ABSTRACT
Enterococcus faecalis, an opportunistic pathogen that causes severe community-acquired and nosocomial infections, has been reported to resist phagocyte-mediated killing, which enables its long-term survival in the host. Metabolism, especially carbohydrate metabolism, plays a key role in the battle between pathogens and hosts. However, the function of carbohydrate metabolism in the long-term survival of E. faecalis in phagocytes has rarely been reported. In this study, we utilized transposon insertion sequencing (TIS) to investigate the function of carbohydrate metabolism during the survival of E. faecalis in RAW264.7 cells. The TIS results showed that the fitness of carbohydrate metabolism-related mutants, especially those associated with fructose and mannose metabolism, were significantly enhanced, suggesting that the attenuation of carbohydrate metabolism promotes the survival of E. faecalis in macrophages. The results of our investigation indicated that macrophages responded to carbohydrate metabolism of E. faecalis and polarized to M1 macrophages to increase nitric oxide (NO) production, leading to the enhancement of macrophage-mediated killing to E. faecalis. Meanwhile, E. faecalis automatically decreased carbohydrate metabolism to escape from the immune clearance of macrophages during intracellular survival. The shift of primary carbon resources for macrophages affected the ability to clear intracellular E. faecalis. In summary, the results of the present study demonstrated that carbohydrate metabolism affects the macrophage-mediated killing of E. faecalis.
IMPORTANCE E. faecalis has become a major pathogen leading to a variety of infections around the world. The metabolic interaction between E. faecalis and its host is important during infection but is rarely investigated. We used transposon insertion sequencing coupled with transcriptome sequencing to explore the metabolic interaction between E. faecalis and macrophages and uncovered that the shift of carbohydrate metabolism dramatically affected the inflammatory response of macrophages. In addition, E. faecalis attenuated carbohydrate metabolism to avoid the activation of the immune response of macrophages. This study provides new insights for the reason why E. faecalis is capable of long-term survival in macrophages and may facilitate the development of novel strategies to treat infectious diseases.
KEYWORDS: Enterococcus faecalis, transposon insertion sequencing, carbohydrate metabolism, macrophage-mediated killing
INTRODUCTION
Enterococcus faecalis is an opportunistic pathogen that primarily colonizes the oral cavity and intestinal tract (1). Due to the high growth rate and strong adaptability of this bacterium to various conditions, such as in vivo environments (2), extreme conditions (3), and the stress of various antibiotics (4), E. faecalis has become a major pathogen leading to a variety of community and nosocomial infections (1). According to clinical reports, E. faecalis is responsible for 7% to 15% of wound infections (5), 5% to 8% of bacteremia (6), 5% to 26% of endocarditis (2), and more than 80% of secondary root canal infections (7). Among the different severe infections caused by E. faecalis, bacteremia and endocarditis are the two most life-threatening infections with a morality rate of more than 17% (2). During infection, innate immunity is first activated to defend against pathogenic invaders (8). Leukocytes, especially macrophages, are recruited to infectious sites to phagocytose and eliminate pathogens (9), which is followed by the delivery of pathogen-associated molecular patterns (PAMPs) from pathogens to activate adaptive immunity to further clear the invading pathogens (9, 10). However, E. faecalis is capable of resisting the phagocyte-mediated killing, delaying the activation of adaptive immunity, promoting its survival inside host cells (11–13), and finally leading to chronic inflammation or systemic infection that is difficult to be cured by antibiotics (2). Therefore, elucidating the pathogenic mechanism by which E. faecalis resists phagocyte-mediated killing is the key to combating E. faecalis-associated infectious diseases.
Metabolic interactions between pathogens and hosts have recently been reported to occur during infection. The host enhances glycolysis for rapid ATP production to trigger an immune response and polarization of macrophages to M1 (14), increases arginine and metabolism for nitric oxide (NO) production to kill bacterial invaders (15), and limits iron acquisition by pathogens (16). On the other hand, pathogens coordinate their own metabolism to adapt to host conditions. For example, Mycobacterium tuberculosis not only decreases primary metabolism and enters a dormant status after successfully colonizing macrophages (17) but also utilizes the energy metabolism of host cells to increase its fitness in host cells (18–20). However, the metabolic interactions between E. faecalis and macrophages have yet to be investigated.
Transposon insertion sequencing (TIS) has been widely used to identify the genetic determinants of bacterial survival or growth under various conditions (21, 22). In the present study, we used TIS to identify the key metabolic genes or pathways affecting E. faecalis survival in macrophages. The results showed that a number of metabolism-related mutants, especially those involved in fructose and mannose metabolism, promoted E. faecalis survival in macrophages. Subsequently, the function of carbohydrate metabolism during E. faecalis survival in macrophages was validated and investigated.
RESULTS
E. faecalis resists macrophage-mediated killing.
E. faecalis has been reported to resist macrophage-mediated killing (11, 12). To better characterize macrophage-mediated killing of E. faecalis, the survival dynamics of the E. faecalis strains OG1RF and ATCC 33186 in RAW264.7 macrophages were monitored. Twenty-four hours postinfection (h.p.i.), the survival of Lactobacillus lactis and Escherichia coli in RAW264.7 cells dramatically decreased by 137-fold and 70-fold, respectively (see Fig. S1A in the supplemental material). In contrast, E. faecalis strains OG1RF and ATCC 33186 in RAW264.7 cells survived well, exhibiting a reduction of less than 2-fold (Fig. S1A). At 48 and 72 h.p.i., L. lactis and E. coli were nearly completely cleared, while E. faecalis strains OG1RF and ATCC 33186 maintained a high level of survival in RAW264.7 cells (Fig. S1A), suggesting that E. faecalis resisted macrophage-mediated killing to promote its survival. To observe the survival of bacteria in macrophages, transmission electron microscopy (TEM) was performed, and the results showed that bacteria located in phagosomes after internalization and the bacterial counts of the E. faecalis strains OG1RF and ATCC 33186 in infected cells were significantly higher than those of L. lactis and E. coli in infected cells after 24 h of infection (Fig. S1B), which was in keeping with the results shown in Fig. S1A.
E. faecalis resists macrophage-mediated killing. (A) RAW264.7 cells were infected with the E. faecalis strains OG1RF and 33186, Lactobacillus lactis, and E. coli DH5α at an MOI of 10. Cells were washed and further incubated with DMEM supplemented with 10% FBS containing vancomycin and gentamycin to kill extracellular bacteria 2 h.p.i. The number of viable bacteria within host cells was counted at 2, 24, 48, and 72 h postinfection. More than three replicates were performed. (B) Characterization of bacterium-containing phagosomes by TEM. RAW264.7 cells were infected with the above strains at an MOI of 10. After 24 h of infection, cells were fixed and processed for imaging by transmission electron microscopy. Representative images are shown. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A comprehensive analysis of metabolism-related genetic determinants of E. faecalis resistance to macrophage-mediated killing.
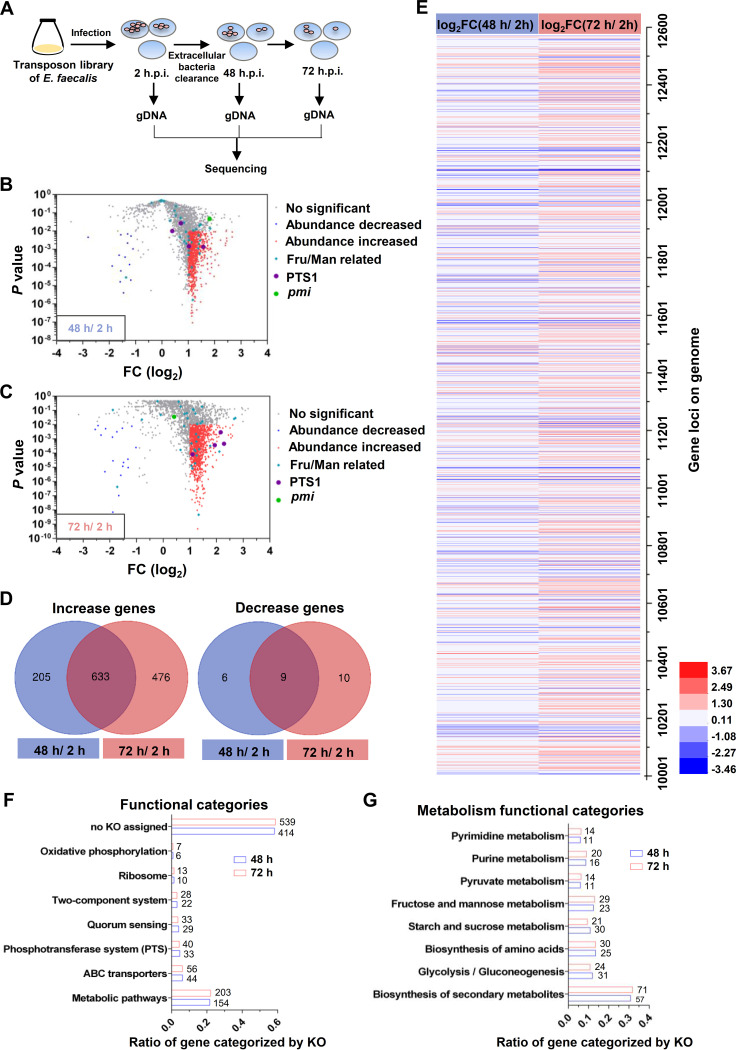
To comprehensively assess the metabolic determinants enabling E. faecalis to resist macrophage-mediated killing, transposon insertion sequencing (TIS) was performed using the transposon insertion mutant library of E. faecalis OG1RF. A schematic diagram of the TIS pipeline employed to identify E. faecalis genetic determinants of resistance to macrophage-mediated killing is shown in Fig. 1A. After RAW264.7 cells were colonized for 2, 48, and 72 h, the transposon insertion mutants were harvested and inoculated into brain heart infusion (BHI) medium for a short incubation period (approximately 2 h) to reduce interference from dead bacteria; this step was followed by genomic DNA (gDNA) extraction and next-generation sequencing (Fig. 1A). The replicates showed a high correlation (see Fig. S2A in the supplemental material). Analysis of the sequencing results by AL-Artemis (23) showed that the transposon mutant library contained 20,852 different mutants (see Table S1 in the supplemental material). Compared with internalized mutants (2 h.p.i.), 853 mutants exhibited a significant difference in abundance (log2 fold change [FC] of ≥1.0 or ≤−1.0; P < 0.01) after 48 h of survival in RAW264.7 cells, including 838 and 15 mutants with abundance increases and decreases, respectively (Fig. 1B and Table S1). After 72 h of survival in RAW264.7 cells, 1,128 mutants showed a significant difference in abundance (log2 fold change of ≥1.0 or ≤−1.0; P < 0.01), including 1,109 and 19 mutants with abundance increases and decreases, respectively (Fig. 1C and Table S1). For the mutants with significant differences, 9 and 633 mutants with abundance decreases or increases, respectively, were enriched in both the 48-h and 72-h groups (Fig. 1D). A heatmap was also generated to display the abundance change for the mutants in the library, and many mutants in the 48-h group showed a similar change as those in the 72-h group (Fig. 1E).
FIG 1.
Fitness screening for E. faecalis survival in macrophages by transposon insertion sequencing (TIS). (A) Schematic diagram of genetic determinant screening. Intracellular bacteria were harvested at 2, 48, and 72 h postinfection (h.p.i.), followed by DNA extraction for sequencing. (B and C) Scatterplots of the fold change (log2 output/input) and corresponding P values of mutants after survival within RAW264.7 cells for 48 h (B) or 72 h (C). The blue and red dots represent mutants with survival decreased (P < 0.01; log2 FC, ≤−1.0) and survival increased (P < 0.01; log2 FC, ≥1.0), respectively. The blue triangles represent fructose and mannose metabolism-related mutants. The purple and green dots represent PTS1 and pmi, respectively. Three or two independent biological replicates were performed. (D) Venn diagram depicting the genes with significant differences after 48 or 72 h of infection. (E) Heatmap of the genes with abundances decreased or increased relative to the input (2 h.p.i.). (F and G) KEGG orthology classification of genes with significant differences in fitness. The percentages shown are the fraction of the total number of depleted genes represented by the number of depleted genes/category. The number of depleted genes in each category is shown.
(A) Correlation of experimental replicates from the TIS results. The sequencing data from experimental replicates of input libraries (n = 3) and output libraries recovered after survival in RAW264.7 cells for 48 h (n = 2) and 72 h (n = 3) are shown. (B) Transcriptome profiles depicting the expression of fructose and mannose metabolism-associated genes at log and stationary phases in BHI medium. Three independent biological replicates were performed. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of transposon insertion mutant library screened for genetic determinates for E. faecalis survival in RAW264.7 cells. Download Table S1, XLS file, 0.8 MB (833.5KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Attenuation of carbohydrate metabolism promoting E. faecalis survival in macrophages.
The genes with transposon insertions resulting in significant changes in abundance in the 48-h and 72-h groups were further assessed by KEGG orthology (KO) analysis to determine their functions. A total of 154 and 203 genes in the 48-h and 72-h groups were identified as being involved in metabolic pathways (Fig. 1F), including 147 (95.4%) and 197 (97.0%) genes with abundance increases in the 2 groups, respectively. These results imply that the attenuation of metabolism may strongly contribute to E. faecalis survival in RAW264.7 cells. Furthermore, these metabolic pathways were primarily involved in pathways such as biosynthesis of secondary metabolites, glycolysis/gluconeogenesis, biosynthesis of amino acids, and fructose and mannose metabolism (23 and 29 genes in the 48- and 72-h groups, respectively) (Fig. 1G). Some genes in the fructose and mannose metabolism category also overlapped with genes in the phosphotransferase system (PTS) category, which also affected E. faecalis survival in RAW264.7 cells (Fig. 1F and G). According to KEGG analyses, multiple PTSs are carried by E. faecalis, hinting that PTS may play key roles for E. faecalis survival in vivo or/and in vitro. In addition, fructose and mannose metabolism have been reported to play important roles during infection (24–26). Therefore, we investigated fructose and mannose metabolism and their implication on the survival of E. faecalis in macrophages.
PTS1 and Pmi regulate carbohydrate metabolism in E. faecalis.
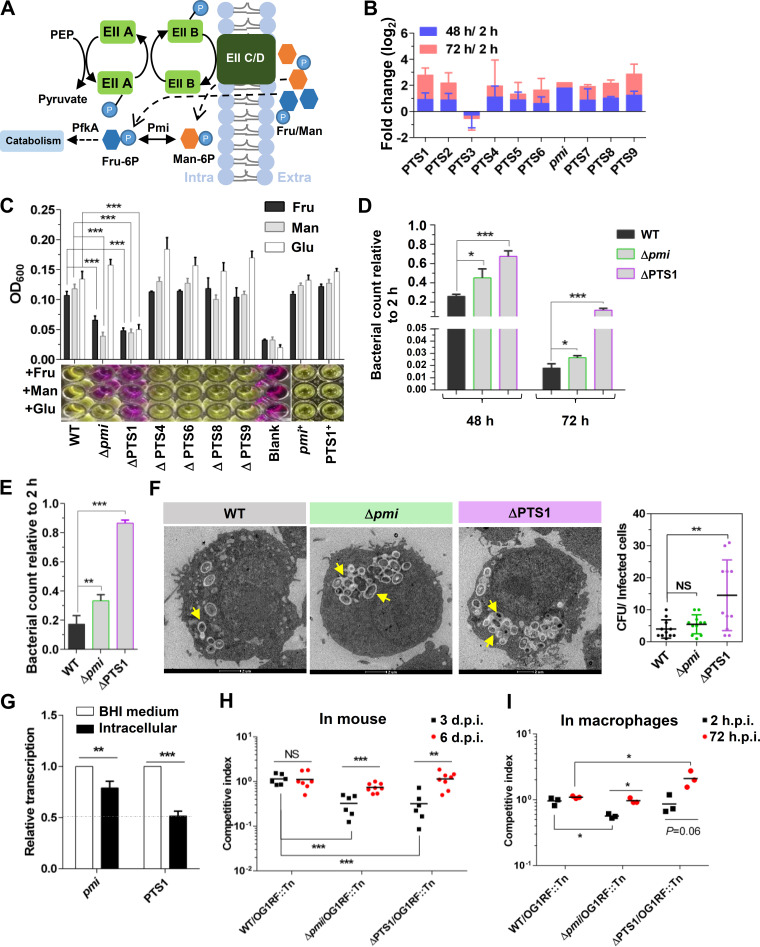
Fructose and mannose transport and metabolism in E. faecalis are depicted in Fig. 2A, where the conversion of phosphoenolpyruvate (PEP) to pyruvate releases phosphoric acid, which is gradually transferred to EIIC/D to phosphorylate extracellular fructose or mannose to form fructose-6-phosphate (fru-6P) or mannose-6-P (man-6P) (Fig. 2A) (25, 27). Next, fructose-6-phosphate or mannose-6-phosphate is transferred into cells by EIIC/D, followed by the isomerization of mannose-6-phosphate into fructose-6-phosphate by mannose-6-phosphate isomerase (Pmi) (Fig. 2A). Fructose-6-phosphate is further metabolized by 6-phosphofructokinase (PfkA) to enter glycolysis and the citrate cycle for ATP and secondary metabolite production (Fig. 2A). Notably, E. faecalis carries one copy of pmi and nine different kinds of fructose and mannose phosphotransferase systems (PTSs; PTS1 to PTS9), implying that E. faecalis may be strongly dependent on fructose and mannose metabolism to support growth or survival. Most of these fructose and mannose metabolism-related mutants in the library showed different degrees of survival enhancement in RAW264.7 cells after 48 or 72 h of infection, except a PTS3 (OG1RF_10547-10549) mutant, which showed a slight decrease (Fig. 2B).
FIG 2.
Carbohydrate metabolism affects E. faecalis survival in macrophages. (A) Schematic diagram of fructose (fru) and mannose (man) transport and metabolism in E. faecalis. (B) The fold change (log2) of fructose and mannose metabolism-related genes after survival in RAW264.7 cells for 48 h (blue) or 72 h (red). The initial data of this histogram are from the TIS results. (C) Growth of E. faecalis strains in DMEM. Sugar-free DMEM supplemented with fructose, mannose, or glucose was used to cultivate the strains. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student’s t test). (D and E) The survival of E. faecalis strains in RAW264.7 cells (D) or primary peritoneal macrophages (E) after 48 or 72 h of infection. The viable bacterial number at the indicated time was counted by plating. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student's t test). (F) Transmission electron microscopy (TEM) results showing the survival of the E. faecalis WT, Δpmi, and ΔPTS1 strains in RAW264.7 cells at 48 h.p.i. The yellow arrows indicate E. faecalis survival in phagosomes. The bacterial counts in infected cells were calculated. Representative images are shown. NS, no significant difference; **, P < 0.01 (based on one-way ANOVA). (G) qRT-PCR results showing the expression of PTS1 and pmi in RAW264.7 cells. The E. faecalis WT strain was cultured in BHI medium and survived in RAW264.7 cells for 24 h. n ≥ 3; ***, P < 0.001 (based on Student’s t test). (H andI) The competitive survival of E. faecalis strains in mice (H) and in primary peritoneal macrophages (I). WT, Δpmi, and ΔPTS1 cells were competed 1:1 versus a transposon mutant (insertion in OG1RF_12558 to OG1RF_12559), which was gentamicin resistant and used to distinguish the Δpmi and ΔPTS1 strains. Six or eight mice were used per group. n ≥ 3; **, P < 0.01; ***, P < 0.001; P values determined via ANOVA followed by Bonferroni’s multiple-comparison posttest.
To further investigate fructose and mannose metabolism in E. faecalis, deletions of these genes were engineered. According to the transcriptional profiles, PTS2 (OG1RF_10340-10342), PTS3, and PTS5 (OG1RF_11549-11552) were expressed only at the log phase in BHI medium (Fig. S2B). In addition, both PTS2 and PTS3 consisted of three genes (Fig. S2B), which may be incomplete for transport function. Thus, the in-frame deletions of PTS1 (OG1RF_10018-10021), PTS4 (OG1RF_11510-11513), PTS6 (OG1RF_11613-11616), PTS7 (OG1RF_12259-12262), PTS8 (OG1RF_12399-12402), PTS9 (OG1RF_12476-12479), and pmi were constructed to further investigate the influence of fructose and mannose metabolism on E. faecalis survival in macrophages. All of these deletions were successfully obtained, except for PTS7. According to growth curves, the deletion of pmi and PTS1 decreased the growth rate of E. faecalis in BHI medium (see Fig. S3A in the supplemental material). In addition, the ΔPTS1 strain showed a growth defect in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with fructose, mannose, or glucose as the sole carbon resource (Fig. 2C), and the complementation of PTS1 restored the defective phenotype to that of the wild-type (WT) strain (Fig. 2C). The Δpmi strain exhibited markedly reduced growth in sugar-free DMEM supplemented with fructose or mannose as the sole carbon resource (Fig. 2C) but grew normally with glucose as the sole carbon resource. The complementation of pmi restored the phenotype to that of the WT strain (Fig. 2C). The E. faecalis ΔPTS4, ΔPTS6, ΔPTS8, and ΔPTS9 strains showed no significant growth defects in vitro (Fig. 2C and S3A). These results suggested that PTS1 and Pmi critically regulate carbohydrate (fructose, mannose, and/or glucose) metabolism in E. faecalis.
(A) Growth curve of E. faecalis strains in BHI medium. n ≥ 3; **, P < 0.01 (based on Student’s t-test). (B) The cytotoxicity of RAW264.7 cells after infection with the E. faecalis WT, Δpmi, and ΔPTS1 strains. More than three replicates were performed. (C) The internalization of the E. faecalis WT, Δpmi, and ΔPTS1 strains by RAW264.7 cells at 2 h.p.i. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student’s t-test). (D) The in vitro growth of the E. faecalis WT strain and the mutant with transposon insertion in OG1RF_12558 to OG1RF_12559, named OG1RF::Tn. Three independent biological replicates were performed. (E) RNA-Seq was performed with RAW264.7 cells infected with E. faecalis WT and ΔPTS1 for 48 h, and KEGG orthology categorization of genes showed significantly different expression in RAW264.7 cells. Download FIG S3, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Carbohydrate metabolism influences E. faecalis survival in macrophages.
The mutants of PTS1 and pmi represented carbohydrate metabolism defects or attenuation in E. faecalis and were employed to investigate the function of carbohydrate metabolism during E. faecalis survival of macrophages. Compared with that of the WT strain at 48 or 72 h.p.i., the survival of the Δpmi and ΔPTS1 strains in RAW264.7 cells was significantly enhanced (Fig. 2D). The cytotoxicity of RAW264.7 cells infected by E. faecalis WT, Δpmi, and ΔPTS1 strains exhibited no obvious differences (Fig. S3B), suggesting that the carbohydrate metabolism of E. faecalis has no cytotoxicity to host cells. Moreover, the viable bacterial counts of Δpmi and ΔPTS1 in primary peritoneal macrophages were also significantly greater than that of the WT strain at 48 h.p.i. (Fig. 2E). After the E. faecalis WT, Δpmi, and ΔPTS1 strains were enabled to colonize RAW264.7 cells for 48 h, the survival status of the strains was monitored by TEM, with the results showing that all of the strains were present in phagosomes (Fig. 2F). Notably, more ΔPTS1 than WT bacteria were present in infected cells (Fig. 2F), while no significant increase in the numbers of Δpmi bacteria compared with WT bacteria was observed in infected cells (Fig. 2F), possibly due to the lower internalization of Δpmi than WT (Fig. S3C). Moreover, reverse transcription-quantitative (qRT-PCR) analysis showed that the expression of PTS1 significantly decreased in RAW264.7 cells compared with BHI medium at 24 h of cultivation, and slightly lower expression levels of pmi were observed in RAW264.7 cells than in BHI medium (Fig. 2G). These results suggested that E. faecalis downregulates carbohydrate metabolism to promote its own survival in macrophages.
Furthermore, the influence of carbohydrate metabolism on E. faecalis survival in the host was investigated using mice as models. In this experiment, the mutant with a transposon insertion in OG1RF_12558 to OG1RF_12559 (named OG1RF::Tn), which was gentamicin resistant and exhibited no obvious growth defect in vitro (Fig. S3D), was used to distinguish between other E. faecalis strains. The OG1RF::Tn strain was mixed 1:1 with the E. faecalis WT, Δpmi, or ΔPTS1 strains and inoculated into the skin of the hind legs of mice at a dose of 107 CFU. Three days postinfection, the Δpmi and ΔPTS1 strains showed significantly lower survival than the WT strain (Fig. 2H), which may have been due to the growth defect of the Δpmi and ΔPTS1 strains observed under in vitro conditions (Fig. 2C and Fig. S3A). However, at 6 days postinfection, the Δpmi and ΔPTS1 strains showed, similar to the WT, a competitive index of approximately 1.0, suggesting that the Δpmi and ΔPTS1 strains may resist immune clearance in mice better than the E. faecalis WT strain. To address this possibility, competitive assays of the E. faecalis WT, Δpmi, and ΔPTS1 strains in primary peritoneal macrophages were conducted. The experimental results showed that Δpmi and ΔPTS1 had lower rates of internalization (2 h.p.i.) than OG1RF::Tn but outcompeted OG1RF::Tn at 72 h of infection (Fig. 2I). However, the competitive index of WT to OG1RF::Tn was approximately 1.0 at 2 or 72 h postinfection. The above results suggested that attenuated carbohydrate metabolism facilitated the resistance of E. faecalis to macrophage-mediated killing, enabling longer survival in the host.
Macrophages respond to carbohydrate metabolism of E. faecalis to activate NO production.
The mechanism by which carbohydrate metabolism in E. faecalis affects macrophage-mediated killing was further explored. As shown in Fig. 2F and in other studies, E. faecalis colonized phagosomes of host cells after internalization (11, 12). In macrophages, the acidification and maturation of phagosomes promote the clearance of pathogens (9, 10). Therefore, we assessed the fitness of E. faecalis strains under low pH, reactive oxygen species (ROS), and lysozyme conditions, which were similar to acidified phagosome conditions. Zou and Shankar previously reported that E. faecalis-containing phagosomes have a pH of 5.82 at 3 h.p.i. (12). Compared with the WT strain, the ΔPTS1 strain exhibited significantly enhanced resistance to different acidic conditions, including pH values of 3.0, 4.0, and 5.0, and the complementation of PTS1 restored the sensitivity of this strain to that of the WT strain (see Fig. S4A in the supplemental material). Δpmi showed slightly higher acidic resistance (pH values of 4.0 to 5.0) than the WT strain, and the pmi-complemented strain showed greater sensitivity to acidic conditions (Fig. S4A). In addition, E. faecalis was highly sensitive to pH 3.0 and 4.0 but could survive at pH 5.0 (more than 48 h) (Fig. S4A). Moreover, the deletions of PTS1 and pmi did not enhance the resistance of E. faecalis to ROS or lysozyme (data not shown), and infections with these two strains did not clearly attenuate the acidification of phagosomes (Fig. S4B), the levels of ROS (Fig. S4C), or the activities of cathepsin B (Fig. S4D) in RAW264.7 cells compared with WT infection.
(A) Sensitivity of the E. faecalis WT, Δpmi, pmi+, ΔPTS1, and PTS1+ strains to acidic conditions, as assessed by inoculating 1.5 × 108 CFU of each strain into modified M9 medium supplemented with 200 μM glucose as the sole carbon resource and adjusting the pH of the medium to 3.0, 4.0, and 5.0. At the indicated time points, the viable bacteria were counted by plating. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student’s t-test). (B to D) Flow cytometry analyses of the acidification of phagosomes (B), production of reactive oxygen species (ROS) (C), and activities of cathepsin B (D) after E. faecalis WT, Δpmi, and ΔPTS1 strain infection. The flow cytometry results were analyzed using FlowJo. More than three independent biological replicates were performed, and the MFI and SD are shown. Download FIG S4, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
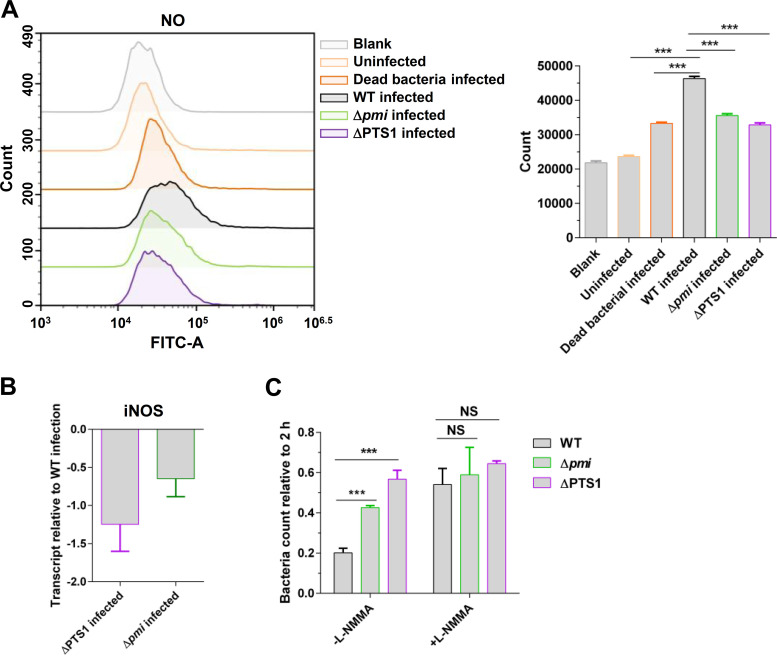
Notably, based on the flow cytometry analysis, compared with E. faecalis WT (mean fluorescence intensity [MFI], 46,367) infection, infection with Δpmi (MFI, 35,624) and ΔPTS1 (MFI, 32,851) significantly decreased the accumulation of nitric oxide (NO) in RAW264.7 cells (Fig. 3A). Meanwhile, we assessed the expression of NO synthase (iNOS) in RAW264.7 cells after infection with E. faecalis WT, Δpmi, and ΔPTS1 strains. Compared with infection by the WT strain, infection by ΔPTS1 and Δpmi decreased the expression of iNOS in RAW264.7 cells (Fig. 3B). The inhibition of NO production in RAW264.7 cells significantly increased the intracellular bacterial counts of E. faecalis WT (FC, 2.68) (Fig. 3C). However, the colonizing advantages of ΔPTS1 and Δpmi in RAW264.7 cells were attenuated compared with WT when the inhibitor (L-NMMA) of NO production was added (Fig. 3C). Taken together, the results described above suggest that macrophages would respond to carbohydrate metabolism of E. faecalis to increase the production of NO.
FIG 3.
Carbohydrate metabolism of E. faecalis promotes NO production in macrophages. (A) Flow cytometry analyses of the level of NO in RAW264.7 cells after infection with the E. faecalis WT, Δpmi, and ΔPTS1 strains. The dead bacteria represent the E. faecalis WT strain killed by heat. The number represents the mean fluorescence intensity (MFI) with standard deviation (SD). (B) qRT-PCR analysis of the transcription of iNOS in RAW264.7 cells. More than three independent biological replicates were performed. (C) The survival of E. faecalis strains in RAW264.7 cells after 48 h of infection. L-NMMA (300 μM) was added to inhibit the production of NO in RAW264.7 cells. The viable bacterial number at the indicated time was counted by plating. n ≥ 3; **, P < 0.01 (based on Student’s t test).
Carbohydrate metabolism of E. faecalis activates the polarization of macrophages to M1 to induce the production of NO.
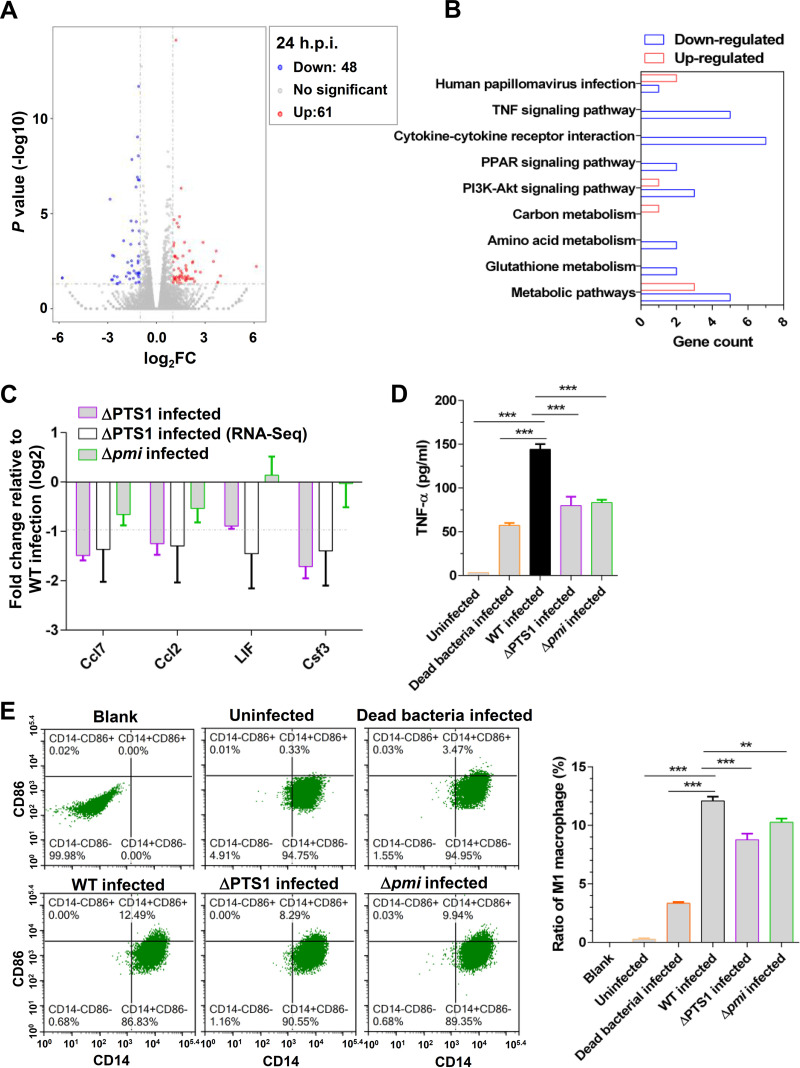
To further investigate the mechanism by which carbohydrate metabolism of E. faecalis activates the production of NO in host cells, transcriptome sequencing (RNA-Seq) was performed with RAW264.7 cells infected by E. faecalis WT and ΔPTS1, which represented defects in carbohydrate metabolism in E. faecalis. The biological replicates of RNA-Seq showed a high correlation (see Table S2 in the supplemental material). Compared with RAW264.7 cells infected by WT E. faecalis, RAW264.7 cells infected by ΔPTS1 elicited the upregulation of 61 genes, and 48 others were downregulated (Fig. 4A). These significantly differentially expressed genes were further analyzed by KEGG orthology, which indicated that the cytokine-cytokine receptor interaction and the tumor necrosis factor (TNF) signaling pathway were clearly upregulated by PTS1 (Fig. 4B). Analysis of qRT-PCR results also showed that PTS1 activated the expression of Ccl7, Ccl2, LIF, and Csf3 (Fig. 4C), which were associated with cytokine-cytokine receptor interactions and the TNF signaling pathway. In addition, infection with Δpmi slightly decreased the expression of Ccl7 and Ccl2 in RAW264.7 cells compared with WT infection (Fig. 4C). Furthermore, the results of enzyme-linked immunosorbent assays (ELISAs) showed that infection with the E. faecalis WT strain notably activated the secretion of TNF-α from RAW264.7 cells (Fig. 4D). However, the secretion of TNF-α from RAW264.7 cells infected by ΔPTS1 and Δpmi was significantly lower than that of cells infected by the WT strain (Fig. 4D). These results were consistent with the RNA-Seq results, suggesting that carbohydrate metabolism in E. faecalis activates inflammatory response in RAW264.7 cells.
FIG 4.
Carbohydrate metabolism in E. faecalis activates the polarization of macrophages to M1 to induce NO production. (A) Volcano plots were utilized to analyze the RNA-Seq results. A total of 61 upregulated genes (red dots) and 48 downregulated genes (blue dots) in RAW264.7 cells infected with the ΔPTS1 strain compared with the WT strain infected at 24 h.p.i. The cutoff was log2 FC of ≥1 or ≤−1, and a P value of <0.01 was set as significant. Two independent biological replicates were performed. (B) Significantly different genes categorized by KO. (C) Validation of RNA-Seq results by qRT-PCR. More than three independent biological replicates were performed. (D) ELISA analyses of the secretion of TNF-α from RAW264.7 cells. n ≥ 3; ***, P < 0.001 (based on Student’s t test). (E) Scatterplots were utilized to analyze the polarization of RAW264.7 cells after infection with the E. faecalis WT, Δpmi, and ΔPTS1 strains. RAW264.7 cells were labeled with CD14 and CD86, which is the hallmark of M1 macrophages. Three independent biological replicates were performed, and representative images are shown.
RNA-Seq results for RAW264.7 cells infected with the E. faecalis WT and ΔPTS1 strains. Download Table S2, XLS file, 13.2 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
It is well known that iNOS is the hallmark of M1 macrophages (28), and the polarization of macrophages to M1 is involved in inflammatory response and NO production (29). Therefore, we assessed the polarization of RAW264.7 cells after E. faecalis WT, Δpmi, and ΔPTS1 strain infection. An analysis of flow cytometry showed that infection with the E. faecalis WT strain induced the polarization of macrophages to M1 (12.49%) (Fig. 4E), but the heat-killed WT bacteria induced polarization to M1 at 3.47% (Fig. 4E). Moreover, viable bacterial infection also induced more NO and TNF-α production in RAW264.7 cells than heat-killed bacterial infection (Fig. 3B and 4D). These results suggest that viable E. faecalis bacteria strongly induce the polarization of macrophages to M1 to promote inflammation and NO production. Besides, infection with ΔPTS1 (8.29%) and Δpmi (9.94%) showed lower polarization of macrophages to M1 than WT infection (Fig. 4E), suggesting that attenuation of carbohydrate metabolism in E. faecalis decreases the polarization of macrophages to M1 to reduce NO production.
Carbohydrate shift affects the killing of E. faecalis by macrophages.
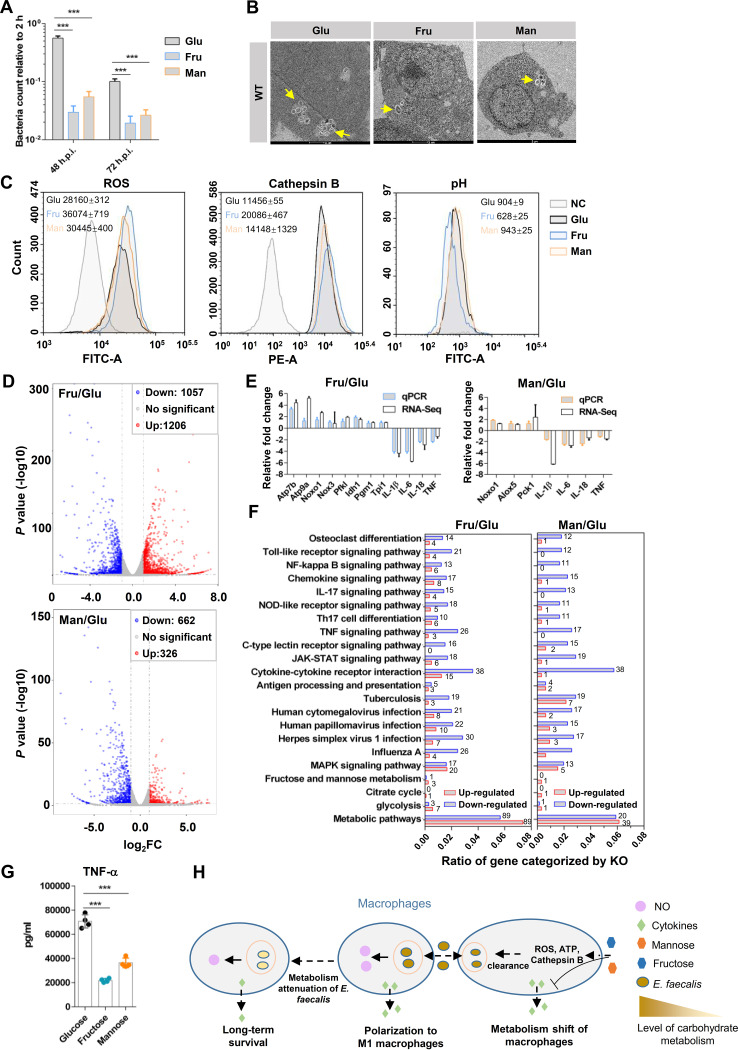
The influence of carbohydrate metabolism on macrophage-mediated killing was also investigated. To manipulate the carbohydrate metabolism of RAW264.7 cells, RAW264.7 cells were cultured in sugar-free DMEM supplemented with glucose, fructose, or mannose as the primary carbon resource after internalization of the E. faecalis WT strain. At 48 or 72 h.p.i., the intracellular bacterial counts in the RAW264.7 cells treated with fructose and mannose were significantly lower than in those of cells treated with glucose (Fig. 5A). The survival status of the E. faecalis WT strains in RAW264.7 cells was monitored by TEM, which showed that the addition of exogenous fructose and mannose dramatically decreased the bacterial counts in RAW264.7 cells compared with that observed in glucose-treated cells (Fig. 5B). These results suggested that fructose and mannose enhance macrophage-mediated killing of E. faecalis. To explore the mechanism governing this phenomenon, the production of intracellular ROS, the activities of cathepsin B, and the acidification of E. faecalis-containing phagosomes were detected by flow cytometry. Compared with RAW264.7 cells treated with glucose and E. faecalis infection, fructose and mannose significantly increased the production of ROS and the activities of cathepsin B in RAW264.7 cells after E. faecalis infection (Fig. 5C). In addition, RAW264.7 cells treated with fructose (MFI, 628) exhibited a lower pH in E. faecalis-containing phagosomes than cells treated with glucose and mannose, but no clear difference in pH between glucose (MFI, 904) and mannose (MFI, 934) was observed (Fig. 5C).
FIG 5.
Carbohydrate metabolism affects macrophage-mediated killing of E. faecalis. (A) Sugar shift affects macrophage-mediated killing of E. faecalis. After E. faecalis was internalized by RAW264.7 cells, the extracellular bacteria were killed by gentamicin and vancomycin, and the medium was simultaneously changed to fresh sugar-free DMEM supplemented with glucose, fructose, or mannose as the primary carbon resource. At 48 and 72 h postinfection, the viable bacteria in host cells were counted by plating. n ≥ 3; ***, P < 0.001 (based on the Student’s t test). (B) TEM analysis showed the status and bacterial number of the E. faecalis WT strain in RAW264.7 cells cultured with sugar-free DMEM supplemented with glucose, fructose, or mannose. Representative images are shown. (C) Flow cytometry analyses of the production of ROS, the activities of cathepsin B, and the acidification of E. faecalis-containing phagosomes in RAW264.7 cells. RAW264.7 cells were treated with different sugars (glucose, fructose, or mannose) and infected with E. faecalis. More than three independent biological replicates were performed. The mean fluorescein intensity (MFI) with standard deviation (SD) was calculated and is shown. (D) Volcano plots were utilized to analyze the RNA-Seq results. RAW264.7 cells treated with glucose, fructose, or mannose were subjected to RNA-Seq. Red dots and blue dots represent upregulation (log2FC, ≥1; P < 0.01) and downregulation (log2 FC, ≤−1; P < 0.01), respectively. Two independent biological replicates were performed. (E) Validation of RNA-Seq results by qRT-PCR. RAW264.7 cells were treated with glucose, fructose, and mannose after E. faecalis infection. The expression of genes in RAW264.7 cells treated with fructose and mannose compared to glucose is shown. Three independent biological replicates were performed. (F) KO categorization of genes with significant differences according to RNA-Seq results. The gene ratio represents the number of genes enriched by one category relative to all the significant genes counted. The number of depleted genes in each category is shown. (G) ELISA analyses of the secretion of TNF-α by RAW264.7 cells treated with glucose, fructose, and mannose after E. faecalis infection. n ≥ 3; ***, P < 0.001 (based on Student’s t test). (H) Schematic diagram shows carbohydrate metabolism coordinating the survival of E. faecalis in macrophages. Macrophages sense the stimulation of carbohydrate metabolism or metabolites from E. faecalis and polarize to M1 to induce the production of NO for bacterial killing. On the other hand, the shift in carbohydrate metabolism affects macrophage-mediated killing of E. faecalis and inflammation.
To further investigate the influence of carbohydrate on macrophage-mediated killing, RNA-Seq was performed (see Table S3 and S4 in the supplemental material). According to the RNA-Seq results, fructose significantly increased the expression levels of 1,206 genes and decreased the expression levels of 1,057 genes (Fig. 5D and Table S3), and mannose significantly increased the expression levels of 326 genes and decreased the expression levels of 662 genes compared with those in RAW264.7 cells treated with glucose and E. faecalis infection (Fig. 5D and Table S4). Among these significantly different genes, ROS-related genes in RAW264.7 cells treated with fructose (Noxo1 log2FC, 2.69; Nox3 log2FC, 1.01) and mannose (Noxo1 log2FC, 1.28; Alox5 log2FC, 1.15; Alox12 log2FC, 1.44) were significantly upregulated compared with glucose-treated cells (Table S3 and S4). ATP synthase-related genes, which promote the acidification of E. faecalis-containing phagosomes, showed significant increases in RAW264.7 cells treated with fructose (Atp7b log2FC, 5.14; Atp9a log2FC, 5.18; Atp6v0d2 log2FC, 2.74) (Table S3). However, the cathepsin B gene (Ctsb) showed no significant change in fructose (log2FC, 0.46) and mannose (log2FC, −0.45) compared with glucose (Table S3 and S4). qRT-PCR analysis was performed to validate the RNA-Seq results and showed that fructose significantly increased the expression of ROS-related genes (Noxo1 and Nox3) and ATP synthesis-related genes (Atp7b and Atp9a) in RAW264.7 cells compared with glucose (Fig. 5E), which was consistent with the RNA-Seq results. In addition, mannose significantly increased the expression levels of Noxo1 and Alox5, of which both promoted the production of ROS in RAW264.7 cells.
RNA-Seq for RAW264.7 cells treated with glucose and fructose. Download Table S3, XLS file, 13.1 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-Seq for RAW264.7 cells treated with glucose and mannose. Download Table S4, XLS file, 13.1 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
According to the analysis of RNA-Seq results based on KEGG orthology, fructose and mannose significantly upregulated energy metabolism, such as fructose and mannose metabolism, citrate cycle, glycolysis, and other metabolic pathways, compared with RAW264.7 cells treated with glucose and infected with E. faecalis (Fig. 5E and F). In addition, fructose and mannose dramatically downregulated the expression levels of innate immunity components, including the Toll-like receptor signaling pathway, NF-κB signaling pathway, chemokine signaling pathway, interleukin-17 (IL-17) signaling pathway, and TNF signaling pathway (Fig. 5E to G), which implies that fructose and mannose promote macrophage-mediated killing independent of inflammatory responses. The results described above showed that carbohydrate metabolism affects the ability of macrophages to mediate killing of E. faecalis and simultaneously represses the activation of inflammation compared with glucose treatment.
DISCUSSION
E. faecalis resists immune clearance by the host and causes severe infectious diseases worldwide. Although several factors have been reported to contribute to the resistance mechanisms of E. faecalis against host immunity, such as the enterococcal polysaccharide antigen (EPA) (30), the impact of metabolites or metabolic pathways on the ability of E. faecalis to resist macrophage-mediated killing has rarely been reported. According to the results of transposon insertion sequencing, a total of 15 and 19 mutants with decreased fitness in RAW264.7 cells were identified from the 48- and 72-h groups, respectively, of which 9 overlapped (Fig. 1D). Among these nine mutants, a mutation in rpoN, which was screened out for causing a fitness reduction (log2FC, −2.18) (Table S1), has been reported to promote the killing of phagocytes by Pseudomonas aeruginosa (31). The other genes with disruptions that caused fitness reductions included those encoding an acetyltransferase (log2FC, −1.66), aspartate aminotransferase (log2FC, −1.59), and a LysR family transcriptional regulator (log2FC, −1.73) (Table S1), of which all will be further investigated in the future. In addition, mutants in OG1RF_11714, which encodes a polysaccharide antigen EPA-related protein, glycosyltransferase, displayed an abundance reduction after 48 h (log2FC, −0.713) or 72 h (log2FC, −0.829) of survival in RAW264.7 cells (Table S1), which was consistent with the results reported by Smith (30).
Bacteria depend on the phosphotransferase system (PTS) to sense and transport extracellular carbohydrates. The PTS has been also reported to affect virulence gene expression and infection in Streptococcus sp., Bacillus anthracis, and Vibrio cholerae (32–34), and mannose metabolites can also act as signaling molecules to coordinate virulence gene expression and influence infection (25). Surface components, including teichoic acids, a capsule, and enterococcal polysaccharide antigen, play key roles for Enterococcus sp. escape from phagocytosis (35–37). In addition to affecting cellular resistance to low pH (Fig. 4A), carbohydrate metabolism may also mediate virulence factor expression or surface components to affect E. faecalis resistance to macrophage-mediated killing, which warrants further investigation.
Metabolic interactions are important in the battle between pathogens and hosts (38–40). When sensing bacterial invasion, the host reprograms metabolism to aerobic glycolysis, which is called the Warburg effect (41). The metabolism shift leads to the accumulation of succinate, which induces inflammation (42). Therefore, some bacterial pathogens need to coordinate their own metabolism to escape from immune activation of the host. M. tuberculosis enters a dormant-like status after colonizing host cells, enhancing their ability to resist immune clearance or avoid immune activation (43). Our results demonstrate that E. faecalis attenuates carbohydrate metabolism to escape from the activation of host immunity and facilitates its own survival in macrophages (Fig. 5H). Fan et al. also showed that the gluconate metabolism of E. faecalis activates the inflammation of macrophages (44). However, how carbohydrate metabolism of E. faecalis triggers immune activation has not been investigated to date. To figure out this question, the transcriptomes of RAW264.7 cells infected with the E. faecalis WT and ΔPTS1 strains were performed (48 h.p.i.) and showed that PTS1 affected the energy metabolism of RAW264.7 cells, including the insulin signaling pathway, pyruvate metabolism, fructose and mannose metabolism, pentose phosphate pathway, citrate cycle, insulin resistance, phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway, and AMP-activated protein kinase (AMPK) signaling pathway (Fig. S3E). Some investigations have also shown that the enhancement of glycolysis and pentose phosphate pathway activity in macrophages promotes rapid energy production and the polarization of macrophages to M1, thereby leading to rapid antibacterial and inflammatory responses (45). Thus, the metabolites of E. faecalis may trigger the host to reprogram its own metabolism for the immune response.
Compared with glucose, fructose and mannose (especially fructose) have been shown promoting the macrophage-mediated killing by increasing the production of ROS, the activities of cathepsin B, and the acidification of phagosomes (Fig. 5C), which is independent of inflammatory response (Fig. 5E and F). Thus, we speculate that the shift of metabolism may affect nutrition obtained by E. faecalis in macrophages. According to TEM images (Fig. 2F, 5B, and S1B), some E. faecalis bacteria were dividing in host cells after 24 or 48 h of survival, implying that E. faecalis may replicate in macrophages. The shift of carbohydrates for macrophage cultivation may affect the nutrient acquisition of E. faecalis during intracellular survival. Xian et al. reported that Ralstonia solanacearum hijacks host metabolites to support its own replication (38). Thus, whether E. faecalis hijacks host carbohydrates as nutrients to promote survival warrants further study.
In summary, the results of the present study helped to elucidate the mechanism by which carbohydrate metabolism mediates the macrophage-mediated killing of E. faecalis (Fig. 5H), which may provide important strategies for the treatment of chronic inflammatory and infectious diseases caused by E. faecalis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are described in Table S5 and oligonucleotides in Table S6 in the supplemental material. E. faecalis strains were routinely grown at 37°C or 42°C (inducing transposition) in brain heart infusion (BHI) broth or agar. E. coli and L. lactis were grown in Luria-Bertani (LB) broth or agar, and erythromycin (Erm) was added at a final concentration of 200 μg/ml to select pTetH or pGhost derivatives. For complementation assays, erythromycin was used at a final concentration of 30 μg/ml to select positive colonies and anhydrotetracycline (Tet) was used at a final concentration of 10 ng/ml to induce gene expression.
Bacterial strains and plasmids used in this study. Download Table S5, XLS file, 0.03 MB (35.5KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S6, XLS file, 0.02 MB (23KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transposon insertion sequencing and mapping.
The construction of the transposon insertion library of E. faecalis was described by Wei et al. (46). High-throughput sequencing and analysis were previously described (21, 47). Briefly, genomic DNA was extracted and fragmented by sonication, followed by end repairing, A-tailing, and addition of adaptors and P5/P7 sequences by PCR. Three or two library replicates of inputs and outputs were applied to high-throughput sequencing on the Illumina HiSeq 2500 platform. The sequencing results were processed with adapter trimming, mapping to the genome, and tallying as previously described (21). The read counts for each locus were normalized among the libraries according to sequencing depth. The fold change of each locus was generated with the output compared with the input read counts. Conditional essential genes were determined using the HMM module of EL-ARTIST (23). Functional classification is based on the KEGG (https://www.genome.jp/kegg/).
Construction of deletion and complementation.
The in-frame deletion of E. faecalis OG1RF was constructed by allele exchange using the procedure previously described (30). pGhost9 derivatives, which were replicated in a temperature-dependent manner in host cells, were electroporated, and transformants were selected at 28°C on BHI agar plates with erythromycin (30 μg/ml). Then, E. faecalis harboring the pGhost9 derivative was grown at a nonpermissive temperature (42°C) in the presence of erythromycin to induce single crossover recombination. The second recombination event happened after 7 serial subcultures at 28°C without erythromycin, and the double crossover mutation was identified by PCR and sequencing.
For complementation, DNA fragments coding PTS1 and pmi were amplified by PCR using the oligonucleotides described in Table S6. PCR products were ligated to pTetH, which were digested by NcoI and BamHI in advance. The constructed pTetH derivatives were transferred in E. faecalis cells by electroporation, and positive colonies were screened by erythromycin (30 μg/ml) and sequenced. For complementation assays, pTetH derivatives were anhydrotetracycline-inducible expression.
Infection of macrophages and the intracellular bacterial counting.
The RAW264.7 cells and primary peritoneal macrophages were used as the cell model, and the cell culture and infection procedures were followed as described by Zou (12). Briefly, cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. A total of 2.0 × 105 cells were seeded into 24-well plates and then infected by E. faecalis strains at a multiple of infection (MOI) of 10 or 20, following immediately by centrifugation at 300 × g for 10 min to facilitate bacterial sinking on RAW264.7 cells. At 2 hour postinfection, the extracellular bacteria were then removed by washing with sterile phosphate-buffered saline (PBS), followed addition of vancomycin (16 μg/ml) and gentamicin (150 μg/ml) to kill the extracellular bacteria. Fresh culture medium was changed every 24 h. At the indicated time points, the macrophages were lysed by 4°C 0.1% Triton X-100. The intracellular bacteria were quantified by serial dilution and plate counting. NO inhibitor (L-NMMA) was added if necessary.
Transmission electron microscopy.
A total of 1 × 106 RAW264.7 cells were seeded into 6-well plates to grow overnight and then infected with E. faecalis strains, E. coli, or L. lactis at an MOI of 20 for 2 h at 37°C. The extracellular bacteria were washed twice with sterile PBS and killed by vancomycin and gentamicin after 2 h postinfection. At the indicated time points, 5% glutaraldehyde was added to fix infected RAW264.7 cells. The fixed cells were dehydrated and embedded in Epon/Araldite, cured, and sectioned. The sections were placed on glow-discharged 150-mesh Cu grids, which were imaged on a Talos L120C transmission electron microscope (TEM). A total of 11 infected cells were random picked up to statistically calculate the average count of intracellular bacteria.
E. faecalis adaption to acidic conditions.
To test the sensitivity of E. faecalis strains to acidic conditions, the M9 medium supplemented with 200 μM glucose as the sole carbon resource was adjusted to pH 3.0, 4.0, and 5.0. Thus, E. faecalis was capable of surviving into the modified M9 medium but could not grow. E. faecalis strains were inoculated into this modified M9 medium (pH 3.0, 4.0, and 5.0) with a dose of 1.5 × 108 CFU. At the setting time points, the viable bacteria in the modified M9 medium were counted by a series of dilutions and plating onto BHI agar plates.
Detection of intraphagosomal acidification.
The detection of phagosome acidification was conducted as described previously (48). First, E. faecalis strains were labeled with fluorescein isothiocyanate (FITC; pH sensitive; Thermo Fisher) and Alexa Fluor 647 (pH insensitive; Thermo Fisher) fluorescent dyes. One milliliter of a E. faecalis suspension was mixed with 25-μl FITC and 25-μl Alexa Fluor 647 dye solutions, and the suspension was incubated in the dark for 1 h at room temperature with continuous shaking. The RAW264.7 cells were infected with E. faecalis strains labeled with FITC and Alexa Fluor 647 for 2 h. The extracellular bacteria were washed three times with sterile PBS, and the cells were continually incubated to the indicated time point. The mean fluorescent intensity of FITC and Alexa Fluor 647 was analyzed with flow cytometry.
Detection of ROS and NO productions and cathepsin B activities.
A total of 4 × 105 RAW264.7 cells were seeded onto 12-well plates and cultured overnight. At 2 h postinfection, RAW264.7 cells were washed three times with PBS, received fresh DMEM medium with 10% FBS, and continued incubating at 37°C for 24 h. At the setting time points, the cells were washed twice with PBS and stained with the MitoSOX Red mitochondrial superoxide indicator (Thermo Fisher) for ROS detection, Magic Red cathepsin B (Bio-Rad) for cathepsin B detection, or 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA; Beyotime Biotechnology) for NO detection. After being washed three times with PBS, the cells were subjected to analysis by flow cytometry. The mean fluorescent intensity was calculated.
Total RNA extraction and qRT-PCR.
The total RNA of RAW264.7 cells or E. faecalis was extracted with a commercial RNA isolation kit (BioFlux). The mRNA was reverse transcribed into cDNA by the PrimeScript RT reagent kit (TaKaRa). The qRT-PCR was performed with a LightCycler 480 II instrument (Roche), and the CT values were analyzed. The relative fold change was calculated with the housekeeping gene gyrB (for E. faecalis) or the gene Gapdh (for RAW264.7 cells) as an internal control.
RNA-Seq and data analysis.
RNA-Seq and data (NCBI accession number PRJNA734659) analysis were conducted as previously described (49). Briefly, total RNA of RAW264.7 cells infected with E. faecalis were extracted with a commercial RNA isolation kit (BioFlux), and mRNA was enriched by magnetic beads. For RNA-Seq, the VAHTS stranded mRNA-Seq library prep kit (Vazyme) was used to construct strand-specific RNA-Seq libraries. After libraries were qualified, the sequencing was conducted on the HiSeq 2500 platform. The Illumina HiSeq sequencer captured the fluorescence signal and converted the optical signal into a sequencing peak through computer software to obtain sequence information on the tested fragments. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was carried out based on RNA-Seq data online (https://www.genome.jp/kegg/tool/map_pathway2.html).
Isolation of primary peritoneal macrophages.
The isolation of primary peritoneal macrophages was performed as previously described (50). Briefly, male C57BL/6 mice (5 weeks old) were each injected with 2 ml aged 3% thioglycolate to induce the production of macrophages. Five days after induction, mice were sacrificed, and primary peritoneal macrophages were isolated from mouse abdomens. After being washed three times with PBS, primary peritoneal macrophages were seeded into 24-well plates and cultured in DMEM supplemented with 10% FBS.
Mouse infection and competitive index detection.
The competitive index of E. faecalis strains in mice was performed as described by Chong (51). Male C57BL/6 mice (6 to 7 weeks old; 20 to 23 g) were anesthetized with isoflurane and hind leg hair removed. The skin was disinfected with 75% ethanol, and then 5 μl of mix bacteria (1.0 × 107 CFU) were inoculated under the skin. At the indicated time points, the mice were euthanized and a 1 by 1-cm piece of skin surrounding the inoculated site was excised into sterile PBS. The excised skins were homogenized, and viable bacteria were counted by diluted plating on BHI agar supplemented with antibiotics. The strain with an intergenic insertion of the transposon between OG1RF_12558 and OG1RF_12559 (named OG1RF::Tn) was used to distinguish Δpmi and ΔPTS1 according to gentamicin resistance.
Ethics statement.
All the procedures mentioned in this study were approved by the Institutional Ethical Review Board of Shanghai Ninth People’s Hospital, School of Medicine, Shanghai Jiao Tong University, and conducted in conformity with institutional guidelines for the care and use of laboratory animals in School of Medicine, Shanghai Jiao Tong University.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism version 6.0. The results were representative of at least three independent experiments (*, P < 0.05; **, P < 0.01; ***; P < 0.001 [Student’s t test or ANOVA]).
Data availability.
Raw sequencing data in FASTQ format are publicly available for download through the Data Repository for NCBI at https://www.ncbi.nlm.nih.gov/sra/PRJNA734659.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (82001033). We thank Shanghai Institute of Precision Medicine for technical assistance.
L.W., W.Z., and J. Liang conceived and designed the experiments. L.W., F.X., J. Wang, and Y.L. performed experiments. L.W., S.R., Z.H., and J. Liang wrote and revised the manuscript.
We declare no conflict of interest.
Contributor Information
Jingping Liang, Email: liangjpdentist@126.com.
Joshua E. Elias, Chan Zuckerberg Biohub
REFERENCES
- 1.Tendolkar PM, Baghdayan AS, Shankar N. 2003. Pathogenic enterococci: new developments in the 21st century. Cell Mol Life Sci 60:2622–2636. doi: 10.1007/s00018-003-3138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahl A, Iversen K, Tonder N, Hoest N, Arpi M, Dalsgaard M, Chehri M, Soerensen LL, Fanoe S, Junge S, Hoest U, Valeur N, Lauridsen TK, Fosbol E, Hoi-Hansen T, Bruun NE. 2019. Prevalence of infective endocarditis in Enterococcus faecalis bacteremia. J Am Coll Cardiol 74:193–201. doi: 10.1016/j.jacc.2019.04.059. [DOI] [PubMed] [Google Scholar]
- 3.Ran S, Liu B, Jiang W, Sun Z, Liang J. 2015. Transcriptome analysis of Enterococcus faecalis in response to alkaline stress. Front Microbiol 6:795. doi: 10.3389/fmicb.2015.00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harten RMV, Willems RJL, Martin NI, Hendrickx APA. 2017. Multidrug-resistant enterococcal infections: new compounds, novel antimicrobial therapies? Trends Microbiol 25:467–479. doi: 10.1016/j.tim.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. 2016. Global guidelines for the prevention of surgical site infection. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Graninger W, Ragette R. 1992. Nosocomial bacteremia due to Enterococcus faecalis without endocarditis. Clin Infect Dis 15:49–57. doi: 10.1093/clinids/15.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Swimberghe RCD, Coenye T, De Moor RJG, Meire MA. 2019. Biofilm model systems for root canal disinfection: a literature review. Int Endod J 52:604–628. doi: 10.1111/iej.13050. [DOI] [PubMed] [Google Scholar]
- 8.Corleis B, Dorhoi A. 2020. Early dynamics of innate immunity during pulmonary tuberculosis. Immunol Lett 221:56–60. doi: 10.1016/j.imlet.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Flannagan RS, Cosío G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 10.Weiss G, Schaible UE. 2015. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentry-Weeks CR, Karkhoff-Schweizer RAnn, Pikis A, Estay M, Keith JM. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect Immun 67:2160–2165. doi: 10.1128/IAI.67.5.2160-2165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J, Shankar N. 2016. The opportunistic pathogen Enterococcus faecalis resists phagosome acidification and autophagy to promote intracellular survival in macrophages. Cell Microbiol 18:831–843. doi: 10.1111/cmi.12556. [DOI] [PubMed] [Google Scholar]
- 13.Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, Winkle WBV, Simon SI. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 67:6067–6075. doi: 10.1128/IAI.67.11.6067-6075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Stunault MI, Bories G, Guinamard RR, Ivanov S. 2018. Metabolism plays a key role during macrophage activation. Mediators Inflamm 2018:2426138. doi: 10.1155/2018/2426138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatima S, Kamble SS, Dwivedi VP, Bhattacharya D, Kumar S, Ranganathan A, Kaer LV, Mohanty S, Das G. 2020. Mycobacterium tuberculosis programs mesenchymal stem cells to establish dormancy and persistence. J Clin Invest 130:655–661. doi: 10.1172/JCI128043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cumming BM, Addicott KW, Adamson JH, Steyn AJ. 2018. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife 7:e39169. doi: 10.7554/eLife.39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borah K, Girardi K, Mendum TA, Lery L, Beste D, Lara FA, Pessolani M, McFadden J. 2019. Intracellular Mycobacterium leprae utilizes host glucose as a carbon source in Schwann cells. mBio 10:e02351-19. doi: 10.1128/mBio.02351-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borah K, Beyß M, Theorell A, Wu H, Basu P, Mendum TA, Nӧh K, Beste DJV, McFadden J. 2019. Intracellular Mycobacterium tuberculosis exploits multiple host nitrogen sources during growth in human macrophages. Cell Rep 29:3580–3591. doi: 10.1016/j.celrep.2019.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opijnen TV, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Chuah S, Chang C, Chen F. 2020. The surface protein fructose-1, 6 bisphosphate aldolase of Klebsiella pneumoniae Serotype K1: role of interaction with neutrophils. Pathogens 9:1009. doi: 10.3390/pathogens9121009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei L, Qiao H, Sit B, Yin K, Yang G, Ma R, Ma J, Yang C, Yao J, Ma Y, Xiao J, Liu X, Zhang Y, Waldor MK, Wang Q. 2019. A bacterial pathogen senses host mannose to coordinate virulence. iScience 20:310–323. doi: 10.1016/j.isci.2019.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altarac S, Papeš D. 2014. Use of d-mannose in prophylaxis of recurrent urinary tract infections (UTIs) in women. BJU Int 113:9–10. doi: 10.1111/bju.12492. [DOI] [PubMed] [Google Scholar]
- 27.Jeckelmann JM, Erni B. 2020. The mannose phosphotransferase system (Man-PTS)—mannose transporter and receptor for bacteriocins and bacteriophages. Biochim Biophys Acta Biomembr 1862:183412. doi: 10.1016/j.bbamem.2020.183412. [DOI] [PubMed] [Google Scholar]
- 28.Edin S, Wikberg ML, Dahlin AM, Rutegård J, Öberg Å, Oldenborg P, Palmqvist R. 2012. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One 7:e47045. doi: 10.1371/journal.pone.0047045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Li J, Chen Y, Zhang S, Feng C, Hou Z, Cai J, Wang Y, Hui R, Lv B, Zhang W. 2019. MicroRNA-216a promotes M1 macrophages polarization and atherosclerosis progression by activating telomerase via the Smad3/NF-κB pathway. Biochim Biophys Acta Mol Basis Dis 1865:1772–1781. doi: 10.1016/j.bbadis.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Smith RE, Bartłomiej S, Piotr S, Natalia H, Prajsnar TK, Gregory B, Thierry F, Justyna K, Herry J, Andrea H, Mike PW, Serror P, Mesnage AS. 2019. Decoration of the enterococcal polysaccharide antigen EPA is essential for virulence. PLoS Pathog 15:e1007730. doi: 10.1371/journal.ppat.1007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong TG, Mekalanos JJ. 2012. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res 40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundar GS, Islam E, Gera K, Le Breton Y, McIver KS. 2017. APTS EII mutant library in Group A Streptococcus identifies a promiscuous man‐family PTS transporter influencing SLS‐mediated hemolysis. Mol Microbiol 103:518–533. doi: 10.1111/mmi.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bier N, Hammerstrom TG, Koehler TM. 2020. Influence of the phosphoenolpyruvate: carbohydrate phosphotransferase system on toxin gene expression and virulence in Bacillus anthracis. Mol Microbiol 113:237–252. doi: 10.1111/mmi.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Millet YA, Chao MC, Sasabe J, Davis BM, Waldor MK. 2015. A genome-wide screen reveals that the Vibrio cholerae phosphoenolpyruvate phosphotransferase system modulates virulence gene expression. Infect Immun 83:3381–3395. doi: 10.1128/IAI.00411-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiss-Liebisch S, Rooijakkers SHM, Beczala A, Sanchez-Carballo P, Kruszynska K, Repp C, Sakinc T, Vinogradov E, Holst O, Huebner J, Theilacker C. 2012. Secondary cell wall polymers of Enterococcus faecalis are critical for resistance to complement activation via mannose-binding lectin. J Biol Chem 287:37769–37777. doi: 10.1074/jbc.M112.358283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng F, Jacques-Palaz KD, Weinstock GM, Murray BE. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect Immun 70:2010–2015. doi: 10.1128/IAI.70.4.2010-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J Bacteriol 191:6203–6210. doi: 10.1128/JB.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xian L, Yu G, Wei Y, Rufian JS, Li Y, Zhuang H, Xue H, Morcillo RJL, Macho AP. 2020. A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe 28:548–557. doi: 10.1016/j.chom.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Saka HA, Valdivia RH. 2010. Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr Opin Microbiol 13:4–10. doi: 10.1016/j.mib.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bäumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heiden MGV, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LAJ. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai W, Wu F, Zhang Y, Fu Y, Liu Z. 2019. The immune escape mechanisms of Mycobacterium tuberculosis. Int J Mol Sci 20:340. doi: 10.3390/ijms20020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan T, Goeser L, Naziripour A, Redinbo MR, Hansen JJ. 2019. Enterococcus faecalis gluconate phosphotransferase system accelerates experimental colitis and bacterial killing by macrophages. Infect Immun 87:e00080-19. doi: 10.1128/IAI.00080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Neill LAJ, Kishton RJ, Rathmell J. 2016. A guide to immunometabolism for immunologists. Nat Rev Immunol 16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei L, Li M, Xia F, Wang J, Ran S, Huang Z, Liang J. 2021. Phosphate transport system mediates the resistance of Enterococcus faecalis to multidrug. Microbiol Res 249:126772. doi: 10.1016/j.micres.2021.126772. [DOI] [PubMed] [Google Scholar]
- 47.Dale JL, Beckman KB, Willett JLE, Nilson JL, Palani NP, Baller JA, Hauge A, Gohl DM, Erickson R, Manias DA, Sadowsky MJ, Dunny GM. 2018. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3:e00062-18. doi: 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokolovska A, Becker CE, Stuart LM. 2012. Measurement of phagocytosis, phagosome acidification, and intracellular killing of Staphylococcus aureus. Curr Protoc Immunol Chapter 14:Unit14.30. doi: 10.1002/0471142735.im1430s99. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Usher MG, Duan SZ, Ivaschenko CY, Frieler RA, Berger S, Schütz G, Lumeng CN, Mortensen RM. 2010. Myeloid mineralocorticoid receptor controls macrophage polarization and cardiovascular hypertrophy and remodeling in mice. J Clin Invest 120:3350–3364. doi: 10.1172/JCI41080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chong KKL, Tay WH, Janela B, Yong AMH, Liew TH, Madden L, Keogh D, Barkham TMS, Ginhoux F, Becker DL, Kline KA. 2017. Enterococcus faecalis modulates immune activation and slows healing during wound infection. J Infect Dis 216:1644–1654. doi: 10.1093/infdis/jix541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. faecalis resists macrophage-mediated killing. (A) RAW264.7 cells were infected with the E. faecalis strains OG1RF and 33186, Lactobacillus lactis, and E. coli DH5α at an MOI of 10. Cells were washed and further incubated with DMEM supplemented with 10% FBS containing vancomycin and gentamycin to kill extracellular bacteria 2 h.p.i. The number of viable bacteria within host cells was counted at 2, 24, 48, and 72 h postinfection. More than three replicates were performed. (B) Characterization of bacterium-containing phagosomes by TEM. RAW264.7 cells were infected with the above strains at an MOI of 10. After 24 h of infection, cells were fixed and processed for imaging by transmission electron microscopy. Representative images are shown. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Correlation of experimental replicates from the TIS results. The sequencing data from experimental replicates of input libraries (n = 3) and output libraries recovered after survival in RAW264.7 cells for 48 h (n = 2) and 72 h (n = 3) are shown. (B) Transcriptome profiles depicting the expression of fructose and mannose metabolism-associated genes at log and stationary phases in BHI medium. Three independent biological replicates were performed. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of transposon insertion mutant library screened for genetic determinates for E. faecalis survival in RAW264.7 cells. Download Table S1, XLS file, 0.8 MB (833.5KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Growth curve of E. faecalis strains in BHI medium. n ≥ 3; **, P < 0.01 (based on Student’s t-test). (B) The cytotoxicity of RAW264.7 cells after infection with the E. faecalis WT, Δpmi, and ΔPTS1 strains. More than three replicates were performed. (C) The internalization of the E. faecalis WT, Δpmi, and ΔPTS1 strains by RAW264.7 cells at 2 h.p.i. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student’s t-test). (D) The in vitro growth of the E. faecalis WT strain and the mutant with transposon insertion in OG1RF_12558 to OG1RF_12559, named OG1RF::Tn. Three independent biological replicates were performed. (E) RNA-Seq was performed with RAW264.7 cells infected with E. faecalis WT and ΔPTS1 for 48 h, and KEGG orthology categorization of genes showed significantly different expression in RAW264.7 cells. Download FIG S3, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Sensitivity of the E. faecalis WT, Δpmi, pmi+, ΔPTS1, and PTS1+ strains to acidic conditions, as assessed by inoculating 1.5 × 108 CFU of each strain into modified M9 medium supplemented with 200 μM glucose as the sole carbon resource and adjusting the pH of the medium to 3.0, 4.0, and 5.0. At the indicated time points, the viable bacteria were counted by plating. n ≥ 3; *, P < 0.05; ***, P < 0.001 (based on Student’s t-test). (B to D) Flow cytometry analyses of the acidification of phagosomes (B), production of reactive oxygen species (ROS) (C), and activities of cathepsin B (D) after E. faecalis WT, Δpmi, and ΔPTS1 strain infection. The flow cytometry results were analyzed using FlowJo. More than three independent biological replicates were performed, and the MFI and SD are shown. Download FIG S4, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-Seq results for RAW264.7 cells infected with the E. faecalis WT and ΔPTS1 strains. Download Table S2, XLS file, 13.2 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-Seq for RAW264.7 cells treated with glucose and fructose. Download Table S3, XLS file, 13.1 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-Seq for RAW264.7 cells treated with glucose and mannose. Download Table S4, XLS file, 13.1 MB (13.5MB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and plasmids used in this study. Download Table S5, XLS file, 0.03 MB (35.5KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S6, XLS file, 0.02 MB (23KB, xls) .
Copyright © 2021 Wei et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw sequencing data in FASTQ format are publicly available for download through the Data Repository for NCBI at https://www.ncbi.nlm.nih.gov/sra/PRJNA734659.