Abstract
The COVID-19 pandemic creates a significant impact on everyone’s life. One of the fundamental movements to cope with this challenge is identifying the COVID-19-affected patients as early as possible. In this paper, we classified COVID-19, Pneumonia, and Healthy cases from the chest X-ray images by applying the transfer learning approach on the pre-trained VGG-19 architecture. We use MongoDB as a database to store the original image and corresponding category. The analysis is performed on a public dataset of 3797 X-ray images, among them COVID-19 affected (1184 images), Pneumonia affected (1294 images), and Healthy (1319 images) (https://www.kaggle.com/tawsifurrahman/covid19-radiography-database/version/3). This research gained an accuracy of 97.11%, average precision of 97%, and average Recall of 97% on the test dataset.
Keywords: Deep learning, Transfer learning, COVID-19, Pneumonia, MongoDB
Introduction
Medical Imaging plays a significant role in the modern healthcare system. Advancements of medical imaging devices, like Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and X-Ray Imaging have become extremely necessary for disease diagnosis. Deep learning plays an essential factor in the medical industry as it can achieve human-level accuracy for image classification and segmentation tasks [1]. Deep learning can perform great on tasks in the fields like computer-aided diagnostics [2], recognition of lesions and tumors [3], and brain–computer interface [4]. Deep learning can easily execute time-consuming tasks, like detect symptoms of COVID-19 and Pneumonia from the chest X-ray images as the lungs of affected patients carry some visual marks of that distinct disease. Newly identified coronavirus causes an infectious disease namely COVID-19 [5]. Many patients who are suffering from COVID-19 can have minor to moderate effects and recover without additional care. An infected person’s cough and sneeze primarily spread COVID-19. Pneumonia is usually caused by viruses, fungi, and bacteria and creates infection in the lungs [6]. Both COVID-19 and pneumonia become life-threatening for those who have chronic diseases. These diseases decrease the body’s oxygen level and create complications for a person to breathe. Patients with serious conditions have to be hospitalized and need to be on ventilator support.
DBMS plays an important role in the medical field to save data for further analysis. Traditional Database Management Systems are inadequate in semi-structured and unstructured data because of not maintaining Big data features, namely velocity, value, variety, validity, veracity, volume, and volatility [7, 8]. Several high-tech companies like LinkedIn, Google, Facebook, and Amazon have customized storage systems to operate with huge data volumes. Not Only SQL (NoSQL) is a distributed database management system, and its demand is increasing to handle big data efficiently. As working with vast amounts of data, big companies develop their NoSQL databases, i.e., Ebay’s MongoDB [9], Google’s Bigtable [10], Facebook’s Cassandra [7].
In this paper, we developed a system using the transfer learning method to classify chest X-ray images into COVID-19, Pneumonia, and Healthy. At first, we applied several image processing techniques on the chest X-ray images, and used pre-trained VGG-19 architecture, and transferred weights for classification purposes. Finally, MongoDB [9] is used as a database to store the original image and corresponding category images as MongoDB needs less data load time and query time than traditional RDMS in the case of BLOB files. Here, our goal is to make a flexible system that can modify in the future. Our system has achieved a higher accuracy using a pre-trained model’s most of the features. Moreover, this work describes the background database and the advantage of using this kind of database to store classified data for further study. By connecting the deep learning model with the database makes this research complete and formulates a new dimension.
The rest of the paper is arranged as follows: “Related Works” describes some related and important recent works. “X‑ray Image Classification” outlines the methodology of this work. The experimental outcome is analyzed in “Performance Analysis”. Finally, a summary of our findings is described in the “Conclusions” section.
Related Works
In recent times, several good works are done utilizing Convolutional Neural Network to solve medical-related problems such as detecting brain tumors from MRI images, breast cancer recognization, classification of diseases using X-ray images [11]. Fukushima originally introduced CNN with the help of the model developed by Wiesel and Hubel [12]. By utilizing CNN, segmentation of neuron membranes was proposed by Ciresan et al. [13]. Wang et al. [14] developed a unique dataset with 108,948 X-ray images of 32,717 patients, which accomplished excellent outcomes utilizing a deep CNN. Rajpurkar et al. [15] developed a deep CNN architecture to identify 14 distinct pathologies from chest X-ray images, and the model contains 121 layers. Zhou et al. [16] applied the transfer learning approach for distinguishing between malignant and benign tumors by utilizing InceptionV3 architecture. Deniz et al. [17] introduced a classifier by utilizing a pre-trained VGG-16 architecture for breast cancer. During the feature extracting procedure, the proposed system used AlexNet architecture. Furthermore, transfer learning was applied to smaller datasets of MRI images, and the time of feature extraction portion utilize pre-trained networks [18]. Yang et al. [19] utilized GoogLeNet and AlexNet for their experiment on glioma grading, and GoogLeNet displayed excellent results for the analysis compare to AlexNet.
Here, we successfully applied the transfer learning approach on the pre-trained VGG-19 architecture. We measured the system’s performance for the classification of COVID-19-affected, Pneumonia-affected, and Healthy people from the chest X-ray images. Furthermore, MongoDB is used as a storage for the original images and classified labels.
X-ray Image Classification
This section includes the following steps: image pre-processing, classification of X-ray images using transfer learning, and finally, description of data storing process in MongoDB database. The entire working process of the system is shown in Fig. 1.
Fig. 1.
Overview of the whole system
Image Pre-processing
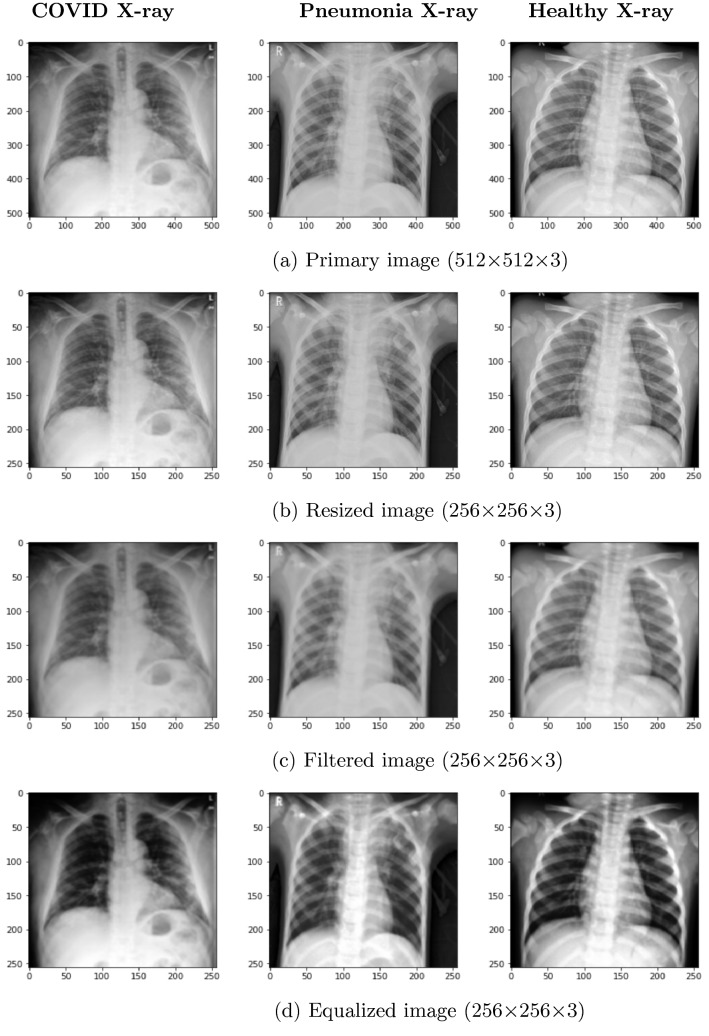
We applied several image processing steps to make the image ready for the experiment. The primary image was 5123 sized. We resize the images into 2563 size. Furthermore, apply a Gaussian filter on the resized image and produce a filtered image.
| 1 |
Equation 1 represents the 2D Gaussian function with a kernel of size 55 is applied. Here, X and Y indicate the length between origin and the horizontal axis and the length between origin and the vertical axis, respectively, and indicates the standard deviation. It removes noise from a resized image. The histogram equalization is applied on every filtered image. This technique enhances the contrast of an image and also spread out values that appear most frequently. These X-ray images are multichannel, and each image has three different (RGB) channels. To utilize the full power of VGG and achieve higher accuracy multi-channel X-Ray images are used, as the VGG-19 [20] model is trained using three channels images. Therefore, we have to separate each channel and apply histogram equalization individually. After that, we merge back every histogram equalized channel and get an equalized image.
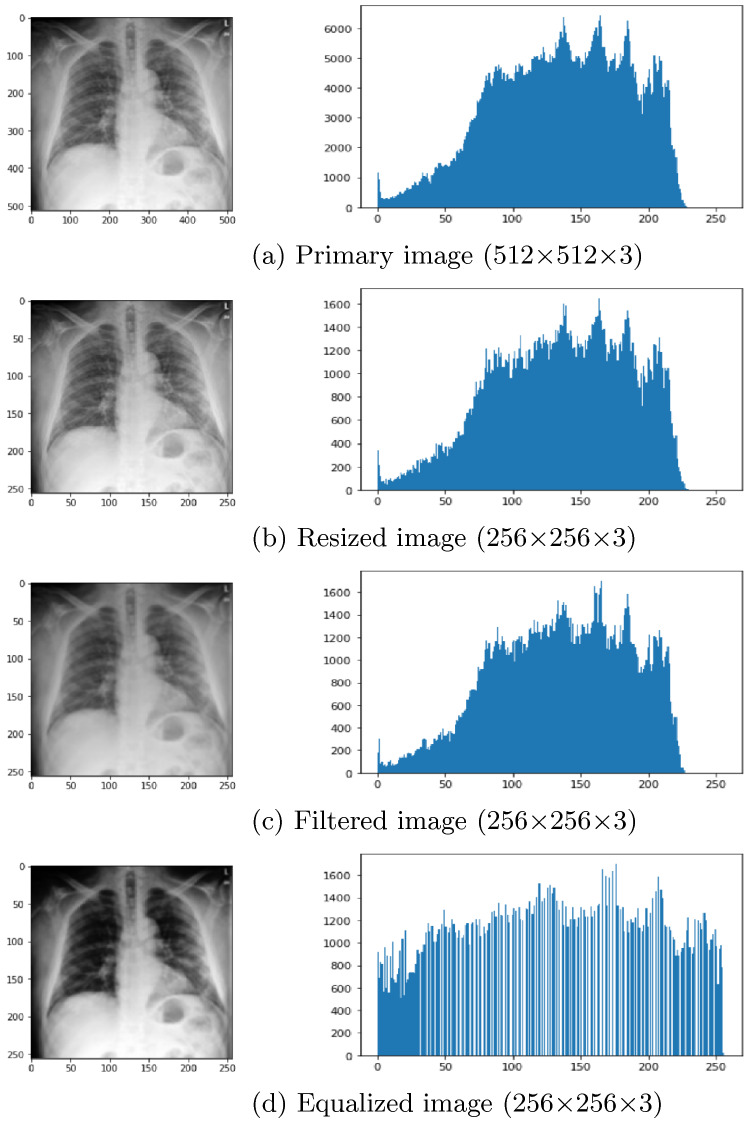
Figure 2b shows the resized image of Fig. 2a. Figure 2c represents the filtered image as it helps to reduce noise and make the image smoother. Also, it enhances the probability for detecting features. Furthermore, Fig. 2d shows the histogram equalized image. Histogram equalization is applied for balancing the contrast of an image by adjusting the intensity distribution. The goal of this method is to provide a linear relationship to the cumulative likelihood function. Figure 3 represents histogram on various pre-processing level outcomes. Figure 3a–c represents the histogram of primary, resized and filtered image, respectively, for an X-ray image. Figure 3d represents the histogram of an equalized image where all the contents are balanced.
Fig. 2.
Image pre-processing result for the system
Fig. 3.
Histogram of various pre-processing level outcomes
Transfer Learning for X-ray Image Classification
Transfer learning is a process to reuse parameters that have been trained before. It implies transferring the pre-trained parameters to a different model. Moreover, pre-trained layers can be applied as a feature extractor.
VGG-19 is a 19-layer deep architecture [20]. It is a CNN model trained on the ImageNet dataset that includes millions of images of 1000 categories. The input layer’s size of VGG-19 is 2243, followed by several filters, each with a kernel size of 33. Therefore, originally it takes three-channel images. It contains multiple convolutional layers. The convolution layer applies several convolution kernels for the extraction of various features.
Transfer Parameters from VGG-19
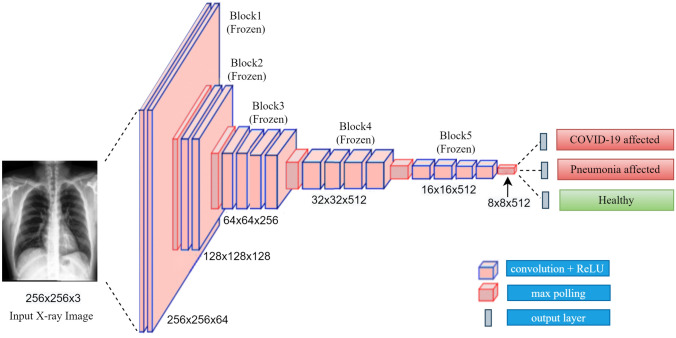
Figure 4 shows the transfer parameters from VGG-19 for the classification task. From the 19 layers of the VGG-19 architecture, we used the first 16 layers. The architecture of the first 16 layers of VGG-19 is not the same as VGG-16 [20]. VGG-16 has fewer convolutional layers than VGG-19. All of these convolutional layers of VGG-19 are also sequentially organized in five blocks. These parameters are considered frozen or non-trainable.
Fig. 4.
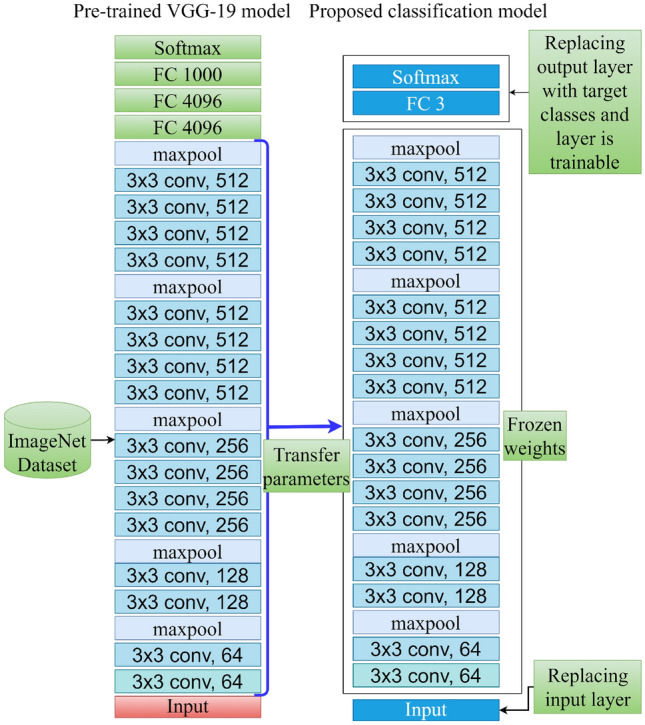
Transfer parameters from VGG-19 for the classification task
The Classification Model
We replace the original input layer of the VGG-19 model with a layer that can take 2563 sized images as input. Instead of the last three layers, added a dense layer with three neurons and used softmax as an activation function. This layer is trainable. Figure 5 represents the entire architecture of the classification model. The overview of the trainable and non-trainable parameters is shown in Table 1.
Fig. 5.
Architecture of the classification model
Table 1.
Parameters for the classification model
| Layers | Parameters |
|---|---|
| Block1 (Non-trainable) | 38,720 |
| Block2 (Non-trainable) | 221,440 |
| Block3 (Non-trainable) | 2,065,408 |
| Block4 (Non-trainable) | 8,259,584 |
| Block5 (Non-trainable) | 9,439,232 |
| Dense (Trainable) | 98,307 |
| Total | 20,122,691 |
| Total trainable | 98,307 |
| Total non-trainable | 20,024,384 |
Mongodb as a Data Storage
In the medical domain, it is essential to store data in a database for further analysis. Figure 6 illustrates the workflow of data storage in the medical domain.
Fig. 6.
Workflow of data storage in the medical field
BLOB file is a collection of binary data. Images, audios, and other types of records are saved as blob files. If an image is converted into a BLOB file, that image becomes compressed and uses less space. Further, we can do several operations as it already been converted into a binary file. Moreover, at the moment of movement of the database not have to bother with the distributing files. For saving an X-ray image in the database, we have to convert it into a BLOB file. The relational database management systems are popular technology in the case of data storage. However, it is difficult to work with BLOB files in the traditional SQL database. Storing BLOB files into an SQL database requires a huge data query process. If the size of an image is small, it can easily store as a BLOB in the SQL DBMS. However, for a large file storing or retrieving BLOB on RDBMS is a costly process and needs large memory. Furthermore, the SQL databases face challenges in the situation like the scalability of the system, and fast data access during complicated operations [21]. It is easy to work with a BLOB file in the MongoDB database as it stores a BLOB file by splitting it into several numbers of chunks. At the time of data retrieval, all the chunks merge into one single file and produce the result. For this reason, MongoDB needs lesser data load time and query time than traditional RDMS. Therefore, we used MongoDB with GridFS interface as a data storage for the system.
After the classification step, the original images and the corresponding label have to store in the MongoDB database. However, MongoDB cannot retain a document file that crosses 16 MB in size. To tackle this problem, the GridFS file storage system is introduced. A combination of MongoDB and GridFS can store image files that are massive in size. Data are separated into two collections in the MongoDB server. These are fs.files and fs.chunks. In fs.files, there are different fields like id, file name, encoding type, MD5, chunk size, length, and upload time. The id field contains a unique key for a file. For this experiment, all the labels are stored in the file name field. The chunk size field determines the volume of a chunk in bytes. The upload time field stands for the time of uploading the file. The server automatically generates MD5, and it represents a check value.
In the case of fs.chunks collection, there are several fields like id, chunk number, file id, and data package. Here id field contains a unique key for a chunk. File id is the same as id in the fs.files for a particular file, and it’s fixed for all the chunks of the same file. The chunk number field represents the chunk’s position, and the data package contains a binary representation of the chunk for an image. Figure 7 represents the data storage for an X-ray image using MongoDB and GridFS interface.
Fig. 7.
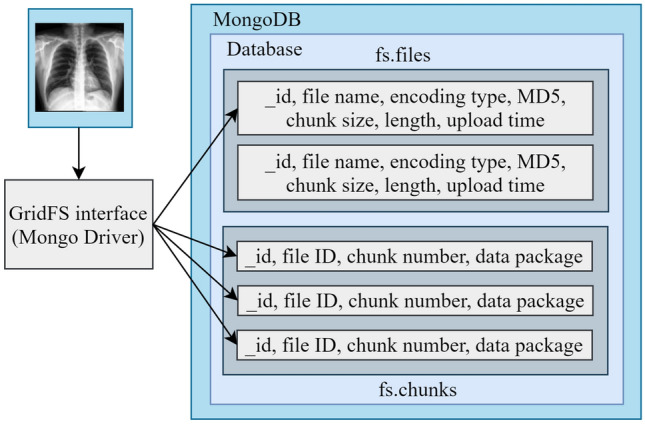
Store images in the MongoDB server
Performance Analysis
The experimental outcomes are illustrated in this Section. We analyze system performance using different performance metrics.
Dataset
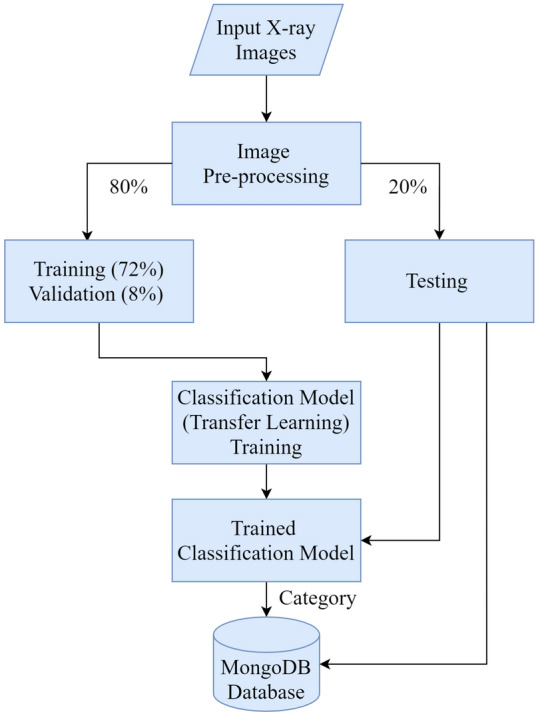
The chest X-ray images obtained from different publicly available sources [22, 23]. We used 3797 images, among these COVID-19 (1,184 images), Pneumonia (1294 images), and Healthy (1319 images). We separated them into three sets from the original dataset and 2733 images, 72% of the dataset, were used for training purposes. 304 images, 8% of the dataset, were used for validation purposes, and the rest of the 760 images, 20% of the dataset, were used for testing purposes. Table 2 represents the summary of the used dataset.
Table 2.
Description of dataset
| Label | Train set | Validation set | Test set | Total |
|---|---|---|---|---|
| COVID-19 | 852 | 94 | 238 | 1184 |
| Pneumonia | 932 | 108 | 254 | 1294 |
| Healthy | 949 | 102 | 268 | 1319 |
Performance Metric
The performance of the classification model is tested using precision, recall, and F1-score. We also use the confusion matrix and ROC curve. We average all the metrics of different classes for validation purposes and give priority base on precision, recall, and F1 score, respectively. During measuring, the following equations are used.
| 2 |
| 3 |
| 4 |
Experimental Setup
We executed all the image pre-processing steps analysis on a computer having an operating system of Windows 10, with NVIDIA GeForce GPU and 8 GB RAM CPU. The rest of the operations, including training and testing of the classification model, were executed on Colab.
Adam optimizer [24] is applied to compile the classification model. Adam optimizer is a powerful optimization algorithm. Moreover, it combines different optimization techniques, including Root Mean Square Propagation, and sparse categorical cross-entropy is used as a loss function. Here, learning rate = 0.001 and momentum parameters = 0.9 & = 0.999. A batch size of 32 and 30 epochs were applied while fitting the training data on the model.
Experimental Results
Figures 8 and 9 represent the accuracy and the loss of the training and validation set, respectively. We got an accuracy of 97.11% while testing the model on the test dataset.
Fig. 8.
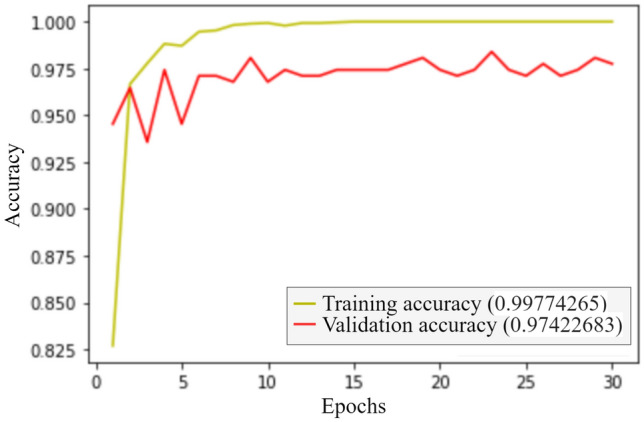
Accuracy curve for the classification process
Fig. 9.
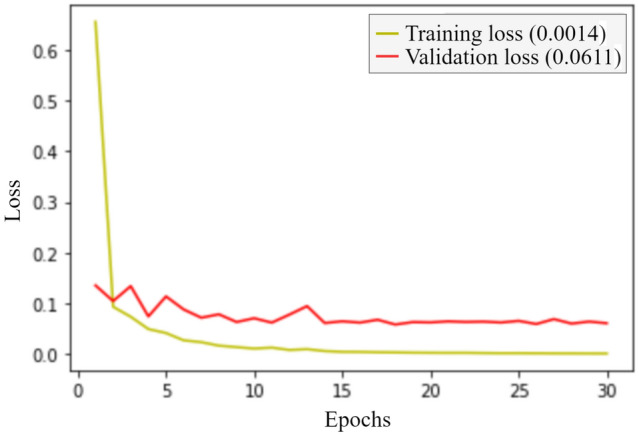
Loss curve for the classification process
Figure 10 shows the confusion matrix for the X-ray image classification model. For testing purposes, 238 COVID-19-affected chest X-ray images as Class 0, 268 images of healthy chest X-ray as Class 1, and 254 images of pneumonia-affected chest X-ray as Class 2 were used. Table 3 represents performance measure indices. The model achieved a precision of 99%, 95%, 97% for Class 0 (COVID-19), Class 1 (Healthy), and Class 2 (Pneumonia), respectively. The average precision is 97%. Figure 11 represents the ROC curve. The overall accuracy grows higher as the curve moves nearer to the top left corner.
Fig. 10.
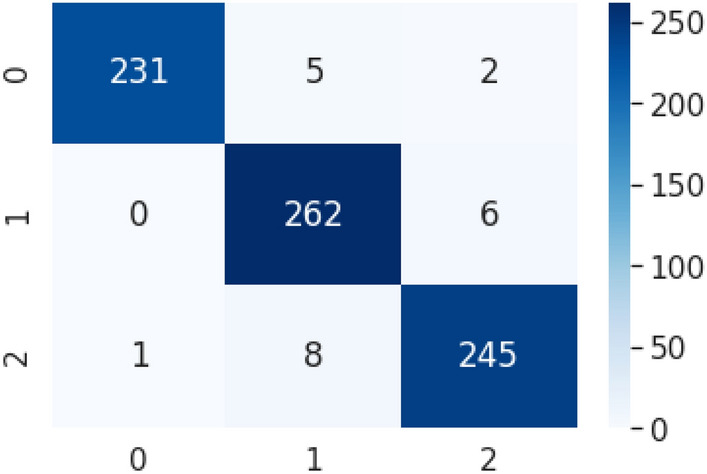
Confusion matrix. Class 0 (COVID-19), Class 1 (Healthy), and Class 2 (Pneumonia)
Table 3.
Performance measure indices
| Class | Precision | Recall | F1-score | Support |
|---|---|---|---|---|
| Class 0 (COVID) | 0.99 | 0.97 | 0.98 | 238 |
| Class 1 (Healthy) | 0.95 | 0.98 | 0.96 | 268 |
| Class 2 (Pneumonia) | 0.97 | 0.96 | 0.96 | 254 |
| Avg./Total | 0.97 | 0.97 | 0.97 | 760 |
Fig. 11.
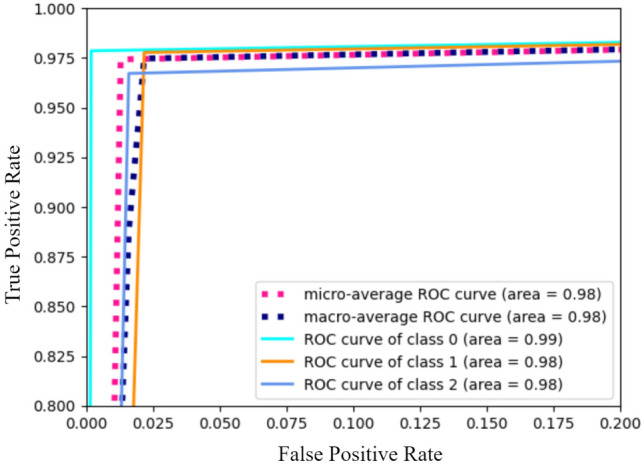
ROC curve of the classification task
Moreover, the recommended system performance is analyzed in Table 4 with related works. This table also describes the methodology behind all the tasks. The proposed method has gained 97.11% accuracy, which is best in all the referenced studies.
Table 4.
Comparison of recently used approaches for COVID-19 detection
| References | Dataset | Methodology | Accuracy (%) |
|---|---|---|---|
| This work | X-ray image | Transfer learning | 97.11 |
| Sethy et al. [25] | X-ray image | Deep learning and SVM | 95.33 |
| Heidari et al. [26] | X-ray image (Same dataset) | Transfer learning | 94.50 |
| Pathak et al. [27] | X-ray image (Same dataset) | Transfer learning | 93.02 |
| Oh et al. [28] | X-ray image | Statistical approach | 88.90 |
| Wang et al. [29] | CT scan | Transfer learning | 85.20 |
| Azemin et al. [30] | X-ray image | Deep learning | 71.90 |
Conclusions
In this paper, our purpose is to develop a deep learning-based method to classify COVID-19- and Pneumonia-affected chest X-ray images by applying transfer learning. Here, we use the pre-trained VGG-19 architecture and transfer its weights and get an excellent accuracy. Moreover, we use MongoDB as a database to store the original image and corresponding category.
Our findings confirm that deep learning can make the diagnostic process more comfortable as it can achieve human-level accuracy for image classification tasks. Deep learning can efficiently execute time-consuming tasks, like detect symptoms of COVID-19 and Pneumonia from the chest X-ray images, and it will be an excellent asset for doctors. Therefore, Deep learning will play an impactful role in saving valuable lives.
Declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Footnotes
This article is part of the topical collection “Computer Aided Methods to Combat COVID-19 Pandemic” guest edited by David Clifton, Matthew Brown, Yuan-Ting Zhang and Tapabrata Chakraborty.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soarov Chakraborty, Email: soarovchakraborty@gmail.com.
Shourav Paul, Email: paulshourav124000@gmail.com.
K. M. Azharul Hasan, Email: az@cse.kuet.ac.bd.
References
- 1.Liu N, Wan L, Zhang Y, Zhou T, Huo H, Fang T. Exploiting convolutional neural networks with deeply local description for remote sensing image classification. IEEE Access. 2018;6:11215–11228. doi: 10.1109/ACCESS.2018.2798799. [DOI] [Google Scholar]
- 2.Asiri N, Hussain M, Al Adel F, Alzaidi N. Deep learning based computer-aided diagnosis systems for diabetic retinopathy: A survey. Artif Intell Med. 2019;99:101701. doi: 10.1016/j.artmed.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Yao L, Wang X, Monaghan J, Mcalpine D, Zhang Y. A survey on deep learning based brain computer interface: recent advances and new frontiers. 2019; arXiv preprint arXiv:190504149 [DOI] [PubMed]
- 5.Velavan TP, Meyer CG. The Covid-19 epidemic. Trop Med Int health. 2020;25(3):278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilani Z, Kwong YD, Levine OS, Deloria-Knoll M, Scott JAG, O'Brien KL, Feikin DR. A literature review and survey of childhood pneumonia etiology studies: 2000–2010. Clin Infect Dis. 2012;54(suppl_2):S102–S108. doi: 10.1093/cid/cir1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakshman A, Malik P. Cassandra: a decentralized structured storage system. ACM SIGOPS Oper Syst Rev. 2010;44(2):35–40. doi: 10.1145/1773912.1773922. [DOI] [Google Scholar]
- 8.Dean J, Ghemawat S. Mapreduce: simplified data processing on large clusters. Commun ACM. 2008;51(1):107–113. doi: 10.1145/1327452.1327492. [DOI] [Google Scholar]
- 9.Chodorow K. MongoDB: the definitive guide: powerful and scalable data storage. Newton: O'Reilly Media Inc.; 2013. [Google Scholar]
- 10.Chang F, Dean J, Ghemawat S, Hsieh WC, Wallach DA, Burrows M, Chandra T, Fikes A, Gruber RE. Bigtable: a distributed storage system for structured data. ACM Trans Comput Syst (TOCS) 2008;26(2):1–26. doi: 10.1145/1365815.1365816. [DOI] [Google Scholar]
- 11.Kallianos K, Mongan J, Antani S, Henry T, Taylor A, Abuya J, Kohli M. How far have we come? artificial intelligence for chest radiograph interpretation. Clin Radiol. 2019;74(5):338–345. doi: 10.1016/j.crad.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima K, Miyake S. Neocognitron: a self-organizing neural network model for a mechanism of visual pattern recognition. In: Arbib MA, Amari SI, editors. Competition and cooperation in neural nets. Berlin: Springer Berlin Heidelberg; 1982. pp. 267–285. [Google Scholar]
- 13.Ciresan D, Giusti A, Gambardella L, Schmidhuber J. Deep neural networks segment neuronal membranes in electron microscopy images. Adv Neural Inf Process Syst. 2012;25:2843–2851. [Google Scholar]
- 14.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, 2017; p 3462–3471. 10.1109/CVPR.2017.369
- 15.Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz CP, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the chexnext algorithm to practicing radiologists. PLoS Med. 2018;15(11):e1002686. doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Zhang Z, Chen YC, Zhao ZY, Yin XD, Jiang HB. A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Transl Oncol. 2019;12(2):292–300. doi: 10.1016/j.tranon.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deniz E, Şengür A, Kadiroğlu Z, Guo Y, Bajaj V, Budak Ü. Transfer learning based histopathologic image classification for breast cancer detection. Health Inf Sci Syst. 2018;6(1):1–7. doi: 10.1007/s13755-018-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed KB, Hall LO, Goldgof DB, Liu R, Gatenby RA. Fine-tuning convolutional deep features for mri based brain tumor classification. In: Medical imaging 2017: Computer-Aided Diagnosis, International Society for optics and photonics, 2017; vol 10134, p 101342E.
- 19.Yang Y, Yan LF, Zhang X, Han Y, Nan HY, Hu YC, Hu B, Yan SL, Zhang J, Cheng DL, et al. Glioma grading on conventional mr images: a deep learning study with transfer learning. Front Neurosci. 2018;12:804. doi: 10.3389/fnins.2018.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014; arXiv preprint arXiv:14091556.
- 21.Chakraborty S, Paul S, Azharul Hasan KM. Performance comparison for data retrieval from nosql and sql databases: a case study for covid-19 genome sequence dataset. In: 2021 2nd International Conference on Robotics, electrical and signal processing techniques (ICREST), 2021; p. 324–328, 10.1109/ICREST51555.2021.9331044.
- 22.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can ai help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 23.Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, Islam MT, Al Maadeed S, Zughaier SM, Khan MS, et al. Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Comput Biol Med. 2021;132:104319. doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014; arXiv preprint arXiv:14126980.
- 25.Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (covid-19) based on deep features and support vector machine. Preprints. 2020;2020:2020030300. [Google Scholar]
- 26.Heidari M, Mirniaharikandehei S, Khuzani AZ, Danala G, Qiu Y, Zheng B. Improving the performance of cnn to predict the likelihood of covid-19 using chest x-ray images with preprocessing algorithms. Int J Med Inf. 2020;144:104284. doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S. Deep transfer learning based classification model for covid-19 disease. Irbm; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh Y, Park S, Ye JC. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, et al. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) Eur Radiol. 2021;31(8):6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Che Azemin MZ, Hassan R, Mohd Tamrin MI, Md Ali MA. Covid-19 deep learning prediction model using publicly available radiologist-adjudicated chest x-ray images as training data: preliminary findings. Int J Biomed Imaging. 2020;2020:1–7. doi: 10.1155/2020/8828855. [DOI] [PMC free article] [PubMed] [Google Scholar]