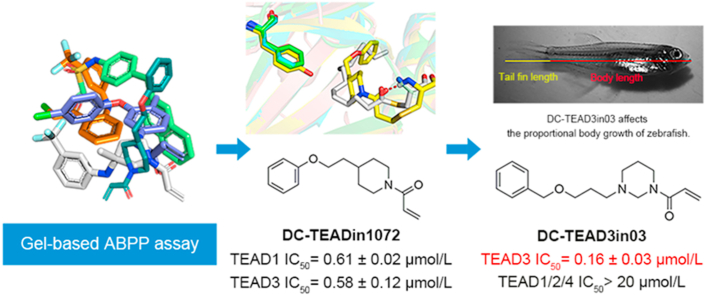
Abstract
The TEA domain (TEAD) family proteins (TEAD1‒4) are essential transcription factors that control cell differentiation and organ size in the Hippo pathway. Although the sequences and structures of TEAD family proteins are highly conserved, each TEAD isoform has unique physiological and pathological functions. Therefore, the development and discovery of subtype selective inhibitors for TEAD protein will provide important chemical probes for the TEAD-related function studies in development and diseases. Here, we identified a novel TEAD1/3 covalent inhibitor (DC-TEADin1072) with biochemical IC50 values of 0.61 ± 0.02 and 0.58 ± 0.12 μmol/L against TEAD1 and TEAD3, respectively. Further chemical optimization based on DC-TEAD in 1072 yielded a selective TEAD3 inhibitor DC-TEAD3in03 with the IC50 value of 0.16 ± 0.03 μmol/L, which shows 100-fold selectivity over other TEAD isoforms in activity-based protein profiling (ABPP) assays. In cells, DC-TEAD3in03 showed selective inhibitory effect on TEAD3 in GAL4-TEAD (1–4) reporter assays with the IC50 value of 1.15 μmol/L. When administered to zebrafish juveniles, experiments showed that DC-TEAD3in03 reduced the growth rate of zebrafish caudal fins, indicating the importance of TEAD3 activity in controlling proportional growth of vertebrate appendages.
KEY WORDS: Hippo pathway, TEAD3, Covalent inhibitor, Palmitoylation inhibitor.
Abbreviations: TEAD, TEA domain family; TEAD3, TEA domain transcription factor 3; YAP, Yes-associated protein; TAZ, transcriptional co-activator with PDZ-binding motif; ABPP, activity-based protein profiling; MS, mass spectrometry; HCC, hepatocellular carcinoma.
Graphical abstract
Through gel-based ABPP palmitoylation assay and follow-up structural optimization, we obtained a selective TEAD3 inhibitor DC-TEAD3in03 that reduced the growth rate of zebrafish caudal fin.
1. Introduction
In the Hippo signaling pathway, the transcription factor TEA domains (TEADs) family proteins (TEAD1‒4) are the most important terminal regulators, which play an important role in development, cell proliferation, cell differentiation, tissue homeostasis, and regeneration1, 2, 3, 4. Although they are widely expressed and evolutionarily conserved, the expression of different subtypes of TEAD proteins (TEAD1‒4) is tissue specific and each isoform performs different functions5. For instance, TEAD1 is reported to be involved in heart development, and knockout of TEAD1 in mouse models can lead to fetal death due to heart defects6. TEAD2 plays a significant role in the development of the nervous system7 while TEAD3 plays an important role in myoblast differentiation, early fiber assembly and other muscle tissue development8, 9, 10. Additionally, TEAD3 was also found to play an important role in regulating cardiac differentiation and epidermal proliferation. Han et al.9 found that in the CVPC (the earlier cardiovascular progenitor cells) stage, YAP interacts with TEAD3 to regulate the cardiac differentiation of human-induced pluripotent stem cells. TEAD1 and TEAD3 jointly regulate epidermal proliferation11. TEAD3 has also been found to be specifically expressed in the placenta and multiple embryonic tissues12. Knockout of TEAD4 in mouse models has been shown to link to fetal death, suggesting that it is crucial for embryo transfer13.
Beyond development, a large number of studies have also shown that the amplification and overactivation of each TEAD plays a context-dependent role in the occurrence and progression of malignant diseases, especially cancers14,15. TEAD1 is reported to be involved in the regulation of prostatic epithelial cell differentiation and epithelial morphogenesis, such as the regulation of cell adhesion to basement membrane16. The overexpression of TEAD1 has been shown to link to poor prognosis of prostate cancer patients17, while the increased expression of TEAD2 mRNA was observed in hepatocellular carcinoma (HCC) patients18. TEAD4 is also highly expressed in a variety of cancers and plays an important role in aspects including epithelial‒mesenchymal transformation (EMT)19, metastasis20, cancer stem cell dynamics21, and chemotherapy resistance while the role of TEAD3 in cancer biology has been less investigated22,23. Besides, many TEAD–YAP target genes, including cell surface receptor tyrosine kinase Axl24, connective tissue growth factor CTGF25, BIRC526 and tumor marker mesothelin are also associated with tumorigenesis27. Therefore, the development of subtype-selective TEAD inhibitors could help understand the pathological role of each TEAD and has great potential in the era of precise medicine.
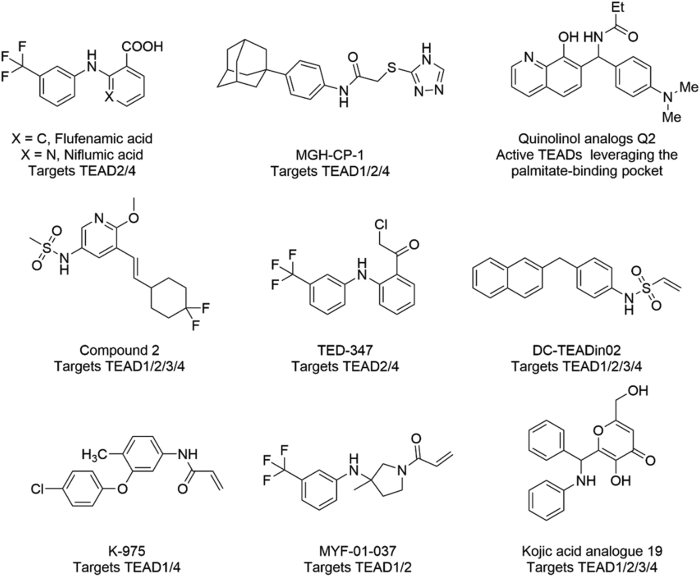
TEAD is a multidomain protein, which is mainly composed of the N-terminal DNA binding domain (DBD) and the C-terminal YAP/TAZ binding domain (YBD) with homology up to 91% between TEAD isoforms3,28,29. TEADs family proteins also contain highly conserved palmitoylation pockets30 (Supporting Information Fig. S1). As the TEAD–YAP protein–protein interaction interface remains challenging to be targeted, more chemistry endeavors have been devoted to the development of the palmitoylation site located in TEAD-YBD in recent years31,32 (Fig. 1). In 2015, the non-steroidal anti-inflammatory drug flufenamic acid was discovered by Pobbati et al. with the KD value of 73 μmol/L33. In 2019, Bum-Erdene et al. discovered a chloromethyl ketone analogue of flufenamic acid TED-347 as a covalent TEAD palmitoylation inhibitor that can allosterically inhibit the TEAD–YAP interaction34. K-975, another covalent TEAD palmitoylation inhibitor, can also directly bind to TEAD protein and inhibit YAP/TAZ–TEAD interaction, and shows strong anti-tumor effect in the preclinical mesothelioma model35. Very recently, Genentech reported a non-covalent compound as a TEAD lipid pocket binder, which inhibited TEAD activity in a dominant-negative manner31. Besides inhibitors, quinolinol compounds have also been found to act as TEAD-dependent transcriptional activators through binding to the lipid pocket36. However, those modulators could target multiple TEADs as reported and the development of subtype-selective TEAD inhibitors lags behind.
Figure 1.
Reported small molecules targeting the palmitate-binding pocket of TEADs.
Previously, our group identified the compound DC-TEADin02, a vinyl sulfonamide derivative, as a pan-TEAD covalent inhibitor through virtual screening37. In the present work, by performing gel-based activity-based protein profiling (ABPP) screening, we identified DC-TEADin1072 as a TEAD1/3 selective inhibitor with the novel chemotype featuring an acrylamide warhead. Through pocket analysis and subsequent rational chemical design, DC-TEAD3in03, a selective, covalent TEAD3 inhibitor was developed with an IC50 value of 0.16 ± 0.03 μmol/L. In GAL4 reporter assays, DC-TEAD3in03 selectively inhibited TEAD3 transcriptional activity. Also as expected, DC-TEAD3in03 significantly increases the content of TEAD3 soluble contents in cellular thermal shift assay (CETSA), which is a reliable indicator for direct target engagement in cells.
In the zebrafish model, DC-TEAD3in03 showed selective reduction of the caudal fin growth rate after 14 days of treatment in 2-month-old juveniles. This provides evidence of TEAD3 activity during proportional growth. Treatment with DC-TEADin1072 on the other hand showed a reduction in both body growth rate and caudal tail growth rate, which suggests TEAD3 redundancy during body development of the zebrafish. These selective inhibitors provide an efficient chemical genetic platform to study the role of TEAD1 and TEAD3 during development.
2. Results and discussion
2.1. Discovery of DC-TEADin1072 as a covalent inhibitor targeting TEAD1 and TEAD3
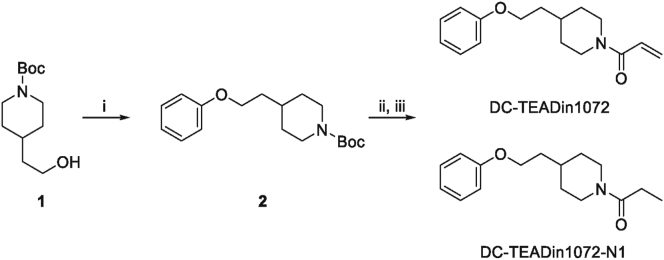
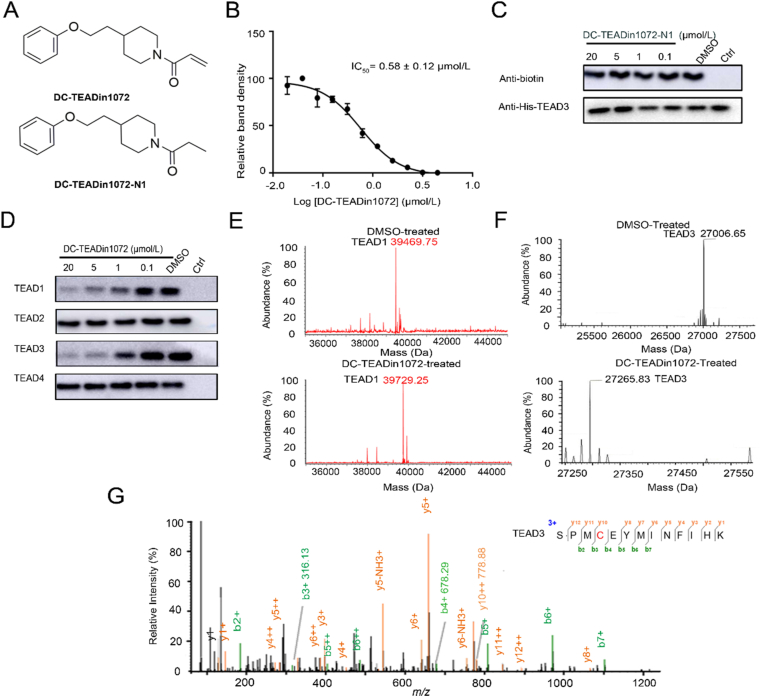
To find compounds bearing novel chemotype against TEAD palmitoylation site, we used gel-based activity-based protein profiling (ABPP) assay to screen in-house covalent compound collections containg 358 compounds with covalent warheads. Compounds with diverse scaffolds were screened with TEAD3-YBD recombinant protein at 50 μmol/L, and hits with >80% inhibition were picked out for futher characterization. Among them, DC-TEADin1072 (synthetic route was shown in Scheme 1) showed inhibitory activity with an IC50 value of 0.58 ± 0.12 μmol/L against TEAD3 (Fig. 2A and B), which was more potent compared with flufenamic acid under the same conditions33 (Supporting Information Fig. S2). We also synthesized a non-covalent counterpart of DC-TEADin1072 (Scheme 1), namely DC-TEADin1072-N1. As expected, DC-TEADin1072-N1 showed minimal inhibition on TEAD palmitoylation (Fig. 2C). Further selectivity profiling indicated that DC-TEADin1072 could also inhibit TEAD1 palmitoylation with an IC50 value of 0.61 ± 0.02 μmol/L against TEAD1 while sparing TEAD2 and TEAD4 homologues (Fig. 2D, Supporting Information Fig. S3). In a mass spectrometry (MS) study, a higher mass peak was detected with the difference in mass by +259.5 Da after the incubation of TEAD1 and TEAD3 protein with excessive DC-TEADin1072 for 24 h at 4 °C, which was in accordance with the molecular weight of DC-TEADin1072 via direct Michael addition (Fig. 2E and F). In addition, the extracted ion chromatogram indicated that the precursor ion of this peptide [SPMC(C16H21NO2) EYMINFIHK] can be only found in compound treated group (Fig. 2G). Collectively, these results confirmed that compound DC-TEADin1072 is a dual TEAD1/3 covalent inhibitor.
Scheme 1.
Synthetic route of DC-TEADin1072 and DC-TEADin1072-N1. Reagents and conditions: (i) phenol, PPh3, DEAD, anhydrous THF, Ar, 0 °C to rt, 8 h. (ii) 4 mol/L HCl in 1,4-dioxane, rt, 2 h. (iii) Acryloyl chloride (propionyl chloride), TEA, DCM, 0 °C, 2 h.
Figure 2.
The discovery of DC-TEADin1072 as a covalent TEAD1 and TEAD3 autopalmitoylation inhibitor. (A) The chemical structures of DC-TEADin1072 and DC-TEADin1072-N1. (B) and (C) The inhibitory activities of DC-TEADin1072 and DC-TEADin1072-N1 in the click-based ABPP assay. The band intensity was quantified in ImageJ (NIH). The experiments were performed in biological triplicates and the data was shown as mean ± SD, n = 3. (D) DC-TEADin1072 selectively inhibited TEAD1 and TEAD3 palmitoylation while sparing TEAD2 and TEAD4. (E)‒(G) Mass spectrometry demonstrated the covalent modification of DC-TEADin1072 on TEAD1 and TEAD3 recombinant protein.
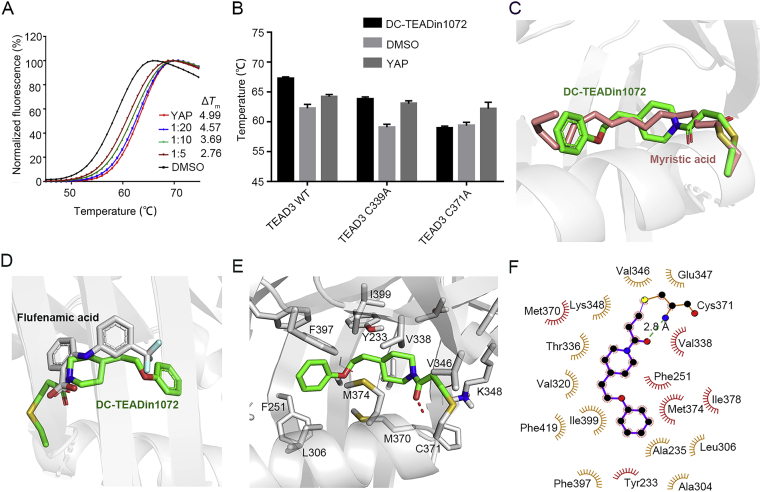
Notably, further differential scanning fluorimetry (DSF) experiments showed that the interactions of wild-type TEAD3-YBD with DC-TEADin1072 resulted in a significant thermal transition with the ΔTm value of ∼5 °C at the ratio of 1:20 (Fig. 3A). To further clarify the specific modification of DC-TEADin1072 on the exact cysteine of TEAD1/3, all cysteines of TEAD1/3-YBD were individually mutated to alanine and subjected to thermal shift evaluation. Compared with other TEAD mutants, the TEAD3-YBD C371A mutant and the TEAD1-YBD C359A mutant significantly abolished the compound effect on protein thermal stability demonstrating selective engagement with Cys371 of TEAD3 and Cys359 of TEAD1 (Fig. 3B, Supporting Information Fig. S4). As it is still controversial whether a small molecule occupying the lipid binding site could abolish the TEAD‒YAP interaction, we also explored the compound effect on TEAD‒YAP interaction in the fluorescence polarization (FP) assay using fluorescein isothiocyanate (FITC)-labeled YAP peptide (61–100). Interestingly, the results indicated that DC-TEADin1072 showed minimal effect on TEAD‒YAP interaction, which is also observed with flufenamic acid and DC-TEADin0237 (Supporting Information Fig. S5).
Figure 3.
Biochemical and biophysical methods demonstrated the direct binding of DC-TEADin1072. (A) TEAD3 thermal shift assays. Synthetic YAP peptide was used as the positive control with the ratio of 1:5. The experiments were performed in biological triplicates. (B) The shift of Tm values of TEAD3-YBD WT and TEAD3-YBD mutants with DC-TEADin1072. (C) and (D) The alignment of DC-TEADin1072 (green) with myristic acid (salmon) (PDB code: 5OAQ)38 and flufenamic acid (grey) (PDB code: 5DQ8)33. (E) and (F) The predicted binding mode of DC-TEADin1072 with TEAD3-YBD in molecular docking studies.
2.2. Binding mode analysis of DC-TEADin1072
To further delineate the binding mode of DC-TEADin1072 and elucidate the mode-of-action (MOA) of the series of compounds, covalent docking was performed using Covalent Docking panel integrated in Maestro 9.137. The results show that DC-TEADin1072 adopts a similar conformation with endogenous substrate myristic acid (Fig. 3C). Additionally, the acrylamide group adjacent to the opening of the pocket forms hydrogen bonds at C371, which shares the similar binding mode to the carboxyl group of flufenamic acid with TEAD2 (Fig. 3D). Molecular docking studies indicate that DC-TEADin1072 could form extensive hydrophobic interaction with V338, F251, M374, M370, V320 and T336 in the hydrophobic groove (Fig. 3E and F). In conclusion, the docking results rationalize the inhibitory activity of DC-TEADin1072 and establish a possible binding mode for further structural modification.
2.3. Development of DC-TEAD3in03 as a selective TEAD3 inhibitor
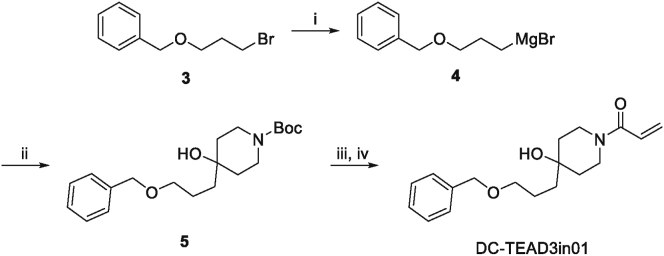
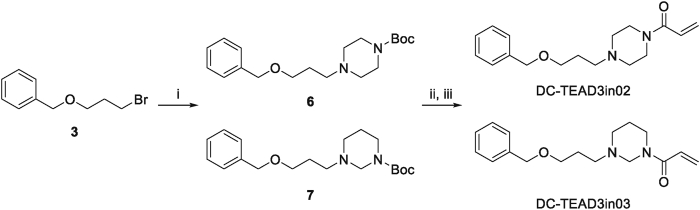
In order to achieve the selectivity within the TEAD family, we compared all the TEADs-YBD crystal structures to see if there is any structural difference to design subtype-selective TEAD inhibitors. Interestingly, we found that there is a unique tyrosine (Y233) in TEAD3, while in other TEAD isoforms, it has been replaced with phenylalanine (Supporting Information Fig. S6). Thus, follow-up medicinal chemistry optimization on the piperidine of DC-TEADin1072 was conducted aiming to design selective inhibitors that could capture this specific hydroxyl group via hydrogen bond. The synthetic routes of DC-TEAD3in01, DC-TEAD3in02 and DC-TEAD3in03 were shown in Scheme 2, Scheme 3.
Scheme 2.
Synthetic route of DC-TEAD3in01. Reagents and conditions: (i) Mg, 1,2-dibromoethane, anhydrous THF, Ar, rt to 60 °C, 2 h. (ii) N-(tert-Butoxycarbonyl)-4-piperidone, anhydrous THF, Ar, 0 °C to rt, 3 h. (iii) 4 mol/L HCl in 1,4-dioxane, rt, 2 h. (iv) Acryloyl chloride, TEA, DCM, 0 °C, 2 h.
Scheme 3.
Synthetic route of DC-TEAD3in02 and DC-TEAD3in03. Reagents and conditions: (i) tert-butyl tetrahydropyrimidine-1(2H)-carboxylate (tert-butyl piperazine-1-carboxylate), K2CO3, DMF, 90 °C, 8 h. (ii) 4 mol/L HCl in 1,4-dioxane, rt, 2 h. (iii) Acryloyl chloride, TEA, DCM, 0 °C, 2 h.
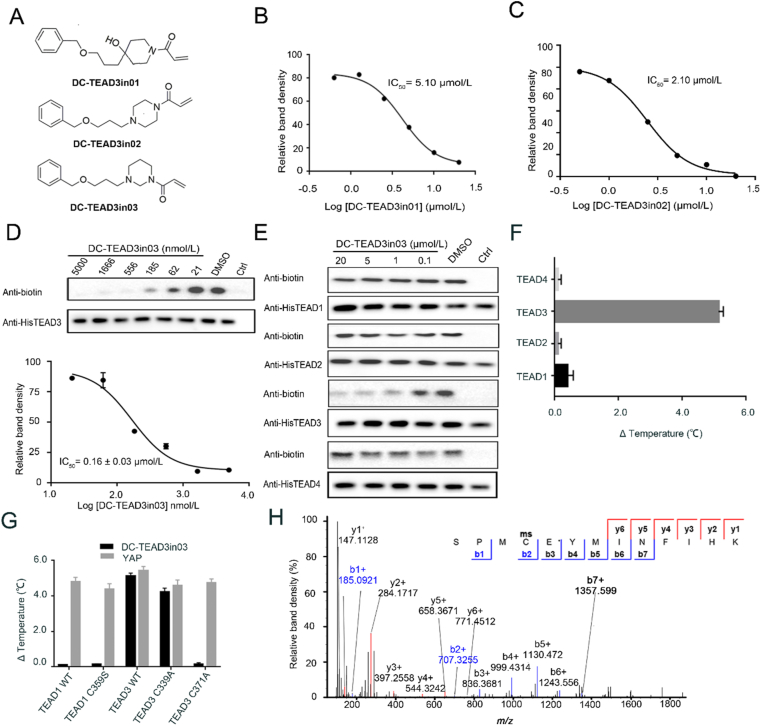
Among this series, DC-TEAD3in01, DC-TEAD3in02 and DC-TEAD3in03 show potent inhibition (Fig. 4A‒D, Supporting Information Fig. S7). Among them, DC-TEAD3in03 shows best inhibitory activity against TEAD3 with the IC50 value of 0.16 ± 0.03 μmol/L and profound selectivity over other TEAD isoforms in both gel-based ABPP and DSF experiments (Fig. 4E and F). Additionally, site mutation studies indicated that DC-TEAD3in03 could significantly improve the thermal stability of TEAD3 protein, while it had minimal effect on widetype TEAD1, TEAD1 C359S and TEAD3 C371A mutants, proving the direct modification site of the compound (Fig. 4G). To further demonstrate the direct interactions of the identified TEAD inhibitors with corresponding TEADs and evaluate the cellular uptake of the compounds, overexpressed TEAD3 from DC-TEAD3in03-treated HEK293T cell samples was enriched and subjected to mass spectrometry. As expected, the peptide containing the exact cysteine was mapped out demonstrating the DC-TEAD3in03 could covalently label TEAD3 in cells (Fig. 4H).
Figure 4.
The biophysical and biochemical characterization of DC-TEAD3in03. (A) The chemical structures of DC-TEAD3in01, DC-TEAD3in02 and DC-TEAD3in03. (B)‒(D) The inhibitory activities of DC-TEAD3in01, DC-TEAD3in02 and DC-TEAD3in03 through CuAAC-based click chemistry. The band intensity was quantified in ImageJ (NIH). The experiments were performed in triplicate and the data was shown as mean ± SD, n = 3. (E) DC-TEAD3in03 selectively inhibited TEAD1 and TEAD3 palmitoylation while sparing TEAD2 and TEAD4. (F) and (G) DC-TEAD3in03 significantly improved the thermostability of TEAD3 but had minimal effect on TEAD1 or corresponding C359S mutant. Synthetic YAP peptide was used as the positive control with the ratio of 1:5. The experiments were performed in triplicate. (H) The modification of transfected-TEAD3 upon 50 μmol/L DC-TEAD3in03 treatment for 6 h was detected by MS experiments.
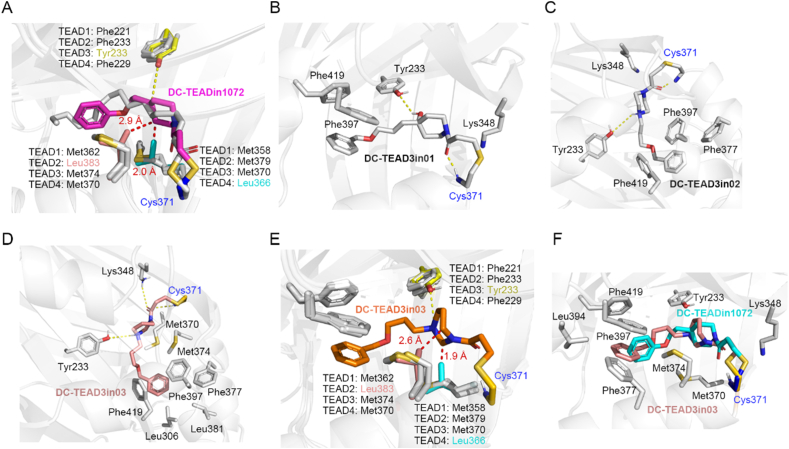
To understand molecular mechanism of the selectivity of those TEAD inhibitors, we revisited the docking result of DC-TEADin1072 and conducted covalent docking for DC-TEAD3in01-03 using covalent docking module in Schrödinger software. The results showed that the L383 of TEAD2 and L366 of TEAD4 have steric clash with DC-TEADin1072, which may account for the selectivity over TEAD2 and TEAD4 (Fig. 5A). The hydroxyl group of DC-TEAD3in01 and the nitrogen atom in piperazine (DC-TEAD3in02) and hexahydropyrimidine moiety (DC-TEAD3in03) could form a hydrogen bond to the unique tyrosine (Y223) of TEAD3, which distinguishes it from TEAD1 (Fig. 5B‒F). To validate the docking model, we also mutated the Y223 to phenylalanine, the activity of DC-TEAD3in03 significantly decreases with the IC50 value close to 18.5 ± 4.9 μmol/L (Supporting Information Fig. S8) in gel-based ABPP assays, which consolidates our docking model.
Figure 5.
The binding mode analysis of DC-TEAD3in03. (A) Binding mode of DC-TEADin1072 (magenta) with TEADs protein (TEAD1: 6IM539, TEAD2: 5EMV30, TEAD3: 5EMW30, TEAD4: 3JUA40). DC-TEADin1072 forms clash (red dash) with Leu383 in TEAD2 and Leu366 in TEAD4, rationalizing the selectivity towards TEAD1 and TEAD3. (B) Binding mode of DC-TEAD3in01 with TEAD3. (C) Binding mode of DC-TEAD3in02 with TEAD3. (D) Binding mode of DC-TEAD3in03 with TEAD3. (E) The clash (red dash) between DC-TEAD3in03 (orange) and TEAD2, TEAD4. (F) Overlap mode of DC-TEADin1072 (cyan) and DC-TEAD3in03 (salmon) with TEAD3.
2.4. DC-TEAD3in03 selectively engaged TEAD3 in cells
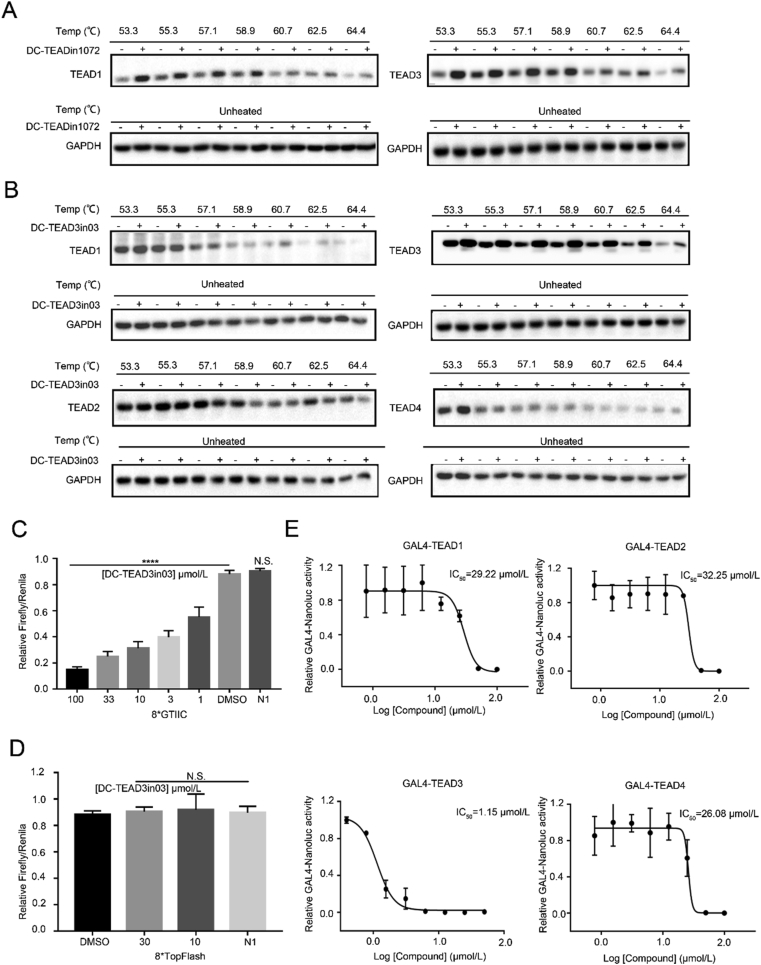
To verify if the observed biochemical selectivity could be translated into cells, we did CETSA experiments to monitor TEAD engagement in HEK293T cells. The results showed that treatment of DC-TEADin1072 could increase the thermal stability of the TEAD1 and TEAD3 while treatment of selective TEAD3 inhibitor DC-TEAD3in03 only increased the soluble fraction of TEAD3 demonstrating its cellular uptake and direct engagement (Fig. 6A and B, Supporting Information Fig. S9).
Figure 6.
DC-TEAD3in03 selectively engaged TEAD3 in cells. (A) HEK293T cells were treated with 50 μmol/L DC-TEADin1072 for 6 h in CETSA experiments to demonstrate the direct binding of DC-TEADin1072 with TEAD1 and TEAD3 in intact cells. (B) HEK293T cells were treated with 50 μmol/L DC-TEAD3in03 for 6 h in CETSA experiments to demonstrate that DC-TEAD3in03 significantly increased the thermal stability of TEAD3 in intact cells. (C) and (D) The effect of DC-TEAD3in03 on TEAD-dependent/independent reporter system. The results shown are mean ± SD of three technical replicates (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, N.S.: P > 0.05), n = 3. (E) The effect of DC-TEAD3in03 on TEAD1, TEAD2, TEAD3, and TEAD4 activities in GAL4 reporter system. HEK293T cells were transfected with a GAL4-LUC reporter, together with expression vectors for GAL4-TEADs constructs as indicated. The results shown are mean of three technical replicates, n = 3.
In order to further evaluate the inhibitory effect of our compounds in cells, we performed TEAD-specific luciferase reporter assays with DC-TEAD3in03 and the negative compound DC-TEADin1072-N1. As expected, the treatment of with DC-TEAD3in03 in cells transfected with the TEAD luciferase reporter led to significant reduction in TEAD reporter activity compared with DC-TEADin1072-N1 while it had minimal effect on the unrelated TOP Flash beta-catenin reporter system, demonstrating the specificity of the compound in the regulation of the Hippo signaling pathway (Fig. 6C and D). In order to simultaneously exclude the interference from different endogenous expression levels of TEAD1‒4 proteins, we expressed each TEAD homologue fused with yeast GAL4 DNA binding domain (TEAD1‒4). The cells were transfected with GAL4-TEAD expression vector together with secreted luciferase reporter vector under the control of promoters containing 9×GAL4 DNA binding elements. At the same time, overexpressing YAP can activate TEAD transcription and significantly enhance luciferase activity. DC-TEAD3in03 was used to treat TEAD1, TEAD2, TEAD3 and TEAD4, respectively. The results showed that DC-TEAD3in03 had selective inhibition on TEAD3 transcrptional activity with its IC50 value of 1.15 μmol/L (Fig. 6E).
2.5. In vivo regulation of proportional growth
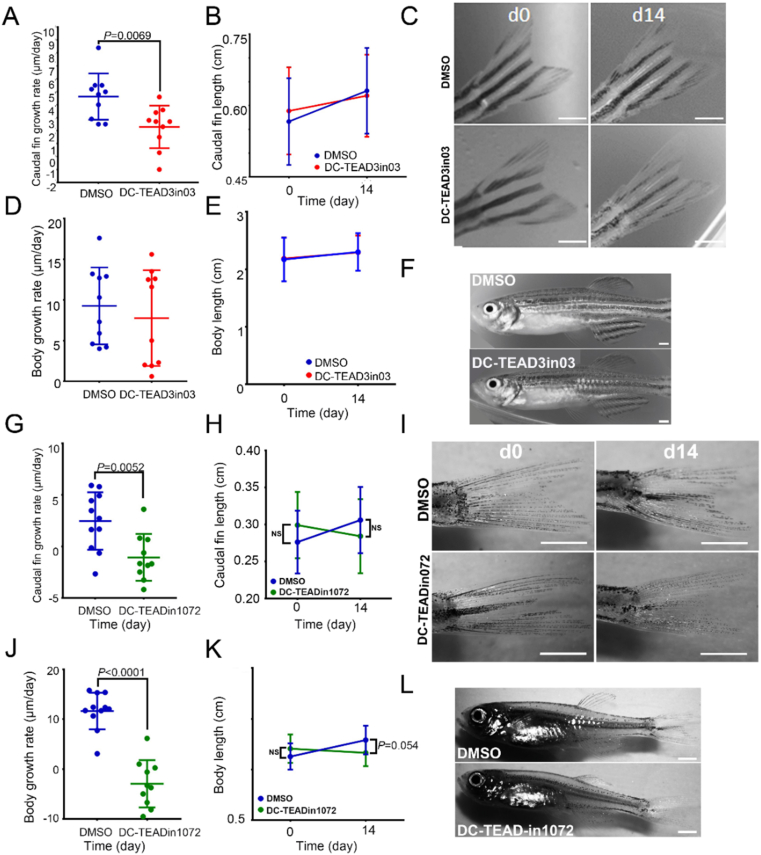
As a downstream effector of the Hippo signal transduction cascade, it has been reported that TEAD1, TEAD2 and TEAD4 regulate the proportional growth of tissues, such as the heart, brain and skull bones during early development, while the role of TEAD3 during development has not been established yet41. To investigate the biological effect of DC-TEAD3in03 on proportional growth of a vertebrate animal, we treated juvenile zebrafish with DC-TEAD3in03 (0.25 μmol/L) for 14 days and evaluated the proportional growth relationship between the animal body and caudal fin. DC-TEAD3in03-treated fish showed a reduction in the growth rate of the caudal fin after 14 days of treatments (Fig. 7A and B). Inspection of the caudal fin showed no ectopic or tumorous growth from the chronic DC-TEAD3in03 treatments (Fig. 7C). Although we observed a significant reduction in the outgrowth of the caudal fin, we did not observe a significant difference between the growth of the overall body rate or lengths (Fig. 7D‒F), indicating that TEAD3 is specifically involved in the proportional growth of appendage tissues. TEAD3 redundancy has been described in keratinocyte growth9. We hypothesized that TEAD3 redundancy is responsible for the observation that the body growth rate did not alter upon DC-TEAD3in03 treatment alone. Therefore, we exposed 2-month old AB wild type juvenile fish to 0.25 μmol/L dual TEAD1/3 inhibitor DC-TEADin1072 for 14 days. We observed that the tail growth speed was reduced compared to DMSO treatment, no differences in tail length and no ectopic aberrations in fin morphology (Fig. 7G‒I), corroborating the findings described in Fig. 6A‒C. The body growth rate decreased significantly upon DC-TEADin1072 treatment and the body length was nearly significantly different (P = 0.054, Fig. 7J and K). No obvious ectopic or tumorous growth were observed in the body of the fish (Fig. 7L). These findings suggest that TEAD1 and TEAD3 have a supplemental role in the proportional body growth during development of juvenile fish.
Figure 7.
DC-TEAD3in03 decreases vertebrate appendage growth during the juvenile growth phase. (A) Growth rates of the caudal fins of DMSO-treated and DC-TEAD3in03-treated zebrafish during the juvenile growth phase (starting at 2 months of age). (B) The average and standard deviation of fin lengths of DMSO- and DC-TEAD3in03-treated fish at the indicated time points. (C) Caudal fins of zebrafish before treatments (d0) and after 14 days of continuous treatment (d14). (D) Growth rates of the bodies of DMSO-treated and DC-TEAD3in03-treated zebrafish during the juvenile growth phase (starting at 2 months of age). (E) Measurements of the body length of untreated and treated zebrafish during the juvenile growth phase (starting at 2 months of age). (F) Images displaying zebrafish bodies after treatment with DMSO and DC-TEAD3in03. (G) Growth rates of the caudal fins of DMSO-treated and DC-TEADin1072-treated zebrafish during the juvenile growth phase (starting at 2 months of age). (H) The average and standard deviation of fin lengths of DMSO- and DC-TEADin1072-treated fish at the indicated time points. (I) Caudal fins of zebrafish before treatments (d0) and after 14 days of continuous treatment (d14). (J) Growth rates of the bodies of DMSO-treated and DC-TEADin1072-treated zebrafish during the juvenile growth phase (starting at 2 months of age). (K) Measurements of the body length of untreated and treated zebrafish during the juvenile growth phase (starting at 2 months of age). (L) Images displaying zebrafish bodies after treatment with DMSO and DC-TEADin1072. Scale bars equal 1 mm.
3. Conclusions
TEADs and its co-activator YAP/TAZ are essential components in Hippo pathway which play a pivotal role in physiological and pathological events. There are four TEAD homologues (TEAD1‒4) in mammalian cells, which showed tissue-specific expression patterns, and each plays a unique role in both development and cancer progression. Thus, the identification of subtype-selective TEAD inhibitors remains of paramount importance, which will allow us to discern the individual contributions of TEAD protein to both normal human physiology and disease. Recent study showed that TEADs could undergo autopalmitoylation which is a more druggable pocket for chemical intervention. However, current inhibitors targeting the palmitoylation sites of TEADs, such as flufenamic acid and TED-347, all act on multiple TEADs. Thus, there is urgent need to develop subtype-selective TEAD inhibitors. Here, starting from the dual TEAD1/3 inhibitor DC-TEADin1072 identified from ABPP-based screening, we obtained a potent, selective TEAD3 covalent inhibitor DC-TEAD3in03 with a novel chemotype. The selectivity was also translated into cell context as validated by TEAD-specific reporter assays and CETSA assays. Our observations that DC-TEAD3in03 specifically reduced the growth rate of the caudal fins without promoting tumorigenesis argues that the inhibitor strictly affects coordinated, proportional growth. To sum up, the study demonstrates for the first time that selective targeting TEAD is achievable which shed light on the development of other TEAD subtype-selective inhibitors beyond TEAD3 and DC-TEAD3in03 could be used an effective chemical tool to investigate the physiological and pathological role of TEAD3 in different contexts.
4. Experimental
4.1. Chemistry
Unless otherwise noted, all reagents and solvents were purchased from commercial suppliers and used without further purification. Reactions were monitored by thin-layer chromatography (TLC) or UPLC‒MS. UPLC‒MS was performed by Waters ultra-performance liquid chromatography (UPLC) H-Class with ACQUITY UPLC BEH C18 reversed-phase column (2.1 mm × 50 mm, 1.7 μm, flow rate = 0.5 mL/min). The analysis process lasted for 5 min with a gradient of 10% acetonitrile to 100% acetonitrile in 0.1% formic acid aqueous solution. Mass analysis was performed by the Waters SQD2 single quadrupole mass detection system. All final compounds were purified by C18 reversed-phase preparative high-performance liquid chromatography (HPLC) column with H2O and CH3CN as eluents and the purity was confirmed to be >95% by UPLC‒MS. 1H NMR and 13C NMR spectra were obtained on BRUKER AVANCE II 400 M or BRUKER AVANCE III 500 M NMR spectrometer. High-resolution electrospray ionization (ESI) mass analysis was performed by an Agilent 1290–6545 UHPLC-QTOF high-resolution mass spectrometer.
4.1.1. tert-Butyl 4-(2-phenoxyethyl)piperidine-1-carboxylate (2)
DEAD (209 mg, 1.2 mmol) was dissolved into a mixture of PPh3 (314 mg, 1.2 mmol) and anhydrous THF (10 mL) under argon at 0 °C and the mixture was allowed to stir for 10 min. Then compound 1 (229 mg, 1.0 mmol) and phenol (113 mg, 1.2 mmol) in anhydrous THF (3 mL) were added. The reaction mixture was stirred in ambient temperature for 8 h. The solvent was removed under vacuum and the residue was purified by flash silica chromatography (gradient 0–10% EA/PE) to afford the title compound 2 (235 mg, yield 77%) as a colorless oil. 1H NMR (400 MHz, chloroform-d) δ 7.33–7.20 (m, 2H), 6.92–6.84 (m, 3H), 4.10 (br, 2H), 4.01 (t, J = 6.0 Hz, 2H), 2.72 (br, 2H), 1.77–1.70 (m, 5H), 1.47 (s, 9H), 1.24–1.08 (m, 2H). LRMS (ESI): m/z Calcd. for C18H27NO3Na [M+Na]+, 328.41, found 328.21.
4.1.2. 1-(4-(2-Phenoxyethyl)piperidin-1-yl)prop-2-en-1-one (DC-TEADin1072)
Compound 2 (100 mg, 0.3 mmol) was added into 4 mol/L HCl in 1,4-dioxane solution (3 mL) and the reaction mixture was allowed to stir at ambient temperature for 2 h. The solvent was removed under vacuum and the residue was used directly in next step without further purification. (ii) the residue obtained in step (i) and TEA (101 mg, 1 mmol) were dissolved in anhydrous DCM (3 mL) at 0 °C, then acryloyl chloride (36 mg, 0.4 mmol) was added dropwise. The reaction was stirred at 0 °C for 2 h then quenched with water. The solvent was removed under vacuum and the residue was purified by reversed-phase preparative HPLC (gradient 5%–95%, CH3CN/H2O with 0.1% TFA) to afford the title compound DC-TEADin1072 (56 mg, yield 72%) as a colorless oil. Purity: 100%. 1H NMR (400 MHz, chloroform-d) δ 7.33–7.23 (m, 2H), 6.94 (t, J = 7.3 Hz, 1H), 6.89 (d, J = 7.8 Hz, 2H), 6.58 (dd, J = 16.8, 10.6 Hz, 1H), 6.25 (dd, J = 16.8, 1.9 Hz, 1H), 5.66 (dd, J = 10.6, 1.9 Hz, 1H), 4.66 (d, J = 13.1 Hz, 1H), 4.07–3.92 (m, 3H), 3.05 (t, J = 12.2 Hz, 1H), 2.64 (t, J = 11.9 Hz, 1H), 1.89–1.78 (m, 3H), 1.78–1.68 (m, 2H), 1.35–1.10 (m, 2H). 13C NMR (126 MHz, CDCl3) δ 164.86, 158.37, 128.98, 127.51, 126.74, 120.22, 113.94, 64.58, 45.64, 41.84, 35.13, 32.64, 32.38, 31.28. HRMS (ESI): m/z Calcd. for C16H22NO2 [M+H]+, 260.1645, found 260.1639.
4.1.3. 1-(4-(2-Phenoxyethyl)piperidin-1-yl)propan-1-one (DC-TEADin1072-N1)
Starting with compound 2 (100 mg, 0.3 mmol), following similar procedures to that for compound DC-TEADin1072 afford the title compound as colorless oil (61 mg, yield 78%). Purity: 99.37%. 1H NMR (400 MHz, chloroform-d) δ 7.31–7.24 (m, 2H), 6.94 (t, J = 7.4 Hz, 1H), 6.91–6.85 (m, 2H), 4.68–4.58 (m, 1H), 4.00 (t, J = 6.1 Hz, 2H), 3.90–3.78 (m, 1H), 3.06–2.93 (m, 1H), 2.62–2.48 (m, 1H), 2.34 (q, J = 7.5 Hz, 2H), 1.85–1.69 (m, 5H), 1.22–1.09 (m, 5H). 13C NMR (126 MHz, CDCl3) δ 171.62, 158.38, 128.97, 120.20, 113.94, 64.62, 45.20, 41.42, 35.19, 32.65, 32.30, 31.35, 26.09, 9.13. HRMS (ESI): m/z Calcd. for C16H24NO2 [M+H]+, 262.1802, found 262.1803.
4.1.4. (3-(Benzyloxy)propyl)magnesium bromide (4)
Polished magnesium strip (162 mg, 6.8 mmol) and anhydrous THF (10 mL) were added into a dried three-necked bottle under argon then degassed the mixture for three times. Compound 3 (1380 mg, 6.0 mmol) in anhydrous THF (2 mL) and 1,2-dibromoethane (10 μL) was added respectively. The mixture was stirred for a while at ambient temperature until reaction initiation was observed and then heated to 60 °C for 2 h. After the reaction completed, the suspension was cooled to ambient temperature and the Grignard reagent was prepared.
4.1.5. tert-Butyl 4-(3-(benzyloxy)propyl)-4-hydroxypiperidine-1-carboxylate (5)
The suspension of compound 4 was cooled to 0 °C and N-(tert-butoxycarbonyl)-4-piperidone (300 mg, 1.5 mmol) in anhydrous THF (2 mL) was added dropwise. The mixture was allowed to stir at ambient temperature for 2 h. While TLC showed the reaction was completed, quenched the reaction with water (10 mL). The mixture was extracted with ethyl acetate (10 mL × 2), and the combined organic layers were dried over anhydrous sodium sulfate and concentrated in vacuum. The residue was purified by flash silica chromatography (gradient 0–5% MeOH/DCM) to afford the title compound 5 (450 mg, yield 86%) as a colorless oil. 1H NMR (400 MHz, chloroform-d) δ 7.39–7.27 (m, 5H), 4.52 (s, 2H), 3.79 (br, 2H), 3.51 (t, J = 5.8 Hz, 2H), 3.15 (br, 2H), 2.46 (s, 1H), 1.77–1.68 (m, 2H), 1.64 (s, 2H), 1.59 (t, J = 7.7 Hz, 2H), 1.49 (s, 2H), 1.45 (s, 9H). LRMS (ESI): m/z Calcd. for C20H31NO4Na [M+Na]+, 372.46, found 372.48.
4.1.6. 1-(4-(3-(Benzyloxy)propyl)-4-hydroxypiperidin-1-yl)prop-2-en-1-one (DC-TEAD3in01)
Starting with compound 5 (105 mg, 0.3 mmol), followed the similar synthetic procedure of compound DC-TEADin1072 to afford the title compound as colorless oil (77 mg, yield 85%). Purity: 100%. 1H NMR (400 MHz, chloroform-d) δ 7.38–7.26 (m, 5H), 6.58 (dd, J = 16.8, 10.6 Hz, 1H), 6.24 (dd, J = 16.8, 1.9 Hz, 1H), 5.65 (dd, J = 10.6, 1.9 Hz, 1H), 4.51 (s, 2H), 4.37 (d, J = 13.1 Hz, 1H), 3.72 (d, J = 13.4 Hz, 1H), 3.56–3.38 (m, 3H), 3.08 (td, J = 12.8, 2.8 Hz, 1H), 2.88 (brs, 1H), 1.78–1.67 (m, 2H), 1.68–1.55 (m, 4H), 1.54–1.39 (m, 2H). 13C NMR (126 MHz, CDCl3) δ 164.78, 137.35, 127.97, 127.44, 127.28, 127.21, 126.75, 72.64, 70.32, 68.28, 41.61, 40.09, 37.75, 37.20, 36.20, 22.78. HRMS (ESI): m/z Calcd. for C18H26NO3 [M+H]+, 304.1907, found 304.1908.
4.1.7. tert-Butyl 4-(3-(benzyloxy)propyl)piperazine-1-carboxylate (6)
Compound 3 (275 mg, 1.2 mmol), tert-butyl piperazine-1-carboxylate (186 mg, 1.0 mmol) and K2CO3 (414 mg, 3.0 mmol) were dissolved in DMF (3 mL) in a thick flask. The reaction mixture was stirred at 90 °C for 8 h. After the mixture cooled to ambient temperature, water (10 mL) was added and extracted with ethyl acetate (8 mL × 2). The combined organic layers were wash with water (10 mL × 2) and saturated saltwater (10 mL × 1) then dried over anhydrous sodium sulfate and concentrated in vacuum. The residue was purified by flash silica chromatography (gradient 0–5% MeOH/DCM) to afford the title compound 6 (298 mg, yield 89%) as a colorless oil. 1H NMR (400 MHz, chloroform-d) δ 7.37–7.26 (m, 5H), 4.49 (s, 2H), 3.51 (t, J = 6.2 Hz, 2H), 3.41 (brs, 4H), 2.43 (t, J = 7.4 Hz, 2H), 2.36 (brs, 4H), 1.79 (td, J = 7.4, 6.2 Hz, 2H), 1.45 (s, 9H). LRMS (ESI): m/z Calcd. for C19H31N2O3 [M+H]+, 335.47, found 335.27.
4.1.8. tert-Butyl 3-(3-(benzyloxy)propyl)tetrahydropyrimidine-1(2H)-carboxylate (7)
Starting with compound tert-butyl tetrahydropyrimidine-1(2H)-carboxylate (186 mg, 1.0 mmol), followed the similar synthetic procedure of compound 6 to afford the title compound as colorless oil (277 mg, yield 83%). 1H NMR (400 MHz, chloroform-d) δ 7.38–7.26 (m, 5H), 4.50 (s, 2H), 4.09 (brs, 2H), 3.53 (t, J = 6.3 Hz, 2H), 3.45 (br, 2H), 2.72 (br, 2H), 2.52 (t, J = 7.4 Hz, 2H), 1.84 (br, 2H), 1.61 (td, J = 7.4, 6.3 Hz, 2H), 1.45 (s, 9H). LRMS (ESI): m/z Calcd. for C19H31N2O3 [M+H]+, 335.47, found 335.57.
4.1.9. 1-(4-(3-(Benzyloxy)propyl)piperazin-1-yl)prop-2-en-1-one (DC-TEAD3in02)
Starting with compound 6 (100 mg, 0.3 mmol), followed the similar synthetic procedure of compound DC-TEADin1072 to afford the title compound as colorless oil (66 mg, yield 76%). Purity: 99.33%. 1H NMR (400 MHz, chloroform-d) δ 7.37–7.24 (m, 5H), 6.54 (dd, J = 16.8, 10.5 Hz, 1H), 6.27 (dd, J = 16.8, 1.9 Hz, 1H), 5.67 (dd, J = 10.5, 1.9 Hz, 1H), 4.49 (s, 2H), 3.76–3.62 (m, 2H), 3.59–3.47 (m, 4H), 2.50–2.39 (m, 6H), 1.85–1.74 (m, 2H). 13C NMR (126 MHz, CDCl3) δ 165.37, 138.55, 128.44, 127.79, 127.68, 127.64, 127.60, 68.43, 55.32, 53.48, 52.80, 45.80, 41.95, 27.14. HRMS (ESI): m/z Calcd. for C17H25N2O2 [M+H]+, 289.1911, found 289.1919.
4.1.10. 1-(3-(3-(Benzyloxy)propyl)tetrahydropyrimidin-1(2H)-yl)prop-2-en-1-one (DC-TEAD3in03)
Starting with compound 7 (100 mg, 0.3 mmol), followed the similar synthetic procedure of compound DC-TEADin1072 to afford the title compound as colorless oil (72 mg, yield 83%). Purity: 100%. Compound DC-TEAD3in03 was obtained as a mixture of non-separable E/Z stereoisomers and showed separate signals in 13C NMR spectrum due to hindered rotation causing by the tertiary amide motif42. 1H NMR (400 MHz, methanol-d4) δ 7.40–7.21 (m, 5H), 6.84–6.64 (m, 1H), 6.29–6.13 (m, 1H), 5.83–5.67 (m, 1H), 4.49 (s, 2H), 4.34 (s, 1H), 4.28 (s, 1H), 3.71–3.65 (m, 2H), 3.54 (q, J = 5.9 Hz, 2H), 2.81 (q, J = 7.7 Hz, 2H), 2.65–2.51 (m, 2H), 1.90–1.75 (m, 2H), 1.75–1.63 (m, 2H). 13C NMR (126 MHz, methanol-d4) δ (E/Z stereoisomers): 167.75, 167.55, 139.81, 129.36, 128.89, 128.83, 128.76, 128.64, 73.94, 73.89, 69.24, 69.20, 68.03, 63.79, 53.12, 51.51, 51.16, 46.59, 43.18, 28.40, 28.31, 24.64, 24.00. HRMS (ESI): m/z Calcd. for C17H25N2O2 [M+H]+, 289.1911, found 289.1916.
4.2. Protein expression and purification
The YAP-binding domain (YBD) of human TEAD1 (209–426), TEAD2 (217–447), TEAD3 (219–435), TEAD4 (217–434) and mutants were cloned in pET28a vector with the N-terminal 6x His tag as previously37. The proteins were expressed in Escherichia coli BL21 (DE3) cells strain by induction with 0.4 mmol/L isopropyl β-d-thiogalactopyranoside (IPTG) at 16 °C overnight. The cells expressing 6x His tag proteins were resuspended buffer containing 50 mmol/L Tris pH 8.0, 100 mmol/L NaCl and 20 mmol/L imidazole pH 8.0, then proteins were then purified by using the HisTrap FF 5 mL column (GE Healthcare). A second purification step by size exclusion chromatography using a Superdex™75 10/300 GL column (GE Healthcare) was then performed in buffer containing 20 mmol/L HEPES pH 8.0 and 100 mmol/L NaCl. The proteins were detected by SDS-polyacrylamide gel electrophoresis.
4.3. Compound library
The in-house covalent compound library containing 358 small molecules with covalent warheads was obtained from commercial vendors including TargetMol (USA) and SPECS (https://www.specs.net/). All compounds were dissolved in DMSO prior to application. All the identified hits will be individually checked by LC‒MS.
4.4. In vitro palmitoylation assay
Synthetic compounds with different concentrations were incubated with nmol/L TEAD (1–4)-YBD protein at 37 °C for 2 h, and flufenamic acid (CSNpharm, #CSN12744) was used as a positive control. Then 2.5 μmol/L palmitoyl coenzyme A (Cayman Chemical, No. 15968) was added into the centrifuge tube, and then 30 mi was incubated on ice. The click chemical reaction is then performed at room temperature. Finally, 6x SDS sample loading buffer containing 30 mmol/L EDTA was added to terminate the reaction, and the sample was boiled at 95 °C for 5 min43. The activity of the compound was then analyzed by Western blotting. The relative band density was quantified in ImageJ (NIH).
4.5. Mass spectrum
4.5.1. Sample preparation
For intact mass analysis, protein sample solution was diluted to a concentration of 1 mg/mL in water with 0.1% formic acid. A total of 2 μg protein was injected for each LC/MS run. For drug conjugate sites analysis, sequencing-grade modified trypsin was added to each sample (enzyme to protein ratio 1:50, w/w) and incubated at 37 °C for 16 h 20 μg of the digested peptide mixture was desalted by C18 tip.
4.5.2. LC/MS instruments and bioinformatics analysis
Intact protein was analyzed using an Ultimate 3000 LC coupled with a Orbitrap Fusion mass spectrometer equipped with a HESI ion source (Thermo Fischer Scientific, San Jose, CA, USA). Intact protein samples were separated with an Agilent PLRP-S column (1.0 mm × 50 mm, 5 μm) using a 15-min gradient (hold at 5% B for 2 min, 5%–35% B for 6 min, 35%–80% B for 2 min, 80% hold for 2 min, 80%–5% B for 0.5 min, 5% B for 2.5 min) at a flow rate of 0.300 mL/min. Mobile Phase A was made up of water with 0.1% formic acid, while Mobile Phase B was made up of acetonitrile with 0.1% formic acid. The mass spectrometry instrument parameters were set as the following: the temperature of the heated capillary was set at 350 °C and the source voltage was set at 3.5 kV. Orbitrap full scan automatic gain control target, 1 × 106; maximum injection time, 50 ms.
Enzymatic digestion products of peptide mixture were analyzed on the Easy nano-LC1000 system using a self-packed column (75 μm × 150 mm; 3 μm ReproSil-Pur C18 beads, 120 Å, Dr. Maisch GmbH, Ammerbuch, Germany) at a flow rate of 300 nL/min. The mobile phase A of RP-HPLC was 0.1% formic acid in water, and B was 0.1% formic acid in acetonitrile. The peptides were eluted with a 60-min gradient (1%–100% mobile phase B) into a nano-ESI orbitrap Q-Exactive mass spectrometer. The mass spectrometer was operated in data-dependent mode with each full MS scan (m/z 300–1600) followed by MS/MS for the 10 most intense ions with the parameters: ≥+2 precursor ion charge, 2 Da precursor ion isolation window and 27 normalized collision energy of HCD. Dynamic Exclusion™ was set as 30 s. The full mass and the subsequent MS/MS analysis were scanned in the Orbitrap analyzer with R = 70,000 and R = 17,500, respectively.
The MS data was analyzed via software MaxQuant (http://maxquant.org/, version 1.6.5.0) with oxidation (M) and protein N-term acetylation set as variable modifications. Trypsin/P was selected as the digestive enzyme with maximum two missed cleavages. The false discovery rate (FDR) for peptides and proteins was controlled <1% by Andromeda search engine.
4.6. Protein thermal shift assay
Protein thermal shift assay was performed in QuantStudio 6 Flex Real-Time PCR System. 5 μmol/L protein was mixed with 5 × SYPRO Orange (Sigma–Aldrich, #S5692) before the thermal cycle (ramped from 25 to 95 °C). All compounds were tested in the assay buffer containing 20 mmol/L Hepes pH 8.5, 100 mmol/L NaCl and 2.5% DMSO. The filter was set at ROX with no quencher and passive filter. The protein melting temperature (Tm) was calculated with fluorescence raw data by the Protein Thermal Shift software v.1.3 (Life Technologies). Data were plotted using GraphPad Prism 7.
4.7. Covalent docking
The specific method of covalent docking has been described previously37. The covalent docking studies were performed in Covalent Docking panel in the Maestro program (Maestro, version 9.1; Schrödinger, LLC: New York, 2010) to conduct covalent docking experiments. TEAD3 Cys371 was picked as the nucleophile for covalent reactions (PDB ID: 5EMV). Other parameters were set as the default.
4.8. Fluorescence polarization assay
Fluorescence polarization experiments were used to determine the effects of compounds or DMSO at different concentrations on TEAD–YAP interactions. The experiment was conducted in a 384-well black plate (Corning, #3575) using FP buffer (25 mmol/L Hepes pH 8.0, 100 mmol/L NaCl, 0.1 mg/mL BSA, 1 mmol/L DTT, and 0.01% NP-40). The compound was incubated with 200 nmol/L TEAD3 protein for 1 h. Then FITC-YAP(61-100) peptide of 30 nmol/L was added and incubated at room temperature for 30 min44. The Envision Multiplate reader (PerkinElmer) was used to measure the fluorescence polarization signal at the excitation wavelength of 480 nm and emission wavelength of 520 nm.
4.9. Cell culture
HEK293T cells were bought from Amercian Type Culture Collection (ATCC) and cultured in Dulebcco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin (Invitrogen).
4.10. Western blotting
Cell lysate samples were heated in a 95 °C heat block for 20 min and separated by 12% acrylamide SDS-PAGE in Tris-glycine buffer. The proteins were transferred to nitrocellulose membranes (GE Healthcare) and blocked for 1 h at room temperature in 5% non-fat milk (diluted in TBST). Blots were incubated at 4 °C overnight with primary antibodies: Anti-TEAD4 antibody (Abcam, #ab58310), Anti-TEAD1-3 (Cell Signaling Technology, #8526, #8870, #13224), GAPDH (Cell Signaling Technology, #5174), YAP (Cell Signaling Technology, #14074), DYKDDDDK Tag (Cell Signaling Technology, #14793), streptavidin-HRP (Cell Signaling Technology, cat#3999), His-Tag (Cell Signaling Technology, #12698). After incubation of primary antibodies, blots were incubated with a 1:10,000 dilution of secondary antibody (BBI life, #D110058) for 1 h at room temperature. Images were detected by GE ImageQuant LAS 4000 system with SuperSignal WestDura (Thermo Scientific, # 34076) as the HRP substrate.
4.11. Luciferase assays
For TEAD reporter assay, HEK293T cells were seeded in 24-well plates (Corning) at the density of 2 × 105/well. Then cells were transfected with 200 ng 8xGTIIC-Luc TEADs reporter construct (Addgene, #34615) or TOPFlash beta-catenin reporter (Addgene, #12456) and 20 ng pGL4.75 Renilla construct (Promega, #E6931) as the internal control. The transfected cells were treated with various concentrations of compounds or DMSO. Luciferase signal was measured using dual-luciferase reporter assay kit (Promega, #E1980) according to the manufacturer's guidelines. Data was collected in EnVision Multilabel Plate Reader (PerkinElmer) and plotted in Prism software.
For GAL4-Luc assay, HEK293T cells were transfected with GAL4-Luc Reporter Plasmid (Genomeditech, #GM-021041) and GAL4-TEAD1 (Addgene, #33108), GAL4-TEAD2 (Addgene, #33107), GAL4-TEAD3 (Addgene, #33106) or GAL4-TEAD4 (Addgene, #33105) expression vectors25. In addition, cells were also transfected with a plasmid expressing YAP45. The cells were treated with DC-TEAD3in03 at a specified concentration and cultured at 37 °C for 12 h. Luciferase signal was colleted using EnVision Multilabel Plate Reader (PerkinElmer).
4.12. Cellular thermal shift assay (CETSA)
Intact cell CETSA was performed in HEK293T cells according to the previously described protocol37. The cells were transfected with TEAD1−4 with 3 × FLAG tags at the N-terminal and treated with 50 μmol/L DC-TEADin1072, DC-TEAD3in03 or an equal volume of DMSO at 37 °C for 6 h, respectively. Then the cells were trypsinized and washed with PBS. After several washes, the cells were resuspended in PBS containing freshly added protease inhibitor (Roche) and aliquoted into PCR tubes. Each tube was heated at a specified temperature for 3 min and restored at room temperature for 3 min. The heated cells underwent three cycles of freezing (in liquid nitrogen, 1 min) and thawing (in room temperature water, 1 min). Then cell lysates were centrifuged at 4 °C and 20,000×g for 30 min to separate the soluble components for western blotting analysis. Primary antibodies: DYKDDDDK Tag (Cell Signaling Technology, #14793) and anti-GAPDH rabbit polyclonal antibody (BBI Life Sciences, #D110016) were used for Western blotting.
4.13. Zebrafish husbandry
AB strain fish were raised in an aquarium system with constantly flowing water under a 26 °C standard light–dark cycle46. Experiments used male and female fish equally. Fish experiments were compliant to the general animal welfare guidelines and protocols (20200903003) approved by legally authorized animal welfare committees (ShanghaiTech University, ShanghaiTech Animal Welfare Committee, Shanghai, China).
4.14. Treatment of juvenile fish
AB wild type juvenile fish (2 months old, 10 per dish) were placed in 30 mL sterilized fish facility water in 90 mm dishes. The water either contained a TEAD3 inhibitor (final concentration of 0.25 μmol/L) or the appropriate dilution of DMSO. The water was continuously aerated with two needles connected to a benchtop pump. The water was refreshed once a day and the fish were fed twice a day. Before the experiment, each fish of each group was imaged using a Zeiss Stemi 508 stereomicroscope equipped with a Zeiss Axiocam 503 color camera controlled by ZEN Blue software (Zeiss) to assess the lengths of the caudal fins and fish bodies. The length of the fish was analyzed by ImageJ (NIH). Fin length was measured from the base of the fin through the middle of the fin, parallel to the middle ray of the fin, to a virtual line connecting the tips of the lobes to average the length of both lobes. Body length was measured from the most anterior point of the fish jaw to the base of the caudal fin.
Acknowledgments
We thank the Institutional Technology Service Center of Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China) for technical assistance in mass spectrometry experiments and analysis. We are grateful to National Centre for Protein Science Shanghai (Protein Expression and Purification system) for their instrument support and technical assistance. We also gratefully acknowledge the financial supports from the National Natural Science Foundation of China (91853205, 81625022, 81821005 to Cheng Luo; 81973166, 21702218, 91753207 to Bing Zhou), Wong Education to Cheng Luo and Bing Zhou, the Department of Science and Technology of Fujian Province (2019T3029 to Cheng Luo, China), the Science and Technology Commission of Shanghai Municipality (19XD1404700 and 18431907100 to Cheng Luo, China) and National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (2018ZX09711002-008 to Cheng Luo, 2018ZX09711002-008-005 to Huijin Feng, 2018ZX09711002-006 to Bing Zhou, China).
Author contributions
Conception of the hypothesis: Cheng Luo; supervision and design: Christopher L. Antos, Bing Zhou and Cheng Luo; Development of methodology: Tian Lu, Yong Li, Wenchao Lu and TWGM Spitters.; Acquisition of data: Tian Lu, Yong Li, Wenchao Lu, TWGM Spitters, Jun Wang, Xueyu Fang, Simian Cai and Huan Xiong. Analysis and interpretation of data: Tian Lu, Yong Li, Wenchao Lu, TWGM Spitters, Jun Wang, Xueyu Fang, Simian Cai, Jing Gao, Yanting Zhou and Zhe Duan. Writing, review, and/or revision of the manuscript: Tian Lu, Yong Li, Wenchao Lu, TWGM Spitters and Christopher L. Antos. Administrative, technical, or material support: Jing Gao, Yanting Zhou, Zhe Duan, Huan Xiong, Liping Liu, Qi Li, Bing Zhou, Hualiang Jiang and Huijin Feng. All authors discuss the study. Study supervision: Christopher L. Antos, Bing Zhou, and Cheng Luo. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no competing financial and non-financial interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.04.015.
Contributor Information
Bing Zhou, Email: zhoubing@simm.ac.cn.
Christopher L. Antos, Email: clantos@shanghaitech.edu.cn.
Cheng Luo, Email: cluo@simm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nouri K., Azad T., Ling M., Janse van Rensburg H.J., Pipchuk A., Shen H. Identification of celastrol as a novel yap-tead inhibitor for cancer therapy by high throughput screening with ultrasensitive yap/taz-tead biosensors. Cancers (Basel) 2019;11:1596–1614. doi: 10.3390/cancers11101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janse van Rensburg H.J., Yang X. The roles of the hippo pathway in cancer metastasis. Cell Signal. 2016;28:1761–1772. doi: 10.1016/j.cellsig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Huh H.D., Kim D.H., Jeong H.S., Park H.W. Regulation of tead transcription factors in cancer biology. Cells. 2019;8:600–622. doi: 10.3390/cells8060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin K.C., Park H.W., Guan K.L. Regulation of the hippo pathway transcription factor tead. Trends Biochem Sci. 2017;42:862–872. doi: 10.1016/j.tibs.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhtar T., Breda J., Grison A., Karimaddini Z., Grobecker P., Iber D. Tead transcription factors differentially regulate cortical development. Sci Rep. 2020;10:4625–4649. doi: 10.1038/s41598-020-61490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsika R.W., Ma L., Kehat I., Schramm C., Simmer G., Morgan B. Tead-1 overexpression in the mouse heart promotes an age-dependent heart dysfunction. J Biol Chem. 2010;285:13721–13735. doi: 10.1074/jbc.M109.063057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko K.J., Kohn M.J., Liu C., DePamphilis M.L. Transcription factor tead2 is involved in neural tube closure. Genesis. 2007;45:577–587. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Z., Yu Y., Cai B., Xu Z., Bao Z., Zhang Y. Yap/tead3 signal mediates cardiac lineage commitment of human-induced pluripotent stem cells. J Cell Physiol. 2020;235:2753–2760. doi: 10.1002/jcp.29179. [DOI] [PubMed] [Google Scholar]
- 9.Li J., Tiwari M., Xu X., Chen Y., Tamayo P., Sen G.L. Tead1 and tead3 play redundant roles in the regulation of human epidermal proliferation. J Invest Dermatol. 2020;140:2081–2084. e4. doi: 10.1016/j.jid.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figeac N., Mohamed A.D., Sun C., Schonfelder M., Matallanas D., Garcia-Munoz A. Vgll3 operates via tead1, tead3 and tead4 to influence myogenesis in skeletal muscle. J Cell Sci. 2019;132:jcs225946. doi: 10.1242/jcs.225946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z., Zhang H., Zhang Y., Liao L., Zhou W., Zhang F. Covalent inhibitors allosterically block the activation of rho family proteins and suppress cancer cell invasion. Adv Sci (Weinh) 2020;7:2000098. doi: 10.1002/advs.202000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquemin P., Chen Z., Martial J.A., Davidson I. Genomic structure and chromosomal mapping of the mouse transcription factor tef-5 (tead3) gene. Mamm Genome. 1999;10:632–634. doi: 10.1007/s003359901059. [DOI] [PubMed] [Google Scholar]
- 13.Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Crawford J.J., Bronner S.M., Zbieg J.R. Hippo pathway inhibition by blocking the yap/taz-tead interface: a patent review. Expert Opin Ther Pat. 2018;28:867–873. doi: 10.1080/13543776.2018.1549226. [DOI] [PubMed] [Google Scholar]
- 15.Elisi G.M., Santucci M., D'Arca D., Lauriola A., Marverti G., Losi L. Repurposing of drugs targeting yap-tead functions. Cancers (Basel) 2018:10. doi: 10.3390/cancers10090329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen T., Li Y., Zhu S., Yu J., Zhang B., Chen X. Yap1 plays a key role of the conversion of normal fibroblasts into cancer-associated fibroblasts that contribute to prostate cancer progression. J Exp Clin Cancer Res. 2020;39:36. doi: 10.1186/s13046-020-1542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight J.F., Shepherd C.J., Rizzo S., Brewer D., Jhavar S., Dodson A.R. Tead1 and c-cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99:1849–1858. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo J.S., Cho S.Y., Rou W.S., Kim J.S., Kang S.H., Lee E.S. Tead2 as a novel prognostic factor for hepatocellular carcinoma. Oncol Rep. 2020;43:1785–1796. doi: 10.3892/or.2020.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Li J., Wu Y., Ge H., Song Y., Wang D. Tead4 overexpression promotes epithelial-mesenchymal transition and associates with aggressiveness and adverse prognosis in head neck squamous cell carcinoma. Cancer Cell Int. 2018;18:178–192. doi: 10.1186/s12935-018-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Wang G., Yang Y., Mei Z., Liang Z., Cui A. Increased tead4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a yap-independent manner. Oncogene. 2016;35:2789–2800. doi: 10.1038/onc.2015.342. [DOI] [PubMed] [Google Scholar]
- 21.Giraud J., Molina-Castro S., Seeneevassen L., Sifre E., Izotte J., Tiffon C. Verteporfin targeting yap1/taz-tead transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int J Cancer. 2020;146:2255–2267. doi: 10.1002/ijc.32667. [DOI] [PubMed] [Google Scholar]
- 22.Chen M., Huang B., Zhu L., Chen K., Liu M., Zhong C. Structural and functional overview of tead4 in cancer biology. Onco Targets Ther. 2020;13:9865–9874. doi: 10.2147/OTT.S266649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L., Yuan L., Sun Y., Wang P., Zhang H., Feng X. Glucocorticoid receptor signaling activates tead4 to promote breast cancer progression. Cancer Res. 2019;79:4399–4411. doi: 10.1158/0008-5472.CAN-19-0012. [DOI] [PubMed] [Google Scholar]
- 24.Xu M.Z., Chan S.W., Liu A.M., Wong K.F., Fan S.T., Chen J. Axl receptor kinase is a mediator of yap-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30:1229–1240. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B., Ye X., Yu J., Li L., Li W., Li S. Tead mediates yap-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L., Li Y., Kim S.M., Bossuyt W., Liu P., Qiu Q. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hucl T., Brody J.R., Gallmeier E., Iacobuzio-Donahue C.A., Farrance I.K., Kern S.E. High cancer-specific expression of mesothelin (msln) is attributable to an upstream enhancer containing a transcription enhancer factor dependent mcat motif. Cancer Res. 2007;67:9055–9065. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- 28.Li Z., Zhao B., Wang P., Chen F., Dong Z., Yang H. Structural insights into the yap and tead complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaan H.Y.K., Chan S.W., Tan S.K.J., Guo F., Lim C.J., Hong W. Crystal structure of taz–tead complex reveals a distinct interaction mode from that of yap–tead complex. Sci Rep. 2017;7:2035–2046. doi: 10.1038/s41598-017-02219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noland C.L., Gierke S., Schnier P.D., Murray J., Sandoval W.N., Sagolla M. Palmitoylation of tead transcription factors is required for their stability and function in hippo pathway signaling. Structure. 2016;24:179–186. doi: 10.1016/j.str.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Holden J.K., Crawford J.J., Noland C.L., Schmidt S., Zbieg J.R., Lacap J.A. Small molecule dysregulation of tead lipidation induces a dominant-negative inhibition of hippo pathway signaling. Cell Rep. 2020;31:107809–107829. doi: 10.1016/j.celrep.2020.107809. [DOI] [PubMed] [Google Scholar]
- 32.Chan P., Han X., Zheng B., DeRan M., Yu J., Jarugumilli G.K. Autopalmitoylation of tead proteins regulates transcriptional output of the hippo pathway. Nat Chem Biol. 2016;12:282–289. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pobbati A.V., Han X., Hung A.W., Weiguang S., Huda N., Chen G.Y. Targeting the central pocket in human transcription factor tead as a potential cancer therapeutic strategy. Structure. 2015;23:2076–2086. doi: 10.1016/j.str.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bum-Erdene K., Zhou D.H., Gonzalez-Gutierrez G., Ghozayel M.K., Si Y.B., Xu D. Small-molecule covalent modification of conserved cysteine leads to allosteric inhibition of the tead center dot yap protein‒protein interaction. Cell Chem Biol. 2019;26:378–389. doi: 10.1016/j.chembiol.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Kaneda A., Seike T., Uemori T., Myojo K., Aida K., Danjo T. Discovery of a first-in-class tead inhibitor which directly inhibits yap/taz-tead protein‒protein interaction and shows a potent anti-tumor effect in malignant pleural mesothelioma. Cancer Res. 2019;79:3086. [Google Scholar]
- 36.Pobbati A.V., Mejuch T., Chakraborty S., Karatas H., Bharath S.R., Gueret S.M. Identification of quinolinols as activators of tead-dependent transcription. ACS Chem Biol. 2019;14:2909–2921. doi: 10.1021/acschembio.9b00786. [DOI] [PubMed] [Google Scholar]
- 37.Lu W., Wang J., Li Y., Tao H., Xiong H., Lian F. Discovery and biological evaluation of vinylsulfonamide derivatives as highly potent, covalent tead autopalmitoylation inhibitors. Eur J Med Chem. 2019;184:111767. doi: 10.1016/j.ejmech.2019.111767. [DOI] [PubMed] [Google Scholar]
- 38.Mesrouze Y., Bokhovchuk F., Meyerhofer M., Fontana P., Zimmermann C., Martin T. Dissection of the interaction between the intrinsically disordered yap protein and the transcription factor tead. Elife. 2017;6 doi: 10.7554/eLife.25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon J.H., Lee J., Lee J.Y., Shin Y.S., Kim D.E., Min J.S. Study on the 2-phenylchroman-4-one derivatives and their anti-MERS-CoV activities. Bull Korean Chem Soc. 2019;40:906–909. doi: 10.1002/bkcs.11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L., Chan S.W., Zhang X., Walsh M., Lim C.J., Hong W. Structural basis of yap recognition by tead4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holden J.K., Cunningham C.N. Targeting the hippo pathway and cancer through the tead family of transcription factors. Cancers (Basel) 2018;10 doi: 10.3390/cancers10030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bisceglia J.Á., Mollo M.C., Orelli L.R. E/z equilibrium in tertiary amides. Part 2: N-acyl-N′-arylhexahydropyrimidines. J Mol Struct. 2010;966:79–84. [Google Scholar]
- 43.Zheng B., DeRan M., Li X., Liao X., Fukata M., Wu X. 2-Bromopalmitate analogues as activity-based probes to explore palmitoyl acyltransferases. J Am Chem Soc. 2013;135:7082–7085. doi: 10.1021/ja311416v. [DOI] [PubMed] [Google Scholar]
- 44.Karatas H., Akbarzadeh M., Adihou H., Hahne G., Pobbati A.V., Yihui Ng E. Discovery of covalent inhibitors targeting the transcriptional enhanced associate domain central pocket. J Med Chem. 2020;63:11972–11989. doi: 10.1021/acs.jmedchem.0c01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith S.A., Sessions R.B., Shoemark D.K., Williams C., Ebrahimighaei R., McNeill M.C. Antiproliferative and antimigratory effects of a novel yap‒tead interaction inhibitor identified using in silico molecular docking. J Med Chem. 2019;62:1291–1305. doi: 10.1021/acs.jmedchem.8b01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerhard G.S., Kauffman E.J., Wang X., Stewart R., Moore J.L., Kasales C.J. Life spans and senescent phenotypes in two strains of zebrafish (danio rerio) Exp Gerontol. 2002;37:1055–1068. doi: 10.1016/s0531-5565(02)00088-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.