ABSTRACT
We describe here the structure and organization of TnCentral (https://tncentral.proteininformationresource.org/ [or the mirror link at https://tncentral.ncc.unesp.br/]), a web resource for prokaryotic transposable elements (TE). TnCentral currently contains ∼400 carefully annotated TE, including transposons from the Tn3, Tn7, Tn402, and Tn554 families; compound transposons; integrons; and associated insertion sequences (IS). These TE carry passenger genes, including genes conferring resistance to over 25 classes of antibiotics and nine types of heavy metal, as well as genes responsible for pathogenesis in plants, toxin/antitoxin gene pairs, transcription factors, and genes involved in metabolism. Each TE has its own entry page, providing details about its transposition genes, passenger genes, and other sequence features required for transposition, as well as a graphical map of all features. TnCentral content can be browsed and queried through text- and sequence-based searches with a graphic output. We describe three use cases, which illustrate how the search interface, results tables, and entry pages can be used to explore and compare TE. TnCentral also includes downloadable software to facilitate user-driven identification, with manual annotation, of certain types of TE in genomic sequences. Through the TnCentral homepage, users can also access TnPedia, which provides comprehensive reviews of the major TE families, including an extensive general section and specialized sections with descriptions of insertion sequence and transposon families. TnCentral and TnPedia are intuitive resources that can be used by clinicians and scientists to assess TE diversity in clinical, veterinary, and environmental samples.
KEYWORDS: mobile genetic elements, genome evolution, antibiotic resistance, virulence, database, insertion sequence, integrons, plasmid-mediated resistance, transposition mechanisms, transposons
INTRODUCTION
Transposable elements (TE) are key facilitators of bacterial evolution and adaptation. They are central players in the emergence of antibiotic and heavy metal resistance and contribute to the transmission of virulence and pathogenic traits. Some TE can capture “passenger genes” (genes not involved in the transposition process) encoding these traits and transmit them to plasmids, where they accumulate and are then transferred within and between bacterial populations by conjugation. TE also contribute significantly to the ongoing reorganization of bacterial genomes, giving rise to new strains that are more adept at proliferating in clinical and agricultural environments, as well as in natural ecosystems.
Understanding TE nature, distribution, and activity is therefore an indispensable part of the struggle to cope with the public health crisis of multiple-antibiotic resistance (ABR) (1, 2). To understand the impact of TE on bacterial populations, it is essential to provide a detailed description and catalog of TE structures and diversity. The simplest TE, known as insertion sequences (IS), have a profound impact on genome organization and function (see references 3, 4, 5, 6, and 7) but do not themselves generally carry integrated passenger genes. There are a large number of significantly more complex TE (Fig. 1), which are arguably even more important in the global emergence of antibiotic resistance (ABR) and other virulence and pathogenicity traits. These are generically called transposons and may carry multiple passenger genes, including some of the most clinically important antibiotic resistance genes. Like IS, these TE are grouped into a number of distinct families with characteristic organizations (3). Their transposition activities facilitate the rapid spread of groups of antibiotic resistance genes and promote their horizontal transfer. Another important aspect of their impact is their ability to assemble passenger genes into resistance clusters (8, 9). While there appears to be a widespread appreciation that mobile plasmids are responsible for the spread of antibiotic resistance, it is less well known that IS and transposons are the conduits that transfer this information between chromosomes and plasmids.
FIG 1.
Structural arrangement of prokaryotic transposable elements. The TE is indicated by a pale-yellow horizontal bar at the top of each section. ORFs are shown as horizontal arrows with arrowheads indicating the direction of expression: purple, transposition-associated genes; red, antibiotic resistance genes; and green, other passenger genes. The inverted terminal repeats found at the ends of the majority of TE are shown as gray arrows, and the direct target repeats generally produced by insertion are indicated by small black arrows. (A) Insertion sequence (IS), a short DNA segment encoding only the mobilization protein (transposase, TnpA), flanked by two imperfect inverted repeats (IRs) and generally containing a short flanking directly repeated duplication (DR) on the target of insertion. (B) The tIS (transporter IS) is structurally similar to an IS but contains passenger genes. These are presently restricted to the IS1595 and IS66 families. (C) Compound transposons are formed by two IS in either a direct or an inverted orientation, flanking a variety of passenger genes, including those for antibiotic resistance. (D) Transposons are more heterogeneous structures and include different sets of transposition-related genes that are specific to each transposon family and multiple antibiotic resistances, virulence, and other passenger genes. This is an example of a Tn3 family transposon with transposon, tnpA, and resolvase genes, tnpR.
There are a number of other bioinformatics resources that cover aspects of prokaryotic TE biology. These include databases for TE passenger genes, such as antibiotic resistance (CARD [10] and ARDB [11]) or toxin/antitoxin gene pairs (TADB [12] and TASmania [13]), as well as the various classes of TE themselves, such as insertion sequences (ISfinder [14]), integrons (INTEGRALL [15]), integrative conjugative elements (ICE; ICEberg [16, 17]), plasmids (PlasmidFinder [18]), or more general databases, which include a variety of these genome components (ACLAME [19–21]). However, there is a need for a resource that collects, compares, and collates detailed information on the various different classes of TE that are responsible for the transmission of medically and economically important passenger genes in an intuitive and accessible way.
Here, we describe TnCentral (https://tncentral.proteininformationresource.org/ [or the mirror link at https://tncentral.ncc.unesp.br/]), a database of detailed structural and functional information on bacterial TE. In addition, TnCentral provides access to TnPedia (https://tnpedia.fcav.unesp.br/), a comprehensive encyclopedia describing the current state of our knowledge of the biology of IS and transposons. Together, TnCentral and TnPedia provide a detailed description of TE diversity with easy-to-understand graphics outputs that are accessible to users without significant bioinformatic knowledge. These databases allow users to rapidly analyze the landscape of TE in genomes (chromosomes and plasmids) isolated from clinical, veterinary, and environmental samples.
RESULTS
TnCentral website content.
As of August 2021, TnCentral contains information on ∼400 TE. About half of these TE are Tn3-family transposons. The remainder are integrons, compound transposons, transposons from the Tn402, Tn554, and Tn7 families, and IS that are associated with TE or are part of compound transposons (see Table S1 in the supplemental material). These include TE with resistance to over 25 different classes of antibiotics and nine different heavy metals. The collection also contains TE that carry a toxin/antitoxin system for bacterial plasmid maintenance (22–24) and TE from xanthomonads carrying genes for plant pathogenicity. Although not considered per se as transposons, we have included the mobile integrons systems because of their important impact in shaping many transposons and their importance in the acquisition and dissemination of ABR.
The table displays the entire collection of TE at present in the database (August 2021), with columns indicating their TnCentral accession numbers, names, synonyms from the literature and/or the Transposon Registry (53), TE type, family group, host organism, and molecular source (e.g., plasmid or chromosome). If no information is provided in the molecular source column, the source is chromosomal or unknown. Download Table S1, XLSX file, 0.04 MB (38.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
TnCentral web portal.
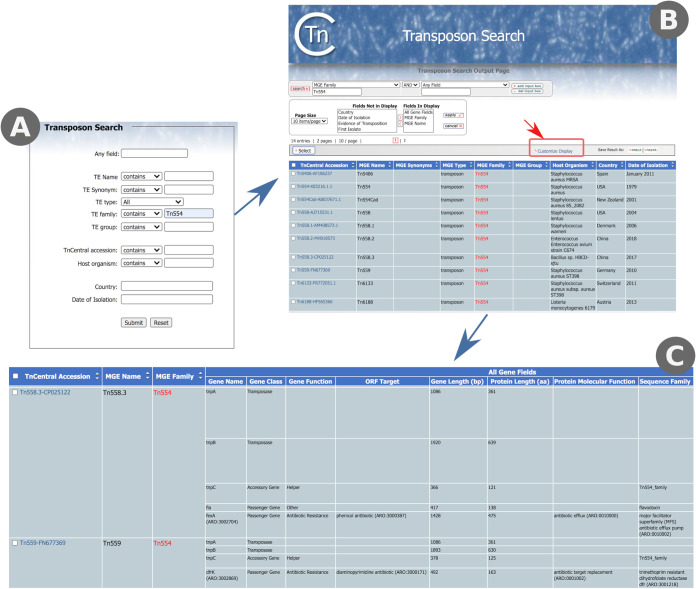
The TnCentral home page is designed to give the user easy access to the contents of TnCentral with a number of options (Fig. 2A), including: TnCentral Search (search of the TnCentral database), Sequence Search (BLAST-like search for sequence similarities in the database), Browse Tn List (view all TE in TnCentral), Tnfinder Software (access to downloadable scripts for identifying potential TE in sequence databases), Documentation (downloadable documentation for TnCentral), For Curators (detailed curation guidelines), TnPedia (TE Encyclopedia), Related Links, and Feedback.
FIG 2.
(A) TnCentral homepage showing clickable links to various TnCentral sections in the box on the left. (B) TnCentral search interface showing search choices for TE on the left and for transposition-related and passenger genes on the left.
TnCentral Search.
The interface provides a variety of search functions divided into two search types: Transposon Search and Gene Search (Fig. 2B).
(i) Transposon Search. The transposon collection can be searched using the transposon name; synonyms, which may have been used in the literature; the type of mobile genetic element (e.g., insertion sequence, transposon, or integron), the family and subgroup to which it belongs, the host organism, country of identification, and date of identification. The latter three search terms are intended for use in epidemiological tracking. These search terms result in a table that can be sorted, customized, and downloaded (see use case 1, below).
(ii) Gene Search. It is also possible to search for TE-associated genes by name, by class (transposase, accessory gene, or passenger gene), or by function (antibiotic resistance or heavy metal resistance) and to retrieve information on the transposons in which they are found (see use case 2, below).
Sequence Search.
Sequence search allows users to perform sequence similarity searches against the TnCentral database using BLAST (25, 26) (see use case 3, below). A text box for entering query sequences is provided. The BLAST tool automatically distinguishes between DNA and protein query sequences. BLAST parameters (e.g., maximum expect value, maximum number of results displayed, and scoring matrix for protein BLAST) can be customized using the menus in the options box, which is located below the query entry box. The page also provides links to several other BLAST interfaces where searches can be initiated. These include the ISfinder (https://isfinder.biotoul.fr/blast.php), NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Comprehensive Antibiotic Resistance (CARD [https://card.mcmaster.ca/analyze/blast]), and Toxin-Antitoxin (TADB [https://bioinfo-mml.sjtu.edu.cn/TADB2/]) databases.
The sequence search results display is currently quite basic, so it is best suited for simple searches such as querying with a transposon sequence to find related transposons in the database or querying with a protein sequence to find transposons in the database that encode the protein. In the future, we plan to enhance the display to facilitate more complex searches such as analyzing the transposon content of a plasmid or genome (see the Discussion). The results table, which is sorted by score, provides the TnCentral Accession for each significant hit hyperlinked to the corresponding entry page, information such as the transposon or protein name, and an alignment column. In the alignment column, the query sequence is represented by the width of the column, and the hits are shown as colored bars positioned according to the portion of the query sequence to which they align. The color of the bar indicates the strength of the match (red indicates strongest; black indicates weakest). Clicking on the alignment column brings up a page that shows the alignment in detail, as well as statistics such as the score and E value.
Browse Tn List.
The browse Tn list option allows the user to browse the entire TnCentral database.
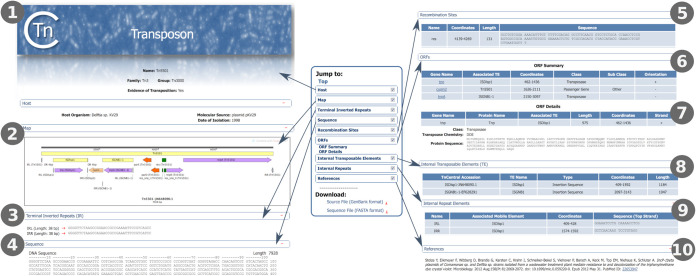
Transposon entry page.
All of the search and browse options provide links to entry pages for each TE (Fig. 3), which provide detailed information about TE features and origins. The page includes various sections: (i) host information (the host species, strain, and plasmid/chromosome in which the transposon was found, as well as the date and geographic location of the isolate) (Fig. 3, section 1); (ii) a graphic representation of the annotated sequence with color-coded features (Fig. 3, section 2); (iii) terminal inverted repeats (Fig. 3, section 3); (iv) the DNA sequence (Fig. 3, section 4); (v) internal recombination sites (e.g., res sites), including their coordinates, length, and DNA sequence (Fig. 3, section 5); (vi) open reading frame (ORF) summary, which includes all protein coding genes in the order in which they appear, 5′–3′, in the TE sequence, the element with which they are associated (important for nested TE in which one TE is inserted into another), their coordinates, their class (e.g., transposase, accessory gene, passenger gene) and subclass (e.g., antibiotic resistance or heavy metal resistance), and their relative orientation within the TE (Fig. 3, section 6); (vii) a detailed ORF description, including the amino acid sequence (Fig. 3, section 7); (viii) if applicable, a table of internal transposable elements (TE inserted in the main element), including the name, type, location, and length (Fig. 3, section 8); (ix) if applicable, a table of internal repeats (i.e., repeat elements, other than the terminal inverted repeats, that are found within the TE), including the associated TE, coordinates, and DNA sequence (Fig. 3, section 9); and (x) bibliographic references with direct links to PubMed (Fig. 3, section 10) (27). Each section can be collapsed using a button on the right-hand side of the section heading. Sections can be viewed either by scrolling down on the page or by clicking on the section name in the menu located on the left side of the page. Sequence files in FASTA and GenBank format can be downloaded using the links on the left side of the page under the menu.
FIG 3.
TnCentral TE entry page. Numbers 1 to 10 indicate various sections of the entry page (see the text for details).
Tnfinder software.
This section provides three user-downloadable scripts written in-house to identify transposons. These scripts help users to screen data sets containing large numbers of genomic sequences using their own servers to identify potential candidate transposons, which can then be manually curated.
The Tn3 Transposon Finder (Tn3_finder) performs the automatic prediction of transposable elements of the Tn3 family in bacteria and archaea. It compares user-provided bacterial and archaeal genome sequences to custom Tn3 transposase and resolvase databases by BLAST alignments. The criteria for identifying potential transposon regions according to similarity, coverage, and distance values can be adjusted by the user. Additional ORFs that might be related to passenger genes are also predicted, and flanking regions can also be retrieved and analyzed. The automatic prediction results are written in report files and pre-annotated GenBank files to help in subsequent manual curation. Tn3_finder allows for the concurrent analysis of multiple genomes by multithreading.
Composite Transposon Finder (TnComp_finder) predicts the putative composite transposons in bacterial and archaeal genomes based on insertion sequence replicas in a relatively short span. It works by comparing nucleotide sequences from bacterial and archaeal genomes to a custom transposon database and identifying duplicated transposons in user-defined genomic regions from BLAST alignments. Similar to Tn3_finder, multithreaded analyses of multiple genomes are available, and the parameters for similarity, coverage, distance, and flanking regions can be adjusted by the user. The results are written in report files and pre-annotated GenBank files to help in subsequent manual curation.
Antibiotic Resistance Gene-associated IS Finder (ISAbR_finder) is an experimental program for the automatic prediction of antibiotic resistance genes associated with known IS elements derived from the ISfinder database and has yet to be tested extensively. It works by comparing IS nucleotide sequences from bacterial and archaeal genomes to a custom antibiotic resistance database based on the parsing of BLAST alignment results, using a number of parameters that can be customized by the user for stricter or more relaxed criteria and allowing multithreaded alignments of multiple genomes. ISAbR_finder also produces report files and pre-annotated GenBank files on which the recommended manual curation should be performed.
Documentation.
This section, which can be downloaded as a .pdf file, provides a short background description of transposons and TnCentral, together with a short description of the curation workflow and planned future developments.
For Curators.
This section provides a detailed description of the curation workflow used to generate the annotated TnCentral data.
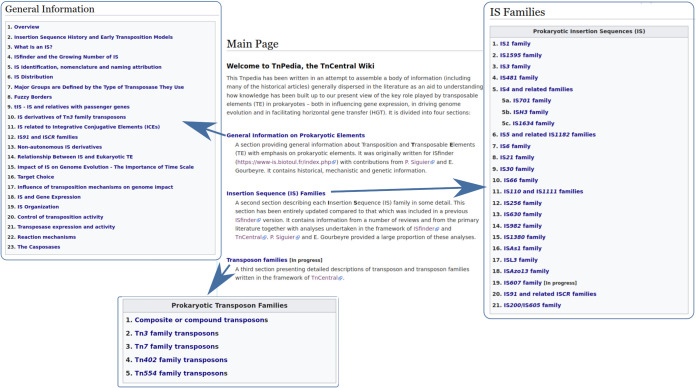
TnPedia.
TnCentral provides access from the homepage to TnPedia, an online knowledge base that contains information concerning transposition in prokaryotes. TnPedia was developed using MediaWiki (https://www.mediawiki.org) and can also be accessed directly (https://tnpedia.fcav.unesp.br/). It is structured into three main sections: general information, IS families, and transposon families (Fig. 4).
FIG 4.
Main sections of TnPedia, a TnCentral-related wiki compiling information on prokaryotic transposable elements. Only three of four sections (general information, IS families, and transposon families) are illustrated. A fourth section, the transposition glossary, is under construction.
The general information section provides a series of clickable sections with an extensive bibliography and direct links to the articles in PubMed. It includes a historical perspective, definitions, and descriptions of a variety of prokaryotic TE, the basic mechanisms involved in their movement, and the enzymes involved in these processes. It also contains information describing their impact on their host genomes and how their activities are controlled.
The IS families section consists of individual chapters describing each of the ∼25 IS families in detail and covers, where possible, the identification of the founding members, their organization, distribution, variability, and phylogenetic relationships; the regulation of their transposition; the impact on their host genomes; and their transposition mechanisms, including genetic, biochemical, and structural studies.
The transposon families section describes each transposon family with information similar to that included in the IS family descriptions but, in addition, includes a detailed description of their structures and the passenger genes that they may carry.
Examples of TnCentral use.
(i) Use case 1: comparing protein coding genes in Tn554 family members. The Tn554 family is a small family restricted to the Firmicutes. Members encode three genes—tnpA, tnpB, and tnpC—involved in transposition (28, 29) (https://tnpedia.fcav.unesp.br/index.php/Transposons_families/Tn554_family). TnpA and TnpB both exhibit a C-terminal motif that shares all of the important catalytic residues of a typical tyrosine site-specific recombinase (28, 29). Tn554-family transposons insert in a sequence-specific way into the DNA repair gene radC (30, 31) and can also be found in a circular form (32–36). To compare the protein coding genes in Tn554 family members side by side, we searched for Tn554 in the TE family field of the transposon search interface (Fig. 5A). Fourteen Tn554 family members were found (only 10 of which are shown in Fig. 5B). In order to perform a side-by-side comparison of the protein coding genes in these TE, we used the customize display option on the search results page, to add the “All Gene Fields” columns, which provide information about the protein coding genes, to the display and to remove several columns (e.g., host organism and country) (Fig. 5B). The results for two of the Tn554 transposons (Tn558.3 and Tn559) are shown in Fig. 5C. Both transposons have the three-part transposition module (tnpA, tnpB, and tnpC) characteristic of the family. However, the two transposons are quite diverse in their passenger genes. Tn558.3 has a gene called fla, which contains a flavodoxin-like domain, and the ABR gene fexA, which confers resistance to phenicol antibiotics. Tn559 has just a single passenger gene, the ABR gene dfrK, which confers resistance to diaminopyrimidine antibiotics. As shown in this example, the flexible search results page makes it easy to compare features across multiple transposons.
FIG 5.
Comparing protein coding genes in Tn554 family members. (A) TnCentral transposon search interface, showing a search for Tn554 in the TE family field. (B) Interface for customizing the columns in the search results display. Clicking on “customize display” (red box) opens the interface. (C) Partial Tn554 family search results after customization to show information on protein coding genes (All Gene Fields).
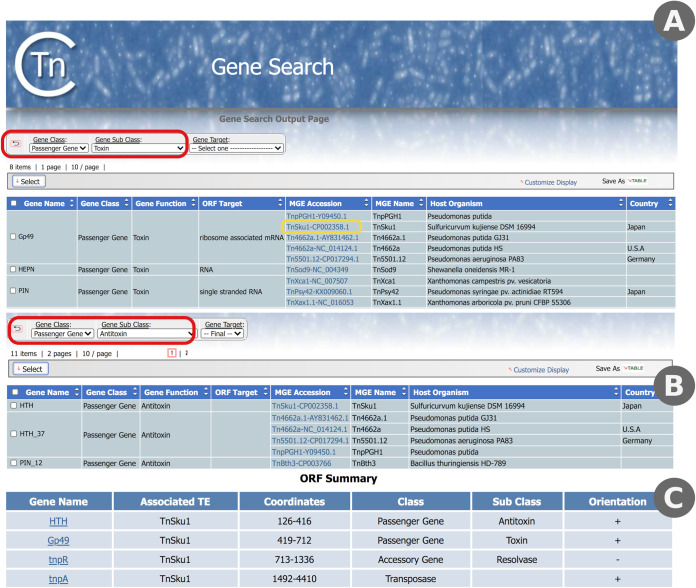
(ii) Use case 2: type II toxin/antitoxin systems in Tn3 transposons. Toxin/antitoxin (TA) systems are implicated in plasmid maintenance in bacterial populations (37). These systems are characterized by a stable toxin and an unstable antitoxin that binds to the toxin and inhibits its lethal effect. Loss of a plasmid carrying a TA system will lead to rapid depletion of the antitoxin, allowing the persistent toxin to kill the cell. Thus, only members of a population that retain the plasmid will survive. Recently, a set of Tn3-family transposons carrying TA systems were characterized and included in the TnCentral database (22). To explore these transposons, we used the TnCentral gene search function, selecting “Passenger Gene” from the gene class pulldown menu and “Toxin” from the gene subclass pulldown menu (Fig. 6A, red box). The search results included eight different toxin genes (Gp49, HEPN, PIN, PIN_3, abiEii, higB, parE, and zeta) found in 43 different transposons. Similarly, transposons carrying antitoxin genes were identified using the gene search function with the gene subclass menu set to “Antitoxin” (Fig. 6B, red box). There were 44 transposons carrying 11 different antitoxin genes. Combinations of toxin and antitoxin genes in individual transposons were examined by going to the ORF summary section of the entry pages for the TA transposons. For example, TnSku1 (Fig. 6B, yellow box; Fig. 6C) has a Gp49 toxin gene and an antitoxin gene containing an HTH domain (referred to as HTH). Most transposons have a single toxin/antitoxin gene pair except for TnXca1, which has two TA pairs, and Tn5501.5, which has a parD antitoxin gene and no toxin gene. The majority of Tn5501 derivatives in TnCentral have a parE toxin gene as well as the parD antitoxin, suggesting that Tn5501.5 may have undergone a deletion in the region containing parE (see Fig. S1 in the supplemental material).
FIG 6.
Exploring toxin/antitoxin genes in TnCentral. (A) Partial results of searching TnCentral for toxin genes. The settings used to obtain these results are shown in the red box. Links to entry pages for the TE carrying the indicated genes are provided in the MGE accession column (e.g., TnSku1-CP002358.1, yellow box). (B) Partial search results for antitoxin genes in TnCentral. Settings are shown in the red box. (C) ORF Summary section of the entry page for TnSku1-CP002358.1, showing the presence of a toxin/antitoxin gene pair (Gp49 toxin/HTH antitoxin).
Maps of Tn5501 and Tn5501.5 showing the loss of parE toxin gene in Tn5501.5. Feature color code: yellow, TE; purple, transposition genes; dark orange, toxin/antitoxin genes; light orange, other open reading frames; grey, repeat elements; green, recombination sites. Maps were created with SnapGene. Download FIG S1, TIF file, 0.3 MB (351.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
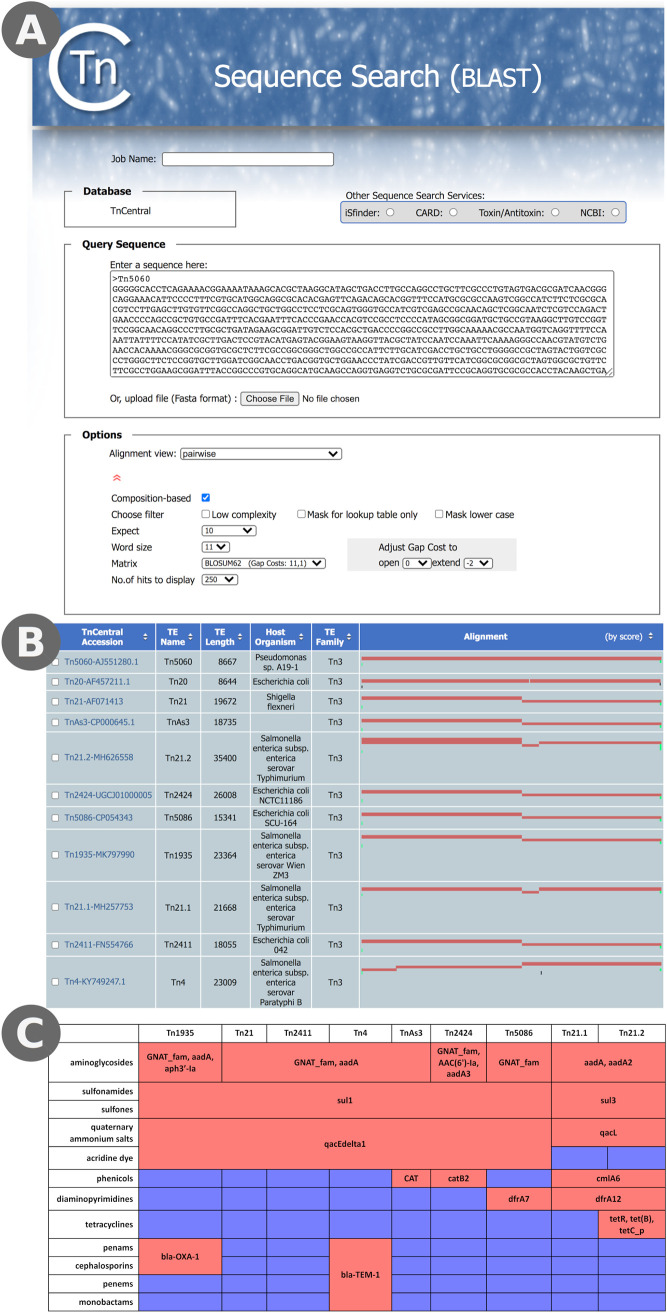
(iii) Use case 3: Tn21 and its relatives. Tn21 is the canonical member of a subfamily of Tn3 transposons that confers a variety of antibiotic resistances (38–40), and several analyses have proposed mechanisms to explain how Tn21 arose from simpler ancestor transposons (see, for example, references 40 and 41). Tn21 has a mercury resistance operon at the 5′ (left) end, a tnpA/tnpR transposition module at the 3′ (right) end, and a transposition-deficient integron (In2) carrying several ABR genes (a GCN5-related N-acetyltransferase [GNAT_fam], sul1, qacEΔ1, and aadA) in the middle (see Fig. S2). These ABR genes confer resistance to aminoglycosides, sulfones, sulfonamides, quaternary ammonium salts, and acridine dye. More recently, a transposon that lacks the integron insertion but is otherwise identical to Tn21 (the hypothetical Tn21 backbone Tn21Δ in reference 40) was discovered (42). This transposon, Tn5060, was proposed to be the ancestor of Tn21 (42). Tn21 also has numerous relatives that carry different combinations of antibiotic resistance genes within and outside the integron. To explore the Tn21 subfamily, we performed a TnCentral sequence search (BLAST) using the putative ancestral Tn5060 sequence (Fig. 7A). In addition to Tn5060 itself, we identified 10 transposons in the database (Tn20, Tn21, Tn21.1, Tn21.2, Tn5086, Tn2411, Tn2424, Tn4, Tn1935, and TnAs3; Fig. S2) that contain all (or nearly all) of the Tn5060 sequence. With the exception of Tn20, which is almost identical to Tn5060 (99.5%), these transposons have two or more discontinuous subregions that align to Tn5060. This suggests that these transposons arose from Tn5060 via the insertion of other sequences. For example, Tn21 has two subregions that align with the Tn5060 sequence: bases 1 to 4633 of Tn5060 align with bases 1 to 4633 of Tn21 (Fig. 7B, left red bar in the alignment column for Tn21) and bases 4629 to 8667 of Tn5060 align with bases 15634 to 19635 of Tn21 (Fig. 7B, right red bar in the alignment column for Tn21). The region of Tn21 from 4633 to 15633 does not align with Tn5060 because it contains an insertion of the In2 integron in the urfM gene (see Fig. S2A and E).
FIG 7.
Analysis of ABR in Tn21 Relatives. (A) TnCentral sequence search using the sequence of Tn5060, the proposed ancestor of Tn21, as a query. The query entry box and the customizable BLAST parameters are shown. The display of the parameters can be hidden/shown by clicking on the red double arrowhead. (B) Sequence search results. The query sequence is represented by the width of the alignment column. The red bars represent regions of the matched transposons that are highly similar to regions of Tn5060. (C) ABR genes and targeted antibiotic classes in Tn21 relatives. Red shading in the table cells indicates that the transposon carries at least one gene targeting the antibiotic class; blue shading indicates that it does not. The ABR genes found in each transposon are indicated in the table cells.
Maps of Tn21 and its relatives. The feature color code is the same as in Fig. S1. Maps were created with SnapGene. Note that the different transposon derivatives are not to scale, but their individual lengths are included. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We compared the antibiotic resistance profiles of the 10 transposons by inspecting their TnCentral entry pages. Tn20, like Tn5060, carries no ABR genes. The other nine transposons carry ABR genes targeting aminoglycosides, sulfones, sulfonamides, and quaternary ammonium salts (Fig. 7C). Other resistances found in a subset of the six include acridine dye (Tn1935, Tn21, Tn2411, Tn4, TnAs3, Tn2424, Tn5086), carbapenams (Tn1935 and Tn4), cephalosporins (Tn1935 and Tn4), carbapenems (Tn4), monobactams (Tn4), phenicols (TnAs3, Tn2424, Tn21.1, Tn21.2), diaminopyrimidines (Tn5086, Tn21.1, Tn21.2), and tetracyclines (Tn21.2). Interestingly, in some cases where the transposons have resistances in common, they are conferred by different genes (Fig. 7C). For example, phenicol resistance is conferred by CAT in TnAs3, catB2 in Tn2424, and cmlA6 in Tn21.1 and Tn21.2. Similarly, sulfonamide and sulfone resistance is conferred by sul1 in all of the antibiotic-resistant family members except for Tn21.1 and Tn21.2, where resistances are conferred by sul3. Thus, even this closely related subfamily of transposons shows diversity in its antibiotic resistance genes. This is partially due to the flexibility of the integron to incorporate new antibiotic resistance gene cassettes but also to insertion of ABR-gene containing elements outside the integron region (e.g., Tn3.1 in Tn4; Fig. S2).
DISCUSSION
Here, we have described TnCentral, a user-friendly resource for exploration of prokaryotic TE. TnCentral provides a flexible search interface, TE-specific entry pages with intuitive graphics and detailed information about TE features, and a BLAST interface that allows users to identify TE that carry features of interest. As shown in the use cases, the flexible search results page makes it easy to compare features across multiple transposons, the detailed entry pages allow exploration of TE passenger genes (such as ABR genes), and the sequence search enables retrieval of TE with related sequences that could be used as a starting point for evolutionary analyses. Moreover, TnCentral provides access to Tnfinder software for locating candidate TE in sequence data and to TnPedia, a comprehensive review of the biology of selected TE families.
As discussed in the introduction, a variety of resources dedicated to aspects of prokaryotic TE biology currently exist. TnCentral’s unique contribution to this universe of resources lies in its coverage of a variety of TE (e.g., different transposon families and compound transposons with their associated IS and integrons) and its detailed focus on both core transposition genes and passenger genes of clinical, environmental, and economic importance. It has the additional feature of providing a clear graphic output for visualizing the often complex structures of TE.
The next step beyond annotation of individual TE is to annotate and visualize the TE content of prokaryotic chromosomes and plasmids. These studies are critical for understanding the propagation of high impact passenger genes, such as those that confer antibiotic resistance. Several tools that address this problem are available. For example, ISsaga (43), which is integrated into ISfinder, annotates IS present in user-provided sequences. Other software suites have been designed specifically to annotate IS in short read raw data (e.g., ISQuest [44], Transposon Insertion Finder [45], ISMapper [46], and panISa [47]) using preassembled libraries of TE and their components, while yet other approaches are based on ab initio prediction (e.g., OASIS [48], ISseeker [49], and ISEscan [50]) or they provide a comparative view of IS mobilization events (e.g., ISCompare [51]). These annotation tools are only as good as their underlying TE databases. ISfinder, which includes nearly 6,000 individual examples of IS classified in distinct families and subfamilies according to their transposition mechanism and structural organization, provides such a rigorous framework for IS, and has been incorporated into a number of annotation pipelines (e.g., ISsaga [43] and MobileElementFinder [52]). However, IS represent only a fraction of prokaryotic TE and, unlike transposons and integrons, they rarely carry passenger genes. We hope that TnCentral will become a benchmark for more complex TE as ISfinder is for IS.
TnCentral is an ongoing project, and we will continue to expand and update the content. In addition to the exporting annotated TE in GenBank format, we plan to make all files available in a SnapGene file format that will allow users to use SnapGene, a commercial software tool (with a free viewer version) for visualizing and documenting nucleotide sequences and their features, to analyze and explore them. We also intend to enhance the visualization of TnCentral Sequence Search (i.e., BLAST) results to better support the analysis of plasmid sequences that may carry a complex complement of TE. For example, we will improve the graphics to show the alignment of multiple hits (i.e., multiple transposons) along the query sequence, enable tooltips that will display the coordinates of the alignment when hovering over a hit in the graphical display, and display the features (e.g., passenger genes or repeat elements) that are included in each hit. Ultimately, we envision that TnCentral could be used to analyze the TE content of a collection of sequences, such as patient, veterinary, and environmental samples from an antibiotic resistance outbreak, to understand TE-driven evolution of the prokaryotic mobilome.
MATERIALS AND METHODS
Curation workflow.
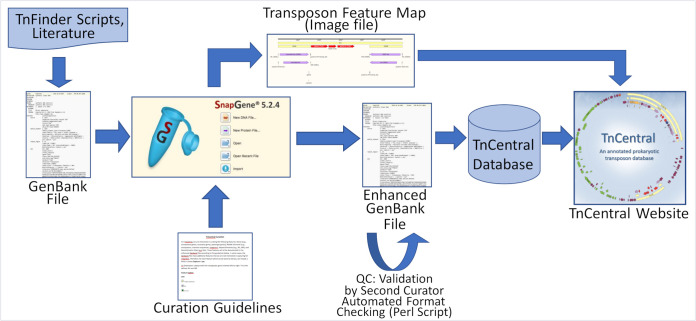
The TnCentral curation workflow is depicted in Fig. 8. Curation is performed by members of the TnCentral development team, as well as by graduate students in bioinformatics courses at Georgetown University Medical Center. TnFinder scripts are run against RefSeq and other sequence databases, and GenBank files potentially containing TE are retrieved. TE sequences are isolated and annotated using SnapGene. Features of interest (i.e., protein coding genes, TE, repeat elements, and recombination sites) are annotated according to detailed curation guidelines (provided in the “For Curators” section of TnCentral). Fully annotated features are saved in a SnapGene custom library. New transposon sequences can be searched against this library, enabling detection of features previously identified in other TEs. All annotated TE files are checked by a second curator. An enhanced GenBank file containing all annotations is exported from SnapGene and checked for common curation formatting errors using a custom Perl script. Detected errors are manually corrected in the SnapGene file, which is then exported as a revised enhanced GenBank file. Information from this GenBank file is used to populate the TnCentral database, which, in turn, serves as the backend for the TnCentral web portal. An image file showing a color-coded map of TE features is also exported from SnapGene and displayed on the TE entry page.
FIG 8.
TnCentral curation workflow (see the text for a description).
Although we have adhered to the standard nomenclature for transposons extracted from the literature, for the many transposons newly identified during TnCentral database-building, we have temporarily used names indicating their source. In all cases, the Transposon Registry (53) accession number is provided as a synonym. There is some ambiguity in the literature concerning class 1 integrons and members of the Tn402 transposon family. Class 1 integrons appear to be derivatives of this transposon family and include members with a range of Tn402 transposition genes with various degrees of completeness. We have therefore elected to include all class 1 integrons as members of the Tn402 family (see Table S1). ISfinder classification is used for the individual IS and, in the case of compound transposons, the group to which they are belong is defined by the flanking IS.
Properties of protein coding genes are annotated with cross-references to database or ontology identifiers whenever possible. Antibiotic resistance gene properties, including gene name, sequence family, antibiotic resistance mechanism, and target drug classes are annotated according to the Antibiotic Resistance Ontology (ARO), as presented in the Comprehensive Antibiotic Resistance Database (CARD) (10). The Pfam (54) and InterPro resources (55) are used to define sequence family information.
TnCentral website implementation.
TE features and sequence information are extracted from the enhanced GenBank files. TE feature information is used for the search and the entry pages, and the TE DNA and protein sequence information are used for the sequence search and display. The extracted data are loaded into the TnCentral database, implemented using MySQL. The website is built on a Linux server with Apache, and the web application is built on Perl CGI. Apache Lucene is used to index the data for flexible and fast search and retrieval. JavaScript is used for the interactive web interface and display. BLAST is used for similarity search.
ACKNOWLEDGMENTS
We thank John Dekker (NIAID, NIH, Bethesda, MD), Fred Dyda and Alison Hickman (NIDDK, NIH, Bethesda, MD), Patricia Siguier (CNRS, Toulouse, France), Susu He (Nanjing University Medical School, Nanjing, China), Laurence van Melderen (Université Libre de Bruxelles, Brussels, Belgium), and Gipsi Lima-Mendez and Bernard Hallet (Université de Louvain la Neuve, Louvain la Neuve, Belgium) for helpful discussions. We also thank Ben Glick (University of Chicago, SnapGene) for his help with the SnapGene software, the student curators at Georgetown University Medical Center for their contributions to the annotation process, and the Protein Information Resource (University of Delaware, Georgetown University Medical Center) for informatics support and by institutional resources. This research was also supported by resources supplied by the Center for Scientific Computing (NCC/GridUNESP) of the São Paulo State University (UNESP).
This project was supported by the U.S. Department of Defense Global Emerging Infections Surveillance Branch (P0020_18_WR). The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained here are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army or the Department of Defense.
This article is dedicated to the memory of Erik Snesrud, who was instrumental in initiating this work but, sadly, was unable to see it completed.
Footnotes
This article is a direct contribution from Michael Chandler, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Alessandra Carattoli, Sapienza University of Rome; Laurent Poirel, University of Fribourg; and Julian Parkhill, Department of Veterinary Medicine.
Citation Ross K, Varani AM, Snesrud E, Huang H, Alvarenga DO, Zhang J, Wu C, McGann P, Chandler M. 2021. TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. mBio 12:e02060-21. https://doi.org/10.1128/mBio.02060-21.
Contributor Information
Mick Chandler, Email: mc2126@georgetown.edu.
Susan Gottesman, National Cancer Institute.
REFERENCES
- 1.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J, Infectious Diseases Society of America . 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Welcome Foundation HM Government UK, London, United Kingdom. [Google Scholar]
- 3.Craig NL (ed). 2015. Mobile DNA III. American Society of Microbiology, Washington, DC. [Google Scholar]
- 4.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. doi: 10.1128/MMBR.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman’s guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 6.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 8.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. 2016. Mechanisms of evolution in high-consequence drug resistance plasmids. mBio 7:e01987-16. doi: 10.1128/mBio.01987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Pop M. 2009. ARDB: Antibiotic Resistance Genes Database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Y, Wei Y, Shen Y, Li X, Zhou H, Tai C, Deng Z, Ou H-Y. 2018. TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res 46:D749–D753. doi: 10.1093/nar/gkx1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akarsu H, Bordes P, Mansour M, Bigot D-J, Genevaux P, Falquet L. 2019. TASmania: a bacterial toxin-antitoxin systems database. PLoS Comput Biol 15:e1006946. doi: 10.1371/journal.pcbi.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 16.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou H-Y. 2012. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Li X, Xie Y, Bi D, Sun J, Li J, Tai C, Deng Z, Ou H-Y. 2019. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res 47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leplae R, Lima-Mendez G, Toussaint A. 2010. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res 38:D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leplae R, Hebrant A, Wodak SJ, Toussaint A. 2004. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res 32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leplae R, Lima-Mendez G, Toussaint A. 2006. A first global analysis of plasmid encoded proteins in the ACLAME database. FEMS Microbiol Rev 30:980–994. doi: 10.1111/j.1574-6976.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- 22.Lima-Mendez G, Oliveira Alvarenga D, Ross K, Hallet B, Van Melderen L, Varani AM, Chandler M. 2020. Toxin-antitoxin gene pairs found in Tn3 family transposons appear to be an integral part of the transposition module. mBio 11:e00452-20. doi: 10.1128/mBio.00452-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szuplewska M, Czarnecki J, Bartosik D. 2014. Autonomous and non-autonomous Tn3-family transposons and their role in the evolution of mobile genetic elements. Mob Genet Elements 4:1–4. doi: 10.1080/2159256X.2014.998537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM. 2016. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol 33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, Holmes JB, Kim S, Kimchi A, Kitts PA, Lathrop S, Lu Z, Madden TL, Marchler-Bauer A, Phan L, Schneider VA, Schoch CL, Pruitt KD, Ostell J. 2019. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 47:D23–D28. doi: 10.1093/nar/gky1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy E, Huwyler L, de Freire Bastos MC. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J 4:3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastos MC, Murphy E. 1988. Transposon Tn554 encodes three products required for transposition. EMBO J 7:2935–2941. doi: 10.1002/j.1460-2075.1988.tb03152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krolewski JJ, Murphy E, Novick RP, Rush MG. 1981. Site specificity of the chromosomal insertion of Staphylococcus aureus transposon Tn554. J Mol Biol 152:19–33. doi: 10.1016/0022-2836(81)90093-0. [DOI] [PubMed] [Google Scholar]
- 31.Murphy E, Reinheimer E, Huwyler L. 1991. Mutational analysis of att554, the target of the site-specific transposon Tn554. Plasmid 26:20–29. doi: 10.1016/0147-619x(91)90033-s. [DOI] [PubMed] [Google Scholar]
- 32.Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin a and related compounds including dalfopristin. Antimicrob Agents Chemother 46:2337–2343. doi: 10.1128/AAC.46.8.2337-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother 49:813–815. doi: 10.1128/AAC.49.2.813-815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:3475–3477. doi: 10.1128/AAC.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Li X-Y, Schwarz S, Yang M, Zhang S-M, Hao W, Du X-D. 2019. Tn6674 is a novel enterococcal optrA-carrying multiresistance transposon of the Tn554 family. Antimicrob Agents Chemother 63:e00809-19. doi: 10.1128/AAC.00809-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution, and function. Crit Rev Biochem Mol Biol 46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 38.De La Cruz F, Grinsted J. 1982. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol 151:222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grinsted J, de la Cruz F, Schmitt R. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24:163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- 40.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. doi: 10.1128/MMBR.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka M, Yamamoto T, Sawai T. 1983. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol 153:1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kholodii G, Mindlin S, Petrova M, Minakhina S. 2003. Tn5060 from the Siberian permafrost is most closely related to the ancestor of Tn21 prior to integron acquisition. FEMS Microbiol Lett 226:251–255. doi: 10.1016/S0378-1097(03)00559-7. [DOI] [PubMed] [Google Scholar]
- 43.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biswas A, Gauthier DT, Ranjan D, Zubair M. 2015. ISQuest: finding insertion sequences in prokaryotic sequence fragment data. Bioinformatics 31:3406–3412. doi: 10.1093/bioinformatics/btv388. [DOI] [PubMed] [Google Scholar]
- 45.Nakagome M, Solovieva E, Takahashi A, Yasue H, Hirochika H, Miyao A. 2014. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics 15:71. doi: 10.1186/1471-2105-15-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treepong P, Guyeux C, Meunier A, Couchoud C, Hocquet D, Valot B. 2018. panISa: ab initio detection of insertion sequences in bacterial genomes from short read sequence data. Bioinformatics 34:3795–3800. doi: 10.1093/bioinformatics/bty479. [DOI] [PubMed] [Google Scholar]
- 48.Robinson DG, Lee M-C, Marx CJ. 2012. OASIS: an automated program for global investigation of bacterial and archaeal insertion sequences. Nucleic Acids Res 40:e174. doi: 10.1093/nar/gks778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams MD, Bishop B, Wright MS. 2016. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb Genom 2:e000062. doi: 10.1099/mgen.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Z, Tang H. 2017. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33:3340–3347. doi: 10.1093/bioinformatics/btx433. [DOI] [PubMed] [Google Scholar]
- 51.Mogro EG, Ambrosis N, Lozano MJ. 2020. Easy identification of insertion sequence mobilization events in related bacterial strains with ISCompare. BioRxiv https://www.biorxiv.org/content/10.1101/2020.10.16.342287v4. [DOI] [PMC free article] [PubMed]
- 52.Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, Petersen TN. 2021. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J Antimicrob Chemother 76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tansirichaiya S, Rahman MA, Roberts AP. 2019. The transposon registry. Mob DNA 10:40. doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong S-Y, Finn RD. 2019. InterPro in 2019: improving coverage, classification, and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table displays the entire collection of TE at present in the database (August 2021), with columns indicating their TnCentral accession numbers, names, synonyms from the literature and/or the Transposon Registry (53), TE type, family group, host organism, and molecular source (e.g., plasmid or chromosome). If no information is provided in the molecular source column, the source is chromosomal or unknown. Download Table S1, XLSX file, 0.04 MB (38.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Maps of Tn5501 and Tn5501.5 showing the loss of parE toxin gene in Tn5501.5. Feature color code: yellow, TE; purple, transposition genes; dark orange, toxin/antitoxin genes; light orange, other open reading frames; grey, repeat elements; green, recombination sites. Maps were created with SnapGene. Download FIG S1, TIF file, 0.3 MB (351.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Maps of Tn21 and its relatives. The feature color code is the same as in Fig. S1. Maps were created with SnapGene. Note that the different transposon derivatives are not to scale, but their individual lengths are included. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.