Abstract
Understanding the glycosylation of the envelope spike (S) protein of SARS-CoV-2 is important in defining the antigenic surface of this key viral target. However, the underlying protein architecture may significantly influence glycan occupancy and processing. There is, therefore, potential for different recombinant fragments of S protein to display divergent glycosylation. Here, we show that the receptor binding domain (RBD), when expressed as a monomer, exhibits O-linked glycosylation, which is not recapitulated in the native-like soluble trimeric protein. We unambiguously assign O-linked glycosylation by homogenizing N-linked glycosylation using the enzymatic inhibitor, kifunensine, and then analyzing the resulting structures by electron-transfer higher-energy collision dissociation (EThcD) in an Orbitrap Eclipse Tribrid instrument. In the native-like trimer, we observe a single unambiguous O-linked glycan at T323, which displays very low occupancy. In contrast, several sites of O-linked glycosylation can be identified when RBD is expressed as a monomer, with T323 being almost completely occupied. We ascribe this effect to the relaxation of steric restraints arising from quaternary protein architecture. Our analytical approach has also highlighted that fragmentation ions arising from trace levels of truncated N-linked glycans can be misassigned as proximal putative O-linked glycan structures, particularly where a paucity of diagnostic fragments were obtained. Overall, we show that in matched expression systems the quaternary protein architecture limits O-linked glycosylation of the spike protein.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the recently emerged zoonotic agent responsible for the COVID-19 pandemic.1−3 SARS-CoV-2 is a member of the Betacoronavirus genera of enveloped RNA viruses, which also includes SARS-CoV-1 and Middle Eastern respiratory syndrome-coronavirus (MERS-CoV).4,5 SARS-CoV-2 tissue tropism is defined by the spike (S) protein.6 The S protein is formed of a trimer of heterodimers (Figure 1A,B) that controls cellular fusion with host cells through the binding of angiotensin-converting enzyme 2 (ACE2) cellular receptors.7−9 The S protein is the major surface transmembrane protein and is the principle antigenic target in current vaccine strategies,10,11 and a key component of serological tests.12,13 It is therefore important that we understand the immunogenic surface of the S protein.
Figure 1.
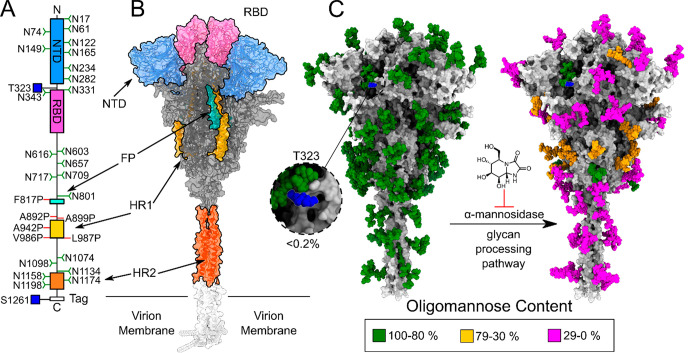
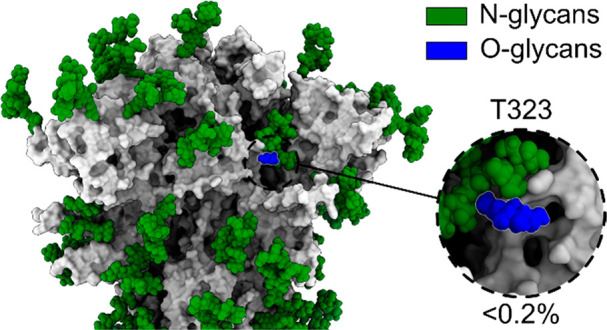
Overview of structure and glycosylation of the S-protein construct, using the full-length S-protein model presented in Allen et al.,31 with the N-terminal domain (NTD) in blue, receptor binding domain (RBD) in pink, the fusion peptide (FP) in cyan, the heptapeptide repeat sequence 1 (HR1) in yellow, and the heptapeptide repeat sequence 2 (HR2) in orange, with oligomannose N-linked glycans in green and the O-linked glycans in blue. (A) Primary structure of the S protein with N- and O-linked glycans and stabilizing protein mutations labeled; (B) full structure of the spike protein, with the NTD, RBD, FP, HR1, and HR2 colored according to the primary structure; (C) spike protein construct with both high mannose and dominant glycans as presented by Watanabe, Allen and co-workers,25 with the Man9GlcNAc2 glycans shown in green, hybrid glycans shown in yellow, and complex glycans shown in magenta and O-linked glycans in blue. Inset: the close-up of the T323 O-glycan on the structure of the high mannose glycoform with the percentage occupancy.
The ectodomains of S proteins of coronaviruses have been shown to be modified with glycans, which can facilitate host-cell binding.14−20 In addition, glycans can obfuscate immunological recognition of the protein surface through glycan shielding,21−23 although not to the same extent as human immunodeficiency virus (HIV).21,24,25
Glycans are carbohydrates added as a post-translational modification, in a process called glycosylation, during glycoprotein secretion.26 Viral spike protein glycosylation occurs via two main mechanisms: the addition of glycans to asparagine residues, termed N-linked glycosylation, or O-linked glycosylation, which typically occurs at serine or threonine residues.27 N-linked glycans are added to Asn residues within the N-linked sequon, N-X-S/T, and so potential N-linked glycosylation sites (PNGS) can be identified from a primary protein sequence. In contrast, O-linked glycans are present in high levels on mucin domains and display a bias toward disordered Ser-, Thr-, Pro-rich (STP) domains, but do not have a conserved sequon.28 This makes O-linked glycans more difficult to study because of their lack of predictability.
The N-linked glycosylation of SARS-CoV-2 S protein has been previously studied, showing high occupancy of 22 PNGS;29−31 however, there are differing reports about the level of occupancy, and localization of, O-linked glycans.25,32−37 In this study we use kifunensine, which inhibits α1,2-mannosidases following the Calnexin/Calreticulin folding cycle,38 to simplify N-linked glycosylation prior to studying the SARS-CoV-2 S protein. The inhibition by kifunensine induces N-linked glycan homogeneity, resulting in the predominant display of Man9GlcNAc2 glycans. Kifunensine has been previously used to study heavily glycosylated glycoproteins in both a crystallographic39 and intact mass spectrometry setting.40 Here, we use kifunensine to simplify bottom-up N-linked fragmentation spectra, lowering the chance of false-positive O-linked glycan identification in several ways.40 Firstly, it simplifies the N-linked glycans while ostensibly leaving the O-linked glycans untouched, preventing possible misassignment of N-linked structures as complex O-glycans, that is, as a complex extended core structure. Secondly, it facilitates initial peptide searches with both N- and O-linked glycan comodifications. Third, it accelerates the manual verification required during joint N- and O-linked glycan searches, allowing better discrimination of misassigned N-glycans.
We study two different forms of prefusion stabilized S protein, the 2P41,42 and HexaPro43 constructs, expressed in the presence of kifunensine, using a bottom-up mass spectrometry (MS) approach with electron-transfer higher-energy collision dissociation (EThcD),44−46 to study O-linked glycosylation. EThcD offers a softer fragmentation approach than pure higher-energy collision dissociation (HCD), preventing dissociation of the glycan from the peptide, which is beneficial for O-linked glycan analysis as there is no conserved sequon.44−46 During data analysis a search of up to three glycan comodifications per peptide, on either N- or O-linked glycan sites, was performed, where the sites can be easily verified or rejected if non-oligomannose-type glycans were observed at the N-linked glycan sites, or if there was incomplete fragmentation between N- and O-linked glycan sites. Using this approach, we observe a single unambiguous O-linked glycan on the S-protein ectodomains at site T323 in the receptor binding domain (RBD), with low occupancy, with our N-linked glycosylation patterns matching previously published data. We considered a glycosylation site unambiguous if it was identified in multiple analytical repeats and had appropriate backbone fragmentation between the O-linked glycan site and any other possible glycan sites. In comparison, monomeric RBD displayed very high occupancy of the T323 site, with greater glycan processing evident. We hypothesize that the higher occupancy and processing of this site is due to a lack of steric hindrance from the quaternary structure of the full trimeric spike. We also explored the impact of kifunensine on O-glycan occupancy and composition on monomeric RBD, observing consistent occupancy between O-linked glycan sites; however, unexpectedly, kifunensine appeared to increase sialylation of O-linked glycans. We find that the use of kifunensine treatment helps reduce misassignments of glycan attachment sites derived from nearby N-glycans as O-glycosylation sites because of poor fragmentation. Elucidation of the pattern of N- and O-linked glycan occupancy are important parameters in defining the antigenic surface of viral glycoproteins.
Methods
Expression and Purification
HEK293F cells were cultured in Freestyle 293 Expression Medium (Fisher Scientific) and maintained at a density of 0.2 × 106 cells/mL at 37 °C, 8% CO2, and 125 rpm shaking. 293F cells were transiently transfected with plasmid containing SARS-CoV-2 S (GenBank ID: MN908947) to generate two forms of soluble SARS-CoV-2 prefusion ectodomain, one with stabilizing proline mutations at residues 986 and 987 (2P)41,42 and another with additional mutations at 817, 892, 899, and 942 (HexaPro).43 Both proteins contained a C-terminal octa-histadine and Strep-tag II. Prior to transfection, two solutions of 10 mL of Opti-MEM (Fisher Scientific) medium were prepared. SARS-CoV-2 S was added to the first solution to give a final concentration of 310 μg/L. To the other solution, 1 mg/mL (pH 7) polyethylenimine (PEI) max reagent was added to generate a ratio of 3:1 PEI max/plasmid DNA. Both solutions were combined and incubated for 30 min at room temperature. Cells were transfected at a density of 1 × 106 cells/mL and incubated for 7 days at 37 °C, 8% CO2, and 125 rpm shaking. At the time of transfection, 20 μM kifunensine was added to elicit oligomannose-type glycans.
Cells were centrifuged at 3041g for 30 min at 4 °C, and supernatant was applied to a 500 mL Stericup-HV sterile vacuum filtration system (Merck) with a pore size of 22 μm. Purification of SARS-CoV-2 spike protein was undertaken using an ÄKTA Pure system (Cytiva). A 5 mL HisTrap FF column (Cytiva) charged with Ni(II) was equilibrated using 10 column volumes (CV) of washing buffer (50 mM Na2PO4, 300 mM NaCl) at pH 7. Supernatant was loaded onto the column at a flow rate of 2 mL/min and washed with 10 CV of washing buffer containing 50 mM imidazole. Protein was eluted in 3 CV of elution buffer (300 mM imidazole in washing buffer) and buffer exchanged to phosphate buffered saline (PBS) using a Vivaspin column (MWCO 100 kDa) (Cytiva).
The nickel purified eluate was concentrated to 1 mL in PBS and injected into a Superdex 200 pg 16/600 column (Cytiva) to isolate trimeric spike protein using size exclusion chromatography (SEC). The column was washed with PBS at 1 mL/min for 2 h, and fractions corresponding to the correct peak on the size exclusion chromatogram were collected and concentrated to 1 mL as above.
Sample Preparation
Three aliquots of each sample were denatured for 1 h in 50 mM Tris/HCl (pH 8.0) containing 6 M urea and 5 mM dithiothreitol (DTT). Next, spike proteins were reduced and alkylated by adding 20 mM iodoacetamide (IAA) and incubated for 1 h in the dark, followed by a 1 h incubation with 20 mM DTT to eliminate residual IAA. The alkylated spike proteins were buffer-exchanged into 50 mM Tris/HCl (pH 8.0) using Vivaspin columns (3 kDa), and two of the aliquots were digested separately overnight using trypsin, chymotrypsin (Mass Spectrometry grade, Promega), or alpha-lytic protease (Sigma-Aldrich) at a ratio of 1:30 (w/w). The next day, the peptides were dried and extracted using C18 Zip-tip (MerckMilipore).
Mass Spectrometry
The peptides were dried again, resuspended in 0.1% formic acid, and analyzed by nanoLC-ESI MS with an Ultimate 3000 HPLC (Thermo Fisher Scientific) system coupled to an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific) using electron-transfer high-energy collision dissociation (EThcD) fragmentation. Peptides were separated using an EasySpray PepMap RSLC C18 column (75 μm × 75 cm). A trapping column (PepMap 100 C18 3 μm × 75 μm × 2 cm) was used in-line with the LC prior to separation with the analytical column. The LC conditions were as follows: 280 min linear gradient consisting of 4–32% acetonitrile in 0.1% formic acid over 260 min followed by 20 min of alternating 76% acetonitrile in 0.1% formic acid and 4% Acn in 0.1% formic acid, used to ensure all the sample had eluted from the column. The flow rate was set to 300 nL/min. The spray voltage was set to 2.7 kV, and the temperature of the heated capillary was set to 40 °C. The ion-transfer tube temperature was set to 275 °C. The scan range was 375–1500 m/z. EThcD reaction kinetics were determined with prior calibration using Pierce FlexMix with supplemental higher energy collision dissociation set to 15%. Precursor and fragment detection were performed using an Orbitrap at a resolution MS1 = 120 000 and MS2 = 30 000. The AGC target for MS1 was set to standard and the injection time set to auto, which involves the system setting the two parameters to maximize sensitivity while maintaining cycle time. Full LC and MS methodology can be extracted from the appropriate raw file using XCalibur FreeStyle software or upon request (MassIVE MSV000087897).
Data Analysis
Glycopeptide fragmentation data were extracted from the raw file using Byos (Version 3.5; Protein Metrics Inc.) and filtered for a score above 100. The glycopeptide fragmentation data were evaluated manually for each glycopeptide; the peptide was scored as true positive when the correct a-, b-, c-, x-, y-, and z-fragment ions were observed along with oxonium ions corresponding to the glycan identified. The MS data were searched using the Protein Metrics 305 N-linked glycan library with sulfated glycans added manually. The relative amounts of each glycan at each site as well as the unoccupied proportion were determined by comparing the extracted chromatographic areas for different glycotypes with an identical peptide sequence. All charge states for a single glycopeptide were summed. The precursor mass tolerance was set at 4 and 10 ppm for fragments. A 1% false discovery rate (FDR) was applied. The relative amounts of each glycan at each site as well as the unoccupied proportion were determined by comparing the extracted ion chromatographic areas (XIC) for different glycopeptides with an identical peptide sequence. Glycans were categorized according to the composition detected. The O-linked glycan occupancy was calculated for each glycopeptide via XIC, and the occupancy was averaged across different glycopeptides across analytical repeats. For N-linked glycosylation, high-intensity glycopeptides (XIC > e10) were selected from analytical repeats to confirm successful kifunensine treatment.
Results and Discussion
High HexaPro N-Linked Glycan Occupancy
Previous publications have identified 22 N-linked glycosylation sites on the S-protein ectodomains.25,33,34 We were able to unambiguously determine the glycosylation state of 21 of the 22 N-linked glycan sites, but did not see signal for the N17 glycan, due to poor N-terminal peptide coverage. With expression in the presence of kifunensine (‘kifunensine treatment’, KT), a majority of oligomannose-type glycans were observed on all sites (Figure 2A) compared to previous publications, which show Spike N-linked glycans consist predominantly of complex-type glycans.25 Over 95% of all sites were observed as being occupied by oligomannose-type glycans, confirming the efficacy of kifunensine (Figure 2B).
Figure 2.
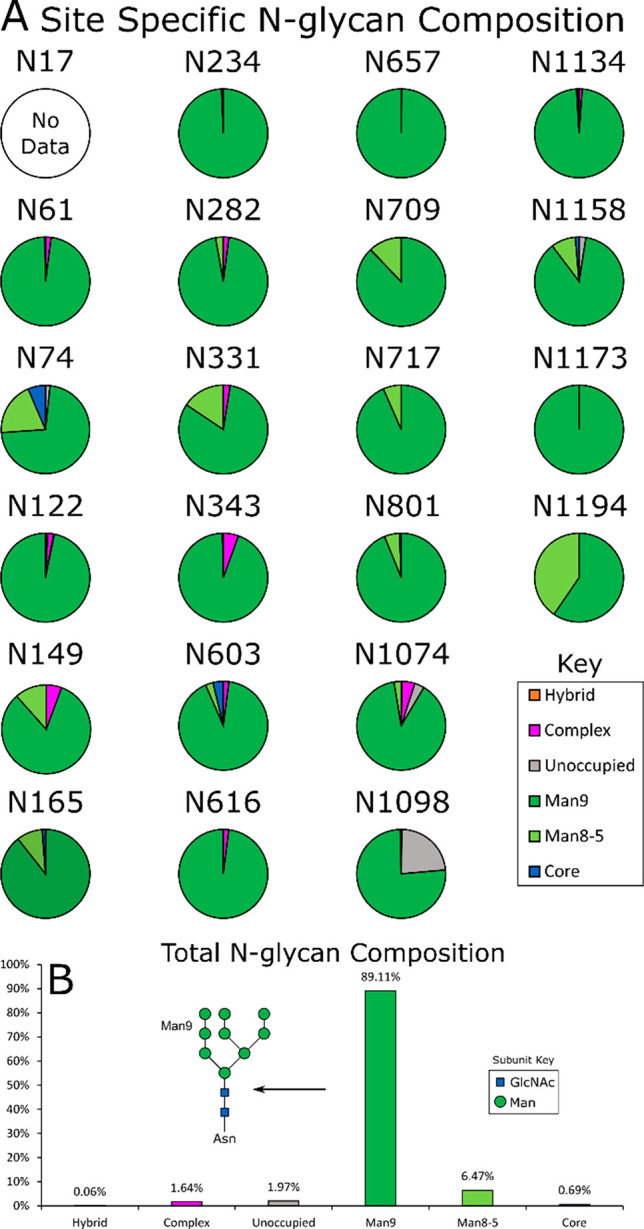
The N-linked glycan composition of HexaPro expressed in the presence of kifunensine: oligomannose-type (green), hybrid-type (orange), complex-type (magenta), unoccupied (gray), and core (blue). (A) Pie charts displaying glycan composition by N-linked glycan site, with a breakdown between Man9GlcNAc2 (Man9; dark green) and other oligomannose-type glycans: Man8-Man5GlcNAc2 (Man8-5; light green). (B) N-linked glycan composition, averaged across all sites.
Only one site of O-linked glycosylation could be unambiguously assigned to the S-protein ectodomain: T323. Low occupancy of the site with a HexNAc(1) (approximately 0.02% occupancy) was observed with fragmentation of either side of the T323 (Figure S2). This single O-glycan site was also observed by Watanabe et al., Zhao et al., and Shajahan et al., although at higher occupancy.25,33,47 The only other unambiguous site of O-linked glycosylation in the present study was a S1261 present on the protein purification tag (approximately 6% occupancy, Figure S2, Table S2).
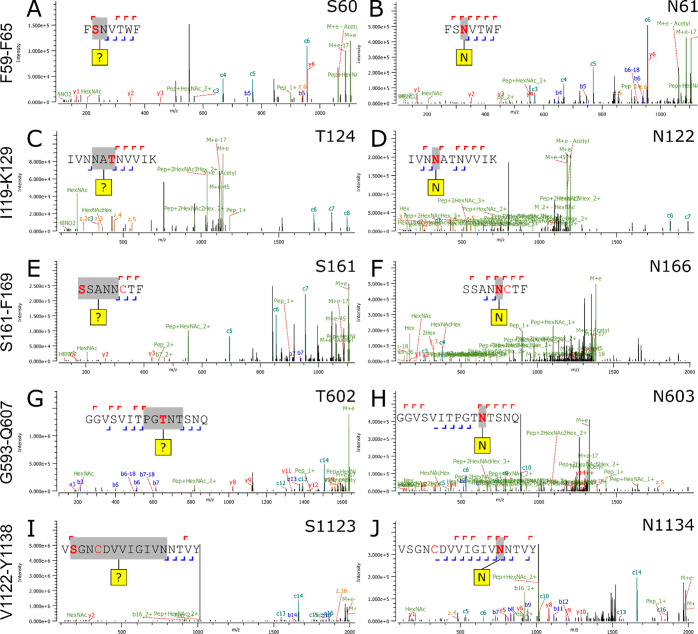
In addition to this, we observed many ambiguous assignments of O-linked glycans during analysis. The majority of these assigned O-linked glycans contained only a single HexNAc within close proximity to previously identified N-linked glycans. Further examination revealed that these sites were ambiguous assignments, where there were no assigned fragment ions between the S/T and the nearby N-glycan (Figure 3). Other modifications could be manually rejected for reasons such as incorrect monoisotopic precursor peak selection, missing isotope peaks, and lack of diagnostic ions.
Figure 3.
(A, C, E, G, I) Example fragmentation spectra of various ambiguous O-linked glycan assignments. (B, D, F, H, J) Corresponding assigned N-glycan, with the glycosylation site and the annotated peptide fragments, with the corresponding assigned glycan site in bold.
Our data is in contrast to recent studies and preprints, suggesting a high number of assigned O-linked glycan sites in the vicinity of N-sequons.32,35,48 A possible reason for these discrepancies could be that the use of PNGase F in these studies, used to remove N-linked glycans, would not remove residual HexNAc(1) remaining from N-linked glycan degradation.49,50 Unambiguous assignment of an O-linked HexNAc(1) in proximity to N-glycan sequons requires a parallel search for N-linked modifications and good fragmentation of the N sequon; otherwise, software pipelines will assign false O-linked glycan sites (Figure 3).
Comparison to 2P Construct
As previous studies have used the 2P stabilized spike construct, which has four fewer proline substitutions compared to the HexaPro construct and exhibits lower expression levels in protein production, we compared glycosylation of HexaPro that of 2P.41,42 We performed an identical O-linked glycan site search on KT 2P. We observed an O-linked HexNAc(1) on T323 at 0.23% occupancy, higher than that observed on HexaPro (Figure 4, Figure S3, Table 1, Table S2). This observation is consistent with the additional proline substitutions increasing the stability of the construct and suppressing the initiation of O-linked glycosylation. No unambiguous O-glycans other than those previously identified were observed.
Figure 4.
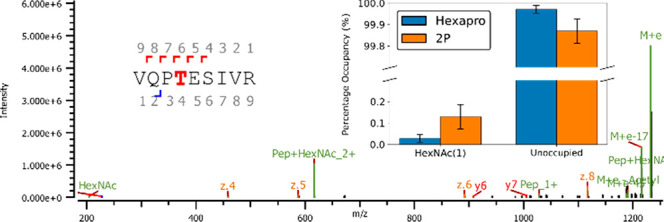
T323 O-glycan site: Example of unambiguous fragmentation spectra with inset: occupancy of T323 with HexNAc(1) in HexaPro (blue) and 2P (orange) constructs. For additional spectra see Figures S2 and S3.
Table 1. Unambiguously Identified O-Linked Glycans by Peptide and Full-Length Construct.
| site | construct | sequence | glycans | score | XIC occupied | XIC unoccupied | occupancy |
|---|---|---|---|---|---|---|---|
| T323 | HexaPro | R.VQPtESIVR.F | HexNAc(1) | 176.05 | 5.91 × 1007 | 5.21 × 1011 | 0.01% |
| T323 | HexaPro | S.NFRVQPtESIV.R | HexNAc(1) | 191.31 | 1.42 × 1008 | 3.00 × 1011 | 0.05% |
| T323 | HexaPro | R.VQPtESIVR.F | HexNAc(1) | 176.14 | 5.32 × 1007 | 6.25 × 1011 | 0.01% |
| T323 | 2P | S.NFRVQPtESIV.R | HexNAc(1) | 170.59 | 3.22 × 1008 | 6.77 × 1010 | 0.47% |
| T323 | 2P | F.RVQPtESIV.R | HexNAc(1) | 192.53 | 1.07 × 1008 | 6.77 × 1010 | 0.16% |
| T323 | 2P | R.VQPtESIVR.F | HexNAc(1) | 197.33 | 5.36 × 1008 | 3.01 × 1011 | 0.18% |
| T323 | 2P | F.RVQPtESIV.R | HexNAc(1) | 171.6 | 3.40 × 1007 | 6.55 × 1010 | 0.05% |
Low-Confidence Ambiguous O-Linked Glycans
As previously stated, we considered a glycosylation site “unambiguous” if it was present on multiple peptides across analytical repeats, and there was sufficient fragmentation data to distinguish it from any other possible O-linked glycan sites. We observed several examples of O-linked glycan sites for which a single peptide was marked as a true positive, at low intensities, including T124, T478, T696/S698/S704 (there was not sufficient fragmentation to localize the O-linked glycan site), and S1175 (Figure S4). We include these ambiguous O-linked glycan sites for transparency; however, we do not have confidence in their validity.
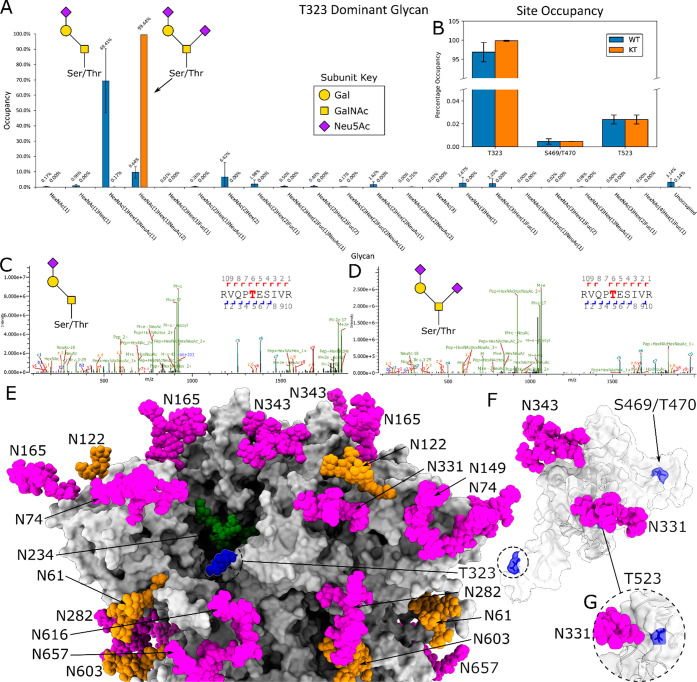
High RBD O-Linked Glycan Occupancy
We explored KT RBD as a comparison to both the HexaPro and 2P constructs, to highlight how the quaternary structure impacts the occupancy on T323. We observed the T323 glycosite at much higher occupancy in the RBD. The dominant glycan in this position is a disialylated core-I (Figure 5A), HexNAc(1)Hex(1)NeuAc(2), representing 99% of total glycan compositions. We also observed unambiguous O-glycan on sites S469/T470 and T523 at occupancies below 1% (Figure 5B). The discrepancy between the occupancy on T323 between full-length S protein is likely due to the fact that T323 is located within a cleft on the full-length trimer, but is more accessible in the monomeric RBD (Figure 5E,F). This is supported by the fact that the N234 glycan (which is in close proximity to T323, Figure 5E) is dominated by oligomannose structures in wild-type (WT) 2P25, and molecular dynamics simulations suggest that this glycan has very low solvent accessibility.31,51 A similar effect is observable with T499 within HIV gp120.52
Figure 5.
WT and KT O-glycan analysis of SARS-CoV-2 RBD: (A) Comparison of glycan composition at site T323 between WT and KT RBD, with the structures of mono- and disialylated core I HexNAc(1)Hex(1) glycans, with WT in blue and KT in orange. (B) Total site occupancy of O-linked glycans at T323, S469/T470, and T523 in RBD. (C, D) Example fragmentation spectra of T323 site mono- and disialylated core-I from (C) WT and (D) KT RBD. (E) S-protein trimer with oligomannose glycans shown in green, hybrid glycans shown in yellow, complex glycans shown in magenta, and O-glycan glycans shown in blue. (F) RBD construct with T323 and S469/T470 and (G) T523 residues in blue, using the full-length S-protein model presented in Allen et al.31
To assess whether kifunensine impacts the occupancy of O-linked glycans, such as through steric hindrance of the glycosylation machinery by the artificial addition of oligomannose-type glycans, we performed a comparison of WT and KT SARS-CoV-2 RBD. We observe the same O-glycan sites (T323, S469/T470, and T523) with similar occupancies (Figure 5B). In comparison to those of KT RBD, the dominant glycans at T323 are mono- and disialylated core-I structures, HexNAc(1)Hex(1)NeuAc(1) and HexNAc(1)Hex(1)NeuAc(2) (Figure 5C,D), representing 80% of total glycan compositions (Figure 5A). Various other O-glycan structures were observed at low levels, with the total occupancy of the T323 site at approximately 97%, comparable to that of KT RBD. Interestingly, we see a distinct shift from mono- to disialylation between WT and KT RBD, and a decrease in overall O-linked glycan complexity at this site (Figure 5A). We hypothesize that the use of kifunensine results in an increase in the overall substrate available for sialyltransferases as kifunensine prevents the sialylation of N-linked glycosylation. In this model, the increase in the sialylation of O-linked glycans caps their further maturation, reducing complexity.53
Conclusion
We use kifunensine to simplify the N-linked glycome of the HexaPro and 2P constructs of the SARS-CoV-2 S protein, observing a single unambiguous O-glycan present on the ectodomain. We and others have previously reported T323 and S325 as O-linked glycosylation sites on the 2P construct; however, we now unambiguously assign T323 as the only readily detectable O-linked glycan site on the ectodomains using the mixture of kifunensine treatment and EThcD. This is in-line with studies on recombinant RBD reporting T323 as the predominate O-linked site.54−56 We also show that kifunensine does not impact the occupancy of O-linked glycans, but may lead to dominance of capped, sialylated O-linked glycans. Further exploration of this effect is required, which we anticipate to be the subject of a future study. Using these techniques, we show that the difference in occupancy of T323 between trimeric S protein and monomeric RBD is likely due to placement of the O-glycan within a cleft on the trimeric protein, imposing steric hindrance on glycosylation machinery.
Our work also highlights that research describing high levels of O-linked glycan sites, which are glycosylated in the absence of proximal N-glycans,32,35,48 may be artifactual in nature. Although it could be envisaged that variations in cell lines and proteins may explain the discrepancies, many of these studies use HEK 293/293F and stabilized proteins.25,29,32,35,47,54,55 It should be noted that these experiments are performed on stabilized constructs in a recombinant setting, and the rules governing O-linked glycosylation of the S protein in the context of natural infection may differ. Additionally, O-linked glycan enrichment may reveal further O-glycan sites that are present at very low levels.36,57
Glossary
Abbreviations
- ACE2
angiotension converting enzyme 2
- FP
fusion peptide
- EThcD
electron-transfer high-energy collision dissociation
- HR1
heptapeptide repeat 1
- HR2
heptapeptide repeat 2
- KT
kifunensine-treated
- NTD
N-terminal domain
- PTM
post-translational modification
- RBD
receptor binding domain
- SARS-CoV-2
severe acute respiratory syndrome coronavirus-2
- S protein
spike protein
- WT
wild type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c01772.
Construct sequences, breakdown of N-link occupancy data, O-glycan fragmentation spectra, and O-glycan peptide tables (PDF)
Author Contributions
# C.E. and J.A. contributed equally to this work. C.E., J.A., M.N.: data collection; C.E., M.N.: sample preparation; C.E., J.A.: data analysis; C.E.: manuscript preparation; C.E., J.A., M.N., M.C.: manuscript revision; M.C.: study design. All authors read and approved the manuscript prior to submission.
Funded by the International AIDS Vaccine Initiative (IAVI) through Grant No. INV-008352/OPP1153692 and the IAVI Neutralizing Antibody Center through the Collaboration for AIDS Vaccine Discovery Grant No. OPP1196345/INV-008813, both funded by the Bill and Melinda Gates Foundation. This work was also supported by the Bright Future Trust, and the University of Southampton Coronavirus Response Fund.
The authors declare no competing financial interest.
Notes
Data are available from the MassIVE database (https://massive.ucsd.edu) with ID MSV000087897, doi:10.25345/C5425B.
Supplementary Material
References
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Yu Y.; Xu J.; Shu H.; Xia J.; Liu H.; Wu Y.; Zhang L.; Yu Z.; Fang M.; Yu T.; Wang Y.; Pan S.; Zou X.; Yuan S.; Shang Y. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020, 8 (5), 475–481. 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J.; Wang D.; Xu W.; Holmes E. C.; Gao G. F.; Wu G.; Chen W.; Shi W.; Tan W. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395 (10224), 565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P. S.The Molecular Biology of Coronaviruses. In Advances in Virus Research; Academic Press: Jan 1, 2006; pp 193–292. 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E.; van Doremalen N.; Falzarano D.; Munster V. J. SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523. 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.; Wang M. L.; Chien C. S.; Yarmishyn A. A.; Yang Y. P.; Lai W. Y.; Luo Y. H.; Lin Y. T.; Chen Y. J.; Chang P. C.; Chiou S. H. Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection. Front. Immunol. 2020, 11, 1022. 10.3389/fimmu.2020.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M.; Marzi A.; Munster V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5 (4), 562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. D.; Watanabe Y.; Chawla H.; Newby M. L.; Crispin M. Subtle Influence of ACE2 Glycan Processing on SARS-CoV-2 Recognition. J. Mol. Biol. 2021, 433 (4), 166762. 10.1016/j.jmb.2020.166762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D.; de Wit E. Next-Generation Vaccine Platforms for COVID-19. Nat. Mater. 2020, 19, 810–812. 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- Dong Y.; Dai T.; Wei Y.; Zhang L.; Zheng M.; Zhou F. A Systematic Review of SARS-CoV-2 Vaccine Candidates. Signal Transduction and Targeted Therapy 2020, 5, 237. 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A.; Faustini S. E.; Perez-Toledo M.; Jossi S.; Aldera E.; Allen J. D.; Al-Taei S.; Backhouse C.; Bosworth A.; Dunbar L. A.; Ebanks D.; Emmanuel B.; Garvey M.; Gray J.; Kidd I. M.; McGinnell G.; McLoughlin D. E.; Morley G.; O’Neill J.; Papakonstantinou D.; Pickles O.; Poxon C.; Richter M.; Walker E. M.; Wanigasooriya K.; Watanabe Y.; Whalley C.; Zielinska A. E.; Crispin M.; Wraith D. C.; Beggs A. D.; Cunningham A. F.; Drayson M. T.; Richter A. G. SARS-CoV-2 Seroprevalence and Asymptomatic Viral Carriage in Healthcare Workers: A Cross-Sectional Study. Thorax 2020, 75 (12), 1089–1094. 10.1136/thoraxjnl-2020-215414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. M.; Faustini S. E.; Williams L. J.; Cunningham A. F.; Drayson M. T.; Shields A. M.; Kay D.; Taylor L.; Plant T.; Huissoon A.; Wallis G.; Beck S.; Jossi S. E.; Perez-Toledo M.; Newby M. L.; Allen J. D.; Crispin M.; Harding S.; Richter A. G. Validation of a Combined ELISA to Detect IgG, IgA and IgM Antibody Responses to SARS-CoV-2 in Mild or Moderate Non-Hospitalised Patients. J. Immunol. Methods 2021, 494, 113046. 10.1016/j.jim.2021.113046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Hulswit R. J. G.; Widjaja I.; Raj V. S.; McBride R.; Peng W.; Widagdo W.; Tortorici M. A.; van Dieren B.; Lang Y.; van Lent J. W. M.; Paulson J. C.; de Haan C. A. M.; de Groot R. J.; van Kuppeveld F. J. M.; Haagmans B. L.; Bosch B.-J. Identification of Sialic Acid-Binding Function for the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (40), E8508–E8517. 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Xiong X.; Park Y. J.; Tortorici M. A.; Snijder J.; Quispe J.; Cameroni E.; Gopal R.; Dai M.; Lanzavecchia A.; Zambon M.; Rey F. A.; Corti D.; Veesler D. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 2019, 176 (5), 1026–1039. e15 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M. A.; Walls A. C.; Lang Y.; Wang C.; Li Z.; Koerhuis D.; Boons G.-J.; Bosch B.-J.; Rey F. A.; de Groot R. J.; Veesler D. Structural Basis for Human Coronavirus Attachment to Sialic Acid Receptors. Nat. Struct. Mol. Biol. 2019, 26 (6), 481–489. 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R. J. G.; Lang Y.; Bakkers M. J. G.; Li W.; Li Z.; Schouten A.; Ophorst B.; van Kuppeveld F. J. M.; Boons G.-J.; Bosch B.-J.; Huizinga E. G.; de Groot R. J. Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (7), 2681–2690. 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. N.; Richards S.-J.; Guy C. S.; Congdon T. R.; Hasan M.; Zwetsloot A. J.; Gallo A.; Lewandowski J. R.; Stansfeld P. J.; Straube A.; Walker M.; Chessa S.; Pergolizzi G.; Dedola S.; Field R. A.; Gibson M. I. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6 (11), 2046–2052. 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morniroli D.; Giannì M. L.; Consales A.; Pietrasanta C.; Mosca F. Human Sialome and Coronavirus Disease-2019 (COVID-19) Pandemic: An Understated Correlation?. Front. Immunol. 2020, 11, 1480. 10.3389/fimmu.2020.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A. B.; Engin E. D.; Engin A. Dual Function of Sialic Acid in Gastrointestinal SARS-CoV-2 Infection. Environ. Toxicol. Pharmacol. 2020, 79, 103436. 10.1016/j.etap.2020.103436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Berndsen Z. T.; Raghwani J.; Seabright G. E.; Allen J. D.; Pybus O. G.; McLellan J. S.; Wilson I. A.; Bowden T. A.; Ward A. B.; Crispin M. Vulnerabilities in Coronavirus Glycan Shields despite Extensive Glycosylation. Nat. Commun. 2020, 11 (1), 2688. 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. J.; Chang Y. C.; Ko T. P.; Draczkowski P.; Chien Y. C.; Chang Y. C.; Wu K. P.; Khoo K. H.; Chang H. W.; Hsu S. T. D. Cryo-EM Analysis of a Feline Coronavirus Spike Protein Reveals a Unique Structure and Camouflaging Glycans. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (3), 1438–1446. 10.1073/pnas.1908898117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.-Y.; Huang H.-Y.; Chen X.; Cheng C.-W.; Wang S.-W.; Shahed-Al-Mahmud M.; Chen T.-H.; Lo J. M.; Liu Y.-M.; Ma H.-H.; Chang Y.-H.; Tsai C.-Y.; Huang P.-Y.; Chang S.-Y.; Chao T.-L.; Kao H.-C.; Tsai Y.-M.; Chen Y.-H.; Chen C.-Y.; Lee K.-C.; Wu C.-Y.; Jan J.-T.; Lin K.-I.; Cheng T.-J. R.; Ma C.; Wong C.-H.. Impact of Glycosylation on SARS-CoV-2 Infection and Broadly Protective Vaccine Design. bioRxiv, May 25, 2021, 2021.05.25.445523. 10.1101/2021.05.25.445523 (accessed 2021-07-30). [DOI] [Google Scholar]
- Watanabe Y.; Bowden T. A.; Wilson I. A.; Crispin M. Exploitation of Glycosylation in Enveloped Virus Pathobiology. Biochim. Biophys. Acta, Gen. Subj. 2019, 1863 (10), 1480–1497. 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science (Washington, DC, U. S.) 2020, 369 (6501), 330 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R.; Kornfeld S. Assembly of Asparagine-Linked Oligosaccharides. Annu. Rev. Biochem. 1985, 54 (1), 631–664. 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Van den Steen P.; Rudd P. M.; Dwek R. A.; Opdenakker G. Concepts and Principles of O-Linked Glycosylation. Crit. Rev. Biochem. Mol. Biol. 1998, 33, 151–208. 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- Nishikawa I.; Nakajima Y.; Ito M.; Fukuchi S.; Homma K.; Nishikawa K. Computational Prediction of O-Linked Glycosylation Sites That Preferentially Map on Intrinsically Disordered Regions of Extracellular Proteins. Int. J. Mol. Sci. 2010, 11 (12), 4991–5008. 10.3390/ijms11124991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Praissman J. L.; Grant O. C.; Cai Y.; Xiao T.; Rosenbalm K. E.; Aoki K.; Kellman B. P.; Bridger R.; Barouch D. H.; Brindley M. A.; Lewis N. E.; Tiemeyer M.; Chen B.; Woods R. J.; Wells L. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28 (4), 586–601. 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Song Y.; Chen Y.; Wu N.; Xu J.; Sun C.; Zhang J.; Weng T.; Zhang Z.; Wu Z.; Cheng L.; Shi D.; Lu X.; Lei J.; Crispin M.; Shi Y.; Li L.; Li S. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183 (3), 730–738. 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. D.; Chawla H.; Samsudin F.; Zuzic L.; Shivgan A. T.; Watanabe Y.; He W.; Callaghan S.; Song G.; Yong P.; Brouwer P. J. M.; Song Y.; Cai Y.; Duyvesteyn H. M. E.; Malinauskas T.; Kint J.; Pino P.; Wurm M. J.; Frank M.; Chen B.; Stuart D. I.; Sanders R. W.; Andrabi R.; Burton D. R.; Li S.; Bond P. J.; Crispin M. Site-Specific Steric Control of SARS-CoV-2 Spike Glycosylation. Biochemistry 2021, 60 (27), 2153–2169. 10.1021/acs.biochem.1c00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhao W.; Mao Y.; Chen Y.; Zhu J.; Hu L.; Gong M.; Cheng J.; Yang H.. Mucin-Type O-Glycosylation Landscapes of SARS-CoV-2 Spike Proteins. bioRxiv, July 30, 2020, 2020.07.29.227785. 10.1101/2020.07.29.227785 (accessed 2021-03-03). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Praissman J. L.; Grant O. C.; Cai Y.; Xiao T.; Rosenbalm K. E.; Aoki K.; Kellman B. P.; Bridger R.; Barouch D. H.; Brindley M. A.; Lewis N. E.; Tiemeyer M.; Chen B.; Woods R. J.; Wells L. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28 (4), 586–601. 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun J.; Vasiljevic S.; Gangadharan B.; Hensen M.; Chandran A. V.; Hill M. L.; Kiappes J. L.; Dwek R. A.; Alonzi D. S.; Struwe W. B.; Zitzmann N. Assessing Antigen Structural Integrity through Glycosylation Analysis of the SARS-CoV-2 Viral Spike. ACS Cent. Sci. 2021, 7 (4), 586–593. 10.1021/acscentsci.1c00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; Thompson A. J.; Wang X.; Søgaard M.; Fougeroux C.; Frank M.; Diedrich J. K.; Yates J. R.; Salanti A.; Vakhrushev S. Y.; Paulson J. C.; Wandall H. H. Site-Specific O-Glycosylation Analysis of SARS-CoV-2 Spike Protein Produced in Insect and Human Cells. Viruses 2021, 13 (4), 551. 10.3390/v13040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.; Chen C.; Yan J.; Zhang X.; Li X.; Liang X. Comprehensive O-Glycosylation Analysis of the SARS-CoV-2 Spike Protein with Biomimetic Trp-Arg Materials. Anal. Chem. 2021, 93, 10444–10452. 10.1021/acs.analchem.0c04634. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Go E. P.; Ding H.; Anang S.; Kappes J. C.; Desaire H.; Sodroski J.; Sodroski J. G.. Analysis of Glycosylation and Disulfide Bonding of Wild-Type SARS-CoV-2 Spike Glycoprotein. bioRxiv April 1, 2021, 2021.04.01.438120., 10.1101/2021.04.01.438120 (accessed 2021-04-12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein A. D.; Tropea J. E.; Mitchell M.; Kaushal G. P. Kifunensine, a Potent Inhibitor of the Glycoprotein Processing Mannosidase I. J. Biol. Chem. 1990, 265 (26), 15599–15605. 10.1016/S0021-9258(18)55439-9. [DOI] [PubMed] [Google Scholar]
- Chang V. T.; Crispin M.; Aricescu A. R.; Harvey D. J.; Nettleship J. E.; Fennelly J. A.; Yu C.; Boles K. S.; Evans E. J.; Stuart D. I.; Dwek R. A.; Jones E. Y.; Owens R. J.; Davis S. J. Glycoprotein Structural Genomics: Solving the Glycosylation Problem. Structure 2007, 15 (3), 267–273. 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W. B.; Stuckmann A.; Behrens A.-J.; Pagel K.; Crispin M. Global N-Glycan Site Occupancy of HIV-1 Gp120 by Metabolic Engineering and High-Resolution Intact Mass Spectrometry. ACS Chem. Biol. 2017, 12 (2), 357–361. 10.1021/acschembio.6b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J.; Wang N.; Corbett K. S.; Wrapp D.; Kirchdoerfer R. N.; Turner H. L.; Cottrell C. A.; Becker M. M.; Wang L.; Shi W.; Kong W. P.; Andres E. L.; Kettenbach A. N.; Denison M. R.; Chappell J. D.; Graham B. S.; Ward A. B.; McLellan J. S. Immunogenicity and Structures of a Rationally Designed Prefusion MERS-CoV Spike Antigen. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (35), E7348–E7357. 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L.; Goldsmith J. A.; Schaub J. M.; DiVenere A. M.; Kuo H.-C.; Javanmardi K.; Le K. C.; Wrapp D.; Lee A. G.; Liu Y.; Chou C.-W.; Byrne P. O.; Hjorth C. K.; Johnson N. V.; Ludes-Meyers J.; Nguyen A. W.; Park J.; Wang N.; Amengor D.; Lavinder J. J.; Ippolito G. C.; Maynard J. A.; Finkelstein I. J.; McLellan J. S. Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes. Science (Washington, DC, U. S.) 2020, 369 (6510), 1501–1505. 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q.; Wang B.; Chen Z.; Urabe G.; Glover M. S.; Shi X.; Guo L. W.; Kent K. C.; Li L. Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glycoproteome Characterization. J. Am. Soc. Mass Spectrom. 2017, 28 (9), 1751–1764. 10.1007/s13361-017-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiding K. R.; Bondt A.; Franc V.; Heck A. J. R. The Benefits of Hybrid Fragmentation Methods for Glycoproteomics. TrAC, Trends Anal. Chem. 2018, 108, 260–268. 10.1016/j.trac.2018.09.007. [DOI] [Google Scholar]
- Riley N. M.; Malaker S. A.; Driessen M. D.; Bertozzi C. R. Optimal Dissociation Methods Differ for N- A Nd O-Glycopeptides. J. Proteome Res. 2020, 19 (8), 3286–3301. 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A.; Supekar N. T.; Gleinich A. S.; Azadi P. Deducing the N- And O-Glycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. Glycobiology 2020, 30 (12), 981–988. 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W.; Li D.; Zhang N.; Bai G.; Yuan K.; Xiao H.; Gao F.; Chen Y.; Wong C. C. L.; Gao G. F. O-Glycosylation Pattern of the SARS-CoV-2 Spike Protein Reveals an “O-Follow-N” Rule. Cell Res. 2021, 1–3. 10.1038/s41422-021-00545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K. Requirements of Cleavage of High Mannose Oligosaccharides in Glycoproteins by Peptide N-Glycosidase F. J. Biol. Chem. 1986, 261 (1), 172–177. 10.1016/S0021-9258(17)42448-3. [DOI] [PubMed] [Google Scholar]
- Cao L.; Diedrich J. K.; Kulp D. W.; Pauthner M.; He L.; Park S.-K. R.; Sok D.; Su C. Y.; Delahunty C. M.; Menis S.; Andrabi R.; Guenaga J.; Georgeson E.; Kubitz M.; Adachi Y.; Burton D. R.; Schief W. R.; Yates III J. R.; Paulson J. C. Global Site-Specific N-Glycosylation Analysis of HIV Envelope Glycoprotein. Nat. Commun. 2017, 8 (1), 14954. 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L.; Gaieb Z.; Goldsmith J. A.; Hjorth C. K.; Dommer A. C.; Harbison A. M.; Fogarty C. A.; Barros E. P.; Taylor B. C.; Mclellan J. S.; Fadda E.; Amaro R. E. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722. 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A.-J.; Harvey D. J.; Milne E.; Cupo A.; Kumar A.; Zitzmann N.; Struwe W. B.; Moore J. P.; Crispin M. Molecular Architecture of the Cleavage-Dependent Mannose Patch on a Soluble HIV-1 Envelope Glycoprotein Trimer. J. Virol. 2017, 91 (2), e01894-16. 10.1128/JVI.01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A.; Cummings R. D.; Esko J. D.; Stanley P.; Hart G. W.; Aebi M.; Darvill A. G.; Kinoshita T.; Packer N. H.; Prestegard J. H.; Schnaar R. L.; Seeberger P. H.. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: 2017. [PubMed] [Google Scholar]
- Gstöttner C.; Zhang T.; Resemann A.; Ruben S.; Pengelley S.; Suckau D.; Welsink T.; Wuhrer M.; Domínguez-Vega E. Structural and Functional Characterization of SARS-CoV-2 RBD Domains Produced in Mammalian Cells. Anal. Chem. 2021, 93, 6839. 10.1021/acs.analchem.1c00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. S.; Mann M. W.; Melby J. A.; Larson E. J.; Zhu Y.; Brasier A. R.; Jin S.; Ge Y. Structural O-Glycoform Heterogeneity of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Revealed by Native Top-Down Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 12014. 10.1021/jacs.1c02713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos A.; Broome S.; Sharov V.; Ziegenfuss C.; Easton R. L.; Panico M.; Dell A.; Morris H. R.; Haslam S. M. Site-Specific Characterization of SARS-CoV-2 Spike Glycoprotein Receptor-Binding Domain. Glycobiology 2021, 31 (3), 181–187. 10.1093/glycob/cwaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Wang D.; Shipman R. D.; Zhu Z.; Liu Y.; Li L. Simultaneous Enrichment and Separation of Neutral and Sialyl Glycopeptides of SARS-CoV-2 Spike Protein Enabled by Dual-Functionalized Ti-IMAC Material. Anal. Bioanal. Chem. 2021, 1–9. 10.1007/s00216-021-03433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.