We read with interest the recently published study by Alito et al. (1) concerning the stability of IS6110 restriction fragment length polymorphism (RFLP) patterns of multidrug-resistant (MDR) Mycobacterium tuberculosis strains. The authors analyzed the IS6110 RFLP patterns and spoligotypes of two groups of MDR M. tuberculosis strains which they discuss to represent two tuberculosis (TB) outbreaks. Within the first group, both IS6110 fingerprint patterns and spoligotypes of the MDR strains have been found to be identical, whereas among the strains of the second group spoligotypes were identical but the IS6110 patterns showed variations. Based on this, the authors conclude that the rate of change of IS6110 RFLP patterns in particular MDR M. tuberculosis strains may be too fast for a reliable interpretation of strain typing results over a period of a few years.
In our recent paper (3), we also elucidated the stability of IS6110 patterns of drug-resistant M. tuberculosis strains by analyzing 165 serial isolates obtained from 56 patients with drug-resistant TB. We did not observe a higher level of instability of IS6110 patterns in these isolates in comparison with the rates of changes described in other studies comprising mainly drug-susceptible isolates (e.g., references 2, 4, and 6). In addition, no particular M. tuberculosis genotypes showing a higher rate of IS6110 changes have been identified. From these data we conclude that the stability of IS6110 patterns in drug-resistant M. tuberculosis strains does not seem to differ from that of drug-susceptible isolates. The IS6110 changes observed, however, occurred only in MDR isolates (37 of the 56 patients were infected with MDR strains), which is likely to be due to the longer time intervals between the times of retrieval of the serial isolates in the patient group with MDR TB (a mean of 300 days for the MDR isolates compared to a mean of 60 days for the resistant but not MDR isolates). Moreover, we have not found a higher instability of IS6110 patterns in several cases of recent transmission of MDR strains.
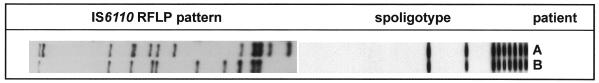
It is well known that the IS6110 patterns of unrelated M. tuberculosis strains generally show a high degree of variability. However, some M. tuberculosis strain families displaying similar IS6110 fingerprint patterns and identical spoligotypes have been described, e.g., the “Beijing family” (5). Hence, the differences in IS6110 patterns among the strains of the second MDR group observed by Alito et al. (1) may be due not to a recent outbreak of a MDR strain showing a higher rate of IS6110 change but to false clustering of strains of an M. tuberculosis family in one fingerprint group. This notion is further supported by the fact that in our German strain collection we found two M. tuberculosis isolates showing the same spoligotype as and an IS6110 pattern similar to those of the strains of the second group of MDR strains described by Alito et al. (1) (Fig. 1).
FIG. 1.
IS6110 RFLP patterns and spoligotypes of two M. tuberculosis strains obtained from two patients living in Germany.
In conclusion, we believe that the data presented by Alito et al. (1) do not give evidence to confirm higher instability of IS6110 patterns among particular MDR M. tuberculosis strains. On the contrary, our results indicate that the evolutionary clock of IS6110 RFLP seems to be identical among drug-susceptible and drug-resistant M. tuberculosis isolates.
REFERENCES
- 1.Alito A, Morcillo N, Scipioni S, Dolmann A, Romano M I, Cataldi A, van Soolingen D. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosisstrains may evolve too fast for reliable use in outbreak investigation. J Clin Microbiol. 1999;37:788–791. doi: 10.1128/jcm.37.3.788-791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cave M D, Eisenach K D, Templeton G, Salfinger M, Mazurek G, Bates J H, Crawford J T. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niemann S, Richter E, Rusch-Gerdes S. Stability of Mycobacterium tuberculosis IS6110restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol. 1999;37:409–412. doi: 10.1128/jcm.37.2.409-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosiscomplex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Soolingen D, Qian L, de Haas P E, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosisin countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosisDNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]