Abstract
The COVID-19 pandemic has exacerbated our society’s tremendous health equity gap. Disadvantaged populations have been disproportionally affected by COVID-19, lacking access to affordable testing, a known effective tool for preventing viral spread, hospitalizations, and deaths. Here, we describe COVID-19 Low-cost Optodiagnostic for Rapid testing (COLOR), a colorimetric biosensor fabricated on cotton swabs using gold nanoparticles modified with human angiotensin-converting enzyme 2 (ACE2), which costs 15¢ to produce and detects SARS-CoV-2 within 5 minutes. COLOR detected very low viral particle loads (limit of detection: 0.154 pg mL−1 of SARS-CoV-2 spike protein) and its color intensity correlated with the cycle threshold (Ct) values obtained using RT-PCR. The performance of COLOR was assessed using 100 nasopharyngeal/oropharyngeal (NP/OP) clinical samples, yielding sensitivity, specificity, and accuracy values of 96%, 84%, and 90%, respectively. In summary, each COLOR test can be manufactured for 15¢ and presents rapid minute-timescale detection of SARS-CoV-2, thus providing a solution to enable high-frequency testing, particularly in low-resource communities.
Keywords: optodiagnostic, low-cost diagnostic, cotton swabs, gold nanoparticles, colorimetric sensor, SARS-CoV-2, COVID-19
Graphical Abstract
INTRODUCTION
COVID-19 has killed over 4.55 million people (1,2) and has disproportionally affected individuals living in resource-limited settings, disadvantaged communities, and low-income countries. High-frequency testing represents an excellent tool for outbreak and viral spread prevention that can only be realized with tests that are accessible, low-cost, and rapid. Such a technology could have a major public health impact broadly, and in low-income countries in particular. However, the most widely used tests today, such as RT-PCR, are slow, relatively expensive, and require highly skilled workers and appropriate lab space.
Colorimetric technologies display properties that may help overcome these issues. They are easy to prepare and use, rapid, portable, and can utilize metallic nanoparticles whose optical properties have been well established and characterized.3 Such tests hold potential for deployment in remote locations, and are amenable for use both at the point-of-care and at home without the need for sophisticated equipment.4 Li et al.5 described a rapid colorimetric test for the simultaneous detection, in human blood, of immunoglobulin M (IgM) and IgG antibodies against SARS-CoV-2, obtaining a sensitivity of 88.66% and specificity of 90.63%. Bartolomeo and collaborators4 developed a colorimetric sensor using gold nanoparticles and antibodies toward three SARS-CoV-2 proteins (e.g., Spike, Envelope, and Membrane proteins), allowing viral detection through optical density measurements that yielded a sensitivity and specificity greater than 95%. However, more affordable technologies using accessible equipment that does not require skilled personnel to operate are needed to enable mass testing at the point of care. Smartphones have been used both as a detector and processor of portable biochemical assays, revolutionizing the field of mobile healthcare by empowering citizens to perform rapid screening tests while obviating the need for expensive equipment or high levels of expertise.6–8
Here, we report COVID-19 Low-cost Optodiagnostic for Rapid testing (COLOR), a SARS-CoV-2 biosensor that uses highly accessible and inexpensive materials. Our method mimics the natural process by which SARS-CoV-2 infects human cells, i.e., through the binding of the viral spike protein to human Angiotensin-Converting Enzyme 2 (ACE2). 9,10 The interaction aforementioned takes place when the receptor binding domain (RBD) from the spike protein subunit S1 binds to ACE2. This is then followed by the fusion of the subunit S2 with the cell membrane. 9,10 The test can be easily assembled for 15¢ using cotton swabs modified with ACE2 to recognize the SARS-CoV-2 spike protein (SP), and gold nanoparticles (AuNano) stabilized with cysteamine (cys) and functionalized with ACE2 (AuNano-cys-ACE2). COLOR enables COVID-19 diagnosis through a simple, minute-timescale color shift that can be detected by the naked eye and whose quantitative results may be obtained using a smartphone camera and a free app. Our technology displayed excellent accuracy (90%), sensitivity (96%), and selectivity (84%) in a panel of 100 clinical patient samples and provides a low-cost solution for COVID-19 testing.
RESULTS AND DISCUSSION
COLOR consists of accessible cotton swabs as a convenient platform for at-home use. The colorimetric biosensor leverages ACE2 immobilized directly onto cotton to ensure selective and rapid recognition of SARS-CoV-2 SP. The AuNano modified with ACE2 enables a visual readout of the results through the formation of a molecular sandwich with SARS-CoV-2, which falls in between the cotton swab-ACE2 and the AuNano-ACE2.
To manufacture COLOR, we first synthesized the AuNano following our previously reported method11 using cysteamine (cys) as a stabilizing agent (Figure 1a). The UV-Vis spectrum obtained for AuNano-cys presented a maximum absorbance at 546 nm (Figure S1), which is characteristic of the plasmonic effect of gold nanoparticles.3 Cys stabilizes AuNano by the covalent binding of thiol (-SH) groups onto the surface of the AuNanos through an Au-S bond. The termination of the amino-functional group of cys provides an electrostatic repulsion due to the protonation state (NH3+), generating free positive charges 12–14 that confer adequate stability to AuNanos when stored at 4 °C for up to 5 days (Figure S2).
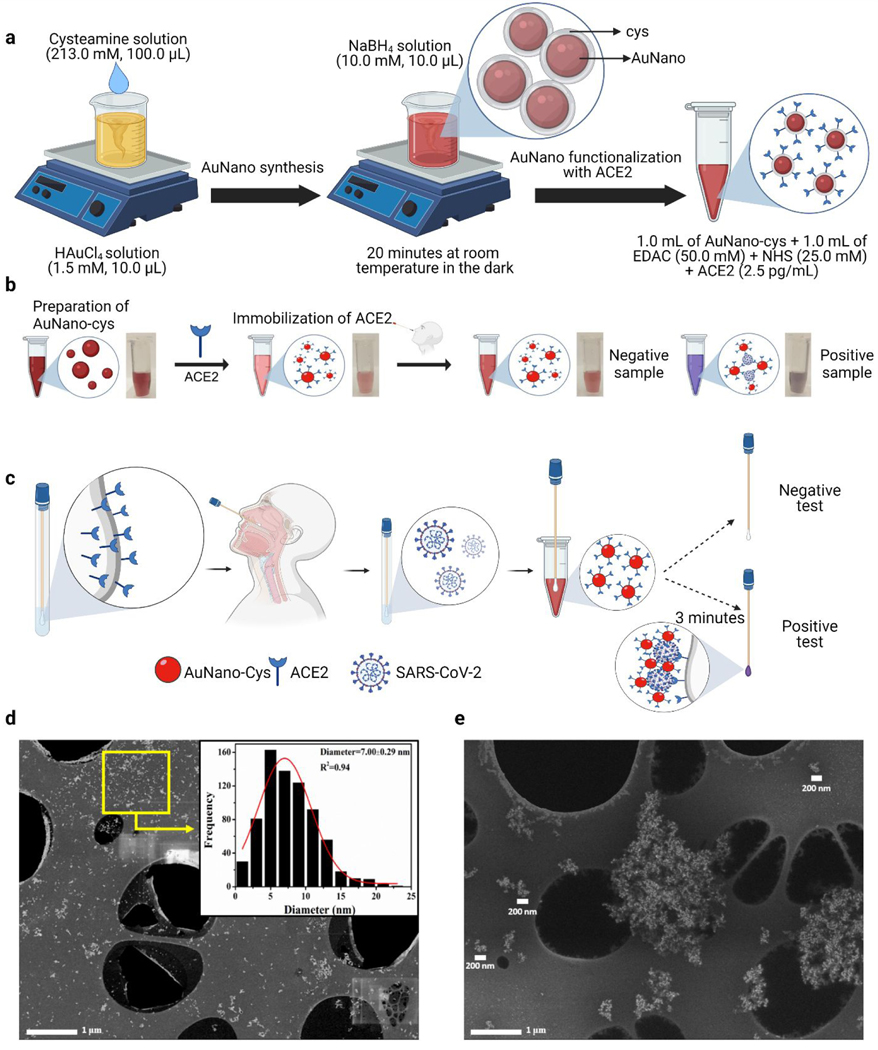
Figure 1. Design, manufacturing, and characterization of COLOR.
(a) Schematic representation of the synthesis steps needed to generate AuNano, including their functionalization with cysteamine (cys) and ACE2. Briefly, cys was added in the presence of chloroauric acid for 20 min at room temperature and protected from light. Sodium borohydride was then added to the solution at room temperature in a light-protected flask. Next, the AuNano-cys were exposed to a mixture of ACE2 and EDAC:NHS for functionalization of the enzyme on the AuNano-cys surface. (b) Steps and photos showing the colorimetric detection of SARS-CoV-2 in an aqueous medium using the synthesized AuNano-cys-ACE2. Proof-of-principle methodology for recognition of the SARS-CoV-2 spike protein through color change (from red to purple, due to the plasmonic effect) of the gold nanoparticles upon aggregation. (c) Schematic representation of the colorimetric steps needed for COVID-19 diagnosis using COLOR. In the presence of SARS-CoV-2, a color shift occurs, and the cotton swabs change color from white to purple after 3 minutes. (d) Morphological characterization of the AuNano-cys solution, which presented a spherical shape and a high dispersion. Inset shows a histogram depicting the size distribution for AuNano-cys with a mean diameter size of 7.00 nm. (e) SEM micrograph of AuNano-cys-ACE2 aggregated in the presence of SARS-CoV-2 SP. Scale bars illustrate that clusters formed between the AuNano-cys-ACE2 and the virus are larger than 200 nm.
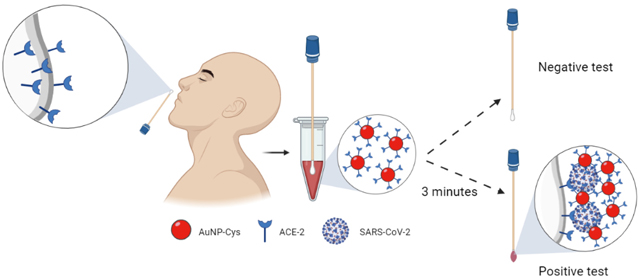
Next, the AuNano-cys were modified with ACE2 using N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC) and N-Hydroxysuccinimide (NHS) to activate the carboxylic functional groups to then covalently bind to amino groups present in the AuNano-cys, generating an amide. This reaction generates a light red solution that maintains its color when exposed to a negative COVID-19 sample (i.e., without SP) in VTM medium (Figure 1c), demonstrating the high stability and selectivity of our modified AuNano-cys-ACE2. On the contrary, when exposed to SARS-CoV-2, the red-colored solution shifts color to purple, resulting from the aggregation of AuNano-cys-ACE2 when in contact with SARS-CoV-2 SP (Figure 1c).
Collectively, the cotton swab modification process consists of three major steps: 1) overnight activation of the cotton surface with a potassium periodate solution containing 1 mL of sulfuric acid at room temperature to allow the formation of aldehyde groups on the cotton’s surface through the conversion of poly-hydroxyl groups; 2) cotton functionalization with ACE2 using EDAC:NHS; and 3) blockage of non-specific sites to ensure ACE2 stabilization by using bovine serum albumin (BSA) (Figure 1c). The subsequent colorimetric detection involves immersion of the cotton swabs into standard SP solutions or clinical SARS-CoV-2 samples. The SP in these samples binds to the surface of the cotton swabs due to its affinity toward ACE2. Subsequently, the cotton swab containing bound SARS-CoV-2 SP was soaked into a AuNano-cys-ACE2 solution for 3 minutes. This step induced aggregation of the AuNano-cys-ACE2 onto the swab surface (positive COVID-19 test), and the red-colored solution turned purple due to the formation of a molecular sandwich consisting of cotton-ACE2/SARS-CoV-2 (or SP alone)/AuNano-cys-ACE2. These results highlight the potential of our synthesized and biofunctionalized AuNano-cys-ACE2 as a functional nanomaterial for diagnosing COVID-19.
We then performed thorough characterization studies of COLOR. Morphological characterization of AuNano-cys-ACE2 dispersion before (Figure 1d) and after (Figure 1e) incubation with SARS-CoV-2 samples was conducted using scanning electron microscopy (SEM). The spherical AuNano-cys dispersion had an averaged size distribution with a diameter of 7.00 ± 0.29 nm and the AuNano-cys-ACE2 had an averaged size distribution with a diameter of 12.71 ± 0.11 nm (Figure S3), which rapidly aggregated when exposed to SARS-CoV-2 due to selective binding with ACE2. Binding to ACE2 yielded the formation of nanostructure aggregates with dimensions higher than 200 nm (Figure 1e), confirming our hypothesis that such an interaction would lead to a color shift (from red-to-purple), observable by the naked eye.
Once our proof-of-principle study was completed, we performed further optimization studies to obtain an optimal analytical signal (greater purple color intensity on cotton swabs), including optimization tests to enhance ACE2 immobilization onto the cotton swabs, assessment of the optimal incubation time of the biosensors in the SP solution, and to enable the binding of the AuNano-cys-ACE2 solution to the cotton-SP complex providing the final color readout. To obtain quantitative results, we photographed the cotton swabs using a white background with ambient lighting (Figure 3b). A distance of 15 cm (perpendicular to the surface where the swabs were placed) was maintained between the smartphone camera (Samsung Galaxy J8 with Android system and camera 10.0.01.77) and the cotton swab biosensor to ensure reproducibility (Figure 3e). All data were plotted as R-R0, where R0 is the red color intensity obtained for the cotton swab coated with AuNP-cys-ACE2 prior to exposure to samples (i.e., the analytical blank), and R is the red color intensity obtained for the cotton swabs coated with AuNP-cys-ACE2 after sample exposure. Thus, using this methodology, we were able to exclude background light interference or any fluctuations on camera focus when the image of the test was captured by our smartphone camera, minimizing errors for the accurate quantitative diagnosis of the samples at the point of care. It is worth noting that R, G, and B parameters provided a similar response in terms of linearity and analytical sensitivity. However, the red color pattern presented a lower relative standard deviation (RSD), and based on these results we selected this parameter to obtain further quantitative results (Figure S4).
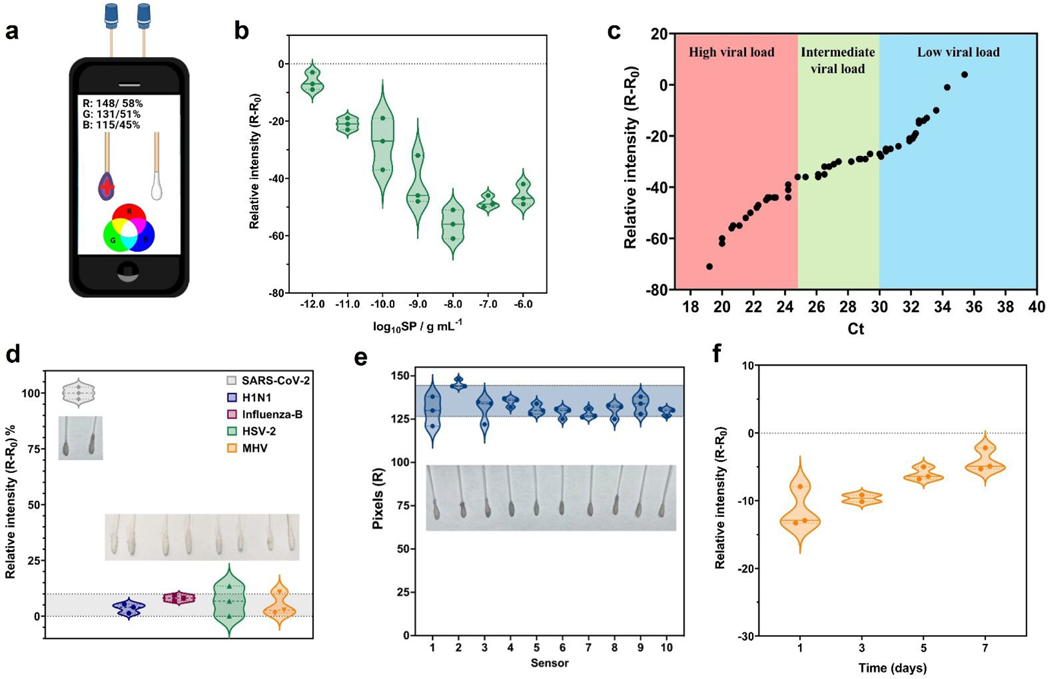
Figure 3. Analytical parameters of COLOR obtained using a free app and a smartphone.
(a) Image of the acquisition process of the color patterns from colorimetric cotton swabs biosensor using RGB app on the smartphone. (b) Analytical curve built using SP at concentrations ranging from 1×10−12 g mL−1 to 1×10−6 g mL−1, with a linear behavior in the concentration ranging from 1×10−12 g mL−1 to 1×10−8 g mL−1 SP. (c) Comparison of the relative red pattern intensity obtained with COLOR including Ct values (ranging from 19.2 Ct to 35.4 Ct) from RT-PCR for all SARS-CoV-2 positive samples tested in this study. (d) Selectivity studies using other coronaviruses and non-coronavirus strains; H1N1-A/California/2009 (105 PFU mL−1); Influenza-B/Colorado (105 PFU mL−1); Herpes simplex virus-2 (108 PFU mL−1) and MHV-murine hepatitis virus (108 PFU mL−1). Each virus was incubated in a final volume of 200 μL for 1 min at room temperature. (e) Reproducibility studies were carried out using 10 modified cotton swabs incubated for 1 min in the presence of 1×10−9 g mL−1 SP, resulting in a relative standard deviation (RSD) of 5.70%. (f) Stability study of COLOR. The cotton swabs were stored at 4 °C in PBS medium (pH = 7.4) over 7 days with the best results obtained until 3 days. Sensitivity values were obtained through analytical curves generated at concentrations ranging between 1×10−12 g mL−1 to 1×10−9 g mL−1 SP.
All experiments were carried out in triplicate (n = 3 biosensors).
The optimization process centered around obtaining the optimal conditions needed to yield the highest analytical sensitivity (angular coefficient) through calibration curves at concentrations ranging from 1×10−11 g mL−1 to 1×10−8 g mL−1 of SP (Figure 2a-c). ACE2 immobilization on the surface of the active cotton swab surface involved covalent anchoring of ACE2 through the formation of an amide bond since the activation process of the cotton swabs with potassium periodate generated aldehyde functional groups confirmed by an increased band signal at 1654 cm−1 using Fourier-transform infrared spectroscopy (FTIR) (Figure S5). The amine terminal group of ACE2 binds to aldehyde functional groups present on the activated cotton surface leading to the formation of amide bonds, which enable a stable anchoring strategy for clinical analyses.15 Collectively, our experiments revealed that the optimal conditions needed for ACE anchoring, and consequently, analytical sensitivity (Figure 2a), involved incubating the cotton swab in an ACE2 solution for 7 hours.
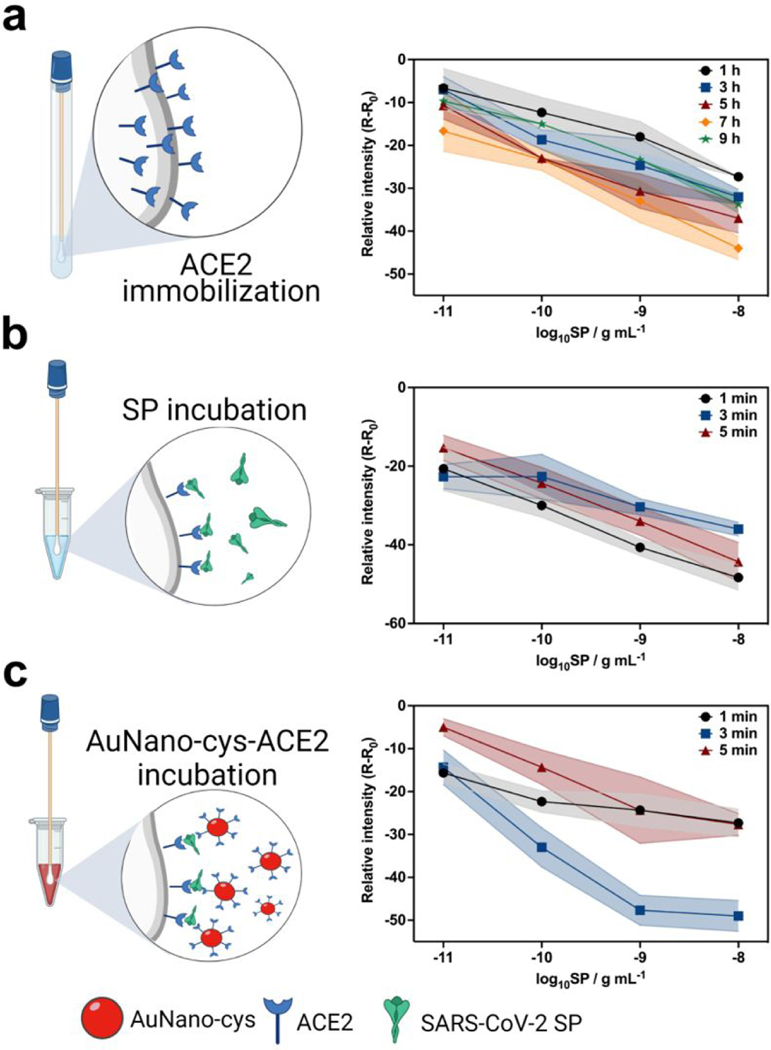
Figure 2. Optimization studies of COLOR.
(a) Study assessing the incubation period needed for functionalization of ACE2 onto the surface of the cotton swabs upon activation of the aldehyde groups with potassium periodate. The optimal incubation period to ensure ACE2 functionalization was determined to be 7 hours (b) Kinetic study of the interaction between the cotton swab-immobilized ACE2 and SP, which revealed an optimal incubation time of 1 min. (c) Study of incubation time between the cotton swab biosensor and AuNano-cys-ACE2, yielding an optimal time of 3 min for best colorimetric results. All results were optimized by evaluating the sensitivity parameters obtained from analytical curves using 1×10−11 g mL−1 to 1×10−8 g mL−1 of SP for 3 different biosensors, color shaded bands represent the uncertainty regions (error bars).
We next assessed the kinetics of the interaction between SP and the ACE2-modified cotton swab for an incubation period ranging from 1 to 5 min (Figure 2b). We selected 1 min as the optimal condition as it yielded excellent sensitivity and analytical frequency (Figure 2b). Finally, the cotton swabs exposed to SP solutions were incubated with AuNano-cys-ACE2, leading to a color change (Figure 2c). The color readout is based on the selective binding of AuNano-cys-ACE2 to the surface of the cotton swab/ACE2/SP complex, leading to aggregation of the modified AuNanos on the functionalized cotton surface, which yields a purple color (Figure 1c). This optimization process was carried out using an incubation period ranging from 1 to 5 min and an optimal sensitivity was obtained after 3 min (Figure 2c). The optimized incubation time of the cotton swabs in the AuNano-cys-ACE2 solution was 3 min (Figure 2c). A lower incubation time yielded a lower sensitivity (low color intensity) due to a restricted time period to allow interactions and sandwich formation between the cotton swab-ACE2-virus-ACE2-AuNano-cys. However, higher incubation times (>5 min) did not provide enhanced sensitivity either, likely due to the fast interaction kinetics of SARS-CoV-2 with ACE2. Additionally, long incubation periods (>5 min) enabled the adsorption of the functionalized nanoparticles onto cotton swabs, reducing the R-R0 value (Figure 2c) which could lead to a false-positive result.
Analytical characterization studies of COLOR
Once all experimental conditions were optimized (Figures 2a-c), we built an analytical curve by exposing the cotton swabs to solutions containing increasing concentrations of SP (10−12 to 10−6 g mL−1 SP) in VTM medium, which provided a visually purple gradient color from 10−12 to 10−8 g mL−1 of SP. Results were quantified using a smartphone camera by analyzing the red color pattern of the photos taken of the cotton swabs (Figure 3b).
The direct use of the red pattern intensity demonstrated a high correlation with the color obtained by the increase in SP concentration, i.e., the red pattern values decreased with increased SP concentration due to the red shift triggered by aggregation of functionalized AuNanos. Thus, the red intensity signals (n = 3 measurements) obtained at each concentration of SARS-CoV-2 SP were plotted as a logarithmic function of the SP concentration, which ranged from 1×10−12 g mL−1 to 1×10−6 g mL−1. A linear behavior was observed in the range of 1×10−12 g mL−1 to 1×10−8 g mL−1 of SP resulting in a sensitivity value of −12.14 ± 0.06 pixels g−1 mL and a linear regression (R2) of 0.99.
The limit of detection (LOD) and limit of quantification (LOQ) were obtained according to the Miller and Miller equation (Eq. 1), 16,17 where the term (y0-yB) is the value of the intercept of the analytical curve after subtraction of the value of the blank solution (i.e., VTM medium), and Sy/x corresponds to the slope error. The LOQ was calculated by multiplying the LOD by a factor of 3.33, as previously described. 18,19 The LOD and LOQ values of the modified cotton swabs were 0.154 pg mL−1 and 0.513 pg mL−1, respectively, underscoring the excellent performance and ultra-sensitive COVID-19 detection capabilities of COLOR. These excellent analytical parameters are similar to those obtained by instrumental methods, such as electrochemical biosensors that also use ACE2 as a biorecognition element for SARS-CoV-2 SP biosensing.20,21 A side-by-side comparison of COLOR’s performance with respect to other optical SARS-CoV-2 detection methods is presented in Table 1. Overall, COLOR displays an extremely low LOD, provides rapid testing time (5 min total; 1 min of incubation with the clinical sample, 30 s first wash with PBS, 3 min incubation with the AuNano-cys-ACE2 solution, and an extra 30 s for the second and final wash with PBS), and is inexpensive to produce with a total cost of 15¢ (8¢ per plastic vial, 1¢ for recombinant ACE2, 1¢ for the cotton swab, and 5¢ for all the other reagents used). The diagnostic time and cost of COLOR are much lower than those of lateral flow immunoassays, the most commonly used technology for high-frequency testing (Table 1).
Table 1.
Side-by-side comparison of the analytical features of different colorimetric-based sensors developed for SARS-CoV-2 detection.
| Sensor | LOD | Target | Technique | Concentration range | Time (min) | Reference |
|---|---|---|---|---|---|---|
| AuNano-cys/ACE2 on cotton swab (COLOR) | 0.154 pg mL−1 | SARS-CoV-2 SP | Colorimetric | 1×10−12 to 1×10−6 g mL−1 | 5 | This work |
| SARS-CoV-2@fAuNPs | Ct value: 36.5 | Antibody (S, E, and M) | Colorimetric | 7.0 to 30.0 Ct | 3 | 4 |
| Lateral flow immunoassay (LFIA) | ND | COVID-19 IgG and IgM antibody | Colorimetric | ND | 15 | 5 |
| AuNPs- RNA | 0.18 ng μL−1 | SARS-CoV-2 ribonucleic acid (RNA) | Colorimetric | 0.2–3 ng μL−1 | 10 | 27 |
| DNAzyme and SARS-CoV-2 RNA | 0.01 ng mL−1 | SARS-CoV-2 RNA | Colorimetric | 101–107 copies mL−1 | ±5 | 28 |
| (MPNs) | 3.2 gene copies μL−1 | SARS-CoV-2 RNA | Fluorescence | 3.2 gene copies μL−1 | 17 | 29 |
| RT-LAMP | 0.75 RNA copies μL−1 | SARS-CoV-2 RNA | Colorimetric | 0.2–47 ng μL−1 | 15–40 | 30 |
| LAMP | 50 RNA copies μL−1 | SARS-CoV-2 RNA | Fluorescence | 50 RNA copies μL−1 | >40 | 31 |
| Lateral flow immunoassay (LFIA) | 0.65 ng mL−1 | Biotinylated antibody | Colorimetric | 0.53 to 0.77 ng mL−1 | ND | 32 |
| Lateral flow immunoassay (LFIA) | 1 pg mL−1 | Protein-anti-SARS-CoV-2 IgM/IgG | Colorimetric | 10–0.001 ng mL−1 | 25 | 33 |
| Lateral flow immunoassay (LFIA) | (IgG) 0.121 U L−1 and (IgM) 0.366 U L−1 | Antibody IgG and IgM | Fluorescent | ND | 15 | 34 |
Antibody (S, E, and M) spike, envelope, and membrane, SARS-CoV-2 (spike, envelope, and membrane), gold nanoparticles (AuNP), functionalized AuNanos (f-AuNPs), ribonucleic acid (RNA), magneto-plasmonic nanoparticles (MPNs); (RT-LAMP) transcription loop-mediated isothermal amplification.
ND: not described.
| (Eq.1) |
To verify the selectivity of COLOR toward SARS-CoV-2, we performed cross-reactivity studies against other viruses. We tested four other viral strains: H1N1 (A/California/2009), Influenza-B/Colorado, Herpes simplex virus-2, and MHV-murine hepatitis virus. Our results revealed no cross-reactivity against any of the viruses tested since all experiments yielded a response lower than 10% of the response obtained by the positive control sample (R-R0), where R and R0 denote the red color intensity for the sample and the analytical blank, respectively (Figure 3d).
Reproducibility assays were conducted to ensure different COLOR tests performed similarly. Each colorimetric device was exposed to 1×10−9 g mL−1 of SP resuspended in a 0.1 mol L−1 PBS solution (pH = 7.4). A relative standard deviation (RSD) of 5.70 % in the analytical signal (red color pattern) was obtained using ten different biosensors (n = 10), indicating excellent reproducibility and suitability for high-frequency testing (Figure 3e). Next, we evaluated the stability of COLOR when stored at 4 °C in PBS medium over 7 days. Analytical curves were generated in 0.1 mol L−1 PBS (pH = 7.4) with concentrations of SP ranging from 1×10−12 g mL−1 to 1×10−9 g mL−1 (Figure 3f). COLOR remained functional after 7 days of storage (Figure 3f). However, the mean sensitivity of the device decreased by 50% when compared to the initial performance when COLOR was used immediately after production (Figure 3f).
Detection of SARS-CoV-2 in clinical samples with COLOR
To demonstrate the translational potential of COLOR, we evaluated its diagnostic efficacy using 100 NP/OP clinical samples (Table 2 and Table S1) containing different viral loads, each of which correlated with a specific cycle threshold (Ct) value as determined by RT-PCR. Ct values of the samples used ranged from 19.2 to 35.4 Ct (Figure 3c). Of note, our colorimetric cotton swabs were able to discriminate high from very low viral loads, and their results correlated with the Ct values obtained by RT-PCR.
Table 2. Detection of SARS-CoV-2 in NP/OP clinical samples with COLOR.
Clinical performance metrics (i.e., sensitivity, specificity, and accuracy) of COLOR using NP/OP samples. Positive and negative values for each clinical sample were obtained by RT-qPCR.
| NP swabs | RT-qPCR |
Sensitivity | Specificity | Accuracy | ||
|---|---|---|---|---|---|---|
| Positive (N=50) | Negative (N=50) | Total (N=100) | ||||
| Positive | 48 | 8 | 56 | 48/50 (96.0%) |
90/100 (90.0%) |
|
| Negative | 2 | 42 | 44 | 42/50 (84.0%) |
||
The clinical samples were collected from inpatients at the Hospital of the University of Pennsylvania (HUP) during January and March of 2021 and heat-inactivated prior to testing. Among the total of 100 samples selected, 50 were confirmed positives for SARS-CoV-2 and the remaining 50 were diagnosed negative for SARS-CoV-2 by RT-PCR. COLOR detected 48 out of 50 SARS-CoV-2 positive samples, and 42 out of 50 SARS-CoV-2 negative samples (Table 2). Based on these results, the sensitivity, specificity, and accuracy of COLOR were 96.0%, 84.0%, and 90%, respectively. In addition, COLOR demonstrated an excellent performance in its ability to diagnose clinical samples with Ct values ranging from 19.2 to 35.4 (Table S1).
Genetic mutations in the SARS-CoV-2 virus have emerged during the COVID-19 pandemic, resulting in variation in the population of circulating viral strains, which may confound the performance of currently available diagnostic methods. As a result, false-negative results may be an issue in molecular tests, particularly if a mutation occurs within the viral genome or antigen assessed by a particular test.22 We expect that the use of ACE2 as a biorecognition element can minimize these errors as long as the virus maintains its interaction with ACE2 through its spike protein. Future studies will evaluate the effect of different mutations (SARS-CoV-2 variants) on COLOR’s performance.
It is also worth noting that COLOR yielded eight false-positive results that can be attributed to the presence of other biomolecules in the complex clinical sample, which likely adsorbed onto the cotton swab surface leading to nanoparticle aggregation, once we used clinical samples without any sample pretreatment step. Note that four out of the eight samples misdiagnosed as positive presented R-R0 values of −11 to −12 (Table S1), which are close to the cut-off value of the method (R-R0 ≤ −10 for positive and R-R0 > −10 for negative results). Selecting a cut-off value is critical for assessing the performance of diagnostic technologies and should consider a balance between sensitivity and selectivity. For example, a method with a high cut-off value can prevent false-positive cases but leads to lower sensitivity. On the other hand, a low cut-off value can increase sensitivity but increases the number of false positives.23 Thus, the cut-off value of COLOR (R-R0 ≤ −10 for positive and R-R0 > −10 for negative results) was selected based on its detectability as determined in the analytical characterization (i.e., the lowest colorimetric readout discernible between the analytical blank and the lowest SARS-CoV-2 spike protein quantified; Figure 3b) and cross-reactivity studies (Figure 3d). We found that a cut-off value of R-R0 ≤ −26 in the case of COLOR reliably diagnoses COVID-19 with no false positives (Table S1).
In follow-up studies, we will assess more clinical samples to verify the influence of sample variability on overall tet performance, as well as strategies to cover the functionalized cotton swabs with a protective membrane to minimize non-specific interactions between interfering molecules present in the biofluid and the cotton surface. COLOR’s accuracy is similar or even superior to most colorimetric lateral flow immunoassays,24 a widely available technology for high-frequency COVID-19 testing. Our test is also substantially more inexpensive and rapid than existing methods (Table 1).
CONCLUSIONS
In this study, we describe COLOR, a low-cost, rapid, and easy-to-assemble COVID-19 test that uses highly accessible materials and provides a visual test readout that can be used to accurately diagnose SARS-CoV-2 infections using a smartphone. Each test costs 15¢ to produce, generates a result within 5 min, is highly sensitive (e.g., LOD of 154 fg mL−1 for SP), and demonstrated 90% accuracy in a study using 100 clinical samples. Collectively, COLOR represents an excellent solution to enable high-frequency testing, particularly in resource-limited populations.
MATERIALS AND METHODS
Materials.
All reagents used in the experiments presented analytical grade. The deionized water (resistivity ≥ 18 MΩ cm at 25 °C) was obtained from a Milli-Q Advantage-0.10 purification system (Millipore). Potassium periodate (KIO4) was obtained by Beantown Chemical. Human angiotensin-converting enzyme 2 (ACE2) Fc Chimera was obtained from GenScript. Spike protein was kindly donated by Scott Hensley from the University of Pennsylvania. N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC) and N-Hydroxysuccinimide (NHS) with a degree of purity ≥98%, gold (III) chloride trihydrate (HAuCl4.3H2O) (99.99%), sodium borohydride (NaBH4) with ≥98% purity, cysteamine hydrochloride (Cys) with 98% purity, phosphate buffer saline powder, pH = 7.4 and glutaraldehyde (25%, v/v) were purchased from Sigma-Aldrich. Cotton swabs from the brand Q-tips were purchased in a drug store (Philadelphia/USA).
Cotton swabs activation.
The cotton surface’s activation was carried out according to the protocol described by Raji et al. 15 First, the cotton swabs were immersed in a 2.0 mmol L−1 potassium periodate (KIO4) solution containing 1.0 mL sulfuric acid overnight at room temperature. Then, the cotton swabs were washed with PBS solution (0.1 mol L−1, pH = 7.4) to remove the excess of oxidizing agent present on their surface, and thereafter, they were kept in PBS buffer prior to functionalization with ACE2. This procedure resulted in the formation of an aldehyde-substituted cotton surface by the conversion of poly-hydroxyl groups, which allowed subsequent anchoring of ACE2. The aldehyde groups present on the activated cotton swabs were confirmed by Fourier-transform infrared spectroscopy (FTIR).
Synthesis of the cysteamine stabilized gold nanoparticles and ACE2 immobilization studies.
The gold nanoparticles were synthesized according to previous studies,14,25 where the AuNanos were stabilized with cys (AuNano-cys) (Figure 1a). Briefly, 100 μL of cysteamine (213.0 mmol L−1) was added to 1.5 mmol L−1 HAuCl4 solution to a final volume of 10.0 mL. The solution was vigorously stirred at 1,200 rpm for 20 minutes at room temperature in a light-protected environment. Subsequently, 10.0 μL of NaBH4 (10.0 mmol L−1) were added to the solution and kept under stirring conditions for an extra 20 minutes in a light-protected flask and at room temperature, until the color of the solution changed from yellow to red. Finally, the solution was stored at 4 °C in a refrigerator for up to 7 days.25,26 Next, 1.0 mL of AuNano-cys solution was modified with ACE2 (2.5 pg mL−1) for 30 minutes at room temperature in 1.0 mL of a solution containing EDAC (50.0 mmol L−1) and NHS (25.0 mmol L−1 in a final volume of 2.0 mL. This protocol enabled the formation of an amide bond between the AuNanos-cys surface and ACE2.
Gold nanoparticle (AuNano) characterization studies.
Morphological characterization of gold nanoparticles stabilized with cysteamine (AuNano-cys) was performed using scanning electron microscopy (SEM). All images were acquired using a JEOL 7500F HRSEM microscope from the Singh Center for Nanotechnology (University of Pennsylvania). The Spectrophotometer measurements were carried out using a Perkin Elmer Multimode Plate Reader spectrophotometer (model EnVision). The absorbance of AuNano-cys was measured at 546 nm at room temperature. The effectiveness of cotton swab activation was evaluated by FTIR spectra obtained using a Perkin Elmer Spectrum 2 equipped with a Diamond UATR 2 detector with a scan range from 4,500 to 500 cm−1 and a total of 32 scans by each measure.
ACE2 immobilization onto the surface of cotton swabs.
To optimize the immobilization of ACE2 on the cotton swab surface, the incubation time of oxidized cotton swabs in 2.5 pg mL−1 ACE2 solution was evaluated at 1, 3, 5, 7, and 9 hours at 4° C. Next, the swabs were washed in 0.1 mol L−1 PBS solution (pH = 7.4) for 30 seconds to remove excess of unbound ACE2 and incubated in 1% (m/m) BSA solution over 30 minutes to block non-modified cotton surface. Subsequently, the swabs were immersed in an SP solution ranged from 10−11 to 10−8 g mL−1 for 5 min, washed to remove excess unbound SP. Finally, the modified swabs containing bound SP were immersed in an AuNano-cys-ACE2 solution for 2 min, followed by rinsing PBS for 30 seconds to remove excess of AuNano. The color shift of the cotton swab surface from white to purple indicates the presence of SP in the solution due to the aggregation of AuNano-cys-ACE2. The best result of incubation time for ACE2 functionalization was 7 hours (Figure 2a).
Analytical optimizations and colorimetric quantification assays of SARS-CoV-2 SP
For quantitative purposes, all colorimetric test images of the cotton swabs were recorded by smartphone camera after 1 minute of the colorimetric test using a smartphone Samsung Galaxy J8 with Android system version 10 0.01.77 at a 15 cm distance (perpendicularly to cotton swabs) in a white background and constant ambient lighting. Colorimetric responses were measured using color pattern intensities (Red, Green, and Blue) by the freeware RGB application to evaluate which parameter provided a better correlation with the color test. The data from digital colorimetry (obtained by smartphone camera) of COLOR were plotted as R-R0, where R0 is the value of parameter R obtained for the cotton swab coated with AuNP-cys-ACE2 before the exposure to samples (analytical blank) and R is the red color intensity obtained for the cotton swabs coated with AuNP-cys-ACE2 after exposure to the samples.
The incubation time of the cotton swab modified with ACE2 in SP solution was optimized. For this, analytical curves were built in the concentration range of 10−11 to 10−8 g mL−1 of SP in 0.1 mol L−1 PBS, varying the incubation time in 1, 3, and 5 minutes. These tests were carried out at room temperature and with the cotton swabs modified with ACE2 at the optimal condition (swab incubated with 1.25 pg mL−1 ACE2 over 7 hours). Subsequently, the swabs were washed in a 0.1 mol L−1 PBS solution (pH = 7.4) for 30 seconds to remove the excess unbound SP. Finally, the swabs were immersed in an AuNano-cys-ACE2 solution for 2 min, followed by rinsing PBS for 30 seconds to remove the excess AuNano-cys. For data processing, the same methodology used to optimize the incubation time of ACE2 was used. Through the results obtained in Figure 2b, it is possible to observe greater detectability for 1 min of incubation.
The last parameter to be optimized was the incubation time of the swabs containing ACE2 and SP in the AuNano-cys-ACE2 solution, i.e., the time required for the swabs to be immersed in a nanoparticle solution for color change. These being the last parameter to be analyzed, the previous steps have already been performed with their respective optimized times (7 hours ACE2 and 1 minute for SP). For this purpose, analytical curves were constructed in the concentrations range from 10−11 to 10−8 g mL−1 SP in 0.1 mol L−1 PBS, varying only the immersion time in AuNP-cys-ACE2 solution in 1, 3, and 5 minutes. Subsequently, the swabs were rinsed with 0.1 mol L−1 of PBS for 30 seconds to remove the excess AuNano-cys-ACE2. For data processing, the same methodology used for the ACE2 incubation time was used. The best result observed was 3 minutes of incubation in the AuNP-cys-ACE2 solution (Figure 2c).
Reproducibility, stability, and cross-reactivity assays.
Reproducibility studies were carried out using 10 different COLOR cotton swabs exposed to 1 ng mL−1 of SP. Color intensity (red pattern) measurements were detected via the RGB app from photos of the cotton swabs taken after 1 min of the test. The stability of the modified cotton swabs was evaluated by the sensitivity value (slope) extracted from analytical curves at an interval range from 1×10−12 to 1×10−6 g mL−1 of SP for 1, 3, 5, and 7 days. Cross-reactivity assays were performed by exposing the modified cotton-swabs to 200 μL of five different viral samples: MHV – murine hepatitis virus at 108 PFU mL−1 (coronavirus), H1N1 – A/California/2009, H3N2 – A/Nicaragua, Influenza B – B/Colorado, HSV2 – herpes simplex virus-2, all at 105 PFU mL−1. After the exposure period of 1 min between COLOR and each virus, the swabs were washed with PBS, exposed to AuNano-cys-ACE2 accordingly to the same protocol described for clinical sample analysis, and analyzed by the RGB app.
RT-PCR analysis
Briefly, the RT-PCR analyses were carried out using the QIAmp DSP Viral RNA Mini Kit (Qiagen) and sample aliquots of 140 μL. For the first step, the virus sample was chemically inactivated under highly denaturing conditions (guanidine thiocyanate) using a biosafety cabinet under BSL-2 enhanced protocols. Next, the presence of RNA in the samples was analyzed in duplicate with the TaqPath™ 1-Step RT-qPCR reagent kit (Life Technologies) using a Quantstudio 7 Flex Genetic Analyzer (ABI). The selection of oligonucleotide primers and probes for the detection of 2019-nCoV was based on the virus nucleocapsid (N) gene. The panel was designed for the specific detection of the 2019-nCoV viral RNA (two primer/probe sets, N1 and N2). Additionally, the primer/probe set to detect the human RNase P gene (RP) in control samples and clinical specimens was also included in the panel as per CDC protocol (2019-nCoVEUA-01). All clinical samples were collected and stored for one week before analysis by RT-PCR and COLOR.
Analysis of clinical samples from patients using COLOR.
One hundred nasopharyngeal/oropharyngeal (NP/OP) clinical samples (50 COVID-19 negative and 50 COVID-19 positive) resuspended in viral transport medium (VTM) were obtained from inpatients after heat-inactivation. All COVID-19 positive samples were confirmed by RT-PCR. We set a cut-off value of −10.0 for relative red pattern changes to determine as positive diagnostic, i.e., positive and negative COLOR results for SARS-CoV-2 infection were R-R0 ≤ −10 and R-R0 > −10, respectively.
Supplementary Material
ACKNOWLEDGMENTS
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). Research reported in this publication was supported by the Nemirovsky Prize and by funds provided by the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. W.R.A. acknowledges funding from Brazilian funding agencies CAPES (88887.479793/2020-00), FAPESP (2018/08782-1), and CNPq (438828/2018-6 and 401256/2020-0) for supporting the research. All figures were prepared using the Biorender drawing toolkit. We thank Scott Hensley, Sara Cherry, Susan Weiss, Ronald Collman, Michael Feldman, Benjamin Abella, and Paul Calahan for donating samples.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The Supporting Information is available free of charge at
Characterization of the synthesized AuNano-cys dispersion by UV-Vis and the stability study of this solution over 5 days; SEM image for AuNano-cys-ACE2 dispersion and the corresponding size histogram distribution; Comparison of RGB parameters for quantification of SARS-CoV-2 SP; FTIR characterization of the activated cotton swabs; table containing the 100 NP/OP clinical samples analyzed. The Supporting Information is available free of charge online.
REFERENCES
- (1).Hui DS; I Azhar E; Madani TA; Ntoumi F; Kock R; Dar O; Ippolito G; Mchugh TD; Memish ZA; Drosten C; Zumla A; Petersen E. The Continuing 2019-NCoV Epidemic Threat of Novel Coronaviruses to Global Health — The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis 2020, 91, 264–266. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu JT; Leung K; Leung GM Nowcasting and Forecasting the Potential Domestic and International Spread of the 2019-NCoV Outbreak Originating in Wuhan, China: A Modeling Study. Obstetrical and Gynecological Survey. 2020, pp 399–400. 10.1097/01.ogx.0000688032.41075.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tang L; Li J. Plasmon-Based Colorimetric Nanosensors for Ultrasensitive Molecular Diagnostics. ACS Sensors 2017, 2 (7), 857–875. 10.1021/acssensors.7b00282. [DOI] [PubMed] [Google Scholar]
- (4).Ventura B. Della; Cennamo M; Minopoli A; Campanile R; Censi SB; Terracciano D; Portella G; Velotta R. Colorimetric Test for Fast Detection of SARS-COV-2 in Nasal and Throat Swabs. ACS Sensors 2020, 5 (10), 3043–3048. 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Li Z; Yi Y; Luo X; Xiong N; Liu Y; Li S; Sun R; Wang Y; Hu B; Chen W; Zhang Y; Wang J; Huang B; Lin Y; Yang J; Cai W; Wang X; Cheng J; Chen Z; Sun K; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol 2020, 92 (9), 1518–1524. 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nelis JLD; Tsagkaris AS; Dillon MJ; Hajslova J; Elliott CT Smartphone-Based Optical Assays in the Food Safety Field. TrAC - Trends Anal. Chem 2020, 129, 115934. 10.1016/j.trac.2020.115934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fan Y; Li J; Guo Y; Xie L; Zhang G. Digital Image Colorimetry on Smartphone for Chemical Analysis: A Review. Meas. J. Int. Meas. Confed 2021, 171 (June 2020), 108829. 10.1016/j.measurement.2020.108829. [DOI] [Google Scholar]
- (8).Roda A; Michelini E; Zangheri M; Di Fusco M; Calabria D; Simoni P. Smartphone-Based Biosensors: A Critical Review and Perspectives. TrAC - Trends Anal. Chem 2016, 79, 317–325. 10.1016/j.trac.2015.10.019. [DOI] [Google Scholar]
- (9).Zhang H; Penninger JM; Li Y; Zhong N; Slutsky AS Angiotensin-Converting Enzyme 2 (ACE2) as a SARS-CoV-2 Receptor: Molecular Mechanisms and Potential Therapeutic Target. Intensive Care Med. 2020, 46 (4), 586–590. 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li F; Li W; Farzan M; Harrison SC Structural Biology: Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science (80-. ). 2005, 309 (5742), 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- (11).Ferreira AL; Lima L. F. De; Moraes AS; Rubira RJG; Constantino CJL; Leite FL; Delgado-silva AO Development of a Novel Biosensor for Creatine Kinase (CK-MB) Using Surface Plasmon Resonance (SPR). Appl. Surf. Sci 2021, 149565. 10.1016/j.apsusc.2021.149565. [DOI] [Google Scholar]
- (12).Farmani MR; Peyman H; Roshanfekr H. Blue Luminescent Graphene Quantum Dot Conjugated Cysteamine Functionalized-Gold Nanoparticles (GQD-AuNPs) for Sensing Hazardous Dye Erythrosine B. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc 2020, 229. 10.1016/j.saa.2019.117960. [DOI] [PubMed] [Google Scholar]
- (13).Liang X; Wei H; Cui Z; Deng J; Zhang Z; You X; Zhang XE Colorimetric Detection of Melamine in Complex Matrices Based on Cysteamine-Modified Gold Nanoparticles. Analyst 2011, 136 (1), 179–183. 10.1039/c0an00432d. [DOI] [PubMed] [Google Scholar]
- (14).Asadzadeh-Firouzabadi A; Zare HR Application of Cysteamine-Capped Gold Nanoparticles for Early Detection of Lung Cancer-Specific MiRNA (MiR-25) in Human Blood Plasma. Anal. Methods 2017, 9 (25), 3852–3861. 10.1039/c7ay01098b. [DOI] [Google Scholar]
- (15).Raji MA; Chinnappan R; Shibl A; Suaifan G; Weber K; Cialla-May D; Popp J; El Shorbagy E; Al-Kattan K; Zourob M. Low-Cost Colorimetric Diagnostic Screening Assay for Methicillin Resistant Staphylococcus Aureus. Talanta 2021, 225 (October 2020), 121946. 10.1016/j.talanta.2020.121946. [DOI] [PubMed] [Google Scholar]
- (16).Miller JC; Miller JN Basic Statistical Methods for Analytical Chemistry: Part I. Statistics of Repeated Measurements: A Review. Analyst 1988, 113 (9), 1351–1356. 10.1039/AN9881301351. [DOI] [Google Scholar]
- (17).Miller JN Basic Statistical Methods for Analytical Chemistry. Part 2. Calibration and Regression Methods. A Review. Analyst 1991, 116 (1), 3–14. 10.1039/AN9911600003. [DOI] [Google Scholar]
- (18).de Lima LF; Maciel CC; Ferreira AL; de Almeida JC; Ferreira M. Nickel (II) Phthalocyanine-Tetrasulfonic-Au Nanoparticles Nanocomposite Film for Tartrazine Electrochemical Sensing. Mater. Lett 2020, 262, 1–5. 10.1016/j.matlet.2019.127186. [DOI] [Google Scholar]
- (19).de Lima LF; Pereira EA; Ferreira M. Electrochemical Sensor for Propylparaben Using Hybrid Layer-By-Layer Films Composed of Gold Nanoparticles, Poly(Ethylene Imine) and Nickel (II) Phthalocyanine Tetrasulfonate. Sensors Actuators, B Chem. 2020, 310 (February), 1–8. 10.1016/j.snb.2020.127893. [DOI] [Google Scholar]
- (20).Torres MDT; de Araujo WR; de Lima LF; Ferreira AL; de la Fuente-Nunez C. Low-Cost Biosensor for Rapid Detection of SARS-CoV-2 at the Point of Care. Matter 2021, 4 (7), 2403–2416. 10.1016/J.MATT.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).de Lima LF; Ferreira AL; Torres MDT; de Araujo WR; de la Fuente-Nunez C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc. Natl. Acad. Sci. U. S. A 2021, 118 (30), 1–9. 10.1073/pnas.2106724118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests | FDA https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed Jul 10, 2021). [Google Scholar]
- (23).Ogawa Taku, Fukumori Tatsuya, Nishihara T. S. Yuji; Okuda Nao, Nishimura Tomoko, Fujikura Hiroyuki N. H.; Imakita Natsuko K. K. Another False-Positive Problem for a SARS-CoV-2 Antigen Test in Japan. J. Clin. Virol 2020, 131 (January), 19–20. 10.1016/j.jcv.2020.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Conklin SE; Martin K; Manabe YC; Schmidt HA; Miller J; Keruly M; Klock E; Kirby CS; Baker OR; Fernandez RE; Eby YJ; Hardick J; Shaw-Saliba K; Rothman RE; Caturegli PP; Redd AD; Tobian AAR; Bloch EM; Benjamin Larman H; Quinn TC; et al. Evaluation of Serological SARS-CoV-2 Lateral Flow Assays for Rapid Point-of-Care Testing. J. Clin. Microbiol 2021, 59 (2). 10.1128/JCM.02020-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sharma AK; Pandey S; Nerthigan Y; Swaminathan N; Wu HF Aggregation of Cysteamine-Capped Gold Nanoparticles in Presence of ATP as an Analytical Tool for Rapid Detection of Creatine Kinase (CK-MM). Anal. Chim. Acta 2018, 1024, 161–168. 10.1016/j.aca.2018.03.027. [DOI] [PubMed] [Google Scholar]
- (26).Alencar WS; Crespilho FN; Martins MVA; Zucolotto V; Oliveira ON; Silva WC Synergistic Interaction between Gold Nanoparticles and Nickel Phthalocyanine in Layer-by-Layer (LbL) Films: Evidence of Constitutional Dynamic Chemistry (CDC). Phys. Chem. Chem. Phys 2009, 11 (25), 5086–5091. 10.1039/b821915j. [DOI] [PubMed] [Google Scholar]
- (27).Moitra P; Alafeef M; Alafeef M; Alafeef M; Dighe K; Frieman MB; Pan D; Pan D; Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14 (6), 7617–7627. 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Anantharaj A; Das SJ; Sharanabasava P; Lodha R; Kabra SK; Sharma TK; Medigeshi GR Visual Detection of SARS-CoV-2 RNA by Conventional PCR-Induced Generation of DNAzyme Sensor. Front. Mol. Biosci 2020, 7 (December), 1–7. 10.3389/fmolb.2020.586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cheong J; Yu H; Lee CY; Lee J; Choi HJ; Lee JH; Lee H; Cheon J. Fast Detection of SARS-CoV-2 RNA via the Integration of Plasmonic Thermocycling and Fluorescence Detection in a Portable Device. Nat. Biomed. Eng 2020, 4 (12), 1159–1167. 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yu L; Wu S; Hao X; Li X; Liu X; Ye S; Han H; Dong X; Li X; Li J; Liu N; Liu J; Zhang W; Pelechano V; Chen WH; Yin X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem 2020, 66 (7), 975–986. 10.1101/2020.02.20.20025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ganguli A; Mostafa A; Berger J; Aydin MY; Sun F; Stewart de Ramirez SA; Valera E; Cunningham BT; King WP; Bashir R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (37), 22727–22735. 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Grant BD; Anderson CE; Williford JR; Alonzo LF; Glukhova VA; Boyle DS; Weigl BH; Nichols KP SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem 2020, 92 (16), 11305–11309. 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- (33).Liu H; Dai E; Xiao R; Zhou Z; Zhang M; Bai Z; Shao Y; Qi K; Tu J; Wang C; Wang S. Development of a SERS-Based Lateral Flow Immunoassay for Rapid and Ultra-Sensitive Detection of Anti-SARS-CoV-2 IgM/IgG in Clinical Samples. Sensors Actuators, B Chem 2021, 329 (July 2020), 129196. 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhang C; Zhou L; Liu H; Zhang S; Tian Y; Huo J; Li F; Zhang Y; Wei B; Xu D; Hu J; Wang J; Cheng Y; Shi W; Xu X; Zhou J; Sang P; Tan X; Wang W; Zhang M; et al. Establishing a High Sensitivity Detection Method for SARS-CoV-2 IgM/IgG and Developing a Clinical Application of This Method. Emerg. Microbes Infect 2020, 9 (1), 2020–2029. 10.1080/22221751.2020.1811161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.