Abstract
The functionality of DNA biomacromolecules has been widely excavated, as therapeutic drugs, carriers, and functionalized modification derivatives. In this study, we developed a series of DNA tetrahedron nanocages (Td), via synchronous conjugating different numbers of i-(X) and therapeutic siRNA on four vertexes of tetrahedral DNA nanocage (aX-Td@bsiRNA, a+b = 4). This i-motif-conjugated Td exhibited good endosomal escape behaviours in A549 tumor cells, and the escape efficiency was affected by the number of i-motif. Furthermore, the downregulating mRNA and protein expression level of epidermal growth factor receptor (EGFR) caused by this siRNA embedded Td were verified in A549 cells. The tumor growth inhibition efficiency of the 2X-Td@2siRNA treated group in tumor-bearing mice was significantly higher than that of non-i-motif-conjugated Td@2siRNA (3.14-fold) and free siRNA (3.63-fold). These results demonstrate a general strategy for endowing DNA nanostructures with endosomal escape behaviours to achieve effective in vivo gene delivery and therapy.
Key words: i-motif, siRNA, Gene delivery, DNA tetrahedron, Endosomal escape, Cancer, Gene silence, Biocompatibility
Graphical abstract
siRNA-loaded DNA tetrahedron nanocages escape from the endosomal degradation and successfully interfere in gene and protein expression level of EGFR in tumor cells.
1. Introduction
As well known, DNA nanostructures, such as intermolecular triplex DNA, t-switch, DNA tweezers, A-motif, and i-motif are prone to pH-sensitive charge-reversal1. Among them, i-motif is a quadruple DNA structure formed by cytosine-rich ssDNA sequences2,3. It can transform its structure in response to the change of pH value4,5, which will contribute to its structural assembly and disassembly. Typically, i-motif can assemble into a quadruple DNA structure in acid and relapse into a linear structure in a neutral or slightly alkaline environment6,7. Specifically, the i-motif exists as a linear DNA structure at physiological pH (pH 7.4). But when the environment turns acidic (pH < 5.0), part of cytosine in the cytosine-rich DNA strand can bind with H+ to form C+. C+ can be paired with unprotonated C to form C:C+, which can form a four-stranded quadruplex structure8,9. At this time, the force between C:C+ is stronger than G–C. Meanwhile, as the pH value reaches a low enough level, all cytosines would be protonated by a large amount of H+. Due to their keen tunability to pH value changes, several reported drug release systems composed of i-motif was demonstrated to release the cargo in response to pH shifts10. Although i-motif nanostructures had shown satisfactory advantages during the delivery of chemotherapy drug11, 12, 13, no research reports on their application in delivering biomacromolecules can be found, such as nucleic acid drugs.
Recently, DNA folding nanostructures had abstracted more and more concern, as biocompatible nanosized delivery system14,15. DNA nanocage demonstrates substantial advantages including high mechanical rigidity16,17, structural stability18,19, and transfection efficiency20, as well as low toxicity and immunogenicity21. Other organic nanomaterials such as lipid nanoparticles and polymers also have excellent transfection ability, but their positive charge will cause cytotoxicity and the formation of “protein corona” which will accelerate elimination in vivo22. Inorganic materials like gold nanoparticles and silica can easily accumulate in the liver and kidney because of their chemical stability while it is hard for them to be degraded in vivo. Therefore, compared with these materials, DNA nanocage possesses better biocompatibility22. It is worth mentioning that DNA nanocage possessed the intrinsic homology with therapeutic siRNA23,24, CpG25, and other nucleic acids26, which facilitated a load of nucleic acids through the design of DNA strands’ sequence. The most important is DNA nanocages can be taken up into cells without transfection reagents. There have been several studies reported that proteins7 and nucleic acids26 can be loaded in DNA nanocages by far. However, nucleic acids, like siRNA have to be delivered to the cytoplasm or nucleus to exert therapeutic effects22,27. In other words, lysosomal escape is one of the key processes for the intracellular disposal of nucleic acid drugs28,29. Though successful delivery of siRNA by DNA nanoparticles had been demonstrated in tumor-bearing mice23,24, there were no reports focusing on the endosomal escape behaviors achieved by the structural modification of DNA nanocage.
The mechanisms of endosomal escape includes29,30: (1) pore formation in the endosomal membrane. Some components like peptides with a high affinity can make the pores in the endosomal membrane. But the dimensions of pores are ∼1–2 nm, which generally limits the efficient release of therapeutic cargo. (2) Proton Sponge Effect and Osmotic Lysis31. The proton sponge effect depends on agents with a high buffering capacity. When these buffers are protonated, the ion balance will be broken, resulting in high osmotic pressure in lysosomes. This can trigger an extensive inflow of ions and water into the endosomes, which leads to the rupture of the endosomal membrane and the release of the entrapped components. (3) fusion in the endosomal membrane, which utilizes fusogenic peptides to release the cargo. In general, virus or bacteria-derived agents, liposomes, polycationic materials (like PEI and poly (amidoamine) (PAMAM) dendrimers), and peptides are used to induce lysosomal escape32. They have potential toxicity, especially viral or bacterial derived agents and polycationic materials33, 34, 35, 36. And the pores caused by peptides are always too small to allow nucleic acids to pass. Therefore, we plan to take advantage of i-motif as a modification onto DNA nanocages. It was supposed that i-motif can consume H+ in the acidic environment after intracellular uptake and being captured by lysosomes. Therefore, it can achieve the proton sponge effect with high osmotic pressure and swelling in lysosomes, resulting in an endosomal escape.
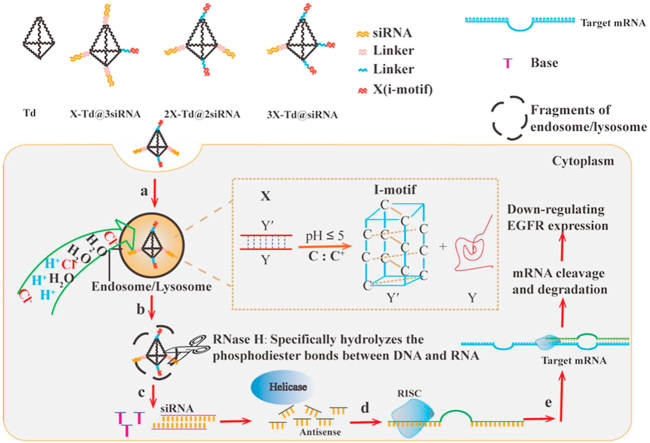
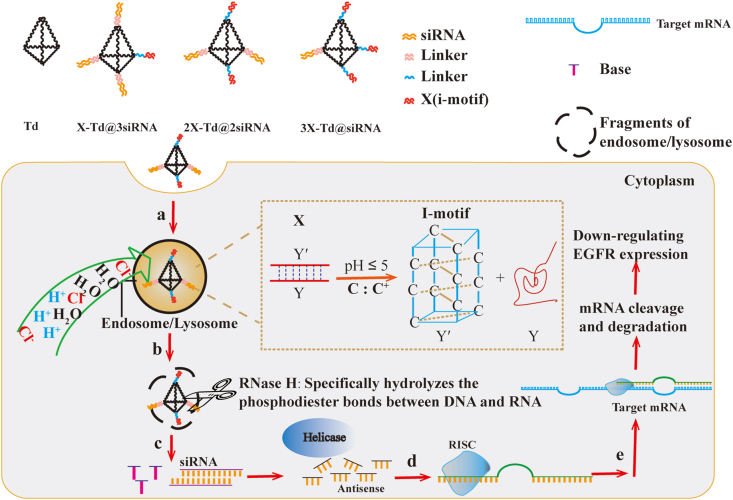
In this study, we developed a series of DNA tetrahedron nanocages (Td), through programmable self-assembly of complementary DNA strands containing different numbers of i-motif DNA structure and therapeutic siRNA, to facilitate the endosomal escape of siRNA after intracellular uptake. As shown in Scheme 1, the selected i-motif DNA structure is consisted of two oligonucleotide strands named Y′ and Y, while Y′ contains four stretches of cytosine rich residues (CCC) and Y is a sequence that is perfectly complementary to Y′. Under the acidic conditions of pH 5.0, the double chains (Y and Y′) will be unwrapped, and Y will be released. Then, the protonated C of Y′ can combine with the unprotonated C to gain a nonclassical base pair (C:C+ base pair), called C-quadruple helix (i-motif). Thus, we supposed that after intracellular uptake into tumor cells, the i-motif-conjugated Td could cause internalization of H+ and Cl‒ into endosomes which makes endosomes swell and crack into fragments. Furthermore, the anti-tumor therapeutic siRNA were simultaneously conjugated on the remaining vertexes of Td to gain these nanostructures (aX-Td@bsiRNA, a+b = 4). Once the endosomes cracked, the aX-Td@bsiRNA nanostructures would successfully escape from endosomes and get distributed in the cytoplasm. Meanwhile, the Dicer enzyme would efficiently cleave the double-stranded siRNA into the therapeutic segment and let them release to generate gene silence.
Scheme 1.
Schematic illustration of the process of siRNA-loaded DNA tetrahedron nanocages with endosomal escape function interfering EGFR gene and protein expression level in tumor cells.
2. Materials and methods
2.1. Chemicals and materials
All specific sequences for assembly of Tds are shown in Supporting Information Table S1. All the DNA and RNA sequences were synthesized by Takara Bio, Inc. (Dalian, China). DNA sequences were designed by computer program RNA STRUCTURE 5.7. Agarose (biochemical grade) was bought from Sinopharm Chemical Reagent Co., Ltd. Gel red, DNase I, and RNase A were obtained from the KeyGEN Institute of Biotechnology (Jiangsu, China). Lyso-Tracker Green, 4% paraformaldehyde, and DAPI were purchased from Beyotime Biotechnology. 5-Diphenyl-2-H-tetrazolium bromide (MTT, Sigma–Aldrich, USA) were used as received. Lipofectamine®3000 (lipo3000) was obtained from Invitrogen. Polyvinylidene difluoride (PVDF) membranes and the enhanced chemiluminescence (ECL) reagents were obtained from Millipore (USA). The primary antibodies used in this study were rabbit anti-EGFR (Abways, Shanghai, China) and rabbit anti-β-actin (Abways, Shanghai, China). Horseradish peroxidase-conjugated AffiniPure goat anti-rabbit IgG secondary antibody was bought from Abways Technology (Shanghai, China). SYBR® Premix Ex Taq™ II was purchased from Takara Bio, Inc. (Dalian, China). All of the other chemicals and reagents were analytic grade and used without further purification.
2.2. Preparation of aX-Td@bsiRNA
Briefly, aX-Td@bsiRNA was assembled by mixing A1, B1, C1, D1, X, antisense, sense (or sense-Cy5) sequences at specific molar concentrations in TM buffer (10 mmol/L Tris-HCl, 5 mmol/L MgCl2, pH 8.0). Then, seven oligonucleotides were annealed by heating the mixture to 95 °C for 8 min and then cooled to 4 °C for 20 min by PCR machine (Eppendrof, Mastercylenexus GSX1, Germany). This i-motif-conjugated Td (aX-Td) was obtained under the same conditions using A1 (or A), B1 (or B), C1 (or C), D (or D-Cy5) and X at specific molar concentrations.
2.3. Agarose gel electrophoresis (AGE)
The assembled structures were verified on 2% agarose gel electrophoresis in TBE buffer (0.045 mol/L Tris-boric acid, 0.001 mol/L EDTA, pH 8.3). Gels stained with Gel red were run with 100 V at room temperature for 1 h. After that, the images were finally visualized using ChemiDoc XRS+™ System with Image Lab Software (BIO-RAD, USA).
2.4. Circular dichroism (CD) spectra
The CD spectra of 2X-Td@2siRNA and single strands of 2X-Td@2siRNA at 0.3 μmol/L were measured at 25 °C in TM buffer (10 mmol/L Tris-HCl, 5 mmol/L MgCl2, pH 8.0) scanning from 200 to 400 nm using a JASCO J810 CD spectrometer. The TM buffer was used to automatically correct the baseline throughout the entire experiment.
2.5. Atomic force microscopy (AFM) imaging
The 30 μL of each sample (0.4 μmol/L) in an aqueous buffer was dispersed onto the surface of mica, and then dried with compressed air for 24 h. Then, samples were fulfilled by means of the tapping mode using the atomic force microscopy (Nano Scope IIIa, Veeco, USA).
2.6. Transmission electron microscopy (TEM) imaging
Here, Td (3 μmol/L) and aX-Td@bsiRNA (3 μmol/L) were dispersed onto holey carbon film on copper grids and analyzed by TEM (H-600, Hitachi, Japan) after being stained negatively with phosphotungstic acid (2%).
2.7. Dynamic light scattering (DLS)
The hydrodynamic sizes and zeta potentials of Td, X-Td@3siRNA, 2X-Td@2siRNA, 3X-Td@siRNA (1.7 μmol/L) at pH 5.5 and 7.4 were measured by Dynamic Light Scattering (DLS) Analyzer (Zetasizer Nano-ZS 90, Malvern Instruments, UK).
2.8. In vitro stability of 2X-Td@2siRNA
For the stability test, 2X-Td@2siRNA solutions (0.4 μmol/L), free siRNA (0.4 μmol/L), single strands of 2X-Td@2siRNA (0.4 μmol/L) were incubated at 37 °C with 75% inactivated fetal bovine serum (FBS, Gibco, USA) and nuclease solution (100U DNase I and 5 mg/mL RNase A) at pre-determined time intervals, respectively. Then the results were probed by 2% Agarose Gel Electrophoresis. Besides, 2X-Td@2siRNA was placed evenly in 1.5 mL of centrifugation tubes containing pH 5.5 and pH 7.4 PBS solution, respectively. Both tubes were incubated under 37 °C, and the size changes of 2X-Td@2siRNA were measured by DLS at pre-determined time points.
2.9. Cell lines and cell culture
The non-small cell lung cancer cell line (A549) was reported to overexpress EGFR37. A549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 media (Gibco, USA) (pH 7.2±0.2) containing 10% fetal bovine serum (FBS), penicillin (50 UI/mL) and streptomycin (50 UI/mL) at 37 °C in a humidified atmosphere with 5% CO2.
2.10. Cell viability assays
To detect the cytotoxicity of aX-Td, A549 cells were cultured in 96-well plates at a density of 2500 cells/well. After being cultured for 24 h, cells were treated with different concentrations of aX-Td for 48 h. Subsequently, the cells were incubated with 20 μL of methyl thiazolyl tetrazolium (0.5 mg/mL) for 4 h. Then, DMSO was added to dissolve the crystallizations. The absorbance at 570 nm was determined.
2.11. In vitro cellular uptake study
To evaluate cellular uptake of aX-Td, flow cytometry was used. A549 cells were seeded into 24-well plates at a density of 2 × 104 cells/well cultured with RPMI 1640 complete media for 24 h. After which, complete media was removed and 200 μL aX-Td resuspended in serum-free media were added. After 4 h incubation, cells were rinsed with cold PBS twice and then isolated from the plates by means of trypsin. Intracellular uptake was analyzed by a flow cytometer (Miltenyi, MACSQuant Analyzer 10, USA). At least 10,000 cells of each sample were analyzed. For this experiment, strand D was labeled with Cy5 for DNA detection inside the cells.
The cellular uptake of aX-Td@bsiRNA was investigated in the same way, in which siRNA loaded in lipofectamine®3000 (lipo3000@siRNA) was set as positive control and free siRNA was set as a negative control. Furthermore, we performed a confocal laser scanning microscopy (LSM700, Carl Zeiss, Germany) to detect the cellular uptake of aX-Td@bsiRNA. A549 cells were seeded into Petri dishes at a density of 2 × 105 cells/dish until sub-confluence and then incubated with Cy5-labeled aX-Td@bsiRNA, lipo3000@siRNA, and free siRNA (0.3 μmol/L) for 10 h. Nuclei were stained with DAPI for 15 min at room temperature.
2.12. Endosomal escape behavior of aX-Td and aX-Td@bsiRNA
To investigate the endosomal escape behavior of aX-Td, A549 cells were seeded into Petri dishes at a density of 2 × 105 cells/dish and then incubated in serum-free medium for different times at the presence of Cy5-labeled aX-Td (0.3 μmol/L). After being incubated at pre-determined times, cells were gently rinsed with PBS and dyed with Lyso-Tracker Green (1:5000 dilution) for 2 h at 37 °C. Subsequently, cells were fixed for 10 min with 4% paraformaldehyde and then incubated for 15 min with DAPI at room temperature. Finally, cells were dispersed in PBS and assessed by confocal laser scanning microscopy (LSM700, Carl Zeiss, Germany).
Endosomal escape behavior of aX-Td@bsiRNA after 10 h incubation was also investigated in the same way, in which lipo3000@siRNA was set as positive control and free siRNA was set as a negative control. The fluorescent images were acquired with 63 × magnification oil immersion lenses.
2.13. Western blot
To determine EGFR protein expression level, A549 cells were seeded onto 6-well plates at 2 × 105 cells per well for 24 h. After that, aX-Td@bsiRNA, free siRNA, lipo3000@siRNA were resuspended in RPMI 1640 media (0% FBS), then added to each well at 300 nmol/L. The cell lysate was collected by using an ice-cold extraction reagent RIPA lysis buffer. The lysate was then centrifuged at 12,000×g for 5 min at 4 °C and the protein concentration was determined by the bicinchoninic acid (BCA) method. Subsequently, 25 μg of protein samples were first separated by 8% SDS-PAGE and transferred onto the PVDF membrane. Then, 5% powdered non-fat milk in Tris-buffered saline with Tween-20 (TBST) was used to block the membrane at 25 °C for 1 h. After which, the membrane was stained with primary EGFR antibody which was added at a dilution of 1:1000 at 4 °C overnight and then incubated with the secondary antibody which was horseradish peroxidase-conjugated AffiniPure Goat anti-rabbit IgG at a dilution of 1:12,500 for 1.5 h at 25 °C. The expression of β-actin was detected as the internal standard for the loading control. β-Actin primary antibody at a dilution of 1:6500 was used. To visualize the protein bands, the ECL reagents were used. The Western blot was developed on a gel imager (ChemiDoc XRS + ™, Bio-Rad, USA).
2.14. Real-time quantitative PCR (RT-qPCR)
To detect EGFR gene expression level, cells were harvested after being treated with aX-Td@bsiRNA, free siRNA, and lipo3000@siRNA for 48 h. Total RNA was extracted from cells using RNeasy Mini Kit (QIAGEN, Germany), then reverse-transcribed with HiScript® Q Select RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech Co., Ltd., Nanjing, China) to obtain cDNAs. Real-time PCR was performed using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus), ROX Plus (TAKARA, Dalian, China) and amplified with Applied Biosystems Stepone™ (Thermo Fisher Scientific, USA). All samples’ EGFR mRNA expression levels were normalized by β-actin amplification. Primer sequences used for EGFR (forward 5ʹ-AGACGCAGATAGTCGCCCAAAG-3ʹ, reverse 5ʹ-TCCATCAGGGCACGGTAGAAG-3ʹ) were designed and synthesized by TAKARA (Dalian, China). β-Actin (forward 5ʹ-AAATCGTGCGTGACATTAA-3ʹ, reverse 5ʹ-CTCGTCATACTCCTGCTTG-3ʹ)38 was synthesized by TAKARA (Dalian, China).
2.15. Antitumor activity and safety in vivo
In order to study antitumor activity in vivo, an orthotopic 4T1 breast tumor model was established. BALB/c mice (5 weeks old, female) were provided by Qinglong Mountain (approval number: SYXK 2017-0001). All mice had free access to chow and water. All live animalsʹ experiments complied with the relevant laws and institutional guidelines and the Animal Experimentation Ethics Committee of China Pharmaceutical University approved the experiments (approval number: SYXK 2018-0019). Briefly, 1 × 106 4T1 cells in PBS (pH 7.4) were subcutaneously injected into the fifth pair of breasts on the left of each mouse. When the tumor size reached ∼100 mm3, the mice were randomly divided into 5 groups (5 mice in each group). Then, these mice were administered (i.v.) with 200 μL of saline (control), Td (naked tetrahedron), 2X-Td@2siRNA, Td@2siRNA (siRNA loaded in Td without endosomal escape capacity), and free siRNA (0.8 mg/kg siRNA, 27.8 nmol/kg Td) every other day for 12 days, respectively. Tumor size was measured using digital calipers and the volume (mm3) was calculated according to the formula (a2b)/2, where a and b were the respective shortest and longest diameter of the tumor. Relative tumor volume (%) was calculated according to the formula (Vx‒V0)/V0, where Vx is the mean volume on Day x and V0 is the mean volume on the first day of treatment (Day 0). Bodyweight was also recorded every day. Relative body weight (%) was calculated according to the formula (Wx‒W0)/W0, where Wx is the mean body weight on Day x and W0 is the mean body weight on the first day of treatment (Day 0). On Day 12, mice were sacrificed by cervical dislocation. Simultaneously, the tumor mass was harvested, weighed, and photographed. Then, major organs (livers, spleens, hearts, lungs, and kidneys) and tumors were collected to be fixed in 10% formalin and embedded in paraffin. Next, five micrometer-thick sections were prepared and stained with H&E. The sections of tumors were also prepared for immunohistochemistry conducted by superbio Technology (Nanjing, China).
2.16. Statistical analysis
All the data were expressed as mean ± standard deviation (SD) obtained from at least three samples. Statistical significance was analyzed using one-way analysis of variance (for multiple-group analysis) and two-tailed (for two-group comparison) Student's t-test. Differences with P < 0.05 were defined as statistically significant.
3. Results and discussion
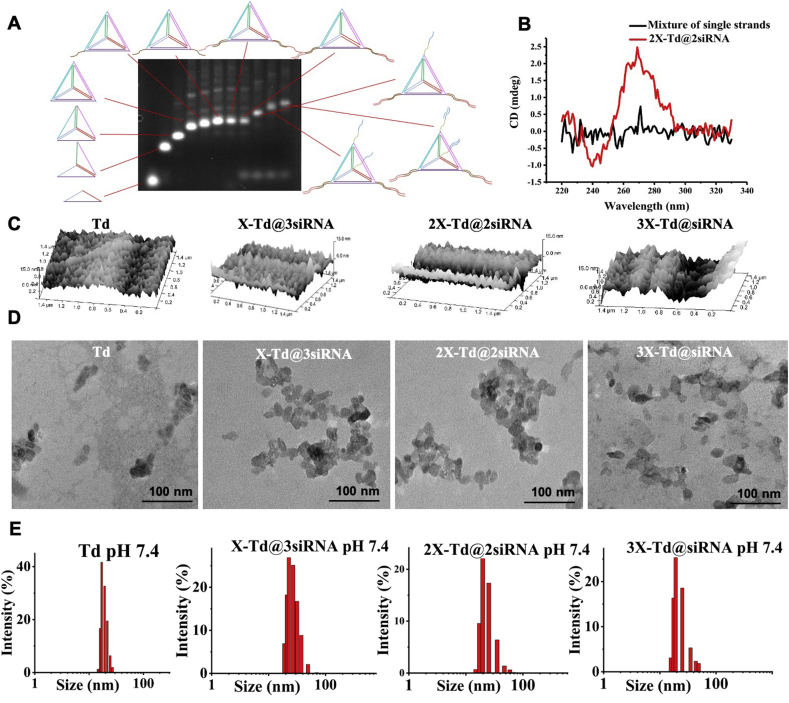
Generally, four designed single-strand DNAs (ssDNA) were utilized to prepare the DNA tetrahedrons by Td self-assembly. In this study, we utilized the original ssDNAs which were first reported by Turberfield and co-workers as the basic Td backbones39,40. Specifically, the strategy for aX-Td@bsiRNA formation was displayed in Table S1. And sequences used to synthesize aX-Td@bsiRNA were described in Supporting Information Table S2. The successful synthesis of aX-Td@bsiRNA was confirmed by 2% agarose gel electrophoresis (AGE, Fig. 1A). The lanes from left to right are A, AB, ABC, ABCD (Td), AB1CD, AB1CD1, AB1CD1+antisense, AB1CD1+antisense + sense, A1B1CD1+antisense + sense, A1B1C1D1+antisense + sense, A1B1C1D1+antisense + sense + X (2X-Td@2siRNA), respectively. As shown in Fig. 1A, there were very sharp bands in each lane and the migration rate slowed down as the molecular weight increases. Td moved more slowly than the ssDNA and other DNA combinations. 2X-Td@2siRNA migrated more slowly than Td because of the extra X and siRNA. These results suggest that Td and 2X-Td@2siRNA were successfully synthesized via an efficient complementary base pairing. Moreover, circular dichroism (CD) spectra were utilized to verify the formation of double strands. Three characteristics of B-form duplex DNA structure were observed in Fig. 1B. They were a positive band around 270 nm, a negative band around 240 nm, and a crossover near 258 nm respectively, which indicated that 2X-Td@2siRNA was synthesized successfully4.
Figure 1.
Characterization of Tds. (A) 2% AGE analysis of Td and 2X-Td@2siRNA. (B) CD spectra of 2X-Td@2siRNA (double strands, red curve) and mixture of 2X-Td@2siRNA strands (single strands, black curve). (C) AFM images of aX-Td@bsiRNA. (D) TEM images of aX-Td@bsiRNA by negative staining with 2% aqueous phosphotungstic acid. The scale bar is 100 nm. And hydrodynamic diameter of aX-Td@bsiRNA from DLS measurements.
Then, the size and zeta potential of Tds were determined by dynamic light scattering (DLS) at both physiological and endosomal pH environments. The results are displayed in Supporting Information Table S3, the sizes of Td, X-Td@3siRNA, 2X-Td@2siRNA, and 3X-Td@siRNA at pH 7.4 are 19.62±2.36, 26.46±3.50, 27.35±2.88, and 26.16±8.46 nm, respectively, which are similar to that at pH 5.5, revealing that DNA tetrahedrons could still keep their integral structures even in endosomal acidic condition. The zeta potentials were all negative at both pH values, indicating that the modification of X and siRNA did not influence the charge of the DNA nanocage skeleton. Furthermore, atomic force microscopy (AFM) images (Fig. 1C) and transmission electron microscopy (TEM) images (Fig. 1D) show the morphology of DNA tetrahedrons, and a high consistency was observed between aX-Td@bsiRNA and Td. Also, it can be seen from the TEM images that the DNA tetrahedron has a particle size of about 20 nm. And the two-dimensional morphology of DNA tetrahedral nanocage is a triangular or irregular quadrilateral, which is consistent with the two-dimensional structure of the tetrahedron. These results indicate that the synchronous conjugation of i-motif DNA and therapeutic siRNA did not influence the formation of the three-dimensional structure formation of Td.
To verify Td could improve the stability of siRNA at the presence of FBS and nucleases, free siRNA, 2X-Td@2siRNA, and single strands of 2X-Td@2siRNA were incubated at 37 °C with 75% inactivated FBS and nuclease solution (100 U DNase I and 5 mg/mL RNase A) for different time. From Supporting Information Figs. S1A and S1B images, it could be found that 2X-Td@2siRNA can maintain its structural integrity for 30 min in nucleases and 120 min in 75% FBS, while the degradation of DNA single strands and free siRNA started from the beginning of incubation. These results indicated that the rigid structure of the self-assembled Td could protect siRNA from rapid degradation41, 42, 43, 44 by the nucleases and FBS, whereas the DNA single-strand without a rigid structure did not possess this function. Furthermore, the in vitro stability of 2X-Td@2siRNA was investigated in pH 5.5 PBS and pH 7.4 PBS over time. After incubation in pH 5.5 PBS at 37 °C for 25 min (Supporting Information Fig. S2A), the particle size changed from 23.44±1.73 nm to 53.74±10.20 nm, showing a trend of continuous increase over time. While in pH 7.4 PBS (Fig. S2B), the particle size reached 63.12±14.53 nm after incubation at 37 °C for 25 min. Hence, no significant increase in size could be observed. These results indicate that the ion composition, concentration, and pH value of these test buffer would influence the protonation and stretch of the DNA chains, resulting in the size change of assembled DNA nanostructures.
To further analyze the cytotoxicity of aX-Td toward A549 cells, an MTT assay was carried out. Supporting Information Fig. S3A gives no significant change in cytotoxicity as the concentration of DNA increased. And the cell viability of aX-Td was all above 80% at different concentrations. In other words, Tds had no obvious cytotoxicity. As shown in Fig. S3B, there was no significantly different cell viability could be found between those groups treated with free siRNA, X-Td@3siRNA, 2X-Td@2siRNA, and 3X-Td@siRNA. Namely, aX-Td@bsiRNA did not show remarkable anti-cancer effects in vitro after 48 h treatment.
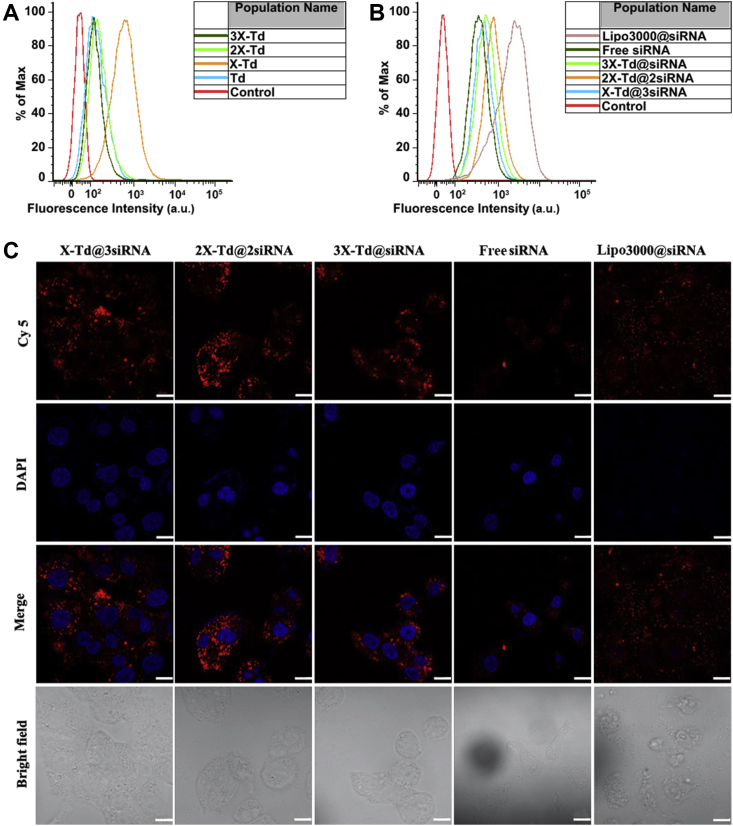
To investigate the cellular uptake efficiency of aX-Td, Td frameworks were labeled by Cy 5. The results from the flow cytometer are shown in Fig. 2. As shown in Fig. 2A, after incubated with aX-Td and Td for 4 h, the cellular uptake of X-Td was significantly higher than other groups and there was no significant difference between the other three groups. DNA tetrahedrons was proved to carry negative charges, thus they must overcome electrostatic repulsion with cell membranes to be taken into cells45. When one of the Td's facets is faced up with the membrane surface, it will form a “face attack” which has high repulsion with cell membranes resulting in difficulties for Td internalization. Generally, when a “face attack” occurred, Td will rotate to produce an angle with cell membranes and go through a “corner attack”, which will decrease the repulsion46,47. When the four vertices of Td were not fully occupied, one X modification will reduce the occurrence of the “face attack” of Td, which is advantageous to Td internalization, thereby enhancing its cellular uptake. However, as the number of X increasing, the appearance of “corner attack” decreased, especially 3X-Td. Therefore, the cellular uptake of 2X-Td and 3X-Td was lower than that of X-Td.
Figure 2.
Cellular uptake in A549 cells of empty cages (aX-Td) after 4 h analysed by flow cytometer (A), siRNA-loaded in Td (aX-Td@bsiRNA) after 10 h with lipo3000@siRNA as a positive control and free siRNA as a negative control analysed by flow cytometer (B) and confocal laser scanning microscopy (C). Scale bar = 10 μm.
To analyze the cellular uptake of aX-Td@bsiRNA (Fig. 2B), siRNA was labeled with Cy 5. Since the i-motif content in different nanostructures is diverse, we adjusted the dosage of nanostructures including i-motif, to guarantee the consistency of siRNA dosage in both in vivo and in vitro experiments. Namely, the ratio of i-motif to siRNA was fixed as 1:3, 1:1, and 3:1 for X-Td@3siRNA, 2X-Td@2siRNA, and 3X-Td@siRNA, respectively. When the dose of siRNA changes, the amount of X-Td will change accordingly. Simultaneously, siRNA loaded in lipofectamine®3000 (lipo3000@siRNA) was set as a positive control and free siRNA was set as a negative control. Flow cytometry results showed that the cellular uptake of lipo3000@siRNA was the strongest, followed by 2X-Td@2siRNA. However, there was no significant difference between 3X-Td@siRNA and X-Td@3siRNA. It's worth noting that free siRNA had the lowest cellular uptake. Similarly, the results were also confirmed by Confocal laser scanning microscopy images (Fig. 2C). There was little free siRNA localized in the cell, indicating siRNA had poor ability into cells. Contrary to free siRNA, aX-Td@bsiRNA showed stronger red fluorescence intensity, revealing that there were more Cy 5 labeled siRNA taken into cells. These results indicate that the conjugation of siRNA on the vertex of aX-Td would benefit for their intracellular uptake into A549 cells.
At the same time, as shown in bright-field images (Fig. 2), the morphology of cells in aX-Td@bsiRNA was complete, which was contrary to that in lipo3000@siRNA. What we need to pay attention to is that there was no obvious cell morphology observed in the lipo3000@siRNA group (Fig. 2), and the cells were destroyed into fragments due to the toxicity of lipo3000. Based on the above results, it was revealed that aX-Td could significantly improve the cellular uptake of free siRNA with low toxicity compared to lipo3000.
After incubation with aX-Td@bsiRNA, the fluorescence intensity of 2X-Td@2siRNA was stronger than that of 3X-Td@siRNA and X-Td@3siRNA, while there was no significant difference between X-Td@3siRNA and 3X-Td@siRNA. It may be due to electrostatic repulsion with cell membranes. When three same ligands (X or siRNA) are modified on the vertex of Td, it would like the three legs of a tripod. Under this circumstance, it would increase the emergence of invalid “face attack” which had high repulsion. However, two ligands modified Td (2X-Td@2siRNA) was easier to produce an angle with cell membranes that would decrease the electrostatic repulsion, thereby enhancing its cellular uptake.
To further identify the endosomal escape capacity of aX-Td and aX-Td@bsiRNA, subcellular colocalization studies were conducted. Tds were labeled with Cy5 (red), nuclei were dyed with DAPI (blue) and endosomes were stained with Lyso Tracker Green (green). The capacity of endosomal escape was represented by the overlap (yellow or orange) of the fluorescence signals of Cy five and Lyso Tracker Green. First, we investigated the endosomal escape behavior of empty vectors modified with different numbers of X. As displayed in Supporting Information Figs. S4A–S4D, the intensity of red fluorescence was getting stronger as time prolonged which indicated that Tds were all entering in cells and the modification of X would not impede the cellular uptake of Td. When treated with aX-Td, the red fluorescence intensity of X-Td was always the strongest indicating that one X could enhance Td's cellular uptake, which was consistent with the flow cytometry results.
Additionally, after treated for 6 h, the overlap fluorescence of Td was most distinct in merged pictures and the red fluorescence signal of Td remained punctate, which meant Td was still entrapped in endosomes. The Pearson coefficient of the overlap fluorescence in Td (0.673±0.06, Supporting Information Fig. S4E) was more than 0.5, which also implied Td colocalized with endosomes. Contrary to Td without modified with X, obvious endosomal escape was observed in all aX-Td groups, revealing that X could endow Td endosomal escape capacity. As shown in Fig. S4E, the Pearson coefficient demonstrated that as the number of X modified on Td increasing, the overlap fluorescence decreased significantly. The overlap fluorescence appeared in X-Td group's merged pictures after 4 h incubation, indicating that X-Td was endocytosed into endosomes. After 6 h incubation, there was still some orange fluorescence observed in X-Td group. But the difference was the intensity of red fluorescence gets stronger, which demonstrated that a part of X-Td had escaped from endosomes. In 2X-Td group, the red fluorescence started to appear after 4 h incubation in merged pictures, indicating that there was some 2X-Td escaped from endosomes into the cytoplasm. After 6 h, the red fluorescence became stronger and there was no orange fluorescence detected, which suggested that most of 2X-Td had escaped. In 3X-Td group, it was distinctly contrary to Td that there were no overlap fluorescence signals detected and the red fluorescence was no longer punctate but diffusely distributed in the cytoplasm, which suggested that 3X-Td had already get rid of endosomes. To be noted, the endosomal escape capacity of 2X-Td was not as good as that of 3X-Td in 4 h, but there was no difference between them in 6 h. Taken together, the endosomal escape capacity of 2X-Td was similar to 3X-Td. This phenomenon could also be seen from the Pearson coefficient, which has no significance between 2X-Td (0.44±0.07) and 3X-Td (0.43±0.08) (Fig. S4E).
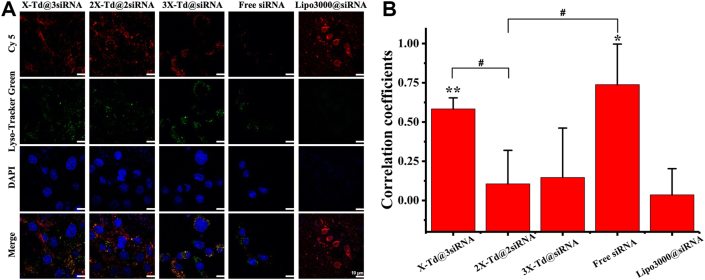
Then, we observed the endosomal escape behavior of aX-Td@bsiRNA with lipo3000@siRNA and free siRNA as controls. As displayed in Fig. 3, compared to free siRNA, there was a higher intensity of red fluorescence observed in aX-Td@bsiRNA treated groups after 10 h incubation, which indicated that siRNA loaded in Td could enhance its cellular uptake. At the same time, there were obvious red fluorescence and little overlap fluorescence observed in the cytoplasm, implying that aX-Td assisted siRNA to get rid of endosomes successfully. The endosomal escape capability of aX-Td@bsiRNA could be explained by the proton sponge effect caused by i-motif. After intracellular uptake and captured by lysosomes, i-motif conjugated on DNA nanocages are prone to consume H+ in the acidic environment, resulting in high osmotic pressure and burst of lysosomes. Moreover, it also demonstrated X did not detach from Td, and aX-Td@bsiRNA still maintain their structural integrity before into endosomes, which is essential to ensure the protonation of C and endosomal escape of siRNA.
Figure 3.
(A) Endosomal escape behavior of aX-Td@bsiRNA incubated for 10 h analysed by confocal laser scanning microscopy. Scale bar = 10 μm. All data are expressed as the mean ± SD, n = 3. (B) The Pearson correlation coefficients between Cy five and endosomes were calculated by Image J software. The significance of the differences (∗P < 0.05 versus lipo3000@siRNA group, ∗∗P < 0.01 versus lipo3000@siRNA group, #P < 0.05 versus 2X-Td@2siRNA group) was evaluated by two-tailed Student’ s t-test.
We could see clearly from Fig. 3A that the overlap fluorescence of lipo3000@siRNA did not exist, indicating lipo3000 had predominant endosomal escape capacity. The Pearson coefficient of the overlap fluorescence also indicated that (0.04±0.17 Fig. 3B). But the cells started to be broken and had no obvious morphology after 10 h, which could be explained by their potential cytotoxicity.
Meanwhile, among aX-Td@bsiRNA, a very large amount of red fluorescent signal appeared in the 2X-Td@2siRNA group, implying that the amount of 2X-Td@2siRNA (The Pearson coefficient of the overlap fluorescence in 2X-Td@2siRNA was 0.11±0.21) escaping from endosomes was larger than that of X-Td@3siRNA (The Pearson coefficient of the overlap fluorescence in X-Td@3siRNA was 0.58±0.07) and 3X-Td@siRNA (The Pearson coefficient of the overlap fluorescence in 3X-Td@siRNA was 0.15±0.31). It should be due to the dual factors that were cellular uptake and endosomal escape capacity. Different from the other four groups, there was almost no red fluorescence in free siRNA, indicating free siRNA had poor cellular uptake and endosomal escape capacity. Overall, Td could significantly improve the cellular uptake of siRNA, and X could endow Td with a certain endosomal escape capacity. In addition, it could be seen that the cytotoxicity of Tds was much smaller than transfection commercial reagent (lipo3000).
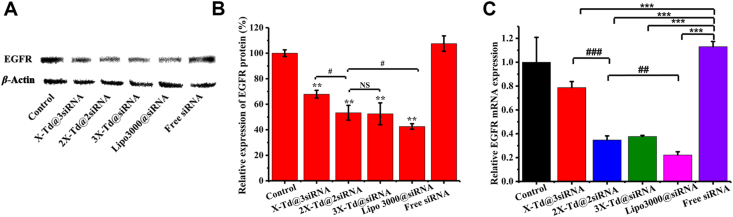
Western blot was used to verify qualitatively if aX-Td@bsiRNA could inhibit the expression of EGFR in A549 cells. As shown in Fig. 4A, the intensity of the free siRNA band had no significant difference from the control, which suggested that free siRNA did not play a role in gene silencing. On the contrary, the intensity of aX-Td@bsiRNA decreased markedly compared to free siRNA, which demonstrated that the efficiency of aX-Td@bsiRNA was superior to that of free siRNA. Moreover, the results showed that the predominant cellular uptake capacity and endosomal escape capacity of aX-Td were beneficial to siRNA in silencing gene expression. Among aX-Td@bsiRNA, the intensity of 2X-Td@2siRNA and 3X-Td@siRNA was lower than that of X-Td@3siRNA, indicating that the gene silencing efficiency of 2X-Td@2siRNA and 3X-Td@siRNA was higher than X-Td@3siRNA. Furthermore, we could also conclude that this is because of the difference in cellular uptake and endosomal escape capacity. But there was no difference between the intensity of 2X-Td@2siRNA and 3X-Td@siRNA, which was shown in Fig. 4B. We combined with the previous results that the endosomal escape capacity of 2X-Td@2siRNA was as good as 3X-Td@siRNA but the cellular uptake of 2X-Td@2siRNA was superior to 3X-Td@siRNA. However, the gene silencing efficiency revealed no difference between 2X-Td@2siRNA and 3X-Td@siRNA indicating that endosomal escape capacity was more important for gene silencing. In addition, the intensity of 2X-Td@2siRNA and 3X-Td@siRNA was similar to that of lipo3000@siRNA, which is to say that 2X-Td@2siRNA and 3X-Td@siRNA could transfect siRNA efficiently and inhibit EGFR expression in vitro effectively even as well as lipo3000@siRNA did.
Figure 4.
In vitro EGFR protein and mRNA expression level treated with siRNA-loaded Tds (aX-Td@bsiRNA) after 48 h analysed by Western blot (A), the semiquantitative analysis by Image J of the protein level (B) and RT-qPCR (C). All data are expressed as the mean ± SD, n = 3. ∗∗∗P < 0.001, ∗∗P < 0.01 versus free siRNA group. #P < 0.05, ##P < 0.01, ###P < 0.001 versus 2X-Td@2siRNA group. The significance of the differences was evaluated by two-tailed Student's t-test.
Furthermore, we detected relative EGFR mRNA expression quantitatively by Real-Time quantitative PCR (RT-qPCR). The relative mRNA expression levels of X-Td@3siRNA, 2X-Td@2siRNA, 3X-Td@siRNA, lipo3000@siRNA, and free siRNA group were 78.75±5.04%, 34.77±3.39%, 37.66±0.92%, 22.19±2.70%, and 113.05±4.31% respectively. Among them, the mRNA expression level in the free siRNA group was not significantly different from that in the control group, indicating that free siRNA did not generate a significant gene silencing. On the contrary, the mRNA expression levels of X-Td@3siRNA, 2X-Td@2siRNA, and 3X-Td@siRNA were significantly decreased compared with free siRNA, indicating that aX-Td@bsiRNA could downregulate the mRNA expression effectively in vitro and cellular uptake was important for siRNA as well as endosomal escape capacity. And among aX-Td@bsiRNA, the mRNA expression level in the 2X-Td@2siRNA and 3X-Td@siRNA groups were lower than X-Td@3siRNA, indicating 2X-Td@2siRNA and 3X-Td@siRNA induce better gene silencing in vitro because of their better endosomal escape capacity. At last, the gene silencing efficiency of 2X-Td@2siRNA and 3X-Td@siRNA was lower than that of lipo3000@siRNA, but the difference was not very distinct. Simultaneously, the previous results showed 2X-Td@2siRNA and 3X-Td@siRNA were safer. In a word, aX-Td@bsiRNA could downregulate EGFR mRNA expression level in A549 cells remarkably, which is consistent with the results of Western blot.
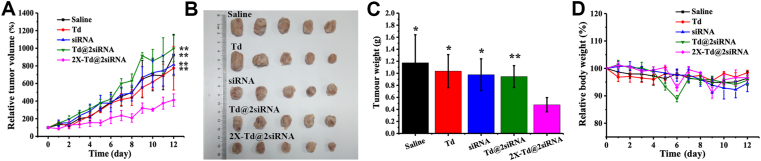
Both in vitro cytology and pharmacodynamic results showed that 2X-Td@2siRNA had better endosomal escape capacity and gene silencing efficiency. Subsequently, antitumor activity and safety of 2X-Td@2siRNA in vivo were investigated with saline, Td, Td@2siRNA (which has no endosomal escape capacity), and free siRNA as controls by orthotopic 4T1 breast tumor model. The result of relative tumor volume (Fig. 5A) shows that the tumor volume in Td and saline groups increased rapidly, and there was no significant difference between the two groups, which revealed that the blank carrier had no antitumor activity. Additionally, the tumors in free siRNA and Td@2siRNA also grew rapidly, indicating that siRNA and Td@2siRNA without good cellular uptake and endosomal escape capacity could not effectively inhibit tumor growth. Different from the other four groups, the tumor growth rate of the 2X-Td@2siRNA group significantly decreased, implying that 2X-Td@2siRNA had good tumor inhibition in vivo.
Figure 5.
In vivo anti-tumor effect. (A) 4T1 tumor growth curves of different groups after injection treatments. (B) Photos of the tumors collected from different groups of mice after 12 days injection treatments. (C) Average weights of the tumors in each treatment group collected after 12 days. (D) Body weights of mice after various injection treatments. ∗P < 0.05, ∗∗P < 0.01 versus 2X-Td@2siRNA. All of the values are shown as means ± SD (n = 5).
Furthermore, the visual image of the mice tumors (Fig. 5B) demonstrates that the tumors in the 2X-Td@2siRNA group were significantly smaller than the other four groups. The average tumor weights (Fig. 5C) of the saline group, the Td group, the free siRNA group, the Td@2siRNA group, and the 2X-Td@2 siRNA group were 1.17, 1.04, 0.98, 0.95, and 0.48 g, respectively. The efficiency of tumor growth inhibition in the 2X-Td@2siRNA group was significantly higher than that in Td@2siRNA (3.14-fold) and free siRNA (3.63-fold). It was further illustrated that 2X-Td@2siRNA had better in vivo antitumor activity.
Besides, the images of immunohistochemistry were used to observe gene silencing efficiency of 2X-Td@2siRNA in vivo qualitatively. The results are shown in Fig. S4A, in which blue represented the nuclei and yellow represented the EGFR protein in the tumor cells. It could be seen clearly that compared to the saline group, the EGFR expression in Td group had no significant decrease, indicating that Td did not exert obvious in vivo gene silencing as same as the saline group. Similarly, EGFR expression level in free siRNA had no significant change compared to the saline group, indicating that free siRNA also did not exert obvious in vivo gene silencing. While in Td@2siRNA group, the EGFR expression level had a slight decrease compared to free siRNA, which implied that the good cellular uptake of Td was advantageous to siRNA in in vivo gene silencing. In the 2X-Td@2siRNA group, the EGFR expression level decreased significantly compared to free siRNA and Td@2siRNA groups, indicating that 2X-Td@2siRNA was superior in in vivo gene silencing. The average optical density of IHC images (Fig. S4B) also reveals the results. Furthermore, it also suggested cellular uptake and endosomal escape capacity had a distinct influence on in vivo gene silencing of siRNA. However, endosomal escape capacity was more essential for siRNA in gene silencing.
The relative body weight (Fig. 5D) shows that there was no significant difference between the five groups from the Day 0 to Day 12 of administration, indicating that the carrier and siRNA-loaded DNA nanocages had no serious toxicity to the body. At the same time, the major organs (hearts, livers, spleens, lungs, and kidneys) of the five groups were observed by stained with H&E (Supporting Information Fig. S5). The images showed that the organs of free siRNA, Td@2siRNA, Td, 2X-Td@2siRNA group had no obvious difference from that of the saline group. It further indicated that the carrier and siRNA-loaded DNA nanocages were safe in vivo.
4. Conclusions
In summary, a series of DNA nanostructures (aX-Td@bsiRNA) gifted with favorable endosomal escape behaviors were developed to achieve efficient siRNA in vivo delivery. This siRNA delivery system can supply the multifunctionality required to address challenges for siRNA in vivo delivery, including protecting siRNA from nuclease-mediated degradation, facilitating its cellular uptake, and promoting endosomal escape. The particle size, AFM, and TEM showed aX-Td@bsiRNA was about 20 nm with negative charges. Besides, compared to free siRNA and DNA single strands, 2X-Td@2siRNA could maintain its complete structure for longer times in FBS (120 min) and nucleases (30 min). Furthermore, Td could significantly improve the cellular uptake of siRNA in A549 cells with lower cytotoxicity and X could help siRNA get rid of being entrapped in endosomes by introducing endosome membrane lysis. Moreover, the results obtained from Western blot and RT-qPCR also indicated that aX-Td@bsiRNA could downregulate EGFR protein and mRNA expression level in A549 markedly, even close to transfection reagent lipo3000. X-Td@3siRNA, 2X-Td@2siRNA, and 3X-Td@siRNA could downregulate EGFR mRNA expression level to 78.75%, 34.77%, and 37.66%, respectively, which were significantly lower than free siRNA. Meanwhile, 2X-Td@2siRNA could inhibit tumor growth effectively in vivo. Compared to free siRNA and Td@2siRNA, 2X-Td@2siRNA could effectively downregulate EGFR expression in 4T1 tumor and showed excellent antitumor efficiency with low systemic toxicity in vivo, thanks to its good stability, effective cellular uptake, and high efficiency in the endosomal escape. All these results represent that DNA tetrahedrons modified with X as promising siRNA-based therapeutics carriers have achieved great progress in further in vivo delivery for gene therapy.
Acknowledgments
This work was supported by Double First-Class Innovation Team of China Pharmaceutical University (CPU2018GY40, China), National Mega-project for Innovative Drugs (2019ZX09721001, China).
Author contributions
Xiu Han and Xiaole Qi designed the research. Xiu Han, Xiang Xu and Ziheng Wu carried out the experiments and performed data analysis. Xiu Han wrote the manuscript. Xiaole Qi revised the manuscript. Zhenghong Wu and Xiaole Qi supervised this research. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.02.009.
Contributor Information
Zhenghong Wu, Email: zhenghongwu66@cpu.edu.cn.
Xiaole Qi, Email: qixiaole523@cpu.edu.cn.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Ma W.J., Yan L.A., He X.X., Qing T.P., Lei Y.L., Qiao Z.Z. Hairpin-contained i-motif based fluorescent ratiometric probe for high-resolution and sensitive response of small pH variations. Anal Chem. 2018;90:1889–1896. doi: 10.1021/acs.analchem.7b03972. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y., Pan X., Li F., Yang D. pH-responsive reversible DNA self-assembly mediated by zwitterion. Chem Res Chin Univ. 2020;36:1–6. [Google Scholar]
- 3.Day H.A., Pavlou P., Waller Z.A.E. I-motif DNA: structure, stability and targeting with ligands. Bioorg Med Chem. 2014;22:4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 4.Liu D.S., Balasubramanian S. A proton-fuelled DNA nanomachine. Angew chem int ed. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 5.Panczyk T., Wolski P. Molecular dynamics analysis of stabilities of the telomeric Watson‒Crick duplex and the associated i-motif as a function of pH and temperature. Biophys Chem. 2018;237:22–30. doi: 10.1016/j.bpc.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Nesterova I.V., Nesterov E.E. Rational design of highly responsive pH sensors based on DNA i-motif. J Am Chem Soc. 2014;136:8843–8846. doi: 10.1021/ja501859w. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.H., Kim K.R., Ahn D.R., Lee J.E., Yang E.G., Kim S.Y. Reversible regulation of enzyme activity by pH-responsive encapsulation in DNA nanocages. ACS Nano. 2017;11:9352–9359. doi: 10.1021/acsnano.7b04766. [DOI] [PubMed] [Google Scholar]
- 8.Bielecka P., Dembska A., Juskowiak B. Monitoring of pH using an i-motif-forming sequence containing a fluorescent cytosine analogue, tC. Molecules. 2019;24:952–965. doi: 10.3390/molecules24050952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepper C.P., Williams M.A.K., Edwards P.J.B., Filichev V.V., Jameson G.B. Effects of pressure and pH on the physical stability of an i-motif DNA structure. ChemPhysChem. 2019;20:1567–1571. doi: 10.1002/cphc.201900145. [DOI] [PubMed] [Google Scholar]
- 10.Lee G.J., Kim T.I. pH-Responsive i-motif conjugated hyaluronic acid/polyethylenimine complexes for drug delivery systems. Pharm Times. 2019;11:247–257. doi: 10.3390/pharmaceutics11050247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Zhang Z., Xu S., Wang F., Shen Y., Huang S. pH, redox and photothermal tri-responsive DNA/polyethylenimine conjugated gold nanorods as nanocarriers for specific intracellular co-release of doxorubicin and chemosensitizer pyronaridine to combat multidrug resistant cancer. Nanomedicine. 2017;13:1785–1795. doi: 10.1016/j.nano.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Shi J., Zhou M. Probing the conformational switch of i-motif DNA using tunable resistive pulse sensing. Biochim Biophys Acta Gen Subj. 2018;1862:2564–2569. doi: 10.1016/j.bbagen.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Fan C., Pei H., Shi J., Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 14.Li H., He B., Liu X., Li J., Liu Q., Dong W. Regulation on toll-like receptor 4 and cell barrier function by RAB26 siRNA-loaded DNA nanovector in pulmonary microvascular endothelial cells. Theranostics. 2017;7:2537–2554. doi: 10.7150/thno.17584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang J.H., Kim K.R., Lee H., Ahn D.R., Ko Y.T. In vitro and in vivo behavior of DNA tetrahedrons as tumor-targeting nanocarriers for doxorubicin delivery. Colloids Surf B Biointerfaces. 2017;157:424–431. doi: 10.1016/j.colsurfb.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Luo X., Chidchob P., Rahbani J.F., Sleiman H.F. Encapsulation of gold nanoparticles into DNA minimal cages for 3D-anisotropic functionalization and assembly. Small. 2018;14:1702660. doi: 10.1002/smll.201702660. [DOI] [PubMed] [Google Scholar]
- 17.Wu D., Wang L., Li W., Xu X., Jiang W. DNA nanostructure-based drug delivery nanosystems in cancer therapy. Int J Pharm. 2017;533:169–178. doi: 10.1016/j.ijpharm.2017.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Corbett S.L., Sharma R., Davies A.G., Walti C. Enhancement of RECA-mediated self-assembly in DNA nanostructures through basepair mismatches and single-strand nicks. Sci Rep. 2017;7:1–9. doi: 10.1038/srep41081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raniolo S., Vindigni G., Ottaviani A., Unida V., Iacovelli F., Manetto A. Selective targeting and degradation of doxorubicin-loaded folate-functionalized DNA nanocages. Nanomedicine. 2018;14:1181–1190. doi: 10.1016/j.nano.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Hu Q., Wang S., Wang L., Gu H., Fan C. DNA nanostructure-based systems for intelligent delivery of therapeutic oligonucleotides. Adv Healthc Mater. 2018;7:1701153. doi: 10.1002/adhm.201701153. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G., Zhang Z., Yang J. DNA tetrahedron delivery enhances doxorubicin-induced apoptosis of HT-29 colon cancer cells. Nanoscale Res Lett. 2017;12:495. doi: 10.1186/s11671-017-2272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williford J.M., Wu J., Ren Y., Archang M.M., Leong K.W., Mao H.Q. Recent advances in nanoparticle-mediated siRNA delivery. Annu Rev Biomed Eng. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 23.Xia K., Kong H., Cui Y., Ren N., Li Q., Ma J. Systematic study in mammalian cells showing no adverse response to tetrahedral DNA nanostructure. ACS Appl Mater Interfaces. 2018;10:15442–15448. doi: 10.1021/acsami.8b02626. [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Jiang Q., He L., Zhan P., Liu Q., Liu S. Self-assembled double-bundle DNA tetrahedron for efficient antisense delivery. ACS Appl Mater Interfaces. 2018;10:23693–23699. doi: 10.1021/acsami.8b07889. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Pei H., Zhu B., Liang L., Wei M., He Y. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Ma W., Zhu Y., Shi S., Li Q., Mao C. Inhibiting methicillin-resistant Staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659. doi: 10.1021/acs.nanolett.8b02166. [DOI] [PubMed] [Google Scholar]
- 27.Mokhtarieh A.A., Lee J., Kim S., Lee M.K. Preparation of siRNA encapsulated nanoliposomes suitable for siRNA delivery by simply discontinuous mixing. Biochim Biophys Acta Biomembr. 2018;1860:1318–1325. doi: 10.1016/j.bbamem.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Ozpolat B., Sood A.K., Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev. 2014;66:110–116. doi: 10.1016/j.addr.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Selby L.I., Cortez-Jugo C.M., Such G.K., Johnston A.P. Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017;9 doi: 10.1002/wnan.1452. [DOI] [PubMed] [Google Scholar]
- 31.Lächelt U., Kos P., Mickler F.M., Herrmann A., Salcher E.E., Rödl W. Fine-tuning of proton sponges by precise diaminoethanes and histidines in pDNA polyplexes. Nanomedicine. 2014;10:35–44. doi: 10.1016/j.nano.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Liang W., Lam J.K. Endosomal escape pathways for non-viral nucleic acid delivery systems. Mol Regul Endo. 2012;17:429–456. [Google Scholar]
- 33.Chen G., Wang Y., Xie R., Gong S. Tumor-targeted pH/redox dual-sensitive unimolecular nanoparticles for efficient siRNA delivery. J Control Release. 2017;259:105–114. doi: 10.1016/j.jconrel.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteban-Fernández de Ávila B., Angell C., Soto F., Lopez-Ramirez M.A., Báez D.F., Xie S. Acoustically propelled nanomotors for intracellular siRNA delivery. ACS Nano. 2016;10:4997–5005. doi: 10.1021/acsnano.6b01415. [DOI] [PubMed] [Google Scholar]
- 35.Wu C., Li J., Wang W., Hammond P.T. Rationally designed polycationic carriers for potent polymeric siRNA-mediated gene silencing. ACS Nano. 2018;12:6504–6514. doi: 10.1021/acsnano.7b08777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y., Lin Y., Chen Z., Hu Q., Liu Y., Yu S. Enhanced endosomal escape by light-fueled liquid-metal transformer. Nano Lett. 2017;17:2138–2145. doi: 10.1021/acs.nanolett.6b04346. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M., Zhang X., Bai C.X., Wei M.Q. Inhibition of epidermal growth factor receptor expression by RNA interference in A549 cells. Acta Pharmacol Sin. 2004;25:61–67. [PubMed] [Google Scholar]
- 38.Shen J., Xu R., Mai J., Kim H.-C., Guo X., Qin G. High capacity nanoporous silicon carrier for systemic delivery of gene silencing therapeutics. ACS Nano. 2013;7:9867–9880. doi: 10.1021/nn4035316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman R.P., Berry R.M., Turberfield A.J. The single-step synthesis of a DNA tetrahedron. Chem Commun. 2004;12:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 40.Goodman R.P., Schaap I.A., Tardin C.F., Erben C.M., Berry R.M., Schmidt C.F. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Sci. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 41.Okholm A.H., Kjems J. DNA nanovehicles and the biological barriers. Adv Drug Deliv Rev. 2016;106:183–191. doi: 10.1016/j.addr.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 42.Bujold K.E., Hsu J.C., Sleiman H.F. Optimized DNA “nanosuitcases” for encapsulation and conditional release of siRNA. J Am Chem Soc. 2016;138:14030–14038. doi: 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- 43.Charoenphol P., Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol Pharm. 2014;11:1721–1725. doi: 10.1021/mp500047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway J.W., McLaughlin C.K., Castor K.J., Sleiman H. DNA nanostructure serum stability: greater than the sum of its parts. Chem Commun. 2013;49:1172–1174. doi: 10.1039/c2cc37556g. [DOI] [PubMed] [Google Scholar]
- 45.Lee D.S., Qian H., Tay C.Y., Leong D.T. Cellular processing and destinies of artificial DNA nanostructures. Chem Soc Rev. 2016;45:4199–4225. doi: 10.1039/c5cs00700c. [DOI] [PubMed] [Google Scholar]
- 46.Ding H., Li J., Chen N., Hu X., Yang X., Guo L. DNA nanostructure-programmed like-charge attraction at the cell-membrane interface. ACS Cent Sci. 2018;4:1344–1351. doi: 10.1021/acscentsci.8b00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chao J., Liu H., Su S., Wang L., Huang W., Fan C. Structural DNA nanotechnology for intelligent drug delivery. Small. 2014;10:4626–4635. doi: 10.1002/smll.201401309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.