ABSTRACT
Hypervirulent Klebsiella pneumoniae (hvKp) bacteria are more virulent than classical K. pneumoniae (cKp) with resultant differences in clinical manifestations and management. It is unclear whether all hvKp isolates share a similar pathogenic potential. This report assessed the utility of siderophore production, mucoviscosity, and murine infection for defining the virulence spectrum of hvKp. Three strain cohorts were identified and defined based on the CD1 mouse subcutaneous (SQ) challenge model: (i) fully virulent hvKp strains (fvhvKp), lethal at a challenge inoculum (CI) of ≤103 CFU; (ii) partially virulent hvKp strains (pvhvKp), lethal at a CI of >103 to 107 CFU; (iii) classical K. pneumoniae, not lethal at a CI of 107 CFU. Quantitative siderophore and mucoviscosity assays differentiated fvhvKp and pvhvKp strains from cKp strains but were unable to differentiate between the fvhvKP and pvhvKP strain cohorts. However, SQ challenge of CD1 mice and intraperitoneal (IP) challenge of CD1 and BALB/c mice, but not C57BL/6 mice, were able to discriminate between an fvhvKp and a pvhvKp strain; SQ challenge of CD1 mice may have the greatest sensitivity. cKp was differentiated from hvKp both by SQ challenge of CD1 mice and IP challenge of all three mouse strains. These data identify a means to define the relative virulence of hvKP strains. It remains unclear whether the observed differences of hvKp virulence in mice translates to human infection. However, these data can be used to sort random collections of K. pneumoniae strains into fvhvKp and pvhvKp strain cohorts and assess for differences in clinical manifestations and outcomes.
IMPORTANCE The pathogenic potential of hvKp strains is primarily mediated by a large virulence plasmid. The minimal set of genes required for the full expression of the hypervirulent phenotype is undefined. A number of reports describe hvKp strains possessing only a portion of the virulence plasmid; the clinical consequences of this are unclear. Therefore, the goal of this report was to determine whether virulence among hvKp strains varied and, if so, how to best identify the relative virulence of hvKp isolates. Data demonstrate hvKp pathogenic potential varies in CD1 and BALB/c murine infection models. In contrast, measurements of siderophore production and mucoviscosity were unable to discriminate the differences in hvKp isolate virulence observed in mice. This information can be used in future studies to determine the mechanisms responsible for differences between fully virulent hvKp and partially virulent hvKp and whether the differences observed in mice translate to disease in humans.
KEYWORDS: Klebsiella pneumoniae, hypervirulent, infection model, mice, mucoviscosity, pathogenic potential, siderophores, virulence
INTRODUCTION
Klebsiella pneumoniae is widely known for its propensity to cause health care-associated and hospital-acquired infections (HAI) and has become increasingly resistant to antimicrobials (1, 2). However, it is less appreciated that two distinct pathotypes are currently circulating, termed classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) (3), and selected epidemiologic, clinical, and genetic features distinguish these pathotypes. cKp is primarily an opportunistic pathogen that causes HAI in individuals with comorbidities; pneumonia, urinary tract infection, or surgical site infection are common infectious syndromes (4). In stark contrast, hvKp usually causes community-acquired infection (CAI), often in otherwise healthy hosts (3). A monomicrobial hepatic abscess in the absence of biliary disease and community-acquired pneumonia are the most commonly recognized syndromes; however, equally canonical features are the development of multiple sites of infection, several of which are unusual for Klebsiella in a healthy host such as meningitis, necrotizing fasciitis, epidural abscess, splenic, prostatic, or brain abscess, and endophthalmitis (3).
Initial studies established that the genotype that conferred hvKp’s hypervirulent phenotype was associated with the presence of a large >180-kDa plasmid on which hvKp-specific virulence factors resided (5–7). Genotypic biomarkers can be used to accurately identify hvKp strains and an inferred increase in virulence relative to cKp strains. The possession of the combination of the iucA, iroB, peg-344, prmpA, and prmpA2 genes which reside on the canonical hvKp virulence plasmid, albeit imperfectly, most accurately predicts whether a strain possesses the hypervirulent phenotype. This result was validated in the outbred CD1 murine infection model, which was highly discriminatory for differentiating hvKp-rich and cKp-rich strain cohorts that were developed based on clinical criteria from human infection (8). Subsequently, a number of studies have described hvKp isolates that possess some, but not all, of these biomarkers and/or plasmids that contain only a portion of the genomic content present in pK2044 or pLVPK (9–13). Hybrid plasmids that consist of antimicrobial resistance genes and a fraction of the hvKp-specific virulence plasmid have also been described (14–16). However, it remains unclear which combination of genetic elements residing on the hvKp-specific virulence plasmid is needed for the full expression of the hypervirulent phenotype, the relative importance of individual elements, and whether these differences affect the pathogenic potential of a given hvKp strain.
Phenotypic markers have also been used as a means to identify the hypervirulent phenotype. These markers include a variety of in vitro assays, infection models, and the clinical syndrome. The string test, which is a qualitative test for mucoviscosity, is not optimally accurate for differentiating hvKp strains from cKp strains (3, 8). Neutrophil and complement-mediated bactericidal activity are also unable to reliably differentiate hvKp from cKp strains (17–20). Likewise, although capsule types have also been proposed as a means to identify hvKp strains, both cKp and hvKp strains can possess a given capsule type (3). However, the production of >30 μg/ml of siderophores in K. pneumoniae has been shown to strongly correlate with lethality in the outbred CD1 murine infection model (8), but this assay is burdensome and not routinely utilized. Alternatively, the phenotype of virulence in infection models is also used. The Galleria mellonella infection model has been commonly employed for this purpose, but it is unable to reliably differentiate hvKp from cKp strains (20–23). Although the clinical syndrome of invasive infection in healthy individuals from the community may be highly suggestive that the offending pathogen is an hvKp isolate, this distinction cannot always be made, especially with the recognition that hvKp can cause health care-associated and nosocomial infection (9, 24, 25). The most accurate phenotypic means to differentiate hvKp from cKp strains has been assessing virulence in mouse infection models such as subcutaneous (SQ) challenge of outbred CD1 mice (8). The accuracy of other mouse strains and challenge routes has been less rigorously assessed.
Data from our group, and others, have suggested that not all hvKp strains are equally virulent in murine infection models (8, 26). Therefore, the goal of this study was to establish whether a virulence spectrum of hypervirulent K. pneumoniae exists, and if so, the value of siderophore production, mucoviscosity, and various murine infection models for identifying differences in hvKP pathogenic potential. The data generated established that a virulence spectrum exists for hvKp strains when assessed in CD1 and BALB/c mice, but not C57BL/6 mice. Unfortunately, quantitative total siderophore production and quantitative mucoviscosity assays were not able to discern the differences in pathogenic potential of hvKp strains identified in mice. Whether the observed differences of hvKp virulence in mice translates to human infection is presently unknown but can now be elucidated in future studies.
RESULTS
Identification of the fvhvKp, pvhvKp, and cKp strain cohorts.
The CD1 subcutaneous challenge (SQ) infection model has been validated for differentiating hvKp from cKp pathotypes, and it is extraordinarily sensitive as evidenced by at least a 5-log-unit window for detecting differences in lethality after bacterial challenge with fully virulent hvKp and cKp strains. Therefore, this model was initially used to assess for potential differences in virulence between hvKp strains (8). Our hvKp strain library (n = 100) and a collection of isolates from the Walter Reed Army Institute of Research Multidrug-Resistant Organism Repository and Surveillance Network (MRSN) that were identified by possessing all of the hvKp-specific biomarkers iucA, iroB, prmpA, prmpA2, and peg-344 (n = 13) underwent screening. The following three K. pneumoniae strain cohorts were defined by their phenotype after SQ challenge of outbred CD1 mice; fully virulent hvKp strains (fvhvKP) were lethal at a challenge inoculum (CI) of ≤103 CFU; partially virulent hvKp strains (pvhvKp) were lethal at a CI >103 to 107 CFU; and classical K. pneumoniae (cKp) were not lethal at a CI of 107 CFU. Of the 100 hvKp strains in our strain library that consists of isolates from otherwise healthy, ambulatory patients with a clinical syndrome of tissue invasive infection (e.g., hepatic and extrahepatic abscesses, necrotizing fasciitis, or endophthalmitis), 11/100 (11%) possessed a pvhvKP phenotype. Of the 13 MRSN isolates, 7/13 (54%) possessed a pvhvKP phenotype. For this study, 15 fvhvKp strains, 14 pvhvKp strains, and 15 cKp strains were chosen for further study. Quantitative lethality data for these strains is listed in Table S1 in the supplemental material. All fvhvKp strains and 13/14 pvhvKp strains (hvKp42 lacks iucA and rmpA) possess all of the hvKp-specific virulence plasmid genes iucA, iroB, prmpA, prmpA2, and peg-344, which have been shown to accurately identify hvKp strains (8), whereas none of the cKp strains possessed these biomarkers (Table S1). hvKp2 (fvhvKp), hvKp94 (pvhvKp), and cKp1 (cKp) were selected for additional detailed studies in CD1, BALB/c, and C57BL/6 mice. These data support that not all hvKp strains share a similar pathogenic potential in mice.
Strains used in this study. Download Table S1, XLSX file, 0.02 MB (19.4KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SQ challenge of CD1 mice is able to differentiate hvKp2 (fvhvKp) from hvKp94 (pvhvKp).
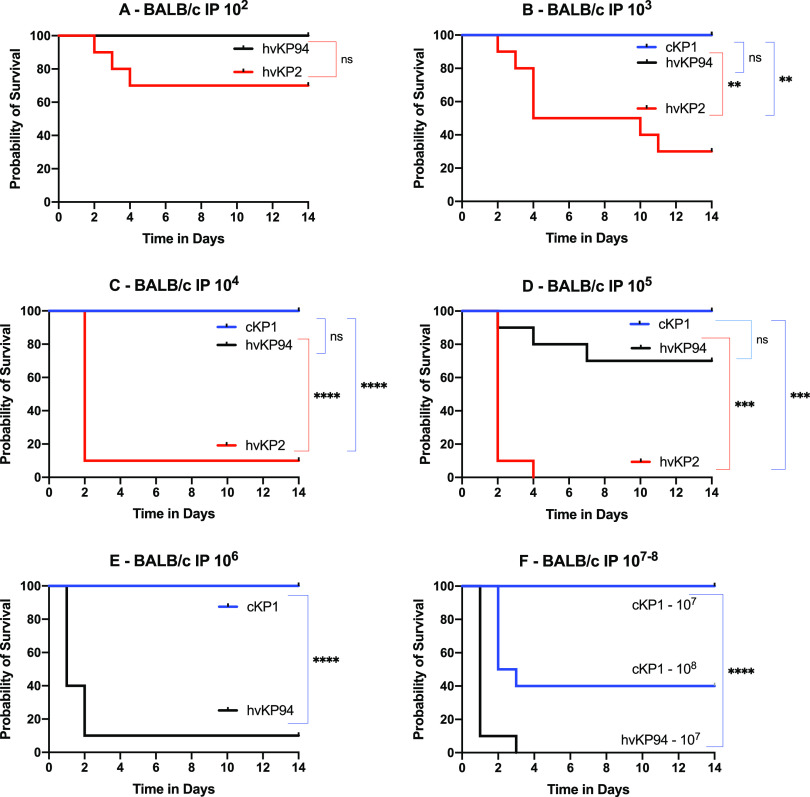
CD1 mice underwent SQ challenge with the fvhvKP strain hvKp2 and the pvhvKp strain hvKp94. hvKp2 was significantly more lethal after SQ challenge with approximately 102 to 103 CFU compared to hvKp94 at similar titers; resultant mortality rates for these CIs were 80% and 100% for hvKP2 compared to 0% and 0% for hvKP94, respectively (Fig. 1A and B and Table 1). SQ challenge with approximately 104 to 107 CFU of hvKp94 resulted in mortality rates of 20%, 70%, 80%, and 80, respectively. Because a 100% mortality rate was predicted based on the 103 CFU challenge data, mice were not challenged with 104-7 CFU of hvKp2 for comparative purposes, since this was deemed inappropriate. Exact titers and P values are listed in Tables S3 and 4, respectively. These data further support that a virulence spectrum exists for hvKp and that SQ challenge of CD1 mice can clearly distinguish between fvhvKp and pvhvKp strains.
FIG 1.
Survival of outbred CD1 mice after subcutaneous (SQ) challenge with hvKp2, hvKp94, and cKp1. (A) The mice were challenged with 2.6 × 102 CFU of hvKp2 and 2.75 × 102 CFU of hvKp94. (B) The mice were challenged with 2.8 × 103 CFU of hvKp2, 3.15 × 103 CFU of hvKp94, and 2.58 × 103 CFU of cKp1. (C) The mice were challenged with 2.85 × 104 CFU of hvKp94, 3.15 × 105 CFU of hvKp94, and 2.85 × 106 CFU of hvKp94. (D) The mice were challenged with 2.48 × 107 CFU of hvKp94, 2.5 × 107 CFU of cKp1, and 3.2 × 108 CFU of cKp1. Strains were grown overnight in LB medium. An in extremis state or death was scored as nonsurvival. Total n = 10 (n = 5 in each of two independent experiments) for each titer for each strain. Values that are significantly different by the log rank (Mantel-Cox) test are indicated as follows: for panel A, ***, P = 0.0003; for panel B, ****, P < 0.0001; and for panel D, ***, P = 0.0003. ns, not significantly different.
TABLE 1.
Mortality rates of CD1, BALB/c, and C57BL/6 mice after challenge with cKp1, hvKp2, and hvKp94
| Mouse | Infectiona | Strain | Mortality rate (%) for mice challenged with the following CFU: |
LD50b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 102 | 103 | 104 | 105 | 106 | 107 | 108 | ||||
| CD1 | SQ | cKp1 | 0 | 0 | 0 | UC | ||||
| hvKp2 | 80 | 100 | UC | |||||||
| hvKp94 | 0 | 0 | 20 | 70 | 80 | 80 | 1.2 × 105 | |||
| CD1 | IP | cKP1 | 0 | 0 | 0 | 0 | 0 | 60 | 2.38 × 108 | |
| hvKp2 | 40 | 60 | 70 | 100 | 1.12 × 103 | |||||
| hvKp94 | 0 | 0 | 0 | 50 | UC | |||||
| BALB/c | IP | cKP1 | 0 | 0 | 0 | 0 | 0 | 60 | 2.61 × 108 | |
| hvKp2 | 30 | 70 | 90 | 100 | 7.98 × 102 | |||||
| hvKp94 | 0 | 0 | 0 | 30 | 90 | 100 | 5.44 × 105 | |||
| C57BL/6 | IP | cKp1 | 0 | 0 | 0 | 0 | 0 | 80 | 2.34 × 108 | |
| hvKp2 | 20 | 40 | 70 | 90 | 4.56 × 103 | |||||
| hvKp94 | 0 | 0 | 50 | 80 | 100 | 3.19 × 104 | ||||
SQ, subcutaneous challenge; IP, intraperitoneal challenge.
LD50, 50% lethal dose; UC, unable to calculate.
Challenge inocula for animal experiments. Download Table S3, XLSX file, 0.01 MB (9.9KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IP challenge of BALB/c and CD1 mice, but not C57BL/6 mice, is able to differentiate hvKp2 (fvhvKp) from hvKp94 (pvhvKp).
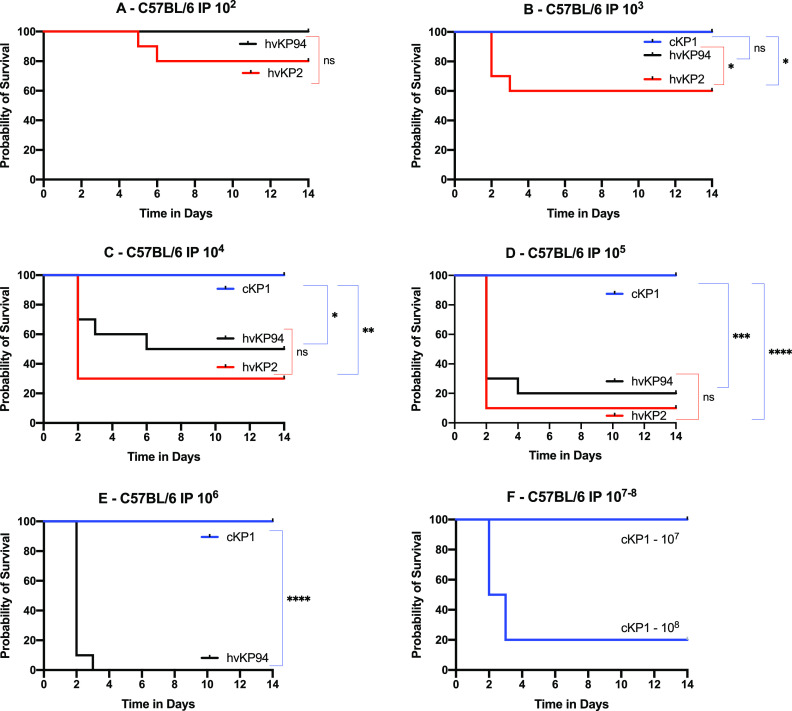
Intraperitoneal (IP) challenge of C57BL/6 or BALB/c mice is commonly used to determine the relative virulence of K. pneumoniae. Therefore, these mouse strains underwent challenge with the fvhvKP strain hvKp2 and the pvhvKp strain hvKp94 to determine whether IP challenge of these mice would be able to differentiate fvhvKp and pvhvKp strains. C57BL/6, BALB/c, and CD1 mice underwent IP challenge with hvKP2 (fvhvKp) and hvKP94 (pvhvKp) (Fig. 2, 3, and 4 and Table 1). When CD1 and BALB/c mice were challenged IP with hvKP2 and hvKP94, significant differences in mortality rates were observed over multiple CIs (Fig. 2 and 3 and Table 1). For CD1 mice, hvKP2 was significantly more lethal after IP challenge with approximately 102 to 105 CFU compared to hvKP94 at similar titers; resultant mortality rates for these CIs were 40%, 60%, 70%, and 100% for hvKP2 compared to 0%, 0%, 0%, and 50% for hvKP94, respectively (Fig. 2A to D and Table 1). For BALB/c mice, hvKP2 trended (P = 0.067) more lethal after IP challenge with approximately 102 CFU and was significantly more lethal with approximately 103 to 105 CFU compared to hvKP94 at similar titers; resultant mortality rates for these CIs were 70%, 90%, and 100% for hvKP2 compared to 0%, 0%, and 30%, for hvKP94, respectively (Fig. 3A to D and Table 1). By contrast, for C57BL/6 mice, a significant difference in lethality was observed only after IP challenge with approximately 103 CFU of hvKp2 compared to hvKp94 at a similar titer; the resultant mortality rates for this CI were 40% and 0%, respectively (Fig. 4B and Table 1). Exact titers and P values are listed in Tables S3 and 4, respectively. These data support that IP challenge of CD1 and BALB/c mice, but not C57BL/c mice, can clearly distinguish between fvhvKp and pvhvKp strains.
FIG 2.
Survival of outbred CD1 mice after intraperitoneal (IP) challenge with hvKp2, hvKp94, and cKp1. (A) The mice were challenged with 2.65 × 102 CFU of hvKp2 and 2.90 × 102 CFU of hvKp94. (B) The mice were challenged with 3.96 × 103 CFU of hvKp2, 2.73 × 103 CFU of hvKp94, and 2.05 × 103 CFU of cKp1. (C) The mice were challenged with 3.96 × 104 CFU of hvKp2, 2.73 × 104 CFU of hvKp94, and 2.05 × 104 CFU of cKp1. (D) The mice were challenged with 3.96 × 105 CFU of hvKp2, 2.90 × 105 CFU of hvKp94, and 1.45 × 105 CFU of cKp1. (E) The mice were challenged with 1.45 × 106 CFU of cKp1, 1.45 × 107 CFU of cKp1, and 2.5 × 108 CFU of cKp1. Strains were grown overnight in LB medium. An in extremis state or death was scored as nonsurvival. Total n = 10 (n = 5 in each of two independent experiments) for each titer for each strain. Values that are significantly different by the log rank (Mantel-Cox) test are indicated as follows: for panel A, *, P = 0.03; for panel B, **, P = 0.004; for panel C, **, P = 0.001; for panel D, *, P = 0.01, ***, P = 0.0001, and ****, P < 0.0001. ns, not significantly different.
FIG 3.
Survival of BALB/c mice after intraperitoneal (IP) challenge with hvKp2, hvKp94, and cKp1. (A) The mice were challenged with 2.60 × 102 CFU of hvKp2 and 2.75 × 102 CFU of hvKp94. (B) The mice were challenged with 2.35 × 103 CFU of hvKp2, 2.80 × 103 CFU of hvKp94, and 1.32 × 103 CFU of cKp1. (C) The mice were challenged with 2.35 × 104 CFU of hvKp2, 2.80 × 104 CFU of hvKp94, and 1.32 × 104 CFU of cKp1. (D) The mice were challenged with 2.35 × 105 CFU of hvKp2, 2.95 × 105 CFU of hvKp94, and 1.32 × 105 CFU of cKp1. (E) The mice were challenged with 2.78 × 106 CFU of hvKp94 and 1.93 × 106 CFU of cKp1. (F) The mice were challenged with .93 × 107 CFU of cKp1 and 2.73 × 108 CFU of cKp1. Strains were grown overnight in LB medium. An in extremis state or death was scored as nonsurvival. Total n = 10 (n = 5 in each of two independent experiments) for each titer for each strain. Values that are significantly different by the log rank (Mantel-Cox) test are indicated as follows: for panel B, **, P = 0.001; for panel C, ****, P < 0.0001; for panel D, ***, P = 0.0001 (cKp1 compared to hvKp2) and ***, P = 0.0002 (hvKp2 compared to hvKp94); for panel E, ****, P < 0.0001; for panel F, ****, P < 0.0001. ns, not significantly different.
FIG 4.
Survival of C57BL/6 mice after intraperitoneal (IP) challenge with hvKp2, hvKp94, and cKp1. (A) The mice were challenged with 2.60 × 102 CFU of hvKp2 and 2.75 × 102 CFU of hvKp94. (B) The mice were challenged with 2.35 × 103 CFU of hvKp2, 2.8 × 103 CFU of hvKp94, and 1.32 × 103 CFU of cKp1. (C) The mice were challenged with 2.35 × 104 CFU of hvKp2, 2.95 × 104 CFU of hvKp94, and 1.32 × 104 CFU of cKp1. (D) The mice were challenged with 2.35 × 105 CFU of hvKp2, 2.95 × 105 CFU of hvKp94, and 1.32 × 105 CFU of cKp1. (E) The mice were challenged with 2.78 × 106 CFU of hvKp94 and 1.93 × 106 CFU of cKp1. (F) The mice were challenged with 1.93 × 107 CFU of cKp1 and 2.73 × 108 CFU of cKp1. Strains were grown overnight in LB medium. An in extremis state or death was scored as nonsurvival. Total n = 10 (n = 5 in each of two independent experiments) for each titer for each strain. Values that are significantly different by the log rank (Mantel-Cox) test are indicated as follows: for panel B, *, P = 0.03; for panel C, *, P = 0.01, **, P = 0.001; for panel D, ***, P = 0.0003, ****, P < 0.0001; for panel E, ****, P = 0.0001. ns, not significantly different.
SQ challenge of CD1 mice may be more sensitive for differentiating fvhvKp from pvhvKp strains compared to IP challenge of CD1 and BALB/c mice.
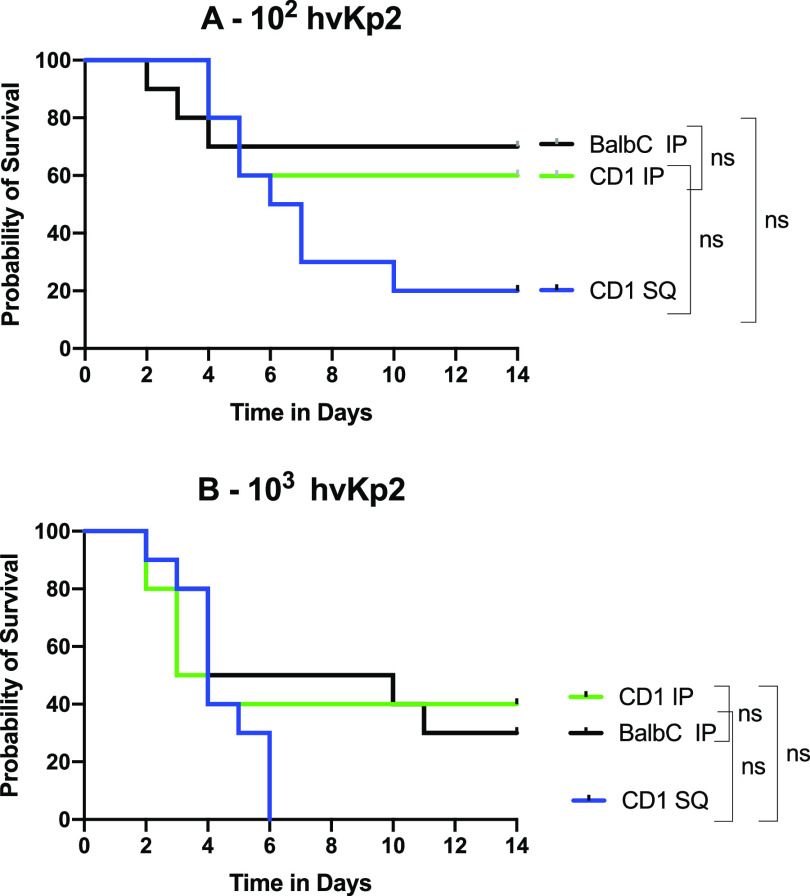
Although both SQ and IP challenge of CD1 mice and IP challenge of BALB/c mice were able to identify significant differences in virulence, as measured by mortality rates between hvKp2 (fvhvKp) and hvKp94 (pvhvKp), SQ challenge was more numerically discerning than IP challenge (Fig. 5 and Table 1). Mortality rates for CD1 mice after SQ challenge with approximately 102 to 103 CFU of hvKP2 were 80% and 100%, respectively (Fig. 5 and Table 1) compared to mortality rates after IP challenge of CD1 mice of 40% (P = 0.158) and 60% (P = 0.381), respectively (Fig. 5 and Table 1), and to mortality rates after IP challenge of BALB/c mice of 30% (P = 0.095) and 70% (P = 0.077), respectively (Fig. 5 and Table 1). No mortality was observed for hvKp94 at similar CIs in CD1 mice challenged SQ and CD1 and BALB/c mice challenged IP. The 100% percent morality rate seen after SQ challenge of CD1 mice with approximately 103 CFU of hvKp2 was not observed after IP challenge in CD1 and BALB/c mice until a CI of approximately 105 CFU was administered (Table 1). Taken together, these data suggest that SQ challenge of CD1 mice may be more sensitive for differentiating fvhvKp from pvhvKp strains than IP challenge of CD1 or BALB/c mice.
FIG 5.
Comparison of CD1 mice challenged SQ with hvKp2 to CD1 and BALB/c mice challenged IP with hvKp2. (A) The CD1 mice were challenged with 2.6 × 102 CFU of hvKp2 SQ and 2.65 × 102 CFU of hvKp2 IP, and the BALB/c mice were IP challenged with 2.6 × 102 CFU of hvKp2. (B) The CD1 mice were challenged with 2.80 × 103 CFU of hvKp2 SQ and 3.96 × 103 CFU of hvKp2 IP, and the BALB/c mice were IP challenged with 2.35 × 103 CFU of hvKp2. Strains were grown overnight in LB medium. An in extremis state or death was scored as nonsurvival. Total n = 10 (n = 5 in each of two independent experiments) for each titer for each strain. ns, not significantly different.
IP challenge of BALB/c, CD1, and C57BL/6 mice or SQ challenge of CD1 mice is able to differentiate hvKp from cKp strains.
In all tested mouse strains and challenge routes, no mortality occurred when challenged with ≤1 × 107 CFU of cKP1 (Fig. 1, 2, 3, and 4 and Table 1). Due to the enhanced virulence of the fvhvK strain hvKp2, lethality was readily noted when low CIs of 102 to 103 CFU were used, whereas for the less virulent pvhvK strain hvKp94, this was not observed consistently until CIs of ≥105 to 106 CFU were used. Interestingly, mortality rates of 60%, 60%, and 80% were observed when CD1, BALB/c, or C57BL/6 mice were challenged IP with approximately 108 CFU of cKP1. By contrast, a 0% mortality rate was observed when CD1 mice were challenged SQ with 3.2 × 108 CFU of cKp1. Whether this was due to a direct bacterial effect and/or the host inflammatory response induced by this high IP CI is unclear. These data support that SQ challenge of CD1 mice or IP challenge of CD1, BALB/c, or C57BL/c mice can distinguish between hvKp and cKp strains.
Quantitative siderophore or mucoviscosity assays are able to accurately differentiate hvKp from cKp strains but are unable to differentiate fvhvKp strains from pvhvKp strains.
Although the murine CD1 SQ challenge and the CD1 and BALB/c IP challenge infection models can accurately differentiate fvhvKp from pvhvKp strains, their use is not pragmatic for the clinical microbiology laboratory or for the study of large numbers of strains. Further, the development of alternatives to the use of animals is desirable when possible. Therefore, we assessed whether the phenotypes of quantitative total siderophore production and mucoviscosity could be used to accurately differentiate fvhvKp from pvhvKp strains.
(i) Quantitative siderophore assay. Previous data have shown that total siderophore production of >30 μg/ml is able to accurately differentiate between hvKp and cKp strains (8). Therefore, the next logical step was to determine whether total siderophore production could differentiate fvhvKp from pvhvKp strains. To accomplish this, three strain cohorts were assessed: (i) fvhvKp strains (n = 15), (ii) pvhvKp strains (n = 14), and (iii) cKp strains (n = 15) (Table S1). The mean siderophore concentration was 315 μg/ml ± 151 μg/ml, 295 μg/ml ± 153 μg/ml, and 20 μg/ml ± 4.1 μg/ml for the fvhvKp, pvhvKp, and cKp strain cohorts, respectively (fvhvKp compared to pvhvKp, P = 0.11; fvhvKp or pvhvKp compared to cKp, P < 0.0001). Results from individual strains are depicted in Fig. 6. All of the fvhvKp strains and 12/14 pvhvKp strains produced siderophore levels of >100 μg/ml. Although 2/14 pvhvKp strains produced siderophore levels of 15.85 ± 2.57 and 34.51 ± 4.9, these data do not support the use of quantitative siderophore measurement as a means to differentiate fvhvKp strains from pvhvKp strains.
FIG 6.
Quantitative siderophore production of cohorts of classical K. pneumoniae (cKp) strains, partially virulent hypervirulent K. pneumoniae (pvhvKp) strains, and fully virulent hypervirulent K. pneumoniae (fvhvKp) strains. (A) cKp strain cohort; (B) pvhvKp strain cohort; (C) fvhvKp strain cohort; (D) All cohorts. Bacterial strains were grown in iron-chelated M9 minimal medium plus Casamino Acids (c-M9-CA). The horizontal line marks 30 μg/ml. Quantitative siderophore production culture supernatants were assessed from culture supernatants using the chromeazurol S dye assay as described previously (36). A total of three independent assays were performed on different days, and the results were reported as means ± SD. ****, P < 0.0001 by Mann-Whitney test. ns, not significantly different.
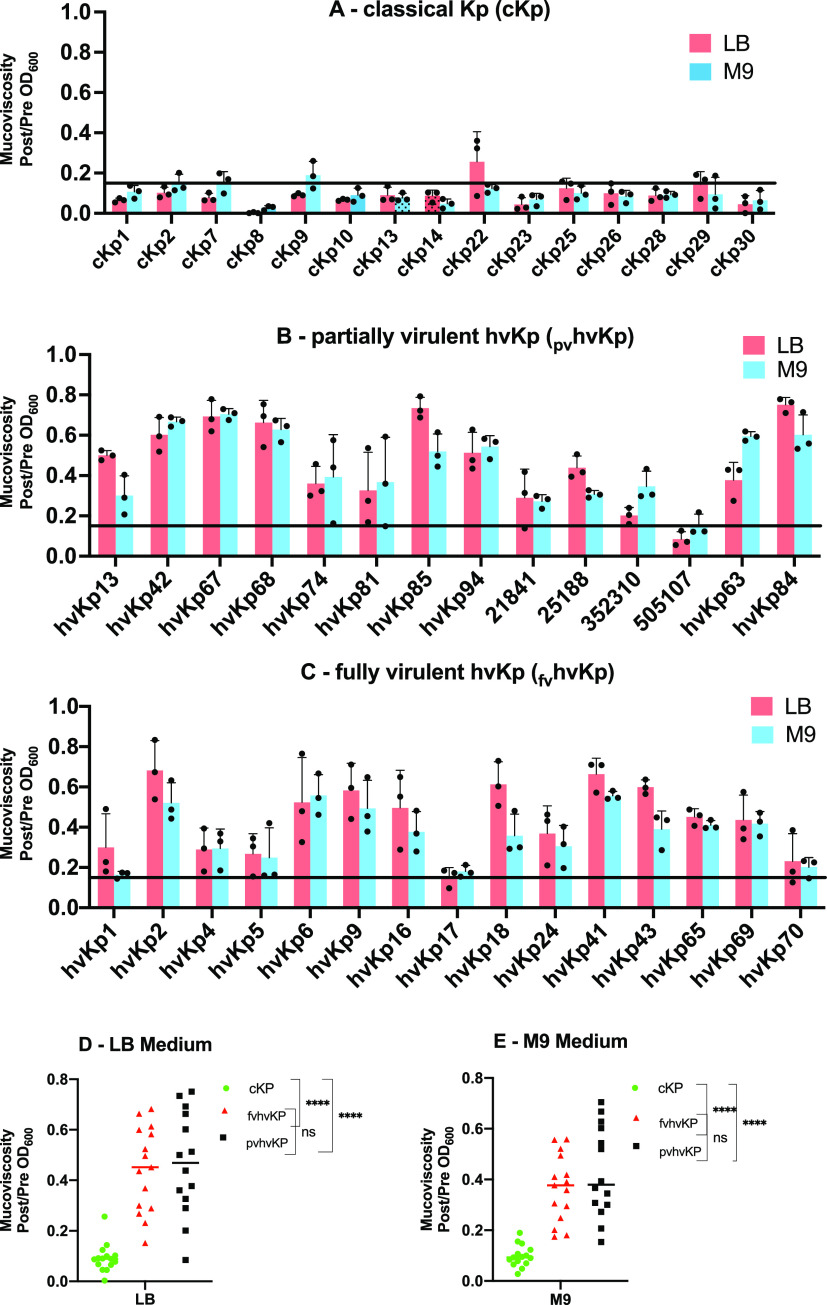
(ii) Quantitative mucoviscosity assay. Mucoviscosity, which is due to RmpA regulated production of RmpD, is a phenotype that is associated with hvKp strains (27–29). Therefore, we performed a quantitative mucoviscosity assay in both lysogeny broth (LB) and c-M9-te (iron-chelated M9 minimum medium plus added trace elements) broth to assess the fvhvKp, pvhvKp, and cKp strain cohorts. The mean mucoviscosity (post/prespin optical density at 600 nm [OD600] ratio) values were 0.44 ± 0.17, 0.47 ± 0.21, and 0.09 ± 0.06 for the fvhvKp, pvhvKp, and cKp strain cohorts, respectively, when grown in LB and 0.37 μg/ml ± 0.13, 0.46 ± 0.18, and 0.10 ± 0.04 when grown in c-M9-te (fvhvKp compared to pvhvKp, P = 0.50; fvhvKp or pvhvKp compared to cKp, P < 0.0001). Results from individual strains are depicted in Fig. 7. All 15 of the fvhvKp and pvhvKp strains had a post/prespin OD600 ratio of >0.15 when grown in c-M9-te broth; 15/15 and 13/14 of the fvhvKp and pvhvKp strains had a post/prespin OD600 ratio of >0.15 when grown in LB. By contrast, 1/15 and 2/15 cKp strains had a post/prespin OD600 ratio of >0.15 when grown in LB and c-M9-te broth, respectively. Interestingly, the mean mucoviscosity values for the index pvhvKp strain hvKp94 were 0.51 ± 0.10 and 0.54 ± 0.05 when grown in LB and c-M9-te broth, respectively, similar to fvhvKp strains. These data demonstrate that the quantitative mucoviscosity assay on average is able to differentiate hvKp from cKp strains but is unable to differentiate fvhvKp from pvhvKp strains.
FIG 7.
Mucoviscosity of cohorts of classical K. pneumoniae (cKp) strains, partially virulent hypervirulent K. pneumoniae (pvhvKp) strains, and fully virulent hypervirulent K. pneumoniae (fvhvKp) strains. Bacterial strains were grown in either lysogeny broth (LB) or iron-chelated M9 minimal medium plus added trace elements (c-M9-te). The culture OD600 was normalized to 1.0 and then centrifuged at 1,000 × g for 5 min. Mucoviscosity was recorded as the post/prespin OD600 ratio. (A) cKp strain cohort; (B) pvhvKp strain cohort; (C) fvhvKp strain cohort; (D) all cohorts grown in LB; (E) all cohorts grown in c-M9-te. The horizontal line marks 0.15. Three independent assays were performed on different days for each strain, and the data were reported as means ± SD. ****, P < 0.0001 by two-way ANOVA.
DISCUSSION
Differentiating cKp from hvKp purely on clinical grounds can be challenging. There is an overlap for selected clinical syndromes, and this becomes more challenging with the recognition that hvKp is causing health care-associated and nosocomial infections similar to cKp (9, 24, 25). Making this distinction has important management implications for fully virulent hvKp strains, for example, vigilance for endophthalmitis, consideration of searching for occult abscess, the use of the largest bore drainage catheter possible, an awareness of propensity for recurrence/relapse, and perhaps hvKp-specific infection control practices (3, 30, 31). Presently, an FDA-approved test for use in the clinical microbiology laboratory is not available to distinguish hvKp strains from cKp strains. However, the presence of iucA, iroB, peg-344, prmpA, and prmpA2 or siderophore production of >30 μg/ml have been shown to accurately differentiate the hvKp and cKp pathotypes (8). Further, albeit limited, data from this report support that a quantitative mucoviscosity assay also holds promise for distinguishing hvKp from cKp. Although impractical for the clinical microbiology laboratory, SQ challenge of CD1 mice is sensitive and accurate for distinguishing the hvKp and cKp pathotypes (8).
Data generated by our group while developing assays to differentiate hvKp from cKp strains, and by others, have suggested that not all hvKp strains are equally virulent in murine infection models (8, 26). Therefore, the goal of this report was to determine whether virulence among hvKp strains varied and, if so, how to best identify the relative virulence of hvKp isolates. Data presented demonstrate that not all hvKp strains share a similar pathogenic potential when assessed by SQ and IP challenge in CD1 mice and by IP challenge of BALB/c mice. Unfortunately, assays that quantitatively measured total siderophore production and mucoviscosity were unable to distinguish between fully and partially virulent hvKp isolates.
It is important to note that it remains unclear whether the observed differences of hvKp virulence in mice translate to human infection. To best answer this question, a random collection of K. pneumoniae strains for which clinical data are available would need to be sorted into fvhvKp, pvhvKp, and cKp strain cohorts and assessed for potential differences in clinical manifestations and outcomes. Key clinical metrics to measure would be the relative ability of each strain cohort to infect multiple sites, metastatically spread, and/or mediate infectious syndromes not usually observed for cKp strains such as central nervous system infection, endophthalmitis, or necrotizing fasciitis. Understanding the clinical behavior of pvhvKp strains in humans could have important implications for management and infection control (3, 30, 31).
The clinical data available for the pvhvKp strains in our collection does not resolve this issue. First, these isolates were not random. Ten isolates were part of the hvKp-rich strain cohort that was used to identify biomarkers to define hvKp (8), and inclusion criteria were based on (i) community acquisition in an otherwise healthy host, (ii) multiple sites of infection or metastatic spread, and (iii) unusual sites of infection for cKp such as endophthalmitis, meningitis, necrotizing fasciitis, nonhepatic abscess (8). Four isolates were from a collection of multidrug-resistant and extremely drug-resistant (MDR-XDR) isolates obtained from military personnel, which were subsequently shown to be hvKp based on the presence of the biomarkers iucA, iroB, prmpA, prmpA2, and peg-344. Nonetheless, some information can be gleaned from this collection. Of 11 pvhvKp strains for which clinical data were available, 10 caused community-acquired infection. Only one of these strains (hvKp42) resulted in multiple sites of infection causing meningitis and brain abscess. However, this strain was from Taiwan and shipped unfrozen to the United States for study. Therefore, a genetic alteration due to handling and storage cannot be excluded. The pvhvKp strain hvKp94 caused prostatic abscesses, but it was likely directly introduced as a result of transrectal biopsy. Therefore, since pvhvKp strains can cause community-acquired infection in otherwise healthy hosts, clinical data support enhanced virulence compared to cKp strains, but it remains unclear whether they are equally virulent to fvhvKp strains and if differences exist, what the management and infection control implications would be, if any.
It is important to note that a knowledge gap exists on the relative virulence of strains having fewer than four or five of iucA, iroB, peg-344, prmpA, and prmpA biomarkers. Twenty-eight of 29 hvKp strains in this study possessed all five of these markers (iucA and rmpA2 were absent in one strain), which is not surprising since they are usually linked on the canonical hvKp virulence plasmid (8). Preliminary data from our group that assessed strains with only one to three of these biomarkers in the CD1 SQ challenge model suggest that these strains are significantly less virulent than the fvhvKp and pvhvKp pathotypes in murine infection models. Additional studies are needed to establish where they stand on the virulence spectrum between fvhvKp and cKp and the potential clinical implications for human disease. Additionally, iutA, iroE, and prmpA can be located within integrative and conjugative elements and may be less reliable markers for the hvKp hypervirulent phenotype since their presence does not necessarily predict the presence of the virulence plasmid (32).
The spectrum of virulence of hvKp strains can be defined via the use of murine infection models. In this study, we established that SQ challenge of CD1 mice or IP challenge of CD1 and BALB/c mice enabled the identification of hvKp strains with decreased virulence that we have termed pvhvKp; SQ challenge of CD1 mice may be the most sensitive model for discerning this difference. Importantly, IP challenge of C57BL/6 mice was poorly discriminatory, so not all mouse models are equal in this regard. Although the mechanisms responsible for these differences were not assessed, one could speculate that outbred mice are more resistant to infection, which in turn increases discriminatory power. Likewise, BALB/c mice possess a TH-2 bias, thereby resulting in increased resistance to extracellular pathogens such as K. pneumoniae, whereas C57BL/6 mice possess a TH-1 bias that results in decreased resistance to extracellular pathogens (18). The route of infection also appears to be important, which may be due to differences in site-specific host defenses. Further, IP challenge results in a more rapid bacteremia due to peritoneal fluid drainage via the lymphatics directly into the bloodstream, which may expedite clearance of lower CIs.
As demonstrated in this study of hvKP94 and other pvhvKp strains, the presence of the combination of iucA, iroB, peg-344, prmpA, and prmpA2 does not necessarily predict the full hypervirulent pathotype, at least as defined in murine infection models (8, 26, 33). One explanation is that other, yet to be identified, virulence genes are needed for maximal virulence, and these genes may not be closely linked with iucA, iroB, peg-344, prmpA, and prmpA2. A genomic comparison of fvhvKp strains with pvhvKp strains may prove to be insightful for delineating the full pantheon of virulence factors requisite for expression of a complete hvKp phenotype.
It is certain that multiple different mechanisms could be responsible for decreased virulence in pvhvKp strains. The first would be the absence or decreased expression of defined virulence factors. The pvhvKp strain hvKp94 produced low levels of total siderophores. It has been previously shown that aerobactin accounts for the majority of siderophores produced by hvKp (34). Preliminary data did not demonstrate a genomic alteration in the aerobactin biosynthetic operon for hvKp94 that precluded production. Therefore, the mechanism(s) responsible for low siderophore production is of interest and may lend insight into the regulation of aerobactin production in hvKp.
This study has several limitations. The mouse breeds assessed were not exhaustive nor were the routes of infection. For example, pulmonary challenge was not studied. However, it was previously shown that lethality was similar after SQ and pulmonary challenge of CD1 mice with the fvhvKp strain hvKp1 (35). For this study, male mice were exclusively used; therefore, any potential gender-based differences would not have been identified. It would also be important to perform the quantitative mucoviscosity assay and in vivo studies on a greater number of fvhvKp, pvhvKp, and cKp strains. Studies on putative hvKp strains that do not possess the complete repertoire set of iucA, iroB, peg-344, prmpA, and prmpA2 would also be of interest.
In summary, this report established that in murine infection models, not all hvKp strains are equally virulent. It remains unclear whether the partially virulent hvKp phenotype observed in mice translates to human disease. This will be important to determine since there are important differences in the management of infections caused by fully virulent hvKp compared to cKp.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study, their origin, sites of infection, and selected genotypic and phenotypic characteristics are listed in Table S1 in the supplemental material. A number of these strains have been previously reported (designated by reference 8 in Table S1) for which quantitative siderophore production and semiquantitative in vivo data were previously reported. For in-depth murine infection studies hvKp2, hvKp94, and cKp1 were used. hvKp2 (ST23, K1; iucA+ iroB+ peg-344+ prmpA+ prmpA2+) was isolated from the blood of a patient who presented from the community in Buffalo, New York, with bacteremia and endophthalmitis (8). hvKp94 (ST23, K1; iucA+ iroB+ peg-344+ prmpA+ prmpA2+) was isolated from the blood of a patient from Buffalo, New York, post-transrectal prostatic biopsy that was complicated by multiple prostatic abscesses. cKp1 (ST2424-1LV, K undefined [not K1, K2, K5, K20, K54, K57]; iucA iroB peg-344 prmpA prmpA2 not present) was isolated from the blood of a patient from Montreal, Quebec, Canada (8). All strains were stored at −80°C prior to use.
Multiplex PCR assay for the identification of selected virulence genes.
Genomic DNA (gDNA) was extracted from 300 μl of a strain grown overnight in lysogeny broth (LB) (5 g yeast extract, 10 g tryptone, 10 g NaCl). Cells underwent centrifugation, and the resultant pellet was resuspended and incubated for 15 min in a Tris-EDTA (TE) lysing buffer containing 0.5% sodium dodecyl sulfate (SDS) and 0.16 mg/ml proteinase K, followed by a 30-min incubation at 37°C with DNase-free RNase (0.033 mg/ml). Next, 36 μl of 5 M sodium chloride was added, and the solution was vortexed vigorously and centrifuged (4°C, 11,500 × g, 10 min) to precipitate protein and cellular debris. The gDNA was extracted using isopropanol, followed by two washes with 70% ethanol before resuspension in 10 mM Tris (pH 8.5). DNA concentration and purity were assessed using a Nanodrop One spectrophotometer with a minimum requirement of a 260/280 ratio of 1.8. Multiplex PCR was performed using 2× Frogga Taq Mix, 12.5 ng gDNA, 0.1 nM each primer and nuclease-free water in a 10-μl reaction mixture. PCR conditions were as follows: 2 min at 95°C; 35 cycles, with 1 cycle consisting of 15 s at 95°C, 30 s at 60/53°C, and 1.5 min at 72°C, with a hold at 4°C. The PCRs were analyzed on a 1.5% agarose gel. Four multiplex PCRs were performed with the following primer pair combinations (Table S2): (i) iucA pair #1, iroB, irp2, and gapA (control for DNA integrity); (ii) peg589 and peg1631; (iii) peg-344 and prmpA; (iv) iucA pair #2, prmpA2 and terB. The hypervirulent strain hvKP1 (8) served as a positive control, and water served as contamination control.
Multiplex PCR primers. Download Table S2, XLSX file, 0.01 MB (10.1KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative siderophore assay.
For the quantitative siderophore assay, strains were grown individually overnight at 37°C in iron-chelated M9 minimal medium containing Casamino Acids (c-M9-CA) (35), and culture supernatants were assessed using the chromeazurol S dye assay as described previously (36). A minimum of three assays were performed, and the results were reported as means ± standard deviations (SD).
Mucoviscosity assay.
Overnight cultures grown in LB broth were used to inoculate 10 ml of either LB or iron-chelated M9 minimum medium plus added trace elements (5 μg/ml CaCl2, 1 μg/ml CoCl2, 20 μg/ml MgCl2, 10 μg/ml MnCl2) (c-M9-te) to a starting OD600 of approximately 0.2. Pilot experiments compared the mucoviscosity of selected strains grown to either an OD600 of 1.0, grown for 6 h, or grown for 24 h. It was determined that the 24-h cultures demonstrated higher mucoviscosity values and allowed greater differentiation between strains; therefore, mucoviscosity was measured after growth for 24 h. Strains were grown shaking (275 rpm) for 24 h at 37°C, and the OD600 was normalized to 1.0 (prespin OD600) with either LB or c-M9-te minimal medium as appropriate. The OD600-adjusted culture (1.5 ml) was then centrifuged at 1,000 × g for 5 min. Seven hundred microliters was carefully withdrawn from the top of the supernatant without disturbing the pellet, and the OD600 was measured (postspin OD600). Mucoviscosity was recorded as the post/prespin OD600 ratio. Three independent assays were performed on different days for each strain, and the data were reported as means ± SD.
Mouse subcutaneous and intraperitoneal challenge infection models.
Animal studies were reviewed and approved by the Veterans Administration Institutional Animal Care Committee and the University at Buffalo-SUNY and were carried out in strict accordance with the recommendations in the guidelines delineated in the "NIH Guide for the Care and Use of Laboratory Animals" (revised 1985) and the "Ethics of Animal Experimentation Statement" (Canadian Council on Animal Care, July, 1980) as monitored by the Institutional Animal Care and Use Committee (IACUC). All efforts were made to minimize suffering. Veterinary care for the animals was supplied by the staff of the Veterans Administration Animal Facility under the direction of a fully licensed veterinarian. BALB/c, C57BL/6, and CD1 male mice, 4 to 6 weeks old, were obtained from Charles River Laboratories and quarantined for 5 days before use. Bacteria were grown overnight in LB and were subsequently serially diluted to the required titers in 1× phosphate-buffered saline (PBS). A 100-μl bacterial suspension was injected either subcutaneously (SQ) or intraperitoneally (IP) using a 0.5-ml insulin syringe. The bacterial titer was enumerated by serial dilutions of the inoculum. The animals were closely monitored for 14 days after challenge for the development of the study endpoints, survival or severe illness (in extremis state)/death, which was recorded as a dichotomous variable. Signs that were monitored and that resulted in immediate euthanasia using methods consistent with the recommendations of the American Veterinary Medical Association Guidelines included hunched posture, ruffled fur, labored breathing, reluctance to move, photophobia, and dehydration. Challenge inocula for all animal experiments are listed in Table S3.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 9.0. A log rank (Mantel-Cox) test was performed to analyze in vivo infection model data. A Mann-Whitney test was used to compare siderophore concentrations between strain cohorts, and a two-way analysis of variance (ANOVA) was performed for the mucoviscosity assays to compare strain cohorts when grown in LB and c-M9-te medium. Bonferroni posttests were used to account for multiple comparisons. A P value of <0.05 was considered statistically significant. P values for all animal experiments are listed in Table S4.
P values for animal experiments. Download Table S4, XLSX file, 0.01 MB (10.8KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by NIH R21 AI123558-01 and 1R21AI141826-01A1 (T. A. Russo) and the Department of Veterans Affairs VA Merit Review (I01 BX004677-01) (T. A. Russo).
The funders had no role in the decision to publish or the preparation of this article.
Contributor Information
Thomas A. Russo, Email: trusso@buffalo.edu.
Vincent B. Young, University of Michigan—Ann Arbor
REFERENCES
- 1.Chen L, Todd R, Kiehlbauch J, Walters M, Kallen A. 2017. Notes from the field: pan-resistant New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae − Washoe County, Nevada, 2016. MMWR Morb Mortal Wkly Rep 66:33. doi: 10.15585/mmwr.mm6601a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA, NISC Comparative Sequencing Program Group . 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 5.Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. 1989. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol 3:1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 6.Nassif X, Sansonetti PJ. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608. doi: 10.1128/IAI.54.3.603-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang HL, Chiang MK, Liou WJ, Chen YT, Peng HL, Chiou CS, Liu KS, Lu MC, Tung KC, Lai YC. 2010. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29:689–698. doi: 10.1007/s10096-010-0915-1. [DOI] [PubMed] [Google Scholar]
- 8.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2017. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/s1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 10.Lam MMC, Wyres KL, Duchene S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, Tournebize R. 2014. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 12:41. doi: 10.1186/1741-7007-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630-15. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, Ren J, Zhu T, Cao Z, Yu Z, Shao C, Shen Z, Ding B, Yuan J, Zhao X, Guo Q, Xu X, Huang J, Wang M. 2016. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 6:165. doi: 10.3389/fcimb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. 2018. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Wai-Chi Chan E, Zhang R, Chen S. 2019. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol 4:2039–2043. doi: 10.1038/s41564-019-0566-7. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Ye L, Chan EW, Zhang R, Chen S. 2020. Tracking recombination events that occur in conjugative virulence plasmid p15WZ-82_Vir during the transmission process. mSystems 5:e00140-20. doi: 10.1128/mSystems.00140-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomakova DK, Hsiao CB, Beanan JM, Olson R, Macdonald U, Keynan Y, Russo TA. 2012. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 18.Gao Q, Shen Z, Qin J, Liu Y, Li M. 2020. Antimicrobial resistance and pathogenicity determination of community-acquired hypervirulent Klebsiella pneumoniae. Microb Drug Resist 26:1195–1200. doi: 10.1089/mdr.2019.0439. [DOI] [PubMed] [Google Scholar]
- 19.Shen P, Berglund B, Chen Y, Zhou Y, Xiao T, Xiao Y, Zhou K. 2020. Hypervirulence markers among non-ST11 strains of carbapenem- and multidrug-resistant Klebsiella pneumoniae isolated from patients with bloodstream infections. Front Microbiol 11:1199. doi: 10.3389/fmicb.2020.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Q, Lan P, Huang D, Hua X, Jiang Y, Zhou J, Yu Y. 2018. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol 18:94. doi: 10.1186/s12866-018-1236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russo TA, MacDonald U. 2020. The Galleria mellonella infection model does not accurately differentiate between hypervirulent and classical Klebsiella pneumoniae. mSphere 5(1):e00850-19. doi: 10.1128/mSphere.00850-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Du P, Zhao J, Li B, Wang C, Sun L, Lu B, Wang Y, Liu Y, Cao B. 2020. Phenotypic and genomic characterization of virulence heterogeneity in multidrug-resistant ST11 Klebsiella pneumoniae during inter-host transmission and evolution. Infect Drug Resist 13:1713–1721. doi: 10.2147/IDR.S243836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Shi J, Zhao Y, Xie Y, Tang Y, Jiang X, Lu Y. 2020. Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur J Clin Microbiol Infect Dis 39:1673–1679. doi: 10.1007/s10096-020-03890-z. [DOI] [PubMed] [Google Scholar]
- 24.Harada S, Aoki K, Yamamoto S, Ishii Y, Sekiya N, Kurai H, Furukawa K, Doi A, Tochitani K, Kubo K, Yamaguchi Y, Narita M, Kamiyama S, Suzuki J, Fukuchi T, Gu Y, Okinaka K, Shiiki S, Hayakawa K, Tachikawa N, Kasahara K, Nakamura T, Yokota K, Komatsu M, Takamiya M, Tateda K, Doi Y. 2019. Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J Clin Microbiol 57:e01206-19. doi: 10.1128/JCM.01206-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. 2020. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence 11:1215–1224. doi: 10.1080/21505594.2020.1809322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyrouthy R, Dalmasso G, Birer A, Robin F, Bonnet R. 2020. Carbapenem resistance conferred by OXA-48 in K2-ST86 hypervirulent Klebsiella pneumoniae, France. Emerg Infect Dis 26:1529–1533. doi: 10.3201/eid2607.191490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker KA, Miller VL. 2020. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol 54:95–102. doi: 10.1016/j.mib.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, Sepúlveda VE, Quinn JD, Miller VL. 2019. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio 10:e00089-19. doi: 10.1128/mBio.00089-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker KA, Treat LP, Sepúlveda VE, Miller VL. 2020. The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. mBio 11:e01750-20. doi: 10.1128/mBio.01750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marr CM, Russo TA. 2018. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther 17:71–73. doi: 10.1080/14787210.2019.1555470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sellick JA, Russo TA. 2018. Getting hypervirulent Klebsiella pneumoniae on the radar screen. Curr Opin Infect Dis 31:341–346. doi: 10.1097/QCO.0000000000000464. [DOI] [PubMed] [Google Scholar]
- 32.Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. 2011. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology (Reading) 157:3446–3457. doi: 10.1099/mic.0.050336-0. [DOI] [PubMed] [Google Scholar]
- 33.Lazareva I, Ageevets V, Sopova J, Lebedeva M, Starkova P, Likholetova D, Lebedeva M, Gostev V, Moiseenko V, Egorenkov V, Navatskaya A, Mitroshina G, Myasnikova E, Tsvetkova I, Lobzin Y, Sidorenko S. 2020. The emergence of hypervirulent bla(NDM-1)-positive Klebsiella pneumoniae sequence type 395 in an oncology hospital. Infect Genet Evol 85:104527. doi: 10.1016/j.meegid.2020.104527. [DOI] [PubMed] [Google Scholar]
- 34.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. 2015. Aerobactin, but not yersiniabactin, salmochelin and enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. 2017. Metabolite transporter PEG344 is required for full virulence of hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect Immun 85:e00093-17. doi: 10.1128/IAI.00093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains used in this study. Download Table S1, XLSX file, 0.02 MB (19.4KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Challenge inocula for animal experiments. Download Table S3, XLSX file, 0.01 MB (9.9KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiplex PCR primers. Download Table S2, XLSX file, 0.01 MB (10.1KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
P values for animal experiments. Download Table S4, XLSX file, 0.01 MB (10.8KB, xlsx) .
Copyright © 2021 Russo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.