Summary:
C. elegans must distinguish pathogenic from nutritious bacterial food sources among the many bacteria it is exposed to in its environment1. Here we show that a single exposure to purified small RNAs isolated from pathogenic Pseudomonas aeruginosa (PA14) is sufficient to induce pathogen avoidance, both in the treated animals and in four subsequent generations of progeny. The RNA interference and piRNA pathways, the germline, and the ASI neuron are required for bacterial small RNA-induced avoidance behavior and transgenerational inheritance. A single P. aeruginosa non-coding RNA, P11, is both necessary and sufficient to convey learned avoidance of PA14, and its C. elegans target, maco-1, is required for avoidance. Our results suggest that this ncRNA-dependent mechanism evolved to survey the worm’s microbial environment, use this information to make appropriate behavioral decisions, and pass this information on to its progeny.
Small RNAs from PA14 induce avoidance
C. elegans’ natural habitat contains many bacterial species; about a third are Pseudomonas, which can be beneficial or detrimental1. Despite their natural attraction to pathogenic Pseudomonas aeruginosa (PA14), C. elegans learn to avoid this pathogen after becoming ill2. We discovered that worms pass this learned avoidance of PA14 to their progeny3, but the nature of the signal conveying the pathogen’ identity was unknown.
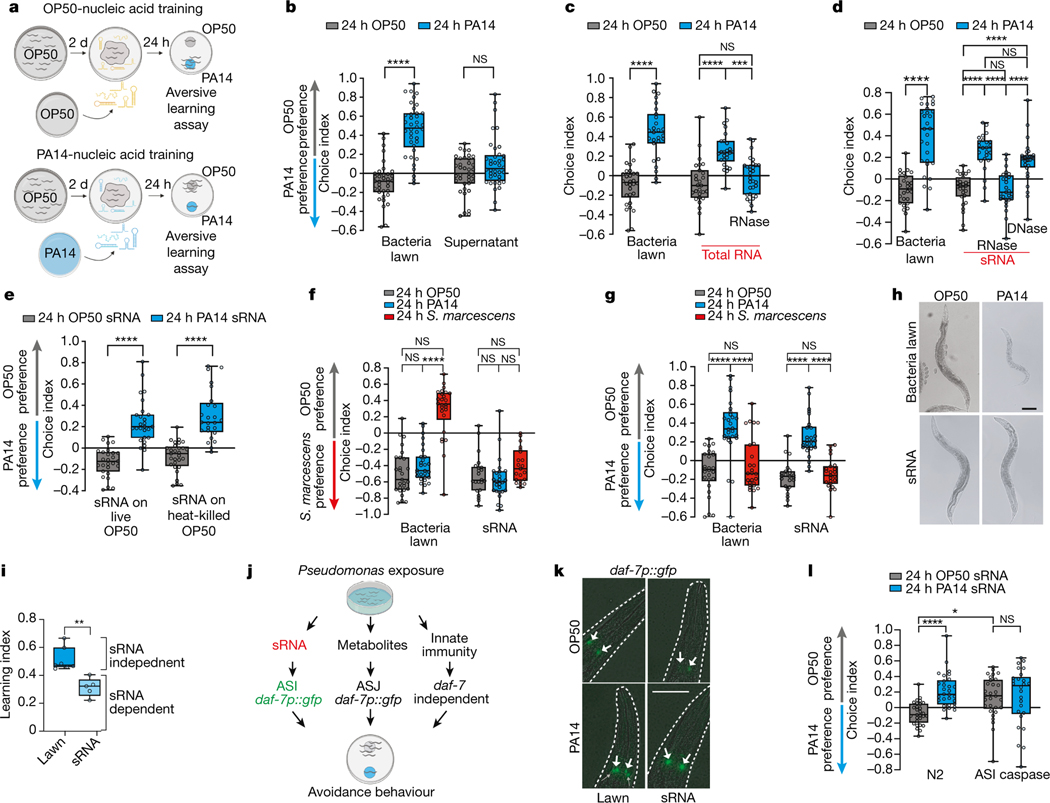
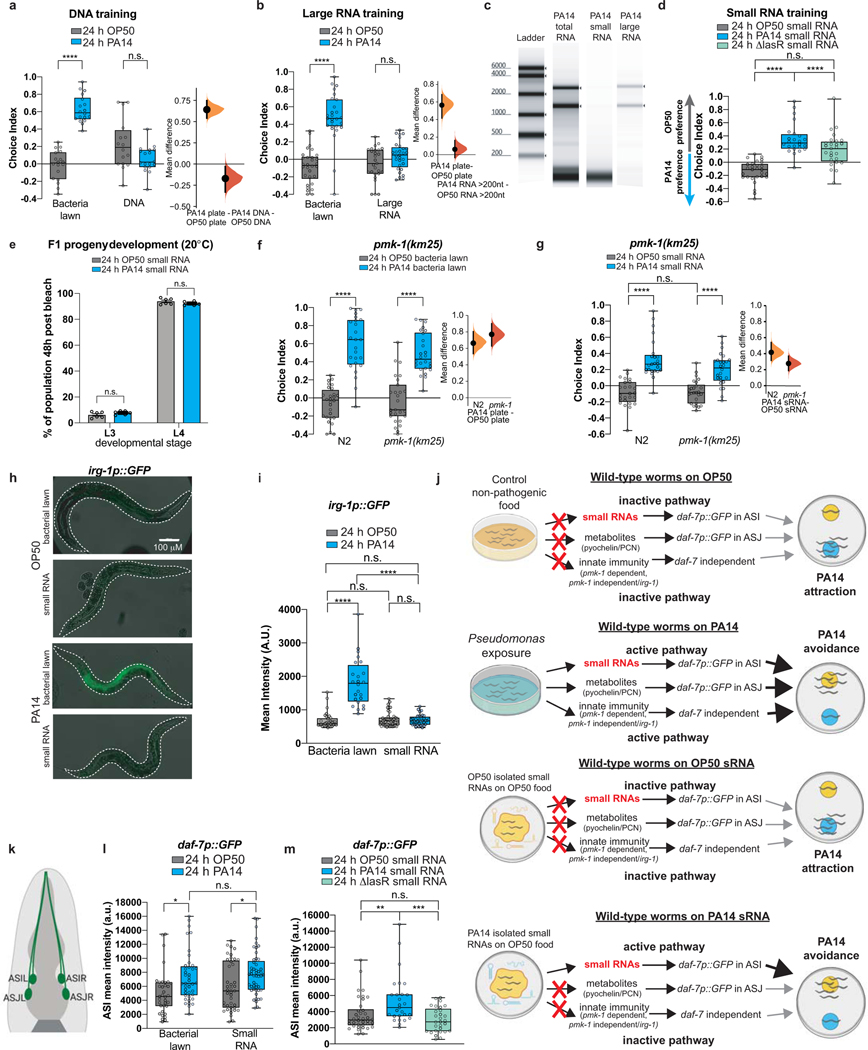
To identify this signal, we trained worms for 24h on non-pathogenic E. coli OP50 spiked with components isolated from pathogenic PA14 or OP50, and then tested avoidance behavior using a standard OP50 vs PA14 bacteria lawn choice assay (Fig. 1a). Although bacterial metabolites can alter worm behavior4, training worms with PA14 supernatant did not induce avoidance learning (Fig. 1b). Next, we tested nucleic acid components (total DNA, total RNA, large RNA (>200nt), small RNA (<200nt) and RNase- and DNase-treated fractions) from pathogenic (25°C, plate-grown) PA14, adding the purified samples to lawns of OP50 for 24h (Fig. 1a, c–d; Extended Data Fig. 1a–b). PA14 total RNA, small RNA (sRNA), and DNase-treated sRNA samples (Fig. 1c–d) induced avoidance of PA14, while DNA and large RNAs had no effect at the concentrations tested (Fig. 1b, Extended Data Fig. 1a–c).
Fig. 1: PA14 small RNAs are sufficient to induce C. elegans pathogen avoidance.
(a) Worms were trained on non-pathogenic E. coli (OP50), PA14 lawns, or OP50 lawns spiked with bacterial components. Choice assays to OP50 vs PA14 bacteria were then performed. Choice index (CI) = (# on OP50 - # on PA14)/(total). (b) Worms exposed to a PA14 lawn (24h) learn to avoid PA14, but PA14 supernatant does not elicit PA14 avoidance. (c) Training with purified PA14 total RNA confers PA14 avoidance; RNase treatment abolishes this effect. (d) Purified PA14 sRNAs (<200 nt) induce PA14 avoidance; RNase treatment abolishes this effect, while DNase treatment does not. (e) Heat killing bacteria does not abolish sRNA learning. (f) Trained worms were tested in an OP50 vs S. marcescens (Sm) choice assay. Sm-lawn training induces Sm avoidance, while exposure to Sm sRNAs does not. PA14 sRNA training does not affect OP50 vs Sm preference. (g) PA14 lawn and PA14 sRNA training induce PA14 avoidance, while Sm bacteria and sRNA training do not affect PA14 preference. (h) Unlike PA14, sRNA exposure does not cause illness. (i) sRNA-induced learning is ~half of that of PA14 lawns. (Learning Index=Average PA14 CI–Average OP50 CI). Each data point = LI from an independent experiment containing ~7–10 choice assay plates with an average of 115 worms/plate. Unpaired, two-tailed student’s t-test. (j) Worms learn to avoid Pseudomonas through several independent mechanisms. (k) ASI daf-7p::GFP expression increases upon PA14 lawn or sRNA exposure (white arrows); expression in ASJ is induced only in PA14 lawn-trained animals (grey arrows). (l) Genetic ablation of ASI neurons abolishes PA14 sRNA-induced learning. Each dot represents an individual choice assay plate (average of 115 worms/plate) from all replicates. Biological replicates (c-h, k, m (3), b(4), i(6)). Box plots: center line = median, box range is 25th - 75th percentile; whiskers denote minimum–maximum values. One-Way (c-d) and Two-Way ANOVA (b, e, f, l), Tukey’s multiple comparison test. *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001, n.s. = not significant. Estimation plots provided in Supplementary information. See Supplemental table 4 for exact sample sizes (n) and p-values.
Worms trained on heat-killed OP50 E. coli bacteria supplemented with purified PA14 sRNAs still learned avoidance (Fig. 1e). sRNAs isolated from the less virulent ΔlasR mutant did not induce PA14 avoidance (Extended Data Fig. 1d). Together, these results suggest that sRNAs that are present in pathogenic bacteria, rather than changes to bacterial metabolism once inside the worm, are responsible for learned pathogenic avoidance.
sRNA-induced avoidance is species-specific
Serratia marcescens exposure also induces avoidance2; however, this avoidance is not passed on to the next generation3, and PA14-induced transgenerational avoidance is species-specific3. Treatment with sRNA isolated from S. marcescens does not induce avoidance of S. marcescens or PA14 (Fig. 1f–g), and sRNA from PA14 does not induce avoidance of S. marcescens (Fig. 1f). Therefore, like transgenerational inheritance of avoidance, sRNA-induced avoidance is a response induced by specific bacteria.
sRNA avoidance is immunity-independent
The fact that purified sRNAs are sufficient to induce P. aeruginosa avoidance in the absence of intact pathogen suggests that sRNA-induced avoidance does not require virulence. Worms treated with sRNAs are healthy and their offspring develop normally (Fig. 1h, Extended Data Fig. 1e). Moreover, the innate immunity regulator pmk-15 is not required for avoidance (Extended Data Fig. 1f–g), consistent with pmk-1’s dispensability for worm aversive behavior6,7. Additionally, the pmk-1-independent innate immune response (irg-1p::GFP)8 is not induced by PA14 sRNA (Extended Data Fig. 1h–i). Thus, mothers learn avoidance through the innate immune response (lawn exposure, metabolites)9 causing half of the avoidance behavior exhibited by lawn-trained worms (Fig. 1i–j, Extended Data Fig. 1j); in a separate mechanism identified here, pathogen-derived bacterial sRNAs induce the other half of the mother’s learned avoidance (Fig. 1i–j, Extended Data Fig. 1j).
ASI daf-7 expression is induced by PA14 sRNA
The TGF-β ligand DAF-7 is induced in ASI and ASJ neurons when exposed to PA144 (Extended Data Fig. 1k), and daf-7 expression increases in the ASI of progeny of PA14-trained mothers3. daf-7p::GFP3,4, which exhibits ASJ fluorescence in response to bacterial metabolites4 and ASI fluorescence in progeny of PA14-trained mothers3, is induced by PA14 sRNA exposure solely in the ASI neurons of trained mothers, resembling F1-F4 expression after PA14 training3 (Fig. 1k, Extended Data Fig. 1l). ΔlasR sRNA did not increase ASI daf-7 levels (Extended Data Fig. 1m). Worms with genetically-ablated ASI neurons exhibit high naïve avoidance of PA14, but sRNAs do not induce further avoidance (Fig. 1l), suggesting that the ASI neuron is required for bacterial sRNA-mediated PA14 avoidance (Fig. 1k), as it is required in the F1 generation for transgenerational inheritance of avoidance3.
sRNA avoidance requires the RNAi pathway
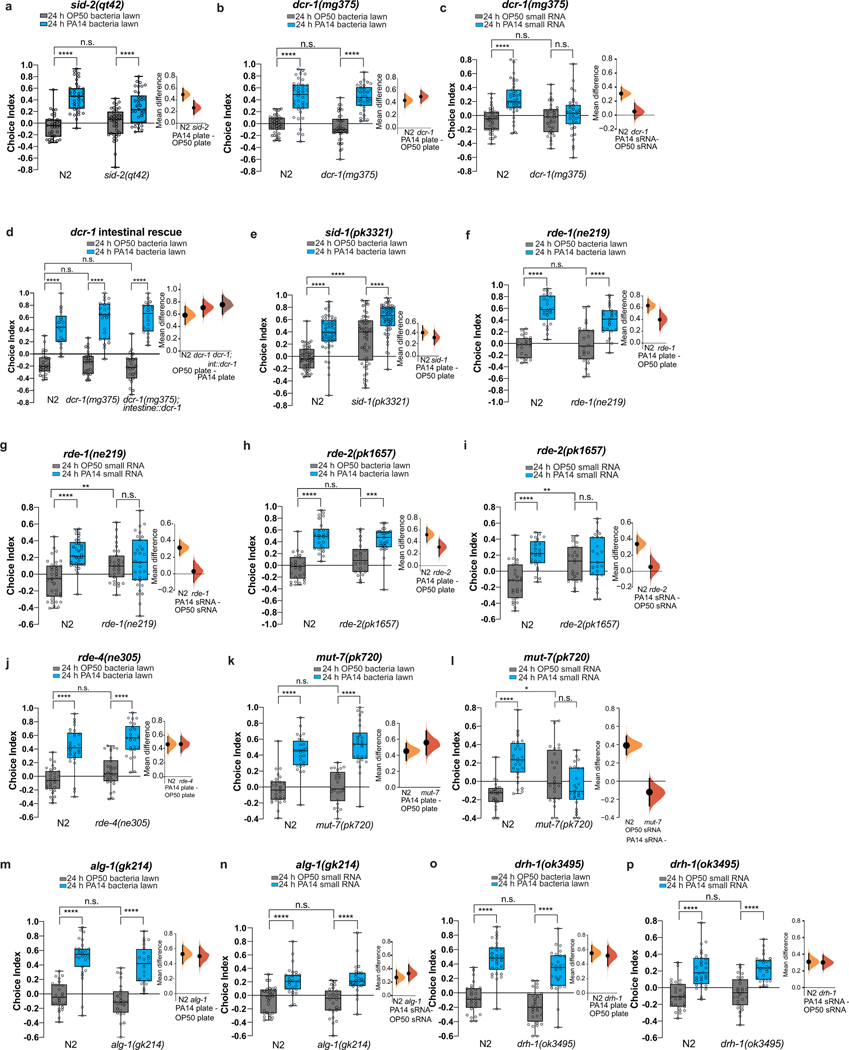
We next wondered whether bacterial sRNA-induced avoidance requires the RNA interference (RNAi) pathway10. While neither the dsRNA transporter SID-211 nor the dsRNA endoribonuclease Dicer/DCR-112,13 is required for avoidance induced by lawn exposure (Extended Data Fig. 2a–b), sRNA-induced avoidance specifically requires SID-2 and Dicer/DCR-112,13 (Fig. 2a–c, Extended Data Fig. 2c).
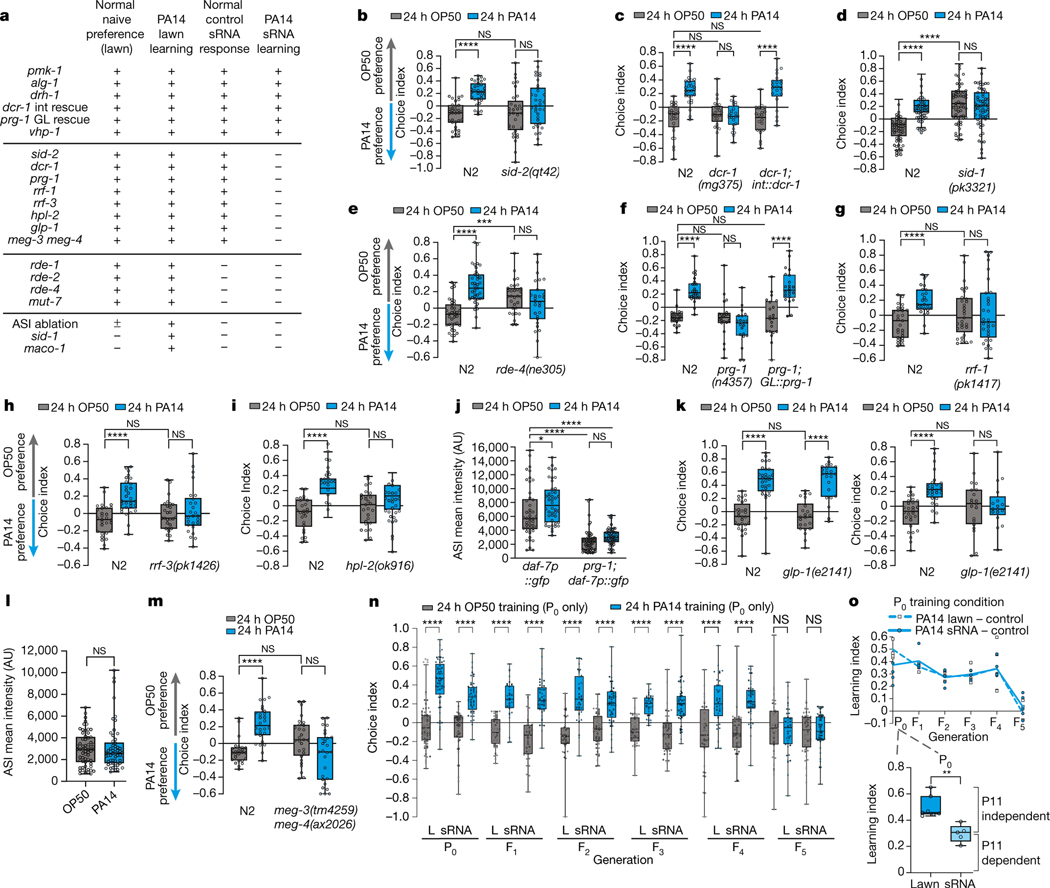
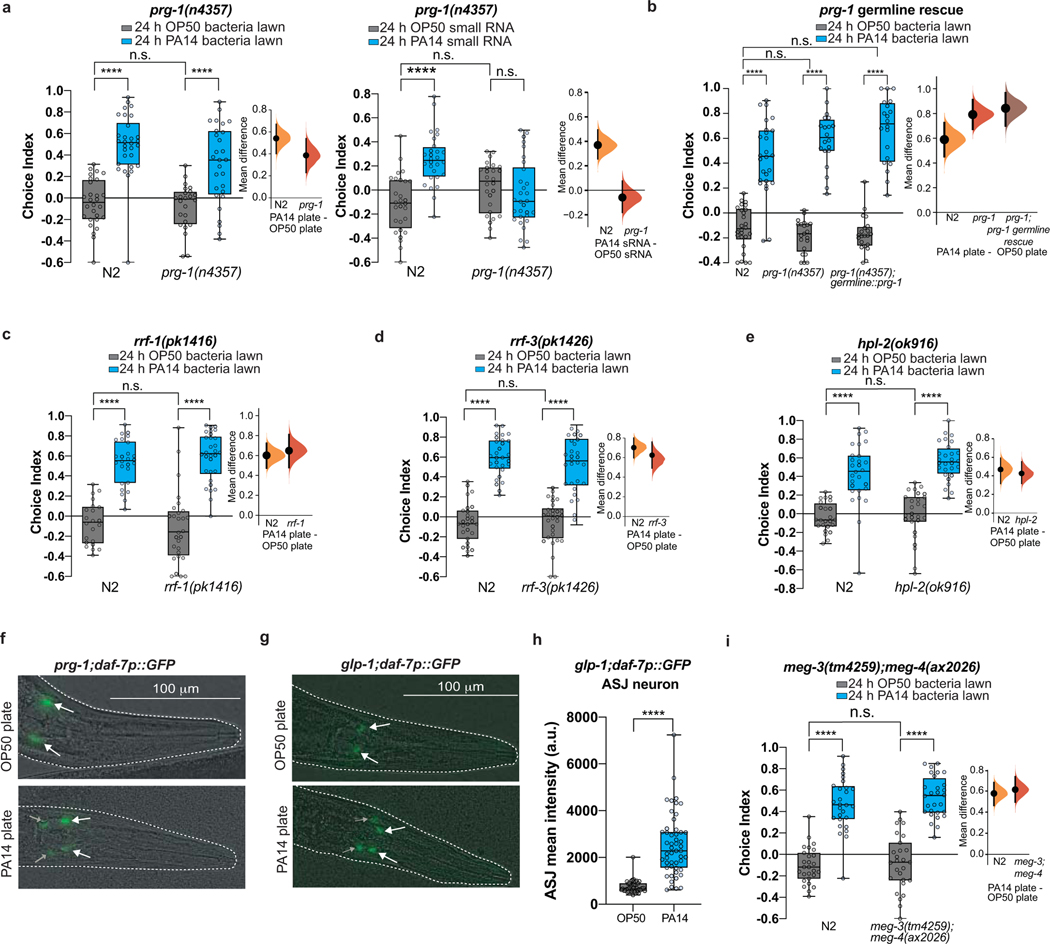
Fig. 2: dsRNA transport, processing machinery, piRNA Piwi/PRG-1 Argonaute pathway, and the germline are required for bacterial sRNA-induced avoidance.
(a) Summary: genes required for naive preference, PA14 lawn learning, and sRNA learning. (+: functions like wild type; -: defective (required for) naïve choice or learning. +/−: statistically borderline). (b) sid-2(qt42) is required for sRNA-induced learning. (c) Intestinal rescue of dcr-1 restores sRNA-mediated learning. (d) sid-1(pk3321) and (e) rde-4(ne305) exhibit abnormal sRNA response. (f) Germline prg-1 rescue restores sRNA-induced learning. (g) rrf-1(pk1417), (h) rrf-3(pk1426), and (i) hpl-2(ok916) mutants are deficient in PA14 sRNA-induced avoidance. (j) prg-1(n4357) mutants do not induce ASI neuron daf-7p::GFP expression upon PA14 training (n= 46, 44, 58, 55 neurons, left to right). (k) Germlineless glp-1(e2141) mutants learn to avoid PA14 after lawn training (left), but are defective in sRNA-induced learning (right). (l) glp-1 mutants do not induce ASI neuron daf-7p::GFP expression (n = OP50(60), PA14(54) neurons, two-sided student’s t-test). (m) Germline P granule mutants exhibit defective sRNA-induced learning. (n) Untrained progeny of PA14 lawn (L) and sRNA-trained mothers avoid PA14 from generation F1-F4; F5 resumes PA14 attraction. (o) Learning index for each generation, mean±SEM. (o, inset, P0; two-sided student’s t-test) Each data point = LI from an independent experiment of ~7–10 choice assay plates; each dot represents an individual choice assay plate (average of 115 worms/plate) from all replicates. (b-k, m) Two-Way ANOVA, Tukey’s multiple comparison test. Biological replicates (c, f-m(3), b, e(4), n(sRNA)(5), d, n(lawn)(6). Box plots: center line = median, box range is 25th - 75th percentile; whiskers denote minimum–maximum values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001, n.s. = not significant. Estimation plots provided in Supplementary information. See Supplemental table 4 for exact sample sizes (n) and p-values.
The dsRNA transporter SID-2 is expressed in the intestine11,14 and is required for sRNA-induced learning (Fig. 2b), while other components, including DCR-1, are expressed more broadly15. Intestinal rescue of dcr-1 restored sRNA-mediated learning (Fig. 2c, a; Extended Data Fig. 2d), suggesting that uptake and processing of bacterial sRNAs by SID-2 and DCR-1, respectively, are initiated in the intestine.
Aberrant sRNA responses in RNAi mutants
While Dicer and SID-2 are required for PA14 sRNA learning, we identified a class of RNAi mutants with aberrant PA14 and/or sRNA responses. sid-116 (systemic RNAi-defective) mutants exhibit high naïve PA14 avoidance (Extended Data Fig. 2e) but still learn to avoid PA14 with lawn training; however, sid-1 is required for sRNA-induced learning (Fig. 2d). By contrast, RNAi-defective (RDE) mutants rde-1/AGO-317, rde-2/mut-818, and rde-419, and mut-720 (an RDE-2 interactor18), have normal naïve PA14 attraction and avoidance after PA14 lawn training, but have an abnormal response to control (OP50) sRNAs, and show no further avoidance upon PA14 sRNA training (Extended Data Fig. 2f–I, Fig. 2e), indicating that they are defective for normal sRNA responses (Fig. 2a). These results are consistent with neuronal rde-4’s reported roles in chemotaxis and the requirement for the rde genes in response to E. coli21,22.
sRNA response is viRNA/miRNA independent
C. elegans processes both exogenous siRNAs and C. elegans-produced microRNAs using different Argonaute homologs. MicroRNA-specific Argonaute alg-1/AGO-123 mutants were competent for PA14 sRNA-induced learning (Fig. 2a, Extended Data Fig. 2m–n) as were mutants of the virus-responsive RNAi component drh-1 (Fig. 2a; Extended Data Fig. 2o–p). Thus, intracellular pathogen response to viral infection is not involved in C. elegans’ sRNA-induced learned avoidance24,25. Moreover, microRNA processing is unaffected in the dcr-1(mg375) mutant26, which is unresponsive to sRNA from PA14 (Fig. 2c). Together, these data suggest that the siRNA pathway, not microRNA or viral processing pathways, mediates bacterial sRNA-induced avoidance learning.
Avoidance requires the piRNA pathway
Piwi/PRG-1 and its downstream components are required for inheritance of learned pathogenic avoidance3, but were not required for maternal pathogen avoidance learning upon PA14 lawn training3 (Extended Data Fig. 3a–e). Therefore, we were surprised that prg-1 mutant mothers were defective in sRNA-induced avoidance response (Fig. 2f, a, Extended Data Fig. 3a). Furthermore, rrf-127, rrf-328 (RDRPs), and hpl-229 (heterochromatin regulator) mutants phenocopied prg-1’s sRNA learning defect (Fig. 2g–i, a; Extended Data Fig. 3c–e). Consistently, prg-1 mutants exposed to PA14 lawns failed to upregulate daf-7 in the ASI, but still induced daf-7 expression in the ASJ neurons (Fig. 2j, Extended Data Fig. 3f).
sRNA avoidance requires a germline
Like wild type, germline-less glp-1(e2141)30 mutants avoid PA14 after training on PA14 (Fig. 2k left) and upregulate daf-7 expression in the ASJ neurons (Extended Data Fig. 3g–h), demonstrating that learning via innate immune pathways does not require a functional germline. However, glp-1 mutants fail to exhibit sRNA-induced avoidance of PA14 (Fig. 2k right, a), and did not increase daf-7p::gfp in ASI neurons (Fig. 2l). Worms with defective germline granules (meg-3;meg-4 mutants)31 are also unable to induce avoidance in response to PA14 sRNA (Fig. 2m, a), despite having normal naïve preferences and lawn learning (Extended Data Fig. 3i). Finally, germline-specific expression of prg-1 fully rescued prg-1’s sRNA-induced PA14 avoidance (Fig. 2f). Thus, while dispensable for innate immunity-induced avoidance, a functional germline is required for sRNA-mediated learned avoidance, and the piRNA pathway acts in the germline to do so. Furthermore, our data suggest that bacteria-derived sRNAs do not act directly in neurons, but rather through an indirect mechanism that first requires uptake by the intestine, then piRNA processing and P granule function in the germline to communicate to the ASI neurons.
sRNA memory is transgenerationally inherited
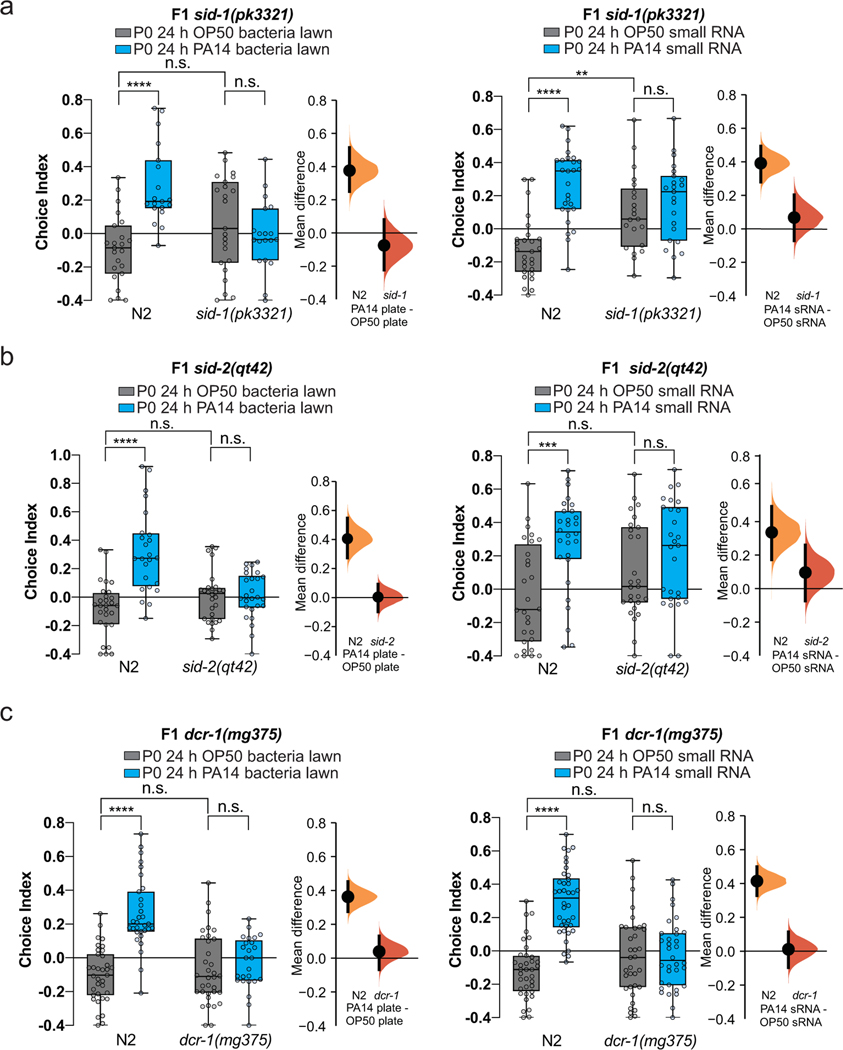
We previously showed that PA14 training induces heritable avoidance behavior persisting through the F4 generation3. Remarkably, a single 24hr exposure of C. elegans to PA14 sRNA induced avoidance of PA14 not only in mothers, but also in the subsequent four generations (Fig. 2n–o), replicating the transgenerationally-inherited avoidance induced by pathogenic bacteria3. This avoidance persisted despite the fact that neither the mothers nor their progeny had ever directly encountered PA14. Transgenerational inheritance of avoidance for both bacterial lawn and sRNA exposure required SID-1, SID-2, and DCR-1 (Extended Data Fig. 4a–c). The similar magnitude of the behavior in progeny of live bacteria- and sRNA-trained animals suggests that the sRNA response is sufficient to fully explain the transgenerational change in behavior (Fig. 2o).
Our data support a model in which mothers respond acutely through the innate immune response9 to pathogen exposure and metabolites, which induces daf-7 expression in the ASJ neuron4, causing half of the avoidance behavior exhibited by trained worms (Fig. 2o inset; 1i, Extended Data Fig. 1j). Separately, pathogenic bacterial sRNAs are taken up by mothers and processed through the canonical RNAi pathway via Dicer activity in the intestine. Subsequent germline signaling involving the piRNA pathway and epigenetic modifiers3 induce daf-7 expression in the ASI neuron, ultimately regulating avoidance of PA14. Only sRNAs are required for transgenerational inheritance of the avoidance behavior signal (Fig. 2n–o; 4g).
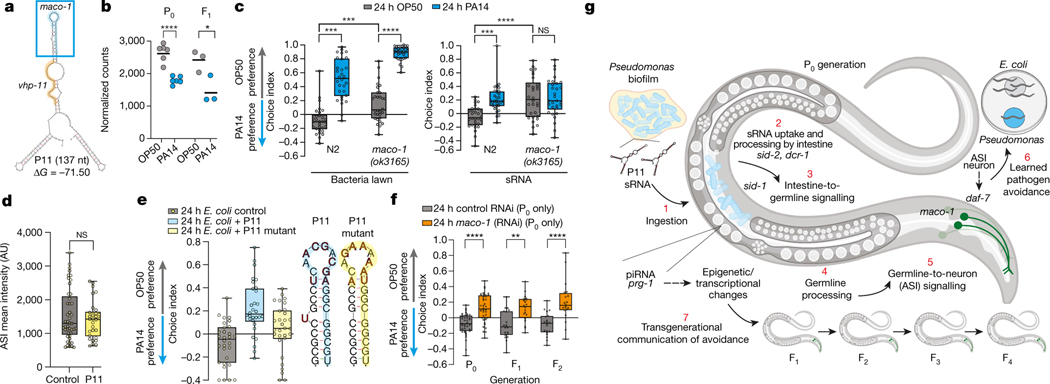
Fig. 4: PA14 P11 sRNA induces avoidance through its target, maco-1.
(a) Predicted secondary structure of wild-type P11 with regions of worm gene homology. (b) maco-1 expression is reduced in PA14-exposed mothers and their naïve F1 progeny3 (mean, DESeq2; P0 p-adj =4.6×10−9, (n=6 replicates/condition), F1 p-adj=0.014, (n=4 replicates/condition). (c, left) maco-1 mutants exhibit high naïve PA14 avoidance, but learning is intact when trained on PA14 lawns. (c, right) maco-1 mutants do not increase PA14 avoidance when trained with PA14 sRNAs. (d) maco-1 mutants do not increase ASI daf-7p::GFP expression upon E. coli+P11 training (n = control(45), P11(30) neurons, two-sided student’s t-test). (e) Wild-type worms fail to avoid PA14 when trained with a P11 mutant in which the maco-1 homology site is altered. Wild-type bases in bold were mutated to disrupt maco-1 homology but retain predicted P11 structure (ΔG = −71.30)46. (f) Mothers trained on maco-1(RNAi) for 24hrs exhibited PA14 avoidance persisting for at least two generations. (g) Model of PA14 P11 sRNA-induced transgenerational learned avoidance. For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. Biological replicates c, e, f(3), d(4). Box plots: center line = median, box range is 25th - 75th percentile; whiskers denote minimum–maximum values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001, n.s. = not significant. Estimation plots provided in Supplementary information. See Supplementary Table 4 for exact sample sizes (n) and p-values.
PA14-P11 sRNA induces avoidance of PA14
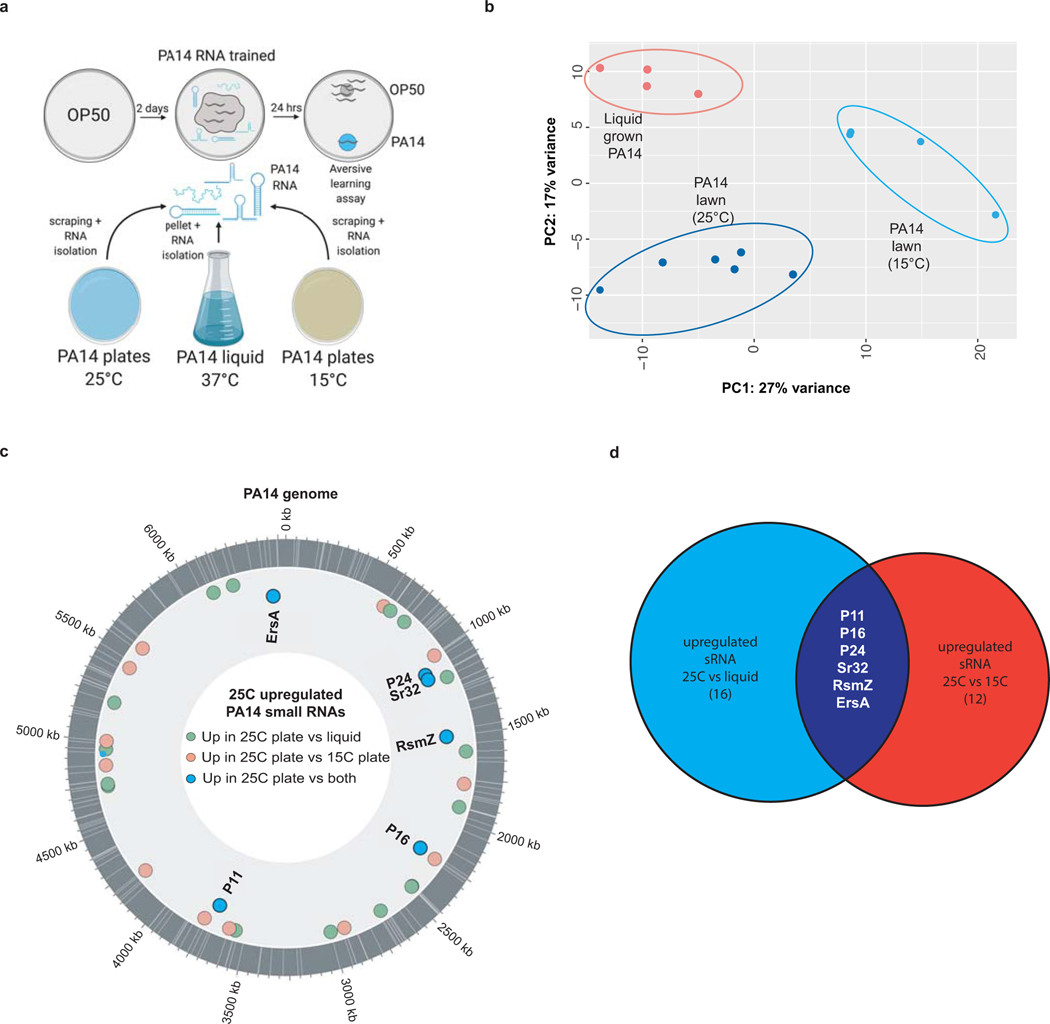
C. elegans utilizes information from pathogenic PA14 sRNA to induce avoidance, despite never having been sick; we wondered whether this was due to a specific sRNA. sRNAs isolated from pathogenic conditions (25°C-plate grown PA14), but not from less-virulent 15°C-grown PA14 and liquid-grown PA14, induced avoidance of PA14 (Fig. 3a–b). Differential expression analysis (Extended Data Fig. 5a–d) revealed 18 and 22 annotated sRNAs32 significantly more highly expressed in 25°C-grown PA14 compared to either 15°C-grown or liquid-grown PA14, respectively (Supplementary Tables 1, 2). Six were upregulated specifically in 25°C samples: P11, P16, P24, PA14sr-032, ErsA, and PrrB/RsmZ (Supplementary Tables 1, 2).
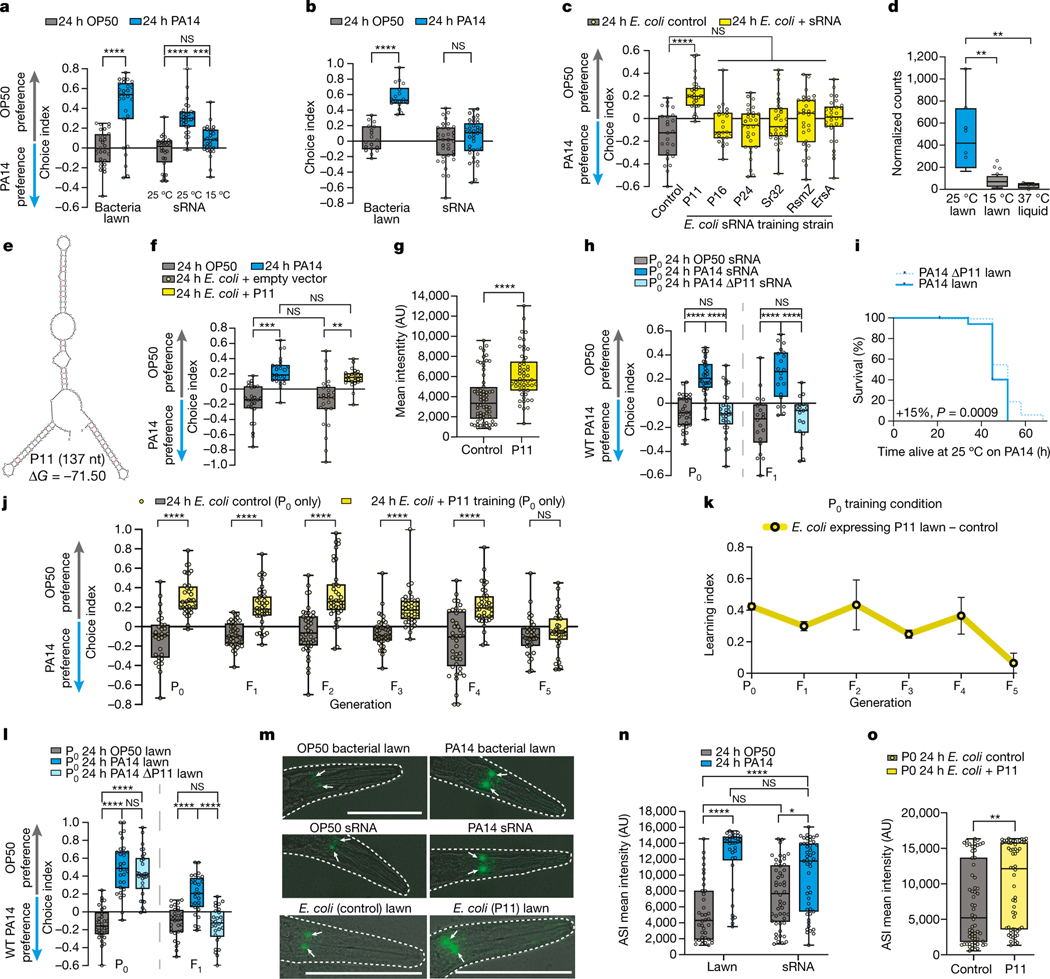
Fig. 3: The PA14 P11 sRNA is required and sufficient for learned avoidance behavior.
(a-b) sRNAs isolated from 15°C-grown (a) or liquid-grown PA14 (b) do not induce PA14 avoidance, while PA14 lawns and sRNA from 25°C plate-grown PA14 do. (c) PA14 preference after training on E. coli expressing PA14 sRNAs (red). (d) DESeq2-normalized counts of P11 expression (25°C vs 15°C, padj=0.006, 6.8x fold change; 25°C vs liquid, padj=0.002, 13.7x fold change; n=25°C(6), 15°C(4), liquid(4) biological replicates). (e) Predicted secondary structure of P11 (ΔG = −71.50)46. (f) sRNAs isolated from PA14 (blue) or E. coli expressing P11 (yellow) induce PA14 avoidance. (g) E. coli+P11 training induces daf-7p::GFP expression in ASI neurons (n = control(65), P11(54) neurons, two-sided student’s t-test). (h) PA14-ΔP11 sRNA does not induce PA14 avoidance in mothers (P0, left), or their progeny (F1, right). (i) Deletion of P11 reduces PA14 pathogenicity. Log-rank (Mantel-Cox test), n=120 animals/condition. (j, k) Mothers exposed to lawns of E. coli+P11 avoid PA14. Untrained progeny of P11-trained mothers avoid PA14 from generation F1-F4. 5th generation resume PA14 attraction. The LI for each generation (k), mean±SEM. (l) Mothers trained on PA14-ΔP11 bacteria avoid PA14 (left), but fail to transmit avoidance (right). (m) ASI daf-7p::GFP expression (white arrows) remains elevated in F1 progeny of PA14-, PA14 sRNA-trained (n, (n= 38, 34, 50, 47 neurons left to right)), and P11-trained mothers (o; (n = control(62), P11(58) neurons, two-sided student’s t-test)). For all learning assays, each dot represents an individual choice assay plate (average of 115 worms/plate) from all replicates. Biological replicates i(2), a, c-d, f-h, l-o(3), b, j, k(4). Box plots: center line = median, box range is 25th - 75th percentile; whiskers denote minimum–maximum values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001, n.s. = not significant. Estimation plots provided in Supplementary information. See Supplemental table 4 for exact sample sizes (n) and p-values.
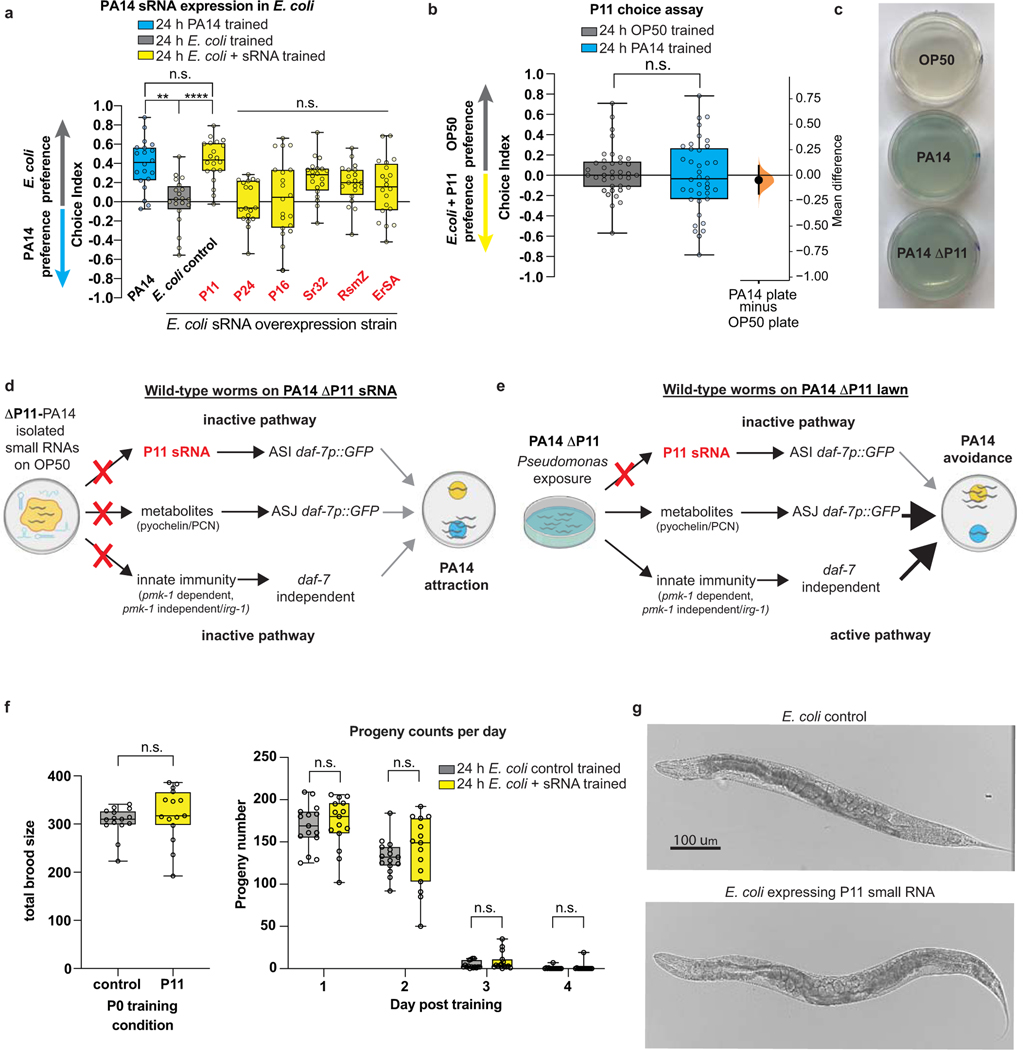
Training worms on E. coli individually expressing each of the six PA14 sRNAs showed that only P11 (“E. coli-P11”) induced PA14 avoidance behavior similar to animals treated with pathogenic PA14 bacteria (Fig. 3c, Extended Data Fig. 6a). (Worms do not avoid E. coli-P11, eliminating P11 itself as the detected component (Extended Data Fig. 6b).) The function of P11 (Fig. 3d–e), a 137-nt non-coding RNA (ncRNA) specific to the Pseudomonas family33, is unknown, but its P. stutzeri ortholog nfiR is required for growth under nitrogen stress conditions34. Worms treated with sRNAs isolated from E. coli-P11 also displayed PA14 avoidance at a similar magnitude to treatment with PA14 sRNA (Fig. 3f). E. coli-P11 treatment induced daf-7 expression in the ASI neuron (Fig. 3g), as with PA14 sRNAs (Fig. 1k). By contrast, mothers treated with sRNAs isolated from PA14 lacking P11 (PA14-ΔP11) (Fig. 3h, Extended Data Fig. 6c–e) did not acquire PA14 avoidance, nor did their progeny (Fig. 3h). Together, these results demonstrate that P11 is both necessary and sufficient for PA14 avoidance learning, despite the fact that exposure to P11 ncRNA does not make worms ill, nor affect reproduction (Extended Data Fig. 6f–g). Thus, C. elegans may have evolved detection of P11, which is both involved in PA14 pathogenesis (Fig. 3i) and upregulated in P. aeruginosa grown in virulent conditions (Fig. 3d), as a biomarker of pathogenic PA14.
P11 induces transgenerational memory
Exposing mothers to E. coli-P11 induced transgenerational inheritance through the F4 generation, similar to that observed with PA14 lawns and PA14 sRNA (Fig. 3j–k). Inherited learned avoidance required P11, as a PA14-ΔP11 lawn failed to caused avoidance in progeny (Fig. 3l). Consistent with sRNAs being required for transgenerational avoidance, PA14 sRNA (Fig. 3m–n) and E. coli-P11 (Fig. 3m, o) exposure to mothers induced daf-7 expression in progeny ASI, similar to PA14 lawn exposure4 (Fig. 3m–n).
MACO-1 is required for sRNA avoidance
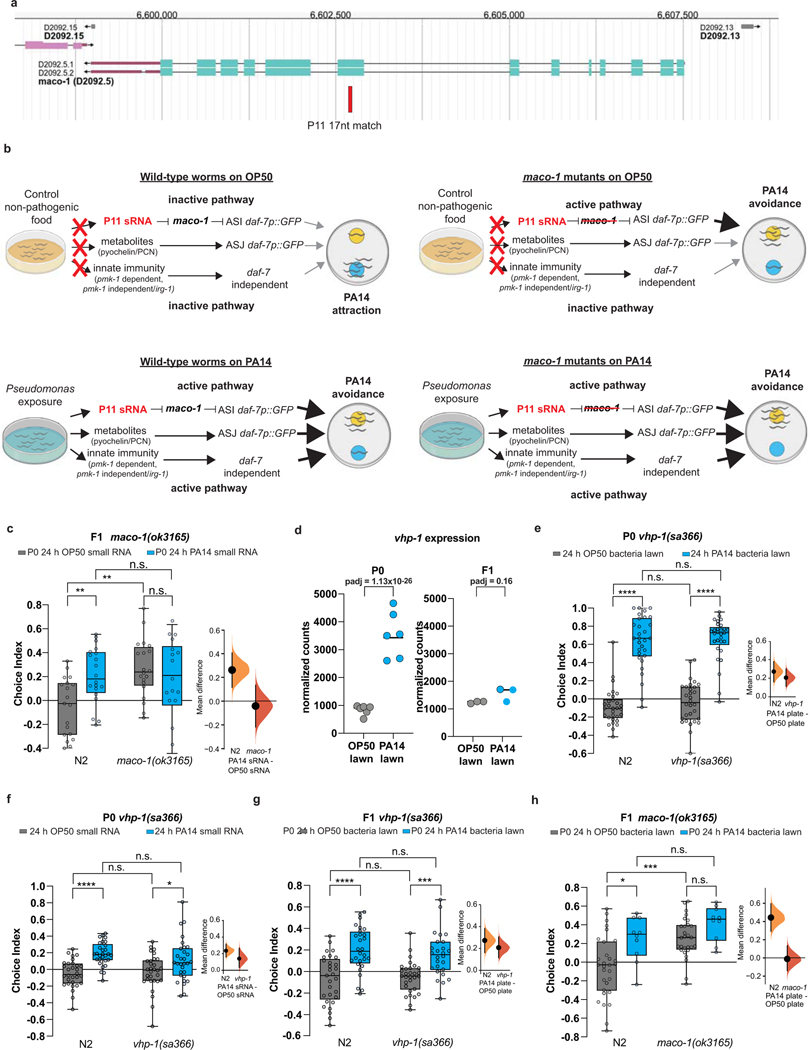
A behavioral response to a specific bacterial sRNA implies the existence of a matching C. elegans sequence. We analyzed the C. elegans genome, including mRNAs and non-coding RNAs, for P11 homology (Table S3). The C. elegans homolog of the mammalian neuronal gene Macoilin35, maco-1, contains the longest perfect match (17nt) to P11 (Fig. 4a, Extended Data Fig. 7a). maco-1 functions in chemotaxis, thermotaxis36, oxygen sensing, and neuronal excitability37. It is expressed in neurons, including the ASI, where it is required for dauer formation38. Upon PA14 exposure, maco-1 expression decreases in mothers and progeny (Fig. 4b). Like ASI mutants, maco-1(ok3165) null mutants exhibit high naïve avoidance of PA14 (Fig. 4c), and do not increase PA14 avoidance upon sRNA treatment (Fig. 4c, right); nor do their progeny (Extended Data Fig. 7b–c). (By contrast, vhp-1, which contains a 16nt match to P11 (Fig. 4a), increased expression upon exposure to PA14, and a null allele (vhp-1(sa366)) did not affect PA14 or sRNA-induced avoidance or inheritance (Extended Data Fig. 7d–g, Fig. 2a).) maco-1 mutants’ innate immune pathways are intact, as maco-1 can learn to avoid PA14 lawns (Fig. 4c, left), but this does not persist in progeny (Extended Data Fig. 7b, h). Expression of daf-7p::gfp in the ASI of maco-1 mutants is not increased upon treatment with E. coli-P11 (Fig. 4d); moreover, training on a P11 mutant that disrupts the perfect match to maco-1 but conserves P11 secondary structure induced no avoidance (Fig. 4e). These results suggest that MACO-1 regulates daf-7 expression in the ASI specifically in response to P11.
Wild-type mothers exposed to E. coli expressing maco-1 dsRNA for 24hrs learned to avoid PA14 (Fig. 4f). Like E. coli-P11, this single exposure to maco-1 RNAi in the P0 generation was sufficient to recapitulate transgenerational memory of PA14 in naïve progeny. Therefore, maco-1 is specifically required for response to PA14 sRNAs, likely through its sequence matching P11.
Together, our data suggest that the ingestion of PA14 and subsequent uptake and Dicer-mediated processing of the PA14 ncRNA P11 in the intestine, followed by further processing and piRNA regulation in the germline, leads to downregulation of maco-1, which in turn is required for upregulation of daf-7 expression in the ASI and subsequent maternal avoidance behavior (Fig. 4g).
Discussion
Here we identified a trans-kingdom signaling system that uses components of the RNAi pathway to “read” bacterial sRNAs. Bacterial sRNAs are an ideal biomarker message, because they are dynamically regulated and reflect bacterial pathogenic state. This sRNA-sensing pathway depends on processing through the germline and subsequent communication to neurons, and is independent of metabolites and innate immunity pathways4. While trans-kingdom signaling has been reported in which sRNAs of a pathogen hijack the host immune system to avoid detection39,40, here we report that an animal host uses a pathogen-produced sRNA to mount a behavioral defense response.
Remarkably, this single exposure of C. elegans to specific bacterial sRNAs causes an avoidance response that is propagated for four additional generations. While other previously-described trans-kingdom signaling systems benefit the pathogen, C. elegans identifies sRNAs of pathogens in order to protect itself and its progeny, and to induce a search for less-pathogenic food sources. This phenomenon may explain why both systemic and transgenerational epigenetic inheritance responses to exogenous RNA exist: to modify the worm’s behavior in response to encounters with naturally abundant, pathogenic bacterial species, which are identified by the worm through unique and decipherable sRNA signatures. This process is distinct from responses that require prolonged, multi-generational bacterial exposure to induce dauer formation41 or infection adaptation42. Rapid, lasting behavioral changes may be more efficient and reproductively advantageous, forcing worms to escape and explore new, potentially safe environments. Passing this avoidance behavior to future generations of progeny may spare them from ever experiencing a prolonged exposure to that pathogen, despite its abundance in the environment. Such a species-specific and plastic response may provide worms with a powerful survival mechanism that is fast-acting and rapidly reversible, a first line of defense against pathogens. This trans-kingdom communication paradigm may represent an adaptive immune memory that prepares future generations for encounters with harmful environmental conditions, allowing them to properly respond to a pathogenic threat without ever experiencing infection and illness.
Materials and Methods
C. elegans and bacterial strains cultivation
Worm strains were provided by the C. elegans Genetics Center (CGC). KU25: pmk-1(km25), AU133: agls17[Pmyo-2::mCherry + Pirg-1::GFP], PY7505: oyIs84 [gpa-4p::TU#813 + gcy-27p::TU#814 + gcy-27p::GFP + unc-122p::DsRed], NL3321: sid-1(pk3321), HC271: ccIs4251 [(pSAK2) myo-3p::GFP::LacZ::NLS + (pSAK4) myo-3p::mitochondrial GFP + dpy-20(+)] I. qtIs3 [myo-2p::GFP dsRNA hairpin]. mIs11 [myo-2p::GFP + pes-10p::GFP + gut-promoter::GFP], YY470: dcr-1(mg375), WM27: rde-1(ne219), NL3531: rde-2(pk1657), WM49: rde-4(ne301), COP2012: alg-1(knu867), RB2519: drh-1(ok3495), CQ636 dcr-1(mg375); vha-6p::dcr-1::dcr-1 3’UTR + [Pmyo-2p::mCherry], MAH23: rrf-1(p1417), NL2099: rrf-3(ok1426), NL917: mut-7(pk204), SX922: prg-1(n4357), RB995: hpl-2(ok916), CQ605 prg-1(n4357); ksIs2 [Pdaf-7p::GFP + rol-6(su1006)], CF1903: glp-1(e2141), CQ640: glp-1(e2141); ksIs2 [Pdaf-7p::GFP + rol-6(su1006)] (strain was made by mating CF1903 with FK181), YL243: unc-119(ed) III; vrls79 [pie-1p::GFP::prg-1 + unc-119(+)], CQ655: prg-1(n4357); unc-119(ed) III; vrls79 [pie-1p::GFP::prg-1 + unc-119(+)] (strain was made by mating SX922 with YL243), JH3225: meg-3(tm4259); meg-4(ax2026), FK181: ksIs2 [Pdaf-7p::GFP + rol-6(su1006)], RB2329: maco-1(ok3165), CQ654 :ksIs2 [Pdaf-7p::GFP + rol-6(su1006)]; maco-1(ok3165) (this strain was made by mating FK181 with RB2329), JT366: vhp-1(sa366). Hermaphrodites were used in all experiments.
Bacterial strains:
P. aeruginosa PA14, and P. aeruginosa ΔlasR were gifts from Z. Gitai. OP50 was provided by the CGC. Control (L4440) and maco-1 RNAi were obtained from the Ahringer library and sequenced verified before use.
General worm maintenance:
Worm strains were maintained at 15°C on High Growth Media (HG) plates (3 g/L NaCl, 20 g/L Bacto-peptone, 30 g/L Bacto-agar in distilled water, with 4 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4, and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving) on E. coli OP50 using standard methods.
General bacterial cultivation:
OP50, P. aeruginosa PA14, and P. aeruginosa ΔlasR were cultured overnight in autoclaved and cooled Luria Broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl in distilled water) shaking (250 rpm) at 37°C. E. coli strains expressing PA14 small RNAs were cultured overnight in Luria Broth supplemented with 0.02% arabinose w/v and 100 μg/mL carbenicillin.
Training plate/worm preparation
Worm preparation:
Eggs from young adult hermaphrodites were obtained by bleaching and subsequently placed onto HG plates and incubated at 20°C for 2 days. Synchronized L4 worms were used in all training experiments. For experiments involving CF1903 (glp-1(e2141)), eggs from mutant and wild-type adult hermaphrodites were obtained by bleaching and placed onto High Growth (HG) plates and left at 25°C for 2 days. Germline loss was confirmed in adult glp-1(e2141) worms raised at 25°C.
Bacteria lawn (25°C) training plate preparation:
Overnight cultures of bacteria (prepared as described above) were diluted in LB to an Optical Density (OD600) = 1 and used to fully cover Nematode Growth Media (NGM) ((3 g/L NaCl, 2.5 g/L Bacto-peptone, 17 g/L Bacto-agar in distilled water, with 1 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4, and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving) plates. For preparation of E. coli expressing PA14 small RNAs, bacteria were seeded on NGM plates supplemented with 0.02% arabinose and 100 μg/mL carbenicillin. All plates were incubated for 2 days at 25°C unless specified otherwise (in separate incubators for control/pathogen seeded plates). On the day of training (i.e., 2 days post bleaching), plates were left to cool on a benchtop for 1 hr to equilibrate to room temperature before the addition of worms. Additionally, for E. coli strains expressing PA14 sRNAs, 200 μL of 0.01% arabinose was spotted onto seeded training plates 1 hr prior to use.
Bacteria lawn (15°C) training plate preparation:
PA14 was prepared by centrifuging 5 mL overnight cultures for 10 minutes at 5000 x g. The supernatant was removed, and the remaining pellet was resuspended in 5 mL of fresh LB. Washed bacteria were used to inoculate (1:500) fresh LB to grow at 15°C for 2 days. Cultures were diluted in LB to an OD600 = 1 and used to seed NGM plates. Plates were incubated at 15°C for 2 days.
DNA/Supernatant/Small RNA training plate preparation:
200 μL of OP50 was spotted in the center of a 10 cm NGM. Plates were incubated at 25°C for 2 days.
Heat-killed bacteria training plate preparation:
One day before plate use, overnight bacteria cultures of OP50 were centrifuged at 5,000 x g for 10 minutes. Following centrifugation, pellets were resuspended in 1/10 volume of LB. Resuspended bacteria pellets were heat shocked at 95°C for 1 hr. Heat shocked bacterial suspensions were left to cool at room temperature for 1 hr, and 200 μL of heat killed bacteria was spotted in the middle of a 10 cm NGM plate supplemented with 100 μg/mL carbenicillin. Plates were incubated at 25°C for 1 day, prior to use. No bacterial growth was observed on heat-killed bacteria plates both before worm training and 24h after worm training.
DNA preparation and training:
Overnight cultures were pelleted at 5,000 x g for 10 minutes at room temperature. DNA was prepared from pelleted bacteria according using the Qiagen DNeasy Blood and Tissue kit and subsequently used fresh. 10 ng of bacterial DNA was placed onto E. coli spots and left to completely dry at room temperature for approximately 1 hr before the addition of worms for training.
Supernatant:
Overnight bacterial cultures (undiluted) were pelleted at 5,000 x g for 10 minutes at room temperature. Supernatant was collected and filtered using a 0.22 μm syringe filter. For worm training plates, 1 mL of filtered supernatant was put onto OP50 spots and left to completely dry at room temperature for approximately 1 hr before the addition of worms for training.
Preparation of bacteria for RNA isolation
Bacteria for RNA collection were prepared as described for training plates (i.e. 2 days on plates at 25°C). Bacterial lawns were collected from the surface of NGM plates using a cell scraper. Briefly, 1 mL of M9 buffer was applied to the surface of the bacterial lawn, and the bacterial suspension following scraping was transferred to a 15 mL conical tube. PA14, ΔlasR, or E. coli expressing PA14 sRNA strains from 10 plates or OP50 from 15 plates was pooled in each tube and pelleted at 5,000 x g for 10 minutes at 4°C. The supernatant was discarded and the pellet was resuspended in 1 mL of Trizol LS for every 100 μL of bacterial pellet recovered. The pellet was resuspended by vortexing and subsequently frozen at −80°C until RNA isolation.
Bacteria RNA isolation
To isolate RNA from bacterial pellets, Trizol lysates were incubated at 65°C for 10 min with occasional vortexing. Debris was pelleted at 7000 x g for 5 min at 4°C. The supernatant was transferred to new tubes containing 1/5 the volume of chloroform. Samples were mixed thoroughly by inverting and centrifuged at 12000 x g for 10 min at 4°C. The aqueous phase was used at input for RNA purification using the mirVana miRNA isolation kit according to the manufacturer’s instructions for total RNA, large RNA (>200 nt), or small RNA (<200 nt) isolation. Purified RNA was used immediately or frozen at −80°C until further use.
RNase treated samples:
For RNase treatment of purified RNA, samples containing 100 μg of RNA were treated with 2.5 μL of an RNAse A (500 U/mL) and RNase T (20,000 U/mL) cocktail for every 50 μL of RNA (RNase Cocktail Enzyme Mix, Ambion). Samples were incubated at room temperature for 20 minutes before adding to worm training plates seeded with OP50. RNase degradation was confirmed using an Agilent 2100 Bioanalyzer. 100 μg of purified small RNA (measured prior to RNAse degradation) treated with RNase was spotted onto plates prior to use for worm training.
DNase treated samples:
For DNase treatment, samples containing 100 μg of purified RNA were treated with 2U of DNase I per 10 μg of RNA using the Invitrogen DNA-free kit according to the manufacturer’s instructions. 100 μg of purified small RNA treated with DNase was spotted onto plates prior to use for worm training.
Total/small RNA/small RNA on heat killed bacteria:
240 μg of total RNA, or 100 μg of small or large, or RNase/DNase-treated small RNA was placed directly onto OP50 spots and left to completely dry at room temperature (~ 1 hr) before use on day of experiment for worm training.
Worm preparation for training
Synchronized L4 worms were washed off plates using M9 and left to pellet on the bench top for approximately 5 minutes. 5 μL of worms were placed onto small RNA-spotted training plates, while 10 μL or 40 μL of worms were plated onto OP50 or E. coli expressing PA14 small RNAs, or pathogen-seeded training plates, respectively. Worms were incubated on training plates at 20°C in separate containers for 24 hr. After 24 hr, worms were washed off plates using M9 and washed an additional 3 times to remove excess bacteria. Worms were tested in an aversive learning assay described below.
Aversive learning assay
Overnight bacterial cultures were diluted in LB to an Optical Density (OD600) = 1, and 25 μL of each bacterial suspension was spotted onto one side of a 60 mm NGM plate and incubated for 2 days at 25°C. After 2 days assay plates were left at room temperature for 1 h before use. Immediately before use, 1 μL of 1M sodium azide was spotted onto each respective bacteria spot to be used as a paralyzing agent during choice assay. To start the assay (modified from 2), worms were washed off training plates in M9 allowed to pellet by gravity, and washed 2 additional times in M9. 5 μL of worms were spotted at the bottom of the assay plate, using a wide orifice tip, midway between the bacterial lawns. Aversive learning assays were incubated at room temperature for 1 hr before manually counting the number of worms on each lawn. Plating a large number worms (>200) on choice assay plates was avoided, since excess worms clump at bacterial spots making it difficult to distinguish animals, and high densities of worms can alter behavior.
In experiments in which F1 and subsequent generations are used: Day 1 worms after from parental (P0) training were bleached and eggs were placed onto HG plates and left for 3 days at 20°C. All animals tested are washed off HG plates with M9 at Day 1. Some of the pooled animals were subjected to an aversive learning assay, while the majority of worms were bleached to obtain eggs, which were then placed onto HG plates left at 20°C for 3 days and used to test F2s.
Imaging and fluorescence quantitation
All images were taken on a Nikon Eclipse Ti microscope. DIC images of whole worms following OP50, or PA14 lawn or small RNA training were imaged at 20X.
Z-stack multi-channel (DIC, GFP) of day 1 adult GFP transgenic worms were imaged every 1 μm at 60X magnification; Maximum Intensity Projections and 3D reconstructions of head neurons were built with Nikon NIS-Elements. To quantify daf-7p::GFP levels, worms were prepared and treated as described for pathogen training. Worms were mounted on agar pads and immobilized using 1 mM levamisole. GFP was imaged at 60X magnification and quantified using NIS-Elements software. Average pixel intensity was measured in each worm by drawing a Bezier outline of the neuron cell body for 2 ASI head neurons and/or 2 ASJ head neurons.
For irg-1p::GFP quantification, whole worms were prepared as described above and imaged at 20x magnification. Average pixel intensity was measured in each worm by drawing a Bezier outline the entire worm.
Brood size assay
L4-stage mothers were trained for 24 h on control (empty vector) or P11-expressing E. coli as described above. After 24 h, 15 individual worms for each condition were transferred to NGM plates seeded with OP50. Worms were transferred to fresh plates every 24 h. Progeny containing plates were incubated at 20°C for 2 days before progeny were counted. Worms were moved daily until progeny production ceased.
Progeny development assay
Mothers were trained on OP50 or PA14 small RNAs as described above. After 24 h of training, mothers were bleached and progeny were transferred to OP50-seeded NGM plates. Plates were incubated at 20°C for 2 days before progeny were assayed for developmental stage.
Small RNA sequencing
Each sample of PA14 small RNA was tested for C. elegans behavior prior to sequencing. The size distribution of small RNA samples was examined on Bioanalyzer 2100 using RNA 6000 Pico chip (Agilent Technologies, CA). For small RNA-seq, around 300 nanograms of small RNA from each sample was first treated with RNA 5’ Pyrophosphohydrolase (New England Biolabs, MA) at 37°C for 30 minutes, then converted to Illumina sequencing libraries using the PrepX RNA-seq library preparation protocol on the automated Apollo 324™ NGS Library Prep System (Takara Bio, CA). Briefly, the treated RNA samples were ligated to 2 different adapters at each end, then reverse transcribed to cDNA and amplified by PCR using different barcoded primers. The libraries were examined on Bioanalyzer DNA High Sensitivity chips (Agilent, CA) for size distribution, quantified by Qubit fluorometer (Invitrogen, CA), then pooled at equal molar amount and sequenced on Illumina NovaSeq 6000 S Prime flowcell as single-end 122 nt reads. The Pass-Filter (PF) reads were used for further analysis.
Small RNA analysis
Pseudomonas aeruginosa (UCBPP-PA14) small RNA stranded reads were trimmed to remove adapters using Cutadapt (v1.16.6). Reads were mapped to the CP000438.1 genome using BWA-MEM. For small RNA analysis, count tables were generated using previously annotated intergenic non-coding RNAs (sRNA)32. Differential gene expression between 25°C plate vs 15°C plate and 25°C plate vs liquid conditions was performed using DESeq2. The Principal component analysis (PCA) plot was generated using DESeq2 on the regularized log transformed counts.
Strain construction
The dcr-1 intestinal rescue strain was made by amplifying 1602 bases upstream of the vha-6 translational start site, and subsequently using PCR to fuse the vha-6 promoter to the dcr-1 cDNA and 3’UTR. Purified PCR products were injected at 10 ng/μL with 1 ng/μL myo-2p::mCherry into dcr-1(mg375) to generate strain CQ636.
E. coli expressing PA14 small RNA strain construction
For cloning the six differentially expressed PA14 sRNA species into E. coli, we utilized the experimentally determined sequences reported by Wurtzel et al. 201243. sRNA sequences were amplified from P. aeruginosa PA14 genomic DNA using the primer pairs described below. The sRNAs were cloned into plasmid, pBAD18-Amp44 which contains an arabinose inducible promoter upstream of a multiple cloning site. Plasmids were transformed into E. coli MG1655. sRNA expression was induced by growing E. coli on NGM plates supplemented with 0.1% arabinose. Proper sRNA production was confirmed for each of the six overexpression strains using RT-PCR. Secondary structure folding predictions for P11 and P11 mutants were performed using Mfold45.
| Primer | Sequence | Restriction Site |
|---|---|---|
|
| ||
| P11-5 | gatacagctagcAATAGCGCAAGTTGGCACG | NheI |
| P11-3 | gatacaaagcttCAGCTTGATCTTCTTCATGCGG | HindIII |
| P16-5 | gatacagctagcGTTCGACGGTGATCGACGAC | NheI |
| P16-3 | gatacaaagcttTCGGCTGTTTCCGTTGGC | HindIII |
| P24-5 | gatacagctagcATTGCGCAATCAAGGTTCTGG | NheI |
| P24-3 | gatacaaagcttGGTAAACGCCGGGTAAAGAATGG | HindIII |
| ersA-5 | gatacagctagcGAATGGCTTCTTGAGCCCTTCG | NheI |
| ersA-3 | gatacaaagcttGCCTGCTGGACGAGGACC | HindIII |
| rsmZ-5 | gatacagctagcGCCGAAGAAGCTTCGCTGG | NheI |
| rsmZ-3 | gatacctctagaCCAAGGCCCGCAAACAGC | XbaI |
| sr32-5 | gatacagaattcGATTCGTGGCTGCACGGG | EcoRI |
| sr32-3 | gatacaaagcttTGCTCCAGAACTACCCCTGG | HindIII |
PA14-ΔP11 mutant strain construction
The unmarked deletion of the P11 ncRNA was constructed by two-step allelic exchange using plasmid pEXG246. Briefly, ~400 bp fragments directly upstream and downstream of the P11 sequence were amplified from gDNA using primer pairs (P11-KO-1, P11-KO-2) and (P11-KO-3, P11-KO-4), respectively. Upstream and downstream fragments were fused together using overlap-extension PCR with primer pair (P11-KO-1, P11-KO-4), and the resulting fragment was cloned into the HindIII site of plasmid pEXG2. The pEXG2 plasmid was integrated into P. aeruginosa PA14 through conjugation with the donor strain E. coli S17. Exconjugants were selected on 30 μg/mL gentamycin and then the mutants of interest were counterselected on 5% sucrose. Proper deletion was confirmed through PCR using primers (P11-seq-5, P11-seq-5).
P11-KO-1) GATACAAAGCTTCCTATGGCGAGATCGAAGCC
P11-KO-2) GCTCCTGCATAAGGTTGGCGCTTGCAGGGATAACGCGGG
P11-KO-3) CCCGCGTTATCCCTGCAAGCGCCAACCTTATGCAGGAGC
P11-KO-4) GATACAAAGCTTGCCGATTGCCGGGTATCG
P11-seq-5) GAAAGACCGCGGGGAGCC
P11-seq-3) CAGTTCTTCCAGGGTACGGACC
E. coli expressing P11 maco-1 homology mutant
To generate the P11 overexpression construct with mutations disrupting the 17-nt of homology to the maco-1 gene, fragments to the left and right of the homology site were amplified from the pBAD18-P11 using primer pairs (P11-macoI-1, P11-macoI-2) and (P11-macoI-3, P11-macoI-4), respectively, and inserted into the NheI/HindIII-cut pBAD18 plasmid by Gibson assembly.
P11-maco-I-1 CTCCATACCCGTTTTTTTGGGCTAGCCATGGAGATAGGCAATAGCGC
P11-maco-I-2 CGCCGCCATTTTTCTGTGGCGGCGCTTTGTTGTCGATTGTCGGG
P11-maco-I-3 CGCCGCCACAGAAAAATGGCGGCGTTTCCCGACCGAACGGGAC
P11-maco-I-4 TCTCATCCGCCAAAACAGCCAAGCTCAGCTTGATCTTCTTCATGCGG
PA14-ΔP11 survival assay
OP50, PA14, or PA14-ΔP11 were grown in liquid culture and diluted as described above. 200 μL of diluted bacteria was spread to completely cover a 6 cm NGM plate. Plates were incubated for 2 days at 25°C to allow bacterial growth. Plates were equilibrated to 20°C before the addition of L4 worms to plates. Assays were performed at 25°C. Assays were counted every 8–10 h until all animals on pathogenic plates died.
Statistical analysis of choice assay data
Populations of animals were raised together under identical conditions and were randomly distributed into treatment conditions. Trained animals were pooled and randomly chosen for choice assays. For all choice assays, each dot represents an individual choice assay plate (~10–300 worms/plate) with all data shown from at least 3 independent replicates (Table S4). Plates were excluded that contained less than 10 total worms per plate. The box extends from the 25th to 75th percentiles, with whiskers from the minimum to the maximum values. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. All figures in the main text and supplement represent pooled data from independent experiments. Results from individual experiments are provided in the statistics supplement. All estimation plots were generated using https://www.estimationstats.com/#/. Additional statistics were generated using Prism 8. Counting of worms on choice assay plates was performed blind with respect to worm genotype and training condition.
Data and materials availability
Sequencing data is available: BioProject PRJNA553700.
All data related to the study that are not provided in the files can be obtained upon reasonable request from the author.
Extended Data
Extended Data 1: PA14 small RNAs induce maternal PA14 avoidance and increased daf-7 expression in the ASI neurons.
(a) Worms exposed to a PA14 bacterial lawn for 24 hr learn to avoid PA14 in a choice assay, while PA14 DNA exposure alone does not induce maternal avoidance of PA14. (b) Training with large RNAs (>200 nt) isolated from bacterial lawns of PA14 is not sufficient for maternal PA14 avoidance. (c) Bioanalyzer results of isolated PA14 total RNA and fractionated small and large RNAs. RNA levels were normalized for worm training. (d) ΔlasR sRNA exposure does not induce PA14 learned avoidance. (e) Development of progeny of PA14 small RNA-trained mothers was not delayed compared to progeny of OP50 trained mothers. n= 6 plates/conditions with 23–142 progeny/plate, mean±SEM. (f) pmk-1(km25) worms learn to avoid PA14 when exposed to PA14 bacteria lawns. (g) pmk-1(km25) is not required for PA14 sRNA-induced pathogenic learning. (h) irg-1p::GFP expression is induced by PA14 bacterial lawn exposure, but not by PA14 sRNAs alone. (i) GFP intensity from (h) was quantified, n = 33, 24, 43, and 35 worms, left to right. (j) Model of PA14 bacteria lawn and small RNA learning. (k) Location of ASI and ASJ neurons. (I) 24 h exposure of worms to a PA14 lawn or PA14 small RNAs increases daf-7p::GFP expression in the ASI neurons. Two-Way ANOVA, Tukey’s multiple comparison test. n = 35, 41, 41, and 49 neurons, left to right. (m) ΔlasR sRNA exposure does not induce changes in daf-7p::GFP ASI expression. One-Way ANOVA, Tukey’s multiple comparison test. n = 38, 27, and 29 neurons, left to right. Biological replicates a(2), b-d, f-i, m(3), l(6). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. One-Way (d, m) and Two-Way ANOVA (a, b, e-g, i, l), Tukey’s multiple comparison test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Extended Data 2: Small RNA and bacterial lawn training of C. elegans RNAi pathway mutants.
(a) Wild-type and sid-2(qt42) worms were trained on OP50 or PA14 bacterial lawns for 24h and tested for learned PA14 avoidance. (b-c) Wild-type and dcr-1(mg375) worms were trained on OP50 or PA14 lawns (b) or OP50 or PA14 sRNA (c). (d) Wild-type, dcr-1(mg375), and dcr-1(mg375);vha-6p::dcr-1 worms were trained on OP50 or PA14 bacterial lawns for 24h and tested for learned PA14 avoidance. Training and choice assays for dcr-1(mg375) and dcr-1(mg375);vha-6p::dcr-1 worms were performed on the same plates. For choice assays, transgenic worms expressing the rescue construct and pharyngeal mCherry were counted using a fluorescence dissecting microscope. Non-transgenic, non-fluorescent dcr-1(mg375) siblings from the same plates were also counted and the results are shown. (e) Wild-type and sid-1(pk3321) worms were trained on OP50 or PA14 bacterial lawns for 24h and tested for learned PA14 avoidance. sid-1 mutants have a constitutively high naïve avoidance, but are able to learn avoidance after training on a PA14 lawn, but not after training on small RNAs. (f, g) Wild-type, rde-1(ne219), (h, i) rde-2(pk1657), or (j) rde-4(ne305) worms were trained on OP50 or PA14 bacterial lawns (f, h, j) or small RNA (g, i) for 24h and tested for learned PA14 avoidance. rde-1, rde-2, and rde-4 mutants are able to learn avoidance following bacteria lawn training, but do not additionally avoid PA14 following small RNA training. (k, l) Wild-type and mut-7(pk720) worms were trained on OP50 or PA14 lawns (k) or sRNA (l) for 24h and tested for learned PA14 avoidance. These mutants (e-l) have high naïve preference after training on small RNAs only. Wild-type and (m, n) alg-1(gk214) or (o, p) drh-1(ok3495) worms were trained on OP50 or PA14 bacterial lawns (m, o) or sRNA (n, p) for 24h and tested for learned PA14 avoidance. Biological replicates d, f, h-p(3), a-c, g(4), e(6). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-Way ANOVA (a-p), Tukey’s multiple comparison test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Extended Data 3: Small RNA and bacterial lawn training of C. elegans small RNA pathway and germline mutants.
(a-e) Wild-type and (a) prg-1(n4357), (b) prg-1 germline rescue, (c) rrf-1(pk1417), (d) rrf-3(pk1426) or (e) hpl-2(ok916) worms were trained on OP50 or PA14 bacterial lawns or sRNA (a, right) for 24h and tested for learned PA14 avoidance. (f-g) Representative images of OP50 or PA14 trained prg-1;daf-7::gfp (f) or glp-1;daf-7::gfp (g) mothers. (ASI neuron = white arrow, ASJ neuron = gray arrow). (h) Germline-less glp-1 mutants induce daf-7p::GFP expression in the ASJ neuron after PA14 lawn exposure. (n = OP50(42), PA14(46) neurons, two-tailed Student’s t-test. (i) Wild-type and meg-3(tm4259);meg-4(ax2026) worms were trained on OP50 or PA14 bacterial lawns for 24h and tested for learned PA14 avoidance. Biological replicates a-i(3). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-Way ANOVA (a-e, i), Tukey’s multiple comparison test. ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Extended Data 4: sid-1, sid-2, and dcr-1 F1 progeny of plate and small RNA trained parents.
(a-c) Progeny of (a) sid-1, (b) sid-2, and (c) dcr-1 are defective in both transgenerational pathogen avoidance following maternal bacterial lawn (left) and PA14 sRNA training (right). Biological replicates a-c(3). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-Way ANOVA (a-c), Tukey’s multiple comparison test. **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Extended Data 5: RNA-seq of small RNAs from PA14.
(a) Small RNA training protocol using RNA isolated from PA14 cultures grown at 25°C or 15°C on plates, or in liquid culture. (b) Principal component analysis of PA14 small RNA sequencing. (c) 25°C upregulated PA14 sRNAs compared to 15°C-grown and liquid-grown PA14 sRNAs. The outermost gray circle represents the PA14 genome, with previously identified sRNAs that were upregulated (DESeq2, adjusted p-value ≤ 0.05) in the 25°C plate condition relative to 15°C plate grown PA14 (pink) or liquid grown PA14 (green) are indicated. Overlapping sRNAs upregulated in the 25°C plate condition relative to both other conditions are noted (blue dots). sRNAs annotated with white lines. (d) Venn diagram showing the number of PA14 sRNAs upregulated at 25°C growth conditions compared to liquid growth (blue circle) or 15°C growth (red circle). The 6 shared upregulated sRNAs are shown.
Extended Data 6: Identification and testing of the differentially regulated PA14 small RNAs.
(a) P11 expressed in E. coli induces PA14 avoidance in a choice assay between PA14 and E. coli strain MG1655. Worms were trained on OP50, PA14, E. coli strain MG1655 containing empty vector (control), or E. coli strain MG1655 expressing PA14 small RNAs (red). Following training, a choice assay was performed between E. coli strain MG1655 and PA14. Similar to Fig 3c, worms trained on PA14 or E. coli expressing P11 exhibited PA14 avoidance behavior. One-way ANOVA with Tukey’s multiple comparison test. (b) PA14 plate-trained worms do not avoid E. coli expressing P11 compared to OP50 in a choice assay. Two-tailed student’s t-test (c) PA14-ΔP11 bacteria grown on NGM plates produce pyocyanin (blue pigment on plates) similar to PA14. (d-e) Model showing induced learning pathways for wild-type worms on isolated PA14-ΔP11 small RNAs (d) or on PA14-ΔP11 bacteria lawns (e). (f) Exposure to E. coli expressing P11 does not affect total brood size (left) (two-tailed student’s t-test), or the number of progeny hatched per day (right), two-way ANOVA with Tukey’s multiple comparison test. N = 15 worms were analyzed per condition. (g) Worms exposed to control (top) or E. coli expressing P11 appear healthy after 24 of training. Biological replicates a(2), b-c, g(3). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. **p ≤ 0.01, ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Extended Data 7: P11 region of homology with maco-1 and behavior of vhp-1 mutants.
(a) Wormbase depiction of the maco-1 genomic locus and the region of P11 homology (red). (b) Model depicting induced learning pathways in wild-type or maco-1 mutant worms as a result of OP50 or PA14 lawn exposure. (c) F1 progeny of OP50 or PA14 sRNA exposed maco-1 mutant mothers were tested for PA14 avoidance behavior. (d) vhp-1expression is increased in PA14-exposed mothers, but not in F1 progeny3. Mean; DESeq2 p-adj values are shown. P0, n=6 replicates/condition; F1, n=4 replicates/condition. (e) The vhp-1(sa336) mutation is a null allele (early stop codon)40. Wild-type, and vhp-1(sa366) worms were trained on OP50 or PA14 bacterial lawns for 24h and tested for learned PA14 avoidance (top). (f) Wild-type, and vhp-1(sa366) worms were trained on OP50 or PA14 small RNA for 24h and tested for learned PA14 avoidance. (g) Progeny of vhp-1(sa366) PA14 lawn trained animals inherit PA14 avoidance behavior. (h) F1 progeny of OP50 or PA14 lawn exposed maco-1 mutant mothers were tested for PA14 avoidance behavior. Biological replicates c, e-h(3). For choice assays, each dot represents an individual choice assay plate (average of 115 worms/plate) with data shown from all replicates. For box plots, center line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-Way ANOVA (c, e-h), Tukey’s multiple comparison test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, n.s. = not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplemental table 4 for exact sample sizes (n) and p-values.
Supplementary Material
Acknowledgments:
We thank the C. elegans Genetics Center for strains; the Genomics Core Facility at Princeton University; W. Wang for helping develop methods to sequence bacterial small RNAs; the Murphy lab for discussion; and Robert Clausen and Jasmine Ashraf for assistance. CTM is the Director of the Glenn Center for Aging Research at Princeton and an HHMI-Simons Faculty Scholar.
Funding: This work was supported by a Pioneer Award to CTM (NIGMS DP1GM119167), the Glenn Foundation for Medical Research (GMFR CNV1001899), the HHMI-Simons Faculty Scholar Program (AWD1005048), T32GM007388 (NIGMS) support of RSM and GDV, and a Pioneer Award to ZG (DP1A1124669).
Footnotes
Competing interests: Authors declare no competing interests.
Supplementary Information is available for this paper.
References:
- 1.Samuel BS, Rowedder H, Braendle C, Félix M-A & Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci 113, E3941–E3949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Lu H. & Bargmann CI Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Moore RS, Kaletsky R. & Murphy CT Piwi/PRG-1 Argonaute and TGF-β Mediate Transgenerational Learned Pathogenic Avoidance. Cell 177, 1827–1841.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meisel JD, Panda O, Mahanti P, Schroeder FC & Kim DH Chemosensation of Bacterial Secondary Metabolites Modulates Neuroendocrine Signaling and Behavior of C. elegans. Cell 159, 267–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DH et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Melo JA & Ruvkun G. Inactivation of Conserved C. elegans Genes Engages Pathogen- and Xenobiotic-Associated Defenses. Cell 149, 452–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K. & Mylonakis E. An Intestine-Derived Neuropeptide Controls Avoidance Behavior in Caenorhabditis elegans. Cell Rep. 20, 2501–2512 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Estes KA, Dunbar TL, Powell JR, Ausubel FM & Troemel ER bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci 107, 2153–2158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troemel ER et al. p38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. elegans. PLoS Genet. 2, e183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghildiyal M. & Zamore PD Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winston WM, Sutherlin M, Wright AJ, Feinberg EH & Hunter CP Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci 104, 10565–10570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E, Caudy AA, Hammond SM & Hannon GJ Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Ketting RF Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwan DL, Weisman AS & Hunter CP Uptake of Extracellular Double-Stranded RNA by SID-2. Mol. Cell 47, 746–754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaletsky R. et al. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLOS Genet. 14, e1007559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winston WM Systemic RNAi in C. elegans Requires the Putative Transmembrane Protein SID-1. Science 295, 2456–2459 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Tabara H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Tops BBJ et al. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33, 347–355 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabara H, Yigit E, Siomi H. & Mello CC The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861–871 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Ketting RF, Haverkamp THA, van Luenen HGAM & Plasterk RHA mut-7 of C. elegans, Required for Transposon Silencing and RNA Interference, Is a Homolog of Werner Syndrome Helicase and RNaseD. Cell 99, 133–141 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Posner R. et al. Neuronal Small RNAs Control Behavior Transgenerationally. Cell 177, 1814–1826.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H. et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grishok A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Sowa JN et al. The Caenorhabditis elegans RIG-I Homolog DRH-1 Mediates the Intracellular Pathogen Response upon Viral Infection. J. Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashe A. et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welker NC et al. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893–903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoki K, Moriguchi H, Yoshioka T, Okawa K. & Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smardon A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol 10, 169–178 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Couteau F, Guerry F, Müller F. & Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3, 235–241 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin J. & Kimble J. glp-1 Is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51, 589–599 (1987). [DOI] [PubMed] [Google Scholar]
- 31.Ouyang JPT et al. P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cell 50, 716–728.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurtzel O. et al. The Single-Nucleotide Resolution Transcriptome of Pseudomonas aeruginosa Grown in Body Temperature. PLoS Pathog. 8, e1002945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livny J. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 34, 3484–3493 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan Y. et al. NfiR, a New Regulatory Noncoding RNA (ncRNA), Is Required in Concert with the NfiS ncRNA for Optimal Expression of Nitrogenase Genes in Pseudomonas stutzeri A1501. Appl. Environ. Microbiol 85, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuvbachieva A. et al. Identification of a novel brain-specific and reelin-regulated gene that encodes a protein colocalized with synapsin. Eur. J. Neurosci 20, 603–610 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Miyara A. et al. Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in Caenorhabditis elegans. PLoS Genet. 7, e1001384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arellano-Carbajal F. et al. Macoilin, a Conserved Nervous System–Specific ER Membrane Protein That Regulates Neuronal Excitability. PLoS Genet. 7, e1001341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neal SJ et al. A Forward Genetic Screen for Molecules Involved in Pheromone-Induced Dauer Formation in Caenorhabditis elegans. G3amp58 GenesGenomesGenetics 6, 1475–1487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudzik C, Hou Y, Ma W. & Axtell MJ Exchange of Small Regulatory RNAs between Plants and Their Pests. Plant Physiol. 182, 51–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiberg A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palominos MF et al. Transgenerational Diapause as an Avoidance Strategy against Bacterial Pathogens in Caenorhabditis elegans. mBio 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton NO et al. Cysteine synthases CYSL-1 and CYSL-2 mediate C. elegans heritable adaptation to P. vranovensis infection. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambers JR & Sauer K. The MerR-Like Regulator BrlR Impairs Pseudomonas aeruginosa Biofilm Tolerance to Colistin by Repressing PhoPQ. J. Bacteriol 195, 4678–4688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzman LM, Belin D, Carson MJ & Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hmelo LR, Borlee BR, Almbald H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, & Harrison JJ Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc 10, 1820–1841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho J, Tumkaya T, Aryal S, Choi H. & Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nat. Methods 16, 565–566 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.