Summary
Astrocytes extensively infiltrate the neuropil to regulate critical aspects of synaptic development and function. This process is regulated by transcellular interactions between astrocytes and neurons via cell adhesion molecules. How astrocytes coordinate developmental processes amongst each other to parse out the synaptic neuropil and form non-overlapping territories is unknown. Here we identify a molecular mechanism regulating astrocyte-astrocyte interactions during development to coordinate astrocyte morphogenesis and gap junction coupling. We show that hepaCAM, a disease-linked, astrocyte-enriched cell adhesion molecule, regulates astrocyte competition for territory and morphological complexity in the developing mouse cortex. Furthermore, conditional deletion of Hepacam from developing astrocytes significantly impairs gap junction coupling between astrocytes and disrupts the balance between synaptic excitation and inhibition. Mutations in HEPACAM cause megalencephalic leukoencephalopathy with subcortical cysts in humans. Therefore, our findings suggest that disruption of astrocyte self-organization mechanisms could be an underlying cause of neural pathology.
eTOC Summary
How astrocytes coordinate their interactions with different cells and establish non-overlapping territories is unknown. Baldwin et al., show that hepaCAM controls astrocyte morphogenesis and competition for territory. Loss of hepaCAM from astrocytes alters localization of Connexin 43, impairs gap junction coupling, and disrupts the balance between synaptic excitation and inhibition.
Introduction
Astrocytes, a major glial cell type in the brain, form elaborately branched arbors to directly interact with many different cells and structures (Allen and Lyons, 2018) and control a wide variety of physiological processes that are fundamentally important for proper brain development and function, including synapse formation, regulation of neurotransmission, and establishment of the blood-brain barrier (Baldwin and Eroglu, 2017; Chung et al., 2015). Despite these vital functions of astrocytes in the brain, the cellular and molecular mechanisms that drive the development of astrocyte morphological complexity and regulate astrocyte interactions with other cells are poorly understood. Furthermore, astrocyte dysfunction is implicated in many neurodevelopmental disorders, including autism and schizophrenia (Blanco-Suarez et al., 2016; Sloan and Barres, 2014); however, the extent to which disruptions to astrocyte development and function drive disease pathogenesis is unclear.
Astrocytes establish distinct non-overlapping territories to tile the brain, an evolutionarily conserved phenomenon that is present from fruit flies to mammals (Bushong et al., 2002; Oberheim et al., 2006; Stork et al., 2014). The molecular mechanisms driving astrocyte tiling are unknown, but may be mediated through contact inhibition via astrocyte-astrocyte interactions (Distler et al., 1991). Astrocyte tiling is disrupted in disease states and following injury, suggesting that establishment of astrocytic domains is important for normal brain function (Myer et al., 2006; Oberheim et al., 2008). Astrocyte-astrocyte interactions also mediate astrocyte proliferation and maturation (Li et al., 2019), and cell-cell communication (Giaume et al., 2010). Astrocytes are extensively coupled through gap junctions, and disruptions to gap junction coupling are observed in many disease and injury states (Mayorquin et al., 2018). Collectively, these studies suggest that the ability of astrocytes to balance discrete anatomical domains and a functionally coupled network is important for normal brain function.
Astrocytes express cell adhesion molecules that may participate in cell-cell interactions (Zhang et al., 2016). Indeed, we found that astrocyte-expressed neuroligins mediate astrocyte-neuron interactions to synchronize astrocyte morphogenesis with synaptogenesis (Stogsdill et al., 2017). However, how astrocytes coordinate their development with neighboring astrocytes and balance these contacts with synaptic contacts, is unknown. We hypothesized that specific cell-adhesion molecules expressed at astrocyte-astrocyte cell junctions might mediate interactions between neighboring astrocytes to regulate astrocyte development and function. Furthermore, we postulated that dysfunction of astrocytic cell adhesions could facilitate the pathogenesis of neurodevelopmental brain disorders.
To test this hypothesis, we investigated the function of hepatocyte cell adhesion molecule (hepaCAM) in astrocyte and brain development. HepaCAM, also known as GlialCAM, is an astrocyte-enriched cell adhesion molecule that was first identified in human cancer cell lines, participating in cell-cell and cell-ECM interactions to regulate cell migration and proliferation (Lee et al., 2009; Moh et al., 2005). In cultured astrocytes, hepaCAM forms homophilic cis and trans interactions to facilitate the targeting of transmembrane proteins to cell-cell junctions, including MLC1, a membrane protein of unknown function, and the chloride channel ClC-2, for which hepaCAM also serves as an auxiliary subunit (Capdevila-Nortes et al., 2015; Elorza-Vidal et al., 2020; Jeworutzki et al., 2012). Mutations to the extracellular domain of hepaCAM cause megalencephalic leukoencephalopathy with subcortical cysts (MLC), a developmental and progressive brain disorder characterized by early-onset macrocephaly, white matter swelling, intellectual disability and epilepsy (Lopez-Hernandez et al., 2011a; van der Knaap et al., 2012).
Despite the causal link between hepaCAM mutations and human neurological disorders, the function of hepaCAM in astrocytes during brain development remains poorly understood.
Results
HepaCAM regulates astrocyte morphogenesis in vitro and in vivo
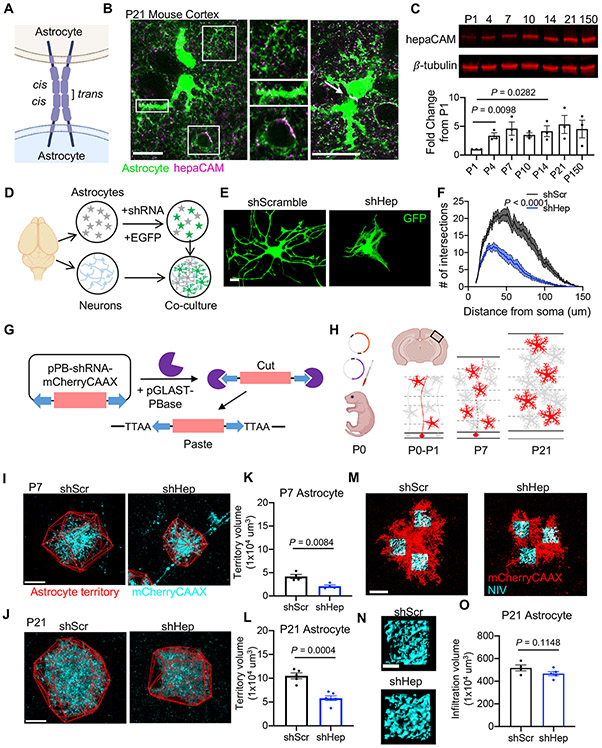
In the mouse primary visual cortex (V1) at postnatal day 21 (P21), we found strong hepaCAM expression in astrocytes with concentrated expression at astrocyte-astrocyte contacts (Figure 1A and 1B). HepaCAM protein expression is evident at postnatal day 4 (P4) and increases during postnatal development (Figure 1C). To investigate the function of hepaCAM in astrocyte development, we used primary rat astrocyte and neuron co-cultures to examine astrocyte morphogenesis following knockdown of hepaCAM with short hairpin RNA (shRNA) (Figures 1D and S1A-S1C). Compared to astrocytes transfected with a scrambled shRNA control (shScr), hepaCAM knockdown (shHep) significantly reduced astrocyte branching complexity (Figures 1E and 1F) and astrocyte length (shScr: 2274.2 ± 44.2 μm vs. shHep: 810.8 ± 41.5 μm, P < 0.0001). Additionally, the twisted and overlapping branches of hepaCAM knockdown astrocytes presented a distinct phenotype from knockdown of astrocytic neuroligins (Stogsdill et al., 2017) such as neuroligin 1 (NL1) (Figures S1E and S1F) and NL2 (Figures S1G and S1H). Moreover, the combined knockdown of NL2 and hepaCAM further reduced astrocyte complexity, suggesting that hepaCAM and NL2 regulate astrocyte morphogenesis through independent pathways (Figures S1G and S1H).
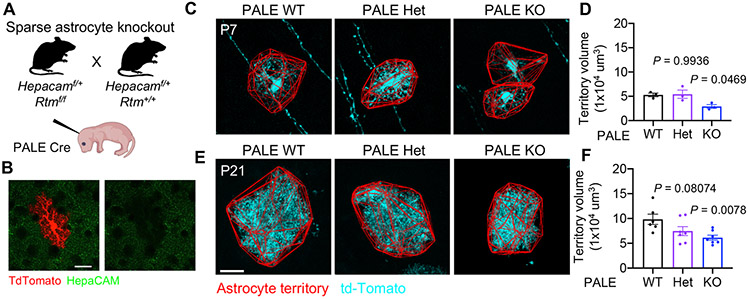
Figure 1: HepaCAM regulates astrocyte morphogenesis in vitro and in vivo.
(A) hepaCAM at cell-cell junctions in homophilic cis and trans interactions. (B) hepaCAM (magenta) in the visual cortex of Aldh1L1-eGFP mice at P21. Scale bar, 20 μm. (C) Western blot analysis of hepaCAM protein expression in mouse cortex. Quantification of hepaCAM protein expression normalized to β-tubulin as a fold change from P1. n = 3 mice/timepoint. Mean ± s.e.m. One-way ANOVA, Tukey’s post-test. (D) Schematic of astrocyte-neuron co-culture assay. (E) Rat astrocytes transfected with EGFP and scrambled shRNA (shScr), or shRNA targeting Hepacam (shHep) and co-cultured with cortical neurons. Scale bar, 20 μm. (F) Quantification of astrocyte branching complexity. Data are mean ± s.e.m. n = 4 independent experiments, 20 cells/condition/experiment. ANCOVA, Tukey’s post-test. (G) Adaptation of Piggybac transposon system. Co-expression of pPB-shRNA-mCherryCAAX and pGLAST-PBase yields genomic integration of shRNA and mCherry-CAAX in developing astrocytes. (H) Overview of Postnatal Astrocyte Labeling by Electroporation (PALE) with Piggybac plasmids. (I and J) Images of V1 L5 astrocytes at P7 (I) and P21 (J) expressing mCherry-CAAX (cyan) and shScr or shHep. Astrocyte territory in red. Scale bar, 20 μm. (K and L) Average territory volumes of P7 (K) and P21 (L) astrocytes. n = 4 mice/condition, 2-8 cells/mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. (M) Images of V1 L5 astrocytes at P21 expressing mCherry-CAAX (red) and shRNA. Scale bar, 20 μm. (N) NIV reconstructions (cyan). Scale bar, 5 μm. (O) Average NIV. 3 ROIs/cell, 4-12 cells/mouse, 4 (shScr) or 5(shHep) mice. Data points, mouse averages. Bars are mean ± s.e.m. Nested t test. See also Figure S1.
Neuroligins mediate astrocyte arborization in response to neuronal contact via interactions with neuronal neurexins (Stogsdill et al., 2017). In cultured cells, hepaCAM forms homophilic trans-interactions at cell-cell junctions (Capdevila-Nortes et al., 2015). While hepaCAM is highly enriched in astrocytes, neurons express detectable amounts of hepaCAM mRNA (Zhang et al., 2014). To test whether hepaCAM controls astrocyte morphology via homophilic interactions between astrocytes and neurons, we silenced hepaCAM expression only in neurons and found that neuronal contact-induced astrocyte morphogenesis was not altered, suggesting that astrocytic hepaCAM controls astrocyte morphogenesis in a cell-autonomous manner (Figures S1I-S1K).
To determine whether hepaCAM regulates astrocyte morphogenesis in vivo, we used Postnatal Astrocyte Labeling by Electroporation (PALE) combined with the PiggyBac transposon system (Chen et al., 2014) to introduce shRNA and a membrane-targeted mCherry (mCherry-CAAX) into a sparse population of astrocytes in the mouse cortex at P0 (Figures 1G and 1H). At P7, we found that Layer 5 astrocytes expressing shHep had a significantly smaller territory volume compared to shScr astrocytes (Figures 1I and 1K). At P21, following a period of astrocyte growth and extensive elaboration, the territory volume of shHep astrocytes remained significantly smaller than shScr astrocytes (Figures 1J and 1L). The infiltration of fine astrocyte processes into the neuropil (neuropil infiltration volume, NIV) was unchanged at P21 (Figures 1M-1O). Taken together, these results show that hepaCAM is required for proper astrocyte territory growth and morphogenesis in the developing mouse cortex.
HepaCAM regulates astrocyte morphogenesis via its extracellular and intracellular domains
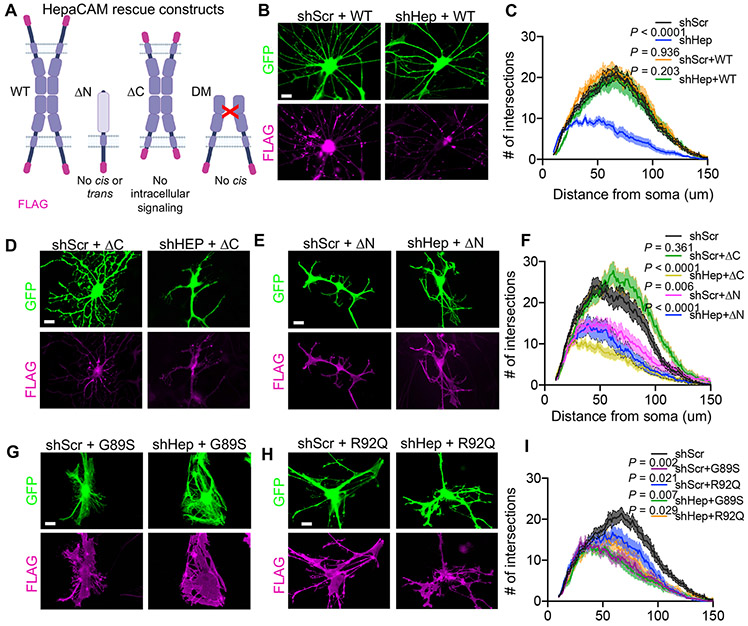
To investigate the molecular mechanisms through which hepaCAM regulates astrocyte morphogenesis, we performed rescue experiments in vitro by expressing mutated versions of an shRNA-resistant human hepaCAM protein with a C-terminal FLAG tag (Figure 2A). Co-expression of wild-type (WT) human hepaCAM with shHep fully rescued astrocyte morphological complexity (Figures 2B and 2C), whereas expression of hepaCAM lacking its intracellular domain (ICD) (ΔC) did not (Figures 2D and 2F). Additionally, expression of WT hepaCAM caused a noticeable straightening of branches not observed in control conditions or with expression of hepaCAM ΔC. Replacing the extracellular domain (ECD) of hepaCAM with the ECD of the human IL-2 receptor, (ΔN) disrupts hepaCAM-mediated cis- and trans-interactions in vitro (Capdevila-Nortes et al., 2015). Co-expression of hepaCAM ΔN with shHep was unable to rescue astrocyte morphological complexity (Figures 2E and 2F), indicating that interactions mediated by the hepaCAM ECD are necessary for proper astrocyte morphogenesis. Furthermore, overexpression of hepaCAM ΔN with shScr significantly reduced astrocyte complexity, indicating that the ICD of hepaCAM, when uncoupled from its ECD, may have a dominant-negative function (Figures 2E and 2F). Collectively, these results demonstrate that both the intracellular and extracellular domains of hepaCAM are necessary for proper astrocyte morphogenesis and suggest that hepaCAM regulates astrocyte morphogenesis in vitro by coupling cell adhesion to intracellular signaling mechanisms.
Figure 2: HepaCAM regulates astrocyte morphogenesis via extracellular and intracellular domains.
(A) shRNA-resistant human hepaCAM rescue constructs with C-terminal FLAG tag (magenta). (B, D, E, G, H) Astrocytes (green) expressing shRNA and FLAG-tagged hepaCAM rescue constructs (magenta) and co-cultured with neurons. Scale bar, 20 μm. (C, F, I) Quantification of astrocyte branching complexity. n = 3 independent experiments, 20 cells/condition/experiment. Data are mean ± s.e.m. ANCOVA, Tukey’s post-test.
Single amino acid substitutions in the extracellular IgV domain of hepaCAM cause MLC in humans (Lopez-Hernandez et al., 2011a). To determine whether disease-causing mutations to hepaCAM impair astrocyte morphogenesis in vitro, we transfected astrocytes with mutant versions of hepaCAM carrying either a dominant (G89S) (Figure 2G) or recessive (R92Q) (Figure 2H) MLC-causing mutation. Both of these mutations impair hepaCAM cis dimerization and localization of hepaCAM to cell-cell junctions (Elorza-Vidal et al., 2020). In astrocytes co-cultured with neurons, hepaCAM mutants were expressed broadly throughout the cell (Figure 2G and 2H), in contrast to the restricted distribution of WT and endogenous hepaCAM (Figure 2B), suggesting that these mutations cause a loss of specific targeting and/or retention at cell-cell junctions. Over-expression of hepaCAM disease mutants (DM) in shScr or shHep astrocytes significantly reduced astrocyte branching complexity (Figure 2I) and substantially altered astrocyte branching organization, causing individual astrocytes to branch onto themselves and form web-like structures (Figures 2G and 2H). These results show that disease-causing mutations in hepaCAM impair astrocyte morphogenesis in a dominant-negative fashion in vitro and suggest that hepaCAM cis interaction as well as proper hepaCAM localization are required for astrocyte morphogenesis.
Connexin 43 knockdown phenocopies hepaCAM knockdown in vitro and in vivo
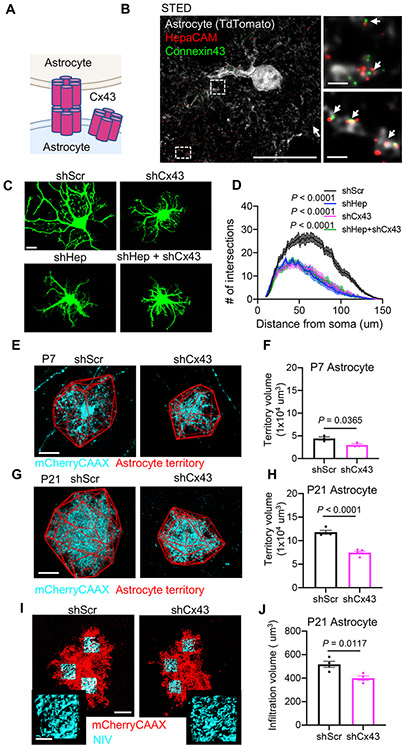
Previous studies found that hepaCAM is required to localize transmembrane proteins ClC2 and MLC1 to cell-cell junctions (Capdevila-Nortes et al., 2013; Hoegg-Beiler et al., 2014; Jeworutzki et al., 2012). Recently hepaCAM was also found to be important for stabilizing the gap junction protein Connexin 43 (Cx43) at cell-cell junctions in vitro (Wu et al., 2016), a finding which we confirmed in astrocyte-only cultures from Hepacam knockout mice (Figure S2A). Cx43 forms gap junction channels between neighboring astrocytes, allowing for the exchange of ions and small molecules (Figure 3A). Independently of channel activity, connexins also function as cell adhesion molecules to control cell-cell interactions and cell migration (Elias et al., 2007; Pannasch et al., 2014), processes also regulated by hepaCAM (Lopez-Hernandez et al., 2011b; Moh et al., 2009). Given the overlapping functions of hepaCAM and Cx43, we wondered whether these two molecules might work together in regulating astrocyte morphogenesis.
Figure 3: Connexin 43 knockdown phenocopies hepaCAM knockdown in vitro and in vivo.
(A) Cx43 organization in astrocytes. (B) Three-color stimulated emission depletion (STED) image of a V1 TdTomato+ astrocyte (grey) at P21. Scale bar, 20 μm (left) and 1 μm (right). (C) Astrocytes expressing shRNAs targeting hepaCAM (shHep), Cx43 (shCx43), or a scrambled control (shScr). Scale bar, 20 μm. (D) Quantification of astrocyte branching complexity. Data are mean ± s.e.m. n = 3 independent experiments, 20 cells/condition/experiment. ANCOVA, Tukey’s post-test. (E and G) V1 L5 astrocytes at P7 (E) and P21 (G) expressing mCherry-CAAX (cyan) and shScr or shCx43. Astrocyte territory in red. Scale bar, 20 μm. (F and H) Average territory volumes of P7 (F) and P21 (H) astrocytes. n = 3 mice/condition, 5-13 cells/mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. (I) V1 L5 astrocytes at P21 expressing mCherry-CAAX (red) and shRNA with NIV reconstructions (cyan). Scale bars, 20 μm (inset 5 μm). (J) Average NIV of shScr and shHep astrocytes. 3 ROIs/cell, 3-12 cells/mouse, 4 mice/condition. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. See also Figures S2 and S3.
Using stimulated emission depletion (STED) and confocal microscopy, we found hepaCAM and Cx43 frequently co-localized at astrocyte processes in the mouse cortex (Figure 3B, S2B, and S2C). To determine whether hepaCAM and Cx43 interact in the mouse brain, we performed native affinity purifications followed by liquid chromatography-coupled high-resolution mass spectrometry (AP-MS) with two different hepaCAM antibodies and observed co-immunoprecipitation between hepaCAM and Cx43 (Figure S2D and S2E). To probe the functional relationship between hepaCAM and Cx43 in regulating astrocyte morphogenesis, we used an shRNA to knockdown Cx43 (shCx43) in vitro and in vivo (Figures S3A and S3B). In astrocytes co-cultured with neurons, Cx43 knockdown significantly reduced astrocyte morphological complexity similar to hepaCAM knockdown and the combined knockdown of hepaCAM and Cx43 (shHep + shCx43) did not further reduce astrocyte complexity (Figures 3C and 3D), suggesting that hepaCAM and Cx43 function in the same pathway to regulate astrocyte morphogenesis in vitro. Next, we used PALE combined with the PiggyBac transposon system to knockdown Cx43 in sparse populations of astrocytes in the developing mouse visual cortex. Similar to hepaCAM knockdown, shCx43 astrocytes had a significantly smaller territory volume at P7 (Figures 3E and 3F). At P21, both the territory volume and neuropil infiltration volume of shCx43 astrocytes were significantly reduced (Figures 3G-3J), showing that Cx43 knockdown impairs astrocyte morphogenesis in vivo.
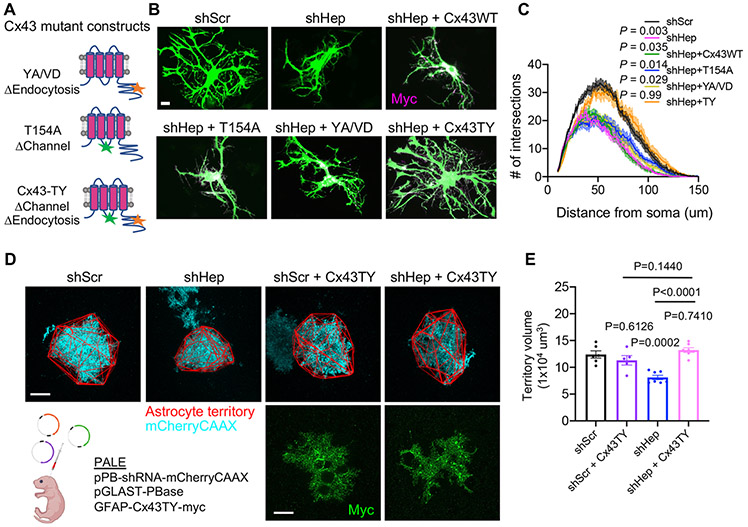
HepaCAM stabilizes Cx43 to regulate astrocyte morphogenesis through a channel-independent mechanism
Because membrane localization of Cx43 was disrupted in astrocyte cultures from Hepacam KO mice (Figure S2A), we hypothesized that hepaCAM might regulate astrocyte morphogenesis by controlling the localization and/or stability of Cx43. To test this, we attempted to rescue the morphology of shHep astrocytes by introducing different shRNA-resistant Cx43 mutant constructs (Figure 4A). Wild-type Cx43 with a C-terminal Myc tag (Cx43-WT) significantly increased the complexity and total process length of astrocytes transfected with shCx43 (Figures S3C-S3E), yet was unable to rescue the complexity of shHep astrocytes (Figures 4B and 4C). Cx43 is a dynamically regulated protein with a half-life of under two hours (Beardslee et al., 1998). To test whether stabilizing Cx43 at the membrane could overcome the requirement for hepaCAM and rescue astrocyte morphological complexity, we used previously described Cx43 mutations to disrupt endocytosis, Cx43-Y286A/V289D (Cx43-YA/VD) (Johnson et al., 2013) (Figure 4A). Similar to Cx43-WT, Cx43-YA/VD significantly increased the complexity of shCx43 astrocytes (Figures S3G and S3J) but failed to increase the complexity of shHep astrocytes (Figures 4B and 4C). A previous study suggested that Cx43 overexpression may produce unwanted side effects due to excess channel function and that Cx43 may also have channel-independent adhesive functions (Elias et al., 2007). To test whether inhibiting channel function might enable endocytosis-deficient Cx43 to exert its adhesive properties while avoiding potential toxicity due to excess channel function, we generated Cx43-TY (Figure 4A) which lacks both the endocytosis motif and the previously characterized T154A mutation that blocks channel function (Beahm et al., 2006). Both Cx43-T154A and Cx43-TY rescued the complexity of shCx43 astrocytes (Figures S3F, S3J, S3H, and S3K), indicating that channel function is not required for Cx43-mediated control of astrocyte morphogenesis. In contrast to the punctate expression of Cx43-WT and Cx43-YA/VD in astrocytes co-cultured with neurons, Cx43-T154A and Cx43-TY were distributed throughout the cell (Figure S3L). The T154A mutation was previously shown to increase the invasive properties of human metastatic melanoma cells (Zucker et al., 2013). Therefore, in addition to blocking channel function, the T154A mutation may also enhance adhesive function of Cx43.
Figure 4: HepaCAM stabilizes Cx43 to regulate astrocyte morphogenesis through a channel-independent mechanism.
(A) Connexin 43 rescue constructs resistant to shRNA and mutated to disrupt channel function (T154A), endocytosis (YA/VD), or both channel function and endocytosis (TY). (B) Astrocytes (green) expressing shScr, shHep, or shHep and Myc-tagged Cx43 rescue constructs (magenta). Scale bar, 20 μm. (C) Quantification of astrocyte branching complexity. n = 3 independent experiments, 20 cells/condition/experiment. Mean ± s.e.m. ANCOVA, Tukey’s post-test. (D) Images of V1 L5 astrocytes at P21 expressing mCherry-CAAX (cyan), shScr or shHep, and Cx43-TY-myc (green). Astrocyte territory in red. Scale bar, 20 μm. (E) Territory volumes of shScr and shHep astrocytes without or with Cx43-TY expression. n= 5-7 mice/condition, between 1-6 cells per mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested one-way ANOVA, Tukey’s post-test. See also Figure S3.
Expression of Cx43-TY in shHep astrocytes fully restored the morphological complexity of shHep astrocytes to control levels, whereas the T154A mutant alone did not (Figure 4B and 4C), indicating that stabilizing Cx43 at the membrane, in the absence of Cx43 channel activity, is able to overcome loss of hepaCAM with respect to astrocyte morphogenesis. This effect was specific to hepaCAM and not due to a general increase in astrocyte adhesion as Cx43-TY did not rescue astrocyte morphological complexity in shNL2 astrocytes (Figure S3M and S3N). To determine whether Cx43-TY could rescue astrocyte morphogenesis in hepaCAM knockdown astrocytes in vivo, we performed PALE to express shRNA, mCherry-CAAX, and Cx43-TY (Figure 4D). Indeed, expression of Cx43-TY in shHep astrocytes rescued astrocyte territory volume to shScr control levels (Figure 4D and 4E). Collectively, these experiments reveal hepaCAM and Cx43 as important regulators of astrocyte morphogenesis in vitro and in vivo and suggest that hepaCAM controls astrocyte morphogenesis by regulating the surface stabilization of Cx43. These findings also demonstrate that Cx43 controls astrocyte morphogenesis via a channel-independent mechanism that is likely mediated through its adhesive properties.
HepaCAM regulates astrocyte competition for territory
Next, to determine the roles of astrocytic hepaCAM in brain development and function, we generated a transgenic mouse line to conditionally delete Hepacam from all astrocytes (Figures S4A and S4B). To test the cell autonomous effects of deleting one or both copies of Hepacam, we used PALE to introduce Cre into a sparse population of cortical astrocytes in Hepacam+/+, Hepacamflox/+, and Hepacamflox/flox mice at P0 (PALE WT, PALE Het, and PALE KO, respectively) (Figure 5A and 5B). These mice also expressed one copy of the Gt(ROSA)26Sortm14(CAG-tdTomato)Hze (RTM) transgene to label Cre positive (Cre+) cells with td-Tomato. Similar to hepaCAM knockdown with shRNA, Hepacam PALE KO astrocytes had a smaller territory volume than PALE WT astrocytes at P7 and P21, but not at P60 (Figures 5C-5F, S4D and S4E).
Figure 5: Sparse deletion of one or both alleles of Hepacam reduces astrocyte territory volume.
(A) Strategy for sparse deletion of Hepacam from cortical astrocytes. (B) Successful deletion of hepaCAM (green) from td-Tomato+ (red) astrocytes in Hepacamf/f RTM mice at P21. Scale bar, 20 μm. (C and E) V1 L5 astrocytes from P7 (C) and P21 (E) Cre+ astrocytes (cyan) from Hepacam+/+ (PALE WT), Hepacamf/+ (PALE Het), and Hepacamf/f (PALE KO) mice. (D and F) Territory volume of td-Tomato+/Cre PALE astrocytes at P7 (D) and P21 (F). P7 n = 3 mice/genotype, 4-8 cells/mouse. P21 n = 6 WT, 7 Het, and 7 KO mice, 4-12 cells/mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested one-way ANOVA, Tukey’s post-test. See also Figure S4.
To determine the developmental and physiological consequences of deleting Hepacam from all astrocytes in the mouse cortex, we crossed Hepacamflox/flox;RTM mice with Aldh1L1-cre/ERT2 mice (Srinivasan et al., 2016) and administered tamoxifen at P2 and P3 (Figure 6A). This protocol efficiently and specifically targeted astrocytes in the mouse cortex (Figure 6B) with 95.6% ± 0.8% (mean ± s.e.m.) of astrocytes (16,511 cells counted from 15 animals), and only 0.56% ± 0.06% of neurons (92,555 cells counted from 15 animals), expressing td-Tomato. We confirmed successful deletion of hepaCAM via immunohistochemistry and Western blot of mouse cortical lysates at P21 (Figures 6B and S4C). HepaCAM protein expression was reduced by 86% in the cortex of Hepacam astrocyte conditional knockout mice (cKO: Hepacamf/f, RTMf/+, CreTg/0) compared to wild type (WT: Hepacam+/+, RTMf/+, CreTg/0), indicating that a majority of hepaCAM protein is expressed in astrocytes. HepaCAM protein expression was not decreased in conditional heterozygous mice (cHet: Hepacamf/+, RTMf/+, CreTg/0) (Figure S5C). At P21, we found no difference in the brain volume or cortical thickness between Hepacam WT, cHet, and cKO mice (Figures S5A and S5B). To count the number of different cell types in the cortex and determine the specificity of Cre-mediated recombination, we performed automated image segmentation based on nuclear marker labeling (Figures S5C and S6A-S6C). Hepacam deletion from astrocytes did not alter the number or density of neurons (NeuN+), astrocytes (Sox9+/Olig2−), Olig2+ cells, or interneurons in the visual cortex at P21(Figures S5D-S5F, S6D, and S6E).
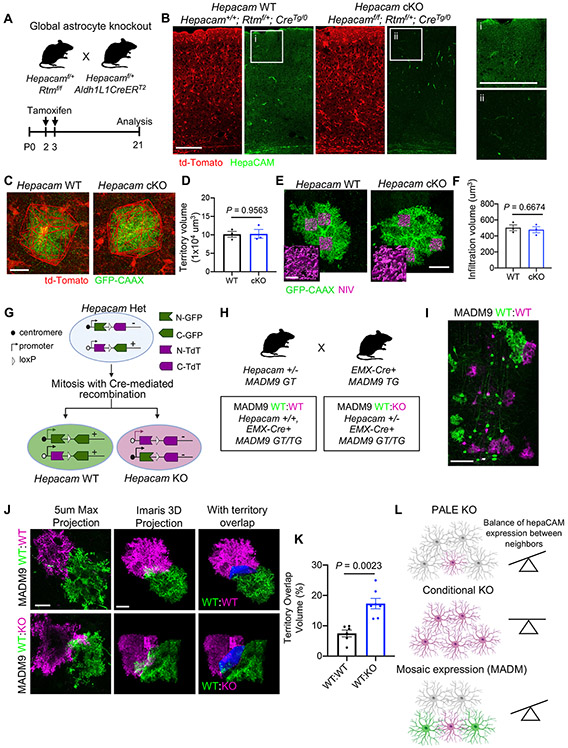
Figure 6: HepaCAM regulates astrocyte competition for territory.
(A) Strategy for conditional deletion of Hepacam from developing astrocytes. (B) Visual cortex of Hepacam WT (Hepacam+/+; Rtmf/+; Aldh1L1CreERT2/(Tg/0)) and Hepacam cKO (Hepacamf/f; Rtmf/+; Aldh1L1CreERT2/(Tg/0)) mice at P21. Minimal hepaCAM expression observed in Hepacam cKO mice (ii) compared to WT mice (i). Scale bar, 100 μm. (C) V1 L5 astrocytes from Hepacam WT and Hepacam cKO mice at P21 expressing GFP-CAAX. Cre+ astrocytes express td-Tomato (red). (D) Average territory volume. n = 4 mice per genotype, 5-8 cells per mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. (E) GFP-CAAX-expressing astrocytes (green) and NIV (magenta) from P21 Hepacam WT and Hepacam cKO mice. Scale bar, 20 μm (inset, 5 μm). (F) Average NIV. n = 4 mice per genotype, 5-8 cells per mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. (G) Strategy for combining Hepacam mutant mice with mosaic analysis with double markers (MADM) chromosome 9 mice. (H) Breeding scheme to generate MADM9 WT:WT and Hepacam MADM9 WT:KO mice. (I) Example image of mosaic labeling in the mouse cortex in MADM9 WT:WT mice with EMX-Cre. Scale bar, 100 μm. (J) Left: Max projection of 5 μm of a z-stack confocal image from MADM9 WT:WT (green and magenta cells both WT) and MADM9 WT:KO (green cells WT, magenta cells KO). Middle: Imaris 3D projection image of entire z-stack image. Right: region of territory overlap between magenta and green neighboring astrocytes (blue). Scale bar, 20 μm. (K) Quantification of the percentage of territory volume overlap between neighboring astrocytes. n = 6 WT:WT and 7 WT:KO mice, 1-2 astrocyte pairs per mouse. Data points are mouse averages. Bars are mean ± s.e.m. Nested t test. (L) Summary of deletion strategies and effect on astrocyte territory. See also Figure S5, S6, and S7.
Because sparse deletion of Hepacam decreased astrocyte territory volume, but we found no change in cell number or cortex size in Hepacam cKO mice, we hypothesized that hepaCAM might participate in a self-recognition mechanism that regulates astrocyte competition for territory. In this scenario, a difference in hepaCAM expression between neighboring astrocytes would result in altered astrocyte territory volume (Figures 5C-5F), but deletion of Hepacam from all astrocytes would leave territory volume unchanged. To test this possibility, we sparsely labeled astrocytes in the cortex of Hepacam WT and cKO mice with GFP-CAAX by injecting PHP.eB2 serotype Adeno-associated virus (AAV) expressing GFP-CAAX under control of the human minimal GFAP promoter (GfaABC1D) (Figures 6C-6F). At P21, we found no difference in the average territory volume between Hepacam WT and cKO astrocytes (Figures 6C-6D). Furthermore, deletion of Hepacam from astrocytes (Figures 6E-6F) or neurons (Figure S7A-S7D) did not alter astrocyte NIV (Figure S7C and S7D). Collectively, these findings suggest that astrocytes may compete for territory through a hepaCAM-based self-recognition mechanism that is dependent on expression of hepaCAM by neighboring astrocytes.
To test this hypothesis, we sparsely overexpressed hepaCAM protein in Hepacam WT, Het, or KO mice using PALE (Figure S7E). Despite successful hepaCAM overexpression (OE) (Figure S7F), astrocyte territory volume did not increase beyond WT levels (Figure S7F and S7G). The inability of hepaCAM-OE to increase territory volume in Hepacam KO mice could be due to a requirement for hepaCAM transcellular interactions and lack of hepaCAM in neighboring cells. However, this would not explain the inability of hepaCAM-OE to increase territory volume in Hepacam Het and WT mice. An alternative explanation could be a saturation effect, not allowing us to increase the surface hepaCAM levels in WT and Het mice. To overcome these potential limitations, we used mosaic analysis with double markers (MADM). MADM enables the generation of genetic mosaic mice in which a small fraction of genetically defined cells can be fluorescently labeled and at the same time the dosage of a candidate gene of interest may be modulated (e.g. GFP+ WT cells and Td-Tomato+ KO cells)(Contreras et al., 2020; Zong et al., 2005). We introduced the Hepacam mutation into the MADM paradigm to produce sparse populations of GFP+ Hepacam WT cells and Td-Tomato+ Hepacam KO cells (Figure 6G). As a control, we used MADM mice with two WT Hepacam alleles (Figure 6H and 6I). In genetically mosaic mice, we found that WT GFP+ and KO Td-tomato+ astrocytes displayed a significantly higher degree of territory overlap compared to two neighboring WT astrocytes (Figure 6J and 6K). Collectively, these results show that hepaCAM is required for the proper recognition and establishment of territories between neighboring astrocytes (Figure 6L).
Deletion of Hepacam from astrocytes disrupts Cx43 localization and gap junction coupling
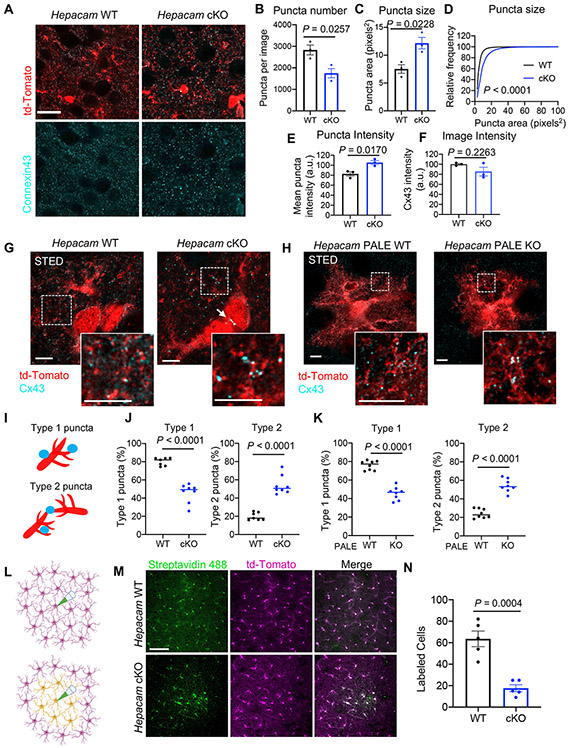
Because we found a functional link between hepaCAM and Cx43 in vitro (Figure 3), we next investigated Cx43 localization in Hepacam cKO mice. Hepacam cKO mice had reduced Cx43 puncta density (Figure 7A and 7B) and increased puncta size and intensity at P21 (Figure 7C-7E), though the overall intensity of Cx43 signal per image was unchanged (Figure 7F). Increased Cx43 puncta size was also observed at P60 (Figure S7H-S7J). Using STED microscopy, we found that many of the smaller and dimmer Cx43 puncta found in WT cortices were absent in Hepacam cKOs (Figure 7G). Moreover, a significantly higher percentage of Cx43 puncta in Hepacam cKO mice were localized to points of astrocyte-astrocyte contact (Figure 7I and 7J), including at points of soma-soma contacts (Figure 7G, arrow), something we never observed in WT mice. While this change in distribution of Cx43 to astrocyte-astrocyte contacts in Hepacam cKO mice is different than the change in sub-cellular localization that we observed in vitro (Figure S2A), this difference could be attributed to the distinctive properties of astrocytes cultured by themselves in the absence of neurons, and astrocytes grown in their native environment of the mouse brain. We observed similar changes to Cx43 localization in Hepacam PALE KO astrocytes (Figure 7H and 7K) indicating that hepaCAM controls Cx43 localization in a cell-autonomous manner in vivo. Depletion of Cx43 from individual astrocytes using PALE did not cause any obvious changes in the distribution or localization of hepaCAM (Figure S7K).
Figure 7: Deletion of Hepacam from astrocytes disrupts Cx43 localization and gap junction coupling.
(A) Confocal images of Cx43 (cyan) expression in Cre+ td-Tomato-expressing astrocytes (red) in L5 of the V1 cortex of Hepacam WT and cKO mice at P21. Scale bar, 20 μm. (B) Average number of Cx43 puncta per image and (C) Cx43 puncta size. Paired two-tailed t-test. (D) Cumulative probability distribution of Cx43 puncta size. Kolmogorov-Smirnov test. (E) Average intensity of individual Cx43 puncta and (F) average intensity of Cx43 signal per image. Paired two-tailed t test. (B-F) 5 images/section, 3 sections/brain, from 3 sex-matched littermate pairs. Data points are mouse averages. Bars are mean ± s.e.m. (G) STED microscopy images of Cx43 expression from Hepacam WT and cKO mice at P21. Scale bar, 5 μm. (H) STED images of Cx43 expression in sparsely transfected Hepacam PALE WT or PALE KO astrocytes from L5 of V1 at P21. Scale bar, 5 μm. (I) Schematic of classification of Cx43 puncta as Type 1 (on branches) or Type 2 (between branches). (J) Quantification of Cx43 puncta localization in Hepacam WT vs cKO mice and (K) PALE WT vs PALE KO astrocytes. 4 neuropil-containing ROI/image, 8 images/genotype from 3 sex-matched littermate pairs. Data are mean ± s.e.m. Unpaired two-tailed t test. (L) Overview of gap junction coupling assay. Neurobiotin was loaded into a single td-Tomato+ astrocyte (magenta) in acute cortical slices for 30 min. (M) Images of cortical slices stained with Streptavidin 488 (green) to detect neurobiotin-labeled cells. Scale bar, 100 μm. (N) Average number of labeled cells per slice. n = 5 slices from at least 3 mice per condition. Data are mean ± s.e.m. Unpaired, two-tailed t test. See also Figure S7.
To determine whether altered Cx43 localization and puncta size in Hepacam cKO mice corresponded to changes in astrocytic gap junction coupling, we injected neurobiotin into individual astrocytes in L5 visual cortex in acute slices from Hepacam WT and cKO mice (Figure 7I). Compared to WT mice, gap junction coupling between cortical astrocytes in Hepacam cKO mice was significantly impaired (Figures 7M and 7N). Based on the changes we observed in Cx43 localization, the decrease in neurobiotin-labeled cells in Hepacam cKO mice could be a result of increased coupling between an individual astrocyte’s own processes at the expense of coupling between neighboring astrocytes (Figure S7L). Alternatively, loss of hepaCAM could impair Cx43 channel function in addition to altering its localization. Collectively these results demonstrate that astrocytic hepaCAM regulates the sub-cellular localization of Cx43 and indicate that hepaCAM is required for proper astrocyte-astrocyte self-organization and gap junction coupling in the mouse cortex.
Astrocytic hepaCAM regulates inhibitory synapse formation and function
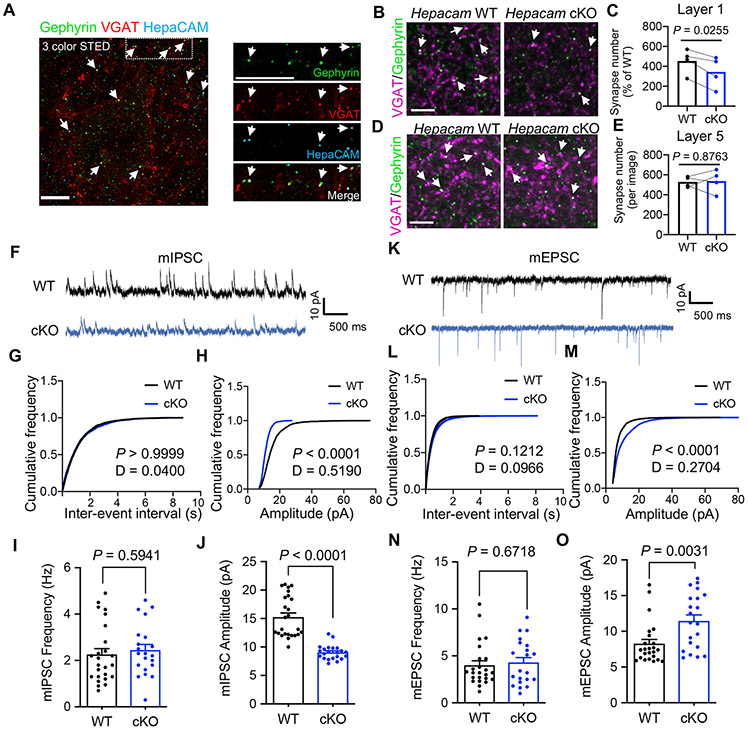
In addition to concentration at sites of astrocyte-astrocyte contacts (Figure 1B), we found hepaCAM puncta in close proximity to or co-localized with 49.01% ± 1.2% of inhibitory synapses in the mouse visual cortex (Figure 8A, arrows). In contrast, hepaCAM puncta were only co-localized with 21.3% ± 0.7% of excitatory intracortical and 5.6% ± 1.1% of excitatory thalamocortical synapses (Figures S8A and S8B). To determine whether hepaCAM in astrocytes is required for proper synaptogenesis in the developing brain, we quantified synapse density in the visual cortex of WT and Hepacam cKO mice at P21. In Hepacam cKO mice we found a marked decrease in the density of inhibitory synapses in Layer 1, but not in Layer 5 (Figures 8B-8E), a phenotype that persisted at P60 (Figure S8G and S8H). However, we found no difference in the density of excitatory intracortical or thalamocortical synapses (Figures S8C-S8F).
Figure 8: Astrocytic hepaCAM regulates inhibitory synapse formation and function.
(A) Three-color STED image of L5 V1 at P21 showing co-localization (arrows) of hepaCAM (cyan) with inhibitory pre- (VGAT, red) and post-synaptic (Gephryin, green) markers. Scale bar, 5 μm. (B and D) Images of inhibitory synapses (VGAT (magenta) and Gephyrin (green)) in L1 (B) and L5 (D) of V1 cortex. Scale bar, 5 μm. (C and E) Average number of inhibitory synapses per image. n = 5 images/section, 3 sections/mouse, 4 sex-matched littermate pairs. Data points represent mouse averages. Bars are mean ± s.e.m. Paired two-tailed t test. (F) mIPSC traces from L5 pyramidal neurons in acute V1 slices from Hepacam WT and cKO mice. (G and H) Representative cumulative distributions of mIPSC frequency (G) and amplitude (H) from Hepacam WT and cKO pyramidal neurons. Kolmogorov-Smirnov test (I and J) Average mIPSC frequency (I) and amplitude (J). n = 25 WT and 22 cKO neurons from 3 mice per genotype. Data are mean ± s.e.m. Unpaired, two-tailed t test. (K) mEPSC traces from L5 pyramidal neurons in acute V1 slices from hepaCAM WT and cKO mice. (L and M) Representative cumulative distributions of mEPSC frequency (L) and amplitude (M) from Hepacam WT and cKO pyramidal neurons. Kolmogorov-Smirnov test. (N and O) Average neuron mEPSC frequency (N) and amplitude (O). n = 24 WT and 21 cKO neurons from 3 mice per genotype. Data are mean ± s.e.m. Unpaired, two-tailed t test. See also Figure S8.
To determine whether astrocytic hepaCAM is required for proper synaptic function, we performed whole-cell patch-clamp recordings of miniature inhibitory and excitatory postsynaptic currents (mIPSCs and mEPSCs, respectively) in L5 pyramidal neurons of the visual cortex in acute brain slices from Hepacam WT and cKO mice at P21. We chose to record L5 pyramidal neurons because these cells possess large dendritic trees that project to all layers of the cortex, interacting with astrocytes in all of these layers (Harris and Shepherd, 2015). In neurons from Hepacam cKO mice, found a strong decrease in the amplitude of mIPSCs compared to WT, with no change in frequency (Figures 8F-8J). Conversely, mEPSCs was increased in Hepacam cKO mice compared to WT mice, with no change in the frequency (Figures 8K-8O). Together, these results show that hepaCAM in astrocytes plays a critical role in regulating the strength of both inhibitory and excitatory synapses in the mouse cortex. Furthermore, these results indicate that loss of hepaCAM from astrocytes disrupts the balance between synaptic excitation and inhibition.
Discussion
HepaCAM regulates astrocyte self-organization, coupling, and synaptic function
During brain development, astrocytes establish and balance numerous contacts with synapses, neighboring astrocytes, and other brain cells and structures. Here we show that hepaCAM is a key molecular player in this process, regulating astrocyte self-organization, morphogenesis, and gap junction coupling. To coordinate these processes, hepaCAM regulates the stability and subcellular localization of Cx43. While loss of Hepacam from astrocytes in vivo impairs gap junction coupling (Figure 7M and 7N), we find that the ability of Cx43 to regulate astrocyte morphogenesis in vitro and territory volume in vivo is independent of channel function (Figure 4B-4E). Thus, while changes to astrocyte competition for territory and gap junction coupling in Hepacam cKO mice both result from changes to Cx43 localization and/or function, these phenotypes can occur independently of one another. Untangling the molecular properties of the hepaCAM and Cx43 interactions in these distinct, yet related aspects of astrocyte development is an important topic for future study. Finally, we show that loss of Hepacam from astrocytes impairs both excitatory and inhibitory synaptic function, highlighting the causative role of astrocyte dysfunction in synaptic dysfunction.
How does hepaCAM regulate synaptic function?
HepaCAM was recently identified as a component of the inhibitory postsynaptic proteome in neurons (Uezu et al., 2016). Could astrocytic hepaCAM control inhibitory synapse formation and function via transcellular homophilic interactions with neuronal hepaCAM at inhibitory post synapses? Future studies are needed to test this possibility. Alternatively, synaptic changes that occur in Hepacam cKO mice could be due to altered Cx43 localization. Cx43 regulates the firing rate of orexin neurons in the lateral hypothalamic area (Clasadonte et al., 2017), and Cx43 and Cx30 both regulate excitatory synaptic activity in the hippocampus (Chever et al., 2014; Pannasch et al., 2014). However, deletion of Cx43 or Cx30 from hippocampal astrocytes causes a significant decrease in mEPSC amplitude and frequency (Chever et al., 2014; Pannasch et al., 2014), whereas deletion of Hepacam from cortical astrocytes increased mEPSC amplitude. While these differences in phenotype could suggest independent roles of hepaCAM and Cx43 at synapses, they could also be attributed to differences in brain region or developmental deletion strategies. A role for Cx43 at inhibitory synapses has not yet been described. Using STED microscopy, we found that Cx43 puncta were infrequently localized with inhibitory synapses (9.91% ± 0.5% of inhibitory synapses), and we observed no changes in this frequency in Hepacam cKO mice (8.24% ± 0.1%, P = 0.16, unpaired two-tailed t test). Therefore, the function of hepaCAM at inhibitory synapses may be independent of Cx43 and could instead be related to hepaCAM’s function as an auxiliary chloride channel subunit, or possibly a role for hepaCAM in localizing other transmembrane proteins to inhibitory synapses. Testing these possibilities will require further investigation into the function of hepaCAM at inhibitory synapses.
Distinct roles of different astrocytic cell adhesion molecules in astrocyte morphogenesis and synaptogenesis
While we have found that loss of different cell adhesion molecules, including hepaCAM and neuroligins, reduce astrocyte morphological complexity in vitro and in vivo, further mechanistic studies revealed that loss of these molecules have distinct effects on the development and function of astrocytes and synapses. For example, while deletion of NL2 from astrocytes decreases excitatory synapse number, deletion of Hepacam decreases inhibitory synapse number.
Where NL2 knockout decreases synaptic excitation and increases inhibition (Stogsdill et al., 2017), deletion of Hepacam does the reverse. In fact, the synaptic phenotypes of Hepacam cKO mice more reminiscent of our findings in NrCAM KO astrocytes, despite the fact that NrCAM KO astrocytes have increased morphological complexity (Takano et al., 2020). The stark contrast between the synaptic and morphometric phenotypes of these three mouse models highlights the complex role that astrocytes play in precisely regulating the formation and function of different types of synapses using distinct molecular players.
Role of hepaCAM dysfunction in the pathogenesis of neurological disorders
The synaptic and gap junction coupling phenotypes in Hepacam cKO mice have important implications for understanding the pathophysiology of neurological disorders, in particular MLC. The changes in synaptic strength in Hepacam cKO mice suggest enhanced excitatory synaptic activity, which could underly the epilepsy phenotypes observed in humans with MLC (Shao et al., 2019; Yalcinkaya et al., 2003). Changes in synapse formation and function could also drive cognitive decline, intellectual disability, and autism spectrum disorder observed in MLC patients (Lopez-Hernandez et al., 2011a). While we did not observe a change in brain volume at P21 in Hepacam cKO mice, MLC patients have increased brain volume, and studies in mouse models of MLC suggest that defects in brain water and ion homeostasis are responsible for macrocephaly, white matter swelling and vacuolization (Bugiani et al., 2017; Dubey et al., 2015). Additional studies are required to determine how the developmental defects we observed in Hepacam cKO mice contribute to the emergence of these characteristic white matter phenotypes. Furthermore, understanding whether disease-causing mutations in hepaCAM impair Cx43 function will be necessary to determine if targeting Cx43 is a useful therapeutic strategy to treat MLC. In conclusion, we have identified hepaCAM as a molecular link connecting astrocyte-astrocyte and astrocyte-synapse interaction, with direct implications for astrocyte dysfunction as a driving cause of neuropathology.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Katherine Baldwin (ktbaldwin@med.unc.edu).
Materials availability
The reagents generated in this study are available without restriction.
Data and code availability
The datasets generated during this study are available from the lead contact upon request. The code generated during this study is available here: https://github.com/ErogluLab/CellCounts
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All mice were used in accordance with the Institutional Animal Care and Use Committee (IACUC) and the Duke Division of Laboratory Animal Resources (DLAR) oversight (IACUC Protocol Numbers A173-14-07 and A147-17-06). All mice were housed under typical day/night conditions of 12-hours cycles. Aldh1L1-EGFP (RRID:MMRRC_011015-UCD) mice were obtained through MMRRC. Aldh1l1-Cre/ERT2 BAC transgenic (RRID:IMSR_JAX:029655), ROSA-td-Tomato Ai14 (RTM) (RRID:IMSR_JAX:007914), FLPo (RRID:IMSR_JAX:012930), CMV-Cre (RRID:IMSR_JAX:006054) and EMX1-Cre (RRID: IMSR_JAX: 005628) lines were obtained through Jackson Laboratory. Wild type CD1 mice used for PALE were purchased through Charles River Laboratories (RRFD:IMSR_CRL:022). Hepacam germline knockout mice used for co-immunoprecipitation experiments were described previously (Hoegg-Beiler et al., 2014). MADM9-GT and MADM9-TG mice were described previously (Contreras et al., 2020). Hepacam conditional knockout mice were generated in collaboration with the Duke Transgenic Mouse Facility using homologous recombination. Briefly, a bacterial artificial chromosome (BAC) with homology to the Hepacam genomic locus was generated to insert loxP sites before Exon 2 and after Exon 4. BACs were injected into mouse ES cells (G4 -129S6B6F1) and positive clones were selected for injection into pseudo-pregnant females. Chimeric offspring males were mated to C57BL/6J females (JAX #000664) and offspring with germline transmission of the conditional allele were identified via PCR amplification of genomic DNA using the following primers (WT band 443 bp; floxed band 532 bp): Forward 5’- TACCCAGCCAATAAGGAGTAGAC-3’ and reverse 5’- CATTGCCTGTCTTCTGCACTT-3’. Mice with successful germline transmission were mated with Flpo transgenic mice on C57BL/6J background to remove the Neo selection cassette. Successfully recombined offspring were confirmed by PCR and mated either to C57BL/6J, RTM, CMV-Cre, or Aldh1l1-CreERT2 BAC transgenic mice to generate mice breeding pairs for experiments. Genotyping primer sequences are listed in Table S1.
Mice were used for experiments at P21, or as specified in the text and figure legends. For all experiments, littermate pairs of the same sex were randomly assigned to experimental groups based on genotype. For all experiments, mice of both sexes were included in analysis, and we did not observe any influence or association of sex on the experimental outcomes. Criteria for inclusion, exclusion, and randomization is listed for each experiment in specific subsections of the Methods Details section.
Primary Cultures
Cortical Neuron Isolation and Culture
Purified (glia-free) rat cortical neurons were prepared as described previously (Stogsdill et al., 2017). Briefly, cortices from P1 rat pups of both sexes (Sprague Dawley, Charles River Laboratories, SD-001) were micro-dissected, digested in papain (~7.5 units/mL) at 33°C for 45 minutes, triturated in low and high ovomucoid solutions, resuspended in panning buffer (DPBS (Gibco 14287) supplemented with BSA and insulin) and passed through a 20 μm mesh filter (Elko Filtering 03-20/14). Filtered cells were incubated on negative panning dishes coated with Bandeiraea Simplicifolia Lectin 1 (x2), followed by goat anti-mouse IgG+IgM (H+L) (Jackson ImmunoResearch 115-005-044), and goat anti-rat IgG+IgM (H+L) (Jackson ImmunoResearch 112-005-044) antibodies, then incubated on positive panning dishes coated with mouse anti-L1 (ASCS4, Developmental Studies Hybridoma Bank, Univ. Iowa) to bind cortical neurons. Adherent cells were collected by forceful pipetting with a P1000 pipette. Isolated neurons were pelleted (11 min at 200 g) and resuspended in serum-free neuron growth media (NGM; Neurobasal, B27 supplement, 2 mM L-Glutamine, 100 U/mL Pen/Strep, 1 mM sodium pyruvate, 4.2 μg/mL Forskolin, 50 ng/mL BDNF, and 10 ng/mL CNTF). 70,000 neurons were plated onto 12 mm glass coverslips coated with 10 μg/mL poly-D-lysine (PDL, Sigma P6407) and 2 μg/mL laminin and incubated at 37°C in 10% CO2. On day in-vitro (DIV) 2, half of the media was replaced with NGM Plus (Neurobasal Plus, B27 Plus, 100 U/mL Pen/Strep, 1 mM sodium pyruvate, 4.2 μg/mL Forskolin, 50 ng/mL, BDNF, and 10 ng/mL CNTF) and AraC (10 μM) was added to stop the growth of proliferating contaminating cells. On DIV 3, all of the media was replaced with NGM Plus. In experiments involving lentivirus infection, 100 μL of supernatant containing lentivirus plus polybrene (1 μg/mL) was added to the AraC NGM mixture on DIV 2 and completely washed out on DIV 3 and replaced with NGM Plus containing 100 ng/mL BDNF. Neurons were fed on DIV 6 and DIV 9 by replacing half of the media with NGM Plus.
Cortical Astrocyte Isolation and Culture
Rat cortical astrocytes were prepared as described previously (Stogsdill et al., 2017). P1 rat cortices from both sexes were micro-dissected, papain digested, triturated in low and high ovomucoid solutions, and resuspended in astrocyte growth media (AGM: DMEM (Gibco 11960), 10% FBS, 10 μM, hydrocortisone, 100 U/mL Pen/Strep, 2 mM L-Glutamine, 5 μg/mL Insulin, 1 mM Na Pyruvate, 5 μg/mL N-Acetyl-L-cysteine). Between 15-20 million cells were plated on 75 mm2 flasks (non-ventilated cap) coated with poly-D-lysine and incubated at 37°C in 10% CO2. On DIV 3, removal of non-astrocyte cells was performed by forcefully shaking closed flasks by hand for 10-15 seconds until only an adherent monolayer of astrocytes remained. AraC was added to the media from DIV 5 to DIV 7 to eliminate contaminating fibroblasts. On DIV 7, astrocytes were trypsinized (0.05% Trypsin-EDTA) and plated into 12-well or 6-well dishes. On DIV 8, cultured rat astrocytes were transfected with shRNA and/or expression plasmids using Lipofectamine LTX with Plus Reagent (Thermo Scientific) per the manufacturer’s protocol. Briefly, 1 μg (12-well) or 2 μg (6-well) total DNA was diluted in Opti-MEM containing Plus Reagent, mixed with Opti-MEM containing LTX (1:2 DNA to LTX) and incubated for 30 minutes. The transfection solution was added to astrocyte cultures and incubated at 37°C for 3 hours. On DIV 10, astrocytes were trypsinized, resuspended in NGM plus, plated (20,000 cells per well) onto DIV 10 neurons, and co-cultured for 48 hours.
Cell Lines
HEK293T
HEK293T cells used to produce lentivirus and adeno-associated virus were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/mL Pen/Strep, 2 mM L-Glutamine, and 1 mM sodium pyruvate. Cell were incubated at 37°C in 5% CO2 and passaged every 2-3 days.
METHOD DETAILS
shRNA and cDNA Plasmids
shRNA Plasmids
pLKO.1 Puro plasmids containing shRNA (pLKO.1-shRNA) against mouse/rat hepaCAM (shHep_1: TRCN0000126619; GCCCAATTTAGAGGCCACTTA, and shHep_2: TRCN0000126622; CGAGACCGTATCCGGCTCTTT), mouse/rat Neuroligin 1 (TRCN0000032019; CCCAACACTATACCAGGGATT ), and mouse/rat Connexin 43 (TRCN0000068474; AACTTCTTGATTTCAATCTGC) were obtained from the RNAi Consortium (TRC) via Dharmacon. In experiments where only one hepaCAM shRNA was used, shHep or shHepacam refers to shHep_2. A scrambled shRNA sequence was generated (GATACGAGACTCCTAATTCGC) and cloned into the pLKO. 1 TRC cloning vector (Moffat et al., 2006) according to Addgene protocols (https://www.addgene.org/tools/protocols/plko/). To generate pLKO.1 shRNA plasmids that express EGFP (pLKO.1-shRNA-EGFP), CAG-EGFP was removed from pLenLox-shNL1-CAG-EGFP (Chih et al., 2006) and inserted between Kpn1 and Spel sites in pLKO.1 Puro, replacing the puromycin resistance gene. pLKO.1 shRNA mCherry plasmids were generated by replacing EGFP with mCherry between KpnI and NheI sites.
PiggyBac plasmids
pPBCAG-EGFP and pGLAST-PBase were a gift from Dr. Joseph Loturco (Chen and LoTurco, 2012). To generate pPBCAG-mCherry-CAAX, mCherry-CAAX was inserted between XmaI and NotI restrictions sites to replace EGFP. To insert the hU6 promoter and shRNA in pPBCAG-mCherry-CAAX, a DNA fragment containing hU6 and shRNA was amplified from pLKO.1-shRNA using Phusion High-Fidelity DNA Polymerase (NEB) with primers that introduced SpeI restriction sites (Forward Primer: GGACTAGTCAGGCCCGAAGGAATAGAAG; Reverse Primer: GGACTAGTGCCAAAGTGGATCTCTGCTG). PCR products were purified, digested with SpeI, and ligated into pPBCAG-mCherry-CAAX at the SpeI restriction site. An analytical digest with EcoRI followed by sequencing was used to confirm the orientation of the inserted DNA fragment.
AAV plasmids
pZac2.1-GfaABC1D-Lck-GCaMP6f was a gift from Dr. Baljit Khakh (Addgene plasmid #52924). GFP-CAAX was cloned into pZac2.1-GfaABC1D-Lck-GCaMP6f between XmaI and NotI restriction sites to generate pZac2.1-GfaABC1D-GFP-CAAX. mCherry-CAAX was cloned into pZac2.1-GfaABC1D-Lck-GCaMP6f between XmaI and NotI restriction site to generate pZac2.1-GfaABC1D-mCherry-CAAX.
HepaCAM plasmids
pcDNA3.1 expression plasmids encoding human hepaCAM with a C-terminal FLAG tag and various deletions or point mutations (ΔC, ΔN, G89S, and R92Q) were described previously (Capdevila-Nortes et al., 2015). The QuikChange Lightning Site Directed Mutagenesis Kit (Agilent) Site-directed mutagenesis was used to replace the FLAG tag with an HA tag for in vivo studies. Mutagenesis primers for hepaCAM WT, ΔN G89S, and R92Q are available in Table S2. HepaCAM-WT-HA was then cloned into pZac2.1-GfaABC1D-Lck-GCaMP6f using EcoRI and NotI.
Connexin 43 plasmids
pcDNA-Cx43-Myc (Addgene plasmid # 60693) and pcDNA-Cx43-YA/VD-Myc (Addgene plasmid # 60701) were gifts from Parmender Mehta (Johnson et al., 2013). Site-directed mutagenesis was used to confer shRNA-resistance to these plasmids (Cx43_RM_Fwd: TGCACCTGAAGCAAATAGAGATTAAGAAGTTCAAGTACGGGATTG; Cx43_RM_Rev: CAATCCCGTACTTGAACTTCTTAATCTCTATTTGCTTCAGGTGCA). To generate the T154A mutation plasmid (Cx43-T154A), site-directed mutagenesis was performed on shRNA-resistant Cx43-Myc (T154A_Fwd: GGCTTGCTGAGAGCCTACATCATCAGCATCCT; T154A_Rev: TGATGATGTAGGCTCTCAGCAAGCCGCCCCTC). To generate a Cx43 construct with both YA/VD and T154A mutations, the same T154A mutagenesis primers were used to perform mutagenesis on shRNA resistant pcDNA-Cx43-YA/VD-Myc. All mutations were confirmed by sequencing. To generate pZac2.1-GfaABC1D-Cx43-TY-Myc for in vivo use, pcDNA-Cx43-TY-Myc was digested with EcoRI and XhoI and inserted in pZac2.1-GfaABC1D at EcoRI and SalI sites.
Lentivirus Production and Transduction
Lentiviruses containing shRNA targeting vectors were produced to test knockdown efficiency of shRNA constructs in cultured primary astrocytes or to bulk transduce neurons with shRNA and GFP. To produce lentivirus, HEK293T cells were transfected with a pLKO.1 shRNA Puro targeting plasmid (for astrocyte transduction) or a pLKO.1 shRNA GFP plasmid (for neuron transduction), an envelope plasmid (VSVG) and a packaging plasmid (dR8.91) using X-tremeGENE (Roche). One day after transfection, the media was replaced with AGM (for astrocyte transduction) or NGM lacking B27 (for neuron transduction), and media containing lentivirus was collected on days 2 and 3 post-transfection. To test the knockdown efficiency of shRNAs in astrocytes, rat or mouse primary astrocytes at DIV 7 were plated in 6-well dishes in 2 mL of AGM. On DIV 8, 1 mL of AGM was removed, and 500 μL of fresh AGM was added along with 500 μL of lentivirus-containing media and 1 μg/mL polybrene. Cultures were treated with puromycin (1 μg/mL) from DIV 10-15 to select for transduced cells. Cells were lysed at DIV 15 for protein extraction and Western blot analysis.
Protein Extraction and Western Blotting
Protein was extracted from cultured rat and mouse astrocytes using membrane solubilization buffer (MSB: 25 mM Tris pH 7.4, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 0.5% NP-40, and protease inhibitors). Cultured astrocytes were washed twice with ice-cold TBS containing 1 mM CaCl2 and 1 mM MgCl2 and incubated on ice in membrane solubilization buffer for 20 minutes with occasional agitation. Cell lysates were collected, vortexed briefly, and centrifuged at 4°C at high speed for 10 minutes to pellet non-solubilized material. The supernatant was collected and stored at −80°C. To collect protein from CD1 WT mice at different developmental ages, mice P7 or younger were euthanized via rapid decapitation, and mice older than P7 were euthanized using chamber CO2 administration. Cortices were micro-dissected and flash frozen in liquid nitrogen. Frozen brains were powderized with liquid nitrogen and lysed directly with Tissue Protein Extraction Reagent (T-PER; Thermo) containing protease inhibitors (Roche), spun at max speed at 4°C for 15 minutes, and supernatant collected and stored at −80°C.
To collect protein from Hepacam WT, cHet, and cKO mice at P21, mice were anesthetized with 200 mg/kg tribromoethanol (avertin) and perfused with TBS/Heparin, cortices rapidly dissected, and flash frozen in liquid nitrogen. Brains were homogenized in 1 mL of Lysis Buffer R (150 mM NaCl, 50 mM Tris, 1 mM EDTA, pH 7.5 containing protease inhibitors (Roche) 1 mM Na3VO4, 20 mM NaF, and 10 mM beta-glycerophosphate) using a Potter-Elvehjem PTFE pestle and glass tube (Sigma). Brain homogenate was collected and combined with equal volume of modified RIPA buffer lacking SDS (M-RIPA: 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 2% NP40, 2% deoxycholate, pH 7.5 containing protease inhibitors (Roche) 1 mM Na3VO4, 20 mM NaF, and 10 mM beta-glycerophosphate) and incubated with rotation at 4°C for 20 minutes. Lysate was centrifuged at max speed at 4°C for 10 minutes, supernatant collected and stored at −80°C.
Pierce BSA Protein Assay Kit (Thermo Fisher) was used to determine protein concentration, and lysates were mixed with 2x Laemmli Sample Buffer (BioRad) containing 5% β-ME and incubated at 45°C for 45 minutes to denature proteins. Either 10 μg (cultured astrocyte lysates) or 50 μg (brain lysates) of protein was loaded into 4-15% gradient pre-cast gels (Bio-Rad) and run at 150 V for 1 hour. Proteins were transferred at 100 V to PVDF membrane (Millipore) for 1 hour, blocked in Odyssey Blocking Buffer (LI-COR) and incubated in primary antibody overnight at 4°C (see list below). The next day, membranes were washed with PBS, incubated in LI-COR secondary antibodies, washed in PBS, dried for >1 hour and imaged on an Odyssey Infrared Imaging system using Image Studio software. Protein expression was quantified using Image Studio Lite software.
Immunocytochemistry
Astrocyte-neuron co-cultures on glass coverslips were fixed on DIV 12 with warm 4% PFA for 7 minutes, washed 3 times with PBS, blocked in a blocking buffer containing 50% normal goat serum (NGS) and 0.4% Triton-X 100 for 30 minutes, and washed in PBS. Samples were then incubated overnight at 4°C in primary antibodies diluted in blocking buffer containing 10% NGS, washed with PBS, incubated in Alexa-Fluor conjugated secondary antibodies (Life Technologies) for 2 hours at room temperature, and washed again in PBS. Coverslips were mounted onto glass slides (VWR Scientific) with Vectashield mounting media containing DAPI (Vector Labs), sealed with nail polish, and imaged on an AxioImager M1 (Zeiss) fluorescence microscope. Images of healthy astrocytes with strong expression of fluorescent markers that did not overlap with other fluorescent astrocytes were acquired at 40x magnification in red, green, and/or DAPI channels using a CCD camera. Astrocyte morphological complexity was analyzed in FIJI using the Sholl analysis plugin (Ferreira et al., 2014) as described previously (Stogsdill et al., 2017). Sholl analysis curves were statistically compared using an analysis of co-variance (ANCOVA) with a Tukey’s post-hoc test. Astrocyte process length was measured in ImageJ using the NeuroAnatomy SNT plugin (Arshadi et al., 2020). For analysis of sub-cellular localization of Cx43 mutant constructs, single optical section confocal images were acquired using an Olympus FV 3000 microscope with a 60x objective. The individual acquiring the images was always blinded to the experimental condition. In all cases, a minimum of 3 independent experiments were performed, with each independent experiment consisting of a primary cell culture obtained from a different litter of mixed sex animals. The exact number independent experiments are indicated in the figure legend for each experiment. At least 20 cells were imaged per condition per experiment. Experiments in which the peak of the Sholl analysis curve for the control condition was greater than or equal to 20 were included in analysis. In cases where the peak of the control condition was less than 20, these cultures were deemed to be in suboptimal health and were excluded. Astrocytes that contained a single nucleus as revealed by DAPI stain, expressed fluorescent markers and fusion proteins as required by the experiment, and were not overlapping with other labeled astrocytes, were selected for imaging.
Postnatal Astrocyte Labeling by Electroporation (PALE)
Late P0/early P1 mice were sedated by hypothermia until anesthetized and 1 μL of plasmid DNA mixed with Fast Green Dye was injected into the lateral ventricle of one hemisphere using a pulled glass pipette (Drummond). For shRNA knockdown experiments in wild type CD1 mice, the 1 μL of DNA contained 1.2 ug of pGLAST-PBase and 0.8 ug of pPB-shRNA-mCherryCAAX. For Hepacam flox/RTM mice, the 1 μL of DNA contained 2 ug of pCAG-Cre. For PALE-mediated overexpression of Cx43-TY, 0.7 ug pGLAST-PBase, 0.5 ug pPB-shRNA-mCherryCAAX, and 0.8 ug pZac2.1-gfaABC1D-Cx43-Ty-myc were injected in a total volume of 1 μL. For hepaCAM overexpression, 1 ug of pZac2.1-gfaABC1D-hepaCAM-HA and 1 ug of pZac2.1-gfaABC1D-mCherry-CAAX were injected in a total volume of 1 μL. Following DNA injection, electrodes were oriented with the positive terminal above the visual cortex and the negative terminal below the chin, and 5 discrete 50 msec pulses of 100 V spaced 950 msec apart were applied. Pups were recovered on a heating pad, returned to their home cage, and monitored until collection at P7, P21, or P60. In the case of plasmid expression in CD1 mice, assignment to experimental groups was randomly determined for each litter. For Hepacam flox/RTM mice, assignment to experimental groups was based on genotype. The number of replicates for each experiment is indicated in the figure legends. All animals that appeared healthy at the time of collection were processed for data collection. All brains were examined for presence of electroporated cells. Due to the precise timing needed to label astrocytes shortly after birth and the sparse nature of this procedure, some brains did not have successful labeling of astrocytes. Brains with no labeled astrocytes were excluded from the study.
Tamoxifen administration
Tamoxifen (Sigma) was dissolved in corn oil (Sigma) at a concentration of 10 mg/mL and further diluted in corn oil to 1.25 mg/mL (for P2 injection) and 2.5 mg/mL (for P3 injection). At P2 and P3, 40 μL of the respective tamoxifen solution was injected into the milk spot using an insulin syringe, for a dose of 0.05 mg at P2 and 0.1 mg at P3.
Adeno-associated virus (AAV) production and administration
Purified AAV was produced as described previously (Uezu et al., 2016). Briefly, HEK293T cells were transfected with pAd-DELTA F6, serotype plasmid AAV PHP.eB, and AAV plasmid (pZac2.1-gfaABC1D-GFP-CAAX). Three days after transfection, cells were collected in 15 mM NaCl, 5 mM Tris-HCl, pH 8.5 and lysed with repeat freeze-thaw cycles followed by treatment with Benzonase (Novagen 70664) at 37°C for 30 minutes. Lysed cells were pelleted by centrifugation and the supernatant, containing AAV was applied to an Optiprep density gradient (Sigma D1556, 15%, 25%, 40% and 60%) and centrifuged at 67,000 rpm using a Beckman Ti-70 rotor for 1 hour. The AAV-enriched fraction was isolated from between 40% and 60% iodixanol solution and concentrated by repeated washes with sterile PBS in an Amicon Ultra-15 filtration unit (NMWL: 100 kDa, Millipore UFC910008) to a final volume of ~100 μL and aliquoted for storage at −80°C. For labeling of astrocytes, AAV containing pZac2.1-GfaABC1D-GFP-CAAX was injected intracortically into Hepacam WT and cKO mice at P2. For neuronal knockout of Hepacam combined with labeling of astrocytes, AAV containing pZac2.1-GfaABC1D-GFP-CAAX and AAV containing pZac2.1-Syn1-Cre were injected intracortically into Hepacam WT or Hepacam flox/flox mice at P2. Briefly, pups were anesthetized by hypothermia, 1 μL of concentrated AAV was injected bilaterally into the cortex (at a depth of 2 mm) using a Hamilton syringe, and pups were recovered on a heating pad and monitored until collection at P21.
Immunohistochemistry
Sample Preparation
Mice used for immunohistochemistry were anesthetized with 200 mg/kg tribromoethanol (avertin) and perfused with TBS/Heparin and 4% PFA. Brains were collected and post-fixed in 4% PFA overnight, cryoprotected in 30% sucrose, frozen in a solution containing 2 parts 30% sucrose and 1 part O.C.T. (TissueTek) and stored at −80°C. Floating coronal tissue sections of 20 μm, 40 μm or 100 μm thickness were collected and stored in a 1:1 mixture of TBS/glycerol at −20°C. For immunostaining, sections were washed in 1x TBS containing 0.2% Triton-X 100 (TBST), blocked in 10% NGS diluted in TBST, and incubated in primary antibody overnight (20 μm) or 2-3 nights (> 20 μm) at 4°C with shaking. Following primary incubation, sections were washed in TBST, incubated in Alexa-Fluor conjugated secondary antibodies diluted 1:200 (Life Technologies) for 2-3 hours at room temperature, washed with TBST, and mounted onto glass slides using a homemade mounting media (90% Glycerol, 20 mM Tris pH 8.0, 0.5% n-Propyl gallate) and sealed with nail polish. For primary antibodies produced in mouse, isotype specific secondary antibodies were always used (e.g. goat anti-mouse IgG1) to avoid excessive background staining. For DAPI staining, DAPI (1:50,000) was added to the secondary antibody solution for the final 10 minutes of incubation. Images were acquired on Olympus FV 3000 microscope or a Zeiss 780 (astrocyte territory experiments, only). For STED microscopy, samples were prepared as described above, but with secondary antibody concentrations at 1:100 and using STED-optimized fluorophores when possible (Oregon Green 488, Alexa Fluor 594, Atto647N). STED images were acquired on a Leica SP8 STED microscope using a white light excitation laser, 775 depletion laser (red and far red channels), and 560 depletion laser (green channel). STED images were deconvolved using Huygens Professional software. The researcher acquiring the images was blinded to the experimental group.
Astrocyte territory volume analysis
To assess the territory volume of individual astrocytes in the developing mouse cortex, 100 μm-thick floating sections containing V1 astrocytes labeled sparsely via PALE with mCherry-CAAX, td-Tomato, or GFP-CAAX were collected. High magnification images containing an entire astrocyte (50-60 μm Z-stack) were acquired on the Zeiss 780 upright microscope with the 63x objective. Criteria for data inclusion required that the entirety of the astrocyte could be captured within a single brain section, and that the astrocyte was located in layer 5 of the visual cortex. Astrocytes in which the entire astrocyte could not be captured within the section or were located in other layers or outside of the visual cortex were excluded from the study and not imaged. Imaged astrocytes were analyzed using Imaris Bitplane software as described previously (Stogsdill et al., 2017). Briefly, surface reconstructions were generated and the Imaris Xtensions “Visualize Surface Spots” and “Convex Hull” were used to create an additional surface render representing the territory of the astrocyte. The volume of each territory was recorded, and astrocyte territory sizes from biological replicates were analyzed across experimental conditions using a nested t-test or a nested one-way ANOVA followed by Tukey’s post-hoc test. The number of animals and cells/animal analyzed are specified in the figure legend for each experiment.
For analysis of territory overlap volume in MADM mice, brains from WT MADM or Hepacam Het MADM mice were collected. In total, the brains from 53 mice were collected and processed for this experiment, and the cortices were examined for the presence of red and green neighboring astrocytes. Due to the rare nature of this event, many brains did not contain red and green neighboring astrocytes. All instances of red and green neighboring astrocytes that occurred in the cortex were imaged and analyzed. To perform the analysis, convex hulls were generated for neighboring GFP+ and td-Tomato+ positive astrocytes. The convex hull for each territory was used to create a mask of the original fluorescent channel (e.g. convex hull for GFP+ cell was used to create a mask for the GFP channel). The co-loc tool in Imaris was then used to generate a co-localization channel representing the area of overlap of the masked GFP and Td-tomato channels. Finally, a convex hull was generated for the co-localization surface reconstruction, and the volume of this convex hull was recorded as the territory overlap volume. Territory overlap volume measurements were normalized to the territory volume of WT (green) astrocyte to control for variations in the percentage of each astrocytes captured in the image.
Neuropil infiltration volume analysis
To measure the extent of astrocyte infiltration into the neuropil, 60x high magnification Z-stack images with 2x optical zoom were acquired from mCherry-CAAX or GFP-CAAX labeled astrocytes in 40 μm brain sections. Criteria for inclusion required the astrocyte to be located within layer 5 of the visual cortex, express the fluorescent label, and have at least 15μm in the z-dimension contained within the section. Astrocytes that did not meet these criteria were excluded from the study and not imaged. For each astrocyte, three ROIs (12.65 μm x 12.65 μm x 10 μm) containing the neuropil, and devoid of cell soma, large branches, and end feet were chosen and reconstructed using the surface tool in Imaris. The surface volume of each ROI was calculated and the three data points from each astrocyte were averaged. Data from biological replicates were analyzed using a nested t test or nested one-way ANOVA. The number of animals and cells/animal analyzed are specified in the figure legend for each experiment.
Cell counting imaging and analysis
For the cell counting assays described in Extended Data Figures 5 and 6, tile scan images containing the visual cortex from P21 Hepacam WT, cHet, and cKO mice were rapidly acquired on an Olympus FV 3000 using the resonant scanner and 20x objective. Stitched images were digitally restored to improve resolution using a trained content-aware image restoration (CARE) network (Weigert et al., 2018). For each brain section, an ROI of equal size (700 pixels2; 870.08 μm2) spanning L1 through L6 of the visual cortex was selected for analysis of cell number. For each nuclear marker, labeled nuclei were identified using a machine-learning based method for imaged segmentation (U-Net) (Falk et al., 2019). Full source code for this method is available here: https://github.com/ErogluLab/CellCounts. Following segmentation, images were inspected for accuracy, and any missed cells were segmented manually. Co-localization of two nuclear markers was determined in ImageJ by overlaying individually segmented channels and creating a mask of the overlapping regions. For Sox9 and Olig2 cell counts, 3 sections per brain from 5 sex-matched littermate groups were analyzed. For NeuN cell counts, 3 sections per brain from 5 sex-matched littermate groups were analyzed. Differences in cell number between genotype were analyzed using one-way ANOVA. For the cell counting assays described in Extended Data Figure 7, tile scan images containing the visual cortex from P21 Aldh1l1-eGFP mice were acquired on the Olympus FV 3000 using the Galvano scanner and a 30x objective. The number of cells labeled with GFP and/or nuclear markers was quantified by hand using the cell counter tool in ImageJ. 3 sections per brain from 3 independent animals were analyzed. All mice that appeared healthy at the time of collection (P21) were included in this study. No data was excluded.
Synapse imaging and analysis
Staining, image acquisition, and analysis of synaptic density were performed as described previously (Risher et al., 2018; Stogsdill et al., 2017) with adjustments. Synaptic staining was performed in coronal sections (20 μm thick) containing the visual cortex from P21 Hepacam WT and cKO mice. To label pre and postsynaptic proteins, the following antibody combinations were used: VGluT1 and PSD95 (excitatory, intracortical), VGluT2 and PSD95 (excitatory, thalamocortical), and VGAT and Gephyrin (inhibitory). High magnification 60x objective plus 1.64x optical zoom Z-stack images containing 15 optical sections spaced 0.34 μm apart were obtained using the Olympus FV 3000 inverted confocal microscope. Each Z-stack was converted into 5 maximum projection images (MPI) by condensing three consecutive optical sections using ImageJ. Synapses were identified by the colocalization of pre and postsynaptic puncta. The number of co-localized synaptic puncta of excitatory intracortical (VGluT1/PSD95), excitatory thalamocortical (VGluT2/PSD95), and inhibitory (VGAT/Gephyrin) synapses were obtained using the ImageJ plugin Puncta Analyzer(Ippolito and Eroglu, 2010). For all analyses 15 MPIs were analyzed per mouse (5 MPI/tissue section, 3 tissue sections/mouse). Between 4 and 5 sex-matched littermate pairs were analyzed for each synaptic staining combination, as indicated in the figure legends. All mice that appeared healthy at the time of collection (P21) were included in this study. No data was excluded.
Connexin 43 puncta analysis
Images for confocal analysis of Connexin 43 puncta were collected using the parameters described above for synaptic imaging. Co-localization of Cx43 and hepaCAM puncta was analyzed using the ImageJ plugin Puncta Analyzer as described above. Number for Cx43 puncta number, size, and intensity were also extracted using the Puncta Analyzer. Average intensity of Cx43 puncta per image was analyzed in ImageJ by obtaining measurements for integrated density. For all analyses 15 MPIs were analyzed per mouse (5 MPI/tissue section, 3 tissue sections/mouse) from 3 sex-matched littermate pairs. To quantify changes in Cx43 subcellular localization between Hepacam WT and cKO mice, STED microscopy images of Cx43 and td-Tomato were acquired with 93x objective using 2x zoom at a resolution of 3504 x 3504 pixels, for 6 optical sections spaced 0.18 μm apart. Images were deconvolved using Huygens Professional software and maximum projection images representing 1.0 μm in the z-direction were generated for analysis. Cx43 puncta localization was manually characterized as Type 1 or Type 2 (see Figure 7I) in reference to td-Tomato signal in 4 ROIs (8 μm x 8 μm) per image. A total of 8 cells from at least 3 mice were collected for each experimental condition, as described in the figure legend.
HepaCAM co-immunoprecipitation from mouse brain
Brain membranes from wildtype (WT) and Hepacam knockout (KO) mice were solubilized with ComplexioLyte 47 as described previously(Lopez-Hernandez et al., 2011a) and subject to co-immunoprecipitation with two different antibodies against hepaCAM, a mouse monoclonal (Ab1) and a rabbit polyclonal (Ab2) (Lopez-Hernandez et al., 2011a), and a non-specific control IgG. Mass spectrometric analysis was carried out as described previously (Schwenk et al., 2014). Peptides obtained from tryptic in-gel digests were dissolved in 0.5% trifluoroacetic acid and loaded onto a C18 PepMap100 precolumn (particle size 5 mm; Dionex/Thermo Scientific, Germany) with 0.5% (v/v) acetic acid using an UltiMate 3000 HPLC (Dionex/Thermo Scientific, Germany). Bound peptides were eluted and separated with an aqueous-organic gradient (from 0.5% (v/v) acetic acid to 0.5% (v/v) acetic acid in 80% (v/v) acetonitrile, 80 min total) in a PicoTip emitter (i.d. 75 mm; tip 8 mm; New Objective, USA) manually packed with ReproSilPur 120 ODS-3 (C18; particle size 3 mm; Dr. Maisch HPLC, Germany) and electrosprayed (2.3 kV; transfer capillary temperature 250°C) into an LTQ Orbitrap XL tandem mass spectrometer with the described settings.
Peak lists were extracted from fragment ion spectra using the “msconvert.exe” tool (ProteoWizard; http://proteowizard.sourceforge.net/; v3.0.6906; Mascot generic format with filter options “peakPicking true 1-” and “threshold count 500 most-intense”) and the precursor m/z values were shifted by the median m/z offset of all peptides assigned to proteins in a preliminary database search with 15 ppm. Corrected peak lists were searched with Mascot 2.6 (Matrix Science, UK) against the UniProtKB/Swiss-Prot database release 2019-1, mouse, rat, human). Acetyl (Protein N-term), Carbamidomethyl (C), Gln->pyro-Glu (N-term Q), Glu->pyro-Glu (N-term E), Oxidation (M), Phospho (ST), Phospho (Y), Propionamide (C) and up to one missed tryptic cleavage were allowed as variable modifications. Peptide and fragment mass tolerance were set to ± 5 ppm and ± 0.8 Da, respectively. The expected value cut-off for peptide assignment was set to 0.5. Proteins either representing exogenous contaminations (e.g., keratins, trypsin, IgG chains) or identified by only one specific peptide were not considered.
Label-free quantification of proteins was carried out as described previously(Schmidt et al., 2017). Peptide signal intensities (peak volumes, PVs) from MS full scans were determined and offline mass calibrated using MaxQuant v1.6.3.3 (http://www.maxquant.org). PV peak elution times in evaluated datasets were pairwise aligned using LOESS regression. PVs were then assigned to peptides based on their m/z and elution time obtained either directly from MS/MS-based identification or inferred indirectly using in-house developed software (matching tolerances of ± 2 ppm and ± 1 min). Molecular abundances of proteins were estimated using the abundancenormspec score defined as the sum of all assigned and protein isoform-specific PVs divided by the number of MS-accessible protein isoform-specific amino acids (Bildl et al., 2012).
Gap junction coupling assay
Four to six-week old Hepacams WT and Hepacam cKO mice were anesthetized with 200 mg/kg tribromoethanol (avertin), decapitated, their brains dissected and placed in ice-cold cutting solution containing (in mM): 222 sucrose, 2.6 KCl, 27 NaHCO3, 1.5 NaH2PO4, 2 CaCl2, and 2 MgSO4, bubbled with 95% O2:5% CO2 (pH 7.4). Sagittal brain slices (350 μm) containing the visual cortex were cut using a vibratome (Leica VT100S) filled with ice-cold cutting solution. After sectioning, slices were recovered for 45 minutes at 33°C in oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM): 123 NaCl, 3 KCl, 26 NaHCO3, 1 NaH2PO4, 10 glucose, 2 CaCl2, and 2 MgSO4, bubbled with 95% O2:5% CO2 (pH 7.4). After recovery, slices were placed in a recording chamber under an upright microscope (Olympus BX61W1, 40x water-dipping objective with 0.5x C-mount adaptor) and continuously perfused with oxygenated ACSF at 2.5 mL/min. All experiments were carried out at room temperature (20–22°C). Astrocytes in the primary visual cortex were identified initially on morphological criteria under epifluorescence for RFP reactivity in transgenic mice and patched onto under infrared-differential interference contrast microscopy.
Pipettes (from borosilicate capillaries, World Precision Instruments TW 150F-4) had resistance of 5-8 MΩ when filled with an internal solution containing the following (in mM): 123 K-methylsulfate, 10 KCl, 10 HEPES, 5 NaCl, 2.5 Mg-ATP, 0.3 Na-GTP (pH 7.4, 290 mOsm). Neurobiotin (2 mg/mL; Vector Laboratories, SP-1120) was also added fresh to the internal solution for post-hoc immunohistochemical staining and evaluation of gap junction coupling. Patch-clamp recordings were performed with an EPC 10 USB (HEKA Elektronik). Data was filtered at 3 kHz and sampled at 10 kHz with PatchMaster V2x90.4. Electrical membrane properties of astrocytes were measured in voltage-clamp mode by applying a series of voltage pulses from −80 mV to + 80 mV (40 ms, 10 mV increments) from a holding potential of −80 mV. A linear current-voltage (I-V) relationship was observed based on real-time analysis on the PatchMaster software for each patched cell. There was also a lack of action potential firing in response to depolarizing current steps (current-clamp mode, 600 ms, 10 pA increments). After the electrical properties of the patch cell were verified to show typical astrocyte membrane properties, neurobiotin was allowed to diffuse passively for 30 minutes in current-clamp mode, after which the slice was immediately fixed in cold 4% PFA overnight. Slices were then washed with PBS containing 0.3% Triton-X 100 (PBST) and incubated in Streptavidin 488 at 1:500 (Life Technologies) for 2 nights at 4°C followed by washing in PBST and mounting in homemade mounting media (described above). Z-stack images spanning at least 60 μm in thickness were acquired on the Olympus FV 3000 using a 10x objective with 2x zoom for quantification. A second image was then taken with a 30x objective for a higher quality figure image. Quantification of neurobiotin-labeled cells was performed in ImageJ using the Cell Counting tool. Slices in which the patched cell remained healthy for the duration of the 30 minutes in current-clamp mode were included in this study. Unhealthy slices, or slices in which the cell died before the end of the 30 minute time period were excluded. Imaging and analysis were performed on slices where the patched cell could be visually identified by fluorescent labeling. Slices in which no fluorescence was overserved were excluded from the study.
Electrophysiology
P21 Hepacam WT and Hepacam cKO mice of both sexes were anesthetized with urethane (1.5-2.0 g/kg, i.p.) and decapitated. Brains were transferred to ice-cold modified artificial cerebrospinal fluid (aCSF) containing (in mM): 240 Sucrose, 25 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2 and 3.5 MgCl2, equilibrated with 95% O2:5% CO2. (pH 7.4). Brain slices (300 μm thick, sagittal) containing the V1 visual cortex were cut in ice-cold aCSF using a VT1200S vibratome. Slices were incubated for 1 hour at RT in aCSF containing (in mM): NaCl 126, KCl 3, MgCl2 1.3, CaCl2 2.5, NaHCO3 26, NH2PO4 1.25 and glucose 11. Slices were perfused with aCSF (3-4 mL/min) and lamina V neurons were recorded under voltage-clamp mode. Glass pipette electrodes (4-6 MΩ resistance) were pulled from borosilicate capillaries (World Precision Instruments, Inc.) using a P-97 Flaming/Brown micropipette puller (Sutter Instrument Co.). Miniature EPSCs (mEPSCs) were recorded at −70 mV holding potential in the presence of 1 μM tetrodotoxin (TTX) and 100 μM picrotoxin. Pipettes were filled with an internal solution (in mM): 135 K-gluconate, 5 KCl, 0.5 CaCl2, 2 MgCl2, 5 EGTA, 5 HEPES, 5 Mg-ATP (adjusted to pH 7.3). For mIPSCs recordings, pipettes were filled with an internal solution (in mM): Cs2SO4 110, CaCl2 0.5, MgCl2 2, EGTA 5, HEPES 5, Mg-ATP 5, tetraethylammonium (TEA)-Cl 5 (adjusted to pH 7.3). The mIPSCs were recorded at 0 mV holding potential, and 1 μM tetrodotoxin, 100 μM AP5 and 10 μM CNQX were added to the perfusion solution. For all recordings, series resistance was closely monitored, and whole-cell recordings were conducted with a pipette access resistance of less than 20 MΩ, and the recording data were included for those cells that showed less than 20% change in resistance during data acquisition. Synaptic currents were recorded with an Axopatch 700B amplifier, filtered at 3 kHz, digitized at 5 kHz. The data were stored with a personal computer using pCLAMP 10.3 software. mIPSCs or mEPSCs events were detected and analyzed using Mini Analysis Program ver. 6.0.3. A minimum of 6 cells was recorded from each brain, from three pairs of mice. For each brain slice, td-Tomato positive cells were viewed to ensure similar levels of Cre+ cells. The individuals performing the cellular recordings were always blinded to the genotypes of the mice.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed in GraphPAD Prism 8 or 9 with the exception of ANCOVA analysis for in vitro co-culture experiments which was performed using XLSTAT (Addinsoft). Exact value of n, what n represents, and specific statistical tests for each experiment are indicated in the figure legend for each experiment. All data are represented as mean ± standard error of the mean and individual data points are shown for all data, where applicable. Exact P-values are listed in the figure for each experiment. Sample sizes were determined based on previous experience for each experiment to yield high power to detect specific effects. No statistical methods were used to predetermine sample size. All experimental animals that appeared healthy at the time of tissue collection were processed for data collection. Specific details for inclusion, exclusion, and randomization are included in the specific subsections of the Method Details section.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Connexin 43 | Cell Signaling | Cat# 3512, RRID:AB_2294590 |
| FLAG (M2) | Sigma | Cat# F1804, RRID:AB_262044 |
| GAD-67 | Millipore | MAB5406, RRID:AB_2278725 |
| Gephyrin | Synaptic Systems | Cat# 147 002, RRID:AB_2619838 |
| GFAP | Sigma | Cat# G3893, RRID:AB_477010 |
| GFP | Millipore | Cat# AB16901, RRID:AB_90890 |
| GFP | Aves Labs | GFP-1020, RRID:AB_10000240 |
| Hepacam | R&D Systems | Cat# MAB4108, RRID:AB_2117687 |
| Hepacam | Lopez-Hernandez et al., 2011 | N/A |
| HA | Roche | 11867423001, RRID:AB_390918 |
| L1 (ASCS4) | DSHB | Cat# ascs4, RRID:AB_528349 |
| Myc (4A6) | Millipore | Cat# 05-724, RRID:AB_309938 |
| NeuN (A60) | Millipore | Cat# MAB377, RRID:AB_2298772 |
| Olig2 (211F1.1) | Millipore | Cat# MABN50, RRID:AB_10807410 |
| PSD95 | Thermo Fisher | Cat# 51-6900, RRID:AB_2533914 |
| RFP (rabbit) | Rockland | Cat# 600-401-379, RRID:AB_2209751 |
| RFP (5F8) | Chromotek | Cat# 5f8-100, RRID:AB_2336064 |
| RFP (chicken) | Rockland | 600-901-379, RRID:AB_10704808 |
| Sox9 | Millipore | Cat# AB5535, RRID:AB_2239761 |
| Tubulin | LI-COR Biosciences | Cat# 926-42211, RRID:AB_1850029 |
| VGAT | Synaptic Systems | Cat# 131 004, RRID:AB_887873 |
| VGluT1 | Millipore | Cat# AB5905, RRID:AB_2301751 |
| VGluT2 | Synaptic Systems | Cat# 135 404, RRID:AB_887884 |
| Bacterial and Virus Strains | ||
| AAVPHP.eB-gfaABC1D-GFPCAAX | This paper | N/A |
| AAVPHP.eB-Syn1-Cre | This paper | N/A |
| One Shot STBL3 | Thermo Fisher | Cat# C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AraC | Sigma | Cat# C1768 |
| B27 | Gibco | Cat# 17504044 |
| B27 Plus | Gibco | Cat# A3582801 |
| BSA | Sigma | Cat# A4161 |
| BDNF | PeproTech | Cat# 450-02 |
| CNTF | PeproTech | Cat# 450-13 |
| DMEM | Gibco | Cat# 11960 |
| DPBS | Gibco | Cat# 14287 |
| Fetal Bovine Serum | Thermo Fisher | Cat# 10-437-028 |
| Forskolin | Sigma | Cat# F6886 |
| L-Glutamine | Gibco | Cat# 25030-081 |
| Hydrocortisone | Sigma | Cat# H0888-5G |
| Insulin | Sigma | Cat# I1882 |
| Lipofectamine LTX | Thermo Fisher | Cat# 15338100 |
| Neurobasal | Gibco | Cat# 21103049 |
| Neurobasal Plus | Gibco | Cat# A3582901 |
| Neurobiotin | Vector Laboratories | Cat# SP-1120-50 |
| Odyssey Blocking Buffer | LI-COR Biosciences | Cat# 927-40000 |
| Opti-MEM | Thermo Fisher | Cat# 11058021 |
| Optiprep | Sigma | Cat# D1556-250ML |
| Pen/Strep | Gibco | Cat# 15140 |
| Poly-D-Lysine | Sigma | Cat# P6407 |
| Sodium Pyruvate | Gibco | Cat# 11360-070 |
| Streptavidin 488 conjugate | Life Technologies | Cat# S11223 |
| Tamoxifen | Sigma | Cat# T5648 |
| Triton-X-100 | Roche | Cat# 11332481001 |
| X-tremeGENE | Roche | Cat# 06366236001 |
| Critical Commercial Assays | ||
| Endo-Free Maxi Prep Kit | Qiagen | Cat# 12362 |
| Quikchange Lightning Mutagenesis | Agilent | Cat# 210518 |
| Experimental Models: Cell Lines | ||
| HEK 293T | ATCC | CRL-11268 |
| Rat primary cortical neurons | This paper | N/A |
| Rat primary cortical astrocytes | This paper | N/A |
| Mouse primary cortical astrocytes | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Aldh1L1-EGFP | MMRRC | RRID:MMRRC_011015-UCD |
| Mouse: B6;FVB-Tg(Aldh1l1-cre/ERT2)1Khakh/J | Jackson | RRID:IMSR_JAX:029655 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson | RRID:IMSR_JAX:007914 |
| Mouse: B6.129S4-Gt(ROSA)26Sortm2(FLP*)Sor/J | Jackson | RRID:IMSR_JAX:012930 |
| Mouse: Hepacam flox | This paper | N/A |
| Mouse: Hepacam germline KO (for co-IP) | Hoegg-Beiler et al., 2014 | N/A |
| Mouse: Hepacam germline KO (for MADM) | This paper | N/A |
| Mouse: CD1 | Charles River | RRID:IMSR_CRL:022 |
| Mouse: EMX1-Cre | Jackson | RRID:IMSR_JAX:005628 |
| Mouse: MADM-9 GT | Contreras et al, 2020 | N/A |
| Mouse: MADM-9 TG | Contreras et al, 2020 | N/A |
| Mouse: B6.C-Tg(CMV-cre)1Cgn/J | Jackson | RRID:IMSR_JAX:006054 |
| Rat: Sprague-Dawley | Charles River | 001 |
| Oligonucleotides | ||
| Genotyping Primers | This paper | Table S1 |
| Mutagenesis Primers | This paper | Table S2 |
| Recombinant DNA | ||
| pLKO.1 Puro (TRC Cloning Vector) | Moffat et al., 2006 | RRID:Addgene_10878 |
| pLKO.1-shHepacam_1 | Dharmacon | TRCN0000126619 |
| pLKO.1-shHepacam_2 | Dharmacon | TRCN0000126622 |
| pLKO.1-shConnexin43 | Dharmacon | TRCN0000068474 |
| pLKO.1-shNL1 | Dharmacon | TRCN0000032019 |
| pLKO.1-shScramble | This paper | N/A |
| pLKO.1-shScramble-GFP | This paper | N/A |
| pLKO.1-shHepacam_1-GFP | This paper | N/A |
| pLKO.1-shHepacam_2-GFP | This paper | N/A |
| pLKO.1-shConnexin43-GFP | This paper | N/A |
| pLKO.1-shNL1-GFP | This paper | N/A |
| pLKO.1-shNL2-GFP | Stogsdill et al, 2017 | N/A |
| pPB-CAG-EGFP | Chen and LoTurco, 2012 | N/A |
| pGLAST-PBase | Chen and LoTurco, 2012 | N/A |
| pPB-hU6-shRNA-mCherry-CAAX | This paper | N/A |
| pcDNA-Hepacam-FLAG | Capdevila-Nortes et al., 2015 | N/A |
| pcDNA-HepacamΔC-FLAG | Capdevila-Nortes et al., 2015 | N/A |
| pcDNA-HepacamΔN-FLAG | Capdevila-Nortes et al., 2015 | N/A |
| pcDNA-Hepacam-G89S-FLAG | Capdevila-Nortes et al., 2015 | N/A |
| pcDNA-Hepacam-R92Q-FLAG | Capdevila-Nortes et al., 2015 | N/A |
| pcDNA-Hepacam-HA | This paper | N/A |
| pZac2.1-Hepacam-HA | This paper | N/A |
| pcDNA-Cx43-Myc | Johnson, et al., 2013 | RRID:Addgene_60693 |
| pcDNA-Cx43-YA/VD-Myc | Johnson, et al., 2013 | RRID:Addgene_60701 |
| pcDNA-Cx43-RM-Myc (shRNA-resistance mutant) | This paper | N/A |
| pcDNA-Cx43-RM-T154A-Myc | This paper | N/A |
| pcDNA-Cx43-RM-YA/VD-Myc | This paper | N/A |
| pcDNA-Cx43-RM-TY-Myc | This paper | N/A |
| pZac2.1-GfaABC1D-Cx43-TY-Myc | This paper | N/A |
| pZac2.1-GfaABC1D-Lck-GCaMP6f | Baljit Khakh Lab | RRID:Addgene_52924 |
| pMD2.G-VSVG | Moffat et al., 2006 | RRID:Addgene_12259 |
| pCMV-dR8.91 | Moffat et al., 2006 | N/A |
| pZac2.1-GfaABC1D-GFP-CAAX | This paper | N/A |
| pZac2.1-GfaABC1D-mCherry-CAAX | This paper | N/A |
| pZac2.1-Syn1-Cre | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPAD | https://www.graphpad.com/scientific-software/prism/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Puncta Analyzer | Ippolito and Eroglu, 2010 | N/A |
| Imaris 8.2.4 | BitPlane | https://imaris.oxinst.com/packages |
| TIBCO Statistica Software (Version 13.3) | TIBCO | www.tibco.com |
| pCLAMP 10.3 | Molecular Devices | www.moleculardevices.com |
| Mini Analysis Program ver. 6.0.3 | Synaptosoft | http://www.synaptosoft.com/MiniAnalysis/ |
| XLSTAT | Addinsoft | https://www.xlstat.com/en/ |
| Image Studio | LICOR | www.licor.com |
| Other | ||
| Digital restoration and image segmentation | This paper | https://github.com/ErogluLab/CellCounts |
Highlights.
HepaCAM regulates astrocyte competition for territory in the mouse cortex
Loss of astrocytic hepaCAM alters Connexin 43 localization and gap junction coupling
Connexin 43 regulates astrocyte morphology through channel-independent mechanisms
Loss of astrocytic hepaCAM decreases synaptic inhibition and increases excitation
Acknowledgments:
This work was supported by the National Institutes of Health (R01 DA047258 and R01 NS102237 C.E., F32 NS100392 K.T.B.) and Holland Trice Brain Research Award to (C.E.). K.T.B. was supported by postdoctoral fellowships from the Foerster-Bernstein Family and The Hartwell Foundation. Hippenmeyer lab was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovations programme (No. 725780 LinPro) to S.H. We thank the Duke Light Microscopy Core Facility, the Duke Transgenic Mouse Facility, Dr. U. Schulte for assistance with proteomic experiments, and Dr. D. Silver for critical review of the manuscript. Cartoon elements of figure panels were created with BioRender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Allen NJ, and Lyons DA (2018). Glia as architects of central nervous system formation and function. Science 362, 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshadi C, Eddison M, Günther U, Harrington KIS, and Ferreira TA (2020). SNT: A Unifying Toolbox for Quantification of Neuronal Anatomy. bioRxiv, 2020.2007.2013.179325 [DOI] [PubMed] [Google Scholar]
- Baldwin KT, and Eroglu C (2017). Molecular mechanisms of astrocyte-induced synaptogenesis. Curr Opin Neurobiol 45, 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beahm DL, Oshima A, Gaietta GM, Hand GM, Smock AE, Zucker SN, Toloue MM, Chandrasekhar A, Nicholson BJ, and Sosinsky GE (2006). Mutation of a conserved threonine in the third transmembrane helix of alpha- and beta-connexins creates a dominant-negative closed gap junction channel. J Biol Chem 281, 7994–8009. [DOI] [PubMed] [Google Scholar]
- Beardslee MA, Laing JG, Beyer EC, and Saffitz JE (1998). Rapid turnover of connexin43 in the adult rat heart. Circ Res 83, 629–635. [DOI] [PubMed] [Google Scholar]
- Bildl W, Haupt A, Muller CS, Biniossek ML, Thumfart JO, Huber B, Fakler B, and Schulte U (2012). Extending the dynamic range of label-free mass spectrometric quantification of affinity purifications. Mol Cell Proteomics 11, M111 007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Suarez E, Caldwell AL, and Allen NJ (2016). Role of astrocyte-synapse interactions in CNS disorders. J Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiani M, Dubey M, Breur M, Postma NL, Dekker MP, Ter Braak T, Boschert U, Abbink TEM, Mansvelder HD, Min R, et al. (2017). Megalencephalic leukoencephalopathy with cysts: the Glialcam-null mouse model. Ann Clin Transl Neurol 4, 450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, and Ellisman MH (2002). Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 22, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila-Nortes X, Jeworutzki E, Elorza-Vidal X, Barrallo-Gimeno A, Pusch M, and Estevez R (2015). Structural determinants of interaction, trafficking and function in the ClC-2/MLC1 subunit GlialCAM involved in leukodystrophy. J Physiol 593, 4165–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila-Nortes X, Lopez-Hernandez T, Apaja PM, Lopez de Heredia M, Sirisi S, Callejo G, Arnedo T, Nunes V, Lukacs GL, Gasull X, et al. (2013). Insights into MLC pathogenesis: GlialCAM is an MLC1 chaperone required for proper activation of volume-regulated anion currents. Hum Mol Genet 22, 4405–4416. [DOI] [PubMed] [Google Scholar]
- Chen F, and LoTurco J (2012). A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J Neurosci Methods 207, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Maher BJ, and LoTurco JJ (2014). piggyBac transposon-mediated cellular transgenesis in mammalian forebrain by in utero electroporation. Cold Spring Harb Protoc 2014, 741–749. [DOI] [PubMed] [Google Scholar]
- Chever O, Pannasch U, Ezan P, and Rouach N (2014). Astroglial connexin 43 sustains glutamatergic synaptic efficacy. Philos Trans R Soc Lond B Biol Sci 369, 20130596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, and Scheiffele P (2006). Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 51, 171–178. [DOI] [PubMed] [Google Scholar]
- Chung WS, Allen NJ, and Eroglu C (2015). Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol 7, a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasadonte J, Scemes E, Wang Z, Boison D, and Haydon PG (2017). Connexin 43-Mediated Astroglial Metabolic Networks Contribute to the Regulation of the Sleep-Wake Cycle. Neuron 95, 1365–1380 e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Davaatseren A, Amberg N, Hansen AH, Sonntag J, Andersen L, Bernthaler T, Heger A, Johnson R, Schwarz LA, et al. (2020). A Genome-wide Library of MADM Mice for Single-Cell Genetic Mosaic Analysis. bioRxiv, 2020.2006.2005.136192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler C, Dreher Z, and Stone J (1991). Contact spacing among astrocytes in the central nervous system: an hypothesis of their structural role. Glia 4, 484–494. [DOI] [PubMed] [Google Scholar]
- Dubey M, Bugiani M, Ridder MC, Postma NL, Brouwers E, Polder E, Jacobs JG, Baayen JC, Klooster J, Kamermans M, et al. (2015). Mice with megalencephalic leukoencephalopathy with cysts: a developmental angle. Ann Neurol 77, 114–131. [DOI] [PubMed] [Google Scholar]
- Elias LA, Wang DD, and Kriegstein AR (2007). Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448, 901–907. [DOI] [PubMed] [Google Scholar]
- Elorza-Vidal X, Xicoy-Espaulella E, Pla-Casillanis A, Alonso-Gardon M, Gaitan-Penas H, Engel-Pizcueta C, Fernandez-Recio J, and Estevez R (2020). Structural basis for the dominant or recessive character of GLIALCAM mutations found in leukodystrophies. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk T, Mai D, Bensch R, Cicek O, Abdulkadir A, Marrakchi Y, Bohm A, Deubner J, Jackel Z, Seiwald K, et al. (2019). U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 16, 67–70. [DOI] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjostrom PJ, and van Meyel DJ (2014). Neuronal morphometry directly from bitmap images. Nat Methods 11, 982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaume C, Koulakoff A, Roux L, Holcman D, and Rouach N (2010). Astroglial networks: a step further in neuroglial and gliovascular interactions. Nat Rev Neurosci 11, 87–99. [DOI] [PubMed] [Google Scholar]
- Harris KD, and Shepherd GM (2015). The neocortical circuit: themes and variations. Nat Neurosci 18, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg-Beiler MB, Sirisi S, Orozco IJ, Ferrer I, Hohensee S, Auberson M, Godde K, Vilches C, de Heredia ML, Nunes V, et al. (2014). Disrupting MLC1 and GlialCAM and ClC-2 interactions in leukodystrophy entails glial chloride channel dysfunction. Nat Commun 5, 3475. [DOI] [PubMed] [Google Scholar]
- Ippolito DM, and Eroglu C (2010). Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E, Lopez-Hernandez T, Capdevila-Nortes X, Sirisi S, Bengtsson L, Montolio M, Zifarelli G, Arnedo T, Muller CS, Schulte U, et al. (2012). GlialCAM, a protein defective in a leukodystrophy, serves as a ClC-2 Cl(−) channel auxiliary subunit. Neuron 73, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Mitra S, Katoch P, Kelsey LS, Johnson KR, and Mehta PP (2013). Phosphorylation on Ser-279 and Ser-282 of connexin43 regulates endocytosis and gap junction assembly in pancreatic cancer cells. Mol Biol Cell 24, 715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LH, Moh MC, Zhang T, and Shen S (2009). The immunoglobulin-like cell adhesion molecule hepaCAM induces differentiation of human glioblastoma U373-MG cells. J Cell Biochem 107, 1129–1138. [DOI] [PubMed] [Google Scholar]
- Li J, Khankan RR, Caneda C, Godoy MI, Haney MS, Krawczyk MC, Bassik MC, Sloan SA, and Zhang Y (2019). Astrocyte-to-astrocyte contact and a positive feedback loop of growth factor signaling regulate astrocyte maturation. Glia 67, 1571–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Hernandez T, Ridder MC, Montolio M, Capdevila-Nortes X, Polder E, Sirisi S, Duarri A, Schulte U, Fakler B, Nunes V, et al. (2011a). Mutant GlialCAM causes megalencephalic leukoencephalopathy with subcortical cysts, benign familial macrocephaly, and macrocephaly with retardation and autism. Am J Hum Genet 88, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Hernandez T, Sirisi S, Capdevila-Nortes X, Montolio M, Fernandez-Duenas V, Scheper GC, van der Knaap MS, Casquero P, Ciruela F, Ferrer I, et al. (2011b). Molecular mechanisms of MLC1 and GLIALCAM mutations in megalencephalic leukoencephalopathy with subcortical cysts. Hum Mol Genet 20, 3266–3277. [DOI] [PubMed] [Google Scholar]
- Mayorquin LC, Rodriguez AV, Sutachan JJ, and Albarracin SL (2018). Connexin-Mediated Functional and Metabolic Coupling Between Astrocytes and Neurons. Front Mol Neurosci 11, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Moh MC, Tian Q, Zhang T, Lee LH, and Shen S (2009). The immunoglobulin-like cell adhesion molecule hepaCAM modulates cell adhesion and motility through direct interaction with the actin cytoskeleton. J Cell Physiol 219, 382–391. [DOI] [PubMed] [Google Scholar]
- Moh MC, Zhang C, Luo C, Lee LH, and Shen S (2005). Structural and functional analyses of a novel ig-like cell adhesion molecule, hepaCAM, in the human breast carcinoma MCF7 cells. J Biol Chem 280, 27366–27374. [DOI] [PubMed] [Google Scholar]
- Myer DJ, Gurkoff GG, Lee SM, Hovda DA, and Sofroniew MV (2006). Essential protective roles of reactive astrocytes in traumatic brain injury. Brain 129, 2761–2772. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Tian GF, Han X, Peng W, Takano T, Ransom B, and Nedergaard M (2008). Loss of astrocytic domain organization in the epileptic brain. J Neurosci 28, 3264–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Wang X, Goldman S, and Nedergaard M (2006). Astrocytic complexity distinguishes the human brain. Trends Neurosci 29, 547–553. [DOI] [PubMed] [Google Scholar]
- Pannasch U, Freche D, Dallerac G, Ghezali G, Escartin C, Ezan P, Cohen-Salmon M, Benchenane K, Abudara V, Dufour A, et al. (2014). Connexin 30 sets synaptic strength by controlling astroglial synapse invasion. Nat Neurosci 17, 549–558. [DOI] [PubMed] [Google Scholar]
- Risher WC, Kim N, Koh S, Choi JE, Mitev P, Spence EF, Pilaz LJ, Wang D, Feng G, Silver DL, et al. (2018). Thrombospondin receptor alpha2delta-1 promotes synaptogenesis and spinogenesis via postsynaptic Rac1. J Cell Biol 217, 3747–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N, Kollewe A, Constantin CE, Henrich S, Ritzau-Jost A, Bildl W, Saalbach A, Hallermann S, Kulik A, Fakler B, et al. (2017). Neuroplastin and Basigin Are Essential Auxiliary Subunits of Plasma Membrane Ca(2+)-ATPases and Key Regulators of Ca(2+) Clearance. Neuron 96, 827–838 e829. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B, and Schulte U (2014). Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84, 41–54. [DOI] [PubMed] [Google Scholar]
- Shao LR, Habela CW, and Stafstrom CE (2019). Pediatric Epilepsy Mechanisms: Expanding the Paradigm of Excitation/Inhibition Imbalance. Children (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, and Barres BA (2014). Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr Opin Neurobiol 27, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, and Khakh BS (2016). New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR, and Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, and Freeman MR (2014). Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83, 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cornwell A, Li J, Peng S, Osorio MJ, Aalling N, Wang S, Benraiss A, Lou N, Goldman SA, et al. (2017). SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J Neurosci 37, 4493–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Wallace JT, Baldwin KT, Purkey AM, Uezu A, Courtland JL, Soderblom EJ, Shimogori T, Maness PF, Eroglu C, et al. (2020). Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, and Soderling SH (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Boor I, and Estevez R (2012). Megalencephalic leukoencephalopathy with subcortical cysts: chronic white matter oedema due to a defect in brain ion and water homoeostasis. Lancet Neurol 11, 973–985. [DOI] [PubMed] [Google Scholar]
- Weigert M, Schmidt U, Boothe T, Muller A, Dibrov A, Jain A, Wilhelm B, Schmidt D, Broaddus C, Culley S, et al. (2018). Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat Methods 15, 1090–1097. [DOI] [PubMed] [Google Scholar]
- Wu M, Moh MC, and Schwarz H (2016). HepaCAM associates with connexin 43 and enhances its localization in cellular junctions. Sci Rep 6, 36218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcinkaya C, Yuksel A, Comu S, Kilic G, Cokar O, and Dervent A (2003). Epilepsy in vacuolating megalencephalic leukoencephalopathy with subcortical cysts. Seizure 12, 388–396. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, and Luo L (2005). Mosaic analysis with double markers in mice. Cell 121, 479–492. [DOI] [PubMed] [Google Scholar]
- Zucker SN, Bancroft TA, Place DE, Des Soye B, Bagati A, and Berezney R (2013). A dominant negative Cx43 mutant differentially affects tumorigenic and invasive properties in human metastatic melanoma cells. J Cell Physiol 228, 853–859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available from the lead contact upon request. The code generated during this study is available here: https://github.com/ErogluLab/CellCounts