Abstract
10-Deacetylbaccatin III (10-DAB) C10 acetylation is an indispensable procedure for Taxol semi-synthesis, which often requires harsh conditions. 10-Deacetylbaccatin III-10-β-O-acetyltransferase (DBAT) catalyzes the acetylation but acetyl-CoA supply remains a key limiting factor. Here we refactored the innate biosynthetic pathway of acetyl-CoA in Escherichia coli and obtained a chassis with acetyl-CoA productivity over three times higher than that of the host cell. Then, we constructed a microbial cell factory by introducing DBAT gene into this chassis for efficiently converting 10-DAB into baccatin III. We found that baccatin III could be efficiently deacetylated into 10-DAB by DBAT with CoASH and K+ under alkaline condition. Thus, we fed acetic acid to the engineered strain both for serving as a substrate of acetyl-CoA biosynthesis and for alleviating the deacetylation of baccatin III. The fermentation conditions were optimized and the baccatin III titers reached 2, 3 and 4.6 g/L, respectively, in a 3-L bioreactor culture when 2, 3 and 6 g/L of 10-DAB were supplied. Our study provides an environment-friendly approach for the large scale 10-DAB acetylation without addition of acetyl-CoA in the industrial Taxol semi-synthesis. The finding of DBAT deacetylase activity may broaden its application in the structural modification of pharmaceutically important lead compounds.
KEY WORDS: Microbial cell factory, Biosynthetic pathway of acetyl-CoA, Bioconversion of 10-deacetylbaccatin III to baccatin III, 10-Deacetylbaccatin III-10-β-O-acetyltransferase, Deacetylation of baccatin III, High-cell-density fermentation, Taxol semi-synthesis
Abbreviations: aceE, pyruvate decarboxynase; aceF, dihydrolipoamide transacetylase; ACS, acetyl-CoA synthetase; DBAT, 10-deacetylbaccatin III-10-β-O-acetyltransferase; E. coli, Escherichia coli; GAPD, glyceraldehyde-3-phosphate dehydrogenase; lpdA, lipoamide dehydrogenase; PANK, pantothenate kinase; PDH, pyruvate dehydrogenase complex; PGK, 1,3-bisphosphoglycerate kinase; Se/acsL641P, acetyl-CoA synthetaseL641P of Salmonella enterica
Graphical abstract
To acetylate 10-deacetylbaccatin III for producing baccatin III, we refactored acetyl-CoA biosynthetic pathway and combined acetyltransferase DBAT in Escherichia coli. By supplying 6 g/L 10-DAB, baccatin III reached 4.6 g/L.
1. Introduction
Taxol (generic name: paclitaxel) is one of the three most extensively applied anticancer chemotherapeutic agents other than doxorubicin and cisplatin, which has enormous market demand at present1, 2, 3. Currently, the chemical semi-synthesis of Taxol has become one of the major approaches to supply this agent in clinic, which is derived from the more abundant 10-deacetylbaccatin III (10-DAB) (ca. 1 g/kg, 0.1% dry weight) isolated from the needles of Taxus baccata or other yew trees4. The chemical semi-synthetic route of Taxol was firstly reported by Denis et al.5 in 1988 which includes the synthesis and addition of the C10 and C13 side chains to 10-DAB. Typically, the C10-hydroxyl acetylation of 10-DAB is initiated by the C7-hydroxyl protection to produce 7-TES-10-DAB through the acylation of triethylsilane (TES). Thereafter, 7-TES-10-DAB experiences C10-hydroxyl acetylation to produce 7-TES-baccatin III by at least 3 steps with the use of pyridine, triethylsilane and acetyl chloride5. Since then, the acetylation process has been extensively optimized by different labs, in which various catalysts, such as 4-dimethylaminopyridine, 4-dialkylaminopyridine-type nucleophilic organocatalyst, acetic anhydride and Lewis acids (ZnCl2, and CeCl3), were often utilized6, 7, 8. Among these catalysts, ZnCl2 or CeCl3 were considered to be more efficient to directly generate baccatin III with the yields of 93% or 91%, both of which required acetic anhydride as the acetyl donor and reacted in tetrahydrofuran (THF) solution under N28,9. Despite these advances, the nonspecific acetylation of C7-hydroxyl is still a challenge. Additionally, some toxic or hazardous chemicals including pyridine, triethylsilane, acetic anhydride and 4-dimethylaminopyridine are often used, let alone some harsh conditions, such as −45 °C and argon shield.
Enzymatic conversion from 10-DAB to baccatin III is recognized to be superior to conventional chemical methods owing to its highly catalytic efficiency, excellent stereo- and regio-selectivity and the mild reaction conditions. Over the past two decades, the catalytic enzymes involved in this reaction have been widely investigated. Except the C10 deacetylase from Nocardioides luteus SC 1391210, many studies were focused on the 10-deacetylbaccatin III-10-β-O-acetyltransferase (DBAT) from Taxus species11, 12, 13. In fact, both 10-DAB and baccatin III are the consecutive biosynthetic intermediates of Taxol; meanwhile, the conversion from 10-DAB to baccatin III in the yew plant is catalyzed by DBAT alone by using acetyl-CoA as the acetyl donor. DBAT was originally purified from the cell suspension cultures of T. chinensis and characterized with an apparent molecular weight of 71 kDa, an optimum pH of 9 and an optimum temperature of 35 °C14. The coding gene of DBAT was first cloned from T. caspidata and expressed in Escherichia coli11. Then, the recombinant enzyme was characterized to have a molecular weight of 49 kDa and an optimum pH of 7.411, which were quite different from those reported in the aforementioned study. Moreover, the recombinant E. coli, which harbored the DBAT gene (dbat), could utilize the intracellular acetyl-CoA as the acetyl donor and convert the substrate 10-DAB into baccatin III, although the baccatin III titer was only 50 nmol/L12. In spite of its potential applications, little progress has been made for integrating DBAT into industrial Taxol semi-synthetic process. One of the limiting factors is the shortage of acetyl-CoA that is used as the acyl donor in the in vitro 10-DAB acetylation, because this acyl donor is not a bulk commodity. In fact, the commercial prices of acyl/aroyl coenzyme A (CoA) thioesters are so expensive, ranging from USD 200 to 300/5 mg (MilliporeSigma)15. Theoretically, at least 1.35 g of acetyl-CoA is required for the enzymatic synthesis of 1 g of baccatin III from 10-DAB. In our previous study, we carried out the DBAT protein engineering and constructed an in vitro one-pot enzymatic reaction system to manufacture Taxol, with 10-deacetyl Taxol as the substrate while acetyl-CoA as the acetyl donor16. Similarly, the supply of acetyl-CoA also became a limiting factor for further scaling-up. On the other hand, the in vivo 10-DAB acetylation also requires a robust biosynthesis of intracellular acetyl-CoA in addition to high expression of recombinant DBAT.
Here, we refactor the innate biosynthetic pathway of bacterial acetyl-CoA in E. coli BL21(DE3) by the modular engineering strategy. Specifically, this study focuses on the native PDH, PANK, PGK and GAPD, together with the heterologous Se/acsL641P, so as to boost the endogenous production of acetyl-CoA. Based on the high acetyl-CoA producing chassis, we construct a microbial cell factory, represented by the recombinant R10 cell (Scheme 1), by introducing DBAT gene into this chassis to achieve the bioconversion from 10-DAB to baccatin III with the native acetyl-CoA as the acetyl donor. Thereafter, we optimize the production of baccatin III on both shake-flask and 3-L bioreactor scales by feeding 10-DAB and acetic acid. Collectively, findings in this study provide an environmentally friendly approach for the conversion of 10-DAB to baccatin III in the industrial chemical semi-synthesis of Taxol.
Scheme 1.
Profile of acetyl-CoA metabolism in recombinant R10 cell. Except the innate enzymes, the additional PDH, PANK, ACS (Se/acsL641P) and DBAT are driven by strong T7 promoter. PDH, PANK, ACS are involved in acetyl-CoA biosynthesis and DBAT is responsible for 10-DAB acetylation. Glucose and acetate are primary substrates for acetyl-CoA biosynthesis and 10-DAB as the substrate for baccatin III formation. Additionally, under alkaline condition DBAT catalyzes the reverse reaction.
2. Materials and methods
2.1. Strains and plasmids
All strains and plasmids used in this study are listed in Table 1. E. coli Trans_1-T1 (TransGen Biotech Beijing, China) was used for plasmid construction and propagation. BL21(DE3) (TransGen Biotech, Beijing, China) was used for heterologous protein expression and in vivo acetylation of 10-DAB to form baccatin III. The plasmids pCDFDuet-1, pRSFDuet-1 and pACYC184 were purchased from Fenghui biotech (Changsha, China).
Table 1.
The plasmids and strains used in this study.
| Description | Source |
|---|---|
| BL21(DE3) | TransGenBiotech |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA, pACYC184-pgk-gapd | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pRSFDuet-Se/acsL641P | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pRSFDuet-pank | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pRSFDuet-pank-Se/acsL641P | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pACYC184-pgk-gapd | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdApank, pRSFDuet-Se/acsL641P | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pRSFDuet-Se/acsL641P, pEASY-Blunt-E1-gapd | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pRSFDuet-Se/acsL641P, pACYC184-pgk-gapd | This study |
| BL21(DE3)-pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA, pACYC184-pgk-gapd, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpd9A-pgk-gapd, pRSFDuet-Se/acsL641P, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pRSFDuet-pank, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pgk-gapd, pRSFDuet- Se/acsL641P-pank, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pACYC184-pgk-gapd, pCWori-dbat | This study |
| BL21(DE3)-pCDFDuet-acE-acF-lpdA-pank, pRSFDuet-Se/acsL641P, pCWori-dbat | This study |
2.2. Chemicals and media
Q5 High-Fidelity DNA Polymerase, all the restriction enzymes, and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Genomic DNA and plasmid DNA were prepared using TIANamp Bacteria DNA Kit and TIANprep Mini Plasmid Kit (TIANGEN Biotech (Beijing) Co., Ltd.), respectively. 10-DAB and baccatin III standards were purchased from J&K Scientific Ltd. (Beijing, China). Acetyl-coenzyme A and CoASH standards were purchased from Sigma‒Aldrich (St. Louis, MO, USA). Isopropyl β-d-thiogalactopyranoside (IPTG), ampicillin sodium salt, kanamycin sulfate and streptomycin sulfate were purchased from Inalco Spa Milano Italy (Inalco, USA). All other chemicals were of analytical grade unless otherwise indicated.
Luria‒Bertani (LB) medium contained 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl (pH 7.0). Terrific broth (TB) medium contained 12 g/L tryptone, 24 g/L yeast extract, 2.31 g/L KH2PO4, 16.25 g/L K2HPO4·3H2O, 4 mL glycerine (pH 7.0). The complex medium (pH 7.0) was used in high-cell-density fermentation in a 3-L bioreactor, which contained 10 g/L glucose, 1.2 g/L MgSO4·7H2O, 4 g/L (NH4)2SO4, 1.7 g/L citric acid monohydrate, 30 g/L tryptone, 5 g/L yeast extract, 3.5 g/L KH2PO4, 7 g/L K2HPO4·3H2O. All media were autoclaved at 121 °C for 20 min. The glucose feeding solution contained 150 g/L glucose, 1.2 g/L MgSO4·7H2O, 20 g/L tryptone and 10 g/L yeast extract and autoclaved at 115 °C for 20 min. The trace element solution contained 7 g/L CuSO4·5H2O, 0.05 g/L MnSO4·H2O, 0.2 g/L CaCl2, 1 g/L FeSO4·7H2O and 0.1 g/L ZnCl2. The trace element solution and other stock solutions including ampicillin (100 mg/mL), kanamycin (50 mg/mL), streptomycin (40 mg/mL), CoASH (10 mg/mL), IPTG (1 mol/L) and 2 mol/L acetic acid, were sterilized by 0.22 μm membrane filtration.
2.3. Plasmid construction
The primers used to amplify each DNA part are listed in Supporting Information Table S1. The gene encoding the codon-optimized acetyl-CoA synthetase variant (Se/acsL641P) (GenBanK Accession: AAO71645.1) from S. enterica was synthesized by SynBio Research Platform at Tianjin University (Tianjin, China). E. coli BL21(DE3) genomic DNA was used as a template for amplification of pank, pgk, gapd, aceE, aceF and lpdA genes. For overexpression of PDH (aceE, aceF and lpdA complex), two rounds of cloning were performed. First, the aceE gene was inserted into pCDFDuet-1 using restriction sites Sbf I/Not I to construct pCDFDuet-aceE. The aceF, lpdA, pgk and gapd genes were inserted into the Nde I/Pac I restriction sites of pCDFDuet-1 to construct plasmids of pCDFDuet-aceF, pCDFDuet-lpdA, pCDFDuet-pgk and pCDFDuet-gapd, respectively. Secondly, the aceF and lpdA were amplified and then overlapped through OE-PCR. The overlapped fragment was cloned into the Not I/Afl II restriction sites of pCDFDuet-aceE to construct pCDFDuet-aceE-aceF-lpdA (pCDFDuet-pdh). In the same way, pgk, gapd and the overlapped fragment pgk-gapd were cloned into the Pac I/Avr II restriction sites of pCDFDuet-acE-acF-lpdA to construct pCDFDuet-acE-acF-lpdA-pgk and pCDFDuet-acE-acF-lpdA-pgk-gapd17. The gene pank was cloned into the EcoR I/Not I restriction sites of pRSFDuet-1 and the Xho I/Pac I restriction sites of pCDFDuet-acE-acF-lpdA to construct pRSFDuet-pank and pCDFDuet-acE-acF-lpdA-pank, respectively. Se/acsL641P was cloned into the Nde I/Xho I restriction sites of pRSFDuet-pank to construct pRSFDuet-pank-Se/acsL641P. The gapd gene was cloned into the “PCR product” part of pEASY-BluntE1. PEASY-BluntE1-gapd was obtained by colony PCR screening to get the correct insertion. All the recombinant genes were controlled by regulatory elements of T7 promoter, lacO, and ribsome binding site. The plasmid pCWori-dbat was preserved by our laboratory. All clones were screened by restriction digestion analysis and subsequently verified by gene sequencing.
2.4. Metabolic engineering for acetyl-CoA production and 10-DAB acetylation
To refactor acetyl-CoA biosynthetic pathway in E. coli, the plasmids were transformed alone or in different combinations into the BL21(DE3) strain (Fig. 1B). Then positive transformant strains were screened by colony PCR. The selected chassis cells were prepared for competent cells by the canonical method of CaCl2 solution. Subsequently, pCWori-dbat was transformed into the aforementioned competent cells to achieve the refactored E. coli (M1‒M12).
Figure 1.
Modular engineering of acetyl-CoA biosynthetic pathway in E. Coli. (A) Schematic diagram of acetyl-CoA biosynthetic pathway and highlighting the focused enzymes. (B) Construction of combinatorial modules including pseudo-operons. (C) Acetyl-CoA production levels in different chassis cells (A.U., arbitrary unit). The data represent the means ± SD, n = 3.
The dry cell weight (DCW) was determined as follows: The optical cell density at 600 nm (OD600) was measured by a UV‒Visible spectrophotometer (IMPLEN, P300, Germany). 5 mL of the culture broth was subjected to centrifugation at 4500 rpm (1902×g) and 4 °C for 10 min and the cell pellet was washed twice with a 0.1% NaCl solution by centrifugation. The collected cell pellet was then freeze-dried overnight to a constant weight. The correlation between DCW and the cell density (OD600) was estimated based on the lyophilization result.
2.5. Shake-flask fermentation
The engineered strain activated from ‒ 80 °C refrigerator was inoculated in 10 mL LB broth, containing amounts of proper antibiotics (100 μg/mL of ampicillin for pCWori, 50 μg/mL of kanamycin for pRSFDuet-1, 40 μg/mL of streptomycin for pCDFDuet-1), and cultivated in a shake-flask at 220 rpm and 37 °C for 24 h. Then the culture was inoculated into 200 mL TB medium containing amounts of proper antibiotics in the proportion of 1:100 (v/v) and until the OD at 600 nm reached 0.8–1.0. Then IPTG was added to the culture with the final concentration of 1 mmol/L and inducted at 20 °C and 220 rpm for a couple of hours. For acetyl-CoA measurement, the cultures at different intervals were collected. For 10-DAB acetylation, 0.5 mg/mL of 10-DAB was added together with IPTG during the IPTG induction period, and 0.5 mL of each culture at different intervals was collected for analysis.
Alternatively, the IPTG-induced strains were collected at 18 h and concentrated to the cell density (OD600) of 40 in 20 mL fresh TB medium, containing amounts of proper antibiotics, 1 mmol/L IPTG, 10-DAB or 50 mmol/L acetic acid were added to the medium, and the cultures were grown at 30 °C and 220 rpm for 48 h to investigate the baccatin III production. Meanwhile, DBAT expression and the product structure were also determined by SDS-PAGE or MS and NMR spectrometry.
To optimize the fermentation process of R10 strain, the cell density (OD600) was set up at 20, 40 and 60, respectively. The concentrations of 10-DAB were set up at 1, 1.5 and 2 mg/mL, respectively. The influence of 50 mmol/L acetic acid was also investigated by measuring the changes of 10-DAB and baccatin III titers and pH profiles under the conditions of adding or non-adding acetic acid, the R10 cells density (OD600) of 40 and 1.5 g/mL 10-DAB during the IPTG induction periods.
2.6. Analytical methods
To rapidly quench the cell metabolism and detect acetyl-CoA, a simplified method was performed18: 20 mL aliquots of the cultures were harvested by centrifugation at 8000 rpm (6010×g) and 4 °C for 2 min and resuspended with 2 mL–80 °C methanol/water (80:20, v/v). The samples were cooled to −20 °C for at least 20 min and then centrifuged at 14,000 rpm (14,462×g) and 4 °C for 10 min, and the resulting supernatant was used for analysis. Alternatively, the fermentation broth was mixed with one volume of methanol and then centrifuged at 14,000 rpm (14,462×g) and 4 °C for 10 min, and the resulting supernatant was used for analysis of the baccatin III titer, the remaining 10-DAB and glucose.
Acetyl-CoA production was analyzed by HPLC on a Shimadzu system (Shimadzu Corporation, Kyoto, Japan) equipped with LC-20AT pumps, SPD-M20A detector and an OSAKA SODA CAPCELL PAK C18 MGII column (5 μm, 4.6 mm × 150 mm) monitoring at 254 nm and 30 °C. Mobile phases used were solvent A (Na3PO4 buffer solution, pH 5.5) and solvent B (Na3PO4 buffer solution (pH 5.5):acetonitrile = 4:1, v/v) with a flow rate of 1 mL/min. The following gradient was used: 0–5 min, 3%–18% B; 5–7.5 min, 18%–28% B; 7.5–12.5 min, 28%–40% B; 12.5–18 min, 40%–42% B; 18–19 min, 42%–97% B.
The HPLC system was also used to analyze the baccatin III titer, the remaining 10-DAB and glucose in the fermentation broth, and the baccatin III deacetylation. The baccatin III titer and the remaining 10-DAB were detected at 230 nm with the solvent A (water) and solvent B (acetonitrile) as the mobile phases. The flow rate was 1 mL/min by using the following gradient: 0–12 min, 30%–62% B; 12–13 min, 62%–90% B. Identities of the compounds were firstly verified by LC‒MS. Similarly, the deacetylation reactions of baccatin III were analyzed at 230 nm with the solvent A (water) and solvent B (acetonitrile) as the mobile phases. The flow rate was 0.4 mL/min by using the following gradient: 0–40 min, 5%–95% B.
The remaining glucose in the fermentation broth was analyzed on Refractive Index detector (Shimadzu Corporation) and OSAKA SODA CAPCELL PAK NH2 UG80 column (5 μm, 4.6 mm × 250 mm) with the mobile phase of 80% acetonitrile and a flow rate of 1 mL/min.
ESI-MS data were acquired using an LCMS-2020 system (Shimadzu Corporation). The structure of baccatin III was further elucidated by NMR spectrometry which was summarized in Supporting Information Table S2. Both 1H NMR (500 MHz) and 13C NMR (125 MHz) spectra of baccatin III in CDCl3 were measured on the Bruker spectrometer (Varian Mercury, USA). Bioconversion rate was calculated as Eq. (1):
| Bioconversion rate (%) of 10-DAB = (10-DAB HPLC peak area before reaction‒10-DAB HPLC peak area after reaction)/10-DAB HPLC peak area before reaction × 100 | (1) |
where 10-DAB concentration and baccatin III concentration (mg/mL; g/L) were calculated based on the HPLC peak areas using the linear regression equation.
2.7. DBAT expression analysis
During the IPTG induction period, 20 mL of the culture broth was harvested after IPTG induction for 8–48 h, respectively. The samples were pelleted by centrifugation at 8000 rpm (6010×g) and 4 °C, for 2 min. The pellets were resuspended in 50 mmol/L Tris-HCl buffer (pH 7.5) with the cell density (OD600) of 20. The resuspended samples were disrupted by sonication on ice for 2 min to prepare the cell homogenate (5 s per pulse, 6 pulses per min) with a sonotrode of 2 mm in diameter and 39% of power (Sonic-vcx-130, Sonics, USA).
The cell homogenate was centrifuged at 12,000 rpm (10,625×g) and 4 °C, for 40 min, and the supernatant was subjected to SDS-PAGE analysis. The DBAT bands were visualized by Coomassie Brilliant Blue staining. The protein expression levels were semiquantitatively measured based on grayscale scanning by the Quantity One software.
DBAT expression analysis of R10 in the high-cell-density fermentation was nearly the same as the aforementioned method except that the samples were collected from the IPTG induction and baccatin III production stages (from 19 to 54 h of cultivation).
2.8. Observation on the baccatin III stability in different pH environment and the DBAT deacetylase activity
To analyze the stability of baccatin III in different pH environment, 1 mg/mL baccatin III was added to the 100 mmol/L Tris-HCl buffer solution (pH 6.0 or 9.0) and incubated at 30 °C for 48 h. The deacetylase activity of DBAT was analyzed under the alkaline condition with the R10 cells or the purified DBAT enzyme. For the R10 cell reaction, the M0 (control) and R10 cells were respectively cultivated and harvested with the same procedure as mentioned in Shake-flask fermentation section. The cell pellets were washed twice with ddH2O by centrifugation. Then, the cell pellets were resuspended in 100 mmol/L Tris-HCl buffer (pH 9.0) containing 1 mg/mL baccatin III. The final cell density (OD600) was adjusted to 40 (mimicking the shake-flask culture condition) and the reaction system was incubated at 30 °C for 48 h.
For the purified DBAT enzyme reaction, the R0 cells (harboring dbat) were used to prepare the recombinant enzyme16. In addition, the M0 cell homogenate was prepared by using the aforementioned method, which was used as a supplement. Before the enzymatic reaction test, this M0 cell homogenate was checked for its impact on the stability of baccatin III under the alkaline condition (Fig. S3). The enzyme reaction system (200 μL) was prepared with 100 mmol/L Tris-HCl buffer (pH 9.0) which contained 5 mg/mL DBAT and 1 mg/mL baccatin III, together with or without 100 μL M0 cell homogenate (prepared with 100 mmol/L Tris-HCl buffer, pH 9.0). The samples were incubated at 37 °C for 0.5, 1, 2 and 4 h, respectively.
Samples were taken during the incubation period and aliquots of the sample were mixed with one volume of methanol and centrifuged at 14,000 rpm (14,462×g) and 4 °C for 10 min, and the resulting supernatant was used for HPLC analysis.
2.9. Exploring the unknown factors from M0 cells
2.9.1. Identification of the unknown factors
To identify the unknown factors from M0 cells (also present in R10 and other bacterial cells) that improve the deacetylation efficiency of DBAT under alkaline condition, we propagated M0 cells by the high-cell-density fermentation approach as mentioned in Section 2.10. The cell pellets were obtained by centrifugation at 8000 rpm (6010×g), 4 °C for 10 min and washed with ddH2O for 3 times. After lyophilization, the cells were suspended in ddH2O and crushed twice under ultra-high pressure (800 bar) to obtain the cell homogenate. This homogenate was extracted three times with ethyl acetate (EtOAc). The EtOAc layer and the water layer were dried by reduced pressure condensation and lyophilization, respectively, to prepare the EtOAc extract and the water extract.
Effects of the two extracts on DBAT deacetylation efficiency were respectively detected using the similar method as described in Section 2.8, with M0 cell homogenate as the control. Briefly, the EtOAc extract, water extract, and M0 cell homogenate (each 0.2 g) were respectively resuspended in 10 mL, 100 mmol/L Tris-HCl buffer (pH 9.0) to prepare the stock solutions. The enzyme reaction system (200 μL) was prepared with 100 mmol/L Tris-HCl buffer (pH 9.0) which contained 1 mg/mL baccatin III, 1 mg/mL DBAT, and 177 μL of the stock solution. The samples were incubated at 37 °C for 1 h and then analyzed with the same method as described in Section 2.8.
For CoASH detection, each sample (0.2 g) was resuspended in 20 mL of 50% methanol solution (v/v) and centrifuged at 14,000 rpm (14,462×g) and 4 °C for 10 min. The resulting supernatant was used for LC‒MS analysis using the similar method as described in Section 2.6.
Additionally, since the TB medium was used for the bacterial shake-flask fermentation, which contained 2.31 g/L KH2PO4 and 16.25 g/L K2HPO4·3H2O, the K+ was another candidate of the unknown factors that was enriched by the cells.
2.9.2. Influence of CoASH and K+ on the deacetyse activity of DBAT against baccatin III under alkaline condition
The reaction system (contained 1 mg/mL baccatin III and 1 mg/mL DBAT) was the same as the aforementioned method and was divided into 4 groups: (1) control; (2) adding 1 mg/mL CoASH; (3) adding 2.31 mg/mL KH2PO4 and 16.25 mg/mL K2HPO4·3H2O; (4) adding 1 mg/mL CoASH plus 2.31 mg/mL KH2PO4 and 16.25 mg/mL K2HPO4·3H2O.
2.10. Fed-batch high-cell-density fermentation
The R10 strain was used for fermentation in a 3-L bioreactor (Shanghai Baoxing Bio-Engineering Equipment, China) equipped with pH, temperature, and dissolved oxygen (DO) monitors. The single clone was firstly grown in 10 mL LB medium supplemented with 100 μg/mL of ampicillin, 50 μg/mL of kanamycin and 40 μg/mL of streptomycin, in a shake-flask at 37 °C and 220 rpm for 7 h, and then 400 μL of suspensions were transferred into 40 mL of LB medium containing the same concentrations of the aforementioned antibiotics in a 100-mL shake-flask and grown at 37 °C and 220 rpm for 7 h as seed cultures. The 40 mL of seed cultures were then transferred into 1 L of the complex medium with antibiotics (100 μg/mL of ampicillin, 50 μg/mL of kanamycin and 40 μg/mL of streptomycin in final concentrations) and 1 mL of the trace element solution.
The fermentation was divided into three stages: pre-induction stage, IPTG induction stage and baccatin III production stage. The pre-induction stage lasted from 0 to 7 h, and the temperature was set at 37 °C with an air flow rate of 3 L/min. The initial pH was 6.8–6.9, which was adjusted by adding proper amount of 5 mol/L ammonium hydroxide. When the initial glucose was completely depleted as indicated by a sudden increase of dissolved oxygen (DO 39%–51%) and pH 7.7–7.8, a simple exponential feed rate was performed19. The IPTG induction stage began at 7 h and ended at 31 h. IPTG was added to the medium with a final concentration of 1 mmol/L to induce the enzyme expression, and the temperature was lowered to 20 °C. During this stage the glucose feeding solution was constantly fed at a rate of 10 mL/h. The baccatin III production stage lasted from 31 to 55 h. Acetic acid and 10-DAB were added and the temperature was increased to 37 °C. The glucose feeding solution was constantly fed at a rate of 5 mL/h. Additionally, the pH was set up and maintained at 6.0 (30 mmol/L acetic acid), 5.8 (35 mmol/L acetic acid) and 5.5 (50 mmol/L acetic acid), respectively, by feeding acetic acid (4 mol/L stock solution) during the whole production stage. During the whole fermentation process, the DO was maintained at approximately 30% with an agitation cascade (100–900 rpm).
2.11. Purification of baccatin III
The crude baccatin III was isolated from the fermentation broth by ethyl acetate extraction, then purified by column chromatography on silica gel (HG/T2354-2010; Qingdao Haiyang Chemical Co., Ltd., China) eluted with a mobile phase containing petroleum ether-dichloromethane-methanol in the ratio of 2.5:1.5:0.25 (v/v/v) to achieve baccatin III.
3. Results
3.1. Refactoring acetyl-CoA biosynthetic pathway in E. coli
To improve the intracellular acetyl-CoA content in E. coli, we screened some enzymes involved in the biosynthetic pathway (Fig. 1A) for modular engineering (Fig. 1B), so as to maximize the pathway flux from glucose to acetyl-CoA and from pantothenate as well as acetate to acetyl-CoA. A total of 12 module combinations were constructed and respectively transformed into E. coli host. First of all, we introduced pdh (aceE, aceF and lpdA complex) into E. coli host (M0 strain) to construct the M1 strain (Fig. 1B). It was found that the acetyl-CoA concentration in M1 was 1.8-fold higher than that in M0 (Fig. 1C). Based on the M1 recombinant gene cluster, we added pgk alone or pgk combined with gapd (both were involved in the glycolysis metabolic pathway) to generate M2 and M3, respectively. Unexpectedly, the acetyl-CoA concentrations in M2 and M3 were even lower than that in M1. Later, M4 strain was constructed, where the genes pgk and gapd were cloned in a low-copy-number vector pACYC184 (Fig. 1B). The results exhibited that the acetyl-CoA productivity of M4 was improved compared with that of M3. Afterwards, we generated M5 by replacing the pACYC184 plasmid with a high-copy-number vector pRSF that carried the gene Se/acsL641P. Compared with M3, M5 also showed a higher intracellular acetyl-CoA concentration, demonstrating that the heterologous Se/acsL641P of Salmonella enteria enhanced acetyl-CoA production in the recombinant strain (Fig. 1C). Similarly, the gene pank also positively affected acetyl-CoA production, since the acetyl-CoA concentrations in both M6 and M8 surpassed those of their counterparts (M3 and M1, respectively, Fig. 1C). Thereafter, we further combined Se/acsL641P with M6 and M8 modules to construct M7 and M10 strains, respectively. As a result, both M7 and M10 were superior to their counterparts in terms of acetyl-CoA production, with M10 being more robust in the biosynthesis of acetyl-CoA. Actually, the acetyl-CoA production of the chassis M10 was three times higher than that of the host cell (Fig. 1C). However, when pgk and gapd (carried by pACYC184) were simultaneously introduced into M8 and M10, respectively, or when gapd alone [carried by pEASY-Blunt-E1 (pBE1) and driven by T7 promoter] was introduced into M10, the acetyl-CoA production of all strains sharply declined (M9 vs. M8, M11 and M12 vs. M10) (Fig. 1C). Thus, the top three chassis cells were defined as M10, M7 and M8, respectively, as judged based on the acetyl-CoA levels.
3.2. Introduction of gene dbat into the selected chassis cells
The gene dbat of T. cuspidata was introduced into M0, M7, M8 and M10, respectively, yielding the corresponding strains of R0 (control), R7, R8 and R10. Considering that the additional PDH, PGK, GAPD, PANK, and Se/acsL641P might affect cell growth, cell densities (OD600) of the four strains were monitored at 8, 16, 24 and 48 h, respectively. The OD600 levels of R0, R7, R8 and R10 reached 15.4, 7.9, 2.4, and 1.0, at 8 h, respectively. Afterwards, the R0 strain rapidly reached its highest OD600 level of 25 at 16 h and then kept its high level till the end of observation. In contrast, the R8 and R10 strains grew much slower and their OD600 levels only reached 11.8 and 11.1 at 48 h (Fig. 2A), suggesting that the expression of these additional genes surely, at least on the cell growth aspect, increased the metabolic burden of the recombinant cells (Fig. 2A). It seemed that the R7 strain exhibited higher tolerance to the introduced gene burden and grew much faster than R8 and R10 within 24 h, but its cell density was quite close to those of R8 and R10 at 48 h (Fig. 2A). We also examined the influence of additional dbat expression on the intracellular acetyl-CoA level after IPTG induction (Fig. 2B). The acetyl-CoA yields of R0, R7, R8 and R10 reached 0.9, 2.5, 2.4 and 2.3 mg/g DCW (dry cell weight) at 8 h, respectively. Then the acetyl-CoA yields steadily declined but all the recombinant strains kept their apparent advantage against the control on the acetyl-CoA productivity, in which R10 exhibited the highest intracellular acetyl-CoA level followed by R8 and R7, suggesting that the dbat gene expression did not disturb the acetyl-CoA production in the recombinant strains (Fig. 2B). Typically, the adverse relationship between the acetyl-CoA production and the cell growth was observed in R8 and R10 strains. This phenomenon may be attributed to the detrimental effect of the accumulated acetyl-CoA on cell growth20. Furthermore, the bioconversion efficiency from 10-DAB (final concentration: 0.5 mg/mL) to baccatin III was examined (Fig. 2C). Again, R10 showed the highest efficiency in baccatin III production after culturing for 16–48 h, followed by R8 and R7. The highest baccatin III yields were observed among the three recombinants at 24 h, of which, the baccatin III concentration in R10 reached 101.6 μg/g DCW (Fig. 2C). Meanwhile, we detected the DBAT expression level in R10 during the cultivation period (Fig. 2D). As a result, the enzyme was continually expressed, with the highest level being detected at 18 h. Further, the R10 product was subjected to LC‒MS analysis and the [M+H]+ peak of R10 product was 587.3 (Fig. S1), which was consistent with that of standard baccatin III and those reported in previous literature16. Later, the product structure was also elucidated to be baccatin III by NMR spectroscopy (Fig. S2A and B).
Figure 2.
Recombinant screening for bioconversion of 10-DAB to baccatin III. (A) Cell densities of R0, R7, R8 and R10 at different cultivation time. (B) Acetyl-CoA yields of R0, R7, R8 and R10 at different cultivation time. (C) Baccatin III yields of R0, R7, R8 and R10 at different cultivation time. (D) DBAT enzyme level of R10 at different cultivation time (calculated according to the gray scale scanning results). The data represent means ± SD, n = 3 (biological replicates).
3.3. Process optimization and acetic acid supplementation to improve the baccatin III production
We further optimized the substrate (10-DAB) concentration and initial R10 cell density during the cultivation period. Under the shake-flask culture conditions, the 10-DAB concentration of 1.5 mg/mL combined with the initial cell density (OD600) of 40 resulted in the highest baccatin III titer (1.14 mg/mL) after 24 h of cultivation (Fig. 3A). Based on this optimized condition, we further observed the time–course profiles of baccatin III and 10-DAB concentrations and pH changes. As shown in Fig. 3B, the baccatin III titer increased rapidly during the first 10 h of cultivation and peaked at 18 h to 1.3 mg/mL. Later, the titer dropped steadily and was finally 0.73 mg/mL at 48 h (Fig. 3B). With the increase in baccatin III yield, the substrate 10-DAB was consumed from the initial level of 1.5 mg/mL to the lowest concentration of 0.32 mg/mL at 18 h (Fig. 3B). Surprisingly, the amount of 10-DAB showed wave-like uplift after 18 h, reaching the final concentration of 0.91 mg/mL at the end of cultivation (Fig. 3B). Meanwhile, the pH value gradually increased from the initial 7.0 to the final 8.8 at 48 h (Fig. 3B). It means that during the late period a part of baccatin III was converted into 10-DAB along with the pH increase. As exhibited in Scheme 1, acetate could be used as a substrate for acetyl-CoA biosynthesis, and the influence of supplementary acetic acid (50 mmol/L) on the acetyl-CoA production in R10 was analyzed. It was illustrated from Fig. 3C that, acetic acid significantly increased the acetyl-CoA titer at any time point of 8, 16 and 24 h.
Figure 3.
Process optimization and influence of adding acetic acid (50 mmol/L) on the production of acetyl-CoA and baccatin III of R10. (A) Optimization of 10-DAB concentration and initial R10 cell density in shake-flask culture. The initial cell densities (OD600) of 20, 40 and 60 were combined with 10-DAB concentrations of 1.0, 1.5, and 2.0 g/mL, respectively. (B) Time-course profiles of baccatin III and 10-DAB concentrations and pH changes under the optimized condition. (C) Influence of adding acetic acid (HAc) on acetyl-CoA production. (D) Time‒course profiles of baccatin III and 10-DAB concentrations with addition of acetic acid. The data represent the means ± SD, n = 3 (biological replicates).
In addition, the impact of supplementary acetic acid (50 mmol/L) on the baccatin III production in R10 was also analyzed by setting the substrate (10-DAB) concentration to 1.5 mg/mL and the initial R10 cell density (OD600) to 40. The supplementation of acetic acid (Fig. 3D) resulted in a relatively acidic environment throughout the cultivation process (pH 5.7–7.8), and the baccatin III titer reached 1.54 mg/mL at 18 h, which was apparently higher than that in the absence of acetic acid addition (Fig. 3B). Moreover, the high baccatin III titer was maintained for about 10 h (maximum, 1.57 mg/mL) before it declined. In the meantime, the 10-DAB concentration was maintained at an extremely low level after 18 h of cultivation (Fig. 3D). It was speculated that baccatin III was unstable under the alkaline environment and easily converted into 10-DAB through deacetylation. Besides being a substrate, feeding acetic acid also maintained the pH environment at a relatively acidic condition, which thus inhibited the deacetylation of baccatin III.
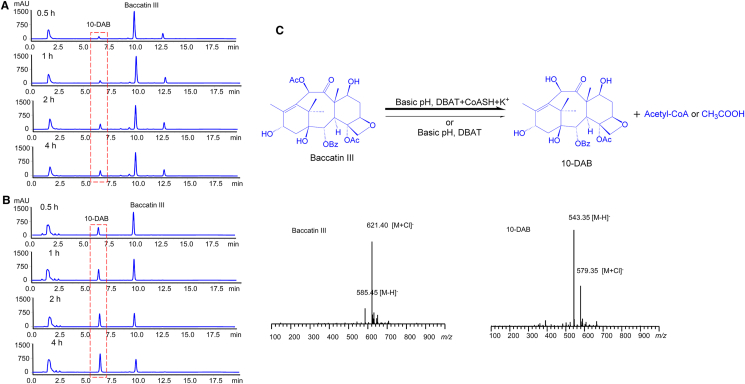
To test our hypothesis, we designed a group of experiments to detect the stability of baccatin III. As shown in Fig. 4, baccatin III was much stable in the pH 6.0 buffer solution during the 48-h observation period (Fig. 4A and Fig. S4C), but its concentration was slowly decreased in the pH 9.0 buffer solution and moreover, 10-DAB was produced simultaneously and slightly accumulated with the extension of incubation time (Fig. 4B and Fig. S4D), indicating that baccatin III was spontaneously deacetylated under alkaline condition. Additionally, the C7 epimerization21 of baccatin III and 10-DAB was also observed under the alkaline condition (Fig. S4D). When the R10 cells (harboring gene dbat) were added into the pH 9.0 solution, the conversion rate of baccatin III to 10-DAB significantly increased (Fig. 4D and Fig. S4F) and the 10-DAB concentration was two-fold higher than that of baccatin III at 12 h. As a control, the M0 cells (not harboring gene dbat) did not exhibit such an effect under the same alkaline condition (Fig. 4C and Fig. S4E). According to such results, the sharp increase in 10-DAB concentration and the simultaneous decrease in baccatin III production were mainly driven by DBAT under alkaline condition.
Figure 4.
Stability of baccatin III under different conditions. (A) Baccatin III in 100 mmol/L Tris-HCl buffer, pH 6.0. (B) Baccatin III in 100 mmol/L Tris-HCl buffer, pH 9.0. (C) Baccatin III in 100 mmol/L Tris-HCl buffer plus M0 cells (as a control of D) (OD600 = 40), pH 9.0. (D) Baccatin III in 100 mmol/L Tris-HCl buffer plus R10 cells (OD600 = 40), pH 9.0.
The DBAT deacetylase activity was further demonstrated by using the purified enzyme assisted by the M0 cell (not harboring dbat) homogenate. When DBAT was added to the alkaline solution, 10-DAB could be detected as early as 0.5 h incubation (Fig. 5A). This conversion from baccatin III to 10-DAB could be significantly enhanced and the concentration of 10-DAB even surpassed that of baccatin III at 4 h when the M0 cell (not harboring dbat) homogenate was added to the aforementioned reaction system, suggesting that some unknown factors are required for the efficient enzymatic deacetylation under alkaline condition (Fig. 5B).
Figure 5.
Deacetylase activity of purified DBAT on baccatin III within 4 h. (A) Baccatin III in 100 mmol/L Tris-HCl buffer with DBAT, pH 9.0. (B) Baccatin III in 100 mmol/L Tris-HCl buffer with DBAT and M0 cell homogenate, pH 9.0. (C) Proposed deacetylation reaction and MS spectra of baccatin III and 10-DAB.
To explore the unknown factors, we propagated the M0 cells by high-cell-density fermentation and extracted the M0 cell homogenate with ethyl acetate and water (Fig. S5A). Both the organic fraction and aqueous fraction were subjected to the enzymatic reaction under alkaline condition, with the M0 cell homogenate as the positive control. We found that only the aqueous fraction could apparently promote the enzymatic deacetylation within 1 h, and the 10-DAB titer was even higher than that of the positive control (Fig. S5B). Coincidently, we identified that CoASH was enriched in the aqueous fraction Fig. S5C and D). Then, we demonstrated that DBAT could efficiently transfer the acetyl group of baccatin III to CoASH and resulted in the formation of 10-DAB and acetyl-CoA, since the 10-DAB titer was 20 times higher than that of the control (Fig. S6). As the TB medium was used for the shake-flask culture, which contained K+ salts, we also checked the influence of K+ on the enzymatic deacetylation. The results indicated that this ion could also improve the deacetylation and resulted in the 6-fold increase on the 10-DAB titer (Fig. S6). The additive effect of CoASH and K+ was also observed, which led to the 50-fold increase on the 10-DAB titer (Fig. S6).
Taken together, the addition of a proper amount of acetic acid into the culture medium not only improved the acetyl-CoA biosynthesis and subsequently increased the baccatin III production in R10, but also inhibited or alleviated the enzymatic deacetylation and epimerization of baccatin III.
3.4. Production of baccatin III by high-cell-density fermentation
We applied the fed-batch high-cell-density fermentation approach to the engineered R10 strain in a 3-L bioreactor with the complex medium (initial pH value of 6.8). The results are summarized in Fig. 6 and more fermentation parameters are displayed in Fig. S7. Briefly, the whole process was divided into three stages, namely, pre-induction stage, IPTG induction stage and baccatin III production stage (Fig. 6). Firstly, we set 10-DAB concentration at 2.0 g/L and acetic acid concentrations at 30 mmol/L (initial pH 6.0), 50 mmol/L (initial pH 5.5), and 65 mmol/L (initial pH 5.0), respectively, at the beginning of the production stage (Fig. 6A‒C). As shown in Fig. 6A‒D, the profiles of acetyl-CoA production, cell density and DBAT expression in these three groups were quite similar during the whole cultivation period, with the only exception that the cell density of 65 mmol/L (pH 5.0) group slightly declined during the production stage. The acetyl-CoA concentrations peaked at the end of IPTG induction and then sharply declined because acetyl-CoA was consumed to form baccatin III. The baccatin III titers increased sharply with the decrease in 10-DAB concentrations until the inflection points at 6 h of production stage. Then, baccatin III production reached a plateau and, at the end of the production stage the baccatin III titers arrived at 1.2 g/L (the 30 mmol/L acetic acid group), 1.6 g/L (the 50 mmol/L acetic acid group) and 0.4 g/L (the 65 mmol/L acetic acid group), respectively (Fig. 6A‒C and E). It seemed that the excessively low pH value (the 65 mmol/L acetic acid group) influenced glucose utilization (Fig. S7) and thereby affected the baccatin III production (Fig. 6C and E). Secondly, we set the initial acetic acid concentrations at 30 mmol/L, 35 mmol/L and 50 mmol/L at the beginning of the production stage, and maintained the pH values at 6.0 (30 mmol/L), 5.8 (35 mmol/L) and 5.5 (50 mmol/L), respectively, by the addition of acetic acid during the whole production stage. Typically, the highest yield of 2.0 g/L baccatin III was achieved in the pH 5.8 group, followed by 1.5 g/L in the pH 6.0 group, and 1 g/L in the pH 5.5 group (Fig. 6F). Then, we defined the pH at 5.8 throughout the whole production stage and set the 10-DAB concentrations at 2.0, 3.0 and 6.0 g/L, respectively. As a result, the baccatin III titers reached 2 g/L (conversion rate, 98%), 3 g/L (conversion rate, 97%) and 4.6 g/L (conversion rate, 78%), respectively, when 2, 3 and 6 g/L 10-DAB were added to the fermenter (Fig. 6G).
Figure 6.
Optimization of baccatin III production by fed-batch fermentation. The fermentation was carried out in a 3-L bioreactor with 2.0 g/L 10-DAB and different concentrations of acetic acid supplemented at the beginning of production stage (a–f). (A) 30 mmol/L acetic acid (initial pH 6.0). (B) 50 mmol/L acetic acid (initial pH 5.5). (C) 65 mmol/L acetic acid (initial pH 5.0). (D) DBAT expression under different initial acetic acid concentrations. (E) Baccatin III titers and 10-DAB conversion rates under different initial acetic acid (HAc) concentrations at 55 h of cultivation. (F) Baccatin III titers and 10-DAB conversion rates under different maintaining pH at 55 h of cultivation. (G) Baccatin III titers and 10-DAB conversion rates under different 10-DAB concentrations and the same maintaining pH (5.8) at 55 h of cultivation. The data represent the means ± SD, n = 3.
In addition, a preliminary purification was carried out by using the fermentation broth of the 3 g/L 10-DAB group as the material. A total of 7.5 g extract was obtained by ethyl acetate extraction. From this extract, 2.59 g purified baccatin III (HPLC purity > 98%) was acquired by column chromatography on the silica gel, with the recovery rate of 86% (the theoretical amount of baccatin III was 3 g, detected by HPLC).
4. Discussion
The DBAT-catalyzed C10-hydroxyl acetylation of 10-DAB during the chemical semi-synthesis of Taxol not only shortens the reaction route, but also decreases the use of hazardous reagents and partially omits the harsh reaction conditions. However, the enzymatic reaction is highly dependent on acetyl-CoA, a specific acetyl donor and a non-bulk and expensive commodity. Therefore, it is impractical to use the commercial acetyl-CoA for the large-scale acetylation of 10-DAB. This bottleneck can be eliminated by the metabolic engineering or synthetic biological approach. The present study aimed to boost the innate biosynthetic pathway of bacterial acetyl-CoA by the modular engineering strategy.
Refactoring metabolic pathway by introducing heterologous enzymes may be associated with a potential risk to increase metabolic burden to host cells due to the redistribution of metabolic flux, accumulation of cellular toxic intermediates or undesirable physiological changes22,23. There is also a trade-off relationship between efficient cell growth and target chemical productivity, economic viability and theoretical feasibility20. Hence, this study only focused on the pyruvate catabolic pathway and coenzyme A (CoASH) anabolic pathway (Fig. 1A) while avoiding excessive interference with the engineered bacterium. Firstly, we introduced the innate PDH (aceE, aceF and lpdA complex), GAPD and PGK enzymes to boost the glycolysis pathway by the modular engineering approach24,25. We found that only over-expression of the additional PDH apparently elevated the cytosolic acetyl-CoA concentration. In fact, over-expression of the additional GAPD and/or PGK had negative effects on acetyl-CoA production (Fig. 1B and C), suggesting that the pyruvate pool controlled by innate GAPD and PGK is enough to guarantee the downstream PDH-catalyzed biosynthesis of acetyl-CoA. In addition, redundant carbon flux flowing into this pathway for pyruvate synthesis might increase the cell metabolic burden and adversely affect acetyl-CoA formation. Additionally, the cell metabolic burden also impacted the cell growth (Fig. 2A). This phenomenon was partially consistent with the previous report where PDH exhibited more additive effects on malonyl-CoA and acetyl-CoA production26, but was contrary to the synergistic effect on malonyl-CoA (downstream acetyl-CoA product) production among PGK, GAPD and PDH24. The possible reason might be that the metabolic network of the latter was changed since the downstream genes (ΔfumC and ΔsucC) involved in acetyl-CoA catabolism were knocked out.
The CoASH anabolic pathway begins with pantothenate, which includes PANK and ACS enzymes (Fig. 1A), and the acetyl group of acetyl-CoA is derived from acetate (Scheme 1 and Fig. 1A). Typically, PANK is the rate-limiting step in CoASH biosynthesis27. Multiple copies of pank gene have been found to up-regulate the CoASH level by 2.7-fold28. ACS is responsible for activating acetate and ligating the acetyl group to CoASH. The L641P mutation of ACS (Se/acsL641P) from Salmonella enterica was reported to maintain the active state of the acetyl CoA synthetase and elevate the production of acetyl CoA as well as the corresponding downstream products26,29. As observed in this study, both the additional PANK and the heterologous Se/acsL641P remarkably promoted the production of cytosolic acetyl-CoA. The combination of PDH, PANK and Se/ascL641P (M10 chassis cell) achieved the highest cytosolic concentration of acetyl-CoA with the productivity over three times higher than that of the host cell (Fig. 1B and C). Meanwhile, the high acetyl-CoA production level was maintained by the corresponding R10 (harboring the additional dbat) throughout the entire culture process (Fig. 2B), which was beneficial to the 10-DAB acetylation/baccatin III production (Fig. 2C). In addition, the continuous DBAT expression in R10 (Fig. 2D) guaranteed the bioconversion process from 10-DAB to baccatin III. Therefore, the baccatin III titer reached 1.14 mg/mL after 24 h of cultivation at the 10-DAB concentration of 1.5 mg/mL and the initial cell density (OD600) of 40 (Fig. 3A).
In the process of recombinant screening, we observed that the baccatin III yield declined sharply at the late cultivation stage (Fig. 2C). Later, it was found that such sharp decrease was coupled with the simultaneous increase in 10-DAB concentration, and the whole process was related to the increase in environmental pH from the initial 7.0 to 8.8 at 48 h (Fig. 3B), suggesting that baccatin III was deacetylated to form 10-DAB under alkaline condition. As a substrate for the biosynthesis of acetyl-CoA, additional acetic acid greatly elevated the acetyl-CoA titer (Fig. 3C), which was in accordance with the other report22 and thereby guaranteed the supply of acetyl donor for the acetylation of 10-DAB. Further, the addition of acetic acid also stabilized the reaction environment at a relatively acidic condition, which overruled the deacetylation and epimerization of baccatin III and thus benefited the baccatin III production (Fig. 3D). In fact, such a dual-effect markedly elevated the baccatin III titer to the maximum 1.57 mg/mL under the shake-flask culture condition and substantially suppressed its deacetylation (Fig. 3D).
Afterwards we found that the deacetylation of baccatin III was mainly catalyzed by DBAT under alkaline condition assisted by some unknown factors (Fig. S4 and Fig. 5). These unknown factors were then demonstrated at least to be CoASH and K+, of which CoASH should be the primary factor that acts as an acetyl group acceptor in the deacetylation of baccatin III, while K+ could further improve the deacetylation. The dual function of DBAT referred to both acetylation of 10-DAB and deacetylation of baccatin III is much like that of the C-10 deacetylase from the soil microorganism Nocardioides lureus. The microbial enzyme could not only hydrolyze the C10 acetyl group of baccatin III to form 10-DAB at the optimum pH of 8, but also catalyze the acetylation of 10-DAB to baccatin III by using vinyl acetate as the acetyl donor at the optimum pH of 7 with the yield of 0.66 g/L (to our knowledge, this is also the highest yield among all the enzymatic acetylation reports to date)10,30. Whether the two enzymes share the same deacetylation mechanism needs to be further investigated.
To overcome the drawback of slow growth and further increase the productivity of baccatin III, the high-cell-density fermentation of R10 was performed in a 3-L bioreactor based on the optimized shake-flask culture conditions (Fig. 3D). We divided the whole process into three stages. The pre-induction stage aimed to increase the cell density (OD600) to 20. While the IPTG induction stage aimed to induce the expression of other genes, including those involved in the acetyl-CoA biosynthetic pathway and DBAT expression. At the production stage, 10-DAB and acetic acid were simultaneously added into the culture medium. Under the optimized conditions, when the 10-DAB concentrations were set at 2, 3 and 6 g/L, the baccatin III titers reached 2 g/L (conversion rate, 98%), 3 g/L (conversion rate, 97%) and 4.6 g/L (conversion rate, 78%), respectively, which has great practical implications for further industrial application in the semi-synthesis of Taxol (Fig. 6G). To the best of our knowledge, these are the highest baccatin III yields among all literature reports. Particularly, our results provide more choices for the industrial purpose, for example, to pursue the highest conversion rate or the highest yield (titer). In the future, the production cost may be further reduced by replacing the expensive IPTG with a cheaper inducer, such as D-galactose. Typically, it should be a good alternative to construct the heterologous chromosomal integrative and constitutive expression system.
5. Conclusions
In this study, we refactored the innate biosynthetic pathway of acetyl-CoA in E. coli. Then, we constructed a microbial cell factory by introducing DBAT gene into this chassis for bioconversion of 10-DAB to baccatin III with the native acetyl-CoA as the acetyl donor. We found that DBAT could efficiently deacetylate baccatin III assisted by the CoASH and K+ in alkaline environment. Feeding acetic acid not only serves as a substrate but greatly alleviates the deacetylation and epimerization of baccatin III. The engineered strain was performed for high-cell-density fermentation and baccatin III titers reached 2, 3 and 4.6 g/L, respectively, when 2, 3 and 6 g/L of 10-DAB was supplied. Our study provides an environmentally friendly approach for the conversion of 10-DAB to baccatin III in the industrial chemical semi-synthesis of Taxol. The finding of DBAT deacetylase activity may broaden its application in the structural modification of pharmaceutically important lead compounds.
Acknowledgments
We thank Yingjin Yuan's lab (Tianjin University, China) for synthesizing the gene encoding the codon-optimized acetyl-CoA synthetase variant (GenBanK Accession: AAO71645.1). This work was supported by the National Key Research and Development Program of China (grant Nos. 2018YFA0901900 and 2020YFA0908003), the Drug Innovation Major Project (grant No. 2018ZX09711001-006-001, China), the National Natural Science Foundation of China (grant No. 81573325), the CAMS Innovation Fund for Medical Sciences (CIFMS; (grant No. 2017-I2M-2-004; 2019-I2M-1-005, China), and PUMC Disciplinary Development of Synthetic Biology (201920100801, China).
Author contributions
Hao Wang, Bo-Yong Zhang designed and performed the experiments and drafted the manuscript. Ting Ging, Tian-Jiao Chen, Jing-Jing Chen and Jin-Ling Yang discussed the experimental results, commented on and proofread the manuscript. Ping Zhu conceived, designed and supervised the study and revised the manuscript. All authors approved the final manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.03.029.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sofias A.M., Dunne M., Storm G., Allen C. The battle of "nano" paclitaxel. Adv Drug Deliv Rev. 2017;122:20–30. doi: 10.1016/j.addr.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Chen J.J., Liang X., Wang F., Wen Y.H., Chen T.J., Liu W.C. Combinatorial mutation on the beta-glycosidase specific to 7-β-xylosyltaxanes and increasing the mutated enzyme production by engineering the recombinant yeast. Acta Pharm Sin B. 2019;9:626–638. doi: 10.1016/j.apsb.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J., Wang Y., Zhou S., Li J., Wang J., Chi D. Remote loading paclitaxel-doxorubicin prodrug into liposomes for cancer combination therapy. Acta Pharm Sin B. 2020;10:1730–1740. doi: 10.1016/j.apsb.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W.C., Gong T., Zhu P. Advances in exploring alternative Taxol sources. RSC Adv. 2016;6:48800–48809. [Google Scholar]
- 5.Denis J.N., Greene A.E., Guenard D., Guerittevoegelein F., Mangatal L., Potier P. Highly efficient, practical approach to natural taxol. J Am Chem Soc. 1988;110:5917–5919. [Google Scholar]
- 6.Damen E.W.P., Braamer L., Scheeren H.W. Lanthanide trifluoromethanesulfonate catalysed selective acylation of 10-deacetylbaccatin III. Tetrahedron Lett. 1998;39:6081–6082. [Google Scholar]
- 7.Yanagi M., Ninomiya R., Ueda Y., Furuta T., Yamada T., Sunazuka T. Organocatalytic site-selective acylation of 10-deacetylbaccatin III. Chem Pharm Bull (Tokyo) 2016;64:907–912. doi: 10.1248/cpb.c16-00037. [DOI] [PubMed] [Google Scholar]
- 8.Holton R.A., Zhang Z., Clarke P.A., Nadizadeh H., Procter D.J. Selective protection of the C7 and C10 hydroxyl groups in 10-deacetyl baccatin III. Tetrahedron Lett. 1998;39:2883–2886. [Google Scholar]
- 9.Holton R.A., Zhang Z., Clark P.A. inventors. Florida State University, assignee. Process for selective derivatization of taxanes. US Patent 6706896 B1. Mar 16. 2004 [Google Scholar]
- 10.Patel R.N., Banerjee A., Nanduri V. Enzymatic acetylation of 10-deacetylbaccatin III to baccatin III by C-10 deacetylase from Nocardioides luteus SC 13913. Enzym Microb Technol. 2000;27:371–375. doi: 10.1016/s0141-0229(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 11.Walker K., Croteau R. Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:583–587. doi: 10.1073/pnas.97.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loncaric C., Merriweather E., Walker K.D. Profiling a Taxol pathway 10β-acetyltransferase: assessment of the specificity and the production of baccatin III by in vivo acetylation in E. coli. Chem Biol. 2006;13:309–317. doi: 10.1016/j.chembiol.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 13.You L.F., Huang J.J., Lin S.L., Wei T., Zheng Q.W., Jiang B.H. In vitro enzymatic synthesis of baccatin III with novel and cheap acetyl donors by the recombinant taxoid 10β-O-acetyl transferase. Biocatal Biotransform. 2019;37:239–245. [Google Scholar]
- 14.Menhard B., Zenk M.H. Purification and characterization of acetyl coenzyme a: 10-hydroxytaxane O-acetyltransferase from cell suspension cultures of Taxus chinensis. Phytochemistry. 1999;50:763–774. doi: 10.1016/s0031-9422(98)00674-8. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan S.A., Nawarathne I.N., Walker K.D. CoA recycling by a benzoate coenzyme A ligase in cascade reactions with aroyltransferases to biocatalyze paclitaxel analogs. Arch Biochem Biophys. 2020;683:108276–108286. doi: 10.1016/j.abb.2020.108276. [DOI] [PubMed] [Google Scholar]
- 16.Li B.J., Wang H., Gong T., Chen J.J., Chen T.J., Yang J.L. Improving 10-deacetylbaccatin III-10-β-O-acetyltransferase catalytic fitness for Taxol production. Nat Commun. 2017;8:15544. doi: 10.1038/ncomms15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu P., Vansiri A., Bhan N., Koffas M.A. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol. 2012;1:256–266. doi: 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- 18.Gao X., Gao F., Liu D., Zhang H., Nie X., Yang C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ Sci. 2016;9:1400–1411. [Google Scholar]
- 19.Kang A., Mendez-Perez D., Goh E.B., Baidoo E.E.K., Benites V.T., Beller H.R. Optimization of the IPP-bypass mevalonate pathway and fed-batch fermentation for the production of isoprenol in Escherichia coli. Metab Eng. 2019;56:85–96. doi: 10.1016/j.ymben.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Soma Y., Yamaji T., Matsuda F., Hanai T. Synthetic metabolic bypass for a metabolic toggle switch enhances acetyl-CoA supply for isopropanol production by Escherichia coli. J Biosci Bioeng. 2017;123:625–633. doi: 10.1016/j.jbiosc.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Jiaher T., Valentino J.S. Degradation of paclitaxel and related compounds in aqueous solutions I: epimerization. J Pharm Sci. 2008;97:1224–1235. doi: 10.1002/jps.21112. [DOI] [PubMed] [Google Scholar]
- 22.Wu G., Yan Q., Jones J.A., Tang Y.J., Fong S.S., Koffas M.A.G. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016;34:652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Shen X., Yuan Q., Yan Y. Microbial synthesis of pyrogallol using genetically engineered Escherichia coli. Metab Eng. 2018;45:134–141. doi: 10.1016/j.ymben.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Xu P., Ranganathan S., Fowler Z.L., Maranas C.D., Koffas M.A. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Xu P., Gu Q., Wang W., Wong L., Bower A.G., Collins C.H. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 26.Liu W., Zhang B., Jiang R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol Biofuels. 2017;10:41–50. doi: 10.1186/s13068-017-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Begley T.P., Kinsland C., Strauss E. The biosynthesis of coenzyme A in bacteria. Vitam Horm. 2001;61:157–171. doi: 10.1016/s0083-6729(01)61005-7. [DOI] [PubMed] [Google Scholar]
- 28.Song W.J., Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J Bacteriol. 1992;174:6411–6417. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y.Y., Jian X.X., Lv Y.B., Nian K.Q., Gao Q., Chen J. Enhanced squalene biosynthesis in Yarrowia lipolytica based on metabolically engineered acetyl-CoA metabolism. J Biotechnol. 2018;281:106–114. doi: 10.1016/j.jbiotec.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Nanduri V.B., Hanson R.L., LaPorte T.L., Ko R.Y., Patel R.N., Szarka L.J. Fermentation and isolation of C10-deacetylase for the production of 10-deacetylbaccatin III from baccatin III. Biotechnol Bioeng. 1995;48:547–550. doi: 10.1002/bit.260480518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.