ABSTRACT
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had a massive impact on human lives worldwide. While the airborne SARS-CoV-2 primarily affects the lungs, viremia is not uncommon. As placental trophoblasts are directly bathed in maternal blood, they are vulnerable to SARS-CoV-2. Intriguingly, the human fetus is largely spared from SARS-CoV-2 infection. We tested whether the human placenta expresses the main SARS-CoV-2 entry factors angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), and furin and showed that ACE2 and TMPRSS2 are expressed in the trophoblast rather than in other placental villous cells. While furin is expressed in the main placental villous cell types, we surveyed, trophoblasts exhibit the highest expression. In line with the expression of these entry factors, we demonstrated that a SARS-CoV-2 pseudovirus could enter primary human trophoblasts. Mechanisms underlying placental defense against SARS-CoV-2 infection likely involve postentry processing, which may be germane for mitigating interventions against SARS-CoV-2.
IMPORTANCE Pregnant women worldwide have been affected by COVID-19. As the virus is commonly spread to various organs via the bloodstream and because human placental trophoblasts are directly bathed in maternal blood, feto-placental infection by SARS-CoV-2 seems likely. However, despite the heightened risk to pregnant women, thus far the transmission risk of COVID-19 to the feto-placental unit seems extremely low. This has been recently attributed to a negligible expression of SARS-CoV-2 entry factors in the human placenta. We therefore sought to explore the expression of the entry factors ACE2 and TMPRSS2 in the different cell types of human placental villi. Using a combination of transcriptome sequencing (RNA-seq), real-time quantitative PCR (RT-qPCR), in situ hybridization, and immunofluorescence, we found that trophoblasts, but not the other main villous cell types, express ACE2 and TMPRSS2, with a broad expression of furin. Correspondingly, we also showed that primary human trophoblasts are permissive to entry of SARS-CoV-2 pseudovirus particles.
KEYWORDS: placenta, trophoblast, SARS-CoV-2, ACE2, TMPRSS2, furin
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a positive-sense, single-stranded RNA virus, is responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to affect millions of human lives worldwide. There is a pressing and unmet need to understand the pathogenesis of SARS-CoV-2, promote disease prevention, enhance diagnostics, repurpose existing therapeutics and innovate new ones, and provide safe pipelines for the development of effective vaccines. Whereas the pathogenesis of COVID-19 remains incompletely understood, accumulating evidence indicates that the initial viral breaching of the immune defense, followed by robust viral replication in the lungs (1), may lead to a systemic cytokine storm (2) that can ultimately result in multiorgan damage or failure.
Angiotensin-converting enzyme 2 (ACE2) was initially identified as a cell membrane carboxypeptidase that modulates blood pressure, including blood pressure during pregnancy, by producing the vasodilator heptapeptide angiotensin-(1–7) (3–5). The viral spike (S) protein of SARS-CoV-2 uses ACE2 receptors as its binding receptor on the host cell membrane (6), thus enabling the subsequent entry of the virus into host cells after proteolytic cleavage of the S protein by transmembrane protease serine 2 (TMPRSS2). This leads to exposure of a fusogenic peptide that promotes fusion of the viral envelope with the host cell membrane and the ensuing endocytosis and viral replication (1, 7). Notably, unlike SARS-CoV, SARS-CoV-2 appears to require an additional cleavage at the S1/S2 site of the S protein by the proprotein convertase furin to enable viral assembly in host cells and to mediate viral spread to permissive cells (8, 9).
Pregnancy poses an increased risk to certain viral infections, such as cytomegalovirus (CMV), rubella, and Zika virus, leading to major fetal morbidity and mortality (10, 11). Due to the high infectivity of SARS-CoV-2 and its transmission via respiratory secretions or droplets, the transmission of SARS-CoV-2 to pregnant women is inevitable. According to the CDC’s report of a COVID-19-associated hospitalization surveillance network across 13 states until 22 August 2020, approximately 45.5% of pregnant women confirmed as having COVID-19 manifested symptoms leading to admission to an intensive care unit (16.2%) and/or the need for mechanical ventilation (8.5%) (12). The risk is heightened in mothers with preexisting morbid conditions (13, 14). In light of the physiological reduction in lung volume that characterizes pregnancy and based on clinical experience from influenza, SARS-CoV-1, H1N1, and Middle East respiratory syndrome (MERS)-CoV infections, severe respiratory infections may worsen during pregnancy, thus potentially exacerbating maternal morbidity and mortality (15–19). Several reports point to a higher incidence of preterm delivery, premature rupture of the membranes, and other pregnancy complications in SARS-CoV-2-infected mothers (20–22). Notably, these obstetrical complications have been associated with infection by SARS-CoV-1, MERS-CoV, metapneumovirus, respiratory syncytial virus, and influenza virus and largely attributed to maternal respiratory complications and consequent hypoxemia (23–25).
Because (i) human placental trophoblasts are directly bathed in maternal blood and (ii) a high fraction of the cardiac output reaches the human placenta, hematogenous spread to the placenta by blood-borne viruses is likely. Some airborne viruses, such as rubella virus, may cause viremia and affect the feto-placental unit. Similarly, the presence of SARS-CoV-2 viral RNA in the blood (RNAemia) has been detected in 8 to 40% of hospitalized COVID-19 patients (26–29), which may explain the hematogenous spread of SARS-CoV-2 to numerous internal organs (30–33). Thus, trophoblasts at the maternal-fetal interface may directly confront circulating SARS-CoV-2 particles in the blood of COVID-19-positive pregnant women.
Unlike rubella virus, CMV, and Zika virus, most data thus far suggest that the incidence of feto-placental transmission of SARS-CoV-2 is very low, even in mothers with severe clinical COVID-19 (34–41). Although there have been reports of SARS-CoV-2 detection in neonatal nasopharyngeal swabs, amniotic fluid, or cord blood, with elevated immunoglobulin M (IgM) levels in the neonatal blood, these have been rare. Some of these assays were followed by a negative test for viral RNA in the neonate (42, 43). In addition, the reliability of the results has been questioned, mainly due to uncertainty regarding the specificity of the IgM assay (35, 44). Similarly, the diagnostic value of detecting SARS-CoV-2 RNA by qPCR in placental biopsy specimens (39–41, 45–47) is questionable in light of the uncertainty regarding contamination with maternal blood.
The potential ability of SARS-CoV-2 to infect human placental trophoblasts depends on the presence of receptors and processing proteins that mediate trophoblast viral entry. Thus far, such data have been scant and relatively inconsistent. SARS-CoV-2 viral proteins, including S and N (nucleocapsid) proteins, were visualized in syncytiotrophoblasts, which are in direct contact with the maternal circulation (45–48). However, a recent analysis of single-cell transcriptome sequencing (RNA-seq) suggested negligible ACE2 and TMPRSS2 expression in the placenta (49). In contrast, earlier studies showed that ACE2 is expressed in the human placenta, especially in syncytiotrophoblasts, the subjacent cytotrophoblasts, and endothelial cells of the umbilical cord (5, 45, 50–58). The expression of placental ACE2 is increased in patients with preeclampsia, suggesting the functional role of ACE2 in countering hypertension at the human uteroplacental interface (5, 50). Less information exists regarding ACE2 expression in the various villous cell types and the expression of TMPRSS2 and furin. We therefore sought to explore the expression of ACE2 and TMPRSS2 in the different cell types of placental villi. The expression of these processing proteins would suggest that trophoblasts are susceptible to SARS-CoV-2 infection and the ensuing transmission to the fetus. Using a combination of RNA-seq, real-time quantitative PCR (RT-qPCR), in situ hybridization-based RNAscope, and immunofluorescence, we found that trophoblasts, but not the other main placental villous cell types, express ACE2 and TMPRSS2 with broad expression of furin. Correspondingly, we also showed that primary human trophoblasts derived from term placentas are permissive to entry of SARS-CoV-2 pseudovirus particles.
RESULTS
The expression of ACE2, TMPRSS2, and furin mRNA in the human placenta and in villous cells.
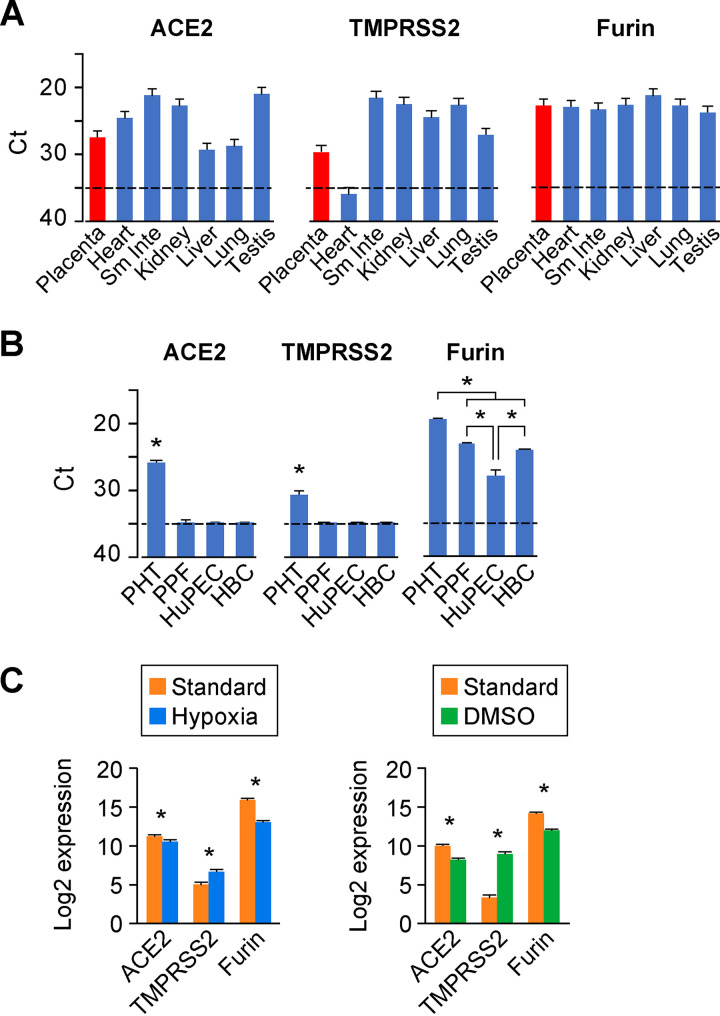
We first used RT-qPCR as a sensitive tool to quantify the expression of three viral entry factors, ACE2, TMPRSS2, and furin, in term human placenta and compared these data to the expression of these transcripts in other human organs, including the heart, small intestine, kidney, liver, lung, and testis. As shown in Fig. 1A, ACE2, TMPRSS2, and furin were clearly expressed in the human placenta, with average threshold cycle (CT) values of 27.5, 29.9, and 22.7, respectively. Intriguingly, the expression level of ACE2 mRNA in the placenta was similar to its expression in the lung, the most common site for SARS-CoV-2 infection in humans. TMPRSS2 levels in most tissues were higher than the placenta, and the expression of furin was fairly comparable among the tissues (Fig. 1A).
FIG 1.
The expression landscape of ACE2, TMPRSS2, and furin in the human placenta and placental cells. (A) The expression of the processing proteins in human organs (Sm Inte, small intestine). Transcripts of ACE2, TMPRSS2, and furin were detected by RT-qPCR using the FirstChoice Human Total RNA Survey Panel. Expression levels are shown by threshold cycle (Ct) values in various human organs. n = 3. (B) The expression of the processing proteins in primary human placental villous cells. Data are derived from cultures of the major primary human placenta cells, including trophoblasts (PHT), fibroblasts (PPF), endothelial cells (HuPEC), and Hofbauer cells (HBC). Expression (mRNA), detected by RT-qPCR, is shown by Ct values. n = 3. *, P < 0.01, ANOVA with post hoc Bonferroni test. For panels A and B, the dashed line represents the assay threshold for the presence of each transcript. Note that lower Ct values indicate higher expression. (C) The expression of the processing proteins in cytotrophoblasts versus syncytiotrophoblasts. Standard/hypoxic conditions (left) and culture in the presence of 1.5% DMSO (right) were performed and analyzed as described in Materials and Methods. RNA-seq data (n = 5) are expressed as log2 value. *, P < 0.01.
To further determine the placental villous cell types that express mRNA of ACE2, TMPRSS2, and furin, we used RT-qPCR to measure these transcripts in the four main villous cell types: primary human trophoblasts (PHT cells), primary placental fibroblasts (PPF cells), primary human placental microvascular endothelial cells (HuPECs), and placental macrophage Hofbauer cells (HBCs). Notably, we routinely culture PHT cells up to 72 h, which has been rigorously tested to ensure their differentiation to the fused multinucleated syncytiotrophoblast phenotype. Interestingly, ACE2 was clearly expressed in PHT cells, whereas expression in PPF, HuPEC, and HBC cells was below the PCR detection threshold. Likewise, TMPRSS2 was also discernibly expressed in PHT cells, but not in the other three cell types. All four main placental cell types expressed furin, with PHT cells exhibiting the highest expression levels (Fig. 1B). These results indicate that, among the main placental villous cell types, only trophoblasts express transcripts for the three key SARS-CoV-2 entry factors. Our data also indicate that furin is ubiquitously expressed in all major cell types of human placenta villi.
We used bulk RNA-seq to validate the expression of ACE2, TMPRSS2, and furin in trophoblasts (Fig. 1C). To profile their expression levels in differentiated syncytiotrophoblasts versus progenitor cytotrophoblasts, we cultured PHT cells in two conditions that are known to hinder the differentiation of cytotrophoblasts to syncytiotrophoblasts in vitro, namely, culture in the presence of 1.5% dimethyl sulfoxide (DMSO) or in hypoxia (<1% O2 for 48 h) (59–62). We found that both conditions resulted in a moderate, yet statistically significant, decrease of ACE2 and furin expression in cytotrophoblasts compared to differentiated syncytiotrophoblasts. Intriguingly, the expression of TMPRSS2 mRNA was significantly increased with exposure to DMSO or hypoxia, compared to the standard condition, which promotes differentiation (Fig. 1C).
The expression of ACE2 and TMPRSS2 proteins in placental villi and in trophoblasts.
Because of the lower and more selective expression pattern of ACE2 and TMPRSS2, we sought to spatially validate the expression of these two transcripts in trophoblasts. As shown in Fig. 2, we performed in situ hybridization using the highly sensitive RNAscope technology. While there were no obvious punctae using the negative-control probe, when using the ACE2 or TMPRSS2 probes, we noted prominent punctae in trophoblasts, located at the outermost layer of the placental villi (Fig. 2, arrows in insets). These results confirmed our findings using RT-qPCR and RNA-seq, with regard to trophoblastic expression of ACE2 and TMPRSS2. To further corroborate our mRNA expression findings, we examined the expression and location of ACE2 and TMPRSS2 proteins in the placental villi and PHT cells using immunofluorescence labeling. As shown in Fig. 3A and B, ACE2 and TMPRSS2 were significantly expressed in the outer, trophoblast layer at the periphery of the placental villi. Furthermore, we used Western blotting to confirm TMPRSS2 expression in cultured PHT cells (Fig. 3C), providing further support to the expression of SARS-CoV-2 processing proteins in human villous trophoblasts.
FIG 2.
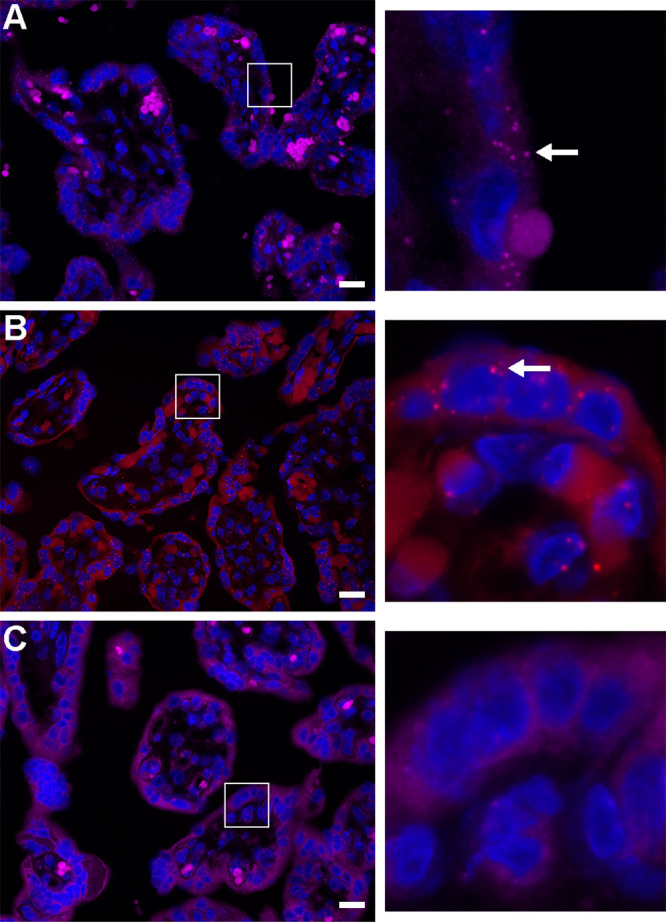
In situ hybridization for the expression of ACE2 and TMPRSS2 mRNA in human placental villi. Paraffin-embedded human placental villi sections were processed as detailed in Materials and Methods. RNAscope probes targeting ACE2 (A), TMPRSS2 (B), or control probe (C) were used according to the manufacturer’s instructions. The hybridized probe signal was amplified using the preamplifier reagent followed by the addition of Opal fluorophores. Both ACE2 (A) and TMPRSS2 (B) show notable punctae (white arrow) in the outermost syncytiotrophoblast layer of the placental villi, compared to the negative control (C). The panels on the right are a magnification of the white squares in panels A to C. The panels represent three independent experiments. Bars, 20 μm.
FIG 3.
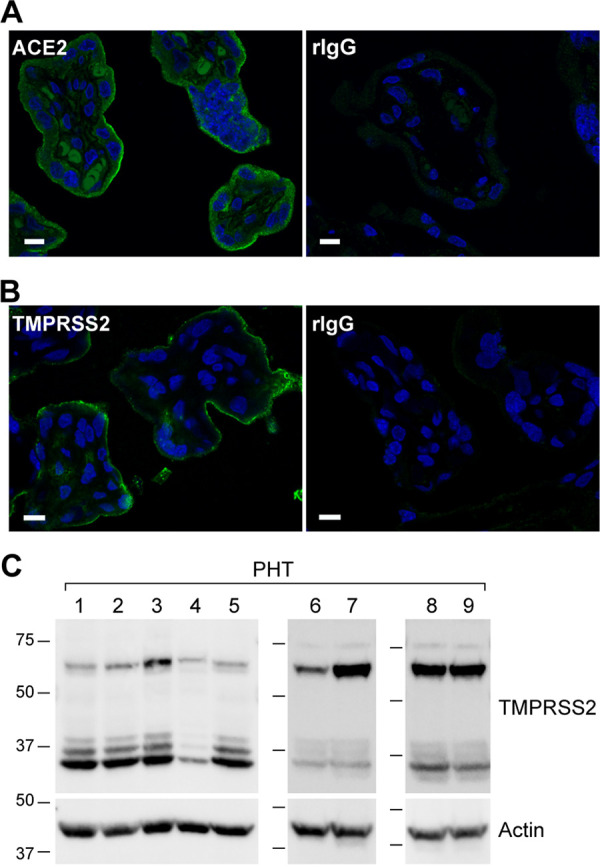
Immunofluorescence localization of ACE2 and TMPRSS2 proteins in the trophoblasts of human placental villi. (A and B) The human placenta villous sections were immunostained with antibodies to ACE2 (A) and TMPRSS2 (B). The concentrations of each pair of specific and control, nonspecific IgG (rIgG), were identical. All image acquisition parameters and intensity scales were identical for each pair of specific and nonspecific antibodies. The panels represent three independent experiments. Bars, 10 μm. (C) Western blot of TMPRSS2 in primary human trophoblasts (PHTs). Whole-cell lysates of PHT cells (60 μg), derived from nine independent placentas, were resolved on SDS-PAGs and transferred to PVDF membranes that were blotted in three independent Western immunoblots using a mouse anti-TMPRSS2 antibody as detailed in Materials and Methods. Note that TMPRSS2 is expressed as a full-length zymogen protein (∼70 kDa) and is processed by proteolytic cleavage (89, 90), producing three activated mature forms with a smaller size (∼37 kDa), derived from two isoforms.
Whereas ACE2 proteins were enriched in the apical membrane of villous trophoblasts, we noted the presence of cytoplasmic punctae in the trophoblasts. This pattern of ACE2 expression prompted us to investigate whether the ACE2 protein undergoes endocytosis in trophoblasts. To address this possibility, we analyzed the colocalization of ACE2 with the canonical early endosome marker EEA.1 in both human villous cryosections and in cultured PHT cells. We found that ACE2- and EEA.1-demarcated endosomes were expressed in villous trophoblasts, with a lower-level expression in the villous core (Fig. 4A to D). Importantly, cytoplasmic ACE2 punctae partially colocalized with EEA.1 in both placental villi (Fig. 4D) and in cultured PHT cells (Fig. 4E to G), suggesting that trophoblastic ACE2 is endocytosed into early endosomes. To assess this possibility, we incubated PHT cells with an Alexa Fluor 488-conjugated ACE2 antibody that recognizes the extracellular domain of ACE2, and we examined the localization of the ACE2-antibody complex in live PHT cells. As shown in Fig. 4H, ACE2 localized to the plasma membrane and intracellular vesicles (Fig. 4I, arrows), thus directly demonstrating endocytosis of ACE2 in trophoblasts. It should be noted that both immunofluorescence and live-cell imaging revealed considerable heterogeneity in the level of ACE2 expression, pattern of localization and endocytosis within the population of PHT cells, likely due to the diverse differentiation state of cells. Overall, our results indicate that the trophoblasts at the outmost layer of the human placental villi express SARS-CoV-2 entry factors, including ACE2, TMPRSS2, and furin, and that these cells are likely capable of ACE2 endocytosis.
FIG 4.
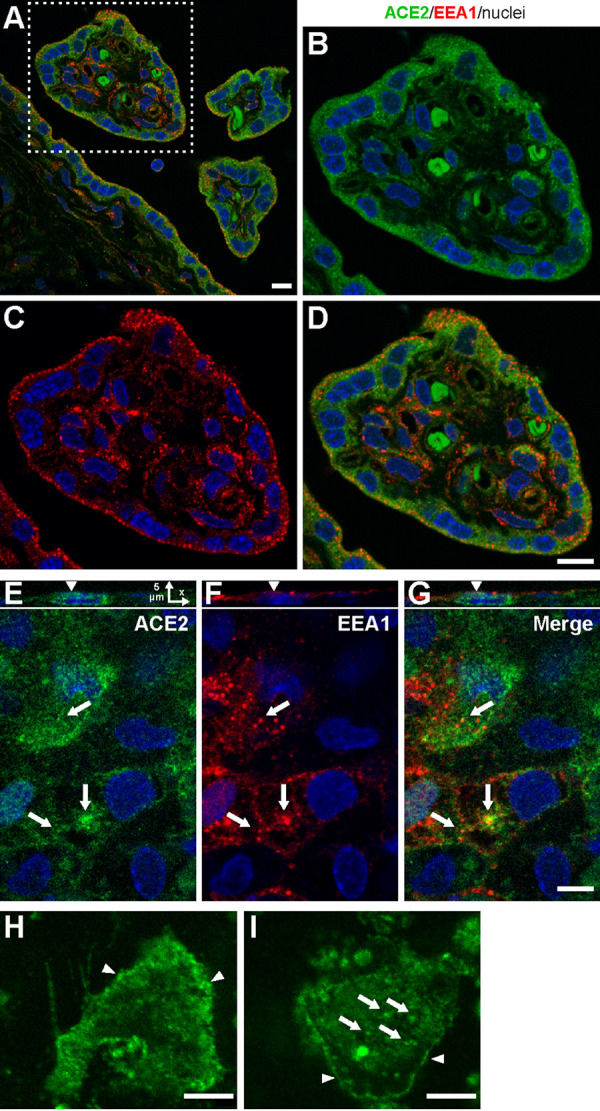
The expression of ACE2 and early endosomal EEA.1 in human placental villi and cultured PHT cells. Placenta crysections were immunolabeled for ACE2 (green) and EEA.1 (red, marker of early/sorting endosomes), and z stacks of x-y confocal images were acquired. (A) Merged image is shown. (B to D) High-magnification ACE2 and EEA.1 images of a cross section of an individual terminal villus, corresponding to the area marked by the square outlined by a white dashed line in panel A. (E to G) Fixed PHT cells were permeabilized and costained for ACE2 and EEA.1 antibodies. x-z images in panel E show a single cross section of the three-dimensional (3-D) image. Arrows in panels E to G show examples of ACE2 localization in EEA.1-containing endosomes. Individual x-y confocal images of 3-D image z stacks are shown. Arrows point at endosomes. (H and I) Representative examples of live-cell images of PHT cells incubated with the Alexa Fluor 488-conjugated ACE2 antibody for 90 min at 37°C. The arrowheads in panel H show ACE2 localization on the plasma membrane, and the arrows in panel I denote the ACE2 cytoplasmic punctae following endocytosis. The panels represent three independent experiments. Bars, 10 μm.
SARS-CoV-2 pseudovirus can enter PHT cells.
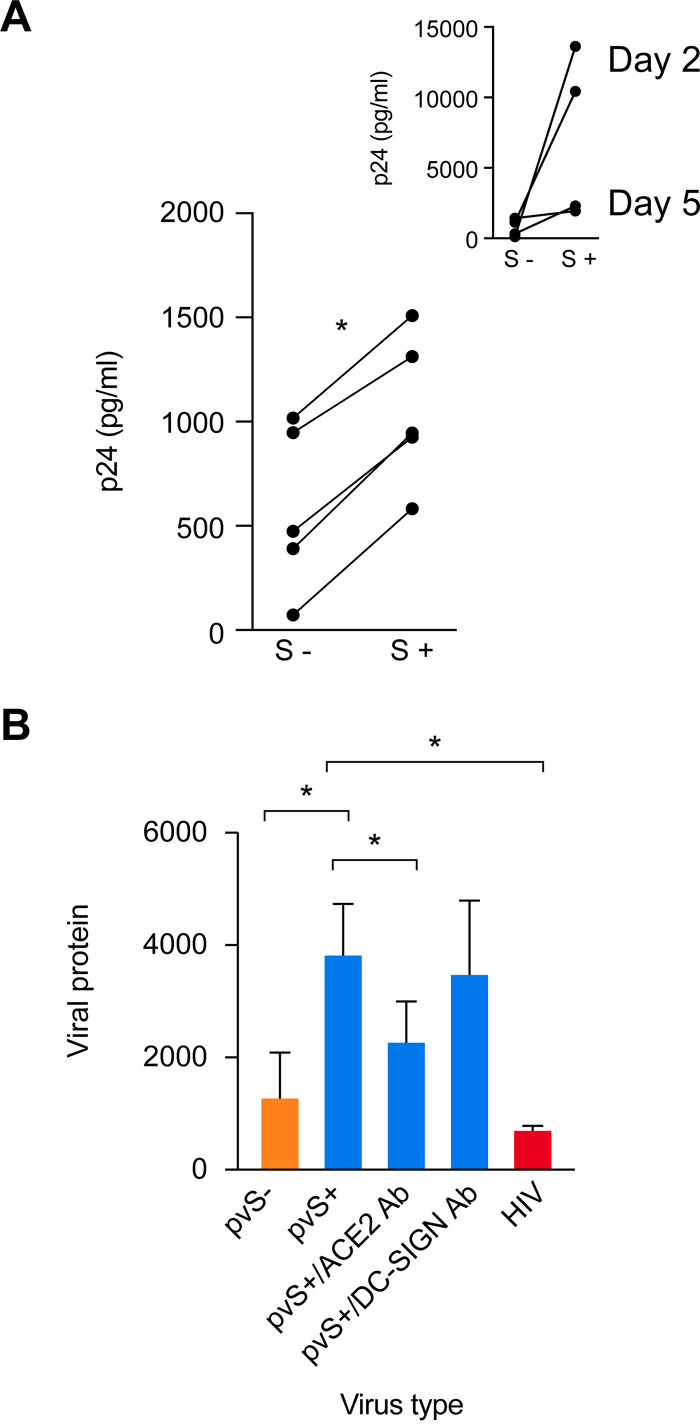
In light of our findings that the three key SARS-CoV-2 entry factors, ACE2, TMPRSS2, and furin, are expressed in villous trophoblasts, we assessed the susceptibility of PHT cells to entry of replication-deficient lentiviral particles pseudotyped with SARS-CoV-2 viral proteins (pvSARS-CoV-2) membrane (M), nucleocapsid (N), and envelope (E), with (SMNE) or without (MNE) the spike (S) protein. We examined the amount of intracellular lentiviral p24 capsid proteins present in pvSARS-CoV-2 viruses as a measure of cellular viral entry. As expected, we detected robust expression of p24 in HEK293T cells overexpressing ACE2 following 2 days of infection with S-protein-expressing pvSARS-CoV-2, with some decline at day 5 (Fig. 5A, inset), confirming the S-protein dependency of pseudovirus entry. Importantly, PHT cells were infected by pvSARS-CoV-2 SMNE at a significantly higher level than by pvSARS-CoV-2 MNE (Fig. 5A). Overall, the degree of PHT cell infection was markedly lower than that using 293T cells. We confirmed the specificity of pvSARS-CoV-2 S+ entry into PHT cells by preincubating the cells with anti-ACE2 antibody, demonstrating that this significantly reduced viral entry, measured by intracellular p24. This reduction was not observed when we used preincubation with anti-DC-SIGN, which is irrelevant for SARS-CoV-2 entry (Fig. 5B). As expected, PHT cells infected with pvSARS-CoV-2 S- or infected with inactivated HIV with conformationally intact gp160 displayed markedly lower level of viral entry. Overall, these results demonstrate the importance of ACE2 and spike protein for pseudovirus entry into PHT cells. These results are consistent with our expression data, suggesting that human trophoblasts are permissive to pvSARS-CoV-2 entry.
FIG 5.
The ACE2-dependent entry of SARS-CoV-2 pseudovirus into cultured PHT cells. (A) PHT (five donors) and 293T-ACE2 cells inoculated with SARS-CoV-2 protein pseudotyped lentiviruses containing SMNE proteins (spike, membrane, nucleocapsid, and envelope) or MNE proteins, as detailed in Materials and Methods. Cells, harvested on day 2 postinoculation with the virus, were washed three times with PBS and trypsinized to remove adherent virus, and the cell pellet was lysed to release intracellular p24. Shown is p24 content (in picograms per milliliter) in cell lysates. *, P < 0.01 (paired t test). (Inset) 293T cells expressing ACE2, assessed at 2 days or 5 days (n = 2 for each) and serving as a positive control. (B) PHT were preincubated or not with 20 μg/ml of anti-ACE-2 or anti-DC-SIGN antibody (Ab) and then inoculated with pvSARS-CoV-2 S+. The PHT cells were also inoculated with pseudovirus lacking S protein (pvS-) or with inactivated HIV-1 as negative controls. Cells were harvested on day 2 postinoculation, and p24 content (in picograms per milliliter) was measured in cell lysates, as detailed in Materials and Methods. *, P < 0.05 (ANOVA and Tukey test, n = 5).
DISCUSSION
Several lines of evidence support our conclusion that villous placental trophoblasts from term human pregnancies express ACE2, TMPRSS2, and furin. (i) We detected the transcripts for ACE2, TMPRSS2, and furin in term placental trophoblasts by RT-qPCR and RNA-seq. (ii) We used RNAscope to visualize intracellular ACE2 and TMPRSS2 mRNA punctae in trophoblasts of the term placental villi. (iii) We identified ACE2 and TMPRSS2 proteins in the term placental villi and in cultured term PHT cells. Whereas furin is expressed in all four cell types we examined, ACE2 and TMPRSS2 are predominantly expressed in the trophoblast layer. Functionally, we demonstrated that cultured PHT cells are permissive to the entry of pvSARS-CoV-2 that express the S protein. Together, our results indicate that the maternal blood-facing villous trophoblasts express the processing proteins that enable SARS-CoV-2 cell entry.
The expression level of ACE2 varies during placental development. While it is expressed in trophoblasts across the various times of gestation surveyed (45, 54, 55, 63, 64), placental ACE2 transcript levels decline from early pregnancy to term (56, 57). The susceptibility of early gestation trophoblasts to SARS-CoV-2 infection remains unclear, and scant data exist regarding feto-placental transmission of SARS-CoV-2 in early pregnancy (65). Interestingly, we found that PHT cell differentiation, which is enhanced as pregnancy progresses, is associated with higher ACE2 and furin expression, but not TMPRSS2 expression. This might have added to the variable level ACE2 protein expression in trophoblasts. The implications of these findings to feto-placental susceptibility to SARS-CoV-2 infection remain to be defined.
TMPRSS2 is obligatory for cleavage of the viral S2 subunit, thus exposing the fusion peptide that facilitates SARS-CoV-2 cell entry and subsequent viral replication (7, 8). Although others have noted that the expression of TMPRSS2 might be below the detection threshold (45, 49), using several sensitive technologies, we were able to unequivocally document TMPRSS2 mRNA and protein expression in trophoblasts, but not in other villous cell types. Placental expression of furin endopeptidase has previously been demonstrated, where it was shown to be required for proteolytic activation of two fusogenic proteins, syncytin-1 and syncytin-2, which are essential for trophoblast fusion (66–69). Unlike the closely related coronavirus SARS-CoV, the S protein of SARS-CoV-2 has evolved to harbor a proteolytic cleavage site of furin, leading to the production of two subunits, S1 and S2, in SARS-CoV-2 permissive cells (70). Importantly, furin-mediated processing of the S protein is critical for SARS-CoV-2-induced cytopathic effects on host cells, cell-cell fusion-mediated viral spread, and SARS-CoV-2 production (8, 9, 71).
Following the initial interaction of SARS-CoV-2 with its receptor, ACE2, on the plasma membrane (6), SARS-CoV-2 usurps the endocytic pathway, culminating in release of its viral genome for viral propagation and assembly of progeny virions. Blocking endocytosis impairs SARS-CoV-2 replication (70, 72), suggesting that proper spatiotemporal trafficking and sorting of SARS-CoV-2 within the endosomal network is required for viral replication in host cells. In line with this finding, we showed that ACE2 protein was colocalized with early endosomes, suggesting that ACE2 and its interacting SARS-CoV-2 are likely to enter endocytic pathways in human trophoblasts. Notwithstanding this observation, current epidemiological data suggest that the feto-placental unit is largely resistant to SARS-CoV-2 infection. Based on these observations, it is likely that SARS-CoV-2 enters trophoblasts, albeit at a lower level than permissive lung epithelial cells. Thereby, we infer that SARS-CoV-2 replication in the human placenta may be limited by the failure to activate postentry pathways, such as endosomal escape or the lysosomal deacidification pathways (73). In line with this intriguing possibility, a recent report demonstrated that interferon-induced transmembrane proteins (IFITM) family member IFITM3, an endosomal protein, restricts SARS-CoV-2 replication via its amphipathic helix domain, which is intimately connected with the endocytosis pathway (74).
Several studies have associated placental histological lesions with COVID-19 infection during pregnancy. While some of these histopathological patterns, such as intervillositis with macrophage or neutrophil infiltration (46–48), are known to accompany other placental viral infections, other lesions are part of maternal or fetal malperfusion lesions (39, 46, 47, 75–78). Whether or not these lesions are caused by SARS-CoV-2 infection or by some other process that enables SARS-CoV-2 villous entry at the site of damage or are a mere coincidence remains to be established.
Current data support the notion that transplacental fetal transmission of SARS-CoV-2 infection is a rare event (12). Our data imply that placental resistance to SARS-CoV-2 infection cannot be assumed on the basis of deficient expression of the SARS-CoV-2 attachment and processing proteins ACE2 and TMPRSS2, as suggested by Pique-Regi et al. (49), but rather is due to post-viral-entry events. Deeper insight into trophoblastic immune response (79) and endocytic pathways will be essential to deciphering the mechanisms underlying resistance to SARS-CoV-2 infection at the maternal-fetal interface.
MATERIALS AND METHODS
Cell culture and reagents.
The institutional review board at the University of Pittsburgh approved all placental procurement, dispersal, and experimental protocols used in these studies. PHT cells were dispersed from term placentas, using a modification of previously published trypsin-DNase-dispase/Percoll protocols (59, 80) and maintained up to 72 h in Dulbecco modified Eagle medium (DMEM; Corning, New York, NY) containing 10% bovine growth serum (BGS; HyClone, Logan, UT) and 1% penicillin-streptomycin (Sigma-Aldrich, St. Louis, MO) at 37°C in a 5% CO2 air atmosphere. PPF cells were isolated by means of our routine standard placental cell isolation procedures and further purified using a mouse anti-CD9 antibody (catalog no. BS3022; Bioworld, St. Louis Park, MN) conjugated to magnetic beads. PPF cell purity was validated by flow cytometry using a mouse antivimentin antibody (clone V9) (catalog no. M0725; Dako, Carpinteria, CA). PPF cells were cultured in DMEM with 10% BGS and antibiotics. HBCs were isolated after our initial trypsin and DNase isolation of PHT cells, following the technique detailed by Tang et al. (81). The purity of HBC cells was validated using the selective marker CD163, with negative signal for CD90, expressed by PPF and other nonmacrophage cell types (81). HuPEC cells were isolated using a previously described protocol, with modifications (82). The identity of HuPEC cells was confirmed via flow cytometry, with positive expression of CD31, endoglin (CD105), and melanoma cell adhesion molecule (CD146) and a lack of CD90 and protein tyrosine phosphatase receptor type C (CD45).
RNA extraction, cDNA synthesis, and real-time quantitative PCR (RT-qPCR) or transcriptome sequencing (RNA-seq).
For mRNA analysis, total RNA was extracted using TRI reagent (Thermo Fisher, Waltham, MA) according to the manufacturer’s protocol. RNA samples were further purified with on-column RNase-free DNase (Qiagen, Germantown, MD). We purchased a FirstChoice Human Total RNA Survey Panel from Thermo Fisher (catalog no. AM6000), which consisted of total RNA from various healthy human tissues, including placenta, heart, intestine, kidney, liver, lung, and testis. Total RNA was reverse transcribed using a HiCapacity cDNA synthesis kit (Thermo Fisher) according to the manufacturer’s protocol. Quantitative PCR was performed using SYBR Select in the ViiA 7 system (Thermo Fisher), and results were presented as threshold cycle (CT) values, or calculated using the 2−ΔCT method (83) and normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Note that GAPDH was within 1 CT value across the human tissues and villous cells. All primer sequences were checked for specificity using BLAST (Basic Local Alignment Search Tool) and are presented in Table S1 in the supplemental material. Dissociation curves were run on all reactions to ensure amplification of a single product with the appropriate melting temperature. Control samples of H2O were included in each qPCR experiment.
Primers used in the studies. Download Table S1, PDF file, 0.02 MB (19.9KB, pdf) .
Copyright © 2021 Ouyang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA libraries of primary human trophoblasts were constructed by different sequencing facilities and sequenced on an Illumina next-generation instrument. After trimming, the RNA sequencing libraries were aligned to human reference genome GRCh38, using the RNA-seq alignment tool STAR (84). The number of reads per gene was calculated for each RNA-seq library, also using STAR. We then used the negative binomial test implemented in the R package DESeq2 (85) to identify ACE2, TMPRSS2, and furin genes that were differentially expressed either between the standard and hypoxia conditions or between the standard and 1.5% DMSO conditions (see also “Statistical analysis” below).
In situ hybridization of ACE2 and TMPRSS2 using RNAscope technology.
Fluorescence in situ hybridization was performed using the RNAscope Fluorescent Multiplex kit (catalog no. 323135; Advanced Cell Diagnostics, Newark, CA) according to the manufacturer’s instructions with minor modifications. Briefly, targeting ACE2 (catalog no. 848151-C3) and TMPRSS2 (catalog no. 470341-C1) probes were purchased from Advanced Cell Diagnostics. Paraffin-embedded sections were deparaffinized twice with xylene, each for 5 min, followed by two 100% ethanol washes for 2 min each. After that, tissues were exposed to hydrogen peroxide for 10 min, heated in kit-provided target retrieval buffer for 15 min at 100°C, and then digested by the provided proteinase plus. After digestion, the probes were added and incubated at 40°C for 2 h. After the probes were rinsed, the signal was amplified using the preamplifier reagent, followed by the addition of Opal fluorophores. The reaction was then blocked, the slides were rinsed and mounted with 4′,6-diamidino-2-phenylindole (DAPI)-Vectashield (Vector labs, Burlingame, CA). Sections were imaged with a Zeiss AxioImager M2 microscope with 40× lens and processed using Zeiss Efficient Navigation microscope software (Carl Zeiss AG, Oberkochen, Germany).
Immunofluorescence of ACE2 and TMPRSS2 in the human placenta.
Cryosections of placental villi were washed and then processed in 1% hydrogen peroxide for 10 min to inhibit endogenous peroxidase. The tissues were then washed, permeabilized with 0.1% Triton X-100 for 1 h, and preincubated with a blocking buffer containing 10% healthy or normal donkey serum (NDS) (catalog no. D9663; Sigma-Aldrich), 3% bovine serum albumin (BSA) (catalog no. A2153; Sigma-Aldrich), 0.1% Triton X-100 (catalog no. X100-500ML; Sigma-Aldrich) in Tris-buffered saline for 1 h at room temperature (RT), followed by incubation with primary antibodies at 4°C in phosphate-buffered saline (PBS) containing 10% NDS, 0.1% Triton X-100, and 3% BSA for 48 h. The following primary antibodies were used: ACE2 (2 μg/ml) (ab15348; ABCAM, Cambridge, MA), TMPRSS2 (1 μg/ml) (ab109131; ABCAM), and normal rabbit IgG (2 μg/ml) for comparison to ACE2, 1 μg/ml for comparison to TMPRSS2 (sc-2027; Santa Cruz Biotechnology, Dallas, TX). After three washes in PBS, the sections were incubated with the corresponding secondary antibodies for 1 h at RT. Nuclei were stained with Hoechst 33342 (catalog no. 62249; Thermo Fisher). All sections were mounted with ProLong Gold Antifade mounting medium (catalog no. P36930; Thermo Fisher).
PHT cells were fixed with freshly prepared 4% paraformaldehyde for 15 min at RT and permeabilized with 0.1% Triton X-100 in PBS containing 0.2% BSA for 3 min at RT. Immunolabeling was then performed with mouse monoclonal antibody ACE2 (E-11; 2 μg/ml) (sc-390851; Santa Cruz Biotechnology) and rabbit polyclonal antibody to EEA.1 (2 mg/ml) (AB-2900; ABCAM) followed by the corresponding secondary antibodies. The nuclei were stained with Hoechst 33342, and cells were mounted with ProLong Gold as described above. To visualize ACE2 traffic in living cells, PHT cells were incubated with Alexa Fluor 488-conjugated ACE2 antibody (4 μg/ml) (sc-390851; Santa Cruz Biotechnology) for 90 min at 37°C, and live-cell imaging was performed at RT.
To obtain high-resolution three-dimensional images of placenta slices and PHT cells, a z stack of confocal images was acquired using a spinning-disk confocal system based on a Zeiss Axio Observer Z1 inverted fluorescence microscope, equipped with a 63× Plan Apo PH, 1.4-numerical-aperture (NA) objective, computer-controlled spherical aberration correction unit, Yokogawa CSU-W1, Vector photomanipulation module, Photometrics Evolve (used for imaging PHT cells), and Hamamatsu Orca-Flash4.0 complementary metal oxide semiconductor (CMOS) (used for imaging of placenta sections) cameras, environmental chamber, piezo stage controller and lasers (405, 445, 488, 515, 561, and 640 nm), all controlled by SlideBook 6 software (Intelligent Imaging Innovations, Denver, CO). Typically, 20 to 30 serial two-dimensional confocal images were recorded at 400-nm intervals. Colocalization of ACE2 with EEA.1 was determined by identifying clear overlapping structures that could be followed in multiple z planes.
Western immunoblotting.
Cells were lysed on ice for 30 min in lysis buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% Triton X-100) supplemented with protease inhibitor cocktail mini EDTA-free and PhoSTOP (Roche, Mannheim, Germany) and centrifuged at 16,000 × g at 4°C for 10 min to remove cell debris. Lysate concentrations were determined with Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher) using a Versa Max microplate reader (Molecular Devices, San Jose, CA). Protein samples (60 μg) were resolved on a sodium dodecyl sulfate-polyacrylamide gel (SDS-PAG), and transferred onto 0.2-μm polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA), using standard procedures. The PVDF membranes were immunoblotted with a mouse monoclonal anti-actin antibody (MAB1501; Sigma) (diluted 1:20,000) and a mouse anti-TMPRSS2 antibody (sc-515727; Santa Cruz Biotechnology) (diluted 1:500). The blots were processed for chemiluminescence using a WesternBright Sirius kit (catalog no. K-12043-D20; Advansta, San Jose, CA) and imaged with the ChemiDoc system (Bio-Rad).
Pseudotyped virus entry of primary human trophoblasts.
PHT cells and 293T-ACE2 (Integral Molecular, Philadelphia, PA) were seeded in 12-well plates at a density of 1.3 × 106 cells/well for PHT (five donors) and 3 × 105 cells/well for 293T-ACE2. Cells were infected with pvSARS-CoV-2 lentiviruses (100 ng/ml based on the p24 content) (Virongy, Manassas, VA) containing SMNE proteins (spike, membrane, nucleocapsid, and envelope), MNE proteins, S protein only, or no SARS-CoV-2 proteins, as well as aldrithiol-inactivated HIV-1 (86) as a negative control. For antibody inhibition experiments, PHT cells were preexposed to 20 μg/ml anti-ACE2 (catalog no. AF933; R&D Systems, Minneapolis, MN ) or anti-DC-SIGN (R&D catalog no. MAB161) for 30 min at 37°C before inoculation with the viruses (1, 7). Cells were harvested at day 2 and day 5 postinfection, and the cells were washed three times with 2 ml of PBS followed by 0.25% trypsin-EDTA for 10 min to release the cells from plastic and to remove any virus adhering to them. The cells were centrifuged at 400 × g for 5 min to pellet cells, which was lysed with 1% Triton X-100 for 15 min at 37°C and centrifuged at 2,000 × g for 5 min. The lysate supernatant was collected, and p24 was measured in the cell lysate using a cytometric bead assay (87). Values were adjusted to normalize for volume differences (1 ml for each fraction).
Statistical analysis.
The main statistical analyses were performed with Prism software (GraphPad, San Diego, CA). Statistical significance for multiple comparison was calculated by one-way analysis of variance (ANOVA) and Tukey post hoc test or by two-tailed paired or unpaired t test, where appropriate. Significance was determined as P < 0.05. Values are presented as means ± standard deviations (SD), derived from at least three independent experiments, as indicated in each figure legend. For statistical analysis of the RNA-seq-based expression of ACE2, TMPRSS2, and furin among the various experimental groups, the P values of differential expression tests were adjusted using Benjamini and Hochberg’s method (88) to control the false discovery rate. In the plots, the expression of the genes was first normalized by the median of ratio method implemented in DESeq2 (85) and then log2 transformed.
Data availability.
RNA-seq data have been deposited with the NCBI BioProject, accession numbers PRJNA674312, PRJNA674329, and PRJNA674366.
ACKNOWLEDGMENTS
We thank Tiffany Coon for technical assistance, Lori Rideout for assistance during manuscript preparation, and Bruce Campbell for editing.
The project was supported by grants from the U.S. National Institutes of Health (NIH) R37-HD086916 (Y.S.) and R01-HL119846 (E.J.S.), the 25 Club of Magee-Womens Hospital (Y.S.), and the Margaret Ritchie R. Battle Family Charitable Fund (Y.S.). The work of L.M. and W.F. was funded by the NICHD Intramural Program.
Y. Sadovsky is a consultant at Illumina, Inc. The other authors report no conflicts.
Contributor Information
Yingshi Ouyang, Email: ouyangy@mwri.magee.edu.
Alexander Sorkin, Email: sorkin@pitt.edu.
Yoel Sadovsky, Email: ysadovsky@mwri.magee.edu.
Matthew B. Frieman, University of Maryland School of Medicine
REFERENCES
- 1.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, HCA Lung Biological Network . 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, Rajagopal S, Pai AR, Kutty S. 2020. Cytokine storm in COVID-19-immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM Consortium Position Paper. Front Immunol 11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosnihan KB, Neves LA, Joyner J, Averill DB, Chappell MC, Sarao R, Penninger J, Ferrario CM. 2003. Enhanced renal immunocytochemical expression of ANG-(1–7) and ACE2 during pregnancy. Hypertension 42:749–753. doi: 10.1161/01.HYP.0000085220.53285.11. [DOI] [PubMed] [Google Scholar]
- 4.Brosnihan KB, Neves LA, Anton L, Joyner J, Valdes G, Merrill DC. 2004. Enhanced expression of Ang-(1-7) during pregnancy. Braz J Med Biol Res 37:1255–1262. doi: 10.1590/s0100-879x2004000800017. [DOI] [PubMed] [Google Scholar]
- 5.Valdés G, Neves LA, Anton L, Corthorn J, Chacón C, Germain AM, Merrill DC, Ferrario CM, Sarao R, Penninger J, Brosnihan KB. 2006. Distribution of angiotensin-(1-7) and ACE2 in human placentas of normal and pathological pregnancies. Placenta 27:200–207. doi: 10.1016/j.placenta.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Klenk HD, Garten W, Steinmetzer T, Böttcher-Friebertshäuser E. 2020. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kleine-Weber H, Pöhlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora N, Sadovsky Y, Dermody TS, Coyne CB. 2017. Microbial vertical transmission during human pregnancy. Cell Host Microbe 21:561–567. doi: 10.1016/j.chom.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira L. 2018. Congenital viral infection: traversing the uterine-placental interface. Annu Rev Virol 5:273–299. doi: 10.1146/annurev-virology-092917-043236. [DOI] [PubMed] [Google Scholar]
- 12.Delahoy MJ, Whitaker M, O’Halloran A, Chai SJ, Kirley PD, Alden N, Kawasaki B, Meek J, Yousey-Hindes K, Anderson EJ, Openo KP, Monroe ML, Ryan PA, Fox K, Kim S, Lynfield R, Siebman S, Davis SS, Sosin DM, Barney G, Muse A, Bennett NM, Felsen CB, Billing LM, Shiltz J, Sutton M, West N, Schaffner W, Talbot HK, George A, Spencer M, Ellington S, Galang RR, Gilboa SM, Tong VT, Piasecki A, Brammer L, Fry AM, Hall AJ, Wortham JM, Kim L, Garg S, COVID-NET Surveillance Team . 2020. Characteristics and maternal and birth outcomes of hospitalized pregnant women with laboratory-confirmed COVID-19 − COVID-NET, 13 states, March 1-August 22, 2020. MMWR Morb Mortal Wkly Rep 69:1347–1354. doi: 10.15585/mmwr.mm6938e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panagiotakopoulos L, Myers TR, Gee J, Lipkind HS, Kharbanda EO, Ryan DS, Williams JTB, Naleway AL, Klein NP, Hambidge SJ, Jacobsen SJ, Glanz JM, Jackson LA, Shimabukuro TT, Weintraub ES. 2020. SARS-CoV-2 infection among hospitalized pregnant women: reasons for admission and pregnancy characteristics − eight U.S. health care centers, March 1-May 30, 2020. MMWR Morb Mortal Wkly Rep 69:1355–1359. doi: 10.15585/mmwr.mm6938e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tartof SY, Qian L, Hong V, Wei R, Nadjafi RF, Fischer H, Li Z, Shaw SF, Caparosa SL, Nau CL, Saxena T, Rieg GK, Ackerson BK, Sharp AL, Skarbinski J, Naik TK, Murali SB. 2020. Obesity and mortality among patients diagnosed with COVID-19: results from an integrated health care organization. Ann Intern Med 173:773–781. doi: 10.7326/M20-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breslin N, Baptiste C, Gyamfi-Bannerman C, Miller R, Martinez R, Bernstein K, Ring L, Landau R, Purisch S, Friedman AM, Fuchs K, Sutton D, Andrikopoulou M, Rupley D, Sheen JJ, Aubey J, Zork N, Moroz L, Mourad M, Wapner R, Simpson LL, D’Alton ME, Goffman D. 2020. Coronavirus disease 2019 infection among asymptomatic and symptomatic pregnant women: two weeks of confirmed presentations to an affiliated pair of New York City hospitals. Am J Obstet Gynecol MFM 2:100118. doi: 10.1016/j.ajogmf.2020.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Wang LL, Zhao SJ, Kwak-Kim J, Mor G, Liao AH. 2020. Why are pregnant women susceptible to COVID-19? An immunological viewpoint. J Reprod Immunol 139:103122. doi: 10.1016/j.jri.2020.103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson DJ, Honein MA, Rasmussen SA, Williams JL, Swerdlow DL, Biggerstaff MS, Lindstrom S, Louie JK, Christ CM, Bohm SR, Fonseca VP, Ritger KA, Kuhles DJ, Eggers P, Bruce H, Davidson HA, Lutterloh E, Harris ML, Burke C, Cocoros N, Finelli L, MacFarlane KF, Shu B, Olsen SJ, Novel Influenza A (H1N1) Pregnancy Working Group . 2009. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet 374:451–458. doi: 10.1016/S0140-6736(09)61304-0. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen SA, Jamieson DJ, Bresee JS. 2008. Pandemic influenza and pregnant women. Emerg Infect Dis 14:95–100. doi: 10.3201/eid1401.070667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong SF, Chow KM, Leung TN, Ng WF, Ng TK, Shek CC, Ng PC, Lam PW, Ho LC, To WW, Lai ST, Yan WW, Tan PY. 2004. Pregnancy and perinatal outcomes of women with severe acute respiratory syndrome. Am J Obstet Gynecol 191:292–297. doi: 10.1016/j.ajog.2003.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shek CC, Ng PC, Fung GP, Cheng FW, Chan PK, Peiris MJ, Lee KH, Wong SF, Cheung HM, Li AM, Hon EK, Yeung CK, Chow CB, Tam JS, Chiu MC, Fok TF. 2003. Infants born to mothers with severe acute respiratory syndrome. Pediatrics 112:e254. doi: 10.1542/peds.112.4.e254. [DOI] [PubMed] [Google Scholar]
- 21.Bellos I, Pandita A, Panza R. 2021. Maternal and perinatal outcomes in pregnant women infected by SARS-CoV-2: a meta-analysis. Eur J Obstet Gynecol Reprod Biol 256:194–204. doi: 10.1016/j.ejogrb.2020.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karimi L, Vahedian-Azimi A, Makvandi S, Sahebkar A. 2021. A systematic review of 571 pregnancies affected by COVID-19. Adv Exp Med Biol 1321:287–298. doi: 10.1007/978-3-030-59261-5_26. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz DA. 2020. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmission of SARS-CoV-2: maternal coronavirus infections and pregnancy outcomes. Arch Pathol Lab Med 144:799–805. doi: 10.5858/arpa.2020-0901-SA. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz DA, Dhaliwal A. 2020. Infections in pregnancy with COVID-19 and other respiratory RNA virus diseases are rarely, if ever, transmitted to the fetus: experiences with coronaviruses, HPIV, hMPV RSV, and influenza. Arch Pathol Lab Med doi: 10.5858/arpa.2020-0211-SA. [DOI] [PubMed] [Google Scholar]
- 25.Yawn DH, Pyeatte JC, Joseph JM, Eichler SL, Garcia-Bunuel R. 1971. Transplacental transfer of influenza virus. JAMA 216:1022–1023. doi: 10.1001/jama.1971.03180320064016. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, Li L, He R, Tan Y, Deng X, Gao M, Tang G, Zhao L, Wang J, Fan Q, Wen C, Tong Y, Tang Y, Hu F, Li F, Tang X. 2020. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 9:469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng OT, Marimuthu K, Ang LW, Mak TM, Lau SK, Anderson DE, Chan KS, Tan TY, Ng TY, Cui L, Said Z, Kurupatham L, Chen MI, Chan M, Vasoo S, Wang LF, Tan BH, Lin RTP, Lee VJM, Leo YS, Lye DC, Singapore 2019 Novel Coronavirus Outbreak Research Team . 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corman VM, Rabenau HF, Adams O, Oberle D, Funk MB, Keller-Stanislawski B, Timm J, Drosten C, Ciesek S. 2020. SARS-CoV-2 asymptomatic and symptomatic patients and risk for transfusion transmission. Transfusion 60:1119–1122. doi: 10.1111/trf.15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. 2020. Extrapulmonary manifestations of COVID-19. Nat Med 26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guadarrama-Ortiz P, Choreño-Parra JA, Sánchez-Martínez CM, Pacheco-Sánchez FJ, Rodríguez-Nava AI, García-Quintero G. 2020. Neurological aspects of SARS-CoV-2 infection: mechanisms and manifestations. Front Neurol 11:1039. doi: 10.3389/fneur.2020.01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulati A, Pomeranz C, Qamar Z, Thomas S, Frisch D, George G, Summer R, DeSimone J, Sundaram B. 2020. A comprehensive review of manifestations of novel coronaviruses in the context of deadly COVID-19 global pandemic. Am J Med Sci 360:5–34. doi: 10.1016/j.amjms.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schröder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. 2020. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaigham M, Andersson O. 2020. Maternal and perinatal outcomes with COVID-19: a systematic review of 108 pregnancies. Acta Obstet Gynecol Scand 99:823–829. doi: 10.1111/aogs.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamouroux A, Attie-Bitach T, Martinovic J, Leruez-Ville M, Ville Y. 2020. Evidence for and against vertical transmission for severe acute respiratory syndrome coronavirus 2. Am J Obstet Gynecol 223:91-e1–91.e4. doi: 10.1016/j.ajog.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karimi-Zarchi M, Neamatzadeh H, Dastgheib SA, Abbasi H, Mirjalili SR, Behforouz A, Ferdosian F, Bahrami R. 2020. Vertical transmission of coronavirus disease 19 (COVID-19) from infected pregnant mothers to neonates: a review. Fetal Pediatr Pathol 39:246–250. doi: 10.1080/15513815.2020.1747120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juan J, Gil MM, Rong Z, Zhang Y, Yang H, Poon LC. 2020. Effect of coronavirus disease 2019 (COVID-19) on maternal, perinatal and neonatal outcome: systematic review. Ultrasound Obstet Gynecol 56:15–27. doi: 10.1002/uog.22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, Li J, Zhao D, Xu D, Gong Q, Liao J, Yang H, Hou W, Zhang Y. 2020. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet 395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosier H, Farhadian SF, Morotti RA, Deshmukh U, Lu-Culligan A, Campbell KH, Yasumoto Y, Vogels CB, Casanovas-Massana A, Vijayakumar P, Geng B, Odio CD, Fournier J, Brito AF, Fauver JR, Liu F, Alpert T, Tal R, Szigeti-Buck K, Perincheri S, Larsen CP, Gariepy AM, Aguilar G, Fardelmann KL, Harigopal M, Taylor HS, Pettker CM, Wyllie AL, Dela Cruz CS, Ring AM, Grubaugh ND, Ko AI, Horvath TL, Iwasaki A, Reddy UM, Lipkind HS. 2020. SARS-CoV-2 infection of the placenta. J Clin Invest 130:4947−4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Algarroba GN, Rekawek P, Vahanian SA, Khullar P, Palaia T, Peltier MR, Chavez MR, Vintzileos AM. 2020. Visualization of SARS-CoV-2 virus invading the human placenta using electron microscopy. Am J Obstet Gynecol 223:275–278. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penfield CA, Brubaker SG, Limaye MA, Lighter J, Ratner AJ, Thomas KM, Meyer J, Roman AS. 2020. Detection of SARS-COV-2 in placental and fetal membrane samples. Am J Obstet Gynecol MFM 2:100133. doi: 10.1016/j.ajogmf.2020.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong L, Tian J, He S, Zhu C, Wang J, Liu C, Yang J. 2020. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA 323:1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng H, Xu C, Fan J, Tang Y, Deng Q, Zhang W, Long X. 2020. Antibodies in infants born to mothers with COVID-19 pneumonia. JAMA 323:1848–1849. doi: 10.1001/jama.2020.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimberlin DW, Stagno S. 2020. Can SARS-CoV-2 infection be acquired in utero? More definitive evidence is needed. JAMA 323:1788−1789. doi: 10.1001/jama.2020.4868. [DOI] [PubMed] [Google Scholar]
- 45.Taglauer E, Benarroch Y, Rop K, Barnett E, Sabharwal V, Yarrington C, Wachman EM. 2020. Consistent localization of SARS-CoV-2 spike glycoprotein and ACE2 over TMPRSS2 predominance in placental villi of 15 COVID-19 positive maternal-fetal dyads. Placenta 100:69–74. doi: 10.1016/j.placenta.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patane L, Morotti D, Giunta MR, Sigismondi C, Piccoli MG, Frigerio L, Mangili G, Arosio M, Cornolti G. 2020. Vertical transmission of coronavirus disease 2019: severe acute respiratory syndrome coronavirus 2 RNA on the fetal side of the placenta in pregnancies with coronavirus disease 2019-positive mothers and neonates at birth. Am J Obstet Gynecol MFM 2:100145. doi: 10.1016/j.ajogmf.2020.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vivanti AJ, Vauloup-Fellous C, Prevot S, Zupan V, Suffee C, Do Cao J, Benachi A, De Luca D. 2020. Transplacental transmission of SARS-CoV-2 infection. Nat Commun 11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Facchetti F, Bugatti M, Drera E, Tripodo C, Sartori E, Cancila V, Papaccio M, Castellani R, Casola S, Boniotti MB, Cavadini P, Lavazza A. 2020. SARS-CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of placenta. EBioMedicine 59:102951. doi: 10.1016/j.ebiom.2020.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pique-Regi R, Romero R, Tarca AL, Luca F, Xu Y, Alazizi A, Leng Y, Hsu CD, Gomez-Lopez N. 2020. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife 9:e58716. doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anton L, Merrill DC, Neves LA, Stovall K, Gallagher PE, Diz DI, Moorefield C, Gruver C, Ferrario CM, Brosnihan KB. 2008. Activation of local chorionic villi angiotensin II levels but not angiotensin (1–7) in preeclampsia. Hypertension 51:1066–1072. doi: 10.1161/HYPERTENSIONAHA.107.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hecht JL, Quade B, Deshpande V, Mino-Kenudson M, Ting DT, Desai N, Dygulska B, Heyman T, Salafia C, Shen D, Bates SV, Roberts DJ. 2020. SARS-CoV-2 can infect the placenta and is not associated with specific placental histopathology: a series of 19 placentas from COVID-19-positive mothers. Mod Pathol 33:2092–2103. doi: 10.1038/s41379-020-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Yuan P, Yang M, Yan Z, Kong S, Yan J, Liu X, Chen Y, Qiao J, Yan L. 2020. SARS-CoV-2 entry factors: ACE2 and TMPRSS2 are expressed in peri-implantation embryos and the maternal-fetal interface. Engineering (Beijing) 6:1162–1169. doi: 10.1016/j.eng.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloise E, Zhang J, Nakpu J, Hamada H, Dunk CE, Li S, Imperio GE, Nadeem L, Kibschull M, Lye P, Matthews SG, Lye SJ. 2020. Expression of severe acute respiratory syndrome coronavirus 2 cell entry genes, angiotensin-converting enzyme 2 and transmembrane protease serine 2, in the placenta across gestation and at the maternal-fetal interface in pregnancies complicated by preterm birth or preeclampsia. Am J Obstet Gynecol 224:298.e1−298.e8. doi: 10.1016/j.ajog.2020.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Chen L, Zhang J, Xiong C, Li X. 2020. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One 15:e0230295. doi: 10.1371/journal.pone.0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komine-Aizawa S, Takada K, Hayakawa S. 2020. Placental barrier against COVID-19. Placenta 99:45–49. doi: 10.1016/j.placenta.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pringle KG, Tadros MA, Callister RJ, Lumbers ER. 2011. The expression and localization of the human placental prorenin/renin-angiotensin system throughout pregnancy: roles in trophoblast invasion and angiogenesis? Placenta 32:956–962. doi: 10.1016/j.placenta.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 57.Hanna N, Hanna M, Sharma S. 2020. Is pregnancy an immunological contributor to severe or controlled COVID-19 disease? Am J Reprod Immunol 84:e13317. doi: 10.1111/aji.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh M, Bansal V, Feschotte C. 2020. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep 32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson DM, Johnson RD, Smith SD, Anteby EY, Sadovsky Y. 1999. Hypoxia limits differentiation and up-regulates expression and activity of prostaglandin H synthase 2 in cultured trophoblast from term human placenta. Am J Obstet Gynecol 180:896–902. doi: 10.1016/s0002-9378(99)70661-7. [DOI] [PubMed] [Google Scholar]
- 60.Thirkill TL, Douglas GC. 1997. Differentiation of human trophoblast cells in vitro is inhibited by dimethylsulfoxide. J Cell Biochem 65:460–468. doi:. [DOI] [PubMed] [Google Scholar]
- 61.Yusuf K, Smith SD, Sadovsky Y, Nelson DM. 2002. Trophoblast differentiation modulates the activity of caspases in primary cultures of term human trophoblasts. Pediatr Res 52:411–415. doi: 10.1203/00006450-200209000-00018. [DOI] [PubMed] [Google Scholar]
- 62.Oh SY, Chu T, Sadovsky Y. 2011. The timing and duration of hypoxia determine gene expression patterns in cultured human trophoblasts. Placenta 32:1004–1009. doi: 10.1016/j.placenta.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Cui D, Liu Y, Jiang X, Ding C, Poon LC, Wang H, Yang H. 2020. Single-cell RNA expression profiling of ACE2 and TMPRSS2 in the human trophectoderm and placenta. Ultrasound Obstet Gynecol 57:248−256. doi: 10.1002/uog.22186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashary N, Bhide A, Chakraborty P, Colaco S, Mishra A, Chhabria K, Jolly MK, Modi D. 2020. Single-Cell RNA-seq identifies cell subsets in human placenta that highly expresses factors driving pathogenesis of SARS-CoV-2. Front Cell Dev Biol 8:783. doi: 10.3389/fcell.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shende P, Gaikwad P, Gandhewar M, Ukey P, Bhide A, Patel V, Bhagat S, Bhor V, Mahale S, Gajbhiye R, Modi D. 2020. Persistence of SARS-CoV-2 in the first trimester placenta leading to vertical transmission and fetal demise from an asymptomatic mother. medRxiv 10.1101/2020.08.18.20177121. [DOI] [PMC free article] [PubMed]
- 66.Chang C, Chen PT, Chang GD, Huang CJ, Chen H. 2004. Functional characterization of the placental fusogenic membrane protein syncytin. Biol Reprod 71:1956–1962. doi: 10.1095/biolreprod.104.033340. [DOI] [PubMed] [Google Scholar]
- 67.Chen CP, Chen LF, Yang SR, Chen CY, Ko CC, Chang GD, Chen H. 2008. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol Reprod 79:815–823. doi: 10.1095/biolreprod.108.069765. [DOI] [PubMed] [Google Scholar]
- 68.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 69.Blaise S, de Parseval N, Bénit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A 100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng YW, Chao TL, Li CL, Chiu MF, Kao HC, Wang SH, Pang YH, Lin CH, Tsai YM, Lee WH, Tao MH, Ho TC, Wu PY, Jang LT, Chen PJ, Chang SY, Yeh SH. 2020. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep 33:108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang YL, Chou YY, Rothlauf PW, Liu Z, Soh TK, Cureton D, Case JB, Chen RE, Diamond MS, Whelan SPJ, Kirchhausen T. 2020. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci US A 117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu V, Fehr AR, Perlman S, Achar SR, Straus MR, Whittaker GR, de Haan CAM, Kehrl J, Altan-Bonnet G, Altan-Bonnet N. 2020. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi G, Kenney AD, Kudryashova E, Zhang L, Hall-Stoodley L, Robinson RT, Kudryashov DS, Compton AA, Yount JS. 2020. Opposing activities of IFITM proteins in SARS-CoV-2 infection. bioRxiv doi: 10.1101/2020.08.11.246678. [DOI] [PMC free article] [PubMed]
- 75.Shanes ED, Mithal LB, Otero S, Azad HA, Miller ES, Goldstein JA. 2020. Placental pathology in COVID-19. Am J Clin Pathol 154:23–32. doi: 10.1093/ajcp/aqaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baergen RN, Heller DS. 2020. Placental pathology in COVID-19 positive mothers: preliminary findings. Pediatr Dev Pathol 23:177–180. doi: 10.1177/1093526620925569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mulvey JJ, Magro CM, Ma LX, Nuovo GJ, Baergen RN. 2020. Analysis of complement deposition and viral RNA in placentas of COVID-19 patients. Ann Diagn Pathol 46:151530. doi: 10.1016/j.anndiagpath.2020.151530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharps MC, Hayes DJL, Lee S, Zou Z, Brady CA, Almoghrabi Y, Kerby A, Tamber KK, Jones CJ, Adams Waldorf KM, Heazell AEP. 2020. A structured review of placental morphology and histopathological lesions associated with SARS-CoV-2 infection. Placenta 101:13–29. doi: 10.1016/j.placenta.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Sinai Immunology Review Project . 2020. Immunology of COVID-19: current state of the science. Immunity 52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF, III.. 1986. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 118:1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- 81.Tang Z, Tadesse S, Norwitz E, Mor G, Abrahams VM, Guller S. 2011. Isolation of Hofbauer cells from human term placentas with high yield and purity. Am J Reprod Immunol 66:336–348. doi: 10.1111/j.1600-0897.2011.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su EJ, Lin ZH, Zeine R, Yin P, Reierstad S, Innes JE, Bulun SE. 2009. Estrogen receptor-beta mediates cyclooxygenase-2 expression and vascular prostanoid levels in human placental villous endothelial cells. Am J Obstet Gynecol 200:427.e1–427.e8. doi: 10.1016/j.ajog.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 83.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 84.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rossio JL, Esser MT, Suryanarayana K, Schneider DK, Bess JW, Jr, Vasquez GM, Wiltrout TA, Chertova E, Grimes MK, Sattentau Q, Arthur LO, Henderson LE, Lifson JD. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J Virol 72:7992–8001. doi: 10.1128/JVI.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Biancotto A, Brichacek B, Chen SS, Fitzgerald W, Lisco A, Vanpouille C, Margolis L, Grivel JC. 2009. A highly sensitive and dynamic immunofluorescent cytometric bead assay for the detection of HIV-1 p24. J Virol Methods 157:98–101. doi: 10.1016/j.jviromet.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 89.Zmora P, Moldenhauer AS, Hofmann-Winkler H, Pöhlmann S. 2015. TMPRSS2 Isoform 1 activates respiratory viruses and is expressed in viral target cells. PLoS One 10:e0138380. doi: 10.1371/journal.pone.0138380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Böttcher-Friebertshäuser E, Stein DA, Klenk HD, Garten W. 2011. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J Virol 85:1554–1562. doi: 10.1128/JVI.01294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in the studies. Download Table S1, PDF file, 0.02 MB (19.9KB, pdf) .
Copyright © 2021 Ouyang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq data have been deposited with the NCBI BioProject, accession numbers PRJNA674312, PRJNA674329, and PRJNA674366.