grubbe W.S., Rasor B.J.. et al. (2020) In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat. Chem. Biol., 16, 912–919.doi: 10.1038/s41589-020-0559-0. [DOI] [PubMed] [Google Scholar]
- 23. Pardee K., Green A.A., Ferrante T., Cameron D.E., Daleykeyser A., Yin P. and Collins J.J. (2014) Paper-based synthetic gene networks. Cell, 159, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D., Ferrante T., McSorley F.R., Furuta Y., Vernet A.. et al. (2016) Portable, on-demand biomolecular manufacturing. Cell, 167, 248–259.e12.doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 25. Benítez-Mateos A.I., Llarena I., Sánchezsánchez-Iglesias A. and Lópezlópez-Gallego F. (2018) Expanding one-pot cell-free protein synthesis and immobilization for on-demand manufacturing of biomaterials. ACS Synth. Biol., 7, 875–884.doi: 10.1021/acssynbio.7b00383. [DOI] [PubMed] [Google Scholar]
- 26. Karig D.K., Bessling S., Thielen P., Zhang S. and Wolfe J. (2017) Preservation of protein expression systems at elevated temperatures for portable therapeutic production. J. R. Soc. Interface, 14, 20161039.doi: 10.1098/rsif.2016.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith M.T., Berkheimer S.D., Werner C.J. and Bundy B.C. (2014) Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques, 56, 186–193.doi: 10.2144/000114158. [DOI] [PubMed] [Google Scholar]
- 28. Sun Z.Z., Hayes C.A., Shin J., Caschera F., Murray R.M. and Noireaux V. (2013) Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp., 79, e50762.doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levine M.Z., Gregorio N.E., Jewett M.C., Watts K.R. and Oza J.P. (2019) Escherichia coli-based cell-free protein synthesis: protocols for a robust, flexible, and accessible platform technology. J. Vis. Exp., 144, e58882.doi: 10.3791/58882. [DOI] [PubMed] [Google Scholar]
- 30. Kwon Y.C. and Jewett M.C. (2015) High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep., 5, 8663.doi: 10.1038/srep08663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang A., Nguyen P.Q., Stark J.C., Takahashi M.K., Donghia N., Ferrante T., Dy A.J., Hsu K.J., Dubner R.S., Pardee K.. et al. (2018) BiobitsTM explorer: a modular synthetic biology education kit. Sci. Adv., 4, eaat5105.doi: 10.1126/sciadv.aat5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiba C.H., Knirsch M.C., Rodrigues Azzoni A., Moreira A.R. and Stephano M.A.. Review cell-free protein synthesis: advances on production process for biopharmaceuticals and immunobiological products. Biotechniques, 70, 126–133. [DOI] [PubMed] [Google Scholar]
- 33. Zhang L., Guo W. and Lu Y. (2020) Advances in cell-free biosensors: principle, mechanism, and applications. Biotechnol. J., 15, 2000187. [DOI] [PubMed] [Google Scholar]
- 34. Kai L. and Schwille P. (2019) Cell-free protein synthesis and its perspectives for assembling cells from the bottom-up. Adv. Biosyst., 3, 1800322. [DOI] [PubMed] [Google Scholar]
- 35. Gregorio N.E., Levine M.Z. and Oza J.P. (2019) A user’s guide to cell-free protein synthesis. Methods Protoc., 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin J. and Noireaux V. (2012) An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol., 1, 29–41. [DOI] [PubMed] [Google Scholar]
- 37. Shin J. and Noireaux V. (2010) Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J. Biol. Eng., 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garamella J., Marshall R., Rustad M. and Noireaux V. (2016) The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol., 5, 344–355. [DOI] [PubMed] [Google Scholar]
- 39. Caschera F. and Noireaux V. (2014) Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie, 99, 162–168. [DOI] [PubMed] [Google Scholar]
- 40. Tayar A.M., Karzbrun E., Noireaux V. and Bar-Ziv R.H. (2015) Propagating gene expression fronts in a one-dimensional coupled system of artificial cells. Nat. Phys., 11, 1037–1041. [Google Scholar]
- 41. Lehr F.O.-X., Hanst M., Vogel M., Kremer J., G̈ H.U., Suess B. and Koeppl H. (2019) Cell-free prototyping of AND-logic gates based on heterogeneous RNA activators. ACS Synth. Biol., 8, 2163–2173.doi: 10.1021/acssynbio.9b00238. [DOI] [PubMed] [Google Scholar]
- 42. Rustad M., Eastlund A., Jardine P. and Noireaux V. (2018) Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth. Biol., 3, ysy002.doi: 10.1093/synbio/ysy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rustad M., Eastlund A., Marshall R., Jardine P. and Noireaux V. (2017) Synthesis of infectious bacteriophages in an E. coli-based cell-free expression system. J. Vis. Exp., 126, e56144.doi: 10.3791/56144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shin J., Jardine P. and Noireaux V. (2012) Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth. Biol., 1, 408–413. [DOI] [PubMed] [Google Scholar]
- 45. Chemla Y., Ozer E., Schlesinger O., Noireaux V. and Alfonta L. (2015) Genetically expanded cell-free protein synthesis using endogenous pyrrolysyl orthogonal translation system. Biotechnol. Bioeng., 112, 1663–1672. [DOI] [PubMed] [Google Scholar]
- 46. Vonshak O., Divon Y., Förste S., Garenne D., Noireaux V., Lipowsky R., Rudorf S., Daube S.S. and Bar-Ziv R.H. (2020) Programming multi-protein assembly by gene-brush patterns and two-dimensional compartment geometry. Nat. Nanotechnol., 15, 783–791.doi: 10.1038/s41565-020-0720-7. [DOI] [PubMed] [Google Scholar]
- 47. Shin Noireaux V.J. (2011) An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol., 1, 29–41. [DOI] [PubMed] [Google Scholar]
- 48. Garamella J., Garenne D. and Noireaux V. (2019) TXTL-based approach to synthetic cells. Methods Enzymol., 15, 783–791.doi: 10.1016/bs.mie.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 49. Garenne D. and Noireaux V. (2020) Analysis of cytoplasmic and membrane molecular crowding in genetically programmed synthetic cells. Biomacromolecules, 21, 2808–2817. [DOI] [PubMed] [Google Scholar]
- 50. Garenne D., Libchaber A. and Noireaux V. (2020) Membrane molecular crowding enhances MreB polymerization to shape synthetic cells from spheres to rods. Proc. Natl. Acad. Sci. USA, 117, 1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caschera F. and Noireaux V. (2016) Compartmentalization of an all-E. coli cell-free expression system for the construction of a minimal cell. Artif. Life, 22, 185–195. [DOI] [PubMed] [Google Scholar]
- 52. Majumder S., Garamella J., Wang Y.-L., Denies M., Noireaux V. and Liu A.P. (2017) Cell-sized mechanosensitive and biosensing compartment programmed with DNA. Chem. Commun., 53, 7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garamella J., Majumder S., Liu A.P. and Noireaux V. (2019) An adaptive synthetic cell based on mechanosensing, biosensing, and inducible gene circuits. ACS Synth. Biol., 8, 1913–1920. [DOI] [PubMed] [Google Scholar]
- 54. Adamala K.P., Martin-Alarcon D.A., Guthrie-Honea K.R. and Boyden E.S. (2017) Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem., 9, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Greiss F., Daube S.S., Noireaux V. and Bar-Ziv R. (2020) From deterministic to fuzzy decision-making in artificial cells. Nat. Commun., 11, 5648.doi: 10.1038/s41467-020-19395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Caschera F. and Noireaux V. (2015) Preparation of amino acid mixtures for cell-free expression systems. Biotechniques, 58, 40–43. [DOI] [PubMed] [Google Scholar]
- 57. Marshall R., Maxwell C.S., Collins S.P., Beisel C.L. and Noireaux V. (2017) Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription–translation systems. Biotechnol. Bioeng., 114, 2137–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin J. and Noireaux V. (2010) Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J. Biol. Eng., 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamane T., Ikeda Y., Nagasaka T. and Nakano H. (2005) Enhanced cell-free protein synthesis using a S30 extract from Escherichia coli grown rapidly at 42 degrees C in an amino acid enriched medium. Biotechnol. Prog., 21, 608–613. [DOI] [PubMed] [Google Scholar]
- 60. Olins P.O., Devine C.S., Rangwala S.H. and Kavka K.S. (1988) The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene, 73, 227–235. [DOI] [PubMed] [Google Scholar]
- 61. Karu A.E., Sakaki Y., Echols H. and Linn S. (1975) The gamma protein specified by bacteriophage gamma. Structure and inhibitory activity for the recBC enzyme of Escherichia coli. J. Biol. Chem., 250, 7377–7387. [PubMed] [Google Scholar]
- 62. Caschera F. and Noireaux V. (2014) Integration of biological parts toward the synthesis of a minimal cell. Curr. Opin. Chem. Biol., 22, 85–91. [DOI] [PubMed] [Google Scholar]
- 63. Stano P., Kuruma Y., Souza T.P. and Luisi P.L. (2010) Biosynthesis of proteins inside liposomes. Methods Mol. Biol., 606, 127–145. [DOI] [PubMed] [Google Scholar]
- 64. Ayoubi-Joshaghani M.H., Dianat-Moghadam H., Seidi K., Jahanban-Esfahalan A., Zare P. and Jahanban-Esfahlan R. (2020) Cell-free protein synthesis: the transition from batch reactions to minimal cells and microfluidic devices. Biotechnol. Bioeng., 117, 1204–1229. [DOI] [PubMed] [Google Scholar]
- 65. Gaut N.J. and Adamala K.P. (2021) Reconstituting natural cell elements in synthetic cells. Adv. Biol., 5, e2000188.doi: 10.1002/adbi.202000188. [DOI] [PubMed] [Google Scholar]
- 66. Shim J., Zhou C., Gong T., Iserlis D.A., Linjawi H.A., Wong M., Pan T. and Tan C. (2021) Building protein networks in synthetic systems from the bottom-up. Biotechnol. Adv., 49, 107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stano P. and Luisi P.L. (2013) Semi-synthetic minimal cells: origin and recent developments. Curr. Opin. Biotechnol., 24, 633–638. [DOI] [PubMed] [Google Scholar]
- 68. Lentini R., Martín N.Y., Forlin M., Belmonte L., Fontana J., Cornella M., Martini L., Tamburini S., Bentley W.E., Jousson O.. et al. (2017) Two-way chemical communication between artificial and natural cells. ACS Cent. Sci., 3, 117–123.doi: 10.1021/acscentsci.6b00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lentini R., Santero S.P., Chizzolini F., Cecchi D., Fontana J., Marchioretto M., Del Bianco C., Terrell J.L., Spencer A.C., Martini L.. et al. (2014) Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun., 5, 4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Godino E., López J.N., Zarguit I., Doerr A., Jimenez M., Rivas G. and Danelon C. (2020) Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. bioRxiv. Commun. Biol., 3, 539.doi: 10.1101/2020.03.29.009639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Van Nies P., Westerlaken I., Blanken D., Salas M., Mencía M. and Danelon C. (2018) Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat. Commun., 9, 1583.doi: 10.1038/s41467-018-03926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Berhanu S., Ueda T. and Kuruma Y. (2019) Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun., 10, 1325.doi: 10.1038/s41467-019-09147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nishimura K., Matsuura T., Nishimura K., Sunami T., Suzuki H. and Yomo T. (2012) Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir, 28, 8426–8432. [DOI] [PubMed] [Google Scholar]
- 74. Sunami T., Matsuura T., Suzuki H. and Yomo T. (2010) Synthesis of functional proteins within liposomes. Methods. Mol. Biol., 607, 243–256.doi: 10.1007/978-1-60327-331-2_20. [DOI] [PubMed] [Google Scholar]
- 75. Noireaux V., Bar-Ziv R., Godefroy J., Salman H. and Libchaber A. (2005) Toward an artificial cell based on gene expression in vesicles. Phys. Biol., 2, P1–P8. [DOI] [PubMed] [Google Scholar]
- 76. Noireaux V. and Libchaber A. (2004) A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA, 101, 17669–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Godino E., López J.N., Foschepoth D., Cleij C., Doerr A., Castellà C.F. and Danelon C. (2019) De novo synthesized Min proteins drive oscillatory liposome deformation and regulate FtsA-FtsZ cytoskeletal patterns. Nat. Commun., 10, 4969.doi: 10.1038/s41467-019-12932-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Blanken D., Foschepoth D., Serrão A.C. and Danelon C. (2020) Genetically controlled membrane synthesis in liposomes. Nat. Commun., 11, 4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dupin A. and Simmel F.C.. Signaling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits.doi: 10.1038/s41557-018-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Furusato T., Horie F., Matsubayashi H.T., Amikura K., Kuruma Y. and Ueda T. (2018) De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth. Biol., 7, 953–961. [DOI] [PubMed] [Google Scholar]
- 81. Song L., Hobaugh M.R., Shustak C., Cheley S., Bayley H. and Gouaux J.E. (1996) Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science, 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on reasonable request.


