Abstract
Objectives:
To apply deep learning algorithms using a conventional convolutional neural network (CNN) and a recurrent CNN to differentiate three breast cancer molecular subtypes on MRI.
Methods:
A total of 244 patients were analyzed, 99 in Training Dataset scanned at 1.5T, 83 in Testing-1 and 62 in Testing-2 scanned at 3T. Patients were classified into 3 subtypes based on hormonal receptor (HR) and HER2 receptor: (HR+/HER2−), HER2+ and triple negative (TN). Only images acquired in the DCE sequence were used in the analysis. The smallest bounding box covering tumor ROI was used as the input for deep learning to develop the model in the Training dataset, by using a conventional CNN and the convolutional long short term memory (CLSTM). Then, transfer learning was applied to re-tune the model using Testing-1(2) and evaluated in Testing-2(1).
Results:
In the Training dataset, the mean accuracy evaluated using 10-fold cross-validation was higher by using CLSTM (0.91) than CNN (0.79). When the developed model was applied to the independent Testing datasets, the accuracy was 0.4–0.5. With transfer learning by re-tuning parameters in Testing-1, the mean accuracy reached 0.91 by CNN and 0.83 by CLSTM, and improved accuracy in Testing-2 from 0.47 to 0.78 by CNN and from 0.39 to 0.74 by CLSTM. Overall, transfer learning could improve the classification accuracy by greater than 30%.
Conclusions:
The recurrent network using CLSTM could track changes in signal intensity during DCE acquisition, and achieved a higher accuracy compared to conventional CNN during training. For datasets acquired using different settings, transfer learning can be applied to re-tune the model and improve accuracy.
Keywords: Breast Neoplasms, Magnetic Resonance Imaging, Machine Learning, Receptors, Estrogen, Receptor, ErbB-2
INTRODUCTION
Breast cancer is a heterogeneous group of disease with different phenotypes, and each subtype has different treatment strategy and prognosis. In the standard clinical practice, the status of the hormonal receptor (HR) and human epidermal growth factor receptor 2 (HER2) are evaluated to decide the appropriate treatments, including the use of hormonal therapy and HER2 targeting therapy. Microarray studies have shown that the morphological and clinical heterogeneity of breast cancer has a molecular basis [1]. Breast MRI can accurately reveal the 3-dimensional high spatial-resolution features of the disease, and is a well-established imaging modality routinely used for diagnosis, pre-operative staging and surgical planning [2]. With technological advances in imaging analysis, computer-aided diagnosis (CAD) and radiomics provide efficient methods to extract quantitative features for diagnosis, and they can also be used for molecular subtype differentiation [3–7]. While most studies extract imaging features from the tumor, it has been shown that features extracted from the peri-tumoral parenchyma outside the tumor also contain useful information [7,8].
After quantitative features were extracted, various classification methods including logistic regression [4,7,8], support vector machine (SVM) [5,8], naïve Bayes model [9] and artificial neural network [10] that could deal with a large number of parameters were applied to build the classification model. While these methods have yielded promising results, since they relied on pre-determined imaging features, the results were dependent on the choice of computer algorithms as well as the contrast variations and image quality. As such, the developed model might be specific to the analyzed dataset and not generally applicable. In the last several years, deep learning using the Convolutional Neural Network (CNN) have been applied for diagnosis and classification of breast lesions on MRI. In contrast to CAD and radiomics that extract specific features to carry out the classification task, CNN uses the raw image and performs the end-to-end learning for classification. The methods have been used for differentiation of benign and malignant lesions and achieved a high accuracy [11–13]. They have also been used for multi-class molecular subtype differentiation, which was a much more challenging task compared to diagnosis and in general had a lower accuracy [14–16]. More sophisticated deep learning networks that can fully utilize all information contained in multi-parametric MRI may help.
The purpose of this study was to apply deep learning networks to differentiate three breast cancer molecular subtypes on MRI, including HR positive and HER2 negative (HR+/HER2−), HR negative and HER2 negative (i.e. triple negative, TN) and HER2 positive (HER2+). The smallest bounding box containing the tumor and the proximal peri-tumor tissue was used as the input. A conventional CNN and a recurrent network using convolutional long short-term memory (CLSTM) that could consider the temporal information in DCE-MRI were applied, and the obtained results were compared. An independent testing dataset acquired using a different MR scanner from another hospital was used to evaluate the applicability of the model developed from the training dataset. Then, the model was re-tuned by transfer learning to investigate its utility for general implementation in different clinical settings.
MATERIALS AND METHODS
Patients
This was a retrospective study by retrieving patients who received breast MRI from two different institutions for analysis. The inclusion criteria were consecutive patients receiving MRI for diagnosis of suspicious lesions or pre-operative staging, and who had surgery with histologically confirmed cancer and molecular subtypes. Only cases presenting as mass lesions with a clear boundary were further selected for this study, in order to minimize the uncertainty in the defined tumor area. The exclusion criteria were patients receiving neoadjuvant treatment such as chemotherapy or hormonal therapy. The training dataset was obtained from one hospital from Aug 2013 to Dec 2014 performed on a Siemens 1.5T system, with a total of 99 patients, including 65 HR+/HER2− (66%), 24 HER2+ (24%), 10 TN (10%) cancers. The mean age was 48 years old (range 22 to 75). The independent testing cases were collected from another hospital performed on a GE 3T system. The testing dataset-1 was collected from Jan 2017 to May 2018, with a total of 83 patients, 54 HR+/HER2− (65%), 19 HER2+ (23%), 10 TN (12%), and mean age of 51 (range 24 to 82). The testing dataset-2 included later cases collected from June to Dec 2018, with a total of 62 patients, 37 HR+/HER2− (60%), 15 HER2+ (24%), 10 TN (16%), and mean age of 49 (range 33 to 72). The study was approved by the Institutional Review Board and the requirement of informed consent was waived.
Histopathological Analysis
The molecular subtypes were obtained from the medical record, based on the examination results of immunohistochemical staining and FISH from the surgical specimen. The tumor size was also obtained from the histological examination result of the surgical specimen. Of the 99 cases in the training dataset, the mean tumor size was 2.6 cm (range 0.4 to 5.0 cm). Of the 83 cases in testing dataset-1, the mean tumor size was 2.0 cm (range 0.7 to 3.5 cm). Of the 62 cases in testing dataset-2, the mean tumor size was 2.1 cm (range 0.5 to 5.3 cm). The tumor size and the distribution of three molecular subtypes in these three datasets were comparable.
MR Imaging Protocol
Only the dynamic-contrast-enhanced (DCE) images were used for analysis. The training dataset was scanned on a 1.5 Tesla scanner (Siemens Magneton Skyra, Erlangen, Germany) with a 16-channel Sentinelle breast coil. DCE-MRI was acquired using a fat-suppressed three-dimensional fast low angle shot (3D-FLASH) sequence with one pre-contrast and four post-contrast frames, with TR/TE=4.50/1.82 msec, flip angle=12°, field of view=32×32 cm, matrix size=512×512 and slice thickness=1.5 mm. The spatial resolution was 0.6×0.6×1.5 mm, and the temporal resolution was 180 seconds for each DCE frame. The contrast medium 0.1 mmol/kg Omniscan® (GE Healthcare, New Jersey, USA,) was administered at the beginning of the second acquisition. The testing dataset was done on a 3T scanner (GE SIGNA HDx, Milwaukee, WI) using a dedicated 8-channel bilateral breast coil. The DCE images were acquired using the volume imaging for breast assessment (VIBRANT) sequence also with fat-suppression, with TR/TE=5/2 msec, flip angle=10°, field of view=34×34 cm, matrix size=416×416 and slice thickness=1.2 mm. The DCE series consisted of one pre-contrast and five post-contrast frames. The spatial resolution was 0.8×0.8×1.2 mm, and the temporal resolution was 130 seconds for each DCE frame. The contrast agent, 0.1 mmol/kg Magnevist® (Bayer Schering Pharma, Berlin, Germany), was injected after the pre-contrast images were acquired.
3D Tumor Segmentation
The tumor was segmented on the contrast enhancement maps generated by subtracting pre-contrast images from post-contrast images taken at the 2nd DCE frame, using the fuzzy-C-means (FCM) clustering algorithm [13]. The 2nd DCE frame could best show the tumor boundary relative to adjacent tissues, thus was chosen for analysis. Some operator input was needed, which was performed by two radiologists (JHC and JZ), with 15 and 8 years of experience interpreting breast MRI, respectively. The range of slices containing the tumor was decided, and then a rectangle box covering the lesion shown on maximum intensity projection (MIP) was drawn. On each slice, FCM was applied to determine the tumor pixels, and then three dimensional connected-component labeling and hole filling was applied to finalize the tumor ROI. Figures 1 and 2 show DCE images from two patients, with the segmented tumor ROI. Since only mass lesions with a clear boundary were included in this study, the segmentation could be done with computer algorithms, without the need of manual correction. After segmentation, tumor ROI’s on all slices were projected together, and the smallest square bounding box covering them was determined as the input for deep learning analysis, as illustrated in [10].
Figure 1:
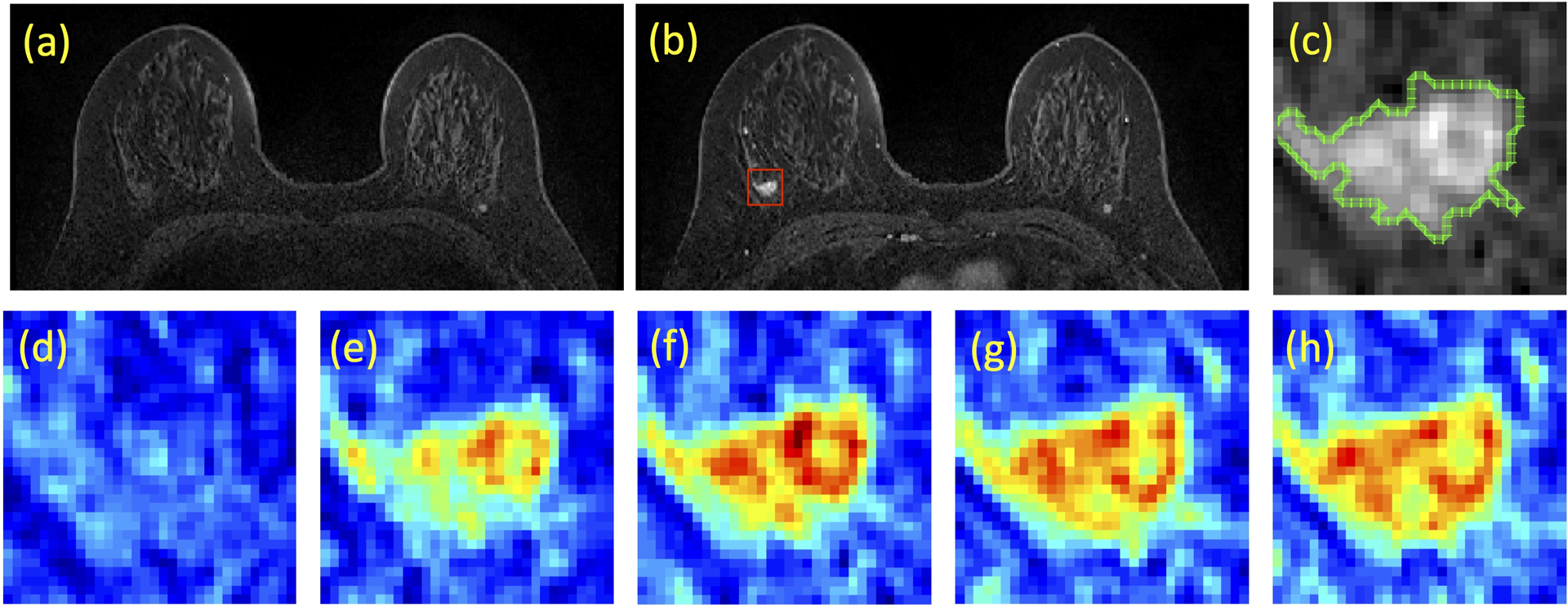
A case example from a 53-year-old woman with triple negative breast cancer in the right breast. (a) Pre-contrast image, (b) Post-contrast image, (c) The zoom-in image of the lesion with outlined tumor boundary obtained from segmentation. The square box is centered at the centroid of the tumor. (d-h) Color-coded DCE images at 5 time frames, one pre-contrast and 4 post-contrast, normalized using the same signal intensity scales.
Figure 2:
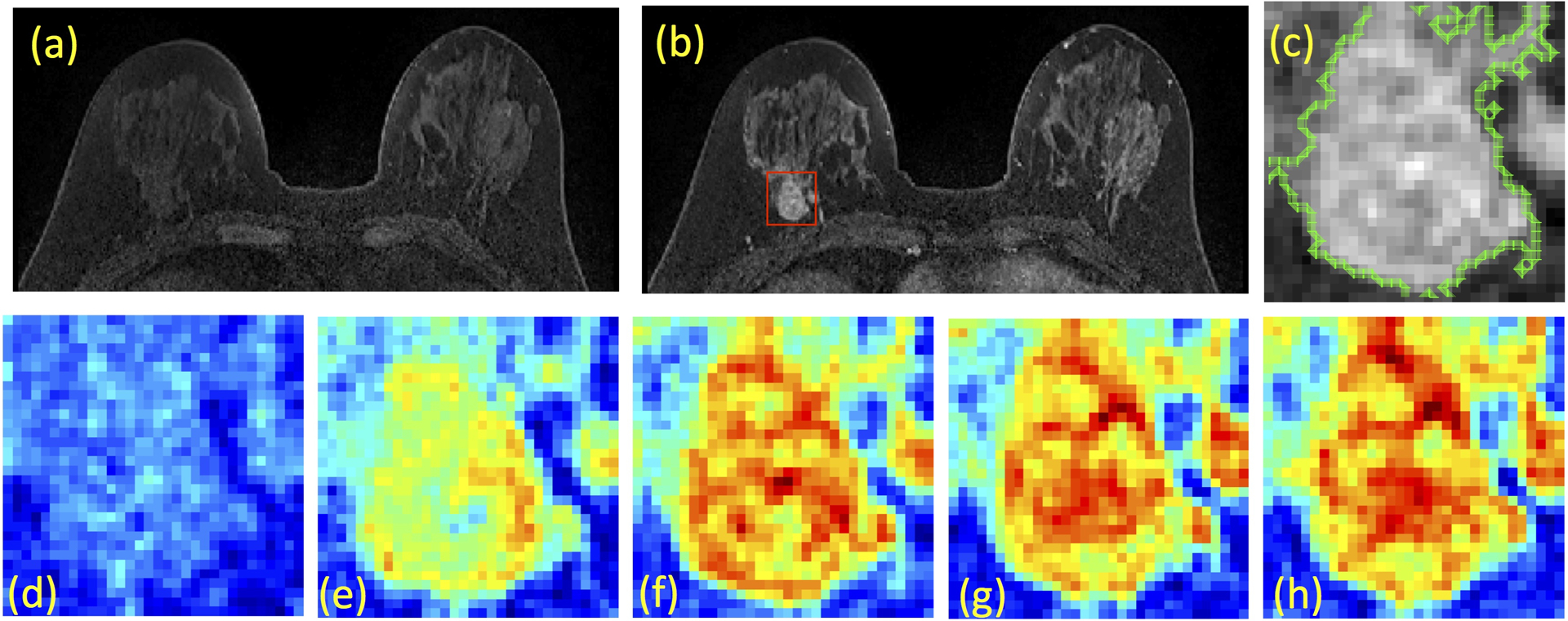
A case example from a 48-year-old woman with Hormonal-positive and HER2-negative breast cancer in the right breast. (a) Pre-contrast image, (b) Post-contrast image, (c) The zoom-in image of the lesion with outlined tumor boundary obtained from segmentation. The square box is centered at the centroid of the tumor. (d-h) Color-coded DCE images at 5 time frames, one pre-contrast and 4 post-contrast, normalized using the same signal intensity scales. Although this patient has moderate breast parenchymal enhancement (BPE), the lesion boundary is clearly visible and can be segmented with computer algorithms.
Deep Learning Networks
For deep learning, each slice was used as an independent input. That is, if a lesion contained 10 slices, all 10 slices were used as input. The cropped frame was resized to 32 × 32. In the training and testing dataset, the images were normalized in the same way to mean=0 and standard deviation=1, so their differences could be handled by standardization. The entire set of DCE images were normalized together so the change of signal intensity could be considered.
The conventional CNN architecture is shown in Figure 3. The DCE images were directly used as inputs, not the subtraction images. For CNN, all 5 sets of pre- and post-contrast images were put in together, with the input size of 32×32×5. Detailed methods using this CNN were reported in Chang et al. [17]. In brief, the architecture used 7 layers and the size of convolution kernel was 3×3. The stride number of the 2nd, 4th and 6th convolution layers in the output transformation was 2, which reduced the spatial resolution to one fourth the size of the input feature map. Instead of max-pooling, this allowed the network to learn down-sampling parameters and facilitated gradient preservation during back-propagation [17]. After each convolution layer, we used rectified linear units (ReLU), which could lead to faster training and sparse representations. The training was implemented using the Adam optimizer. In the training dataset, the parameters were initialized using the heuristic approach with the “He initialization method” [18]. L2 regularization was implemented to prevent over-fitting by limiting the squared magnitude of the kernel weights. Additionally, an early stopping strategy was used to control over-fitting, in which the same echo number was applied to all folds in cross validation. The learning rate for the Adam optimizer was fixed to 0.001 [19].
Figure 3:
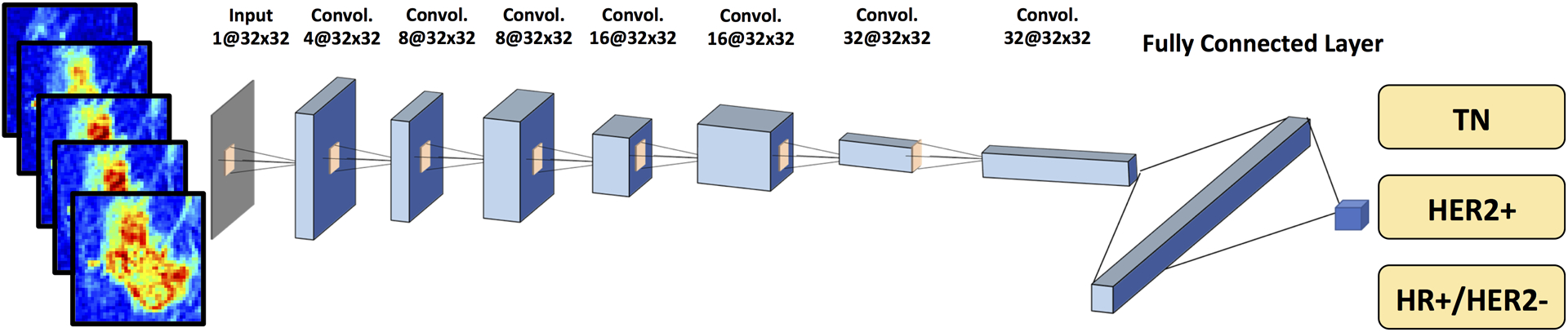
Diagram of convolutional neural network (CNN) architecture. The architecture uses 7 serial convolutional 3 × 3 filters followed by the ReLU nonlinear activation function. Dropout at 50% is applied to all convolutional and fully-connected layers after the second layer. Feature maps are down sampled to 25% of the previous layer by convolutions with a stride length of two. The number of the input channels is 5. The number of activation channels in deeper layers is progressively increased from 8 to 16 to 32 to 64. Softmax is used as the activation function of the last fully connected layer.
Another network, the convolutional long short term memory (CLSTM), was applied to track the temporal information of the changed signal intensity in the DCE time sequence [20], by inputting the 5 DCE datasets into the network one by one, shown in Figure 4. CLSTM is a recurrent neural network (RNN) and has convolutional layers to implement the input transformations and recurrent transformations. This architect can extract spatial features as well as temporal features from a series of images acquired in chronologic order. The same input box used in conventional CNN was used for CLSTM, but the size became 32×32×1 instead of 32×32×5. The output was the three subtypes, and the accuracy was calculated using cases that were correctly predicted to the HR+/HER2−, HER2+, and TN groups.
Figure 4:
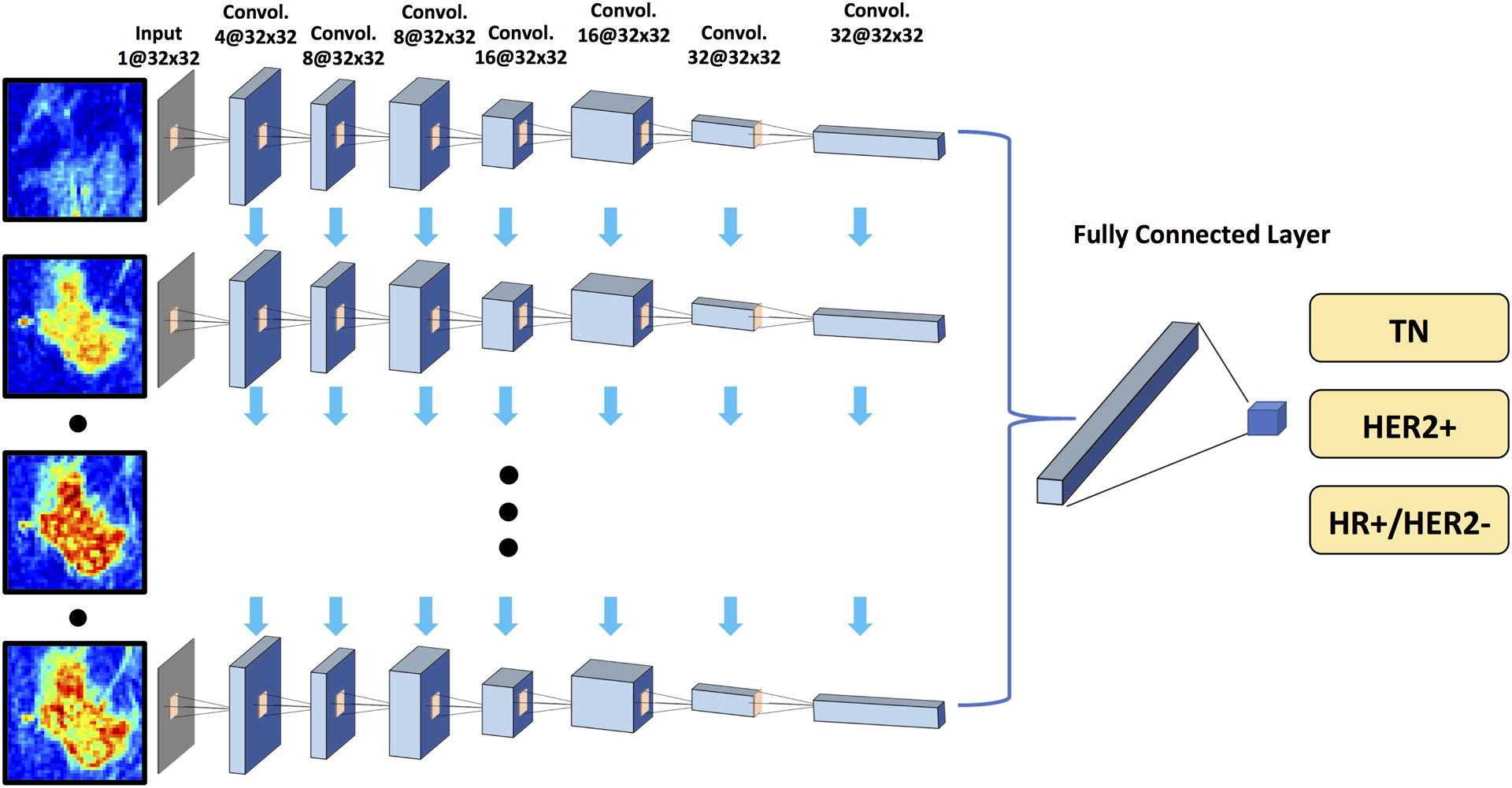
Diagram of convolutional long short term memory network (CLSTM) architecture. The architecture uses 7 serial convolutional LSTM layers via 3×3 filters followed by the ReLU nonlinear activation function. Five sets of pre-contrast and post-contrast DCE images are used as inputs. The configuration of the dropout and down sampling are the same as in Figure 3. The number of the input channels is one. Five sets of pre-contrast and post-contrast DCE images are used as inputs, by adding them one by one into the CLSTM network. The number of activation channels in deeper layers is progressively increased from 4 to 8 to 16 to 32. The last dense layer is obtained by flattening the convolutional output feature maps from all states. Softmax is used as the activation function of the last fully connected layer.
Model Evaluation and Transfer Learning
The first model was developed using the training dataset with 10-fold cross-validation. Each case had one chance to be included in the validation group. The results were pooled together, and the range and mean accuracy obtained using CNN and CLSTM were reported. In addition to 3-way subtype classification, the binary classification was performed to generate ROC curves, and the results obtained using CNN and CLSTM were statistically compared by the DeLong test.
After the model was developed, it was directly applied to the testing dataset-1 and dataset-2 for evaluation. Then, in order to consider datasets acquired using different settings, transfer learning was applied to fine-tune the parameters and develop another model specific to the testing dataset. In the transfer learning, instead of using the random He initialization method during the back-propagation process, the pre-trained model developed using the training dataset was used as the basis, that is, the weights of the trained network from the training dataset were used as the initial value. The transfer learning was done using the testing-1 cases for training with 10-fold cross-validation, and evaluated on testing-2; and then reversely done using testing-2 for training and evaluated on testing-1. This alternative approach could be used to evaluate the robustness of the transfer learning method.
RESULTS
Prediction Accuracy Using CNN and CLSTM
All results are listed in Table 1. When using the conventional CNN, the mean prediction accuracy in the training dataset obtained using 10-fold cross-validation was 0.79 (range 0.73–0.89). When using CLSTM that considered the temporal information in the DCE series, the mean prediction accuracy in the training dataset was improved to 0.91 (range 0.83–0.95). When the developed classification model was directly applied to the testing datasets, the accuracy was much lower. In Testing-1, the accuracy was 0.52 using CNN model and 0.44 using CLSTM model. In Testing-2, the accuracy was 0.47 using CNN model and 0.39 using CLSTM model. These results showed that the developed model from the training dataset acquired using a different scanner could not be applied to the testing dataset.
Table 1.
Accuracy to classify three molecular subtypes in Training and Testing datasets using CNN and CLSTM
| Dataset | Process | CNN | CLSTM |
|---|---|---|---|
| Training Dataset | Initial Training* | 0.73–0.89 (0.79) | 0.83–0.95 (0.91) |
| Testing Dataset-1 | Testing Using the First Trained Model | 0.52 | 0.44 |
| Second Training Using Transfer Learning* | 0.85–0.95 (0.91) | 0.79–0.88 (0.83) | |
| Testing Using the Second Model from Transfer Learning of Dataset-2 | 0.82 | 0.76 | |
| Testing Dataset-2 | Testing Using the First Model | 0.47 | 0.39 |
| Second Training Using Transfer Learning* | 0.82–0.89 (0.85) | 0.74–0.87 (0.82) | |
| Testing Using the Second Model from Transfer Learning of Dataset-1 | 0.78 | 0.74 |
The accuracy in the training process is evaluated using 10-fold cross-validation, and the range (mean) is shown
Binary Prediction Accuracy
In addition to 3-way classification in the training dataset, the binary prediction was performed to differentiate HR+/HER2− vs. others; TN vs. non-TN; and HER2+ vs. HER2−. The ROC curves obtained using CNN and CLSTM are shown in Figure 5. The accuracy, sensitivity, specificity and AUC are summarized in Table 2. The results were in general consistent with the 3-way classification performance, showing a higher accuracy when using CLSTM than CNN, but not reaching the significance level. For HR+/HER2− vs. others, AUC was 0.86 for CNN and 0.92 for CLSTM (p=0.14). For TN vs. non-TN, AUC was 0.84 for CNN and 0.89 for CLSTM (p=0.64). For HER2+ vs. HER2− was 0.90 for CNN and 0.93 for CLSTM (p=0.60).
Figure 5:
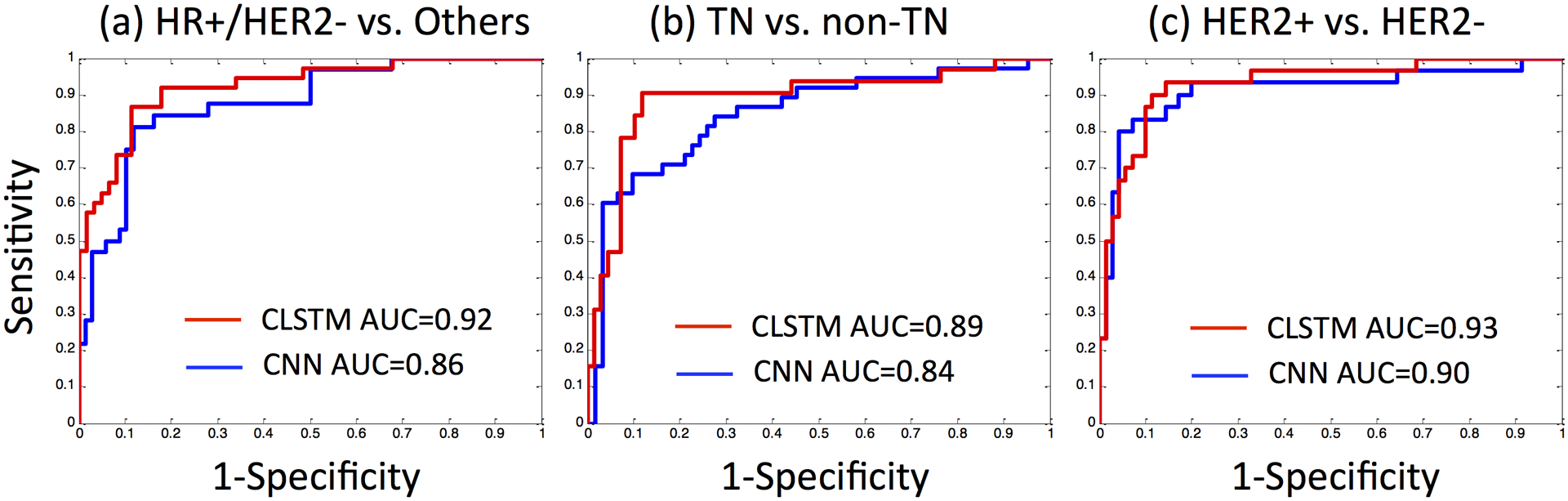
The ROC curves for binary molecular subtype classification in the Training dataset obtained using CNN and CLSTM. (a) HR+/HER2− vs. others, (b) TN vs. non-TN, (c) HER2+ vs. HER2−.
Table 2.
Binary molecular subtype classification performance in the Training dataset using CNN and CLSTM
| CNN | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| HR+/HER2− (N=65) vs. others (N=34) | 0.81 | 0.79 | 0.82 | 0.86 |
| TN (N=10) vs. non-TN (N=89) | 0.76 | 0.71 | 0.79 | 0.84 |
| HER2+ (N=24) vs. HER2− (N=75) | 0.80 | 0.73 | 0.83 | 0.90 |
| CLSTM | ||||
| HR+/HER2− (N=65) vs. others (N=34) | 0.90 | 0.89 | 0.91 | 0.92 |
| TN (N=10) vs. non-TN (N=89) | 0.89 | 0.82 | 0.92 | 0.89 |
| HER2+ (N=24) vs. HER2− (N=75) | 0.92 | 0.90 | 0.93 | 0.93 |
Prediction Accuracy with Transfer Learning
By using the initial trained model as the basis, the parameters were re-tuned in the testing datasets using transfer learning, also evaluated using 10-fold cross-validation. When using CNN, the mean accuracy in re-training of Testing-1 was 0.91 (range 0.85–0.95), and that could be applied to Testing-2 to improve accuracy from 0.47 to 0.78. When using CLSTM, the re-training mean accuracy in Testing-1 was 0.83 (range 0.79–0.88), and that also greatly improved accuracy in Testing-2 from 0.39 to 0.74. Similarly, when using the Testing-2 for re-training, the developed model could be applied to Testing-1 and improved the accuracy from 0.52 to 0.82 using CNN, and from 0.44 to 0.76 using CLSTM. The improvement is summarized in Table 1. The second model developed using transfer learning could improve accuracy by 0.31 and 0.30 using CNN, and 0.35 and 0.32 using CLSTM, overall greater than 30%.
DISCUSSION
Machine learning methods, including radiomics and deep learning, have potential to provide a comprehensive evaluation of the heterogeneous tumor known to be associated with underlying tumor biology [21]. In this study, we applied deep learning to predict three breast cancer molecular subtypes: HR+/HER2−, HER2+ and TN breast cancers that have different treatment strategies. A conventional CNN and a recurrent CLSTM network were used. In the training dataset, the CLSTM that could consider the changing signal intensity in the DCE series achieved a higher mean accuracy of 0.91 compared to the mean of 0.79 by using the conventional CNN. In the independent testing, it was clear that the developed models could not be directly applied, but when transfer learning was used, the re-tuned model could significantly improve accuracy. This study elaborates how the AI methods developed using one training dataset can be implemented in a different clinical setting, e.g. images acquired using different protocols, different scanners or in different hospitals. Although the approach using transfer learning was trivial, yet few studies have actually implemented the transfer learning and demonstrated how it worked using well-characterized datasets.
Breast cancer molecular subtypes are very important for choosing the optimal treatments. HER-2 targeting agents, Trastuzumab and Pertuzumab, are included in the treatment for HER-2 positive cancer. Long-term (5–10 years) hormonal therapy such as tamoxifen and aromatase inhibitors are used for HR positive cancer to prevent recurrence. For the TN cancers, they are more aggressive and no targeted therapy, and thus, more aggressive chemotherapy is usually given to achieve a better outcome. While molecular markers can be evaluated from tissues obtained in biopsy or surgery, it is subject to the tissue sampling bias problem. Breast MR images contain rich information, which may be used for differentiation of molecular subtypes, by using images acquired at the time of diagnosis for a thorough assessment of the entire tumor.
For breast DCE-MRI, the pattern of the DCE kinetics (or, signal intensity time curve) is known to provide important information for lesion diagnosis, which can be taken into consideration in deep learning architecture using various strategies [11–13,22,23]. To consider the full spectrum of this time-dependent intensity information, CLSTM was developed to process the DCE images set by set, as in a previous study [24]. The CLSTM is similar to Long Short-Term Memory (LSTM) network reported by Hochreiter et al. [25], which is a Recurrent Neural Network (RNN) used for processing time series and text. The temporal features contained in the time order of the 5 DCE pre- and post-contrast MRI sets can be fully explored, and that achieved a higher accuracy compared to conventional CNN (0.91 vs. 0.79).
Several studies have applied deep learning to differentiate breast cancer subtypes. Ha et al. applied a deep learning method using residual neural network for subtype differentiation, and reached 70% accuracy and AUC of 0.85 [16]. Zhu et al. applied several different CNN architectures, including GoogleNet, VGG and CIFAR, to analyze DCE-MRI and achieved the best accuracy of 0.65 [14]. All these studies only analyzed a single-institutional dataset, and the reported accuracy was comparable to our result obtained with convention CNN in the training dataset. In an extensive literature search, we have not found any study that included a second independent dataset for testing, as done in our study. In addition to MRI or other breast images, H&E stained histologic images also contain rich information, and present a great opportunity for deep learning-based analysis for subtype classification [26,27].
The term “transfer learning” is used broadly, which is often referring to pre-training. Usually, the pipeline of CNN classification contains 2 stages. First, a network is pre-trained by a natural image dataset to obtain the weights of the trainable parameters, e.g. ImageNet, which is a set of network weights pre-trained by a large public natural image dataset. Next, the training dataset in the intended application is used to fine-tune the pre-trained network to achieve the best performance. For example, Nishio et al. [28], applied VGG16 to differentiate benign nodule, primary lung cancer and metastatic lung cancer on lung CT. The network was initialized using ImageNet, then fine-tuned, which showed increased accuracy from 62.3% to 68% with transfer learning. Two other studies by Yuan et al. [29] and Byra et al. [30], also applied a similar strategy using fine-tuned CNN with pre-trained ImageNet, and achieved higher accuracy for prostate and breast lesion classification. Another strategy, as demonstrated in Samala et al. [31], designed a CNN pre-trained by mammography dataset to classify breast lesions on digital breast tomosynthesis (DBT). For clinical implementation, the cases were usually acquired in a different setting, and as demonstrated here, re-tuning of the parameters is necessary to improve accuracy. Many companies are developing AI tools, and usually the product can achieve a high accuracy using training datasets. For field implementation in different hospitals, transfer learning based on the specific datasets collected in each hospital is necessary. In the present study, we split the testing cases based on the time of MRI, which represented a realistic clinical scenario. For example, if an AI software developed by a company is sold to a hospital, it can be re-trained using retrospective datasets, and then applied to prospective cases.
The major limitation was the small case number, particularly for the TN subtype. Unfortunately, this was a common problem for all cancer subtype differentiation studies no matter whether it was based on histology, molecular biomarkers or genetic mutations. For multi-class differentiation to predict breast cancer molecular subtypes, or to predict different primary tumors in metastasis [32], the overall accuracy was a harsh outcome that often resulted in low accuracy, i.e. each case had to be correctly classified into one of several classes to be counted as accurate. For some clinical applications, combining multi-class into binary classification would be sufficient, e.g. to differentiate lung cancer from other primary cancers in patients with spinal or brain metastasis [24,32]. The application of machine learning for medical imaging analysis can be designed according to the available case number and the clinical indications, as well as whether there are appropriate datasets that can be used for pre-training.
In conclusion, we have implemented two deep learning networks, conventional CNN and CLSTM, to classify three breast cancer molecular subtypes that have different treatment strategies. The accuracy in the training dataset could reach 0.8–0.9, but the developed model could not be directly applied to the independent testing dataset acquired in a different hospital using a different scanner. When using part of the testing dataset for re-tuning, the accuracy could be greatly improved by 30%. The results suggest that deep learning can be applied to aid in tumor molecular subtype prediction, and also that transfer learning can be implemented to re-tune the developed model for wide adoption in different clinical settings.
KEY POINTS.
Deep learning can be applied to differentiate breast cancer molecular subtypes.
The recurrent neural network using CLSTM could track the change of signal intensity in DCE images, and achieved a higher accuracy compared to conventional CNN during training.
For datasets acquired using different scanners with different imaging protocols, transfer learning provided an efficient method to re-tune the classification model and improve accuracy.
List of Abbreviations:
- ADC
Apparent Diffusion Coefficient
- AUC
Area Under the ROC Curve
- CAD
Computer-Aided Diagnosis
- CLSTM
Convolutional Long Short Term Memory
- CNN
Convolutional Neural Network
- DCE-MRI
Dynamic Contrast Enhanced Magnetic Resonance Imaging
- FCM
Fuzzy-C-Means
- GLCM
Gray Level Co-occurrence Matrix
- HR
Hormonal Receptor
- LSTM
Long Short-Term Memory
- RNN
Recurrent Neural Network
- ROC
Receiver Operating Characteristic
- ROI
Region Of Interest
- SE
Signal Enhancement
- SVM
Support Vector Machine
- TN
Triple Negative
REFERENCES
- 1.Sandhu R, Parker JS, Jones WD, Livasy CA, Coleman WB (2010) Microarray-based gene expression profiling for molecular classification of breast cancer and identification of new targets for therapy. Lab. Med 41:364–372 [Google Scholar]
- 2.Houssami N, Turner RM, Morrow M (2017) Meta-analysis of pre-operative magnetic resonance imaging (MRI) and surgical treatment for breast cancer. Breast Cancer Res Treat 165(2):273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agner SC, Rosen MA, Englander S et al. (2014) Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology 272:91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang R-F, Chen H-H, Chang Y-C, Huang C-S, Chen J-H, Lo C-M (2016) Quantification of breast tumor heterogeneity for ER status, HER2 status, and TN molecular subtype evaluation on DCE-MRI. Magn Reson Imaging 34:809–819 [DOI] [PubMed] [Google Scholar]
- 5.Sutton EJ, Dashevsky BZ, Oh JH et al. (2016) Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 44:122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Zhu Y, Burnside ES et al. (2016) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan M, Li H, Wang S, Zheng B, Zhang J, Li L (2017) Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PloS One 12:e0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braman NM, Etesami M, Prasanna P et al. (2017) Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma W, Zhao Y, Ji Y et al. (2019) Breast Cancer Molecular Subtype Prediction by Mammographic Radiomic Features. Acad Radiol 26:196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L, Zhang Y, Nie K, et al. (2019) Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn Reson Imaging 61:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C (2019) Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 290:290–297 [DOI] [PubMed] [Google Scholar]
- 12.Antropova N, Huynh B, Li H, Giger ML (2019) Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J Med Imaging (Bellingham) 6(1):011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Zhang Y, Chang KT, et al. (2020) Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J Magn Reson Imaging 51(3):798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA (2019) Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med 109:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie T, Wang Z, Zhao Q, et al. (2019) Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Front Oncol 9:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha R, Mutasa S, Karcich J, et al. (2019) Predicting Breast Cancer Molecular Subtype with MRI Dataset Utilizing Convolutional Neural Network Algorithm. J Digit Imaging 32:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang P, Grinband J, Weinberg B et al. (2018) Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am J Neuroradiol 39(7):1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. Proceedings of the IEEE international conference on computer vision, pp 1026–1034 [Google Scholar]
- 19.Kingma D, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980 [Google Scholar]
- 20.Shi X, Chen Z, Wang H, Yeung D-Y, Wong W-K, Woo W-C (2015) Convolutional LSTM network: A machine learning approach for precipitation nowcasting. NIPS’15: Proceedings of the 28th International Conference on Neural Information Processing Systems, pp 802–810 [Google Scholar]
- 21.Michael KY, Ma J, Fisher J, Kreisberg JF, Raphael BJ, Ideker T (2018) Visible Machine Learning for Biomedicine. Cell 173:1562–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antropova N, Abe H, Giger ML (2018) Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep con- volutional neural networks. J Med Imaging 5:014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haarburger C, Langenberg P, Truhn D, et al. (2018) Transfer learning for breast cancer malignancy classification based on dynamic contrast- enhanced MR images. In: Maier A, Deserno T, Handels H, Maier-Hein K, Palm C, Tolxdorff T, eds. Bild-verarbeitung für die Medizin Berlin: Springer; 2018;216–221 [Google Scholar]
- 24.Lang N, Zhang Y, Zhang E, et al. (2019) Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imaging 64:4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9:1735–1780 [DOI] [PubMed] [Google Scholar]
- 26.Couture HD, Williams LA, Geradts J, et al. (2018) Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaber MI, Song B, Taylor C, et al. (2020) A deep learning image-based intrinsic molecular subtype classifier of breast tumors reveals tumor heterogeneity that may affect survival. Breast Cancer Res 22:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishio M, Sugiyama O, Yakami M, et al. (2018) Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS One 13:e0200721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, Qin W, Buyyounouski M, et al. (2019) Prostate cancer classification with multiparametric MRI transfer learning model. Med Phys 46:756–765 [DOI] [PubMed] [Google Scholar]
- 30.Byra M, Galperin M, Ojeda-Fournier H, et al. (2019) Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med Phys 46:746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samala RK, Heang-Ping Chan, Hadjiiski L, Helvie MA, Richter CD, Cha KH (2019) Breast Cancer Diagnosis in Digital Breast Tomosynthesis: Effects of Training Sample Size on Multi-Stage Transfer Learning Using Deep Neural Nets. IEEE Trans Med Imaging 38:686–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz-Ramón R, Larroza A, Ruiz-España S, Arana E, Moratal D (2018) Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 28:4514–4523 [DOI] [PubMed] [Google Scholar]


