ABSTRACT
Metabolites from the microbiome influence human, animal, and environmental health, but the diversity and functional roles of these compounds have only begun to be elucidated. Comprehensively characterizing these molecules are significant challenges, as it requires expertise in analytical methods, such as mass spectrometry and nuclear magnetic resonance spectroscopy, skills that not many traditional microbiologists or microbial ecologists possess. This creates a gap between microbiome scientists that want to understand the role of microbial metabolites in microbiome systems and the skills required to generate and interpret complex metabolomics data sets. To bridge this gap, microbiome scientists should engage analytical chemists to best understand the underlying chemical principles of the data. Conversely, analytical scientists are encouraged to engage with microbiome scientists to better understand the biological questions being asked with metabolomics and to best communicate its intricacies. Better communication across the chemistry/biology disciplines will further reveal the “dark matter” within microbiomes that maintain healthy humans and environments.
KEYWORDS: analytical, mass spectrometry, metabolome, microbiome
PERSPECTIVE
The chemical language of microbes is immensely diverse. By this very nature, we poorly understand the molecules that make up the text of their metabolic narratives. Many tools are available to decipher the microbe-microbe and microbe-host chemical interactions, with some of the most powerful being mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. These highly advanced and technical instruments are the Rosetta Stones of microbial chemical ecology because they enable translation of these molecular languages. Many impactful discoveries have been made with these tools demonstrating how microbial chemistry promotes health, disease, immunity, and metabolism of xenobiotics in both human and environmental systems (1–5). Evolving technologies in metabolomics enable more comprehensive assessments of the chemicals within a biological system. The aim of microbiome metabolomics is the large-scale quantitative and qualitative characterization of the small molecules (metabolites) present in a microbiome sample, which represent the functional outputs of the microbiome, its host, and/or its environment. However, challenges exist in interpreting the complex and highly technical data that metabolomics analyses provide. These challenges are becoming more pronounced as the capacity to generate in-depth MS and NMR data on microbial metabolomes grows. While it is becoming routine to capture this chemical information with high degrees of mass accuracy and depth, interpretation of its biological meaning, particularly relating to microbiomes, requires extensive insight and validation of the chemical species identified. Chemical ambiguity can be problematic in microbiome science, as even slight deviations in the structure of a compound can modify the function of a microbial metabolite. For example, chemical changes as subtle as unique epimers of bile acids induced by the microbiome can have dramatic effects on host immunity (2). Study of these microbiome-dependent or microbiome-altering metabolites and their biological activities continues to be an active area of research. There are challenges in this field however, because the identities of many microbial metabolites have not yet been elucidated (6–8) and the biological context begins with metabolite identification. Thus, there is a need for in-depth dialogue on metabolomics data between analytical chemists and the microbiome scientists who aim to interpret it from complex microbial communities.
THE CONCEPT OF METABOLITE IDENTIFICATION AND QUANTIFICATION EXISTS ON A SPECTRUM OF CERTAINTY
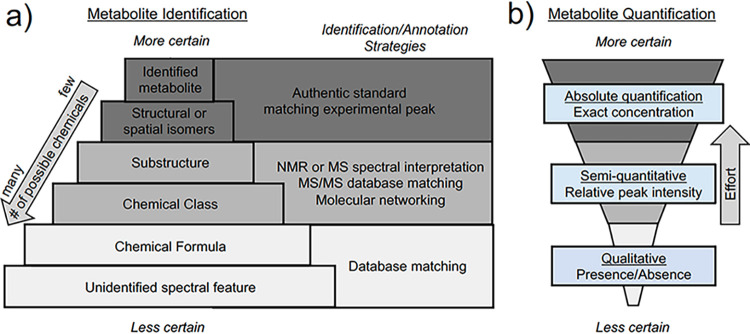
An important concept in analytical chemistry and metabolomics is the spectrum of certainty that exists when annotating and/or quantifying metabolites in a complex biological sample depending on the methods and analytical approaches used. Biologists are cautioned to not assume that a molecule identified from even the best metabolomics informatics pipelines is a known compound without some further validation. Confidence in chemical identification is dependent on the analytical platform, quality of the data, and access to chemical standards. Different analytical platforms (NMR or MS) employ various mechanisms for distinguishing one chemical from another (selectivity) and have differing ability to detect lower abundance chemicals (sensitivity). Depending on the metabolomics approach, metabolite identification can range from highly certain to merely a marginal association, and the biological interpretation will depend on this degree of analytical (un)certainty (Fig. 1). While the microbial dark matter is believed to be highly diverse, it is important to validate that an unknown feature in a metabolomics data set is a true compound derived from biological or environmental origins and not simply chemical or electronic noise due to the measurement itself (9). Traditionally, unambiguous chemical identifications require matching spectral data of an authentic standard with experimentally derived spectra and associated retention times, drift times, or chemical shifts (10). The use of in-house libraries derived from analyses of authentic reference standards (acquired under identical analytical conditions as study samples) is the preferred approach for high confidence identifications. However, authentic standards are usually not available for recently discovered chemicals or metabolic pathways, which limits the ability to validate the identity of these molecules and their associated functions. In some cases, such as in the absence of authentic standards, there is an additional need for the synthesis or isolation and characterization of metabolites to confirm their identity. Unambiguous identification of a single structure may require years of work and many different analytical techniques to rule out all other possibilities. These validation approaches are vital for reproducible microbiome research and dissemination of metabolite data from microbiomes across laboratories.
FIG 1.
The spectrum of uncertainty in identification and quantification of microbial metabolites. (a) Chemical annotations and identifications exist on a spectrum of certainty based on evidence provided and availability of authentic standards. Analysis of experimental samples generates many unidentified spectral features (accurate mass m/z mass spectrometry [MS1]) from which chemical formula can be determined with analysis of fine isotopic distributions. The number of possible chemicals any specific feature can represent decreases as more annotation strategies are applied. Identifying criteria can be obtained by ion fragmentation analyses (tandem mass spectrometry [MS/MS]), retention time analysis and others. Metabolite databases (PubChem, Metlin, HMDB, Massbank, and KEGG) can be used to annotate accurate mass spectral data collected from experimental samples to previously described library spectra (MS1 or MS/MS). Metabolites are not considered identified unless authentic standards are analyzed alongside experimental samples and at least two orthogonal identifying characteristics are matched from authentic chemical standard analysis (e.g., m/z, reverse transcription [RT], MS/MS) compared with experimental samples (10). Identification of metabolites (even with authentic standards) using untargeted metabolomic approaches may not be able to resolve stereoisomers, enantiomers, or exact spatial orientation of complex structures. Elucidation of these may require other targeted analytical approaches to determine exact structures, such as NMR. (b) Similarly, quantifying the concentration of a measured metabolite can be done on a spectrum of accuracy. Absolute quantification can be performed using standard curves prepared from authentic standards to generate units of concentration which can be compared across laboratories. If standards are not available, then raw peak abundances provide relative quantification (which will be variable across platforms) or the ability to perform qualitative analyses based on presence or absence.
Extensive validation of a chemical’s structure with labor-intensive analytical rigor may not always be required depending upon the goal(s) of a microbiome scientist. For example, one can obtain valuable information about a biological system without the need to identify the stereochemistry of a particular chemical group. Instead, annotation at the molecular family level can still be valuable, as one can assess overall chemical shifts from host or environmental perturbations. Microbiome scientists can further calculate diversity measures (both alpha- and beta-diversity) using metabolomics data, whether the metabolites measured are annotated or not, and these metrics can reveal important biological phenomena, such as resistance and resilience of a microbiome system. In fact, the calculation and interpretation of diversity measures, such as the Shannon index, for metabolomics data are a fitting setting to begin this important cross-field discourse. Recent advances in MS data analyses have furthered the biological information that can be mined from spectral data without knowledge of a metabolite annotation, such as molecular networking (11) and the characterization of chemical mass shifts between related molecules (12). Herein lies the need for dialogue between microbiome scientists and analytical chemists to learn from one another about what can be harnessed from metabolomics data and how to best interpret that information.
Similarly, quantification of metabolites in microbiome studies can be done from the level of precise concentrations to relative changes in abundance (Fig. 1). This too must be interpreted appropriately. It is important for the microbiome scientist to know that accurately quantifying a compound comes at the cost of measuring only a few compounds at a time. The thousands of metabolites measured in an untargeted metabolomics experiment will be quantified only in relative abundance across samples. However, this is a concept microbiome scientists are quite familiar with, creating a common place for dialogue and sharing of strategies for the analysis of multivariate and compositional data. Much of the data analyses approaches commonly used in omics studies were developed by ecologists and fine-tuned by microbial ecologists (13, 14). Thus, it is important for microbiome scientists to communicate to the more analytically inclined that many of these approaches can be applied to metabolomics data sets with the potential to enrich their interpretability, but also the data structural challenges that come with them (15). Another important concept in metabolomics is its untargeted or targeted nature. For example, a metabolite or a panel of metabolites identified as important in a discovery study can then be rigorously quantified in a confirmatory or replicating study, reducing the cost by narrowing down the number of targeted metabolite analyses. Parallels exist in microbiome science as well, such as quantitative PCR (qPCR)-based targeting of specific genes for rigorous quantification compared to more exploratory metagenomics methods for characterizing a microbiome’s genetic complement.
BRIDGING THE GAP
Analytical chemists, many of whom work in institutional core centers or academic labs generating metabolomics data, are highly encouraged to transparently discuss the challenges and intricacies of metabolomics data for microbiome samples at the initial stages of a project’s discussion and maintain a continuous dialogue through its completion. This includes not only explaining the spectrum of certainty described above and the per sample cost of targeted and untargeted analyses but also the feasibility of large sample numbers, optimization of metabolite extraction methods, statistical power, and appropriate use of laboratory consumables, some of which that are routine in microbiome science can induce troublesome polymer or salt contamination in analytical instruments. Many of these fruitful dialogues between analytical and microbiome scientists are already occurring, and they should be applauded for bridging this challenging chemical-microbiological gap, but many interactions still exist where the data generated from analytical instruments are not fully harnessed. Analytical chemists, many of whom are highly engaged and interested in the biology being revealed through their instruments, need resources from academic and federal institutions to provide the time and resources to work with microbiome scientists to avoid the unfortunate “data dump” that can lead to either incorrect biological interpretation or wasted effort on data that is never fully analyzed. In turn, many microbiome scientists are well versed in analytical methods, but most do not have the academic or technical training to interpret raw metabolomics data. Thus, educating oneself in the types of metabolomics platforms and instruments used is one step toward improving the dialogue. But perhaps most importantly, microbiome scientists and analytical chemists must work together to develop a project’s specific goals, which may sometimes not require extensive structural characterization. Some analytical chemists may be unaware of the statistical approaches that can reveal biological information from multi-omics data sets without resorting to the labor-intensive procedures required for accurate annotation and quantification of compounds. Diversity indices, machine learning approaches, and multi-omics integration can provide biological insights at the data set scale (16–18). If accurate quantification is required, and it often is, this must be explained to the analytical chemist so they can appropriately design an assay and provide a fair cost estimate.
THE NEED FOR “CHEMINFORMATICIANS” IN MICROBIOME SCIENCE
We propose that research institutions, core centers, and academic labs support and train “cheminformaticians” to bridge the gap between the highly specialized science of metabolomics and the urgent need to understand the role of microbiomes in human and environmental health. The desired dialogue described above is intensive and time-consuming, prohibitively so for many core centers and academic labs. Thus, including and training individuals with experience in interpreting the technical language of metabolomics data but with a microbiome/biological background is highly beneficial. This is akin to the early years of genomics in microbiology, when labs with bench microbiology experience began to need bioinformaticians to help interpret the genomic data that they were generating. We advocate for funding agencies, academic institutions and principal investigators (PIs) to advertise the need for and training of “cheminformaticians” in microbiome science to develop a workforce of analytical language translators who can bridge the gap between microbiome and analytical science. Together, this will lead to new and exciting metabolic narratives about the function of microbiomes.
ACKNOWLEDGMENT
We acknowledge the insights of Dr. Ewy Mathé of the National Center for Advancing Translational Sciences for help with this work.
Contributor Information
Robert A. Quinn, Email: quinnrob@msu.edu.
Jonathan L. Klassen, University of Connecticut
REFERENCES
- 1.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. 2013. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, Zheng Y, Longman RS, Rastinejad F, Devlin AS, Krout MR, Fischbach MA, Littman DR, Huh JR. 2019. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korenblum E, Dong Y, Szymanski J, Panda S, Jozwiak A, Massalha H, Meir S, Rogachev I, Aharoni A. 2020. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc Natl Acad Sci USA 117:3874–3883. doi: 10.1073/pnas.1912130117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vizcaino MI, Crawford JM. 2015. The colibactin warhead crosslinks DNA. Nat Chem 7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva RR, Dorrestein PC, Quinn RA. 2015. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci USA 112:12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peisl BYL, Schymanski EL, Wilmes P. 2018. Dark matter in host-microbiome metabolomics: tackling the unknowns–a review. Anal Chim Acta 1037:13–27. doi: 10.1016/j.aca.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, Downes M, Welch RD, Quinn M, Humphrey G, Panitchpakdi M, Weldon KC, Aksenov A, da Silva R, Avila-Pacheco J, Clish C, Bae S, Mallick H, Franzosa EA, Lloyd-Price J, Bussell R, Thron T, Nelson AT, Wang M, Leszczynski E, Vargas F, Gauglitz JM, Meehan MJ, Gentry E, Arthur TD, Komor AC, Poulsen O, Boland BS, Chang JT, Sandborn WJ, Lim M, Garg N, Lumeng JC, Xavier RJ, Kazmierczak BI, Jain R, Egan M, Rhee KE, Ferguson D, Raffatellu M, Vlamakis H, Haddad GG, et al. 2020. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahieu NG, Huang X, Chen Y-J, Patti GJ. 2014. Credentialing features: a platform to benchmark and optimize untargeted metabolomic methods. Anal Chem 86:9583–9589. doi: 10.1021/ac503092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. 2012. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109:1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann AC, Petras D, Quinn RA, Protsyuk I, Archer FI, Ransome E, Williams GJ, Bailey BA, Vermeij MJA, Alexandrov T, Dorrestein PC, Rohwer FL. 2017. Meta-mass shift chemical profiling of metabolomes from coral reefs. Proc Natl Acad Sci USA 114:11685–11690. doi: 10.1073/pnas.1710248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray J, Curtis J. 1957. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 14.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, Swenson TL, Van Goethem MW, Northen TR, Vazquez-Baeza Y, Wang M, Bokulich NA, Watters A, Song SJ, Bonneau R, Dorrestein PC, Knight R. 2019. Learning representations of microbe–metabolite interactions. Nat Methods 16:1306–1309. doi: 10.1038/s41592-019-0616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM, Price ND. 2019. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol 37:1217–1228. doi: 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- 17.Raghuvanshi R, Vasco K, Vázquez-Baeza Y, Jiang L, Morton JT, Li D, Gonzalez A, DeRight Goldasich L, Humphrey G, Ackermann G, Swafford AD, Conrad D, Knight R, Dorrestein PC, Quinn RA. 2020. High-resolution longitudinal dynamics of the cystic fibrosis sputum microbiome and metabolome through antibiotic therapy. mSystems 5:e00292-20. doi: 10.1128/mSystems.00292-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves CJ, Valdes AM, Spector TD, Menni C. 2018. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]