ABSTRACT
Current epidemics, such as AIDS or flu, and the emergence of new threatening pathogens, such as the one causing the current coronavirus disease 2019 (COVID-19) pandemic, represent major global health challenges. While vaccination is an important part of the arsenal to counter the spread of viral diseases, it presents limitations and needs to be complemented by efficient therapeutic solutions. Intricate knowledge of host-pathogen interactions is a powerful tool to identify host-dependent vulnerabilities that can be exploited to dampen viral replication. Such host-directed antiviral therapies are promising and are less prone to the development of drug-resistant viral strains. Here, we first describe proteomics-based strategies that allow the rapid characterization of host-pathogen interactions. We then discuss how such data can be exploited to help prioritize compounds with potential host-directed antiviral activity that can be tested in preclinical models.
KEYWORDS: host-directed therapies, systems biology, drug repurposing, host-pathogen interactions, proteomics
INTRODUCTION
Viral diseases represent a major cause of mortality across the world and can devastate our global health systems (1). The current coronavirus disease 2019 (COVID-19) pandemic illustrates the need to develop innovative tools that allow the rapid identification of potential therapies when new viral epidemics occur. Approaches exploiting mRNA have allowed the development of effective vaccines against COVID-19 at record speed (2–6). While vaccines are often the most efficient approach to control viral infections in the long term, it needs time to be administered to the general population, does not provide 100% antiviral efficiency, and cannot be administered to every patient (7). Therefore, it is critical to develop antiviral therapies as a measure to treat patients suffering from viral diseases when prevention fails. However, drug development is often a process too slow to be readily deployable to an emerging outbreak of disease. It is therefore key to establish new methods that facilitate the rapid identification of antiviral compounds to treat viral infections in vulnerable patients (8).
To date, most FDA-approved antiviral drugs target viral proteins involved in its replication cycle (9). However, a major challenge of these antiviral compounds is that they facilitate the common emergence of drug-resistant viral strains (10, 11) and are effective only against particular infections, hampering the possibility of pan-viral efficacies and repurposing against emerging new diseases. Viruses rely on the host cellular machinery to ensure their replication. Host-directed therapies (HDTs) take advantage of this dependency and attempt to disrupt the virus replication cycle by inhibiting essential host factors (12–17). Host genes that viruses rely on for survival have a low propensity to mutate within the treatment time frame, and adaptation of the virus to compensatory host factors likely occurs only under long-term selection pressure of a host-directed antiviral. Moreover, different viruses may share dependencies of specific host proteins or functions. Therefore, targeting the host proteins required for viral replication is a viable and innovative strategy that can avoid resistance and lead to potentially broad-spectrum therapeutics as families of viruses often exploit common cellular pathways and processes. However, HDTs require in-depth knowledge of virus-host interactions and their biological significance to virus replication. New approaches are aimed at identifying these cellular host factors, and proteomics approaches provide a powerful tool to elucidate the direct physical interactions between host and pathogen as well as perturbations to the host cells proteome caused by viral infection. Using functional genomics, the anti- or proviral function as well as the endogenous function of the identified host factors can be assessed. For example, the role of host factors on viral replication can be studied by knocking out individual host factors using CRISPR followed by viral infection. Only host factors that are identified as host dependency factors without altering critical endogenous molecular functions are suitable for pharmacological inhibition for HDT. Examples of successful HDT include the use of C-C chemokine receptor type 5 (CCR5) antagonists for the human immunodeficiency virus (HIV), cyclosporine for influenza A virus (IAV) and anti-claudin-1 and antioccludin monoclonal antibodies for hepatitis C virus (HCV). CCR5 antagonists inhibit HIV cell entry by blocking the interaction of the HIV type 1 (HIV-1) gp120 envelope glycoprotein with one of its host coreceptors, CCR5 (18, 19). Cyclosporine targets the interaction between influenza virus protein M1 and the host factor cyclophilin A, increasing the ability of cyclophilin A to inhibit M1 (20). It has also been shown that cyclosporine can inhibit nuclear export of viral RNA (21). HCV can bind to the tight junction protein claudin-1 and occludin to enter cells, and monoclonal antibodies against those two proteins have been shown to decrease HCV infection (22, 23). However, despite these examples, great effort has not been placed on therapeutically targeting the host in an effort to combat infectious disease.
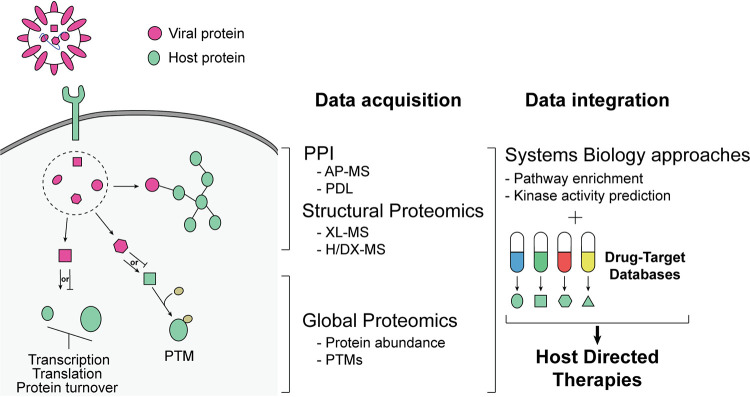
Recently, massive efforts to identify therapeutic strategies to manage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, have been undertaken. Understanding how SARS-CoV-2 interacts with the host cellular machinery and the identification of host dependency factors have led to the identification of promising drugs and investigational new drugs (INDs) that could be repurposed for HDT (24, 25). In this minireview, we will discuss how proteomics and systems biology approaches can streamline the discovery of antiviral host-directed therapies (Fig. 1). We will first give an overview of the most commonly used proteomics approaches to (i) map viral protein interactions with host proteins, (ii) map the global effects of viral infection on cellular signaling, and (iii) determine structure function. We will then describe how such proteomic data can be used to direct drug repurposing or new drug discovery.
FIG 1.
Schematic representation of antiviral host-directed therapy discovery using proteomics and systems biology approaches. AP-MS, affinity purification-mass spectrometry; PDL, proximity-dependent labeling; XL-MS, cross-linking mass spectrometry; H/DX-MS, hydrogen/deuterium exchange mass spectrometry; PTM, posttranslational modification.
PROTEOMICS APPROACHES TO IDENTIFY HOST FACTORS
The identification of putative key host factors is crucial to drive the development of innovative HDTs. Proteomics approaches present many advantages to do so, as they allow the unbiased mapping of protein-protein interactions (PPIs) and signaling perturbations due to infection, and ultimately enable structure-function studies. Coupled with other systems biology approaches, proteomic data are particularly suited to guide the discovery of innovative antiviral therapeutic strategies using HDTs (26). This section briefly gives an overview of mass spectrometry (MS) methods used to study virus-host interactions (reviewed extensively in references 27 and 28).
Proteomics approaches for protein-protein interaction mapping.
Affinity purification-mass spectrometry (AP-MS) and proximity-dependent labeling (PDL) permit the extensive mapping of PPI networks and can uncover host machinery that viral proteins associate with and potentially hijack. Such approaches have successfully been used to map Zika virus, herpesvirus, HIV-1, and SARS-CoV-2 virus-host PPIs and give critical insights into identifying important host factors for those viruses (29–40).
AP-MS is a widely used method to characterize the interactors of epitope-tagged bait proteins (41). Tagged viral proteins are expressed in host cells and purified with their bound interactors that are subsequently identified by MS. AP-MS allows the rapid, quantitative, and unbiased identification of multiple host interactors of a viral protein in a single experiment. While AP-MS presents some limitations, including the potential loss of weak interactions or the difficulty to recover membrane proteins, its ease of use and scalability permit it to generate extensive mapping of PPIs in a very short time. This is exemplified by the timely description of the SARS-CoV-2 interaction network only a few months after the pandemic outbreak (29), an immense undertaking that has been possible due to a massive collaborative effort (42). When designing AP-MS experiments, it is also important to consider that N- or C-terminal tagging can have an effect on protein function, requiring testing the functional status of tagged proteins. Finally, AP-MS also does not discriminate between direct and indirect interactions. However, this can be addressed by coupling AP-MS with cross-linking reagents, which allows the identification of direct interactions (see “Structure MS” section below) (43–45).
PDL uses enzymes fused to the protein of interest to biotinylate interactors in close proximity (46, 47). Two classes of enzymes are mainly used: promiscuous biotin protein ligases (BirA/BioID/TurboID) (48–50) or engineered ascorbic acid peroxidases (APEX) (51–53). In both cases, upon addition of their substrate, proteins in a 10- to 20-nm range are biotinylated and can be subsequently purified using streptavidin resin followed by identification using MS. The main advantages of the PDL approach, compared to AP-MS, resides in its ability to detect transient or weak interactions as well as membrane-bound partners. Moreover, due to the covalent labeling of interacting proteins by biotin, lysis and purification can be performed under stringent conditions to reduce background. PDL can also provide information on the subcellular localization of the identified PPI by the proximal labeling of organelle-specific proteins or specific use of spatial references (54).
Global proteomics approaches.
Viruses trigger global changes in the molecular landscape of infected cells in order to ensure their replication and evasion from the cell’s innate immunity. Measurements of cell signaling rewiring, regulation of protein levels, and changes to transcription upon viral infection provide a holistic understanding of the mechanisms at play during infection.
High-throughput characterization of posttranslational modifications (PTMs) especially offer valuable insight into the biology of viral infections, as PTMs have critical roles in many different aspects of infection that have both proviral (inhibiting interferon response or promoting viral replication and assembly) and antiviral consequences (degrading viral proteins through ubiquitination or inactivating them through changes in PTMs) (55, 56). Various studies also revealed how the dynamic interplay between different PTMs (such as phosphorylation, ubiquitination, and SUMOylation) regulates processes like pathogen-sensing pathways and innate immune signaling, underscoring the importance of characterizing the dynamics of these modifications upon infection (57–60). In particular, phosphoproteomic profiling of infected cells allows identification of changes in kinase activity over the time course of infection (61–63). Kinases represent attractive drug targets, as many kinase-regulating drugs and compounds have been developed for the treatment of various diseases, and numerous studies have shown that host kinases can regulate various steps of the virus replication cycle (16). Recent improvements in phosphopeptide enrichment using ion metal affinity or ion-exchange chromatography coupled to MS now allows for the routine identification of tens of thousands of phosphorylated peptides in a single experiment (64–66). Moreover, computational methods to quantify and localize the phosphorylation sites have also greatly improved, leading to the possibility to infer many kinases’ activity using growing kinase-substrate relationship databases (67–69). Other PTMs such as ubiquitination or acetylation can be assessed using MS (70–73), and an integrative analysis of multiple PTMs with protein abundance can provide insight into the cross talk between different signaling events (74). Changes in PTM abundance upon infection can lead to the identification of pathways or enzymes controlling such PTMs and targetable using HDTs.
Global protein abundance also provides valuable insights as host proteins can be up- or downregulated after virus infection and show deregulation of pathways that could be therapeutically actionable. Classic global proteomics approaches using mass spectrometry have successfully been used to characterize changes in protein levels following infection (74–76). Recent studies combined global proteomics approaches and thermal proteome profiling (TPP) to assess changes in protein levels and activity during SARS-CoV-2 and cytomegalovirus infection (77, 78). Thermal shift assay approaches, such as cellular thermal shift assays (CESTA), could also be used to understand antiviral compound’s molecular consequences and targets in relevant cell models (79).
Finally, viruses induce reorganization of subcellular structure and organelles of infected cells to promote replication. Such spatial changes in protein levels and organization can be probed using the methodologies discussed here and reviewed in detail by Jean Beltran et al. (80). Briefly, organelle fractionation followed by MS (81, 82) or proximity-based biotinylation (83–87) have been used in numerous studies to assess subcellular compartment content and are particularly suited to study how viruses reshape cell proteome landscape.
Structure MS.
Cross-linking mass spectrometry (XL-MS) uses chemical cross-linkers to covalently bridge reactive amino acid residues in close proximity (43, 88, 89). Pairs of linked peptides can then be identified by MS. This technique can therefore provide not only PPI information but structural insights on intra- and intermolecular interaction surfaces as well. Combined with dedicated structural approaches such as cryo-electron microscopy (cryo-EM) and structure predictions through deep learning systems such as AlphaFold (Deepmind), XL-MS facilitates and validates structure determination of challenging protein complexes (90, 91). Cross-linking can also be performed in combination with affinity purification of protein baits in order to determine direct interactions between copurifying proteins and improve AP-MS resolution (92).
Hydrogen/deuterium exchange mass spectrometry (H/DX-MS) allows the study of protein conformation for individual proteins or protein complexes. H/DX-MS measures changes in mass associated with the exchange between deuterium isotopes and hydrogens of the protein backbone amides (93, 94). The rate of the exchange is dependent on the conformational state of the protein and surface accessibility. Therefore, H/DX-MS is useful to probe folding dynamics, allosteric changes, protein conformation, and binding sites (95–99). Applications of H/DX-MS include the characterization of protein structure changes in response to PTMs or characterization of the binding of small molecules to proteins.
DRUG REPURPOSING STRATEGIES FOR DESIGNING ANTIVIRAL THERAPIES
Drug repurposing can offer an expedited timeline to bring host-directed antiviral therapies into clinical settings in a cost-effective and timely manner in comparison to traditional drug discovery, as designing a new compound, characterizing its efficacy, and demonstrating its safety can be a slow and resource-intensive process with a higher risk of failure. Successful applications of repurposing have occasionally been driven by observations of unexpected consequences of drugs, such as the discovery of minoxidil’s effects on hair growth when it was being tested for hypertension and the approval of sildenafil for erectile dysfunction treatment even though it was originally in trial for angina treatment (100, 101). Following advances in various omics technologies and development of computational methodologies, data-driven approaches are also increasingly being used to gain valuable insight into identifying the most suitable candidates for repurposing.
The proteomics approaches described above are suited for repurposing studies, as they enable the identification of host factors and pathways that are hijacked and rewired during infection. Combined with functional genetics to identify host dependency factors, the data obtained provide a rich list of potential targets for therapeutic interventions. Once targets are identified, several databases can be used to determine their druggability and identify existing chemical matter, ranging from preclinical compounds to INDs and FDA-approved drugs. These databases are also used to gain extensive information on relevant drugs, as they catalog their protein targets, structures, chemical properties, or clinical profiles (102). DrugBank is a frequently used database across many studies that make use of drug-target interaction networks and provides detailed information on various properties of more than 14,000 drugs, including their targets, pharmacodynamics, mechanism of action, and toxicity (103). Other valuable resources include DGIdb (the Drug-Gene Interaction Database), ChEMBL, and PharmGKB (the Pharmacogenomics Knowledgebase). DGIdb characterizes the druggable genome by curating information from various drug- and gene-related databases for more than 10,000 drugs and 40,000 genes and annotates their associated drug-target relationships (104). ChEMBL is based on the curation of more than 80,000 publications and offers a broader scope of nearly 2.1 million compounds by providing genomic and chemical data on bioactive drug-like small molecules (105). For studies with a pharmacogenomics focus, PharmGKB is a relevant resource as it curates knowledge on gene-drug associations and effects of genetic variation on drug response (106). Selection of a particular database is typically tailored to specific aims of the approach, and frequently multiple resources are integrated with each other to increase the coverage of drug-gene interaction networks built to connect host factors to relevant drugs.
Several studies have applied proteomics technologies to reveal virus-host interactions and searched for direct interactions between these host factors and drugs, with the aim to identify compounds or drugs that inhibit processes the virus relies on. Studies with this strategy include the work by Dapat et al. on respiratory syncytial virus, where the authors created its interaction network by integrating host factors identified across nine proteomics and seven transcriptomics studies (107). DrugBank was then used to query drugs targeting this network, revealing 177 FDA-approved drugs targeting 78 host proteins belonging to various categories such as anti-infective and anti-cancer agents. An AP-MS study by Gordon et al. identified 332 human proteins interacting with 26 SARS-CoV-2 proteins, revealing various complexes and processes hijacked by the virus, many of which are druggable targets (29). Chemoinformatics approaches and literature search by experts highlighted 69 compounds that target 62 proteins found in the PPI network. Translation inhibitors and molecules that target sigma-1 and sigma-2 receptors emerged as effective antiviral candidates in vitro. The translation inhibitor plitidepsin was subsequently characterized as a potent antiviral in vitro and in vivo with ∼30-fold-higher efficacy than remdesivir (108). Watanabe et al. generated an influenza-human protein interaction network via mass spectrometry and targeted the host factors with small interfering RNAs (siRNAs) to identify those involved in viral replication (109). Querying various drug databases for chemicals targeting these host factors led to the identification of 61 drugs, where further experimental studies highlighted GBF1 inhibitor golgicide A and JAK1 inhibitor ruxolitinib as viable candidates for anti-influenza therapy. Phosphoproteomics approaches (110) have also been successfully used to identify druggable host factors, especially those targeted by kinase inhibitors, which were shown to have antiviral effects against various viruses (111). Dynamics of kinase activity can be inferred computationally following characterization of sites phosphorylated upon infection, and kinases whose activity can explain virus-associated changes in phosphosites are then linked to drugs modulating their activity. This strategy has been applied to various viruses, which led to the nomination of GRK2 inhibitors for influenza (112), JNK1 inhibitors for Japanese encephalitis virus (113), and various inhibitors targeting casein kinase II, p38/MAPK (mitogen-activated protein kinase) signaling, and growth factor receptor signaling for SARS-CoV-2 (61, 114) treatment.
Network-based approaches have also been crucial for nominating drug repurposing candidates by integrating virus-host protein interaction, human protein-protein interaction, and drug-target interaction networks. One key idea behind these approaches is to consider interactions between host factors within a human PPI network and the downstream effects that dysregulation of a protein could cause based on these relationships. These integrative approaches can reveal indirect effects of proteins hijacked by viruses, and searching for drugs that can modulate these downstream effects can present additional druggable opportunities. Network-based studies have especially been valuable for SARS-CoV-2 research, offering resources to identify putative antiviral therapy options in a timely manner. For instance, the study by Zhou et al. generated a coronavirus-host interaction network by integrating various experimental resources that characterized interactions of six different viruses, including SARS-CoV-1 and Middle East respiratory syndrome (MERS)-CoV, to assemble the set of human proteins associated with these viruses (115). This study also generated a drug-target interaction network obtained from various drug databases exemplified above. To identify drugs with the potential to target SARS-CoV-2, Zhou et al. (115) used a “network proximity measure,” which computes distances between the host factors and proteins targeted by a given drug based on the human PPI network’s connectivity and prioritizes drugs whose targets are in close proximity to the host factors.
In another network-based study, Gysi et al. (116) implemented 12 different pipelines that rely on network proximity, network diffusion, and artificial intelligence principles, where they searched for drugs that can target the network of SARS-CoV-2 interactors identified by Gordon et al. (29). Proximity-based approaches focused on the distance between SARS-CoV-2 interactors and targets of drugs to rank drugs based on proximity scores, whereas diffusion methods ranked them based on the network similarity of drug targets and SARS-CoV-2 targets. Graph neural networks were implemented for artificial intelligence-based approaches. After characterizing the results of each individual method, the authors used a multimodal approach to combine different pipelines’ rankings together to generate a consensus list of predictions, taking advantage of the strengths of each individual pipeline. Sadegh et al.’s work offers researchers the opportunity to explore a virus-host interaction network from a drug target identification perspective by developing an online tool called CoVex (Coronavirus Explorer) that provides visualization of virus-host networks, drugs, and their targets (117, 118). They also implemented various network-based algorithms that can build bridges between host factors through neighboring, related proteins and connect these proteins to drugs targeting them, such as multi-Steiner tree, closeness centrality, and degree centrality. Currently, this resource includes PPI networks for SARS-CoV-1 (119, 120) and SARS-CoV-2 (29) with plans to add networks for additional viruses, and it allows the researchers to run custom queries that can start from viral or host proteins of interest to identify a set of drugs that can target them directly or indirectly generating actionable hypotheses on drug repositioning.
Studies highlighted here focused on drug repurposing strategies that have been applied to virus-associated data sets to underscore the utility of these algorithms in identifying antiviral therapies. For additional information on alternative repurposing approaches with a broader scope, we refer readers to several reviews that discuss properties of different algorithms and how various omics technologies and data sources are used for network-based drug repurposing (121–124). Application of these currently available drug repurposing algorithms or development of new approaches specifically tailored for viral proteomics data sets have the potential to identify viable targets for designing effective host-directed antiviral therapies both for viruses that currently lack effective treatments and emergent pathogens.
OPPORTUNITIES FOR PAN-VIRAL THERAPY DESIGN
Drug repurposing studies we discussed up to now have mostly focused on a single virus and characterizing its set of druggable targets. However, when we compare characteristics of different viruses, commonalities between proteins and biological processes that they interact with start to emerge, raising the possibility of identifying druggable host factors shared across multiple viruses (125). These comparisons could focus on a single family and characterize a set of related viruses in detail while taking their sequence homology and evolutionary differences into account. One such study comparing PPI networks of three coronaviruses revealed the set of interactions that are conserved across the three and trends that are shared between more closely related SARS-CoV-1 and SARS-CoV-2 but not identified in MERS-CoV (126). Drugs targeting proteins shared across viruses were also discussed to exemplify how these networks can be used to discover drugs that can be effective against multiple pathogens. Examples of other comparative analyses include ones that go beyond a single family, such as the study by Pichlmair et al., where they characterized human proteins interacting with 70 viral proteins from 30 different viruses using AP-MS (127) and the study by Bösl et al., which built an interaction network of 17 viruses by curating AP-MS and yeast two-hybrid studies (128). Focusing on a broader range of viruses revealed shared biological processes across distinct viruses that illustrate pathways crucial for viral infection and enabled comparisons between different groups of viruses, such as minus-strand single-stranded RNA [ssRNA(−)], plus-strand ssRNA [ssRNA(+)], double-stranded RNA (dsRNA), and double-stranded DNA (dsDNA) viruses. Additionally, Bösl et al. (128) queried DGIdb to identify drugs targeting proteins within this host-virus interaction network, highlighting putative pan-viral therapy options. Cataloging processes that are unique to individual viruses or shared across viruses will generate a valuable resource as we move forward with the design of host-directed antiviral therapies. It will help prioritize drug repurposing opportunities with the potential to be effective broad-acting antivirals and match a specific set of drugs to the most relevant group of viruses based on the intersection between their targets and shared host factors.
CONCLUSION
Proteomics and systems biology approaches are strikingly suited to provide a rapid and useful functional landscape of host-virus interactions. While requiring cross discipline expertise, the current available pipelines using these methods can identify important host factors involved in viral replication. Such insights are critical to develop efficient host-targeted antiviral therapies by allowing us to quickly nominate targetable host functions using repurposed drugs or by developing new compounds able to inhibit specific host-virus protein complex formation or activity. Importantly, as the description of different virus interactions with their host partner grows, it becomes clear that commonalities could be identified and exploited to target essential host factors shared by several viruses.
ACKNOWLEDGMENTS
We thank Manon Eckhardt for her valuable comments.
This work was funded by grants from the National Institutes of Health (P50AI150476, U19AI135972, and R01AI143292); by the Excellence in Research Award (ERA) from the Laboratory for Genomics Research (LGR), a collaboration between University of California, San Francisco (UCSF), University of California, Berkeley (UCB), and GSK (133122P); by funding from F. Hoffmann-La Roche and Vir Biotechnology, and gifts from QCRG philanthropic donors. This work was supported by the Defense Advanced Research Projects Agency (DARPA) under Cooperative Agreement HR0011-19-2-0020.
The views, opinions, and/or findings contained in this material are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government.
The Krogan laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. Nevan Krogan has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, and Interline Therapeutics. He is a shareholder in Tenaya Therapeutics, Maze Therapeutics, and Interline Therapeutics.
Contributor Information
Antoine Forget, Email: antoine.forget@ucsf.edu.
Ileana M. Cristea, Princeton University
REFERENCES
- 1.Bloom DE, Cadarette D. 2019. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol 10:549. doi: 10.3389/fimmu.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardi N, Hogan MJ, Porter FW, Weissman D. 2018. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov 17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, mRNA-1273 Study Group . 2020. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med 383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong W-P, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, et al. 2020. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D, Brachtendorf S, Lörks V, Sikorski J, Hilker R, Becker D, Eller A-K, Grützner J, Boesler C, Rosenbaum C, Kühnle M-C, Luxemburger U, Kemmer-Brück A, Langer D, Bexon M, Bolte S, Karikó K, Palanche T, Fischer B, Schultz A, Shi P-Y, Fontes-Garfias C, Perez JL, Swanson KA, Loschko J, Scully IL, Cutler M, Kalina W, Kyratsous CA, Cooper D, Dormitzer PR, Jansen KU, Türeci Ö. 2020. COVID-19 vaccine BNT162b1 elicits human antibody and T1 T cell responses. Nature 586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 6.Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Şahin U, Jansen KU. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 7.Trovato M, Sartorius R, D’Apice L, Manco R, De Berardinis P. 2020. Viral emerging diseases: challenges in developing vaccination strategies. Front Immunol 11:2130. doi: 10.3389/fimmu.2020.02130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meganck RM, Baric RS. 2021. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat Med 27:401–410. doi: 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri S, Symons JA, Deval J. 2018. Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Res 155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irwin KK, Renzette N, Kowalik TF, Jensen JD. 2016. Antiviral drug resistance as an adaptive process. Virus Evol 2:vew014. doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locarnini S, Bowden S. 2010. Drug resistance in antiviral therapy. Clin Liver Dis 14:439–459. doi: 10.1016/j.cld.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 12.McDougall WM, Perreira JM, Reynolds EC, Brass AL. 2018. CRISPR genetic screens to discover host-virus interactions. Curr Opin Virol 29:87–100. doi: 10.1016/j.coviro.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Gebre M, Nomburg JL, Gewurz BE. 2018. CRISPR-Cas9 genetic analysis of virus-host interactions. Viruses 10:55. doi: 10.3390/v10020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brito AF, Pinney JW. 2017. Protein-protein interactions in virus-host systems. Front Microbiol 8:1557. doi: 10.3389/fmicb.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wakker SI, Fischer MJE, Oosting RS. 2017. New drug-strategies to tackle viral-host interactions for the treatment of influenza virus infections. Eur J Pharmacol 809:178–190. doi: 10.1016/j.ejphar.2017.05.038. [DOI] [PubMed] [Google Scholar]
- 16.Kumar N, Sharma S, Kumar R, Tripathi BN, Barua S, Ly H, Rouse BT. 2020. Host-directed antiviral therapy. Clin Microbiol Rev 33:e00168-19. doi: 10.1128/CMR.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad M, Ranjan K, Brar B, Shah I, Lalmbe U, Manimegalai J, Vashisht B, Gaury M, Kumar P, Khurana SK, Prasad G, Rawat J, Yadav V, Kumar S, Rao R. 2017. Virus-host interactions: new insights and advances in drug development against viral pathogens. Curr Drug Metab 18:942–970. doi: 10.2174/1389200218666170925115132. [DOI] [PubMed] [Google Scholar]
- 18.Roche M, Salimi H, Duncan R, Wilkinson BL, Chikere K, Moore MS, Webb NE, Zappi H, Sterjovski J, Flynn JK, Ellett A, Gray LR, Lee B, Jubb B, Westby M, Ramsland PA, Lewin SR, Payne RJ, Churchill MJ, Gorry PR. 2013. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10:43. doi: 10.1186/1742-4690-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman-Blum SS, Fung HB, Bandres JC. 2008. Maraviroc: a CCR5-receptor antagonist for the treatment of HIV-1 infection. Clin Ther 30:1228–1250. doi: 10.1016/S0149-2918(08)80048-3. [DOI] [PubMed] [Google Scholar]
- 20.Ma C, Li F, Musharrafieh RG, Wang J. 2016. Discovery of cyclosporine A and its analogs as broad-spectrum anti-influenza drugs with a high in vitro genetic barrier of drug resistance. Antiviral Res 133:62–72. doi: 10.1016/j.antiviral.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Zhao Z, Li Z, Xu C, Sun L, Chen J, Liu W. 2012. Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS One 7:e37277. doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu Y, Shirasago Y, Kondoh M, Suzuki T, Wakita T, Hanada K, Yagi K, Fukasawa M. 2018. Monoclonal antibodies against occludin completely prevented hepatitis C virus infection in a mouse model. J Virol 92:e02258-17. doi: 10.1128/JVI.02258-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita M, Iida M, Tada M, Shirasago Y, Fukasawa M, Nagase S, Watari A, Ishii-Watabe A, Yagi K, Kondoh M. 2015. Discovery of anti-claudin-1 antibodies as candidate therapeutics against hepatitis C virus. J Pharmacol Exp Ther 353:112–118. doi: 10.1124/jpet.114.217653. [DOI] [PubMed] [Google Scholar]
- 24.Reuschl A-K, Thorne LG, Zuliani-Alvarez L, Bouhaddou M, Obernier K, Hiatt J, Soucheray M, Turner J, Fabius JM, Nguyen GT, Swaney DL, Rosales R, White KM, Avilés P, Kirby IT, Melnyk JE, Shi Y, Zhang Z, Shokat KM, García-Sastre A, Jolly C, Towers GJ, Krogan NJ. 2021. Host-directed therapies against early-lineage SARS-CoV-2 retain efficacy against B.1.1.7 variant. bioRxiv 10.1101/2021.01.24.427991. [DOI]
- 25.Varona JF, Landete P, Lopez-Martin JA, Estrada V, Paredes R, Guisado-Vasco P, de Orueta LF, Torralba M, Fortún J, Vates R, Barberán J, Clotet B, Ancochea J, Carnevali D, Cabello N, Porras L, Gijón P, Monereo A, Abad D, Zúñiga S, Sola I, Rodon J, Izquierdo-Useros N, Fudio S, Pontes MJ, de Rivas B, Girón de Velasco P, Sopesén B, Nieto A, Gómez J, Avilés P, Lubomirov R, White KM, Rosales R, Yildiz S, Reuschl A-K, Thorne LG, Jolly C, Towers GJ, Zuliani-Alvarez L, Bouhaddou M, Obernier K, Enjuanes L, Fernández-Sousa JM, Plitidepsin – COVID - 19 Study Group, Krogan NJ, Jimeno JM, García-Sastre A. 2021. Plitidepsin has a positive therapeutic index in adult patients with COVID-19 requiring hospitalization. medRxiv 10.1101/2021.05.25.21257505.
- 26.Eckhardt M, Hultquist JF, Kaake RM, Hüttenhain R, Krogan NJ. 2020. A systems approach to infectious disease. Nat Rev Genet 21:339–354. doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas P, Muralidharan M, Krogan NJ, Kaake RM, Hüttenhain R. 2021. Proteomic approaches to study SARS-CoV-2 biology and COVID-19 pathology. J Proteome Res 20:1133–1152. doi: 10.1021/acs.jproteome.0c00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards AL, Eckhardt M, Krogan NJ. 2021. Mass spectrometry-based protein-protein interaction networks for the study of human diseases. Mol Syst Biol 17:e8792. doi: 10.15252/msb.20188792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheerathodi MR, Meckes DG, Jr.. 2020. BioID combined with mass spectrometry to study herpesvirus protein-protein interaction networks. Methods Mol Biol 2060:327–341. doi: 10.1007/978-1-4939-9814-2_19. [DOI] [PubMed] [Google Scholar]
- 31.Coyaud E, Ranadheera C, Cheng D, Gonçalves J, Dyakov BJA, Laurent EMN, St-Germain J, Pelletier L, Gingras A-C, Brumell JH, Kim PK, Safronetz D, Raught B. 2018. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics 17:2242–2255. doi: 10.1074/mcp.TIR118.000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rider MA, Cheerathodi MR, Hurwitz SN, Nkosi D, Howell LA, Tremblay DC, Liu X, Zhu F, Meckes DG, Jr. . 2018. The interactome of EBV LMP1 evaluated by proximity-based BioID approach. Virology 516:55–70. doi: 10.1016/j.virol.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georges AA, Frappier L. 2017. Affinity purification-mass spectroscopy methods for identifying Epstein-Barr virus-host interactions. Methods Mol Biol 1532:79–92. doi: 10.1007/978-1-4939-6655-4_5. [DOI] [PubMed] [Google Scholar]
- 34.Le Sage V, Cinti A, Valiente-Echeverría F, Mouland AJ. 2015. Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol J 12:138. doi: 10.1186/s12985-015-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics 9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristea IM, Carroll J-WN, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem 281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 37.Mayer D, Molawi K, Martínez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, García-Sastre A, Schwemmle M. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res 6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamayoshi S, Noda T, Ebihara H, Goto H, Morikawa Y, Lukashevich IS, Neumann G, Feldmann H, Kawaoka Y. 2008. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 3:168–177. doi: 10.1016/j.chom.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. 2011. Global landscape of HIV-human protein complexes. Nature 481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Jacobs EY, Greco TM, Mohammed KD, Tong T, Keegan S, Binley JM, Cristea IM, Fenyö D, Rout MP, Chait BT, Muesing MA. 2016. HIV-host interactome revealed directly from infected cells. Nat Microbiol 1:16068. doi: 10.1038/nmicrobiol.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang I-F. 2006. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 6:6158–6166. doi: 10.1002/pmic.200600225. [DOI] [PubMed] [Google Scholar]
- 42.Fabius JM, Krogan NJ. 2021. Creating collaboration by breaking down scientific barriers. Cell 184:2271–2275. doi: 10.1016/j.cell.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu C, Huang L. 2018. Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal Chem 90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang S, Hawkridge AM, Johnson KL, Muddiman DC, Prevelige PE, Jr. . 2006. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass spectrometry. J Proteome Res 5:370–377. doi: 10.1021/pr050356f. [DOI] [PubMed] [Google Scholar]
- 45.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han S, Li J, Ting AY. 2018. Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 50:17–23. doi: 10.1016/j.conb.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trinkle-Mulcahy L. 2019. Recent advances in proximity-based labeling methods for interactome mapping. F1000Res 8:F1000 Faculty Rev-135. doi: 10.12688/f1000research.16903.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim DI, Jensen SC, Noble KA, Birendra KC, Roux KH, Motamedchaboki K, Roux KJ. 2016. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. 2016. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc 11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lobingier BT, Hüttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, Krogan NJ. 2017. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169:350–360.e12. doi: 10.1016/j.cell.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar R, Mehta D, Mishra N, Nayak D, Sunil S. 2020. Role of host-mediated post-translational modifications (PTMs) in RNA virus pathogenesis. Int J Mol Sci 22:323. doi: 10.3390/ijms22010323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valerdi KM, Hage A, van Tol S, Rajsbaum R, Giraldo MI. 2021. The role of the host ubiquitin system in promoting replication of emergent viruses. Viruses 13:369. doi: 10.3390/v13030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Y, He C, Wang L, Ge B. 2017. Post-translational regulation of antiviral innate signaling. Eur J Immunol 47:1414–1426. doi: 10.1002/eji.201746959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, Qian C, Cao X. 2016. Post-translational modification control of innate immunity. Immunity 45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 59.Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol 11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 60.Chelbi-Alix MK, Thibault P. 2021. Crosstalk between SUMO and ubiquitin-like proteins: implication for antiviral defense. Front Cell Dev Biol 9:671067. doi: 10.3389/fcell.2021.671067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM, Batra J, Richards AL, Stevenson E, Gordon DE, Rojc A, Obernier K, Fabius JM, Soucheray M, Miorin L, Moreno E, Koh C, Tran QD, Hardy A, Robinot R, Vallet T, Nilsson-Payant BE, Hernandez-Armenta C, Dunham A, Weigang S, Knerr J, Modak M, Quintero D, Zhou Y, Dugourd A, Valdeolivas A, Patil T, Li Q, Hüttenhain R, Cakir M, Muralidharan M, Kim M, Jang G, Tutuncuoglu B, Hiatt J, Guo JZ, Xu J, Bouhaddou S, Mathy CJP, Gaulton A, Manners EJ, Félix E, et al. 2020. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giansanti P, Strating JRPM, Defourny KAY, Cesonyte I, Bottino AMS, Post H, Viktorova EG, Ho VQT, Langereis MA, Belov GA, Nolte-‘t Hoen ENM, Heck AJR, van Kuppeveld FJM. 2020. Dynamic remodelling of the human host cell proteome and phosphoproteome upon enterovirus infection. Nat Commun 11:4332. doi: 10.1038/s41467-020-18168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, Pichlmair A. 2018. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561:253–257. doi: 10.1038/s41586-018-0484-5. [DOI] [PubMed] [Google Scholar]
- 64.Qiu W, Evans CA, Landels A, Pham TK, Wright PC. 2020. Phosphopeptide enrichment for phosphoproteomic analysis - a tutorial and review of novel materials. Anal Chim Acta 1129:158–180. doi: 10.1016/j.aca.2020.04.053. [DOI] [PubMed] [Google Scholar]
- 65.Beltran L, Cutillas PR. 2012. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids 43:1009–1024. doi: 10.1007/s00726-012-1288-9. [DOI] [PubMed] [Google Scholar]
- 66.Fíla J, Honys D. 2012. Enrichment techniques employed in phosphoproteomics. Amino Acids 43:1025–1047. doi: 10.1007/s00726-011-1111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugiyama N, Imamura H, Ishihama Y. 2019. Large-scale discovery of substrates of the human kinome. Sci Rep 9:10503. doi: 10.1038/s41598-019-46385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ochoa D, Jarnuczak AF, Viéitez C, Gehre M, Soucheray M, Mateus A, Kleefeldt AA, Hill A, Garcia-Alonso L, Stein F, Krogan NJ, Savitski MM, Swaney DL, Vizcaíno JA, Noh K-M, Beltrao P. 2020. The functional landscape of the human phosphoproteome. Nat Biotechnol 38:365–373. doi: 10.1038/s41587-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hijazi M, Smith R, Rajeeve V, Bessant C, Cutillas PR. 2020. Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat Biotechnol 38:493–502. doi: 10.1038/s41587-019-0391-9. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Fu Y, Wang Q, Li M, Zhou Z, Dabbagh D, Fu C, Zhang H, Li S, Zhang T, Gong J, Kong X, Zhai W, Su J, Sun J, Zhang Y, Yu X-F, Shao Z, Zhou F, Wu Y, Tan X. 2019. Proteomic profiling of HIV-1 infection of human CD4 T cells identifies PSGL-1 as an HIV restriction factor. Nat Microbiol 4:813–825. doi: 10.1038/s41564-019-0372-2. [DOI] [PubMed] [Google Scholar]
- 71.Fu B, Wang L, Ding H, Schwamborn JC, Li S, Dorf ME. 2015. TRIM32 senses and restricts influenza A virus by ubiquitination of PB1 polymerase. PLoS Pathog 11:e1004960. doi: 10.1371/journal.ppat.1004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murray LA, Sheng X, Cristea IM. 2018. Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat Commun 9:4967. doi: 10.1038/s41467-018-07179-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Zheng H, Zhu J, Dong Q, Wang J, Fan H, Chen Y, Zhang X, Han X, Li Q, Lu J, Tong Y, Chen Z. 2021. Ubiquitin-modified proteome of SARS-CoV-2-infected host cells reveals insights into virus-host interaction and pathogenesis. J Proteome Res 20:2224–2239. doi: 10.1021/acs.jproteome.0c00758. [DOI] [PubMed] [Google Scholar]
- 74.Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, Haas DA, Huang Y, Oubraham L, Wang A, Hamad MS, Piras A, Hansen FM, Tanzer MC, Paron I, Zinzula L, Engleitner T, Reinecke M, Lavacca TM, Ehmann R, Wölfel R, Jores J, Kuster B, Protzer U, Rad R, Ziebuhr J, Thiel V, Scaturro P, Mann M, Pichlmair A. 2021. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 75.Greenwood EJ, Matheson NJ, Wals K, van den Boomen DJ, Antrobus R, Williamson JC, Lehner PJ. 2016. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. Elife 5:e18296. doi: 10.7554/eLife.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, Cinatl J, Münch C. 2020. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selkrig J, Stanifer M, Mateus A, Mitosch K, Barrio-Hernandez I, Rettel M, Kim H, Voogdt CGP, Walch P, Kee C, Kurzawa N, Stein F, Potel C, Jarzab A, Kuster B, Bartenschlager R, Boulant S, Beltrao P, Typas A, Savitski MM. 2021. SARS-CoV-2 infection remodels the host protein thermal stability landscape. Mol Syst Biol 17:e10188. doi: 10.15252/msb.202010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM. 2020. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Commun 11:806. doi: 10.1038/s41467-020-14586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundbäck T, Nordlund P, Martinez Molina D. 2014. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 80.Jean Beltran PM, Cook KC, Cristea IM. 2017. Exploring and exploiting proteome organization during viral infection. J Virol 91:e00268-17. doi: 10.1128/JVI.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Christoforou A, Mulvey CM, Breckels LM, Geladaki A, Hurrell T, Hayward PC, Naake T, Gatto L, Viner R, Martinez Arias A, Lilley KS. 2016. A draft map of the mouse pluripotent stem cell spatial proteome. Nat Commun 7:8992. doi: 10.1038/ncomms9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itzhak DN, Tyanova S, Cox J, Borner GH. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. Elife 5:e16950. doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim BR, Coyaud E, Laurent EMN, St-Germain J, Van de Laar E, Tsao M-S, Raught B, Moghal N. 2017. Identification of the SOX2 interactome by BioID reveals EP300 as a mediator of SOX2-dependent squamous differentiation and lung squamous cell carcinoma growth. Mol Cell Proteomics 16:1864–1888. doi: 10.1074/mcp.M116.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Antonicka H, Lin Z-Y, Janer A, Aaltonen MJ, Weraarpachai W, Gingras A-C, Shoubridge EA. 2020. A high-density human mitochondrial proximity interaction network. Cell Metab 32:479–497.e9. doi: 10.1016/j.cmet.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 85.Redwine WB, DeSantis ME, Hollyer I, Htet ZM, Tran PT, Swanson SK, Florens L, Washburn MP, Reck-Peterson SL. 2017. The human cytoplasmic dynein interactome reveals novel activators of motility. Elife 6:e28257. doi: 10.7554/eLife.28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu L, Doray B, Kornfeld S. 2018. Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc Natl Acad Sci USA 115:8984–8989. doi: 10.1073/pnas.1810291115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoffman AM, Chen Q, Zheng T, Nicchitta CV. 2019. Heterogeneous translational landscape of the endoplasmic reticulum revealed by ribosome proximity labeling and transcriptome analysis. J Biol Chem 294:8942–8958. doi: 10.1074/jbc.RA119.007996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iacobucci C, Götze M, Sinz A. 2020. Cross-linking/mass spectrometry to get a closer view on protein interaction networks. Curr Opin Biotechnol 63:48–53. doi: 10.1016/j.copbio.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 89.Piotrowski C, Sinz A. 2018. Structural investigation of proteins and protein complexes by chemical cross-linking/mass spectrometry. Adv Exp Med Biol 1105:101–121. doi: 10.1007/978-981-13-2200-6_8. [DOI] [PubMed] [Google Scholar]
- 90.Prchal J, Sýs J, Junková P, Lipov J, Ruml T. 2020. Interaction interface of Mason-Pfizer monkey virus matrix and envelope proteins. J Virol 94:e01146-20. doi: 10.1128/JVI.01146-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meyer NL, Hu G, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich DS, Trzynka A, David L, Stagg SM, Chapman MS. 2019. Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR. Elife 8:e44707. doi: 10.7554/eLife.44707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makowski MM, Willems E, Jansen PWTC, Vermeulen M. 2016. Cross-linking immunoprecipitation-MS (xIP-MS): topological analysis of chromatin-associated protein complexes using single affinity purification. Mol Cell Proteomics 15:854–865. doi: 10.1074/mcp.M115.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masson GR, Burke JE, Ahn NG, Anand GS, Borchers C, Brier S, Bou-Assaf GM, Engen JR, Englander SW, Faber J, Garlish R, Griffin PR, Gross ML, Guttman M, Hamuro Y, Heck AJR, Houde D, Iacob RE, Jørgensen TJD, Kaltashov IA, Klinman JP, Konermann L, Man P, Mayne L, Pascal BD, Reichmann D, Skehel M, Snijder J, Strutzenberg TS, Underbakke ES, Wagner C, Wales TE, Walters BT, Weis DD, Wilson DJ, Wintrode PL, Zhang Z, Zheng J, Schriemer DC, Rand KD. 2019. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat Methods 16:595–602. doi: 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Konermann L, Pan J, Liu Y-H. 2011. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev 40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 95.Vadas O, Jenkins ML, Dornan GL, Burke JE. 2017. Using hydrogen-deuterium exchange mass spectrometry to examine protein-membrane interactions. Methods Enzymol 583:143–172. doi: 10.1016/bs.mie.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 96.Harrison RA, Engen JR. 2016. Conformational insight into multi-protein signaling assemblies by hydrogen-deuterium exchange mass spectrometry. Curr Opin Struct Biol 41:187–193. doi: 10.1016/j.sbi.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balasubramaniam D, Komives EA. 2013. Hydrogen-exchange mass spectrometry for the study of intrinsic disorder in proteins. Biochim Biophys Acta 1834:1202–1209. doi: 10.1016/j.bbapap.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. 2011. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics 8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VL, Jr. . 2003. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci USA 100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. 2019. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 101.Ashburn TT, Thor KB. 2004. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 102.Masoudi-Sobhanzadeh Y, Omidi Y, Amanlou M, Masoudi-Nejad A. 2020. Drug databases and their contributions to drug repurposing. Genomics 112:1087–1095. doi: 10.1016/j.ygeno.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 103.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. 2018. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Freshour SL, Kiwala S, Cotto KC, Coffman AC, McMichael JF, Song JJ, Griffith M, Griffith OL, Wagner AH. 2021. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res 49:D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, Magariños MP, Mosquera JF, Mutowo P, Nowotka M, Gordillo-Marañón M, Hunter F, Junco L, Mugumbate G, Rodriguez-Lopez M, Atkinson F, Bosc N, Radoux CJ, Segura-Cabrera A, Hersey A, Leach AR. 2019. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. 2012. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dapat C, Oshitani H. 2016. Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Expert Rev Anti Infect Ther 14:285–297. doi: 10.1586/14787210.2016.1141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.White KM, Rosales R, Yildiz S, Kehrer T, Miorin L, Moreno E, Jangra S, Uccellini MB, Rathnasinghe R, Coughlan L, Martinez-Romero C, Batra J, Rojc A, Bouhaddou M, Fabius JM, Obernier K, Dejosez M, Guillén MJ, Losada A, Avilés P, Schotsaert M, Zwaka T, Vignuzzi M, Shokat KM, Krogan NJ, García-Sastre A. 2021. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watanabe T, Kawakami E, Shoemaker JE, Lopes TJS, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morris MK, Chi A, Melas IN, Alexopoulos LG. 2014. Phosphoproteomics in drug discovery. Drug Discov Today 19:425–432. doi: 10.1016/j.drudis.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 111.Schor S, Einav S. 2018. Repurposing of kinase inhibitors as broad-spectrum antiviral drugs. DNA Cell Biol 37:63–69. doi: 10.1089/dna.2017.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yángüez E, Hunziker A, Dobay MP, Yildiz S, Schading S, Elshina E, Karakus U, Gehrig P, Grossmann J, Dijkman R, Schmolke M, Stertz S. 2018. Phosphoproteomic-based kinase profiling early in influenza virus infection identifies GRK2 as antiviral drug target. Nat Commun 9:3679. doi: 10.1038/s41467-018-06119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ye J, Zhang H, He W, Zhu B, Zhou D, Chen Z, Ashraf U, Wei Y, Liu Z, Fu ZF, Chen H, Cao S. 2016. Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Sci Signal 9:ra98. doi: 10.1126/scisignal.aaf5132. [DOI] [PubMed] [Google Scholar]
- 114.Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J. 2020. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell 80:164–174.e4. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. 2020. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gysi DM, Do Valle Í, Zitnik M, Ameli A, Gan X, Varol O, Sanchez H, Baron RM, Ghiassian D, Loscalzo J, Barabási A-L. 2020. Network medicine framework for identifying drug repurposing opportunities for COVID-19. ArXiv arXiv:2004.07229 [q-bio.MN]. [DOI] [PMC free article] [PubMed]
- 117.Sadegh S, Matschinske J, Blumenthal DB, Galindez G, Kacprowski T, List M, Nasirigerdeh R, Oubounyt M, Pichlmair A, Rose TD, Salgado-Albarrán M, Späth J, Stukalov A, Wenke NK, Yuan K, Pauling JK, Baumbach J. 2020. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat Commun 11:3518. doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Matschinske J, Salgado-Albarrán M, Sadegh S, Bongiovanni D, Baumbach J, Blumenthal DB. 2020. Individuating possibly repurposable drugs and drug targets for COVID-19 treatment through hypothesis-driven systems medicine using CoVex. Assay Drug Dev Technol 18:348–355. doi: 10.1089/adt.2020.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pfefferle S, Schöpf J, Kögl M, Friedel CC, Müller MA, Carbajo-Lozoya J, Stellberger T, von Dall’Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Züst R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel H-J, Schwegmann-Wessels C, Pöhlmann S, Haas J, Drosten C, von Brunn A. 2011. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guirimand T, Delmotte S, Navratil V. 2015. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res 43:D583–D587. doi: 10.1093/nar/gku1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lotfi Shahreza M, Ghadiri N, Mousavi SR, Varshosaz J, Green JR. 2018. A review of network-based approaches to drug repositioning. Brief Bioinform 19:878–892. doi: 10.1093/bib/bbx017. [DOI] [PubMed] [Google Scholar]
- 122.Conte F, Fiscon G, Licursi V, Bizzarri D, D'Antò T, Farina L, Paci P. 2020. A paradigm shift in medicine: a comprehensive review of network-based approaches. Biochim Biophys Acta Gene Regul Mech 1863:194416. doi: 10.1016/j.bbagrm.2019.194416. [DOI] [PubMed] [Google Scholar]
- 123.Wu Z, Wang Y, Chen L. 2013. Network-based drug repositioning. Mol Biosyst 9:1268–1281. doi: 10.1039/c3mb25382a. [DOI] [PubMed] [Google Scholar]
- 124.Alaimo S, Pulvirenti A2. 2019. Network-based drug repositioning: approaches, resources, and research directions. Methods Mol Biol 1903:97–113. doi: 10.1007/978-1-4939-8955-3_6. [DOI] [PubMed] [Google Scholar]
- 125.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, André P, Lotteau V. 2012. New horizons for antiviral drug discovery from virus-host protein interaction networks. Curr Opin Virol 2:606–613. doi: 10.1016/j.coviro.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 126.Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, Jureka AS, Obernier K, Guo JZ, Batra J, Kaake RM, Weckstein AR, Owens TW, Gupta M, Pourmal S, Titus EW, Cakir M, Soucheray M, McGregor M, Cakir Z, Jang G, O’Meara MJ, Tummino TA, Zhang Z, Foussard H, Rojc A, Zhou Y, Kuchenov D, Hüttenhain R, Xu J, Eckhardt M, Swaney DL, Fabius JM, Ummadi M, Tutuncuoglu B, Rathore U, Modak M, Haas P, Haas KM, Naing ZZC, Pulido EH, Shi Y, Barrio-Hernandez I, Memon D, Petsalaki E, Dunham A, Marrero MC, Burke D, Koh C, Vallet T, Silvas JA, et al. 2020. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle C-A, Goncalves A, Bürckstümmer T, Müller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G. 2012. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 128.Bösl K, Ianevski A, Than TT, Andersen PI, Kuivanen S, Teppor M, Zusinaite E, Dumpis U, Vitkauskiene A, Cox RJ, Kallio-Kokko H, Bergqvist A, Tenson T, Merits A, Oksenych V, Bjørås M, Anthonsen MW, Shum D, Kaarbø M, Vapalahti O, Windisch MP, Superti-Furga G, Snijder B, Kainov D, Kandasamy RK. 2019. Common nodes of virus-host interaction revealed through an integrated network analysis. Front Immunol 10:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]