Abstract
Pulmonary fibrosis (PF) is a chronic, progressive, fatal interstitial lung disease with limited available therapeutic strategies. We recently reported that the protein kinase glycogen synthase kinase-3β (GSK-3β) interacts with and inactivates the ubiquitin-editing enzyme A20 to suppress the degradation of the transcription factor CCAAT/enhancer-binding protein beta (C/EBPβ) in alveolar macrophages (AMs), resulting in a profibrotic phenotype of AMs and promoting the development of PF. Here, we showed that chronic lung injury upregulated the stress response protein tribbles homolog 3 (TRIB3), which interacted with GSK-3β and stabilized GSK-3β from ubiquitination and degradation. Elevated GSK-3β expression phosphorylated A20 to inhibit its ubiquitin-editing activity, causing the accumulation of C/EBPβ and the production of several profibrotic factors in AMs and promoting PF development. Activated C/EBPβ, in turn, increased the transcription of TRIB3 and GSK-3β, thereby establishing a positive feedback loop in AMs. The knockdown of TRIB3 expression or the pharmacologic disruption of the TRIB3‒GSK-3β interaction was an effective PF treatment. Our study reveals an intact profibrotic axis of TRIB3‒GSK-3β‒A20‒C/EBPβ in AMs, which represents a target that may provide a promising treatment strategy for PF.
KEY WORDS: E3 ligase, Lung injury, Protein phosphorylation, Protein–protein interaction, Ubiquitination, TRIB3, GSK-3β
Graphical abstract
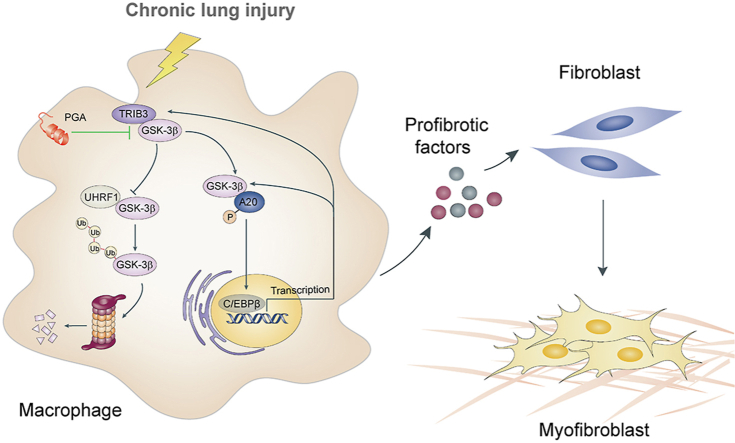
Elevated TRIB3 expression in macrophages positively correlates with the development of pulmonary fibrosis. TRIB3 interaction with GSK-3β obstructs the ubiquitination and degradation of GSK-3β. Targeting TRIB3‒GSK-3β interaction reduces pulmonary fibrosis.
1. Introduction
Pulmonary fibrosis (PF) is a chronic, progressive, fatal interstitial lung disease with forms including idiopathic pulmonary fibrosis (IPF) with unknown causes and others with a known etiology1. Despite increasing research efforts, the pathogenesis of PF remains inconclusive, and no effective intervention is available for this devastating disease2. Recent studies have indicated that alveolar macrophages (AMs) display profibrotic and proinflammatory phenotypes following chronic lung injury and play central roles in the occurrence and progression of PF3, 4, 5, 6. In particular, profibrotic factors released from these AMs activate the transformation of lung fibroblasts into myofibroblasts, the effector cells of PF. However, despite intensive studies on the regulation of AM phenotypes during PF, the precise molecular and cellular mechanisms that modulate these processes remain elusive7.
Our recent study showed that dysfunction of the ubiquitin-editing enzyme A20 in AMs promotes PF by reducing the degradation of the transcription factor CCAAT/enhancer-binding protein beta (C/EBPβ) and increasing the expression of its target genes, thus directing the profibrotic phenotype of AMs8. Glycogen synthase kinase-3β (GSK-3β), whose expression and activity are increased following chronic lung injury, can phosphorylate A20 to suppress its activity. These results show that GSK-3β positively modulates PF by inducing the profibrotic phenotype of AMs to support the differentiation of fibroblasts into myofibroblasts. Therefore, reducing the activity and expression of GSK-3β may suppress the activation of AMs by restoring A20 activity. However, the upstream signal that regulates GSK-3β in AMs during PF progression remains unclear.
The pseudokinase tribbles homolog 3 (TRIB3), a well-known stress and metabolic sensor, is involved in organ fibrogenesis9, 10, 11. For instance, TRIB3 exerts a profibrotic effect in renal tissue and systemic sclerosis via activation of transforming growth factor-β1 (TGF-β1) signaling9,10. Furthermore, stress-induced TRIB3 promotes hepatic fibrosis by affecting the cargo function of sequestosome-1 (SQSTM1), thus causing exosomes to be released from liver cells to activate hepatic stellate cells10. Given that TRIB3 senses various stresses, such as inflammation and hypoxia, we hypothesized that the elevation of TRIB3 expression in AMs following chronic lung injury might drive PF development by triggering an intrinsic regulatory mechanism to direct the profibrotic phenotype of AMs. Here, we reveal a previously unknown profibrotic mechanism by which TRIB3 initiates a positive feedback regulatory loop composed of the protein kinase GSK-3β, the ubiquitin-editing enzyme A20 and the transcription factor C/EBPβ in AMs, suggesting potential therapeutic strategies for treating PF and other fibroproliferative lung diseases.
2. Materials and methods
2.1. Experimental design
Phenotypic, transcriptomic, and functional analyses of alveolar macrophages were conducted to determine the role of TRIB3 in PF development. In addition, we used transgenic mice and protein–protein interaction (PPI) inhibitors to assess the functional outcomes of mechanical suppression of AM activation. The experiments and outcome assessments were carried out in a blinded fashion. A detailed description of the experimental replicates and detailed methods can be found in the following subsections and figure legends.
2.2. In vivo experiments
C57BL/6J mice (6–8 weeks old, 18–20 g) were supplied by Vital River Lab Animal Technology Co., Ltd. (Beijing, China). Trib3 conditional knockout (Trib3fl/fl) mice were obtained from Cyagen Biosciences Inc. (Guangzhou, China). Lyz2Cre mice were supplied by the Model Animal Research Center of Nanjing University (Nanjing, China). A20 conditional deficient (Tnfaip3fl/+) mice were procured from RIKEN (Wako, Japan). Trib3fl/flLyz2Cre mice were generated by intercrossing Trib3fl/fl mice with Lyz2Cre mice. Tnfaip3fl/+Lyz2Cre mice were produced by intercrossing Tnfaip3fl/+ mice with Lyz2Cre mice. Trib3fl/flTnfaip3F/+Lyz2Cre mice were produced by intercrossing Tnfaip3fl/+Lyz2Cre mice with Trib3fl/fl mice. All mice were housed in the animal facilities at the Institute of Materia Medica under specific-pathogen free (SPF) conditions. After earmarking, the mice were randomly grouped, and the sample size was predetermined base on prior experience with similar studies. The mice of n > 6 were used for each condition and genotype. Age and body weight were evenly distributed among groups. The ethical approval for animal experiments was obtained from the Animal Experimentation Ethics Committee of the Chinese Academy of Medical Sciences (approval number: 002802; Beijing, China). All experiments were carried out in compliance with the Institutional Animal Care and Use Committee (Chinese Academy of Medical Sciences) and the ARRIVE guidelines12.
2.3. Human subjects
Frozen sections of human normal and fibrotic lung tissues were supplied by OriGene Inc. (MD, USA). The histologic features of the PF sample (Cat. No. CS502727) were as follows: Hematoxylin and eosin (H&E) staining reveal 45% alveoli, 0% bronchioles, 30% fibrovascular tissue, and 25% diffuse interstitial fibrosis. The individual normal lung tissues with regular alveolar architecture and morphology were analyzed and compared with those of the patient samples. Human bronchoalveolar lavage fluids (BALF) were obtained from the Pulmonary Function Detection Chamber of Beijing Friendship Hospital (Beijing, China). Primary AMs were collected from the BALF of PF patients according to the previous method13. The clinical characteristics of PF patients are summarized in Supporting Information Table S1. The ethical approval for human research was obtained from the Institutional Review Board of Chinese Academy of Medical Sciences and Peking Union Medical College. Written informed consent was signed by all participants. All procedures involving human subjects were carried out in compliance with the principles of the Declaration of Helsinki.
2.4. Animal models
The bleomycin (BLM) induced chronic PF model was produced by repetitive intratracheal spray according to the previous method8. After intraperitoneal (i.p.) anesthesia with 400 mg/kg Avertin (Sigma–Aldrich, USA), the trachea of mice was sprayed 6 times with 1 U/kg BLM (Selleck, China) in 50 μL lipopolysaccharides (LPS)-free phosphate-buffered saline (PBS) using a MicroSprayer (Penn-Century, USA) at an interval of 14 days. All mice were euthanized by an overdose of anesthetics after the final of BLM spray.
The silicosis mouse model was constructed by intratracheally injecting 125 mg/kg aqueous suspensions of silica particles (Sigma–Aldrich). All mice were euthanized on Day 50 following administration of SiO2. The mice in the control group were treated intratracheally with an identical volume of PBS.
For therapeutic experiments, mice were i.p. injected with 1 or 4 mg/kg of peptide every 3 days starting from Day 10 after the final BLM challenge or Day 50 after SiO2 administration for 30 days. All mice were euthanized by an overdose of anesthetics on Day 40 after the final BLM challenge or Day 80 after SiO2 administration.
2.5. Cell lines and primary cultures
HEK293T cells (ATCC, USA, CRL-11268) were cultured in IMDM (Livning, China) containing 10% fetal bovine serum (FBS). Primary human lung fibroblasts (PriCells) were cultured in DMEM/F12 (Gibco, USA) containing 10% FBS. Primary mouse lung fibroblasts (PMLFs) were cultured in DMEM (Livning) with 15% FBS. Primary mouse and human AMs were cultured in RPMI 1640 medium (Gibco) containing 10% FBS. All cells were maintained at 37 °C in a humidified atmosphere with 5% CO2, and were confirmed to be free of mycoplasma contamination.
2.6. Isolation of primary mouse AMs
Mouse AMs were obtained from BALF. First, the mouse lungs were intratracheally injected with 1 mL PBS and then sucked out. After repeating this step for 3 times, the lavage fluids were subjected to centrifugation (300 ×g, 15 min). Next, the pellet was resuspended and cultured in RPMI 1640 medium containing 10% FBS. After 45 min, the culture medium containing the non-adherent cells was discarded. Finally, the attached AMs were rinsed with PBS for 3 times and then used for analysis.
2.7. Isolation of PMLFs
PMLFs were isolated as described previously8. Briefly, mice were euthanized by an overdose of anesthetics and cleaned with 75% ethanol. The lung tissue was collected, washed twice with sterile PBS, and cut into ~1 mm3 pieces. After centrifugation (600 ×g, 5 min), the pellet was resuspended in DMEM medium containing 15% FBS, and cultured at 37 °C in 10-cm culture dishes. After 4–5 days of incubation, the attached fibroblasts were collected for subculturing or other assays.
2.8. Pulmonary function tests
After i.p. anesthesia with 400 mg/kg Avertin, the mice were placed in a flexiVent system (SCIREQ Inc., Montreal, Canada), followed by mechanical ventilation (tidal volume = 10 mL/kg, respiratory rate = 150 breaths/min and positive end-expiratory pressure at 3 cm H2O) for evaluating pulmonary function as described previously14. In addition, a Snapshot-150 perturbation was used to measure dynamic lung compliance.
2.9. Surface plasmon resonance assay
Binding affinity between TRIB3 and the peptide was evaluated using a BIAcore T200 instrument (GE Healthcare, CA, USA) as described previously8. For TRIB3 immobilization with amine-coupling, the CM5 chip was activated using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxy succinimide. TRIB3 protein in PBS was then passed overflow cells until response units were achieved. The activated free binding sites were blocked with ethanolamine. Analyte peptides were injected at a 30 μL/min flow rate and 180 s contact time, followed by 600 s autonomous dissociation time. BIAevaluation software was adopted to calculate the dissociation constant (KD) using the following equation: KD=Kd/Ka, where Kd and Ka are the dissociation rate and the association rate, respectively.
2.10. Assessment of Trib3 transcription activity
To assess Trib3 transcription, TK plasmid and p-Trib3 luciferase reporter were transfected into Cebpb-depleted cells. After transfection, the cells were collected and lysed with lysis buffer. The internal control used was pTK-Renilla. Luciferase activities were then determined using a dual luciferase assay (Promega, WI, USA).
2.11. Gsk3b transcription assay
HEK293T cells were seeded in 12-well plates 24 h before transfection. The Gsk3b-GLuc (HPRM36354-PG04) vectors were obtained from GeneCopoeia, Inc. (Rockville, MD, USA). At 48 h post transfection, the cell culture medium was collected for GLuc and SEAP luminescent assays by using the Secrete-Pair Dual Luminescence Assay Kit according to the manufacturer's instruction (GeneCopoeia).
2.12. Plasmid construction
Myc-tagged GSK-3β and truncating mutants of M1 (amino acids, AAs 1–123), M2 (AAs 124–353), M3 (AAs 124–433), and M4 (AAs 1–353) were cloned into pcDNA3.1-Myc-His(-)-B vector (Invitrogen, USA) by a standard subcloning procedure. TRIB3-FL (AAs 1–359) and its truncations, M1 (AAs 1–179), M2 (AAs 71–315), M3 (AAs 180–358), M4 (AAs 180–315), and M5 (AAs 72–359, Δ180‒315) plasmids were inserted into the pcDNA3.1-N-HA vector using a standard subcloning procedure.
2.13. TRIB3 binding protein identification
The AMs from PF mice were isolated, and the cell lysate was immunoprecipitated with anti-TRIB3 antibody using a Pierce Co-IP Kit (Thermo Fisher Scientific, USA). The peptide fragments were sequenced by mass spectrometry (MS) and then analyzed by Oebiotech Co., Ltd. (Shanghai, China). After protein digestion and gel extraction, the digested products were separated in a 120/60-min gradient elution at 0.600 L/min using the EASY-nLC 1000 system coupled to the Thermo Orbitrap Fusion mass spectrometer. (Thermo Fisher Scientific). The mass of each peptide was identified using a liquid chromatography-tandem mass spectrometry (LC–MS/MS) Q Exactive™ Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The MS/MS data were searched against a Uniprot protein FASTA file using an in-house Proteome Discoverer ver. 1.4 (Thermo Fisher Scientific). The peptide was considered unique if it assembled to a particular protein group.
2.14. Real-time polymerase chain reaction PCR (RT-PCR) and RNA interference
First, total RNA was extracted with TransZol UP (TransGen Biotech, Beijing, China) in accordance with the kit protocol. cDNA synthesis was then performed using One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). Next, RT-PCR was conducted using the KAPA SYBR FAST RT-PCR Master Mix (2×) Kit (Kappa Biosystem, USA) following the kit protocol. RT-PCR primer sequences are shown in Supporting Information Table S2. Trib3 and Uhrf1 siRNAs were synthesized by RIBOBIO (Guangzhou, China) and transfected using Lipofectamine RNA interference MAX Transfection Reagent (Life Technologies, CA, USA) under the kit protocol.
2.15. Immunoblotting, immunostaining and immunohistochemistry
Proteins were extracted from cells and lungs using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China). First, protein concentration was measured by BCA Protein Assay Kit (Applygen Technologies Inc., Beijing, China). Protein lysates were then loaded for SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and proceeded with immunoblot analyses. Finally, signals were recorded using a chemiluminescence imaging system (Tanon 5200; Tanon, Shanghai, China).
For immunofluorescence staining, lung frozen sections or cells on coverslips were first fixed in 4% buffered paraformaldehyde, then permeabilized with 0.5% Triton X-100, blocked with 3% bovine serum albumin (BSA), and finally incubated with specific primary antibody followed by the corresponding secondary antibody. After counterstaining with 4ʹ,6-diamidino-2-phenylindole (DAPI), the images were recorded using a confocal microscope (Olympus Microsystems, CA, USA).
For immunohistochemical staining, the frozen tissue sections were fixed in 4% buffered paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 3% BSA for 30 min at room temperature. Next, the sections were incubated with anti-TRIB3 (1:200, ab137526, Abcam, UK) and anti-alpha-smooth muscle actin (α-SMA, 1:200, BM0002, BOSTER, China) antibody at 4 °C for 24 h, then with the corresponding secondary antibody at room temperature for 30 min. After staining with the freshly prepared 3,3ʹ-diaminobenzidine (DAB) substrate solution (ZSGB-BIO Company, Beijing, China), the sections were subjected to hematoxylin counterstaining, dehydration, and coverslip mounting. The images were recorded using an Olympus DP72 microscope (Olympus Microsystems).
2.16. Co-immunoprecipitation (co-IP)
After lysing with the co-IP buffer [150 mmol/L NaCl, 25 mmol/L Tris-HCI (pH 7.4), 2.5 mmol/L MgCl2, 0.5% NP-40, 0.5 mmol/L EDTA, 5% glycerol], the lysate was incubated with the specific antibodies at 4 °C for 24 h, and then with Protein A/G Plus-Agarose (Santa Cruz, USA) at 4 °C for 2 h. Interaction complexes were detached from the beads by boiling for 10 min, followed by SDS-PAGE and immunoblot analysis.
2.17. Flow cytometry analysis
AMs with indicated treatment were first suspended in 100 μL of PBS. The suspension was then incubated with anti-mouse CD16/32 on ice for 20 min. Next, the cells in all groups, except for the negative control group, were stained with allophycocyanin (APC) anti-mouse F4/80 in the dark on ice for 45 min. After 3 times washing with PBS, the cells were fixed in paraformaldehyde (4%) at room temperature for 15 min, permeabilized with saponin (0.1%), and incubated with fluorescein isothiocyanate (FITC) anti-mouse CD206 in the dark on ice for 45 min. Finally, the cells were detected using a BD FACSVerse™ flow cytometer (BD Biosciences, USA), and analyzed with FCS Express 6 software.
2.18. A20 activity measurement
AMs with indicated treatment were lysed using the co-IP buffer. A20 protein was coimmunoprecipitated with the corresponding antibody (Abcam). After 5 times washing, A20 protein was eluted with the elution buffer obtained from Pierce Co-IP Kit (Thermo Fisher Scientific). The deubiquitinating enzyme (DUB) activities of A20 in AMs were determined using a DUB Activity Kit (StressMarq Biosciences Inc., Canada) per the kit protocol. Fluorescence intensities were recorded using a microplate reader, and 7-amido-4-methylcoumarin (AMC) standard curve was established for data analysis.
2.19. Assessment of lung hydroxyproline
The hydroxyproline content in the right lungs of mice was measured using hydroxyproline content detection kits following with the protocol (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.20. Structured illumination microscopy
Primary AMs were seeded on coverslips 24 h before indicated treatment. After fixing with formalin and permeabilizing in 2% Triton X-100, the cells were quenched with 50 nmol/L glycine in Tris-buffered saline (TBS) for 10 min and then blocked with 3% BSA for 30 min. Subsequently, the coverslips were exposed to the primary antibody at 4 °C for 24 h, and then with the corresponding secondary antibody. After DAPI counterstaining, the slides were sealed with resinene. Images were acquired using a three-dimensional structured illumination microscope (GE healthcare). The images’ intensity plots within a straight line extracted from the immunofluorescence image were analyzed using the Imaris 3D/4D software (Bitplane AG, Switzerland). Adobe Photoshop was used to crop ad process the images. When comparison was performed between images in the same experiment, the levels were equally adjusted, and the ratio between the levels was not altered.
2.21. Chromatin immunoprecipitation (ChIP)
SimpleChIP® Enzymatic Chromatin IP Kit (magnetic beads, Cell Signaling Technology, USA) was used to perform ChIP assays. First, AMs were crosslinked with 1% formaldehyde, washed and lysed. Next, the chromatin was sheared by sonication into fragments. The chromatin fragments were incubated with anti-C/EBPβ or normal rabbit IgG, and then incubated with anti-rabbit IgG Dynabeads. Finally, the purified immunoprecipitated DNA was used for quantitative PCR.
2.22. Three-dimensional collagen gels
PMLFs were suspended in serum-free medium and blended with 3 mg/mL neutralized rat tail type I collagen (ratio = 2:1). Subsequently, the mixture was seeded in 24-well plates at 1 × 105/mL cell density. After collagen coagulating for 1 h at 37 °C, the gel edge was detached from well walls, and 1 mL 10% FBS-containing culture medium was added on the gels. The incubation time depends on cell viability. Finally, the gel images were captured and then analyzed by ImageJ software (National Institutes of Health, USA).
2.23. Statistics
Statistical tests were conducted by GraphPad Prism 5 software (San Diego, CA, USA). All results were described as mean ± standard error of the mean (SEM). The differences between groups were compared by one-way ANOVA or unpaired Student's t-test. The correlation between two variables was determined by Pearson's correlation coefficient. The level of statistical significance was set at P < 0.05. All measurements were performed at least 3 times.
3. Results
3.1. TRIB3 level in AMs are positively correlated with PF development
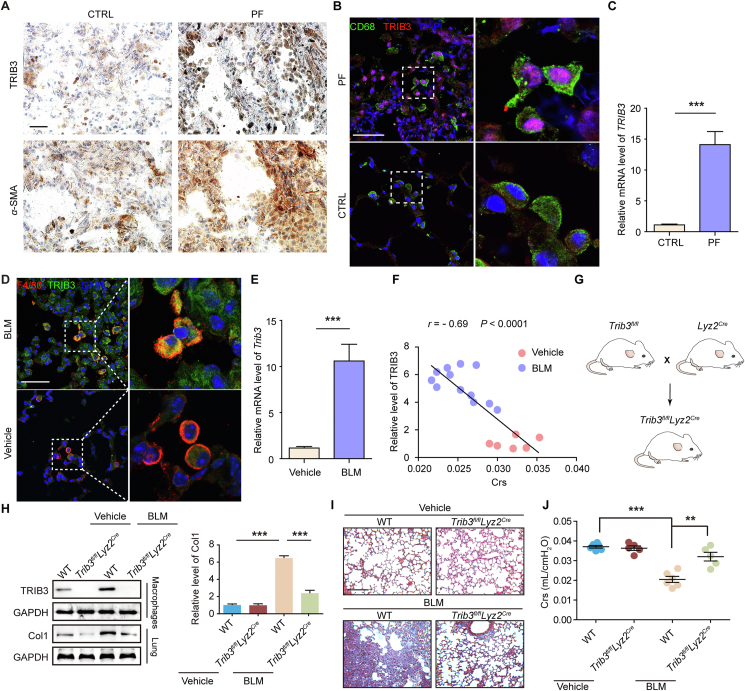
We recently reported that the protein kinase GSK-3β binds to, phosphorylates, and inactivates the ubiquitin-editing enzyme A20 to suppress the degradation of the transcription factor C/EBPβ in AMs, resulting in a profibrotic phenotype of AMs and PF development8. In this study, we wanted to investigate how GSK-3β is positively regulated following chronic lung injury. The expression levels of the stress sensor TRIB3 and α-SMA, a marker of myofibroblasts, were found to be increased in lung sections from PF patients (Fig. 1A). We then examined the cellular source of TRIB3 in lung tissues during PF development by examining the colocalization of TRIB3 with α-SMA, pro-SPC [alveolar epithelial type 2 cell (AEC2) marker], F4/80 (macrophage marker), and VE-cadherin (endothelial cell marker) in fibrotic lung tissues. The results indicated that TRIB3 was significantly upregulated in AMs from patients with PF (Fig. 1B; Supporting Information Fig. S1A and B). In addition, the TRIB3 mRNA level in AMs from patients with PF was markedly higher than that in AMs from age-matched controls (Fig. 1C). Consistent with the data from PF patients, elevated TRIB3 protein expression was observed in AMs from bleomycin bleomycin (BLM)-challenged fibrotic mice (Fig. 1D and Fig. S1C). PCR assays also showed much higher mRNA levels of Trib3 in mice fibrotic lungs than in normal lungs (Fig. 1E). In addition, a negative correlation was found between TRIB3 expression in AMs and lung function in control and PF mice (Fig. 1F). To further confirm whether TRIB3 contributes to PF development, we initially crossed mice containing a floxed Trib3 exon 3 with mice expressing Cre recombinase driven by the Lyz2 promoter, which efficiently targets AMs to produce Trib3fl/flLyz2Cre mice (Fig. 1G and H). These mice were viable and fertile, were born at the expected Mendelian ratio and showed no abnormality during adulthood. The genetic knockout of Trib3 in AMs reduced BLM-induced PF changes, as revealed by the reduction of collagen 1 (Col1) expression (Fig. 1H and I) and the improvement of lung function (Fig. 1J). These findings demonstrate that TRIB3 from AMs is associated with PF progression.
Figure 1.
The elevation of TRIB3 expression in AMs is positively correlated with PF occurrence. (A) Representative immunohistochemical staining of TRIB3 or α-SMA expression in lung tissues from a patient with PF and an age-matched health adult. Scale bar, 50 μm. (B) Confocal image revealing the expression of TRIB3 and CD68 in lung tissue from control or PF patients. Scale bar, 25 μm. (C) The mRNA levels of TRIB3 in AMs from controls or PF patients were determined by RT-PCR (n = 6/group). (D) Confocal image revealing TRIB3 expression in AMs on Day 10 after BLM challenge. Scale bar, 50 μm. (E) The mRNA levels of Trib3 in AMs from control or PF mice were detected by RT-PCR (n = 6/group). (F) Correlation analysis between TRIB3 expression in AMs and respiratory system compliance (Crs) in mice. Each point represents the value of one mouse. Spearman's rank correlation test was employed to determine statistical significance (vehicle, n = 6; BLM, n = 12). (G) Scheme for generating mice lacking Trib3 in the hematopoietic lineage. The Trib3fl/flLyz2Cre mouse model was constructed by intercrossing Trib3fl/fl mice with Lyz2Cre mice. (H) Sample blots showing TRIB3 expression in AMs and Col1 expression in lung tissue. (I) Representative Masson staining of lung tissue after BLM challenge. Scale bar, 200 μm. (J) Crs was measured to determine the changes in fibrosis in the lungs. (C, E) Pooled data from two independently significant experiments. Data are represented as mean ± SEM (n = 5–6 mice, unpaired two-tailed Student's t-test). (G–J) Representatives of two independent experiments. Data are represented as mean ± SEM (n = 5–6 mice, one-way ANOVA). ∗∗P < 0.01, ∗∗∗P < 0.001.
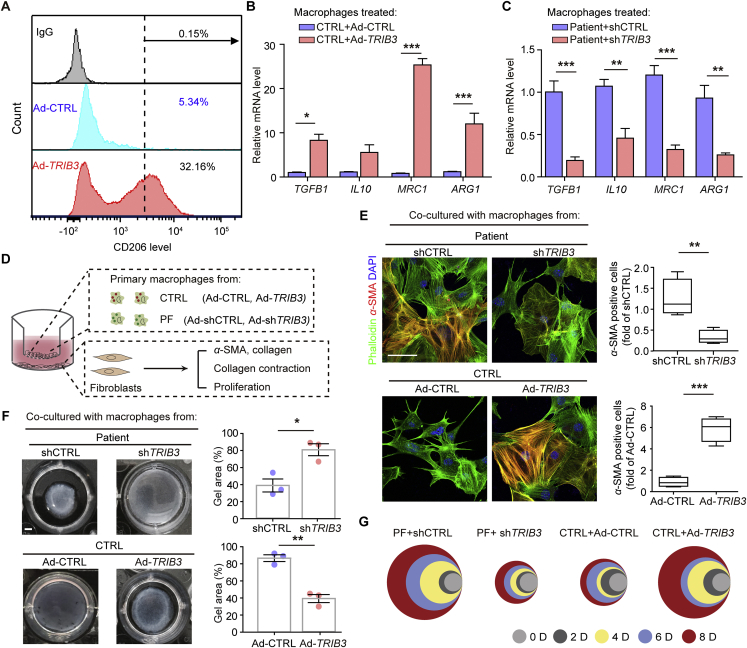
We then determined whether TRIB3 expression could affect the profibrotic phenotype of AMs. The overexpression of TRIB3 in AMs from controls increased the expression of CD206 (Fig. 2A) and the profibrotic genes arginase 1 (ARG1), interleukin 10 (IL10), mannose receptor C-type 1 (MRC1), and TGFB1 (Fig. 2B). In contrast, the depletion of TRIB3 in AMs from PF patients decreased their expression (Fig. 2C). Moreover, in the AM-fibroblast coculture system (Fig. 2D), the silencing of TRIB3 reversed AMs-induced overexpression of α-SMA and Col1 as well as the proliferation and contraction of fibroblasts (Fig. 2D–G). However, TRIB3 overexpression in AMs from controls increased fibroblast activation (Fig. 2D‒G). Similarly, the overexpression of Trib3 induced the profibrotic phenotype of AMs from control mice, whereas the depletion of Trib3 inhibited the profibrotic phenotype of AMs from PF mice (Supporting Information Fig. S2A‒E). These data verify that elevated TRIB3 expression in AMs contributes to PF pathogenesis.
Figure 2.
Elevated TRIB3 expression supports the profibrotic phenotype of AMs in PF patients. (A) Flow cytometric analysis of CD206 expression in TRIB3-overexpressing AMs from control subjects. (B) Quantitative mRNA analysis of target genes in AMs from control subjects treated with the Ad-TRIB3 virus (n = 3). (C) Quantitative mRNA analysis of the target genes in AMs from PF patients transfected with the Ad-shTRIB3 virus (n = 3). (D–G) Ad-CTRL or Ad-TRIB3 viral transfection in AMs from control subjects. Ad-shCTRL or Ad-shTRIB3 viral transfection in AMs from PF patients. The AMs were co-cultured with human primary fibroblasts at different time points (D). The activation of fibroblasts was examined based on α-SMA expression (E) (n = 4), fibroblast contraction in 3D collagen matrices (F) (n = 3), and fibroblast proliferation (G). Different colors represent different time points. The diameter indicates the relative number of cells. Scale bar, 10 μm for E and 2 mm for F. (A–C) and (E–G) are representative of three experiments. Data are represented as mean ± SEM (n = 3–4, unpaired, two-tailed Student's t-test). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
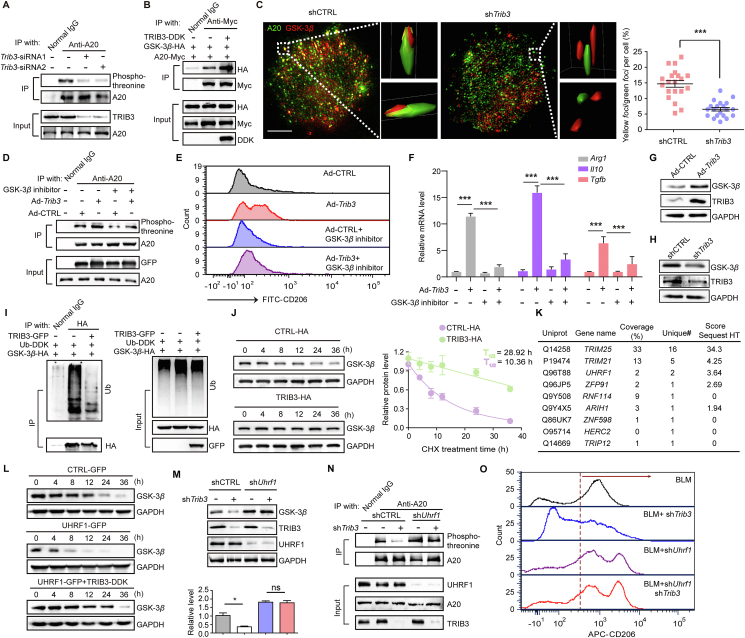
3.2. TRIB3 represses the enzymatic activity of A20 in AMs
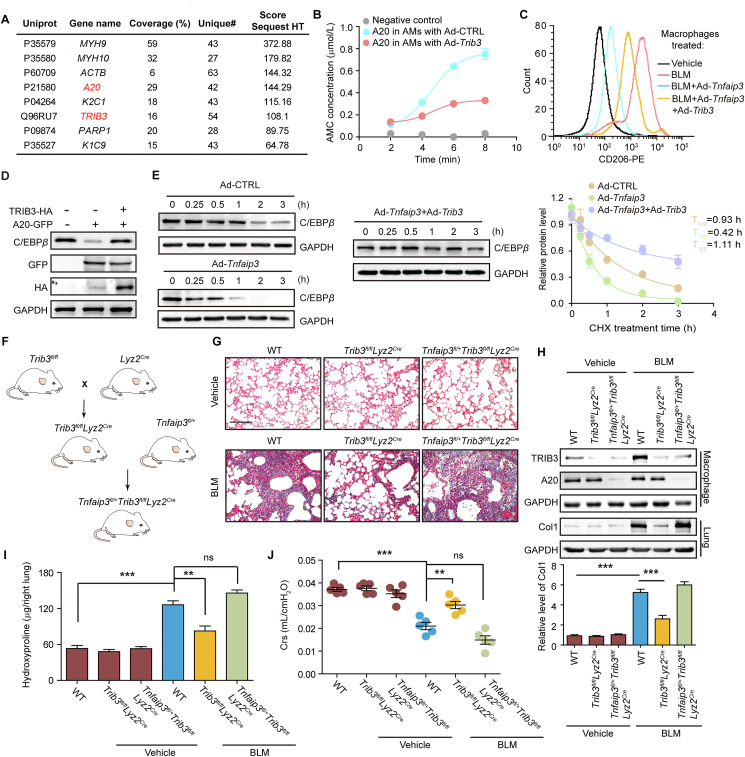
We explored how TRIB3 regulates the phenotype of AMs. Several proteins were screened from the TRIB3-precipitated proteins by combining immunoprecipitation (IP) and mass spectrometry (MS)-based proteomic analysis (Fig. 3A). Among these interacting proteins, A20 attracted our attention due to our recent work identifying its role in AMs and PF development8. Indeed, the activity of A20 was suppressed in AMs transfected with a TRIB3-carrying adenovirus (Fig. 3B). Moreover, the suppression of CD206 expression by A20 was reversed after Trib3 overexpression (Fig. 3C). In line with these results, Trib3 depletion in AMs from BLM mice failed to reduce CD206 expression when Tnfaip3 was silenced (Supporting Information Fig. S3A). Furthermore, the ectopic expression of TRIB3 increased the A20-mediated reduction of the expression of C/EBPβ, which is a substrate of A20 (Fig. 3D). The overexpression of TRIB3 increased the reduced K63-linked ubiquitination but decreased the enhanced K48-linked ubiquitination of C/EBPβ caused by A20 overexpression (Fig. S3B and C). Similarly, the reduced half-life of C/EBPβ in the presence of A20 was prolonged in AMs with TRIB3 overexpression (Fig. 3E). To determine whether TRIB3 contributes to PF pathogenesis by suppressing the activity of A20, we first constructed hematopoietic Trib3-deficient mice (referred to as Trib3fl/flLyz2Cre). These mice were then crossed with mice carrying a floxed Tnfaip3 exon 3 to generate mice with reduced A20 and TRIB3 expression in AMs (referred to as Tnfaip3fl/+Trib3fl/flLyz2Cre mice) (Fig. 3F). Consistent with the above findings, the genetic knockout of Trib3 expression in AMs protected against BLM-induced PF, as indicated by decreases in collagen deposition, Col1 expression, and the hydroxyproline content (Fig. 3G‒I), and improved lung function (Fig. 3J) in these mice. However, the Tnfaip3fl/+Trib3fl/flLyz2Cremice exhibited similar fibrotic changes in the lung to those found in wild-type (WT) mice (Fig. 3G‒J). Additionally, Trib3 depletion suppressed the expression of C/EBPβ in AMs with WT A20 but did not decrease C/EBPβ expression in AMs with A20 depletion (Fig. S3D). These data suggest that TRIB3 exerts its pro-PF effect by impeding A20 activity in AMs.
Figure 3.
TRIB3 supports the profibrotic phenotype of AMs by inhibiting A20 activity and increasing C/EBPβ stability. (A) The TRIB3 protein was precipitated from AMs by the anti-TRIB3 antibody. The immunocomplexes were resolved by SDS-PAGE and detected by silver staining. The protein bands were subjected to MS analysis. (B) AMs collected from control subjects were transfected with the Ad-TRIB3 virus. The activity of A20 immunoprecipitated from these cells was measured. (C) The flow cytometric analysis of CD206 expression in AMs from PF mice transfected with the indicated adenovirus. (D) Sample immunoblots demonstrating the expression of C/EBPβ in AMs after transfection with the indicated adenoviruses. (E) Quantitative analysis of C/EBPβ degradation in the presence of Ad-CTRL, Ad-Tnfaip3, or Ad-Tnfaip3 plus Ad-Trib3. (F) Schematic of representation of the construction triple-transgenic mouse model constructed by (i) intercrossing Trib3-targeted knockout mice with homozygous Lyz2Cre mice to produce Trib3fl/flLyz2Cre transgenic mice and (ii) crossing Trib3fl/flLyz2Cre mice with Trib3-targeted knockout mice to produce triple-transgenic mice (Trib3fl/flTnfaip3fl/+Lyz2Cre mice. (G–J) Analysis of Masson staining (G), Col1 and C/EBPβ expression in AMs (H), hydroxyproline levels in the right lung (I), and Crs (J) were performed to assess the fibrotic changes in lung tissue after BLM challenge. Scale bar, 200 μm. (B–E) Representatives of 2–4 repeats. (G–J) Representative of two individual experiments. Data are represented as mean ± SEM (n = 5 mice in each group, one-way ANOVA). ∗∗P < 0.01, ∗∗∗P < 0.001. ns, not significant.
3.3. TRIB3 promotes A20 phosphorylation by increasing the stability of GSK-3β
Because the phosphorylation of A20 is associated with its ubiquitin-editing activity, we examined whether TRIB3 regulates A20 phosphorylation. The silencing Trib3 expression decreased the threonine phosphorylation of A20 (Fig. 4A). Given that GSK-3β binds to and phosphorylates A20 to suppress its activity, the co-IP assay was conducted to investigate the effect of TRIB3 on the GSK-3β‒A20 interaction. We found that TRIB3 overexpression could enhance the interaction between GSK-3β and A20 (Fig. 4B). Accordingly, the GSK-3β‒A20 interaction was disrupted in Trib3-silenced cells (Fig. 4C and Supporting Information Fig. S4A). However, TRIB3 did not increase the phosphorylation of A20 in the presence of a GSK-3β inhibitor (Fig. 4D). Moreover, the profibrotic phenotype of AMs induced by Trib3 overexpression was suppressed in cells pretreated with a GSK-3β inhibitor (Fig. 4E and F), suggesting that TRIB3 impedes A20 activity via GSK-3β.
Figure 4.
TRIB3 interferes with UHRF1-mediated GSK-3β degradation and enhances A20 phosphorylation. (A) IP assay for detecting A20 phosphorylation after Trib3 deletion. (B) IP assay for evaluating the interaction between A20 and GSK-3β in the presence of TRIB3. (C) Structured illumination microscopy image of Ad-shCTRL- or Ad-shTrib3-treated AMs stained for GSK-3β and A20. The dotted line-bordered square denotes the region processed for 3D surface rendering (insets) (n = 20). Scale bar, 2 μm. (D) IP assay for detecting A20 phosphorylation in AMs after the indicated treatments. (E) The CD206 expression by flow cytometric analysis in AMs after the indicated treatments. (F) Quantitative mRNA analysis of target genes in AMs from control subjects under the indicated treatments (n = 3). (G) Immunoblots revealing GSK-3β expression in AMs from control mice after TRIB3 overexpression. (H) Immunoblot showing GSK-3β expression in AMs from PF mice after TRIB3 depletion. (I) Immunoblot indicating GSK-3β ubiquitination in the presence of TRIB3 in vitro. (J) Quantitative analysis of GSK-3β degradation in the presence of TRIB3. (K) E3 ligases interacting with GSK-3β were identified by MS. (L) Immunoblots showing the degradation of GSK-3β in cells treated with UHRF1 or UHRF1 plus TRIB3. (M) Representative immunoblots of GSK-3β in AMs under the indicated treatments (n = 3). (N) TRIB3 was knocked out in E3-silenced AMs. The phosphorylation of A20 was detected in an IP assay. (O) Flow cytometric analysis of CD206 expression in AMs treated with the indicated adenoviruses. (A, B, D–J, and L–O) Representatives of two individual experiments. (C) Pooled data from two independent experiments. Data are represented as mean ± SEM. ∗∗∗P < 0.001.
We next explored how TRIB3 increases the GSK-3β-mediated phosphorylation of A20. We found that the overexpression of Trib3 increased the protein level of GSK-3β and that the depletion of Trib3 reduced the GSK-3β protein level (Fig. 4G and H). Moreover, the expression of GSK-3β was positively correlated with that of TRIB3 in AMs from PBS- and BLM- challenged mice (Fig. S4B). Intracellular proteins are degraded by the ubiquitin-proteasome system (UPS) or lysosomal autophagy pathways. We found that the UPS inhibitor MG132 but not the autophagy inhibitor 3-MA reversed the Trib3 depletion-induced reduction in GSK-3β expression (Fig. S4C). Indeed, the overexpression of TRIB3 blocked the ubiquitination of GSK-3β and prolonged its half-life (Fig. 4I and J). We then reasoned that TRIB3 might inhibit GSK-3β ubiquitination by suppressing the expression of certain E3 ubiquitin ligases or their interaction with GSK-3β. MS-based proteomic analysis was performed to identify E3 ligases interacting with GSK-3β (Fig. 4K). Tripartite motif containing 21 (TRIM21), TRIM25, and ubiquitin-like with PHD and ring finger domains 1 (UHRF1) were identified as potential E3 ligases of GSK-3β because they increased its ubiquitination (Fig. S4D). Furthermore, the overexpression of TRIB3 suppressed the increase in the ubiquitination of GSK-3β mediated by UHRF1 but not that mediated by TRIM21 or TRIM25 (Fig. S4E). Additionally, the reduced half-life of GSK-3β caused by UHRF1 was prolonged in cells with ectopic expression of TRIB3 (Fig. 4L). The loss of Trib3 failed to reduce the expression of GSK-3β (Fig. 4M), the phosphorylation of A20 (Fig. 4N), and the expression of CD206 (Fig. 4O) in Uhrf1-silenced AMs. These data suggest that TRIB3 disrupts UHRF1-mediated GSK-3β ubiquitination and degradation.
We further investigated how TRIB3 suppresses GSK-3β ubiquitination mediated by UHRF1. The manipulation of TRIB3 expression did not affect the expression of UHRF1 (Fig. S4F). However, the overexpression of TRIB3 disturbed the association of UHRF1 with GSK-3β (Fig. S4G). Immunofluorescence and co-IP assays confirmed that both UHRF1 and TRIB3 could interact with GSK-3β (Supporting Information Fig. S5A‒D). Furthermore, we constructed a series of GSK-3β deletion mutants to confirm their interaction domains. Both TRIB3 and UHRF1 interacted with the N-terminal domain of GSK-3β (Fig. S5E and F). Using TRIB3 deletion mutants, we found that each domain could interact with GSK-3β (Fig. S5G). Hence, the TRIB3‒GSK-3β interaction could interfere with the binding of GSK-3β to UHRF1, likely due to TRIB3 occupying the UHRF1-interacting domain in GSK-3β.
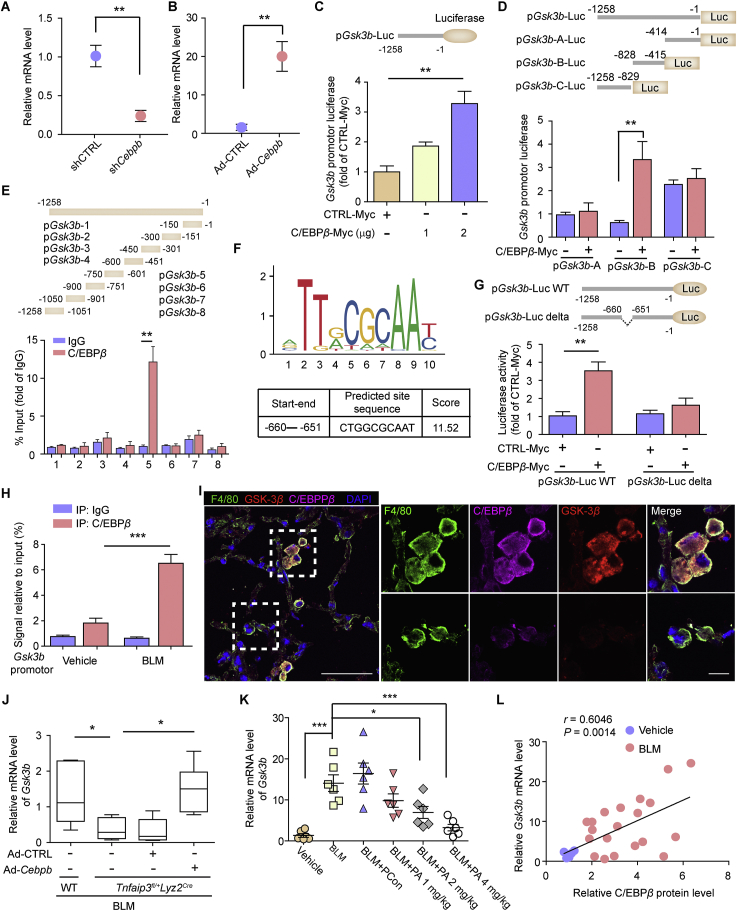
3.4. TRIB3 and GSK-3β transcription elevation induced by C/EBPβ establishes a positive feedback loop in AMs
We have reported that the mRNA levels of Gsk3b, protein coding gene of GSK-3β, are higher in AMs from PF mice than in AMs from control mice8. The increase in TRIB3 expression enhanced the expression of the transcription factor C/EBPβ (Fig. 3D). We thus suspected that the increase in C/EBPβ levels might enhance Gsk3b mRNA levels. Indeed, the ectopic expression of Cebpb, protein coding gene of C/EBPβ, increased the mRNA level of Gsk3b, while the depletion of Cebpb decreased the mRNA level of Gsk3b, suggesting that Gsk3b is a target gene of C/EBPβ (Fig. 5A and B). The overexpression of Cebpb increased Gsk3b promoter-driven luciferase activity (Fig. 5C). Additionally, C/EBPβ increased the transcription of Gsk3b by binding to the B region of the Gsk3b promoter (Fig. 5D). ChIP analysis of C/EBPβ-myc was performed in Cebpb-overexpressing cells, followed by RT-PCR analysis of the Gsk3b promoter. We found that the ChIP products obtained with C/EBPβ-myc were enriched in one conserved promoter region of Gsk3b (−750 to −601), which contained one putative C/EBPβ binding site (Fig. 5E and F). The deletion of this motif prevented the transcription increase induced by Cebpb overexpression (Fig. 5G). Moreover, the amount of C/EBPβ in the Gsk3b promoter region was more remarkable in AMs from PF mice than in AMs from control mice (Fig. 5H). We also found higher Gsk3b levels in AMs with elevated C/EBPβ expression than those with low C/EBPβ levels (Fig. 5I). The in vivo overexpression of Cebpb reversed the reduction in Gsk3b mRNA expression caused by A20 overexpression (Fig. 5J). Moreover, PA, a peptide that shows potent therapeutic efficacy against experimental PF models by accelerating C/EBPβ degradation, also reduced the mRNA expression of Gsk3b in AMs in a dose-dependent manner (Fig. 5K). Notably, a positive correlation between Gsk3b mRNA levels and C/EBPβ protein levels was found in AMs from both control and PF mice (Fig. 5L). Altogether, these findings indicate that the enhanced expression of C/EBPβ in AMs activates GSK-3β transcription during PF.
Figure 5.
C/EBPβ accumulation in profibrotic AMs increases Gsk3b transcription. (A) Quantitative analysis of Gsk3b mRNA expression levels in Cebpb-silenced AMs from PF mice (n = 3). (B) RT-PCR analysis of Gsk3b mRNA levels in Cebpb-overexpressing AMs from control mice (n = 3). (C) Relative expression of Gsk3b promoter-driven luciferase reporters in Cebpb-overexpressing cells (n = 3). (D) Relative expression of the truncated Gsk3b promoter-driven luciferase reporters in Cebpb-overexpressing cells. (E) ChIP assay of Cebpb binding to the Gsk3b promoter (n = 3). (F) Canonical C/EBPβ binding motif in the Gsk3b promoter obtained from JASPAR. (G) The relative expression of the truncated Gsk3b promoter-driven luciferase reporters in Cebpb-overexpressing cells (n = 3). (H) ChIP assay of C/EBPβ binding to the Gsk3b promoter in lung tissues from PBS- and BLM- challenged mice (n = 3). (I) Co-staining of GSK-3β and C/EBPβ in AMs from BLM-challenged mice. Scale bars: 50 μm for the large view and 10 μm for the small view. (J) Quantitative mRNA analysis of Gsk3b expression levels in AMs from the indicated mice. (K) Quantitative mRNA analysis of Gsk3b expression levels in AMs in the presence of PA. (L) Correlation analysis of the Gsk3b mRNA level and C/EBPβ protein level in AMs from PBS- or BLM-challenged mice. (A–E, G–I) Representatives of three individual experiments. (J, K) An independent repeat of two experiments. Data are represented as mean ± SEM (n = 6 mice per group, one-way ANOVA). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Given that TRIB3 is reported to be a target gene of C/EBPβ15, we examined whether C/EBPβ regulates TRIB3 expression in AMs. Indeed, the overexpression of Cebpb increased the mRNA level of Trib3 in AMs, and depletion of Cebpb decreased the Trib3 mRNA level (Supporting Information Fig. S6A and B). The promoter-luciferase activity of Trib3 was reduced in Cebpb-depleted cells (Fig. S6C). Additionally, the amount of C/EBPβ in the promoter region of Trib3 was higher in AMs from PF mice than in AMs from control mice (Fig. S6D). The mRNA level of Trib3 was reduced in AMs overexpressing A20 but was enhanced upon Cebpb overexpression (Fig. S6E). Moreover, the peptide PA reduced the mRNA expression of Trib3 in a dose-dependent manner in PF mice (Fig. S6F). The mRNA level of Trib3 was positively correlated with the protein level of C/EBPβ in AMs from both control and PF mice, suggesting that C/EBPβ transcriptionally induced an increase in TRIB3 expression in AMs during PF (Fig. S6G). These observations indicate that positive feedback mechanisms mediated by the TRIB3‒GSK-3β‒A20‒C/EBPβ axis support the profibrotic phenotype of AMs. TRIB3 increases GSK-3β protein levels by reducing GSK-3β degradation, and GSK-3β interacts with A20 to increase the expression of C/EBPβ, which in turn activates GSK-3β and TRIB3 transcription.
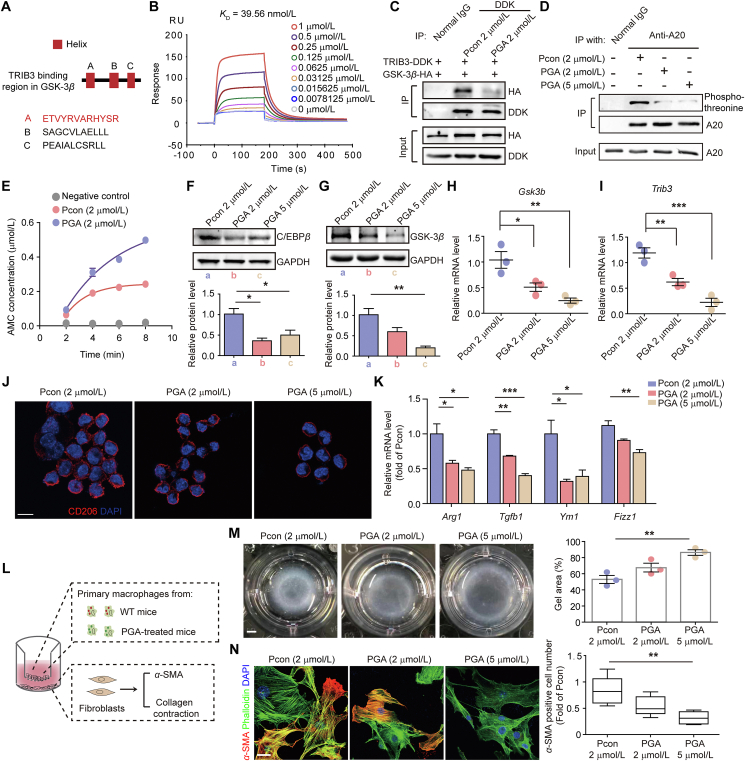
3.5. Disturbing the TRIB3‒GSK-3β interaction reduces PF development
Given that the TRIB3‒GSK-3β interaction inhibits A20 activity and stabilizes C/EBPβ to induce macrophage activation in PF, the disruption of this interaction could serve as a potential strategy for treating PF. The M1 domain of GSK-3β was screened on the I-TASSER server to estimate its secondary structure. Three α-helical peptides (GA, GB, and GC) were identified, among which only the GA peptide exhibited a high affinity for TRIB3 (KD = 39.56 nmol/L, Fig. 6A and B). To verify whether the GA peptide disrupts the TRIB3–GSK-3β interaction in AMs, a cell-penetrating peptide was incorporated into GA through a glycine–glycine linker to synthesize PGA. Pcon, the control peptide, was prepared by incorporating Pep2 into the GB peptide. PGA disrupted the TRIB3‒GSK-3β interaction (Fig. 6C), suppressed A20 phosphorylation (Fig. 6D), and restored the activity of A20 in AMs from PF mice (Fig. 6E). We also found that PGA increased the UHRF1/GSK-3β interaction (Supporting Information Fig. S7A) but showed no effect on the GSK-3β‒A20 interaction (Fig. S7B). The expression of C/EBPβ, the substrate of A20, was decreased in response to PGA (Fig. 6F). The protein and mRNA expression of GSK-3β was impaired in AMs treated with PGA (Fig. 6G and H). Additionally, PGA treatment reduced the mRNA expression of Trib3 (Fig. 6I), and PGA decreased the expression of CD206 (Fig. 6J), and profibrotic phenotype-associated genes (Fig. 6K) in AMs from PF mice. Consequently, PGA inhibited the proliferation and contraction of primary lung fibroblasts (Fig. 6L‒N). These data indicate that PGA suppresses the profibrotic phenotype of AMs by restoring the activity of A20 and reducing the expression of C/EBPβ.
Figure 6.
Disruption of the TRIB3‒GSK-3β interaction suppresses profibrotic AMs by restoring A20 activity. (A) I-TASSER server for analyzing the secondary structures of TRIB3-binding regions in GSK-3β and the amino acid sequences of the helical peptides in GSK-3β. (B) Surface plasmon resonance for analyzing the kinetic interaction between TRIB3 and the α-helical peptide GA. (C) IP assay for determining the interaction between TRIB3 and GSK-3β in the presence of PGA. (D) IP assay for evaluating A20 phosphorylation in the presence of PGA. (E) A20 activity was measured in AMs treated with PGA. (F) Immunoblot analysis of C/EBPβ protein levels in AMs after PGA treatment. The loading control was GAPDH (n = 3). (G) Immunoblot analysis of GSK-3β protein levels in AMs after PGA treatment. The loading control was GAPDH (n = 3). (H) Quantitative mRNA analysis of Gsk3b expression in AMs from PF mice after PGA treatment (n = 3). (I) Quantitative mRNA analysis of Trib3 expression in AMs from PF mice after PGA treatment (n = 3). (J) Immunostaining revealing the expression of CD206 in PGA-treated AMs. (K) Quantitative mRNA analysis of the target genes in AMs from PF mice after different treatments (n = 3). (L) The experimental scheme for determining the cross-talk between AMs from PF mice and primary lung fibroblasts. (M, N) The antifibrotic effect of PGA was verified based on α-SMA expression in fibroblasts (M, n = 3) and fibroblast contraction in 3D collagen matrices (N, n = 4). (B–K, M, and N) Representatives of 2–4 experiments. Data are represented as mean ± SEM (n = 3–4). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bar: 8 μm (J), 2 mm (M), and 50 μm (N).
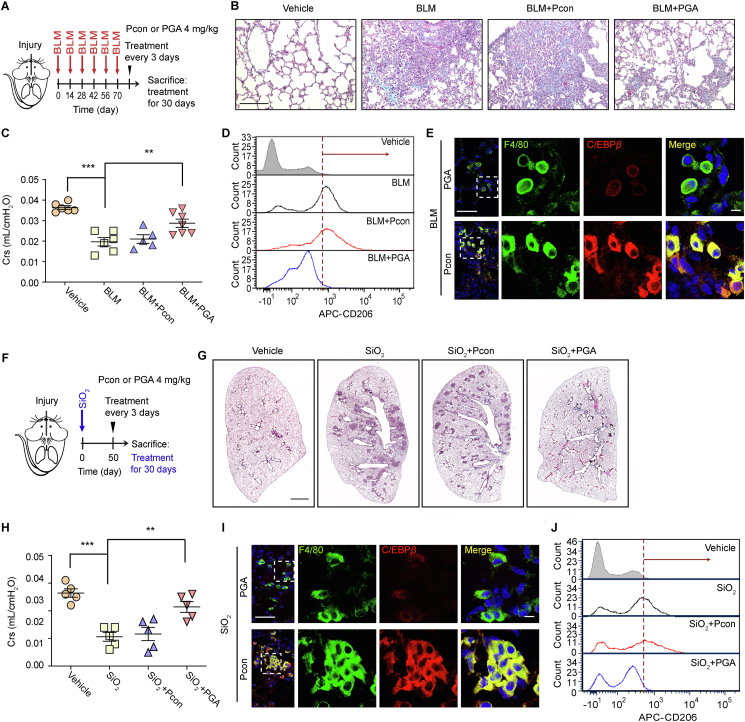
To determine whether PGA can ameliorate PF in vivo, the mice were exposed to PGA or Pcon on Day 10 after the BLM challenge (Fig. 7A). PGA reduced collagen deposition (Fig. 7B) and improved lung function (Fig. 7C). Moreover, PGA decreased the expression of CD206 (Fig. 7D), C/EBPβ (Fig. 7E), and GSK-3β (Fig. S7C) in AMs from fibrotic lung tissue. The mRNA levels of Trib3 and Gsk3b were also decreased in AMs from PGA-treated mice (Fig. S7D and E). A similar antifibrotic effect was observed in PGA-treated SiO2 mice (Fig. 7F), as revealed by a decrease in silicotic nodules in the lungs (Fig. 7G) and improved lung function (Fig. 7H) in these mice. Additionally, the expression of C/EBPβ and CD206 was suppressed in mice treated with PGA (Fig. 7I and J). In addition, combined treatment with low doses of PGA and PA, the peptide that exhibited a therapeutic effect on PF by disturbing the GSK-3β‒A20 interaction in AMs, exerted a potent inhibitory effect on BLM-induced PF, as revealed by reduced collagen deposition (Fig. S7F) and recovered lung function (Fig. S7G). Taken together, these findings imply that TRIB3 is involved in PF pathogenesis via its interaction with GSK-3β and that the peptide that disturbs this interaction is a potent therapeutic option for PF.
Figure 7.
Disruption of the TRIB3‒GSK-3β interaction attenuates lung fibrosis. (A) Schematic diagram of the elucidation of the beneficial effect of PGA in the BLM-induced PF model. All mice were euthanized with an overdose of anesthetic on Day 40 after the final BLM challenge. (B, C) Masson staining (B) and Crs (C) analyses were carried out to assess the changes in lung fibrosis. Scale bar: 200 μm. (D) Flow cytometric analysis of CD206 expression in AMs following PGA treatment. (E) Confocal image demonstrating the expression of C/EBPβ in the AMs of PGA-treated mice. Scale bars: 50 μm for the large view and 10 μm for the small view. (F) Schematic diagram of the elucidation of the beneficial effect of PGA in the silicosis model of PF. Briefly, the silicosis mouse model was constructed by intratracheally administering aqueous suspension of silica particles (125 mg/kg). Mice were administered with PGA twice a week starting from Day 50 after the SiO2 challenge. Then, all mice were euthanized with an overdose of anesthetic on Day 80 after the SiO2 challenge. (G, H) The effect of PGA on mice with SiO2-induced PF. H&E staining (G) and Crs (H) analyses were conducted to assess the fibrotic changes in the lungs of mice after SiO2 administration. Scale bar, 2 mm. (I) Confocal image demonstrating the expression of F4/80 and C/EBPβ in lung tissue from control or silicosis mice after treatment with PGA. Scale bars: 50 μm for the large view and 10 μm for the small view. (J) Flow cytometry revealing the expression of CD206 in AMs from control or silicosis mice after treatment with PGA. (B–E, G–J) Representatives of two independent experiments. Data are represented as mean ± SEM (n = 5–7 mice per group). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
TRIB3 is involved in the regulation of various biological processes that are relevant to fibrotic diseases9, 10, 11. The profibrotic roles of TRIB3 mainly occur in fibroblasts, hepatocytes, or renal mesangial cells. In the current study, TRIB3 was observed to be substantially upregulated in AMs from patients with PF. The overexpression of TRIB3 in AMs increased the expression of profibrotic and immunosuppressive genes. In contrast, the depletion of TRIB3 reduced their expression, indicating that the elevation of TRIB3 expression affects the profibrotic phenotype of AMs. Moreover, the genetic knockout of Trib3 specifically in AMs suppressed BLM-induced lung fibrotic changes. TRIB3 is regulated by diverse stresses, including hypoxia, insulin, nutrient, and endoplasmic reticulum stresses15, 16, 17, 18. Numerous transcription factors are responsible for the regulation of TRIB3 expression15,19, 20, 21. For instance, in response to endoplasmic reticulum stress, the CCAAT/enhancer-binding transcription factors CHOP and ATF4 cooperate to activate TRIB3 promoter activity21. Additionally, the activation of the transcription factor C/EBPβ by insulin increases the transcription of TRIB315,19. We recently reported that C/EBPβ expression is increased in AMs and directs the profibrotic phenotype of AMs following chronic lung injury8. Indeed, the overexpression of Cebpb increased the mRNA level of Trib3 in AMs, whereas the depletion of Cebpb decreased the Trib3 mRNA level. The abundance of C/EBPβ in the promoter region of Trib3 was higher in AMs from PF mice than in AMs from controls. Thus, elevated TRIB3 expression in AMs was transcriptionally induced by the accumulation of the transcription factor C/EBPβ. Conversely, elevated TRIB3 expression increased the protein expression of C/EBPβ by interacting with GSK-3β and inactivating A20 activity, which stabilized C/EBPβ in AMs. Our study reveals that this positive feedback loop may enhance the profibrotic effects of C/EBPβ and TRIB3 following chronic lung injury. In addition to C/EBPβ, elevated ATF4 and CHOP protein levels have also been detected in lung homogenates from patients with IPF22. Thus, further studies are needed to evaluate whether these transcription factors also contribute to the upregulation of TRIB3 in PF.
GSK-3β is involved in the regulation of many cellular activities through the phosphorylation of target proteins23, 24, 25. In recent years, GSK-3β has been shown to be play an essential role in the pathogenesis of fibrotic diseases8,26, 27, 28. However, most studies focus on the change in the kinase activity of GSK-3β under pathological conditions, which is usually controlled by its phosphorylation29. In our previous work, we found that both the activity and expression of GSK-3β were increased in AMs from PF patients, thus supporting the profibrotic phenotype of AMs by suppressing A20 activity. However, the exact mechanism by which GSK-3β is upregulated in AMs during PF is still unknown. Our findings showed that the expression of GSK-3β was upregulated at both the transcriptional and translational levels. Indeed, the mRNA level of Gsk3b was upregulated by C/EBPβ, which acted as a transcription factor and was enriched in the Gsk3b promoter region in the AMs of PF mice. The increase in the protein level of GSK-3β was also related to its increased stability in AMs. Furthermore, the proteasome inhibitor MG132, but not the autophagy inhibitor 3-MA, recovered the reduction of GSK-3β expression caused by TRIB3 depletionin AMs. This result is consistent with a prior report showing that GSK-3β is degraded via the proteasome pathway30. As previously reported, TRIB3 usually acts as a critical modulator of intracellular protein quality control systems31, 32, 33, 34. Indeed, we found that elevated TRIB3 expression in AMs increased the stability of GSK-3β by interfering with its interaction with the E3 ligase UHRF1 and thereby suppressing its degradation. Although additional E3 ligases, such as TRIM21 and TRIM25, have also been identified as enzymes mediating the ubiquitination of GSK-3β, we did not find these proteins to be involved in TRIB3-induced GSK-3β accumulation in AMs. Thus, our work provides insight into the regulatory mechanism of GSK-3β expression in AMs. Further investigation is necessary to investigate the role and expression of UHRF1 in AMs during PF development.
Previously, we found that an α-helix peptide derived from A20 that disrupts the GSK-3β–A20 interaction exerts a protective effect on PF by restoring A20 activity and accelerating C/EBPβ degradation8. Although this peptide has been verified to be effective in treating PF, it does not reduce the expression of GSK-3β which might exert a profibrotic effect by regulating other well-known downstream signals, such as NF-κB. In this study, we designed an additional peptide derived from GSK-3β that disrupted the TRIB3/GSK-3β interaction, increased the UHRF1‒GSK-3β interaction, and thereby accelerated the degradation of GSK-3β. Moreover, this peptide recovered the enzymatic activity of A20 and decreased C/EBPβ expression. Notably, the mRNA expression levels of both Trib3 and Gsk3b were reduced in AMs treated with this peptide. In fact, this peptide seems to be more effective in treating both BLM- and SiO2- challenged PF mice than the peptide derived from A208. The combination of these two peptides at lower doses displayed synergistic therapeutic effects against BLM-induced PF. Because elevated TRIB3 expression was also found in AEC2s following chronic lung injury, the questions of whether the TRIB3‒GSK-3β interaction occurs in AEC2s and whether this PPI inhibitor can interfere with these interactions to prevent PF remain to be explored. Moreover, further studies are needed to verify the target specificity and toxicity of PGA. In addition, the current study focused only on the profibrotic action of TRIB3 in AMs in the context of PF. The TRIB3–GSK-3β interaction may participate in other organ fibrosis diseases, and further investigation may thus be required to demonstrate the broad utility of this peptide.
5. Conclusions
Our study reveals that the overexpression of TRIB3 affects the profibrotic phenotype of AMs and promotes PF development by triggering the complex TRIB3‒GSK-3β‒A20‒C/EBPβ regulatory axis in AMs. These results provide insights into the pathogenesis of PF. Additionally, our study provides a proof-of-concept of a novel treatment strategy for PF by targeting the TRIB3‒GSK-3β interaction.
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2017YFA0205400), the National Natural Science Foundation of China (81530093, 81773781, 81803604, 81874316, 81622010, and 81770800), Chinese Academy of Medical Sciences Central Public-interest Scientific Institution Basal Research Fund (2018PT35004, Molecular Mechanism and Target Discovery of Metabolic Disorder and Tumorigenesis, CAMS Key Lab, China), Beijing Outstanding Young Scientist Program (BJJWZYJH01201910023028, China), and CAMS Innovation Fund for Medical Sciences (2016-I2M-3-008, China).
Author contributions
Conceptualization, Shanshan Liu, Zhuowei Hu, and Bing Cui; Methodology, Xiaoxi Lv, Xupeng Wei, Qiao Li, and Chang Liu; Investigation, Jiali Min, Fang Hua, Xiaowei Zhang, and Ke Li; Writing-original draft, Shanshan Liu; Writing-review and editing, Bing Cui and Zhuowei Hu; Visualization, Pingping Li and Yang Xiao; Funding acquisition, Shanshan Liu, Pingping Li, and Zhuowei Hu; Supervision, Zhuowei Hu and Bing Cui.
Conflicts of interest
The authors have declared no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.06.017.
Contributor Information
Shanshan Liu, Email: shanshan@imm.ac.cn.
Bing Cui, Email: cuibing@imm.ac.cn.
Appendix A Supporting information
The following is the Supplementary data to this article:
References
- 1.Putman R.K., Hatabu H., Araki T., Gudmundsson G., Gao W., Nishino M. Association between interstitial lung abnormalities and all-cause mortality. J Am Med Assoc. 2016;315:672–681. doi: 10.1001/jama.2016.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richeldi L., Collard H.R., Jones M.G. Idiopathic pulmonary fibrosis. Lancet. 2017;389:1941–1952. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 3.Wynn T.A., Vannella K.M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C., Ryan A.J., Murthy S., Carter A.B. Accelerated development of pulmonary fibrosis via Cu,Zn-superoxide dismutase-induced alternative activation of macrophages. J Biol Chem. 2013;288:20745–20757. doi: 10.1074/jbc.M112.410720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson-Casey J.L., Deshane J.S., Ryan A.J., Thannickal V.J., Carter A.B. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity. 2016;44:582–596. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosser D.M., Hamidzadeh K., Goncalves R. Macrophages and the maintenance of homeostasis. Cell Mol Immunol. 2021;18:579–587. doi: 10.1038/s41423-020-00541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spagnolo P., Maher T.M. Clinical trial research in focus: why do so many clinical trials fail in IPF?. Lancet Respir Med. 2017;5:372–374. doi: 10.1016/S2213-2600(17)30122-4. [DOI] [PubMed] [Google Scholar]
- 8.Liu S.S., Lv X.X., Liu C., Qi J., Li Y.X., Wei X.P. Targeting degradation of the transcription factor C/EBPβ reduces lung fibrosis by restoring activity of the ubiquitin-editing enzyme A20 in macrophages. Immunity. 2019;51:522–534. doi: 10.1016/j.immuni.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Tomcik M., Palumbo-Zerr K., Zerr P., Sumova B., Avouac J., Dees C. Tribbles homologue 3 stimulates canonical TGF-signalling to regulate fibroblast activation and tissue fibrosis. Ann Rheum Dis. 2016;75:609–616. doi: 10.1136/annrheumdis-2014-206234. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Sun A., Lv W., Cheng J., Lv S., Liu X. TRB3, up-regulated in kidneys of rats with type1 diabetes, mediates extracellular matrix accumulation in vivo and in vitro. Diabetes Res Clin Pract. 2014;106:101–109. doi: 10.1016/j.diabres.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X.W., Zhou J.C., Peng D., Hua F., Li K., Yu J.J. Disrupting the TRIB3–SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome-mediated HSC activation. Autophagy. 2020;16:782–796. doi: 10.1080/15548627.2019.1635383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilkenny C., Browne W., Cuthill I.C., Emerson M., Altman D.G. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborn-Heaford H.L., Murthy S., Gu L., Larson-Casey J.L., Ryan A.J., Shi L. Targeting the isoprenoid pathway to abrogate progression of pulmonary fibrosis. Free Radic Biol Med. 2015;86:47–56. doi: 10.1016/j.freeradbiomed.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv X.X., Wang X.X., Li K., Wang Z.Y., Li Z., Lv Q. Rupatadine protects against pulmonary fibrosis by attenuating PAF-mediated senescence in rodents. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du K., Ding J. Insulin regulates TRB3 and other stress-responsive gene expression through induction of C/EBP. Mol Endocrinol. 2009;23:475–485. doi: 10.1210/me.2008-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudeau J., Miranda-Saavedra D., Barton G.J., Alessi D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Prudente S., Sesti G., Pandolfi A., Andreozzi F., Consoli A., Trischitta V. The mammalian tribbles homolog TRIB3, glucose homeostasis, and cardiovascular diseases. Endocr Rev. 2012;33:526–546. doi: 10.1210/er.2011-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzer R., Dames S., Tondera D., Klippel A., Kaufmann J. TRB3 is a PI3-kinase dependent indicator for nutrient starvation. Cell Signal. 2006;18:899–909. doi: 10.1016/j.cellsig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Selim E., Frkanec J.T., Cunard R. Fibrates upregulate TRB3 in lymphocytes independent of PPAR by augmenting CCAAT/enhancer-binding protein (C/EBP) expression. Mol Immunol. 2007;44:1218–1229. doi: 10.1016/j.molimm.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Shang Y.Y., Zhong M., Zhang L.P., Guo Z.X., Wang Z.H., Zhang Y. Tribble 3, a novel oxidized low-density lipoprotein-inducible gene, is induced via the activating transcription factor 4-C/EBP homologous protein pathway. Clin Exp Pharmacol Physiol. 2010;37:51–55. doi: 10.1111/j.1440-1681.2009.05229.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van’t Wout E.F.A., Hiemstra P.S., Marciniak S.J. The integrated stress response in lung disease. Am J Respir Cell Mol Biol. 2014;50:1005–1009. doi: 10.1165/rcmb.2014-0019TR. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez C., Perez M., Avila J. GSK3β-mediated phosphorylation of the microtubule-associated protein 2C (MAP2C) prevents microtubule bundling. Eur J Cell Biol. 2000;79:252–260. doi: 10.1078/s0171-9335(04)70028-x. [DOI] [PubMed] [Google Scholar]
- 24.Wakatsuki S., Saitoh F., Araki T. ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B-dependent CRMP2 phosphorylation. Nat Cell Biol. 2011;13:1415–1423. doi: 10.1038/ncb2373. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami F., Suzuki M., Shimada N., Kagiya G., Ohta E., Tamura K. Stimulatory effect of α-synuclein on the tau-phosphorylation by GSK-3β. FEBS J. 2011;278:4895–4904. doi: 10.1111/j.1742-4658.2011.08389.x. [DOI] [PubMed] [Google Scholar]
- 26.Lal H., Ahmad F., Woodgett J., Force T. The GSK-3 family as therapeutic target for myocardial diseases. Circ Res. 2015;116:138–149. doi: 10.1161/CIRCRESAHA.116.303613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S.P., Tao S., Fields T.A., Webb S., Harris R.C., Rao R. Glycogen synthase kinase-3 inhibition attenuates fibroblast activation and development of fibrosis following renal ischemia-reperfusion in mice. Dis Model Mech. 2015;8:931–940. doi: 10.1242/dmm.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergmann C., Akhmetshina A., Dees C., Palumbo K., Zerr P., Beyer C. Inhibition of glycogen synthase kinase 3β induces dermal fibrosis by activation of the canonical Wnt pathway. Ann Rheum Dis. 2011;70:2191–2198. doi: 10.1136/ard.2010.147140. [DOI] [PubMed] [Google Scholar]
- 29.Xi Y.L., Li H.X., Chen C, Liu Y.Q., Lv H.M., Dong S.Q. Baicalin attenuates high fat diet-induced insulin resistance and ectopic fat storage in skeletal muscle, through modulating the protein kinase B/Glycogen synthase kinase 3 beta pathway. Chin J Nat Med. 2016;14:48–55. doi: 10.3724/SP.J.1009.2016.00048. [DOI] [PubMed] [Google Scholar]
- 30.Ougolkov A.V., Fernandez-Zapico M.E., Bilim V.N., Smyrk T.C., Chari S.T., Billadeau D.D. Aberrant nuclear accumulation of glycogen synthase kinase-3β in human pancreatic cancer: association with kinase activity and tumor dedifferentiation. Clin Cancer Res. 2006;12:5074–5081. doi: 10.1158/1078-0432.CCR-06-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua F., Li K., Yu J.J., Lv X.X., Yan J., Zhang X.W. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6:7951. doi: 10.1038/ncomms8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J.M., Sun W., Wang Z.H., Liang X., Hua F., Li K. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat Commun. 2019;10:5720. doi: 10.1038/s41467-019-13700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li K., Wang F., Cao W.B., Lv X.X., Hua F., Cui B. TRIB3 promotes APL progression through stabilization of the oncoprotein PML-RARα and inhibition of p53-mediated senescence. Cancer Cell. 2017;31:697–710. doi: 10.1016/j.ccell.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Li K., Wang F., Yang Z.N., Zhang T.T., Yuan Y.F., Zhao C.X. TRIB3 promotes MYC-associated lymphoma development through suppression of UBE3B-mediated MYC degradation. Nat Commun. 2020;11:6316. doi: 10.1038/s41467-020-20107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.