Abstract
Novel biomarkers for HCC surveillance in cirrhotic patients are urgently needed. Exosomes and their lipid content in particular, represent potentially valuable noninvasive diagnostic biomarkers. We isolated exosomes from plasma of 72 cirrhotic patients, including 31 with HCC. Exosomes and unfractionated plasma were processed for untargeted lipidomics using ultra-high-resolution mass spectrometry. A total of 2,864 lipid species, belonging to 52 classes, were identified. Both exosome fractionation and HCC diagnosis had significant impact on the lipid profiles. Ten lipid classes were enriched in HCC exosomes compared to non-HCC exosomes. Dilysocardiolipins were detected in 35% of the HCC exosomes but in none of the non-HCC exosomes (p<0.001). Cardiolipins and sphingosines had the highest differential effects (fold change of 133.08, q=0.001 and 38.57, q<0.001, respectively). In logistic regression analysis, high abundances of exosomal sphingosines, dilysocardiolipins, lysophosphatidylserines and (O-acyl)-1-hydroxy fatty acids were strongly associated with HCC (OR [95% CI]: 271.1 [14.0–5251.9], p<0.001; 46.5 [2.3–939.9], p=0.012; 14.9 [4.3–51.2), p<0.001; 10.3 [3.2–33.1], p<0.001). Four lipid classes were depleted in HCC exosomes compared to non-HCC exosomes. In logistic regression analysis, lack of detection of sulfatides and acylGlcSitosterol esters was strongly associated with HCC (OR [95% CI]: 215.5 [11.5–4035.9], p<0.001; 26.7 [1.4–528.4], p=0.031). These HCC-associated changes in lipid composition of exosomes reflected alterations in glycerophospholipid metabolism, retrograde endocannabinoid signaling and ferroptosis. In conclusion, this study identified candidate biomarkers for early detection of HCC as well as altered pathways in exosomes that may contribute to tumor development and progression.
Keywords: liver cancer, early detection, screening, exosomes, lipids
Introduction
Liver cancer is the second most common cause of cancer-related deaths worldwide. Hepatocellular carcinoma (HCC) represents 70–90% of all liver cancers. HCC has a poor prognosis with 5 year survival rate below 20% (1) and surveillance in high-risk subjects is a promising approach to reduce mortality. Professional societies such as the American Association for the Study of Liver Diseases, recommend HCC surveillance in patients with cirrhosis (2). Surveillance in cirrhotic patients with ultrasound and serum alpha-fetoprotein (AFP) is commonly used. However, HCC surveillance remains underused in clinical practice, leading to high proportion of late stage detection (3) and both ultrasound and AFP lack sensitivity and specificity (4). Thus, there is a need for innovative approaches to promote HCC surveillance in patients with cirrhosis and for novel blood biomarkers to complement imaging.
Exosomes are membrane-bound nanovesicles (30–150nm) that contain various molecular components such as proteins, lipids and nucleic acids. They are present in all body fluids such as plasma, ascites and urine. Exosomes represent potentially valuable noninvasive diagnostic biomarkers, therapeutic targets and drug carriers (5, 6). They are important players in cancer growth, metastasis and angiogenesis, and therefore important mediators of cancer progression (7–9). Furthermore, exosomes modulate inflammation and downregulate antitumor immunity (10–13). While still in its infancy, the clinical application of exosomes to cancer detection has shown some promise (14, 15). With the rise of interest in exosomes and “omics” studies, databases such as Vesiclepedia have surfaced to track studies and their downstream analyses such as mRNA, miRNA, protein and lipids (16). The most common type of downstream analysis is proteomics followed by mRNA profiling (17–20). More recent “‘omics” studies have also investigated the lipid and metabolite content of exosomes (21–23).
Our study aimed to identify using ultra-high resolution mass spectrometry, differences in lipids in circulating exosomes of patients with cirrhosis and HCC vs. patients with cirrhosis without HCC, evaluate their potential utility as candidate biomarkers for HCC early detection and predict biological alterations affected by these changes.
Methods
STUDY PARTICIPANTS
This study includes 72 participants with cirrhosis (31 with HCC and 41 without HCC), matched by gender, age and etiology (Supplementary Table S1). Participants were recruited from Hepatology and multidisciplinary HCC clinics at Parkland Memorial Health and Hospital System and UT Southwestern Medical Center, using protocols previously described in detail (24, 25). The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center. Informed written consent was obtained from all participants. In brief, cirrhosis was diagnosed histologically, radiographically, or using non-invasive markers of fibrosis (26). All HCC diagnoses were confirmed using the American Association for the Study of Liver Disease criteria and Barcelona Clinic Liver Cancer (BCLC) staging system (27). All HCCs were treatment-naïve at time of recruitment, with blood samples collected typically within 30 days of diagnosis, processed and stored within 4 hours of collection. The same blood draws were used for both clinical lab measurements as part of routine clinical care and exosomes analysis. Heavy alcohol was defined as more than 1 or 2 drinks per day for women and men, respectively. Ascites and hepatic encephalopathy were classified as none, mild or controlled, and severe or uncontrolled. Mild or controlled ascites was defined as small ascites on imaging or adequately treated with diuretics. Mild or controlled hepatic encephalopathy was defined as adequately treated on lactulose and/or rifaximin. Patients requiring admission or other interventions, such as paracentesis, were determined to have severe or uncontrolled hepatic decompensation. MELD and Child Pugh scores were calculated per readily available clinical calculators.
EXOSOMES ISOLATION
Stored aliquots of 500μL EDTA plasma were thawed on ice and subjected to serial centrifugation to remove cellular debris. Before fractionation, 5μL of plasma was collected, snap frozen and stored at −80°C. The plasma samples and a blank sample of phosphate-buffered saline (PBS) were processed as previously described (28). Briefly, samples were diluted with equal parts of PBS and centrifuged in an Optima MAX-XP bench top ultracentrifuge with TLA-55 rotor (Beckman Coulter) in polypropylene tubes at 150,000g at 4°C for 2h. The pellets were washed with PBS and centrifuged again at 150,000g at 4°C for 2h. The resulting pellets were snap frozen and stored for lipidomics profiling at the Proteomics and Metabolomics Core at MD Anderson Cancer Center.
LIPIDOMIC PROFILING
Exosome pellets, unfractionated plasma and blank samples were subjected to Avanti SPLASH® LIPIDOMIX® Mass Spec Standard (330707) in methanol, 0.5μL of 10mM butylated hydroxytoluene in methanol, and 189.5μL of −80°C ethanol and vortexed. The contents of the mixture were then transferred to a Phenomenex Impact Protein Precipitation Plate (CE0–7565) and filtered through using a vacuum manifold. The sample tubes were rinsed with 200μL of ethanol that was subsequently used to elute residual lipids from the protein precipitation plate. The sample was transferred to a glass autosampler vial, dried using a centrifugal vacuum concentrator and reconstituted in 50μL ethanol. The injection volume was 10μL. Mobile phase A (MPA; weak) was 40:60 acetonitrile:0.1% formic acid in 10mM ammonium acetate. Mobile phase B (MPB; strong) was 90:8:2 isopropanol:acetonitrile:0.1% formic acid in 10mM ammonium acetate. The chromatographic method included a Thermo Fisher Scientific Accucore C30 column (2.6μm, 150 × 2.1mm) maintained at 40°C, autosampler tray chilling at 8°C, a mobile phase flowrate of 0.200 mL/min, and a gradient elution program as follows: 0–7 min, 20–55% MPB; 7–8 min, 55–65% MPB; 8–12 min, 65% MPB; 12–30 min, 65–70% MPB; 30–31 min, 70–88% MPB; 31–51 min, 88–95% MPB; 51–53 min, 95–100% MPB; 53–60 min, 100% MPB; 60–60.1 min 100–20% MPB; 60.1–70 min, 20% MPB. A Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer with heated electrospray ionization source was operated in data dependent acquisition mode, in both positive and negative ionization modes, with scan ranges of 150 – 677 and 675 – 1500 m/z. An Orbitrap resolution of 120,000 (FWHM) was used for MS1 acquisition and a spray voltages of 3,600 and −2900 V were used for positive and negative ionization modes, respectively. For MS2 and MS3 fragmentation a hybridized HCD/CID approach was used. Each sample was analyzed in both ionization modes using four 10μL injections making use of the two aforementioned scan ranges. Data were analyzed using Thermo Scientific LipidSearch software (version 4.2.23) and R scripts written in house. The peak areas (area-under-the-curve; AUC) identified in Thermo Scientific LipidSearch software were exported to Microsoft Excel.
STATISTICAL ANALYSIS
Demographic and clinical parameters were compared between HCC and non-HCC patients using two tailed t-test for continuous variables and Fisher test for categorical variables. Lipidomic AUC data were normalized by total signal. AUC peak data were filtered using the blank sample as background and full analysis was performed on analytes identified in at least 20% of the samples. The difference in AUCs between HCC and non-HCC samples was evaluated using Mann-Whitney U test adjusted by Benjamini-Hochberg method (29) to reduce the likelihood of false positives. Principal component analysis (PCA) was performed with the Euclidian-based distances matrix, generated in R using log10-transformed values. Pie graphs, volcano plots, and scatter plots were generated in Graph Prism 8.0.0. The list of lipid classes and lipid species found to be depleted or enriched in HCC exosomes vs non-HCC exosomes were analyzed using Lipid Pathway Enrichment Analysis (LIPEA). To determine the association between abundance of individual lipid classes and HCC, Firth logistic regression (30) was performed using the brglm package in R, with and without adjusting for age, gender and body mass index (BMI). For each lipid class enriched in HCC exosomes, we estimated the odds ratio (OR) and adjusted OR (AOR) for HCC with high abundance (Tertile T3). For each lipid class depleted in HCC exosomes, and often undetected in HCC exosomes, we estimated the OR and AOR for HCC with the lipid class as absent versus detected.
Results
EXOSOME ISOLATION FROM PLASMA OF CIRRHOSIS PATIENTS WITH OR WITHOUT HCC
We collected plasma from 72 patients with cirrhosis, 31 with HCC (HCC) and 41 without HCC (non-HCC). Detailed demographic and clinical parameters of the study participants are provided in Supplementary Table S1. Non-HCC patients were selected so that gender, age and etiology were not statistically different between HCC patients and non-HCC patients. The average age was 62.4 among HCC patients and 59 among non-HCC patients. Hepatitis C virus (HCV) was the most common etiology, representing 45% of HCC patients and 44% of non-HCC patients, followed by alcohol (23% in HCC patients and 24% in non-HCC patients). Child Pugh class A was the most common class, accounting for 58% of HCC patients and 68% of non-HCC patients. The majority of HCC patients (64%) had early stage disease, defined as BCLC stage 0 or A. As expected, patients with HCC had higher AFP levels than non-HCC patients (median 24 ng/mL versus 5 ng/mL, p<0.021).
Exosomes were isolated by ultracentrifugation, the gold standard method for exosome isolation. Unfractionated plasma samples and isolated exosomes were processed for lipidomics by mass spectrometry. While no significant differences in total lipids were detected in HCC versus non-HCC plasma samples (FC=1.13, p=0.21), a decrease in total lipids was observed in HCC exosomes compared to non-HCC exosomes (FC=0.64, p=0.005).
LIPIDOMIC PROFILING OF EXOSOMES
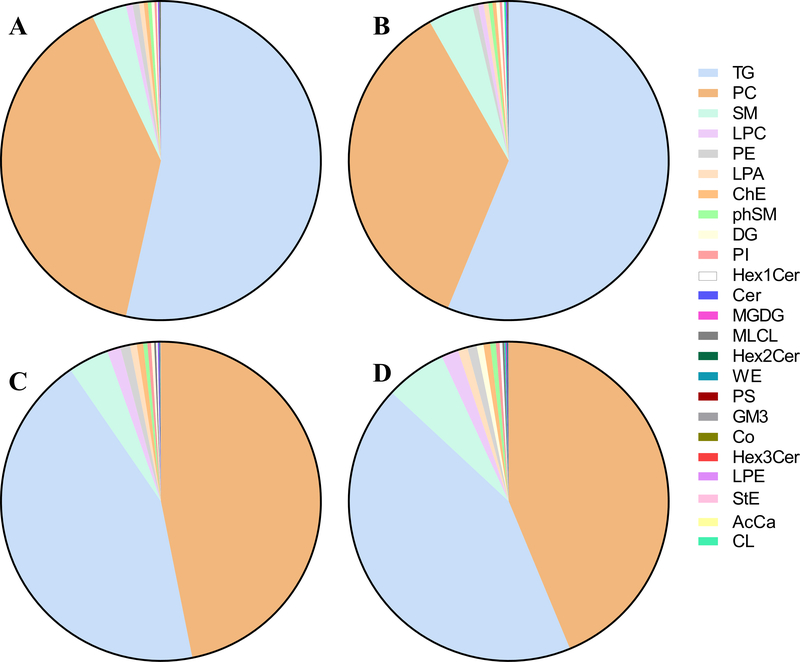
Untargeted lipidomics was performed on all isolated exosomes and unfractionated plasma samples. After filtering to remove signals under background and species detected in less than 20% of the samples, a total of 2,864 lipid species belonging to 52 classes were identified. Among the 2,864 species, 21 were detected only in exosomes and 75 only in plasma. The relative abundances of all 52 classes in exosomes HCC, exosomes non-HCC, plasma HCC and plasma non-HCC, are summarized in Supplementary Table S2. The two most abundant lipid classes were triglyceride (TG) and phosphatidylcholine (PC), with similar abundance of both classes in plasma (ratio TG/PC=0.93–0.98) but an enrichment of TG over PC in exosomes (ratio TG/PC=1.36–1.58). TG represented 53.5%−56.2% of all lipids in exosomes and 43.1%−43.5% in plasma. PC represented 35.5%−39.4% of all lipids in exosomes and 43.7%−46.8% in plasma. The third most abundant class was sphingomyelins (SM) (3.5%−6.2% of all lipids). The next three abundant classes were lysophosphatidylcholines (LysoPC), phosphatidylethanolamines (PE), and lysophosphatidic acids (LPA). Sphingosines phosphate (SPHP) and cyclic phosphatidic acids (cPA) were only detected in unfractionated plasma. Sulfatides (ST) and acylGlcSitosterol esters (AcHexSiE) were only detected in non-HCC plasma and exosomes while dilysocardiolipins (DLCL) were only detected in HCC plasma and exosomes. These results are represented in pie charts in Fig. 1.
Figure 1.
Distribution of lipid classes in plasma and exosomes from HCC and non-HCC patients. Pie diagrams showing the relative abundance of the 20 most abundant lipid classes in: (A) exosomes from non-HCC patients, (B) exosomes from HCC patients, (C) unfractionated plasma samples from the same non-HCC patients, and (D) unfractionated plasma samples from the same HCC patients. AcCa: Acyl carnitines; Cer: Ceramides; ChE: Cholesteryl esters; CL: Cardiolipins; Co: Coenzymes; DG: Diglycerides; GM3: Gangliosides; Hex1Cer: Hexosylceramides; Hex2Cer: Dihexosylceramides; Hex3Cer: Trihexosylceramides; LPA: Lysophosphatidic acids; LPC: Lysophosphatidylcholines; LPE: Lysophosphatidylethanolamines; MGDG: Monogalactosyldiacylglycerols; MLCL: Monolysocardiolipins; PC: Phosphatidylcholines; PE: Phosphatidylethanolamines; phSM: Phytosphingosines; PI: Phosphatidylinositols; PS: Phosphatidylserines; SM: Sphingomyelins; StE: Stigmasteryl esters; TG: Triglycerides; WE: Wax esters.
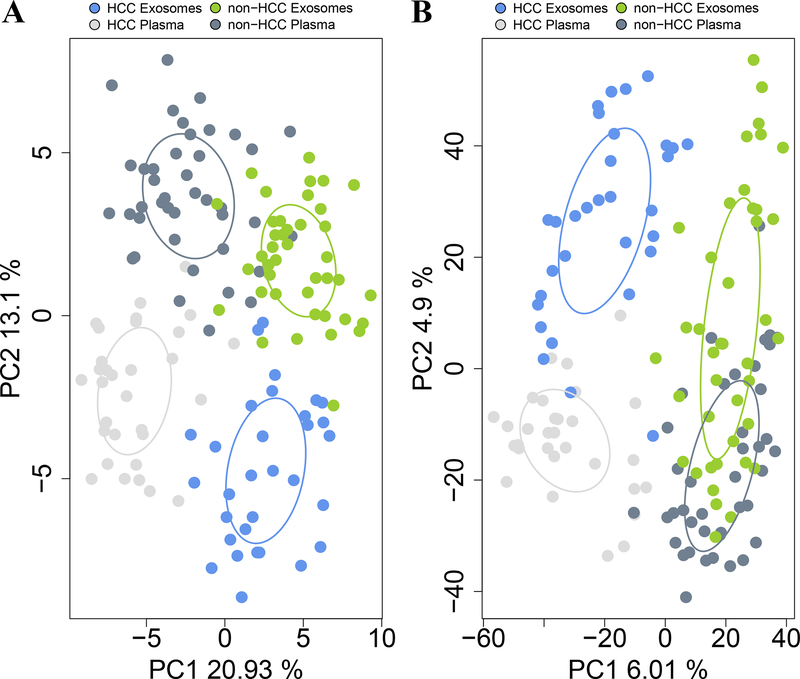
PCA was performed using the relative abundances of lipid classes (Fig. 2A) as well as lipid species (Fig. 2B). Both exosome fractionation and HCC diagnosis had a significant impact on lipid profiles. Lipid classes composition clearly separated exosomes from plasma (p<0.001) as well as HCC from non-HCC (p<0.001), while abundance of lipid species clearly separated HCC from non-HCC (p<0.001) and HCC exosomes from the other three groups (p<0.001).
Figure 2.
Principal Component analyses (PCAs) of plasma and exosomal lipid profiles from HCC and non-HCC patients shows four distinct clusters. PCAs were performed using Euclidean distances, based on (A) log10 relative abundance of detected lipid classes; and (B) log10 relative abundance of detected lipid species. Ellipses were drawn using the SD of point scores. p values were calculated using Permutational Multivariate Analysis of Variance (PERMANOVA) test.
HCC-ASSOCIATED CHANGES IN EXOSOMAL LIPID CLASSES AND SPECIES
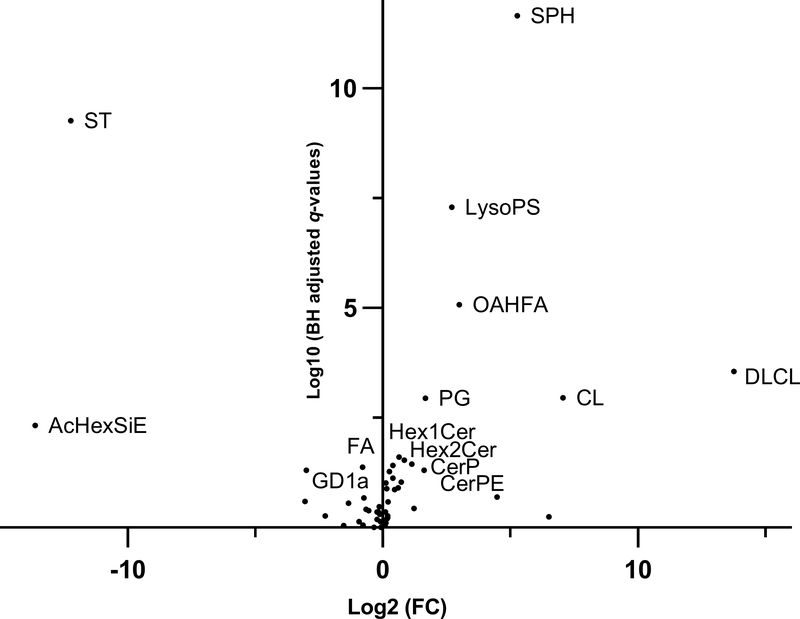
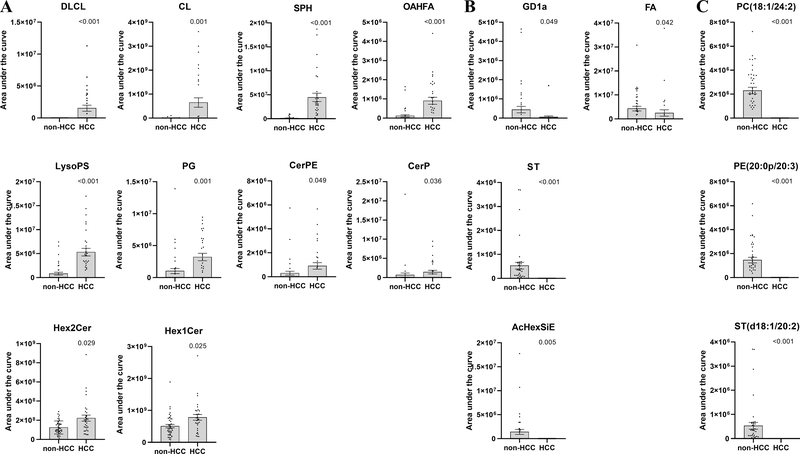
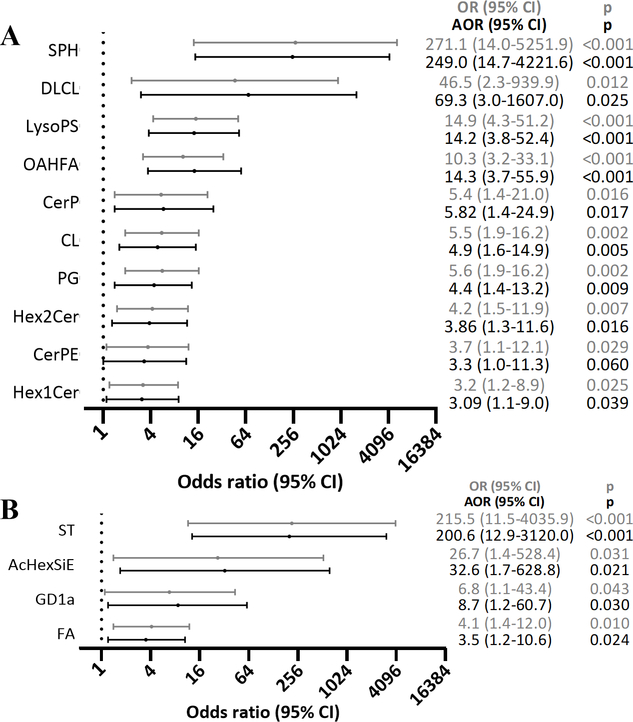
Ten lipid classes were significantly enriched in exosomes from HCC patients compared to exosomes in non-HCC patients (Fig. 3). DLCL were detected in 35% of the HCC exosomes but in none of the non-HCC exosomes (q<0.001). Cardiolipins (CL) and sphingosines (SPH) had the highest differential effects with fold changes of 133.08 (q=0.001) and 38.57 (q<0.001), respectively. The other enriched lipid classes were: (O-acyl)-1-hydroxy fatty acids (OAHFA) (FC=7.94, q<0.001), lysophosphatidylserines (LysoPS) (FC=6.49, q<0.001), phosphatidylglycerols (PG) (FC=3.16, q=0.001), ceramide phosphoethanolamines (CerPE) (FC=3.05, q=0.049), ceramides phosphate (CerP) (FC=2.18, q=0.036), dihexosylceramides (Hex2Cer) (FC=1.78, q=0.029) and hexosylceramides (Hex1Cer) (FC=1.54, q=0.025) (Fig. 4A). Lipid species DLCL(16:0/20:3), CL(18:2/16:0/16:0/24:1), CL(18:2/18:0/18:0/24:1), CL(18:2/16:0/20:4/24:1), SPH(t18:0), OAHFA(18:2/32:0), LysoPS(34:1), PG(18:0/18:2), PG(16:0/18:2), CerPE(d18:1/16:0), CerP(m17:0/22:6), Hex2Cer(d15:0/18:2), Hex2Cer(d14:0/20:4), Hex1Cer(t20:0/18:2), and Hex1Cer(d18:1/22:0) were major contributors of the enrichment of these 10 lipid classes in HCC exosomes. In logistic regression analysis, high abundance (Tertile T3) of SPH, DLCL, LysoPS and OAHFA were strongly associated with HCC (OR [95% CI]: 271.1 [14.0–5251.9], p<0.001; 46.5 [2.3–939.9], p=0.012; 14.9 [4.3–51.2), p<0.001; 10.3 [3.2–33.1], p<0.001). The association remained significant after adjusting for age, gender and BMI (Fig. 5A).
Figure 3.
Volcano plot of lipid classes in HCC exosomes vs non-HCC exosomes. Log10 of Benjamini-Hochberg (BH) adjusted p values (q) and Log2 of fold change (FC) for all lipid classes were fitted onto the plot. Names are shown for lipid classes enriched (FC >1.5, BH q <0.05) or depleted (FC <−1.5, BH q <0.05) in HCC exosomes vs non-HCC exosomes. AcHexSiE: acylGlcSitosterol esters; CerP: ceramides phosphate; CerPE: ceramide phosphoethanolamines; CL: cardiolipins; DLCL: dilysocardiolipins; FA: fatty acids; GD1a: gangliosides; Hex1Cer: hexosylceramides; Hex2Cer: dihexosylceramides; LysoPS: lysophosphatidylserines; OAHFA: (O-acyl)-1-hydroxy fatty acids; PG: phosphatidylglycerols; SPH: sphingosines; ST: sulfatides.
Figure 4.
Scatter plots of selected lipid abundance in HCC exosomes and non-HCC exosomes. (A) Lipid classes enriched in HCC exosomes compared to non-HCC exosomes; (B) Lipid classes depleted in HCC exosomes compared to non-HCC exosomes; (C) Selected lipid species depleted in HCC exosomes compared to non-HCC exosomes. Mean and SEM are shown and significance was determined by Mann-Whitney U test adjusted by Benjamini-Hochberg method. AcHexSiE: acylGlcSitosterol esters; CerP: ceramides phosphate; CerPE: ceramides phosphoethanolamine; CL: cardiolipins; DLCL: dilysocardiolipins; FA: fatty acids; GD1a: gangliosides; Hex1Cer: hexosylceramides; Hex2Cer: dihexosylceramides; LysoPS: lysophosphatidylserines; OAHFA: (O-acyl)-1-hydroxy fatty acids; PG: phosphatidylglycerols; SPH: sphingosines; ST: sulfatides
Figure 5.
Forest plots of lipid classes in HCC exosomes vs non-HCC exosomes. Firth logistic regression was performed to assess the associations, (A) between high (Tertile T3) abundance of enriched lipid classes named in Figure 3 and HCC, and (B) between depletion of the lipid classes named in Figure 3 and HCC. OR: odds ratio; AOR: OR adjusted for age, gender and BMI. AcHexSiE: acylGlcSitosterol esters; CerP: ceramides phosphate; CerPE: ceramide phosphoethanolamines; CL: cardiolipins; DLCL: dilysocardiolipins; FA: fatty acids; GD1a: gangliosides; Hex1Cer: hexosylceramides; Hex2Cer: dihexosylceramides; LysoPS: lysophosphatidylserines; OAHFA: (O-acyl)-1-hydroxy fatty acids; PG: phosphatidylglycerols; SPH: sphingosines; ST: sulfatides.
Four lipid classes were depleted in HCC exosomes compared to non-HCC exosomes (Fig. 3). Abundances of gangliosides (GD1a) and fatty acids (FA) lipid classes were lower in HCC exosomes compared to non-HCC exosomes (FC=−8.05, p=0.049 and FC=−1.75, p=0.042, respectively) (Fig. 4B). Lipid classes ST and AcHexSiE were undetectable in HCC exosomes but detected in 78% and 29% of non-HCC exosomes, respectively. Lipid species FA(20:4), GD1a(d18:1/18:0), GD1a(d18:1/16:0), ST(d18:1/20:2), and AcHexSiE(16:0) were major contributors of the depletion of these four lipid classes in HCC exosomes. In logistic regression analysis, lack of detection of ST and ACHexSiE was strongly associated with HCC (OR [95% CI]: 215.5 [11.5–4035.9], p<0.001; 26.7 [1.4–528.4], p=0.031). The association remained significant after adjusting for age, gender and BMI (Fig. 5B).
Some lipid species were detected in the majority of HCC exosomes but in none of the non-HCC exosomes. These included PC(18:3e/22:4), PC(16:1e/22:6), SM(d14:0/23:1), CerG3GNAc1(t18:0/24:1), WE(26:5/18:0), SPH(t18:0), GM3(d18:1/22:0), TG(25:0/16:0/17:0), MGDG(16:0/21:6), TG(18:0/14:0/16:0), and DG(20:0/16:0). In contrast, the following lipid species were detected in a majority of non-HCC exosomes but in none of the HCC exosomes: PC(20:2e/18:1), PE(16:0/20:4), Hex1Cer(d16:0/26:2), TG(18:1/10:3/18:3), PE(20:0p/18:1), PC(18:1/24:2), LPA(10:0), PE(20:0p/20:3), ST(d18:1/20:2) and SM(t18:1/24:3). Fig. 4C shows depletion of PC(18:1/24:2), PE(20:0p/20:3) and ST(d18:1/20:2).
BIOLOGICAL PATHWAYS ASSOCIATED WITH LIPID CHANGES IN HCC EXOSOMES
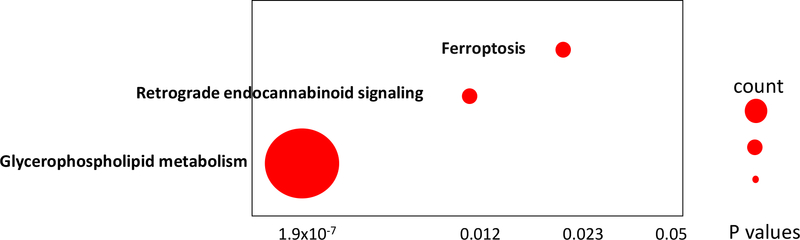
Pathway analysis using LIPEA identified three pathways corresponding to the observed changes in lipid composition in HCC exosomes compared to non-HCC exosomes. These included glycerophospholipid metabolism (p=1.9×10−7), retrograde endocannabinoid signaling (p=0.012) and ferroptosis (p=0.023) (Fig. 6).
Figure 6.
KEGG pathways associated with enriched or depleted lipids in HCC exosomes compared to non-HCC exosomes. These pathways were identified using LIPEA. Each dot represents one pathway and the size of dot depicts the metabolite and lipid counts in our data associated with the pathway.
Discussion
Most studies on the utility of exosomes for diagnosis have focused on proteins and miRNAs. In this study, we used ultra-high resolution mass spectrometry to identify lipid differences between exosomes isolated from cirrhotic patients with and without HCC. An important strength of our approach was that the Orbitrap technology permitted identification of thousands of lipids. A limitation of the workflow, however, is one that plagues the entire metabolomics field − annotation confidence. Although the combination of high scan speed and ultra-high resolution permitted acquisition of MS2 spectra for thousands of lipids, to perform database matching and subsequent annotation, confirmation of those annotations would ultimately require retention time matching, which is not possible using conventional untargeted profiling workflows. Nevertheless, our analysis elucidated a number of novel associations with both exosome isolation and HCC diagnosis having significant impact on lipid profiles.
Ten lipid classes were enriched in exosomes from cirrhotic patients with HCC compared to exosomes from cirrhotic patients without HCC. Among them, SPH had among the highest differential abundance. SPH(t18:0) was the main lipid responsible for this effect. The phosphorylated form of SPH, SPH-1P, has been shown to regulate hepatocyte exosome-dependent liver repair and regeneration (31). Furthermore, exosome adherence and internalization by hepatic stellate cells trigger SPH-1P dependent migration (32). SPH(d18:1)-1P has been proposed as a serum biomarker for HCC in patients with cirrhosis (33) and as a risk marker for HCC in a large population-based cohort (34). A second exosomal lipid class that had strong association with HCC was ST. ST was detected in 78% of non-HCC exosomes but undetectable in HCC exosomes. ST has specific anti-inflammatory and immunomodulatory properties (35). ST reactive-type II NKT cells are immunosuppressive in inflammatory liver diseases and attenuate alcoholic liver disease in mice (36). We also detected LysoPS at significantly higher levels in HCC exosomes compared to non-HCC exosomes. LysoPS(34:1) was the main form responsible for that increase but this specific LysoPS remains largely uncharacterized. Recent studies have revealed important roles for LysoPS signaling in T cell and macrophage functions (37, 38).
In addition to the potential use of the identified analytes in HCC early detection in cirrhotic patients, pathway analysis identified three pathways impacted by the observed changes in lipids and metabolites in HCC exosomes compared to non-HCC exosomes. Greater impact was predicted on glycerophospholipid metabolism. It was recently reported that mTORC2 promotes liver steatosis and HCC in mice, by altering glycerophospholipid synthesis (39). Other pathways included ferroptosis. Interestingly, ferroptosis, a new recognized way of non-apoptosis-regulated cell death characterized by the iron-dependent accumulation of lipid peroxides, shows promise in the therapy of cancer, especially in HCC (40, 41). Whether circulating exosomes by altering glycerophospholipid metabolism or ferroptosis contribute to the development of HCC should be further investigated.
The study has a number of limitations. The results were not independently validated in a separate cohort. The sample size of the discovery cohort was also small. In future studies, it would be valuable to determine the performance of the identified biomarkers by etiology and the complementarity of the identified biomarkers with imaging, ultrasound in particular, in detecting HCC in a prospective cohort of cirrhotic patients.
Altogether, this study identified candidate biomarkers for the detection of early stage HCC in at-risk cirrhosis patients and confirmed the promise of using exosomes as shown in the recently published analysis of purified extracellular vesicles combined to reverse transcription (42). In addition, this study identified pathways altered in HCC exosomes that may contribute to tumor development and progression.
Supplementary Material
Acknowledgments
Financial Support: This research was supported by the Cancer Prevention & Research Institute of Texas grant RP150587 (to LB), NIH grant U01 CA230694 (to AS) and NIH grant R01 CA222900 (to AS). The Metabolomics Core Facility is supported by Cancer Prevention and Research Institute of Texas grant RP130397 and NIH grants S10OD012304-01 and P30CA016672.
Abbreviations:
- AcHexSiE
acylGlcSitosterol esters
- AFP
alpha-fetoprotein
- AOR
adjusted odds ratio
- AUC
area under the curve
- BCLC
Barcelona Clinic Liver Cancer
- BMI
body mass index
- CerP
ceramides phosphate
- CerPE
ceramides phosphoethanolamine
- CI
confidence interval
- CL
cardiolipins
- cPA
cyclic phosphatidic acids
- DLCL
dilysocardiolipins
- FA
fatty acids
- FC
fold change
- GD1a
gangliosides
- GM3
gangliosides
- HCC
hepatocellular carcinoma
- Hex1Cer
hexosylceramides
- Hex2Cer
dihexosylceramides
- MS
mass spectrometry
- LPA
lysophosphatidic acids
- LysoPC
lysophosphatidylcholines
- lysoPS
lysophosphatidylserines
- MGDG
monogalactosyldiacylglycerols
- MPA
mobile phase A
- MPB
mobile phase B
- OAHFA
(O-acyl)-1-hydroxy fatty acids
- OR
odds ratio
- PBS
phosphate-buffered saline
- PC
phosphatidylcholines
- PCA
principal component analysis
- PE
phosphatidylethanolamines
- PG
phosphatidylglycerols
- SM
sphingomyelins
- SPH
sphingosines
- SPHP
sphingosines phosphate
- ST
sulfatides
- TG
triglycerides
- WE
wax esters
Footnotes
Conflict of Interest: Dr. Singal has served on advisory boards or as a consultant for Wako Diagnostics, Glycotest, Exact Sciences, Roche, GRAIL, Genentech, Bayer, Eisai, Exelixis, AstraZeneca, Bristol Myers-Squibb, and TARGET RWE.
References
- 1.Golabi P, Fazel S, Otgonsuren M, Sayiner M, Locklear CT, Younossi ZM. Mortality assessment of patients with hepatocellular carcinoma according to underlying disease and treatment modalities. Medicine (Baltimore) 2017;96:e5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marrero JA. Surveillance for Hepatocellular Carcinoma. Clin Liver Dis 2020;24:611–621. [DOI] [PubMed] [Google Scholar]
- 3.Wolf E, Rich NE, Marrero JA, Parikh ND, Singal AG. Use of Hepatocellular Carcinoma Surveillance in Patients With Cirrhosis: A Systematic Review and Meta-Analysis. Hepatology 2021;73:713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology 2018;154:1706–1718 e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R The biology and function of exosomes in cancer. J Clin Invest 2016;126:1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Z, Li D, Hou S, Zhu X. The cancer exosomes: Clinical implications, applications and challenges. Int J Cancer 2020;146:2946–2959. [DOI] [PubMed] [Google Scholar]
- 7.Ruivo CF, Adem B, Silva M, Melo SA. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res 2017;77:6480–6488. [DOI] [PubMed] [Google Scholar]
- 8.Othman N, Jamal R, Abu N. Cancer-Derived Exosomes as Effectors of Key Inflammation-Related Players. Front Immunol 2019;10:2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest 2016;126:1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbins PD, Dorronsoro A, Booker CN. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J Clin Invest 2016;126:1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurywchak P, Tavormina J, Kalluri R. The emerging roles of exosomes in the modulation of immune responses in cancer. Genome Med 2018;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol 2020;20:209–215. [DOI] [PubMed] [Google Scholar]
- 14.Cheng N, Du D, Wang X, Liu D, Xu W, Luo Y, et al. Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends Biotechnol 2019;37:1236–1254. [DOI] [PubMed] [Google Scholar]
- 15.Gulei D, Petrut B, Tigu AB, Onaciu A, Fischer-Fodor E, Atanasov AG, et al. Exosomes at a glance - common nominators for cancer hallmarks and novel diagnosis tools. Crit Rev Biochem Mol Biol 2018;53:564–577. [DOI] [PubMed] [Google Scholar]
- 16.Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 2019;47:D516–D519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest 2016;126:1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li A, Zhang T, Zheng M, Liu Y, Chen Z. Exosomal proteins as potential markers of tumor diagnosis. J Hematol Oncol 2017;10:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pocsfalvi G, Stanly C, Vilasi A, Fiume I, Capasso G, Turiak L, et al. Mass spectrometry of extracellular vesicles. Mass Spectrom Rev 2016;35:3–21. [DOI] [PubMed] [Google Scholar]
- 21.Luo X, An M, Cuneo KC, Lubman DM, Li L. High-Performance Chemical Isotope Labeling Liquid Chromatography Mass Spectrometry for Exosome Metabolomics. Anal Chem 2018;90:8314–8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penfornis P, Vallabhaneni KC, Whitt J, Pochampally R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer 2016;138:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skotland T, Sagini K, Sandvig K, Llorente A. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev 2020;159:308–321. [DOI] [PubMed] [Google Scholar]
- 24.Yopp AC, Mansour JC, Beg MS, Arenas J, Trimmer C, Reddick M, et al. Establishment of a multidisciplinary hepatocellular carcinoma clinic is associated with improved clinical outcome. Ann Surg Oncol 2014;21:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rich NE, John BV, Parikh ND, Rowe I, Mehta N, Khatri G, et al. Hepatocellular Carcinoma Demonstrates Heterogeneous Growth Patterns in a Multicenter Cohort of Patients With Cirrhosis. Hepatology 2020;72:1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Serag HB, Kanwal F, Feng Z, Marrero JA, Khaderi S, Singal AG, et al. Risk Factors for Cirrhosis in Contemporary Hepatology Practices-Findings From the Texas Hepatocellular Carcinoma Consortium Cohort. Gastroenterology 2020;159:376–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358–380. [DOI] [PubMed] [Google Scholar]
- 28.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3–22. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Statistical Methodology 1995;57:289–300. [Google Scholar]
- 30.Firth D. Bias Reduction of Maximum-Likelihood-Estimates. Biometrika 1993;80:27–38. [Google Scholar]
- 31.Nojima H, Freeman CM, Schuster RM, Japtok L, Kleuser B, Edwards MJ, et al. Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J Hepatol 2016;64:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Ding Q, Yaqoob U, de Assuncao TM, Verma VK, Hirsova P, et al. Exosome Adherence and Internalization by Hepatic Stellate Cells Triggers Sphingosine 1-Phosphate-dependent Migration. J Biol Chem 2015;290:30684–30696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y, Tie C, Wang Y, Bian D, Liu M, Wang T, et al. Upregulation of Serum Sphingosine (d18:1)-1-P Potentially Contributes to Distinguish HCC Including AFP-Negative HCC From Cirrhosis. Front Oncol 2020;10:1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepien M, Keski-Rahkonen P, Kiss A, Robinot N, Duarte-Salles T, Murphy N, et al. Metabolic perturbations prior to hepatocellular carcinoma diagnosis: Findings from a prospective observational cohort study. Int J Cancer 2021;148:609–625. [DOI] [PubMed] [Google Scholar]
- 35.Dasgupta S, Kumar V. Type II NKT cells: a distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics 2016;68:665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maricic I, Sheng H, Marrero I, Seki E, Kisseleva T, Chaturvedi S, et al. Inhibition of type I natural killer T cells by retinoids or following sulfatide-mediated activation of type II natural killer T cells attenuates alcoholic liver disease in mice. Hepatology 2015;61:1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanbhag K, Mhetre A, Khandelwal N, Kamat SS. The Lysophosphatidylserines-An Emerging Class of Signalling Lysophospholipids. J Membr Biol 2020;253:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagida K, Valentine WJ. Druggable Lysophospholipid Signaling Pathways. Adv Exp Med Biol 2020;1274:137–176. [DOI] [PubMed] [Google Scholar]
- 39.Guri Y, Colombi M, Dazert E, Hindupur SK, Roszik J, Moes S, et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 2017;32:807–823 e812. [DOI] [PubMed] [Google Scholar]
- 40.Nie J, Lin B, Zhou M, Wu L, Zheng T. Role of ferroptosis in hepatocellular carcinoma. J Cancer Res Clin Oncol 2018;144:2329–2337. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, et al. Ferroptosis: past, present and future. Cell Death Dis 2020;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun 2020;11:4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.