ABSTRACT
Dietary shifts can have a direct impact on the gut microbiome by preferentially selecting for microbes capable of utilizing the various dietary nutrients. The intake of dietary fiber has decreased precipitously in the last century, while consumption of processed foods has increased. Fiber, or microbiota-accessible carbohydrates (MACs), persist in the digestive tract and can be metabolized by specific bacteria encoding fiber-degrading enzymes. The digestion of MACs results in the accumulation of short-chain fatty acids (SCFAs) and other metabolic by-products that are critical to human health. Here, we implemented a 2-week dietary fiber intervention aiming for 40 to 50 g of fiber per day within the context of a course-based undergraduate research experience (CURE) (n = 20). By coupling shotgun metagenomic sequencing and targeted gas chromatography-mass spectrometry (GC-MS), we found that the dietary intervention significantly altered the composition of individual gut microbiomes, accounting for 8.3% of the longitudinal variability within subjects. Notably, microbial taxa that increased in relative abundance as a result of the diet change included known MAC degraders (i.e., Bifidobacterium and Lactobacillus). We further assessed the genetic diversity within Bifidobacterium, assayed by amplification of the groEL gene. Concomitant with microbial composition changes, we show an increase in the abundance of genes involved in inositol degradation. Despite these changes in gut microbiome composition, we did not detect a consistent shift in SCFA abundance. Collectively, our results demonstrate that on a short-term timescale of 2 weeks, increased fiber intake can induce compositional changes of the gut microbiome, including an increase in MAC-degrading bacteria.
IMPORTANCE A profound decrease in the consumption of dietary fiber in many parts of the world in the last century may be associated with the increasing prevalence of type II diabetes, colon cancer, and other health problems. A typical U.S. diet includes about ∼15 g of fiber per day, far less fiber than the daily recommended allowance. Changes in dietary fiber intake affect human health not only through the uptake of nutrients directly but also indirectly through changes in the microbial community and their associated metabolism. Here, we conducted a 2-week diet intervention in healthy young adults to investigate the impact of fiber consumption on the gut microbiome. Participants increased their average fiber consumption by 25 g/day on average for 2 weeks. The high-fiber diet intervention altered the gut microbiome of the study participants, including increases in known fiber-degrading microbes, such as Bifidobacterium and Lactobacillus.
KEYWORDS: diet intervention, fiber, GroEL, microbiome, metagenomics
INTRODUCTION
The consumption of dietary fiber has declined dramatically in the last century as processed foods have become a larger part of diets in the industrialized world. Preindustrial and modern-day rural societies consume between 60 and 120 g/day fiber, while individuals in the United States consume about half of the daily recommended allowance of 38 g/day for men and 25 g/day for women (1, 2). Declines in fiber intake over the past century have contributed to complications for human health. For example, chronic low-fiber intake has been associated with type 2 diabetes mellitus, heart disease, and colon cancer (3–5). Indeed, a reciprocal diet intervention exchanging African Americans’ low-fiber western diet with rural Africans’ high-fiber diet (increasing, on average, 40 g per day) led to significant decreases in precancerous biomarkers, further providing a link between fiber and human health (6). Furthermore, dietary fiber has been shown to protect against influenza infection (7) and may influence vaccine efficacy (8).
Dietary fiber is a mixture of polysaccharides that resist rapid digestion in the small intestine by endogenous enzymes and persists through the digestive tract into the colon. Once in the colon, fiber can be digested by the resident microbes (1, 9). This is due, in part, to the human genome encoding only 17 enzymes (i.e., glycoside hydrolases) that are capable of digesting carbohydrates (10). Conversely, the resident gut microbial communities collectively encode thousands of diverse enzymes from 152 gene families that can break down dietary fiber (11). In the colon, specialized microbes metabolize recalcitrant carbohydrates and produce fermented by-products, including short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate (12). SCFAs are capable of being absorbed across the human intestinal epithelial cells and have direct impacts on human health (reviewed in reference 13), such as stimulating and maintaining the mucus layer for the gut epithelium (14) and providing an energy source for butyrate-consuming colonocytes (15). SCFAs have also been shown to have immunomodulatory effects, including increased viral protection through altered T-cell metabolism (7) and inhibitory effects on pathogenic bacteria (e.g., Clostridioides difficile) (16).
Understanding the role of dietary fiber in structuring the gut microbiota could provide insights into managing chronic diseases associated with the gut microbiome. Typical diet intervention studies assessing the impact of fiber on gut microbial communities and the production of SCFAs have relied on single fiber supplements (17–19, 89). Fiber supplements such as psyllium husks, inulin, wheat bran, resistant potato starch, and resistant corn starch vary in their efficacy for each individual (17, 20). Individuals might be more or less susceptible to the intervention depending on their initial resident microbial community and its ability to digest a particular fiber supplement. For example, one group investigating the impact of three fermentable fibers on gut microbiome composition and SCFA abundance found no significant effect when study participants consumed 20 to 24 g resistant maize starch per day for 2 weeks (17). However, in addition to the quantity, the variety of dietary fibers may be important. Studies that have increased dietary fiber have previously observed changes in microbiome composition (3, 17, 18), yet results remain mixed on SCFA production (6, 21, 22, 89). Further, the American Gut Project found that individuals who eat more than 30 types of plants in a week have a more diverse gut microbiome (23). Thus, the consumption of a diversity of fiber sources through whole foods may provide more opportunities for an individual’s gut microbiome to respond to the dietary changes and result in more dramatic changes in fiber-degrader abundance and activity in the gut microbiome. The increase of fiber from a diverse set of dietary foods, rather than single fiber supplements, may also contribute to the increased consumption of other micronutrients and vitamins that affect the microbiome as well (24).
In this study, we sought to answer three questions: (i) does a diet rich in fiber from whole foods alter the overall microbiome, (ii) does the intervention alter the abundance and diversity of known fiber degraders (e.g., Bifidobacterium), and (iii) if we observe compositional shifts in the microbiome, do these correspond with metabolic changes in the production of short-chain fatty acids? To address these questions, we developed and employed a course-based undergraduate research experience (CURE) at UC Irvine to assess individual responses to a high-fiber diet (25). Integrating authentic research experiences within laboratory courses to facilitate a deeper understanding of academic and industrial research continues to be a priority for both national education reform and the American Society for Microbiology (25–28). During the intervention, participants were given 10 meals each week from a food service that specializes in providing high-fiber, unprocessed meals. Individuals tracked dietary information of macronutrients for every meal for 3 weeks, with the goal of increasing dietary fiber intake to 50 g/day during a 2-week intervention period. We then compared overall bacterial composition using metagenomic sequencing and assessed the production of volatile SCFAs using mass spectrometry. In addition to the shotgun metagenomic sequencing, we targeted a known fiber degrader, Bifidobacterium, by analyzing its diversity using amplicon sequencing of the groEL marker gene, enabling a unique high-resolution view of the impact of a dietary fiber intervention on a key taxon.
RESULTS
Dietary intervention within the CURE course.
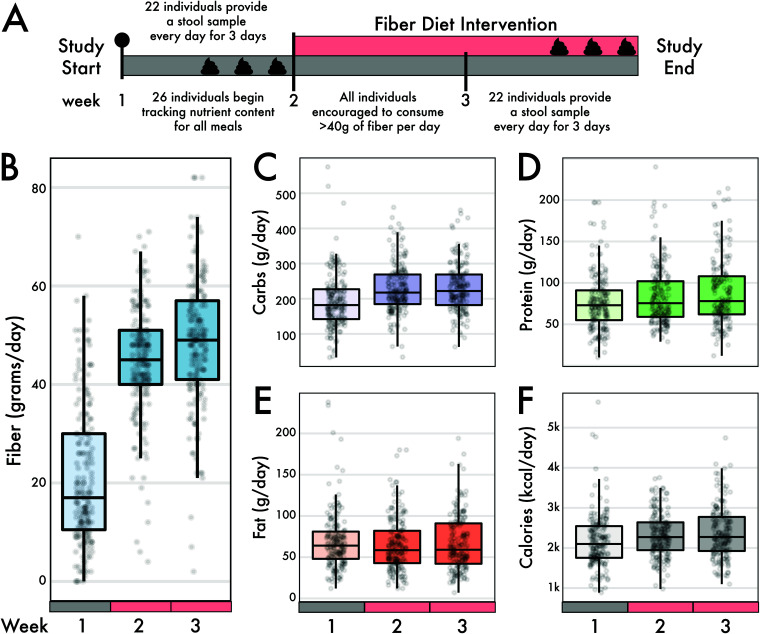
Twenty-six individuals participated in a CURE course at UC Irvine, designed to tandemly investigate pedagogical methods (25) and the role of fiber in the microbiome. We collected nutritional data over 3 weeks from all 26 individuals who initially began the intervention (1 week before and 2 weeks during the dietary intervention) (Fig. 1A). We collated the total amount of macronutrients consumed per day, including fiber, protein, carbohydrates, fats, as well as overall calories (Fig. 1B to F). Additionally, we informally surveyed food items the study participants frequently used to supplement their meal plans, beyond the meals supplied from Thistle, and found that items such as fiber-fortified cereals, lentils or beans, and berries were common (25). For the intervention, subjects increased their average fiber consumption from 21.0 g/day (± 14.2 g/day) before the intervention to 46.4 g/day (± 12.5 g/day) during the intervention (Wilcoxon rank sum test, P < 0.0001) (Fig. 1B). While these dietary shifts increased carbohydrate intake by an average of 84% (36 g) during the intervention (P = 0.013), other macronutrients measured, such as calories, fat, and proteins, did not significantly change (P > 0.05) (Fig. 1C to F).
FIG 1.
Intervention timeline and sample collection. The study subjects began eating their normal diet for 1 week, tracking all their food intake using the MyFitnessPal app. At the end of the week, each individual provided a daily fecal sample on three different days. At the start of week two, subjects started a wholesome high-fiber diet, getting at least 40 g of fiber per day. During week three, subjects were encouraged to get 50 or more grams of fiber per day. At the end of week three, each subject provided a fecal sample on three different days. (B to F) Self-reported macronutrients from individuals using the MyFitnessPal app. Change in macronutrients across the 3-week diet intervention for fiber (B), carbohydrates (C), protein (D), fat (E), and calories (F). Fiber changed the most in magnitude between preintervention intake and during the diet intervention (linear mixed-effects model, P < 0.001). There were modest but significant changes in carbohydrate, protein, and caloric intake, but not fat intake, across the same time interval.
Diet intervention altered gut microbial community composition within individuals.
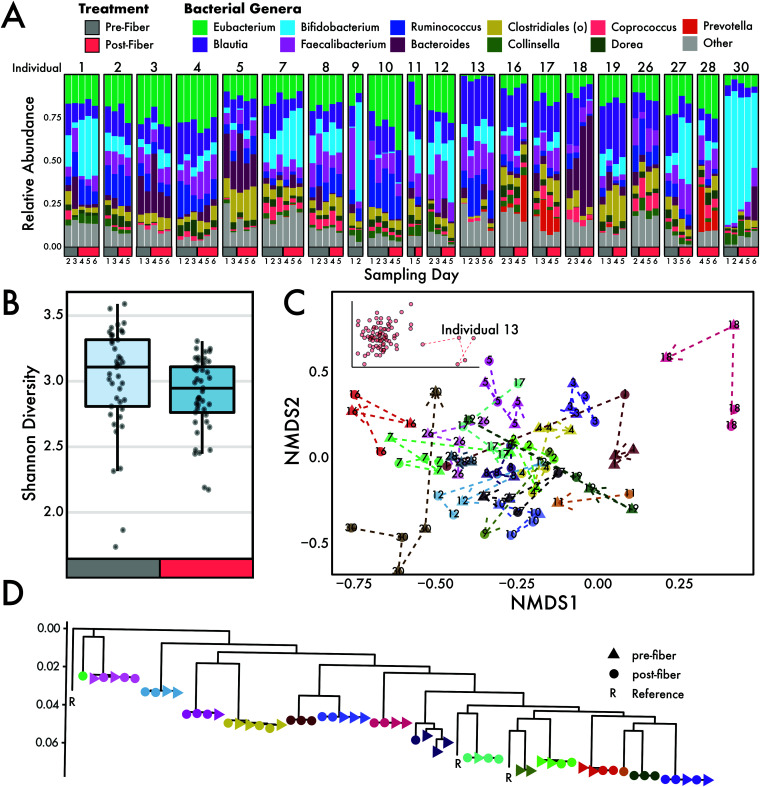
To evaluate whether increased fiber consumption contributed to shifts in the gut microbiome, we characterized the microbial communities from 20 individuals using 86 shotgun metagenomic libraries collected before and after the fiber intervention (Fig. 2A). The alpha diversity of microbial taxa decreased during the high-fiber diet intervention, as measured by the Shannon diversity index (Fig. 2B) (Wilcoxon rank sum test, P < 0.05). Using alternative approaches to assess taxonomy and diversity (see Materials and Methods) showed either no change or supported the decreasing trend of diversity during the intervention period (Fig. S1).
FIG 2.
Microbiome community composition through a dietary fiber intervention. (A) Relative abundances of genera detected in microbiomes from individuals throughout the diet intervention study. (B) Alpha diversity, measured using the Shannon index, changed significantly during the intervention period (Wilcoxon, P < 0.05). (C) NMDS ordination showed that samples from individuals mostly group together. Dotted lines connect the same individual and point toward the final postfiber intervention sample. Samples in this study were highly personalized: the individual explained 78% of the variation in the data. The inset shows an extended version of the NMDS plot that includes individual 13. (D) A phylogeny of Eubacterium rectale strains found in individuals (denoted by color) during the intervention.
Comparisons of diversity measures obtained using different databases for taxonomic assignments. IGG_rich and MIDAS were run using default parameters. IGG-Lenient was run at 25% species quality and 15% marker genes. Download FIG S1, PDF file, 0.1 MB (107.7KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Despite little difference in alpha diversity, beta diversity changed significantly in response to a high-fiber diet. Multivariate analysis of marker gene abundances showed that most of the variation in microbiome composition could be explained by the individual (permutational analysis of variance [PERMANOVA] for main individual effect, R2 = 0.78, P < 0.001) (see Table S2 in the supplemental material). The diet intervention shifted the microbial composition of the entire study cohort significantly (main intervention effect, R2 = 0.014, P < 0.001). Within samples from each individual, the pre- and postdiet intervention samples explain significant variation in the community composition (intervention-by-individual effect, R2 = 0.083, P < 0.001). A linear mixed-effects (LME) model confirmed these results, which identified diet as a significant determinant of an individual’s microbiome composition (LME, P < 0.01). Individual gut microbiome samples grouped together in nonmetric multidimensional space (NMDS; Fig. 2C), further providing support that each individual is associated with a unique microbiome. Some individual gut microbiomes (i.e., individual 13) were more distinct from others (Fig. 2C, inset). Additionally, we used Eubacterium rectale (due to its high coverage in our data) to ask whether the diet intervention had an impact at the strain level. Strains were highly individual specific (PERMANOVA main individual effect, R2 = 0.99, P < 0.001) and did not change in response to increased fiber intake (P > 0.05; Fig. 2D).
PERMANOVA model and term results. Download Table S2, DOCX file, 0.01 MB (13.4KB, docx) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
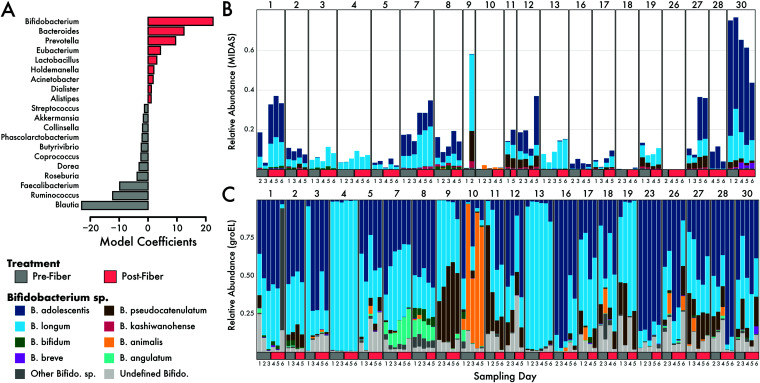
We next parsed the taxonomic data to assess which microbial taxa increased or decreased in response to the diet intervention. One species in the family Lachnospiraceae was significantly negatively associated with increasing fiber intake (Spearman r = −0.43, q = 0.01) (Fig. S2A and B). Coprococcus sp. and Anaerostipes hadrus were both positively associated with increasing fiber intake, but this association was not significant when P values were false discovery rate (FDR) corrected for multiple comparisons (r = 0.32, q = 0.33 for both species) (Fig. S2A). Furthermore, positive linear coefficients of a PERMANOVA model, which detect differences between community compositions due to the diet intervention, included genera such as Bifidobacterium, Bacteroides, and Prevotella (Fig. 3A). Conversely, Blautia and Ruminococcus contributed negative linear coefficients to the PERMANOVA model (Fig. 3A).
FIG 3.
GroEL amplicon analysis of Bifidobacterium during the fiber intervention. (A) Model coefficients of the PERMANOVA analysis (model: species ∼ individual × intervention). Species with high coefficients (positive or negative) were best able to distinguish the pre- versus postdiet intervention groups. Only the top 20 genera are shown. The genus Bifidobacterium had the largest positive coefficient, indicating that it was important to the model for disguising microbiomes before and after the diet intervention. (B and C) Relative abundances of 12 detected species of Bifidobacterium from shotgun metagenomics (B) and groEL amplicon sequencing (C).
(A) Correlations above an r cutoff of >0.2 for microbial abundance and fiber intake. Only Lachnospiraceae bacterium 51870 was significantly negatively correlated at an FDR cutoff of 0.05. (B) Raw spearman correlation of a species of Lachnospiraceae with fiber intake. Download FIG S2, PDF file, 0.05 MB (48.5KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bifidobacterium species were enriched by the diet intervention.
Of the 105 microbial genera detected in this study, Bifidobacterium was the strongest predictor genus for the postintervention microbiomes (Fig. 3A). Indeed, taxonomic analysis of the metagenomic samples identified Bifidobacterium abundances increasing, on average, 1.4-fold between the pre- and postintervention periods (Fig. S3A). Further, we identified several species of Bifidobacterium present within and across individuals, with B. adolescentis being the most abundant species on average (Fig. 3B). When we investigated the taxonomic profiles at the species level, we found that B. adolescentis, B. biavatii, B. breve, B. longum, and B. ruminantium all increased in mean abundance on a high-fiber diet, whereas the other, less abundant species exhibited no change or decreased in abundance (Fig. S3B).
(A) Mean abundance (MIDAS read counts) of the genus Bifidobacterium during the diet intervention. Points are colored by individual. (B) Changes in mean abundance of each species of Bifidobacterium, detected by MIDAS, during the diet intervention period. Download FIG S3, PDF file, 0.10 MB (99KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Given that Bifidobacterium was the strongest predictor genus in the postfiber gut microbiomes, we employed a targeted analysis of the diversity within Bifidobacterium to examine species-level patterns. Specifically, we applied targeted amplicon approaches to amplify the groEL gene, a conserved phylogenetic marker gene to track Bifidobacterium diversity (Fig. S4). Using phylogenetic inference of the groEL gene, we compared the observed Bifidobacterium diversity observed at the community level to our targeted analysis of the groEL gene. Similar to the metagenomic analysis, we found that individuals were largely comprised of B. adolescentis and B. longum, with six other abundant species of Bifidobacterium (Fig. 3C). This analysis also revealed extensive Bifidobacterium diversity within the human gut, detecting 22 species across all individuals.
Bifidobacterium phylogenetic analyses. (A) Multilocus phylogenetic analysis of conserved ribosomal marker genes. (B) Phylogenetic analysis of the groEL gene sequences used for amplicon analyses. The top 8 species observed are shown. Download FIG S4, PDF file, 0.9 MB (925.9KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Since Bifidobacterium species are known to participate in cross-feeding with other gut microbes (reviewed in reference 29), we next assessed the cooccurrence of Bifidobacterium with other genera. Bifidobacterium was positively correlated (r = 0.43, q = 0.001) with an increasing abundance of Lactobacillus and negatively correlated with Roseburia (r = −0.49, q = 0.0002) and Ruminococcus (r = −0.38, q = 0.007) (Fig. S5), suggesting species interactions between these taxa.
Significant (FDR, 0.05) correlations from comparing abundances of 99 different genera with Bifidobacterium. Download FIG S5, PDF file, 0.02 MB (19.1KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes involved in inositol degradation increase on high-fiber diet.
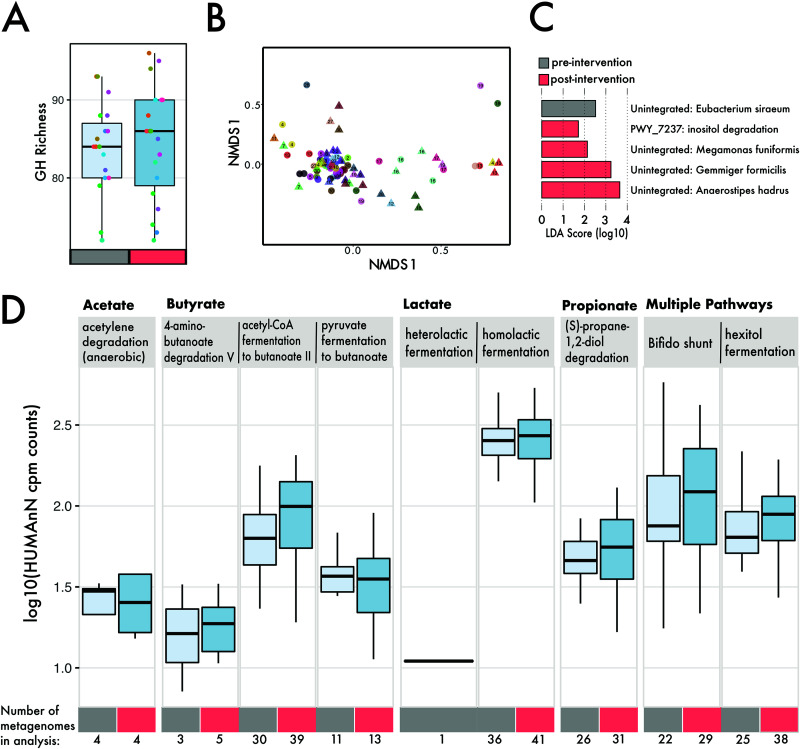
Our results demonstrate that a shift in dietary fiber consumption influenced compositional changes in the gut microbial community. As such, we sought to correlate the observed taxonomic shifts to functional shifts, particularly the enrichment of genes related to carbohydrate degradation. Despite taxonomic shifts at the individual level, we observed no changes in the overall abundance (average number of normalized reads) mapping to gene families for glycoside hydrolases (GH) (Wilcoxon, P = 0.42), carbohydrate esterases (P = 0.58), glycoside transferases (P = 0.73), and polysaccharide lyases (P = 0.77) as a result of the intervention (Fig. S6A). No individual families of GH and polysaccharide lyase CAZy classes changed in abundance during the intervention when corrected for multiple comparisons (Wilcoxon, P > 0.05) (Fig. S6B). Further, the diversity (Fig. 4A) (n = 106 families; ANOVA, P > 0.05) and composition (PERMANOVA, P > 0.05) of GHs detected were indistinguishable between the pre- and postintervention samples. Compositional analysis of all genes identified by HUMAnN revealed no individual signature (PERMANOVA main individual effect, R2 = 0.017, P > 0.05) (Fig. 4B) and no shifts in response to a high-fiber diet (intervention by individual effect, R2 = 0.015, P > 0.05). We performed a linear discriminant analysis to determine if there were pathways that were differentially abundant due to the diet intervention and found inositol degradation (in addition to several unintegrated pathways) to be increased in abundance on a high-fiber diet (Fig. 4C, Fig. S7). For the pathways involved in SCFA metabolism, we found no significant (Wilcoxon, P > 0.05) changes as a result of the high-fiber diet (Fig. 4D).
FIG 4.
Genes involved in carbohydrate degradation and SCFA metabolism within metagenomes. (A) Number of distinct glycoside hydrolase families within individual metagenomes (different-colored circles), separated by preintervention (mean, 83) (gray) and postintervention (mean, 84) (red). (B) NMDS ordination of Euclidean distance matrix based on 19,680 gene features. Shape denotes intervention (triangle, preintervention, circles, postintervention), and individuals are separated based on color. (C) Lefse analysis of pathways that differentiate samples by intervention. (D) Log abundance (copies per million) of pathways involved in SCFA production.
(A) Average abundances (normalized reads) of carbohydrate active enzymes within individuals during the diet intervention period. Each different color point represents an individual, and lines connect the same individual. (B) Log2-transformed GH and polysaccharide lyase gene abundances during the intervention. Download FIG S6, PDF file, 0.2 MB (229.6KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inositol degradation abundance (normalized copies per million) for metagenomes before and after the intervention. Download FIG S7, PDF file, 0.08 MB (83.3KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fecal short-chain fatty acid concentrations were unaltered by the diet intervention.
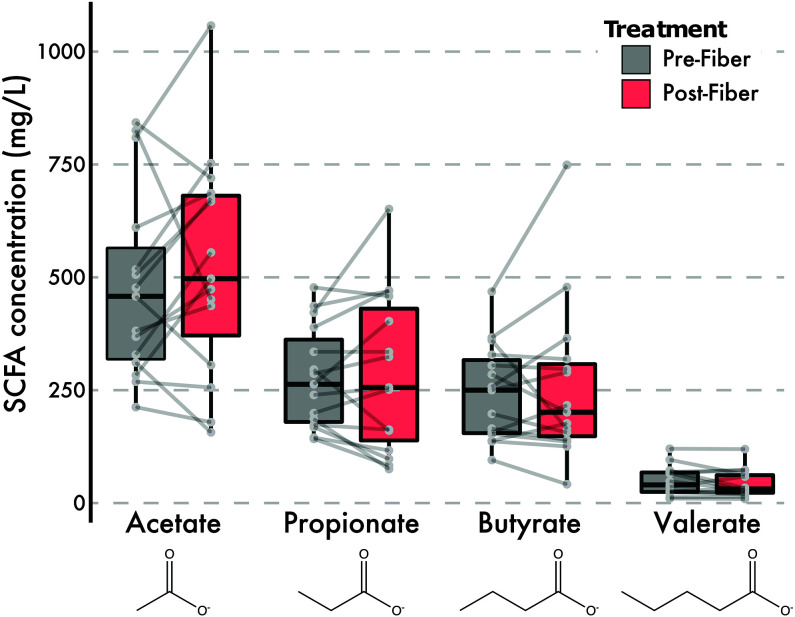
While the presence of genes related to SCFA production provides insights into the functional changes of the microbiome, these genes only reflect the genomic potential to process these pathways. Therefore, we applied a targeted gas chromatography-mass spectrometry (GC-MS) analysis to 149 samples from 18 individuals for the presence of SCFA molecules. Across the intervention period, the average abundance of acetate, propionate, butyrate, and valerate increased (Table 1); however, these increases were not statistically significant (LME, P > 0.05) (Fig. 5). For the eight individuals with samples run in duplicate, three had biological differences that were greater than the technical variation seen in the duplicates (Fig. S8). Acetate had the least technical variation (mean coefficient of variation [CV], 45%), followed by propionate (mean CV, 51%) (Table 1, Fig. S8).
TABLE 1.
Abundances of four SCFAs in samples pre- and postintervention
| SCFA | Preintervention abundance (mg/liter) |
Postintervention abundance (mg/liter) |
||||
|---|---|---|---|---|---|---|
| Mean | SD | CV (%) | Mean | SD | CV (%) | |
| Acetate | 480 | 208 | 43 | 525 | 244 | 47 |
| Propionate | 277 | 112 | 41 | 288 | 175 | 61 |
| Butyrate | 244 | 107 | 44 | 257 | 176 | 68 |
| Valerate | 49 | 31 | 65 | 43 | 31 | 72 |
FIG 5.
GC-FID measurements of fecal volatile SCFAs during intervention. Fecal SCFA abundances, averaged across replicates where applicable, before and after the intervention are shown.
Technical variation of SCFAs seen in a subset of samples run in duplicate. (A) Amount of SCFAs by individual, with color denoting if it was measured during replicate 1 or 2. (B) Normalized (by mean) difference between absolute difference between treatment, subtracted by absolute difference between technical replicates. Larger negative values suggest differences between technical replicates were larger than the differences detected between pre- and postintervention arms. Download FIG S8, PDF file, 0.1 MB (111.6KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We examined the impact of dietary foods, rich in their diversity of fiber, on the human gut microbiome. We expected that an increase in fiber consumption through whole foods consumed would lead to a more generalizable shift of the microbiome, in contrast to previous studies that utilized a single fiber supplement. For instance, a recent meta-analysis (30) found mixed results in how fiber may impact gut microbiome richness and composition. Among papers published before October 2017, only 18% (12 out of 64) of studies (31–42) contained food-based fiber interventions, and most of these studies only modified one aspect of diet (e.g., addition of whole-grain breakfast cereal). One study increased dietary fiber by 40 g from a diverse set of foods during a 5-day period (32). The authors similarly found microbiome composition changes within individuals when they accounted for differences in the subjects’ starting microbiomes. Despite the variation in implementing a fiber intervention, it is becoming increasingly clear that fiber alters the composition of the gut microbiome (17) and the associated microbial changes affect human health (i.e., type 2 diabetes mellitus [3]). A common observation in fiber intervention studies (30) is the specific involvement of the genus Bifidobacterium in response to fiber interventions. However, to our knowledge, no study has documented how fiber impacts the genus at the strain level in the human gut.
Does a diet intervention rich in fiber alter the microbiome?
Past studies have shown that an increase in the diversity of dietary foods could lead to an increase in microbial diversity (24). Moreover, individuals living in rural societies often harbor far greater gut microbial diversity than individuals from western societies (43–45), which may in part be linked to a greater proportion of plant-based polysaccharide intake. However, we did not measure an increase in species diversity (alpha diversity) after subjects consumed >40 g of fiber from a diverse set of foods (Fig. 2B). These results could be attributed to the brevity of the intervention, as the rapid change in dietary composition may result in the loss of microbes poorly adapted to recalcitrant carbohydrates. Similarly, other studies have reported finding no increases in alpha diversity as a result of fiber intake (32, 46–49), which indicates a trade-off where fiber degraders increased while other taxa decreased. Although alpha diversity was unaffected, we did observe a significant impact of the high-fiber diet on microbial community composition (beta diversity) (Fig. 2). The composition of microbial communities within individuals shifted ∼8% during the intervention period. We found changes in communities to be at broader taxonomic levels than the strain level. We could examine strains of E. rectale due to its high coverage in our data, and we showed these strains stayed constant and individual specific during the intervention (Fig. 2D). Future work should determine if this pattern holds up for other species. While we suspect the high-fiber diet treatment played an instrumental role in shifting the microbial composition, we cannot rule out other factors, such as host genetics or nondietary behaviors. As discussed, many food-based fiber interventions have shown mixed results in changing the microbial communities (6, 50). The drastic increase in fiber from a variety of foods may lead to rapid shifts in community composition over the 2-week period. Changes in community composition pre- and postintervention were largely driven by shifts in known fiber degraders, such as Bifidobacterium, Bacteroides, and Prevotella (Fig. 3A).
We expected the taxonomic shifts in the microbiota would be associated with changes in the functional potential of the microbial communities (Fig. 4). While we initially hypothesized that a high-fiber diet would increase the abundance or diversity of carbohydrate active enzymes, we did not detect changes associated with the intervention (see Fig. S6 in the supplemental material). Our findings support a similar result showing no difference in CAZy abundance due to increased fiber intake (49). We acknowledge that sequencing depth is an important consideration in the detection of genes; increasing reads beyond our ∼1.3 million paired-end reads (average per sample; Table S1) may allow for greater detection. However, we did find a notable increase in the abundance of genes mapping to the inositol pathway (Fig. 4C). We suspect that the increased consumption of fiber-fortified cereals and legumes, which contain higher levels of inositol, during the diet intervention allowed for an expansion in organisms capable of breaking down this sugar. There is substantial interest in the role of inositol (specifically phytic acid) in its protective role against colon cancer and other metabolic disorders (51, 52). Next, we assessed whether genes involved in SCFA metabolism changed in abundance during the intervention. Although appreciable cross-feeding between lactate-producing Bifidobacterium spp. and butyrogenic bacteria has been shown (53), we did not find significant increases in genes involved in various SCFA metabolic pathways (Fig. 4D). This further supports our results showing no clear correlations between Bifidobacterium spp. and butyrate producers within our diet intervention (Fig. S5). Indeed, we would not be the first to suggest that these complex trophic interactions require more time to establish (17). Rather, our results suggest that while broad taxonomic shifts occur, these do not correspond to changes in functional potential, and fine-scale (intraspecies) shifts are less susceptible to dietary shifts on short-term timescales.
Sequence statistics, accession IDs, and experimental metadata. Download Table S1, XLSX file, 0.02 MB (25.5KB, xlsx) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Does the intervention alter the abundance and diversity of Bifidobacterium, a known fiber degrader?
Many studies have indicated that bifidobacteria (often identified as the genus Bifidobacterium by fluorescent in situ hybridization probes, PCR, or DNA sequencing) are highly abundant in the gut following increased fiber intake (meta-analysis of 51 studies [30]). The increased abundance of Bifidobacterium is somewhat unsurprising, as they harbor numerous genetic components, such as carbohydrate active enzymes, that make them especially adapted to a fiber-rich diet (54). In one study, both resistant potato starch and inulin increased the relative abundance of Bifidobacterium spp.; however, the 16S amplicon sequencing in this study did not have the resolving power to identify which species of Bifidobacterium were increasing (17). Using a targeted amplicon approach, the groEL gene has been shown to delineate species of Bifidobacterium that otherwise share >99% sequence identity in the 16S rRNA gene, making it a robust marker gene for analyzing within-genus species diversity (55). In our study, the most abundant species of Bifidobacterium were B. adolescentis and B. longum, both of which are efficient degraders of plant-based fructooligosaccharides (FOS) and produce acetate and lactate in the process (56). Mirroring our results, other studies have found selective increases in certain species of Bifidobacterium as a result of carbohydrate intake; for example, in one study, the intake of inulin resulted in a greater increase of B. adolescentis (57). We speculate that on a high-fiber diet, bifidobacteria are the initial members of the community accessing fiber substrates, easily adapted to utilize various FOS, and pivotal to the creation of the initial metabolic cross-feeding networks. Future studies should extend the intervention period to examine the dynamics of longer-term trophic interactions in response to increased dietary fiber intake.
Can we detect diet-induced changes in the abundance of fecal short-chain fatty acids?
While SCFAs did generally increase during the diet intervention, trending toward their naturally occurring gut ratio of 3:1:1 (acetate-propionate-butyrate) (12, 58, 59), we did not observe a statistically significant increase in SCFAs postintervention. Static fecal concentrations of SCFAs may not reflect the total pool of molecules fluxing through a given individual, as the molecules are preferred substrates of the cells lining the gut epithelia (15). It is also possible that the intervention period was too short to observe increases in SCFA abundances.
It should be noted that accurate SCFA measurements are notoriously difficult. Our examination of technical variability within 44 samples from eight individuals showed that technical variation between preintervention replicates or postintervention replicates was greater than the average difference between pre- and postintervention for any given SCFA. One study reported high intrafecal variability of butyrate quantification (coefficient of variation [CV], 38%) before optimizing a freeze-drying method (60). Numerous studies have indicated the benefit of SCFAs to human health (5, 61), yet the heterogeneity in reported acetate, propionate, and butyrate abundances remains high. In one meta-analysis of fiber studies, only butyrate was generally found to increase with fiber intake, yet the heterogeneity of reported results was 70% (I2), similar to other SCFAs analyzed (30). Outside of technical limitations, shifts in microbial community structure are not predictive of changes in static measurements of fecal SCFA abundances (62). The difficulty of finding meaningful correlations between microbiome composition and SCFA abundances likely reflects a failure to measure both circulating and fecal SCFAs across time in conjunction with microbial abundances. Indeed, it has been observed that fecal levels of acetate are inversely related with the rate of its absorption (63). Future studies are needed to confirm whether correlation analysis between fecal SCFAs and microbiome composition is a useful tool to understand the interplay between microbiome, SCFAs, and health.
In sum, our results indicate that gut microbial communities are malleable to an influx in recalcitrant carbohydrates, contributing to significant community and functional shifts in certain metabolic pathways. However, these compositional changes did not correspond to broad functional changes, at least over the short-term timescales for this intervention. Further studies exploring the impact of timing and composition of dietary fiber interventions, particularly while taking into account the starting composition of the gut microbiomes of study participants, are critical for understanding the generalizability of fiber interventions for engineering microbiomes. Increasing fiber intake could have the most impact in contexts where low gut microbial diversity increases the risk of C. difficile infection, such as for nursing home residents and cancer patients or after antibiotic treatment.
MATERIALS AND METHODS
Study design.
Twenty-six UC Irvine students and instructors volunteered for a 3-week high-fiber diet intervention study (Fig. 1A); only 22 individuals elected to provide stool samples for microbiome analyses (from 20 of whom we recovered enough sequence data for analysis; see Table S1 in the supplemental material). The dietary intervention was approved by UC Irvine IRB number 2018-4297. For the first week of the study, all participants consumed their normal diets, tracking all nutritional information using the smartphone application MyFitnessPal (MyFitnessPal, Inc.). Prior to the end of week one, each subject provided three fecal samples from 3 days within the first week. The intervention commenced in week two, when participants were instructed to raise their dietary fiber intake to approximately 40 g per day. To assist with the dietary shifts, we provided 10 meals per week, with ∼15 g of fiber and ∼5.8 unique fruits or vegetables per meal, from the food delivery service Thistle (San Francisco, CA, USA). During week three, subjects were encouraged to further increase fiber intake to ∼50 g of fiber per day. Subjects provided three fecal samples from 3 days during week three, concluding the intervention period. As part of the CURE course, students were educated on human health, dietary information on high-fiber meals, the human gut microbiome, and the quantitative methods for microbiome analyses (from DNA extraction and library preparation to metagenome and statistical analyses), as previously described (25).
Sample collection.
Subjects were given materials to collect fecal samples at home. Each stool sample was split into three 2-ml tubes by the individual and immediately stored in the freezer. When convenient, students transported their anonymized and coded samples using cold packs and insulated boxes to a common laboratory freezer. Upon the conclusion of the intervention period (week 1 or 3), all samples were transported to a −20°C freezer.
DNA extraction and metagenomic library preparation.
To characterize the bacterial community composition of the samples, DNA was extracted with the ZymoBIOMICS 96 DNA kit (product D4309) from Zymo Research using the manufacturer’s suggested protocol. Sequencing libraries were prepared using the Illumina Nextera kit and methods described in Baym et al. (64). Briefly, DNA was diluted to 0.5 ng/μl and added to 0.25 μl of Nextera enzyme and 1.25 μl of tagmentation buffer. This mixture was incubated at 55°C for 10 min and then placed on ice for the remainder of the protocol. Barcodes were added using Phusion polymerase (New England Biolabs), and excess adaptors were cleaned using AMPure XP (Beckman Coulter Life Sciences) magnetic beads. Quality and concentration were assessed using a PicoGreen assay (ThermoFisher), and the distribution of fragment sizes was determined using a Bioanalyzer. These libraries were loaded onto the Illumina Next-Seq 500 at 1.8 pM concentrations and sequenced using Illumina’s mid-output kit for 75-bp paired-end sequencing, resulting in a total of 144,023,583 reads and an average of 1,425,976 reads/sample (maximum, 5,902,966; minimum, 7) (Table S1).
Amplicon library preparation.
To characterize the genetic diversity of Bifidobacterium at a finer genetic scale than could be assayed by metagenomics, we used genus-specific primers to target this group for sequencing (65). Sequencing libraries were prepared by setting up initial 25-μl PCRs with AccuStart II PCR ToughMix (2×), the groEL forward primer (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTCCGATTACGAYCGYGAGAAGCT-3′, 20 μM), and the groEL reverse primer (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCSGCYTCGGTSGTCAGGAACAG-3′, 20 μM). The initial PCR ran for 28 cycles 95°C for 30 s, 60°C for 30 s, and 72°C for 50 s, followed by the addition of 0.5 μl of dual Nextera XT index (Illumina) to each sample, proceeding with an additional 8 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 50 s. Amplicons were pooled based on visual quantification of the bands on an agarose gel and purified using magnetic Speed Beads. The pool was run on a MiSeq PE 300 at the University of California Irvine's Genetic High-Throughput Facility, resulting in a total of 20,052,935 reads and an average of 185,675 reads/sample (maximum, 6,815,601; minimum, 155).
SCFA extraction and measurements.
SCFA extractions were done by following the methods of Zhao et al. (66). One hundred milligrams of fecal material was added to 1 ml of high-performance liquid chromatography (HPLC)-grade water and vortexed for 2 min. Ten microliters of 6N HCl was added to the fecal slurry and vortexed briefly. This mixture was incubated at room temperature for 10 min with occasional shaking. Afterward, the mixture was centrifuged at 14,000 × g for 1 min, and 400 μl of the supernatant was transferred to a new tube, which was then filtered through a 0.22-μm filter. An aliquot (200 μl) of this suspension was then transferred to a glass vial with a 0.2-ml vial insert and stored at −20°C. When running the sample, 10 μl of an internal standard of 10 mM ethyl butyrate was added to the extraction prior to the run. Before running each sample, the instrument was calibrated using a standard comprising 100 mg/liter acetate, propionate, isobutyrate, butyrate, isovalerate, valerate, and ethyl butyrate. Six samples were run on an Agilent 7890A gas chromatograph with dual-column flame ionization detectors. Two microliters per extracted sample was hand injected on a stainless steel column (2 m by 3.2 mm) containing 10% SP-1000 and 1% H3PO4 on 100/120 Chromosorb W AW (Supelco, Inc., Bellefonte, PA, USA). The flow rate of the N2 carrier gas was 26.14 ml/min. Between sets of six samples, the instrument was washed using water and phosphoric acid. Peaks were autointegrated using ChemStation v1.0 on a PC running Windows 2000 (Microsoft). A subset of samples (n = 44 from 8 individuals) were run in duplicate to examine technical variation (see the coefficient of variation [CV] in Table 1), and the average CV was 55%.
Metagenomic sequence analysis.
Raw shotgun metagenome sequences were filtered using Prinseq v0.20.4 (67) to remove sequences that had a mean quality score of 30 or less. Reads from human DNA were also removed by aligning the filtered reads to the human genome (hg38), using Bowtie2 v2.2.7 (68), and keeping the reads that failed to align. A total of 130,755,383 paired-end reads (average of 1,294,607 nonhuman reads/sample) were retained and passed through MIDAS, which assigns taxonomy to short read data using a marker gene approach (69). Species counts per sample represent the average of 100 subsamples, rarefied to 900 sequences per sample using the EcolUtils (v0.1) package in R. Taxonomy was also assessed using IGGsearch (70). To analyze functional differences related to SCFA metabolism between high- and low-fiber treatment groups, HUMAnN3 (71) was used with default parameters. All pathways within the MetaCyc pathway class “Fermentation to Short-Chain Fatty Acids” were searched for within the HUMANnN pathway output, which resulted in nine pathways used for analysis (72). For genes related to carbohydrate breakdown, we translated reads using Prodigal (73) to predict open reading frames (ORFs) and searched all ORFs against the Pfam database (74) with hmmer/3.1b2 (75). Resulting PFAM annotations were then screened against CAZyDB.07202017 (76) with BLAST/2.8.1 (77) using alignments of >70% amino acid identity and 30% coverage. Alpha diversity and PERMANOVA analyses were performed using the Vegan v2.5-6 (78) package in R (79). Nonmetric multidimensional analysis was done using the metaMDS function in Vegan on Bray-Curtis distances. StrainPhlAn (80), under default parameters, was used to analyze strain-level variation within the metagenomes. To root the phylogenetic tree, Prosthecochloris aestuarii (accession no. GCA_000020625) was used, and two reference genomes of Eubacterium rectale (accession no. GCA_000209935 and GCA_001404855).
GroEL amplicon analysis.
We downloaded 780 genomes from the genus Bifidobacterium on the PATRIC database (81). All genomes were screened for completeness by searching for 21 single-copy ribosomal marker genes using Prodigal (73) and HMMer v3.1b2 (75) with an E value of 1 × 10−10. The remaining 578 genomes were used to create a multilocus, concatenated phylogeny of the ribosomal marker genes with ClustalO v1.2.0 (82) to produce a 4,272-amino-acid alignment for phylogenetic analysis using RAxML v8.0.0 (83) under the PROTGAMMABLOSUM62 model for 100 replicates. Next, we parsed the filtered genomes for the groEL gene sequences by using 260 nonredundant gene sequences to build a groEL phylogeny under parameters identical to those of the whole-genome analysis. The groEL amino acid sequences, alignment, and phylogeny were used to construct BLASTp, HMMer, and pplacer reference databases for metagenomic analyses.
For each groEL amplicon library, sequences were quality trimmed and adapters were removed with BBDuk (84) (qtrim = rl trimq = 10 ktrim = r k = 25). Paired-end sequences were merged together with BBMerge (84), and, if paired reads did not overlap, only the forward read was retained. The reads were then searched against the groEL reference databases using BLAT (85) and hmmsearch, respectively. Passed reads were aligned with ClustalO to the pplacer reference package and placed on the groEL reference phylogeny using pplacer v1.1.alpha17 (86). Relative abundance was calculated from the single branch assignments and aggregated at the species level to be normalized by the total number of extracted groEL gene sequences. We show that the phylogenetic relationship between species of Bifidobacterium based on the groEL gene closely reflects a phylogeny based on 21 single-copy marker genes from 578 Bifidobacterium genomes (Fig. S4).
Statistical analysis.
PERMANOVA was conducted on Bray-Curtis dissimilarities at the genus level with 999 permutations using the Adonis test in the vegan package in R (described below). We tested the effect of the intervention (pre- versus postfiber increase), the effect of the individual, and the interaction between these two factors. Genus contributions to significant results from the PERMANOVA model were determined by passing the resulting PERMANOVA object through the coefficients function found in the base Stats package in R. A similar procedure was used to analyze compositional differences between CAZy enzymes and HUMAnN gene predictions in the metagenomes, with permutations on Euclidean distances. Linear mixed-effects modeling, using the nLME package (79) in R, were also conducted for comparison, because they take repeated measures into account. Specifically, to support the PERMANOVA analysis of beta diversity, an LME was performed on the rank-transformed first principal coordinate of a principal coordinate analysis on the Bray-Curtis community dissimilarity matrix. Individual was used as the random effect and the model used the default autoregressive (Lag 1) structure (AR1) for regression across a time series. For the functional analyses, reads analyzed using HUMAnN3 were normalized by copies per million; CAZy reads were normalized to the total number of reads per metagenome and compared using Wilcoxon rank sum test. Gene features for HUMAnN were reduced by analyzing only unstratified data, for which 70% of samples had nonzero reads mapping to each feature. HUMAnN pathway abundances were analyzed in their entirety with stratification and without feature reduction. Lefse (87) was used to determine pathways which may differentiate pre- versus postintervention samples. Wilcoxon rank sum tests were also used to compare nutritional and gene differences between intervention periods when residuals were not normally distributed and reads or macronutrients were averaged out within individuals (by treatment) to account for repeated measures. When normality assumptions of residuals were met (tested using the Shapiro-Wilk test), ANOVAs were used. To assess which taxa were correlated with changing amounts of fiber, all species within each sample (the rarefied species abundance matrix) and fiber were correlated using the Corrr package v0.4.2 (88) in R. To analyze which genera cocorrelate with the genus Bifidobacterium, Spearman correlations were used, and, where appropriate, P values were corrected (q value) for multiple comparisons using a false discovery rate cutoff of 0.05. To assess significance of strains between individuals, cophenetic distances were calculated on the RAxML tree output from StrainPhlAn and passed into the above-described PERMANOVA model.
Data availability.
All scripts are stored on GitHub (https://github.com/aoliver44/Fiber-Analysis). All metagenomic and amplicon sequences are available from NCBI under the BioProject no. PRJNA647720. Metadata linking the shotgun metagenomes and groEL sequences with the appropriate sample ID and intervention can be found in Table S1.
ACKNOWLEDGMENTS
We acknowledge the T32 training grant, which supported Andrew Oliver (1T32AI14134601A1), from UC Irvine’s training program in microbiology and infectious diseases.
We also acknowledge the UCI Microbiome Initiative for supporting the study, Thistle for supporting our provision of high fiber meals, and Heather Maughan for thoughtful edits to the manuscript. We especially thank the students from the course M130L at UC Irvine Spring in 2018.
Contributor Information
Katrine Whiteson, Email: katrine@uci.edu.
Janet K. Jansson, Pacific Northwest National Laboratory
REFERENCES
- 1.Jones JM. 2014. CODEX-aligned dietary fiber definitions help to bridge the fiber gap. Nutr J 13:34. doi: 10.1186/1475-2891-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.US Department of Agriculture, Agricultural Research Service. 2012. Nutrient intakes from food: mean amounts consumed per individual, by gender and age, what we eat in America, NHANES 2009–2010. US Department of Agriculture, Washington, DC. [Google Scholar]
- 3.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Wang X, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C. 2018. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds A, Mann J, Cummings J, Winter N, Mete E, Te Morenga L. 2019. Carbohydrate quality and human health: a series of systematic reviews and meta-analyses. Lancet 393:434–445. doi: 10.1016/S0140-6736(18)31809-9. [DOI] [PubMed] [Google Scholar]
- 5.O’Keefe SJD. 2016. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PGB, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. 2015. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, Marsland BJ. 2018. Dietary fiber confers protection against flu by shaping Ly6c− patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity 48:992–1005. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, Sartor RB, Gewirtz AT, Pulendran B. 2014. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utzschneider KM, Kratz M, Damman CJ, Hullarg M. 2016. Mechanisms linking the gut microbiome and glucose metabolism. J Clin Endocrinol Metab 101:1445–1454. doi: 10.1210/jc.2015-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantarel BL, Lombard V, Henrissat B. 2012. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 12.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. 2013. The influence of diet on the gut microbiota. Pharmacol Res 69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 14.Makki K, Deehan EC, Walter J, Bäckhed F. 2018. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Roediger WEW. 1982. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 83:424–429. doi: 10.1016/S0016-5085(82)80339-9. [DOI] [PubMed] [Google Scholar]
- 16.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. 2019. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10:e02566-18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkataraman A, Sieber JR, Schmidt AW, Waldron C, Theis KR, Schmidt TM. 2016. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome 4:33. doi: 10.1186/s40168-016-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicki CM, Livingston KA, Obin M, Roberts SB, Chung M, Mckeown NM. 2017. Dietary fiber and the human gut microbiota: application of evidence mapping methodology. Nutrients 9:125. doi: 10.3390/nu9020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velázquez M, Davies C, Marett R, Slavin JL, Feirtag JM. 2000. Effect of oligosaccharides and fibre substitutes on short-chain fatty acid production by human faecal microflora. Anaerobe 6:87–92. doi: 10.1006/anae.1999.0318. [DOI] [Google Scholar]
- 21.Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. 2013. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Cuesta-Zuluaga J, Mueller N, Álvarez-Quintero R, Velásquez-Mejía E, Sierra J, Corrales-Agudelo V, Carmona J, Abad J, Escobar J. 2018. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 11:51. doi: 10.3390/nu11010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, Thompson LR, et al. 2018. American gut: an open platform for citizen science microbiome research. mSystems 3:2020. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 25.Sewall JM, Oliver A, Denaro K, Chase AB, Weihe C, Lay M, Martiny JBH, Whiteson K. 2020. Fiber force: a fiber diet intervention in an advanced course-based undergraduate research experience (CURE) course. J Microbiol Biol Educ 21:21.1.40. doi: 10.1128/jmbe.v21i1.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkel S, ASM Task Force on Curriculum Guidelines for Undergraduate Microbiology. 2012. The development of curricular guidelines for introductory microbiology that focus on understanding. J Microbiol Biol Educ 13:32–38. doi: 10.1128/jmbe.v13i1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Research Council. 2003. BIO 2010: transforming undergraduate education for future research biologists. National Research Council, Washington, DC. [Google Scholar]
- 28.American Association for the Advancement of Science. 2009. No TitleVision and change in undergraduate biology education: a call to action: a summary of recommendations made at a national conference organized by the American Association for the Advancement of Science. American Association for the Advancement of Science, Washington, DC. [Google Scholar]
- 29.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, Shanahan ER, Staudacher HM, Campbell KL. 2018. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr 107:965–983. doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- 31.Nemoto H, Ikata K, Arimochi H, Iwasaki T, Ohnishi Y, Kuwahara T, Kataoka K. 2011. Effects of fermented brown rice on the intestinal environments in healthy adult. J Med Investig 58:235–245. doi: 10.2152/jmi.58.235. [DOI] [PubMed] [Google Scholar]
- 32.Tap J, Furet J-P, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, Fontaine E, Doré J, Leclerc M. 2015. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 33.Cooper D, Kable M, Marco M, De Leon A, Rust B, Baker J, Horn W, Burnett D, Keim N. 2017. The effects of moderate whole grain consumption on fasting glucose and lipids, gastrointestinal symptoms, and microbiota. Nutrients 9:173. doi: 10.3390/nu9020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karl JP, Meydani M, Barnett JB, Vanegas SM, Goldin B, Kane A, Rasmussen H, Saltzman E, Vangay P, Knights D, Chen C-YO, Das SK, Jonnalagadda SS, Meydani SN, Roberts SB. 2017. Substituting whole grains for refined grains in a 6-wk randomized trial favorably affects energy-balance metrics in healthy men and postmenopausal women. Am J Clin Nutr 105:589–599. doi: 10.3945/ajcn.116.139683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ampatzoglou A, Atwal KK, Maidens CM, Williams CL, Ross AB, Thielecke F, Jonnalagadda SS, Kennedy OB, Yaqoob P. 2015. Increased whole grain consumption does not affect blood biochemistry, body composition, or gut microbiology in healthy, low-habitual whole grain consumers. J Nutr 145:215–221. doi: 10.3945/jn.114.202176. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho-Wells AL, Helmolz K, Nodet C, Molzer C, Leonard C, McKevith B, Thielecke F, Jackson KG, Tuohy KM. 2010. Determination of the in vivo prebiotic potential of a maize-based whole grain breakfast cereal: a human feeding study. Br J Nutr 104:1353–1356. doi: 10.1017/S0007114510002084. [DOI] [PubMed] [Google Scholar]
- 37.Costabile A, Klinder A, Fava F, Napolitano A, Fogliano V, Leonard C, Gibson GR, Tuohy KM. 2008. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br J Nutr 99:110–120. doi: 10.1017/S0007114507793923. [DOI] [PubMed] [Google Scholar]
- 38.Gråsten SM, Juntunen KS, Mättö J, Mykkänen OT, El-Nezami H, Adlercreutz H, Poutanen KS, Mykkänen HM. 2007. High-fiber rye bread improves bowel function in postmenopausal women but does not cause other putatively positive changes in the metabolic activity of intestinal microbiota. Nutr Res 27:454–461. doi: 10.1016/j.nutres.2007.05.010. [DOI] [Google Scholar]
- 39.Jenkins DJA, Vuksan V, Rao AV, Vidgen E, Kendall CWC, Tariq N, Würsch P, Koellreutter B, Shiwnarain N, Jeffcoat R. 1999. Colonic bacterial activity and serum lipid risk factors for cardiovascular disease. Metabolism 48:264–268. doi: 10.1016/S0026-0495(99)90045-8. [DOI] [PubMed] [Google Scholar]
- 40.Ross AB, Bruce SJ, Blondel-Lubrano A, Oguey-Araymon S, Beaumont M, Bourgeois A, Nielsen-Moennoz C, Vigo M, Fay L-B, Kochhar S, Bibiloni R, Pittet A-C, Emady-Azar S, Grathwohl D, Rezzi S. 2011. A whole-grain cereal-rich diet increases plasma betaine, and tends to decrease total and LDL-cholesterol compared with a refined-grain diet in healthy subjects. Br J Nutr 105:1492–1502. doi: 10.1017/S0007114510005209. [DOI] [PubMed] [Google Scholar]
- 41.Smith SC, Choy R, Johnson SK, Hall RS, Wildeboer-Veloo ACM, Welling GW. 2006. Lupin kernel fiber consumption modifies fecal microbiota in healthy men as determined by rRNA gene fluorescent in situ hybridization. Eur J Nutr 45:335–341. doi: 10.1007/s00394-006-0603-1. [DOI] [PubMed] [Google Scholar]
- 42.Zeng Y, Huang S, Mu G, Zeng X, Zhou X. 2015. Effects of whole grain-bean mixed staple food on intestinal microecology and metabolic parameters of obese people. Chin J Clin Nutr 23:27–34. doi: 10.3760/cma.j.issn.1674-635X.2015.01.006. [DOI] [Google Scholar]
- 43.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallav K, Dowd SE, Villafuerte J, Yang X, Kabbani T, Hansen J, Dennis M, Leffler DA, Newburg DS, Kelly CP. 2014. Effects of polysaccharopeptide from Trametes Versicolor and amoxicillin on the gut microbiome of healthy volunteers. Gut Microbes 5:458–467. doi: 10.4161/gmic.29558. [DOI] [PubMed] [Google Scholar]
- 47.Hooda S, Boler BMV, Serao MCR, Brulc JM, Staeger MA, Boileau TW, Dowd SE, Fahey GC, Swanson KS. 2012. 454 Pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr 142:1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 48.Herman DR, Rhoades N, Mercado J, Argueta P, Lopez U, Flores GE. 2020. Dietary habits of 2- to 9-year-old american children are associated with gut microbiome composition. J Acad Nutr Diet 120:517–534. doi: 10.1016/j.jand.2019.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, Topf M, Gonzalez CG, Robinson JL, Elias JE, Sonnenburg ED, Gardner CD, Sonnenburg JL. 2020. Gut microbiota-targeted diets modulate human immune status. bioRxiv doi: 10.1101/2020.09.30.321448. [DOI] [PMC free article] [PubMed]
- 50.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graf E, Eaton JW. 1985. Dietary suppression of colonic cancer fiber or phytate? Cancer 56:717–718. doi:. [DOI] [PubMed] [Google Scholar]
- 52.Dinicola S, Minini M, Unfer V, Verna R, Cucina A, Bizzarri M. 2017. Nutritional and acquired deficiencies in inositol bioavailability. Int J Mol Sci 18:2187. doi: 10.3390/ijms18102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez CI, Martiny JBH. 2020. Evolutionary relationships among bifidobacteria and their hosts and environments. BMC Genomics 21:26. doi: 10.1186/s12864-019-6435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu L, Lu W, Wang L, Pan M, Zhang H, Zhao J, Chen W. 2017. Assessment of Bifidobacterium species using groEL gene on the basis of Illumina MiSeq high-throughput sequencing. Genes 8:336. doi: 10.3390/genes8110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol 71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. 2008. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 58.Topping DL, Clifton PM. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 59.Cummings JH. 1981. Short chain fatty acids in the human colon. Gut 22:763–779. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueyama J, Oda M, Hirayama M, Sugitate K, Sakui N, Hamada R, Ito M, Saito I, Ohno K. 2020. Freeze-drying enables homogeneous and stable sample preparation for determination of fecal short-chain fatty acids. Anal Biochem 589:113508. doi: 10.1016/j.ab.2019.113508. [DOI] [PubMed] [Google Scholar]
- 61.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. 2016. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol 7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sze MA, Topçuoğlu BD, Lesniak NA, Ruffin MT, Schloss PD. 2019. Fecal short-chain fatty acids are not predictive of colonic tumor status and cannot be predicted based on bacterial community structure. mBio 10:e01454-19. doi: 10.1128/mBio.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogt JA, Wolever TMS. 2003. Fecal acetate is inversely related to acetate absorption from the human rectum and distal colon. J Nutr 133:3145–3148. doi: 10.1093/jn/133.10.3145. [DOI] [PubMed] [Google Scholar]
- 64.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony RK. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036-15. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Junick J, Blaut M. 2012. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol 78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao G, Nyman M, Jönsson JÅ. 2006. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr 20:674–682. doi: 10.1002/bmc.580. [DOI] [PubMed] [Google Scholar]
- 67.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568:505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. 2018. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, Paley S, Subhraveti P, Karp PD. 2018. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res 46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference, p 205–211. In Morishita S, Lee SY, Sakakibara Y (ed), Genome informatics 2009. Imperial College Press, London, UK. doi: 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- 76.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. R package version. https://cran.r-project.org/web/packages/vegan/index.html.
- 79.Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore M, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES, Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N, Theologis A, Yang WH, Hubbell E, Au M, Chung EY, Lashkari D, Lemieux B, Dean C, Lipshutz RJ, Ausubel FM, Davis RW, Oefner PJ, Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES, Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES, Lenné JM, Takan JP, Mgonja MA, Manyasa EO, Kaloki P, Wanyera N, et al. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 80.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure & genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, MacHi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bushnell B. 2015. BBMap. https://sourceforge.net/projects/bbmap/.
- 85.Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsen FA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuhn M, Jackson S, Cimentada J. 2020. corrr: Correlations in R. R package version 0.4.2. https://CRAN.R-project.org/package=corrr.
- 89.Maier TV, Lucio M, Lee LH, VerBerkmoes NC, Brislawn CJ, Bernhardt J, Lamendella R, McDermott JE, Bergeron N, Heinzmann SS, Morton JT, González A, Ackermann G, Knight R, Riedel K, Krauss RM, Schmitt-Kopplin P, Jansson JK. 2017. Impact of dietary resistant starch on the human gut microbiome, metaproteome, and metabolome. mBio 8:e01343-17. doi: 10.1128/mBio.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparisons of diversity measures obtained using different databases for taxonomic assignments. IGG_rich and MIDAS were run using default parameters. IGG-Lenient was run at 25% species quality and 15% marker genes. Download FIG S1, PDF file, 0.1 MB (107.7KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PERMANOVA model and term results. Download Table S2, DOCX file, 0.01 MB (13.4KB, docx) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Correlations above an r cutoff of >0.2 for microbial abundance and fiber intake. Only Lachnospiraceae bacterium 51870 was significantly negatively correlated at an FDR cutoff of 0.05. (B) Raw spearman correlation of a species of Lachnospiraceae with fiber intake. Download FIG S2, PDF file, 0.05 MB (48.5KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Mean abundance (MIDAS read counts) of the genus Bifidobacterium during the diet intervention. Points are colored by individual. (B) Changes in mean abundance of each species of Bifidobacterium, detected by MIDAS, during the diet intervention period. Download FIG S3, PDF file, 0.10 MB (99KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bifidobacterium phylogenetic analyses. (A) Multilocus phylogenetic analysis of conserved ribosomal marker genes. (B) Phylogenetic analysis of the groEL gene sequences used for amplicon analyses. The top 8 species observed are shown. Download FIG S4, PDF file, 0.9 MB (925.9KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significant (FDR, 0.05) correlations from comparing abundances of 99 different genera with Bifidobacterium. Download FIG S5, PDF file, 0.02 MB (19.1KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Average abundances (normalized reads) of carbohydrate active enzymes within individuals during the diet intervention period. Each different color point represents an individual, and lines connect the same individual. (B) Log2-transformed GH and polysaccharide lyase gene abundances during the intervention. Download FIG S6, PDF file, 0.2 MB (229.6KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inositol degradation abundance (normalized copies per million) for metagenomes before and after the intervention. Download FIG S7, PDF file, 0.08 MB (83.3KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Technical variation of SCFAs seen in a subset of samples run in duplicate. (A) Amount of SCFAs by individual, with color denoting if it was measured during replicate 1 or 2. (B) Normalized (by mean) difference between absolute difference between treatment, subtracted by absolute difference between technical replicates. Larger negative values suggest differences between technical replicates were larger than the differences detected between pre- and postintervention arms. Download FIG S8, PDF file, 0.1 MB (111.6KB, pdf) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence statistics, accession IDs, and experimental metadata. Download Table S1, XLSX file, 0.02 MB (25.5KB, xlsx) .
Copyright © 2021 Oliver et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All scripts are stored on GitHub (https://github.com/aoliver44/Fiber-Analysis). All metagenomic and amplicon sequences are available from NCBI under the BioProject no. PRJNA647720. Metadata linking the shotgun metagenomes and groEL sequences with the appropriate sample ID and intervention can be found in Table S1.