Abstract
B-lymphocyte-induced maturation protein (Blimp-1) is a transcriptional repressor that is considered to be a master regulator of terminal B-cell development because it is sufficient to trigger differentiation in the BCL1-cell model. Transcription of the c-myc gene is repressed by Blimp-1 during B-cell differentiation. In this study, we have explored the mechanism by which Blimp-1 represses transcription by using Gal4-fusion protein assays and assays in which Blimp-1 represses the natural c-myc promoter. The results show that Blimp-1 represses the c-myc promoter by an active mechanism that is independent of the adjacently bound activator YY1. Blimp-1 contains two regions that independently associate with histone deacetylase (HDAC) and endogenous Blimp-1 in nuclear extracts binds in vitro to the c-myc Blimp-1 site in a complex containing HDAC. The functional importance of recruiting HDAC for Blimp-1-dependent repression of c-myc transcription is supported by two experiments. First, the HDAC inhibitor tricostatin A inhibits Blimp-1-dependent repression in cotransfection assays. Second, a chromatin immunoprecipitation assay shows that expression of Blimp-1 causes deacetylation of histone H3 associated with the c-myc promoter, and this deacetylation depends on the Blimp-1 binding site in the c-myc promoter.
B-lymphocyte-induced maturation protein (Blimp-1) is a 100-kDa protein which contains five zinc finger motifs. Blimp-1 cDNA was originally isolated in a subtractive screen of the BCL1 B-cell lymphoma cell line following treatment with cytokines interleukin 2 and interleukin 5 (62). This treatment causes BCL1 cells to undergo terminal differentiation, evidenced by altered expression of various mRNAs and cell surface proteins and secretion of immunoglobulin M (62). Since ectopic expression of Blimp-1 alone is sufficient to cause terminal differentiation of BCL1 cells, Blimp-1 is considered to be a “master regulator” of terminal B-cell development. The initial report showed that Blimp-1 expression was limited to mature or terminally differentiated B cells (62).
Multiple differences in gene expression are known to exist between postgerminal center B cells and terminally differentiated plasma cells, the developmental stages thought to be represented by BCL1 cells before and after cytokine treatment. Plasma cells secrete large amounts of immunoglobulin, and in BCL1 cells, J chain is induced upon differentiation to allow secretion of immunoglobulin M (3, 45). Cell surface proteins CD138 (Syndecan-1) and CD47 are also induced upon BCL1 cell differentiation. On the other hand, expression of genes encoding proteins, such as c-Myc (40), CD23 (55), CD22 (61), major histocompatibility complex class II (4, 59), BSAP (Pax-5) (54), early B-cell factor (18), and CIITA (59), is repressed in plasma cells. Since Blimp-1 can initiate the entire developmental cascade in BCL1 cells, it appears that all these genes are either direct targets of Blimp-1 or are regulated by Blimp-1 target genes.
We have previously shown that c-myc is an important target gene of Blimp-1 in BCL1 lymphoma cells (40). c-Myc is required for cell cycle progression through the G0-G1 and S-G2/M transitions (63). c-Myc expression correlates with cell proliferation, being induced upon mitogen stimulation (30, 41, 44, 57) and shut down in quiescent or terminally differentiated cells (14, 23, 37). In addition, overexpression of c-Myc is known to block terminal differentiation in some cell lines (6, 10, 49), suggesting that repression of c-myc is crucial to achieve the nonproliferating state associated with terminal differentiation. Therefore, the fact that Blimp-1 represses c-myc transcription is consistent with the role of Blimp-1 as a master regulator in B-cell terminal differentiation.
The human homolog of Blimp-1, PRDI-BF1, was cloned by its ability to bind the PRDI site in the human beta interferon (IFN-β) promoter (31). PRDI-BF1 was shown to repress the IFN-β promoter, and induction of PRD1-BF1 late in the response to virus infection was postulated to be important for limiting the IFN response (31). Thus, for the only two currently established and physiologically relevant target genes of Blimp-1, c-myc, and IFN-β, Blimp-1 functions as a transcriptional repressor. We wished to analyze the mechanism by which Blimp-1 represses transcription.
Mechanisms of transcriptional repression can be considered in two categories: active repression and passive repression (7, 22, 24, 51). Passive repressors function by interfering with transcriptional activators, either by competing for the same binding site or by masking the function of their activation domains (42, 58). Active repressors repress independently; their activity is not dependent upon interference with specific activators. They may repress transcription by interacting with components of the general transcription machinery, like Tag (19) and even-skipped (25, 35). Alternatively, they may function by recruiting corepressors with intrinsic repression activity. One type of corepressor complex involves recruitment of histone deacetylases (HDACs) (16, 48, 64). Many transcriptional repressors associate with HDACs by bridging proteins that function as corepressors (1, 20, 21, 33, 36, 47, 69). For example, Mad recruits the Sin3 complex that includes mSin3A or -B, HDAC1 or -2, RbAp46 or -48, Ski, and at least two other polypeptides of unknown function, SAP18 and SAP30 (20, 32, 33, 47, 69). The repression complex associating with unliganded nuclear receptors (36, 46), PLZF (8), PLZF-RARα, and Bcl-6 (9) requires the presence of SMRT/NCoR in addition to mSin3 and HDAC. However, YY1 (66) and Rb family proteins Rb (43), p107 (12), and p130 (12) all interact directly with HDAC and no other corepressors are found in their complexes. PLZF and Bcl-6 associate both with SMRT/NcoR and directly with HDAC (9). Recruitment of HDAC to DNA appears to alter nucleosome structure in a local region and inhibit transcription, presumably because acetylation neutralizes the positive charge on lysines in the histone tails and alters intra- and/or internucleosomal structure. Other corepressors, such as Groucho and Kap-1, have also been identified, but their mechanism of action is not yet understood (13).
In this paper, we studied the repression mechanism of Blimp-1 by testing whether Blimp-1 is an active or a passive repressor and by exploring the possible role of HDAC in Blimp-1-dependent repression. On the c-myc promoter, we found that Blimp-1 functions as an active repressor independent of activator YY1, which binds nearby. Also, we found that Blimp-1 associates with HDACs directly, suggesting that it can recruit HDACs to promoters it binds. In addition, inhibition of cellular HDAC activity relieved repression of Blimp-1 on the c-myc promoter as well as the repression of a Gal4-Blimp-1 fusion protein on a thymidine promoter with Gal4 binding sites. Finally, by using a chromatin immunoprecipitation (ChIP) assay, we show that expression of Blimp-1 causes deacetylation of histone H3 at the c-myc promoter. Taken together, these results suggest that Blimp-1-dependent repression involves alteration of local chromatin structure by recruitment of HDAC.
MATERIALS AND METHODS
Plasmids.
Luciferase reporters driven by the c-myc promoter (−1100/+580), mPRF-c-myc promoter (−1100/+580), (27), c-myc promoter (−424/+340), and mYY1-c-myc (−424/+340) (53), the Blimp-1 expression vector pBDP1-F, vector control pBDP1-B (Blimp-1 cDNA in reverse orientation) (62), YY1 expression plasmid pCMV-YY1, and vector control pCMV (53) have been previously described. The expression constructs of hemagglutinin (HA)-tagged full-length Blimp-1 (HA-Blimp-1) and deletion mutants H1 to H4 were cloned by PCR amplification of Blimp-1 cDNA fragments by using pBDP1-F as the template and using the primers shown below, followed by subsequent ligation into the XbaI site on the pCGN vector (29). DNA sequence analysis was performed to confirm the clones.
For HA-Blimp-1, the PCR primers used were 5′GTTCTAGATTTCTCAGATGTTGGAT and 3′CCTCTAGACACGAAACACTTATT. For H1, the PCR primers used were 5′AATCTAGAAAATACATAGTGAACGA and 3′CCTCTAGACACGAAACACTTATT. For H2, the PCR primers used were 5′AATCTAGAAATGGTATCAACAACT and 3′CCTCTAGACACGAAACACTTATT. For H3, the PCR primers used were 5′GTTCTAGATTTCTCAGATGTTGGAT and 3′AATCTAGAAGTTGCCCTTCAGGT. For H4, the PCR primers used were 5′GTTCTAGATTTCTCAGATGTTGGAT, 3′CCTCTAGACACGAAACACTTATT, and 3′GGCTGAAGTTGTTGATACCATTCTCTTCAAACTCGGCCTCTGTC.
pBSK-Blimp-1 full length (B1) was cloned by inserting the XhoI-XhoI Blimp-1 cDNA from pBDP1-F into the XhoI site of the pBluescript SKII(+) (Stratagene). B2 to B5 were cloned by inserting EcoRI-digested Blimp-1 fragments obtained by PCR using H1 to H4 as templates, respectively, and using 5′GTTCTAGAGAATTCACCTCCATAGAA and 3′AATCTAGAGAATTCCCTGAAGTTCTC as primers into the EcoRI site on the pSK vector. B6 was cloned by digesting B4 with SmaI to delete a 500-bp Blimp-1 fragment and religating the remaining plasmid. B7 was made by inserting a Blimp-1 fragment made by PCR using B1 as the template and using the PCR primers 5′AATCTAGAATGAAACAGAATGGCAAGAT and 3′AATCTAGAAGTTGCCCTTCAGGT into the XbaI site of the pSK vector. B8 was made by digesting B6 with PmlI and EcoRI to delete a 300-bp Blimp-1 cDNA fragment and end-filling and religating the remaining plasmid. B9 was made by replacing the 2.1-kb PmlI-PacI Blimp-1 fragment on B1 with the 1.6-kb BsrB1-PacI Blimp-1 fragment cut from Blimp-1 cDNA. B10 was cloned by digesting B9 with EcoNI and EcoRI to delete the 1-kb Blimp-1 fragment and end-filling and religating the remaining plasmid. B11 was made by digesting B9 with AvrII and EcoRI to delete the 1.5-kb Blimp-1 fragment and end-filling and religating the remaining plasmid. B12 was made by inserting the EcoNI-AvrII-digested Blimp-1 fragment made by PCR using B1 as the template and using 5′GAGGCATCCTTACCAAGGAACCTGCT and 3′CAACCTAGGGGAGGGATTGGAGTCCAGTTTTAGAA as primers into EcoNI-AvrII-digested B9. All PCR products used in cloning procedures were confirmed by DNA sequencing. Gal4-Blimp-1 was made by inserting Blimp-1 cDNA resulting from ScaI-BamHI-digested B1, after end-filling, into the SmaI site of the pGal4(1-147) vector (29). Expression plasmids for Blimp-1 fused to Gal4 DNA binding domains (Gal4DBDs) (G1 to G4) were made by inserting the respective PCR fragments obtained by using B1 as the template and using the pairs of primers shown below, after BamHI-XbaI digestion, into the BamHI and XbaI sites of pGal4(1-147). Again, all PCR-generated fragments were confirmed by DNA sequencing. For G1, the PCR primers used were 5′CGGGATCCGTTGGATCTTCTCTTGGA and 3′GTTCTAGAAGGCAGCCAGGTTTTGCTCC. For G2, the PCR primers used were 5′AAAGGATCCGCGTGGTAAGTAAGGAGT and 3′TTCTCTAGACTTTCCGTTTGTGTGAGA. For G3, the PCR primers used were 5′AAAGGATCCTGGCCTATGGGATGGAGA and 3′AAGTCTAGAGGCTGCTGCCACTAAGGA. For G4, the PCR primers used were 5′GCGGATCCAACTGAAGGGCAACTGC and 3′CCTCTAGACACGAAACACTTATT. Gal4-Blimp-1 (amino acids [aa] 557 to 714) and Gal4DBD fused to Blimp-1 with both binding domains deleted were generated by inserting the respective Blimp-1 fragments obtained from B8 and B12 into pGal4(1-147).
Cell lines and culture.
293T human kidney fibroblast cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Gemini). 18-81 pre-B cells were grown in RPMI 1640 medium with 10% heat-inactivated fetal bovine serum and 50 μM β-mercaptoethanol.
Transfections and luciferase assay.
Transient DNA transfections were performed by calcium phosphate precipitation in 293T cells. Briefly, cells were split at a density of 5 × 105 cells/10-cm plate the day before transfection and the cells were fed 3 h before transfection. DNA was added as a Ca3(PO4)2 precipitate (25 mM HEPES, 140 mM NaCl, 750 μM Na2HPO4) to the medium. Twelve hours later, the medium was changed, and 36 h later, cells were harvested. Transient DNA transfections were performed by electroporation in 18-81 pre-B cells. For each transfection, 5 × 106 log-phase cells were collected by centrifugation, washed in RPMI medium, resuspended in 300 μl of the same medium, and transferred to a 0.4-cm electrode gap gene pulser cuvette (BTX). After addition of DNA, the samples were gently shaken and subjected to electroporation at 960 μF and 240 V with a GenePulser apparatus (Bio-Rad). After electroporation, samples were diluted with 10 ml of RPMI culture medium and incubated at 37°C with 5% CO2. For experiments using trichostatin A (TSA), transfected cells were split into two dishes immediately after electroporation: one plate remained untreated and the other was treated with TSA (100 ng/ml). Cells were harvested for the luciferase assay 16 h after electroporation.
For the luciferase assay, 10 ml of transfected cells was harvested and centrifuged at 2,000 rpm (500 × g) at 4°C for 5 min. The cell pellets were lysed (25 mM glycylglycine [pH 7.8], 12 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol [DTT] and 1% Triton) and then centrifuged. One hundred microliters of each supernatant was mixed with 500 μl of luciferase substrate buffer (5 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 15 mM K2HPO4, and 2 mM ATP) and 20 μl of luciferin solution (1 mM luciferin, 25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT), and the luminescence was measured with a Berthold Lumat LB9501 luminometer.
Immunoprecipitation and Western blot analysis.
An aliquot containing 107 transfected 293T cells was washed with 1× phosphate-buffered saline and subsequently lysed in PBS plus 0.1% NP-40 containing protease inhibitors. The lysates were sonicated, clarified by centrifugation, and immunoprecipitated at 4°C with the indicated antibodies and protein A-agarose beads (Santa Cruz). The immunoprecipitations were washed five times with PBS plus 0.1% NP-40 and then resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels, electrotransferred to nitrocellulose membranes, and examined by Western blot analysis. Membranes, after transfer, were blocked in 5% dry milk in PBS for 1 h and then incubated with indicated primary antibodies in PBS plus 2% dry milk for 2 h, followed by incubation with appropriate secondary antibodies in PBS plus 2% dry milk for 1 h. The membranes were washed in PBS plus 0.2% Tween 20 between each step, developed by using an enhanced chemiluminescence detection kit (Pierce), and exposed to X-ray film.
GST fusion proteins and in vitro binding assays.
Glutathione S-transferase (GST) fusion proteins were expressed from the appropriate pGEX recombinant vectors in transformed Escherichia coli XL-1 Blue and were purified by immobilization on a glutathione-agarose matrix as previously described (17) except for two modifications. First, bacteria were grown at 30°C. Second, cells were lysed in a solution containiing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 100 μg of lysozyme/ml, 5 mM DTT, 1.5% N-lauryl sarcosine, the protease inhibitors (each at 2 μg/ml) aprotinin, pepstatin, and leupeptin, and 100 mM phenylmethylsulfonyl fluoride (PMSF). 35S-radiolabeled Blimp-1 proteins were synthesized by a coupled in vitro transcription-translation protocol (TnT; Promega) and incubated with a 50% slurry of the corresponding immobilized GST fusion protein in buffer B (20 mM HEPES [pH 7.6], 100 mM NaCl, 4 mg of dry milk/ml, 5 mM DTT, the protease inhibitors [each at 2 μg/ml] aprotinin, pepstatin, and leupeptin, and 1 mM PMSF) for 2 h at 4°C with gentle rocking. The agarose beads were then washed five times with 1 ml (each time) of PBS plus 0.1% NP-40. Bound proteins were eluted in 20 μl of SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, resolved by SDS-PAGE, and exposed to X-ray film.
EMSA.
P3X nuclear extracts were prepared as previously described (27). Probes were labeled with [γ-32P]ATP. An electrophoretic mobility shift assay (EMSA) was performed as previously described (70). Unlabeled competitor oligonucleotides (50-fold molar excess) or αHDAC1 antibody (Santa Cruz) were incubated with nuclear extracts on ice for 30 min prior to the addition of probe.
ChIP assays.
ChIP assays were performed essentially according to the protocol for the Acetyl-Histone H3 ChIP Assay Kit (Upstate Biotechnology). Twenty-four hours after transfection, 2.5 × 107 18-81 cells were cross-linked by addition of formaldehyde directly into the medium to achieve a final concentration of 1% and incubated for 30 min at 37°C. Formaldehyde was then quenched with 0.125 M glycine. Cells were washed, suspended in PIPES buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40) containing protease inhibitors, pelleted, and resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) with protease inhibitors. The lysates were subsequently subjected to sonication to reduce DNA length to between 200 and 1,000 bp. Samples were then diluted 10-fold by using dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) and precleared by incubating with protein A beads overnight; anti-acetyl histone H3 antibody (Upstate Biotechnology) was added and immunoprecipitation was done overnight at 4°C with rotation. Immunocomplexes were collected with protein A agarose beads and eluted after extensive washing, and cross-links were reversed by heating at 65°C. Samples were subjected to proteinase K treatment. DNA was recovered by phenol-chloroform extraction, ethanol precipitated, and used as a template for PCR (twofold serial dilutions for 25 cycles) using c-myc promoter-specific primers (5′CAACCGTACAGAAAGGGAAAGGACTAGCGC3′ and 5′TCCCTTCCCCACCTCTCTCTATTTTTTTC3′). PCR products were transferred onto a nylon membrane and hybridized with a specific probe. The linearity of the PCR was verified by phosphorimager analysis.
RESULTS
Blimp-1-dependent repression of the c-myc promoter does not involve YY1.
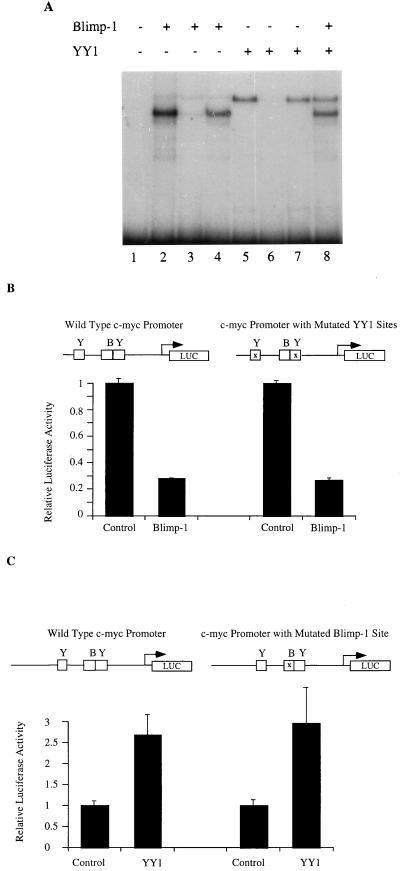
The Blimp-1 binding site, located 290 bp 5′ of the P1 transcriptional start site on the murine c-myc promoter (40), is adjacent to a binding site for YY1, which activates c-myc transcription (52, 53). Earlier studies showed that proteins binding these two sites were able to bind simultaneously and suggested that the proteins binding these sites might bind cooperatively (26, 27). Therefore, we wished to test directly whether the ability of Blimp-1 to repress c-myc transcription involved effects on either the DNA binding or the transcriptional activation properties of YY1. First, an EMSA was used to study whether YY1 and Blimp-1 bind cooperatively on the c-myc promoter. A 213-bp fragment of the c-myc promoter which contains both the Blimp-1 and the 3′ YY1 binding sites was used as a probe in these experiments (26, 53). Recombinant Blimp-1 and YY1, synthesized in vitro, were incubated with the probe. Both Blimp-1 and YY1 alone yielded a single retarded band, shown to represent sequence-specific binding since each was competed away by a specific, but not by a nonspecific, competitor (Fig. 1A, lanes 2 to 5). When Blimp-1 and YY1 were incubated together with the probe under conditions of probe molar excess, both single-protein complexes were observed. However, no slower mobility complex, which would have indicated cooperative binding of Blimp-1 and YY1, was observed (Fig. 1A, lane 8). Therefore, these proteins appear to bind independently on the c-myc promoter.
FIG. 1.
Blimp-1 and YY1 act independently on the c-myc promoter. (A) A 213-bp probe (SmaI-HapII) from the murine c-myc gene (26) encompassing both the Blimp-1 site and the 3′ YY1 site was used with in vitro-translated Blimp-1 (lanes 2 to 4) or YY1 (lanes 5 to 7) or both (lane 8) in the presence of no competitor (lanes 2, 5, and 8), specific competitor (lanes 3 and 6), or nonspecific competitor (lanes 4 and 7) in EMSA. The competitors were added in 50-fold molar excess. (B) One microgram of a luciferase reporter fused with wild-type c-myc promoter (−424/+340) or a c-myc promoter with both two YY1 sites mutated (−424/+340) (53) was transfected into 18-81 cells by electroporation together with 10 μg of a Blimp-1 expression plasmid or control plasmid with Blimp-1 c-DNA inserted in the reverse orientation. The transfected cells were harvested 16 h after transfection, and luciferase activity was then measured. (C) One microgram of a luciferase reporter fused with the wild-type c-myc promoter (−1100/+580) or a promoter containing a mutation in the Blimp-1 site (−1100/+580) (27, 40) was transfected into 18-81 cells by electroporation together with 10 μg of pCMV-YY1, a YY1 expression construct, or a pCMV vector control. Transfection results are averages of three or more independent transfections, and the error bars show 1 standard deviation from the mean.
A transient-transfection assay was used to determine if or how Blimp-1 and YY1 affect one another's regulation of c-myc promoter activity. Cotransfection of a Blimp-1 expression plasmid into 18-81 pre-B cells with a luciferase reporter that is dependent on either a wild-type c-myc promoter fragment or a fragment containing a mutation of both YY1 sites resulted in similar amounts of transcriptional repression (Fig. 1B). Thus, repression of the c-myc promoter by Blimp-1 is independent of the YY1 binding sites or binding of endogenous YY1. Similarly, cotransfections of a YY1 expression plasmid activated a c-myc promoter containing a mutated Blimp-1 binding site as well as it activated a wild-type promoter (Fig. 1C). When YY1 and Blimp-1 expression plasmids were cotransfected together with a reporter dependent on a wild-type c-myc promoter, their activities were additive (data not shown). Thus, we conclude that Blimp-1 represses and YY1 activates the c-myc promoter independently of one another. These data are consistent with the idea that Blimp-1 repression occurs by an active rather than a passive mechanism since it is independent of YY1.
Multiple regions of Blimp-1 are involved in its repression activity.
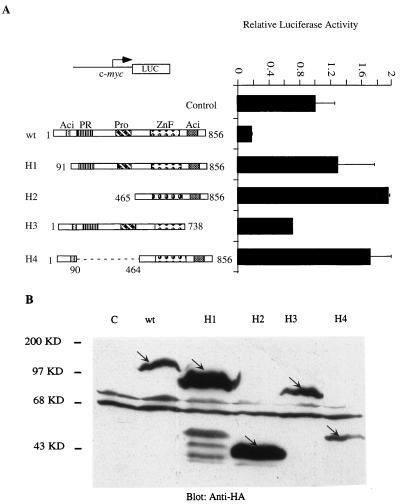
One characteristic of repressors that work by an active mechanism is that DNA binding alone is not sufficient to mediate repression; additional domains of the protein are required. To learn more about the mechanism of Blimp-1 repression, we tested which domains of the protein were required for repression of the c-myc promoter. Expression plasmids for four Blimp-1 mutants were made, in which identifiable structural motifs of Blimp-1 were removed. Blimp-1 contains two acidic regions, one located at each of the N and C termini, a PR region which is homologous to a region on an Rb-associating protein called Riz (5), a proline-rich domain, and a region containing five zinc finger motifs which confer DNA binding (62) (Fig. 2A). Expression of HA-tagged mutant proteins in cells transiently transfected with expression plasmids was monitored by immunoblotting with a monoclonal antibody to the HA tag (Fig. 2B). The repression activity of the Blimp-1 mutants was subsequently tested by cotransfection into 18-81 pre-B cells by using a luciferase reporter dependent on the c-myc promoter (Fig. 2A). Blimp-1 lacking the N-terminal 90 amino acids (H1) failed to repress the c-myc promoter. Also, a larger N-terminal truncation, removing aa 1 to 464 (H2), which includes the N-terminal acidic, PR, and proline-rich domains, also resulted in the loss of repressor activity and showed modest activation. C-terminal truncation of aa 738 to 856, which removed the C-terminal acidic domain (H3), impaired but did not abolish Blimp-1 repression activity. An internal deletion mutant that retained the N-terminal acidic region but lacked the PR and proline-rich domains (H4) showed loss of repression and modest transcriptional activation. These data suggest that multiple regions of Blimp-1, including the N-terminal acidic domain and the region between aa 90 and 464, are required for Blimp-1 to repress the c-myc promoter. The data are also consistent with our supposition that Blimp-1 repression proceeds by an active mechanism, since DNA binding is not sufficient to yield repression.
FIG. 2.
Mutational analysis of Blimp-1 domains required for repression of the c-myc promoter. (A) One microgram of a luciferase reporter driven by the c-myc promoter (−1100/+580) was transfected into 18-81 cells with plasmids encoding full-length HA-Blimp-1 (wt) or various HA-tagged Blimp-1 deletion mutants (H1 to H4) (schematic structures of mutations are shown at left) or a vector with Blimp-1 cDNA inserted in a reverse direction (Control). Cells were harvested 16 h after transfection, and luciferase activity was measured. (B) Ten micrograms of the HA-tagged Blimp-1 deletion constructs (H1 to H4) shown in panel A was transfected into 293T cells. Thirty-six hours later, whole-cell extracts were made and subjected to immunoblotting with a monoclonal antibody recognizing the HA tag (12CA5). The HA-tagged proteins are indicated by arrows. Transfection results are averages of three or more independent transfections, and error bars show 1 standard deviation from the mean.
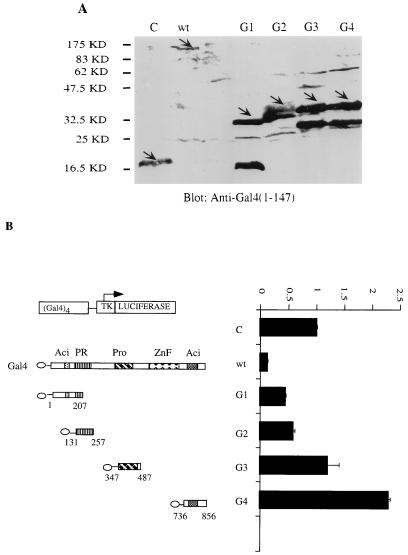
Another characteristic of active repressors is that they can repress transcription in a Gal4-fusion protein assay in which DNA binding is mediated by a heterologous domain. Therefore, the ability of Blimp-1 fused to the Gal4DBD (Gal4DBD-Blimp-1) to repress a synthetic thymidine kinase (tk) promoter containing four Gal4 binding sites, (Gal4)4-tk, was tested. We also fused the Blimp-1 N-terminal acidic domain, the PR domain, the proline-rich domain, and the C-terminal acidic domain with the Gal4DBD and confirmed their expression following transient transfection into 293T cells by immunoblotting with antibody recognizing Gal4DBD (Fig. 3A). Full-length Blimp-1-Gal4DBD repressed the (Gal4)4-tk promoter (Fig. 3B), but not a tk promoter lacking Gal4 binding sites (data not shown), upon cotransfection into 18-81 pre-B cells. However, none of the fusion proteins containing individual Blimp-1 domains showed complete repression (Fig. 3B). Partial repression was observed with the N-terminal acidic and PR domains. Interestingly, the isolated C-terminal acidic domain activated the (Gal4)4-tk promoter (Fig. 3B). Thus, these data also support the notion that Blimp-1 is an active repressor and show that complete repression in this assay requires portions of the Blimp-1 protein not included in the individual domain fusion proteins we tested.
FIG. 3.
Repression by Blimp-1 in a Gal4 fusion protein assay. (A) Ten micrograms of expression plasmids encoding proteins with wild-type Blimp-1 (wt) or different structural motifs of Blimp-1 (G1, N-terminal acidic domain; G2, PR domain; G3, proline-rich domain; G4, C-terminal acidic domain) (schematic diagram of mutations are shown in panel B) fused to the Gal4DBD or, as a control, Gal4DBDs (28) were transfected into 293T cells. Whole-cell extracts made 36 h after transfection were subjected to immunoblotting with a polyclonal antibody recognizing Gal4DBD. Fusion proteins are indicated by arrows. (B) 18-81 cells were transfected with a luciferase reporter fused to a tk promoter which harbors four Gal4 DNA binding sites and with 10 μg of wild-type Blimp-1 (wt) or various Gal4-fusion Blimp-1 domains (G1 to G4) or an empty vector which expressed only Gal4DBD (28). Sixteen hours later, cells were harvested and luciferase activity was measured. Transfection results are averages of three or more independent transfections, and error bars show 1 standard deviation from the mean.
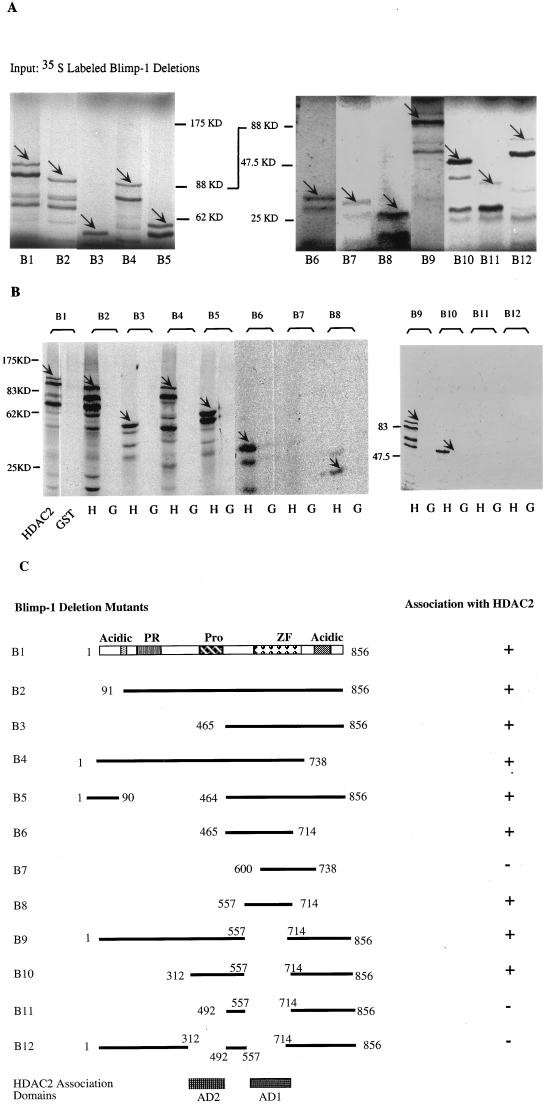
Blimp-1 associates directly with HDAC2 through two independent association domains.
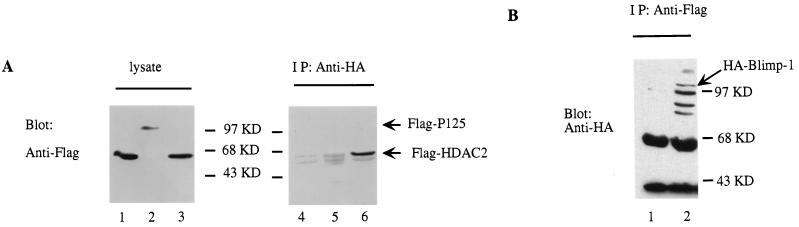
The mechanism of action of some active repressors involves recruitment of HDACs (16, 48, 64). Therefore, we wished to determine if recruitment of HDAC was involved in Blimp-1-dependent repression. First, we used a coimmunoprecipitation assay to determine whether ectopically expressed Blimp-1 and HDAC2 associated in vivo. HA-tagged Blimp-1 and Flag-tagged HDAC2 were expressed in transiently transfected 293T cells. When Blimp-1 was immunoprecipitated with a monoclonal antibody against the HA epitope, HDAC-2 in the immunoprecipitate was easily detected by a monoclonal antibody recognizing the Flag tag (Fig. 4A, lane 6); however, a control Flag-tagged protein was not detected in the immunoprecipitate (Fig. 4A, lane 5). In addition, when HDAC2 was immunoprecipitated with Flag monoclonal antibody, Blimp-1, detected by the HA monoclonal antibody, was present in the immunoprecipitate (Fig. 4B). Similar results were obtained with Flag-tagged HDAC1 in the coimmunoprecipitation assay (data not shown). HDAC family proteins (HDAC1 to -3) are more than 50% homologous and ubiquitously expressed (67); thus, we assume that they function similarly. These data show that Blimp-1 and HDAC1 or HDAC2 are present in the same complex under these experimental conditions.
FIG. 4.
In vivo association between Blimp-1 and HDAC2. (A) 293T cells were cotransfected with an expression plasmid encoding HA-Blimp-1 (lanes 2, 3, 5, and 6) or, as a control, empty vector (lanes 1 and 4) and Flag-HDAC2 (lanes 1, 3, 4, and 6) or an expression plasmid encoding Flag tag control protein Flag-p125 (lanes 2 and 5). Immunoblots were developed with M2 monoclonal antibody to the Flag epitope on either lysates of transfected cells (left) or immunoprecipitates (IP) using monoclonal antibody 12CA5 against the HA epitope from transfected cells (right). Positions of expected Flag-tagged proteins are marked with arrows. (B) 293T cells were cotransfected with HA-Blimp-1 expression plasmids (lanes 1 and 2) and Flag-HDAC2 (lane 2) or an expression construct of Flag tag control protein Flag-p125 (lane 1). Immunoblots were developed with monoclonal 12CA5 HA antibody on immunoprecipitates (IP) using monoclonal M2 Flag antibody from transfected cells.
Most repressors recruit HDACs via bridging molecules (1, 20, 21, 33, 36, 46, 69). However, we were unable to detect association of Blimp-1 with Sin3 by using coimmunoprecipitation and a yeast two-hybrid assay (data not shown) or with NCoR by using a coimmunoprecipitation assay (data not shown). Therefore, we asked if Blimp-1 associated directly with HDAC2 by using a GST fusion protein assay (2). In this experiment, a GST-HDAC2 fusion protein was expressed in bacteria and purified by binding to glutathione agarose beads. 35S-labeled Blimp-1 and mutant forms of Blimp-1, synthesized in vitro, were adjusted to be present at comparable amounts in the reaction mixtures, which were subsequently tested for their ability to associate with the immobilized GST-HDAC2 (Fig. 5A). Blimp-1 was retained by the GST-HDAC2 matrix but not by the control GST matrix (Fig. 5B, lanes B1). Similar results were obtained with GST-HDAC1 (data not shown). We also observed binding of HDAC2 to Blimp-1 by using HA-tagged Blimp-1 that was immunopurified from transiently transfected 293T cells by using beads coated with antibody against the epitope tag (data not shown). These data strongly suggest that Blimp-1 binds directly to HDAC1 and HDAC2; however, we cannot formally rule out the possibility that a tightly bound bridging protein, present in the in vitro synthesis reaction mixture and tightly bound during immunopurification, is involved. The data are also consistent with the hypothesis that Blimp-1 represses transcription by recruiting HDACs.
FIG. 5.
Blimp-1 contains two regions which independently associate with HDAC2. (A) Autoradiograph of [35S]Met-labeled full-length Blimp-1 (B1) and deletion mutants (B2 to B12) synthesized in vitro after resolution on SDS-PAGE gel (panel C, schematic diagram of the various mutations). The in vitro-translated Blimp-1 full-length and deletion mutants are indicated by arrows. (B) Equivalent amounts of glutathione beads with immobilized and purified GST-HDAC2 (lanes H) or GST control (lanes G) were mixed with equivalent amounts of the in vitro-translated Blimp-1 proteins. The beads were washed thoroughly, and bound proteins were eluted with SDS loading buffer before analysis on SDS-PAGE gels. The retained signals of Blimp-1 full-length and deletion mutants are marked by arrows. (C) Summary of Blimp-1 deletion mutations and their abilities to associate with HDAC2 in this assay. The two domains required for HDAC2 binding were shown on the bottom labeled as AD1 and AD2; only AD1 has been shown to be sufficient to mediate the association.
The GST assay was also used to identify the domains of Blimp-1 which were required for association with HDAC2 by testing a series of Blimp-1 truncation and deletion mutants. Mutant B8 (aa 557 to 714), which includes most of the Blimp-1 Zn finger region and 51 amino acids N-terminal to the first zinc finger, was identified as a minimal fragment of Blimp-1, which was sufficient to associate with HDAC2 (Fig. 5B, lanes B6 to B8). This region contains the two zinc finger domains that are essential for sequence-specific DNA binding (31). However, when a Blimp-1 mutant with an internal deletion of this HDAC association domain, B9 (Δaa 557 to 714), was tested, it still associated with HDAC2 (Fig. 5B). This suggested the existence of another, independent HDAC2 association domain in Blimp-1. Therefore, a second series of Blimp-1 mutants, all containing the aa 557-to-714 deletion, was tested for their ability to associate with HDAC2. This study revealed another HDAC2 association domain located between aa 312 and 492, which spans the proline-rich region (Fig. 5B, lanes B10 and B11). A Blimp-1 mutant with both domains deleted, B12, lost its ability to associate with HDAC2, suggesting that there are no more HDAC2 association domains in Blimp-1. These data, summarized in Fig. 5C, identified two domains of Blimp-1 which associate independently with HDAC2 and showed that aa 557 to 714 comprise a minimal HDAC association domain.
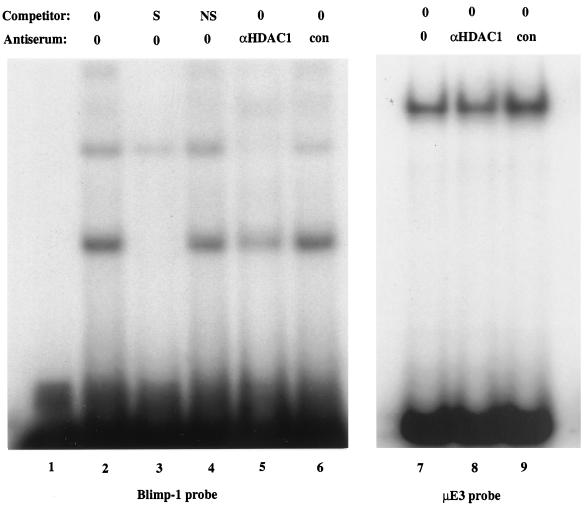
We also wished to determine if Blimp-1 and HDAC associated when the proteins were present at endogenous levels in vivo. Nuclear extracts from P3X plasmacytoma cells, which express Blimp-1, were used in EMSA experiments with an oligonucleotide probe containing the c-myc Blimp-1 site. Two complexes that competed with specific but not with nonspecific oligonucleotide competitors were observed (Fig. 6, lanes 2 to 4). Previous work has shown that antiserum against Blimp-1 ablates all complexes which bind specifically to this site (40). Antibody against HDAC1, but not a control antibody, strongly inhibited formation of both Blimp-1 complexes (Fig. 6, lanes 5 and 6). However, neither anti-HDAC1 nor control antiserum altered an unrelated protein-DNA complex formed on the μE3 site from the immunoglobulin heavy chain intronic enhancer (Fig. 6, lanes 7 to 9), demonstrating that anti-HDAC1 does not ablate protein-DNA complexes nonspecifically. We conclude that endogenous HDAC1, or protein immunologically related to it, is present in these complexes containing DNA and Blimp-1. Ablation of binding by antibodies to HDAC is consistent with our finding that one domain of Blimp-1 which associates with HDAC completely overlaps the zinc finger motifs that confer DNA binding (31). The components of the minor, lower mobility complex are not known; it could contain Blimp-1 associated with two molecules of HDAC, Blimp-1 associated with both HDAC and Groucho (50), or Blimp-1 associated with HDAC and an unidentified protein. The absence of a higher mobility complex binding the probe shows that free Blimp-1, not associated with HDAC, is not detectable in this assay, implying nearly stoichiometric association of Blimp-1 with HDAC. These data show that Blimp-1 and HDAC, present at endogenous concentrations in plasmacytoma cells, associate with one another and that Blimp-1 can recruit HDAC to DNA.
FIG. 6.
Blimp-1 associated with HDAC binds to the c-myc promoter. In lanes 1 to 6, [γ-32P]ATP-labeled oligonucleotides corresponding to the c-myc Blimp-1 binding site (5′CGCGTACAGAAAGGGAAAGGACTAG3′ and 5′CGCGCTAGTCCTTTCCCTTTCTGTA3′) were used with P3X nuclear extracts in the presence or absence of unlabeled oligonucleotide competitors or anti-HDAC1 or control antiserum, as indicated. The competitors were added in 50-fold molar excess. S, specific competitor; NS, nonspecific competitor. In lanes 7 to 9, [γ-32P]ATP-labeled oligonucleotides corresponding to the μE3 site of the immunoglobulin heavy chain enhancer were used with P3X nuclear extracts in the presence of anti-HDAC1 or control antiserum, as indicated.
Blimp-1-dependent repression requires HDAC activity.
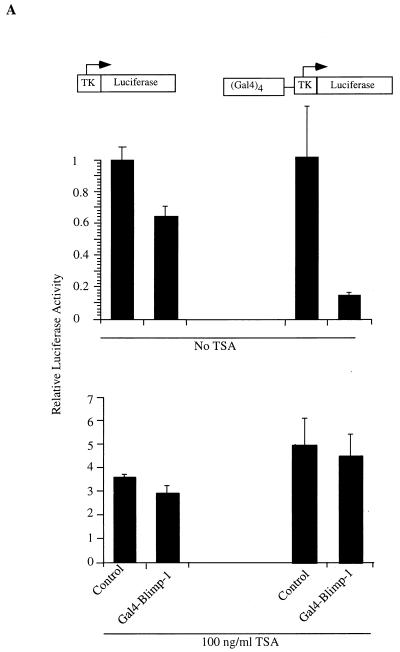
The association between Blimp-1 and HDAC2 suggested that Blimp-1 might repress transcription by recruiting HDACs to the target promoter. To test this hypothesis, we first measured the ability of GAL4DBD-Blimp-1 fusion proteins to repress the (Gal4)4-tk promoter in the presence of TSA, an inhibitor of HDAC activity (60, 68). Full-length Blimp-1 fused to the Gal4DBD repressed the (Gal4)4-tk promoter upon cotransfection into 18-81 pre-B cells in a Gal4 binding site-dependent manner (Fig. 7A). However, in the presence of 100 ng of TSA/ml, Blimp-1-dependent repression was abolished (Fig. 7A).
FIG. 7.
Recruitment of HDAC is sufficient for Blimp-1 repression in the Gal4 assay. (A) Expression plasmids encoding Gal4DBD-Blimp-1 fusion protein (Gal4-Blimp-1) or a vector which expresses only Gal4DBD (Control) were cotransfected into 18-81 cells with the (Gal4)4-tk promoter driving the luciferase gene or tk-Luc (which lacks Gal4DNA binding sites). Where indicated, transfected cells were treated with the HDAC inhibitor TSA at 100 ng/ml after transfection. Cells were harvested 16 h after transfection, and luciferase activities were measured. (B) Ten micrograms of expression plasmid encoding Gal4DBD-Blimp-1 with internal deletions of aa 312 to 492 and aa 557 to 714 (which cannot bind HDAC2; refer to Fig. 5) or Gal4-Blimp-1 containing only aa 557 to 714 (which is sufficient to bind HDAC2; refer to Fig. 5) was cotransfected into 18-81 cells with 2 μg of (Gal4)4-tkLuc. A plasmid which expresses full-length Blimp-1 fused to Gal4DBD (Gal4-Blimp-1) was used as a positive control, and one which expresses Gal4DBD (Control) was used as a vector control. Transfection results are averages of three or more independent transfections, and error bars show 1 standard deviation from the mean.
We also tested whether the association of Blimp-1 with HDAC2 correlated with its ability to repress transcription. Gal4DBD fusion proteins were made with the minimal Blimp-1 fragment (aa 574 to 714), which associates with HDAC2 (Fig. 7B) and the association defective deletion mutant of Blimp-1 (Fig. 5C, 12). Expression of both fusion proteins was confirmed by immunoblotting after transient transfection into 293T cells (data not shown). Upon cotransfection, Gal4DBD-Blimp-1(574-714) repressed the (Gal4)4-tk promoter as well as wild-type Blimp-1 while the association-defective mutant B12 failed to repress transcription (Fig. 7B). Repression by Gal4-Blimp-1 (574-714) was also inhibited in the presence of TSA (data not shown). Thus, Blimp-1-dependent repression in this assay correlates with the ability of Blimp-1 to associate with HDAC2. We conclude that deacetylase activity is required for Blimp-1 repression on the synthetic (Gal4)4-tk promoter and that recruitment of HDAC is one mechanism by which Blimp-1 represses transcription.
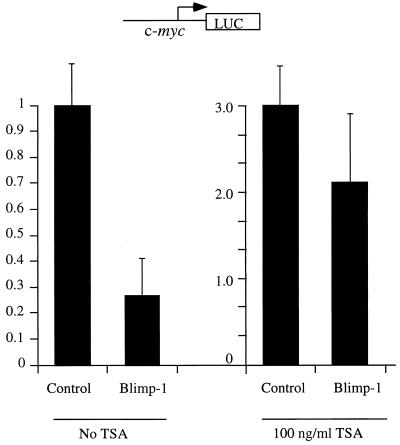
However, a more physiologically relevant question is whether HDAC activity is required for Blimp-1 repression on the c-myc promoter, which is a natural target for Blimp-1 repression during terminal differentiation of B lymphocytes (40). A similar approach using the inhibitor TSA in a cotransfection assay was employed to test this possibility. The results show that treatment of 18-81 pre-B cells with TSA significantly inhibits Blimp-1-dependent repression of the c-myc promoter (Fig. 8). These data suggest that recruitment of HDAC is required for Blimp-1-dependent repression of a natural target, the c-myc promoter.
FIG. 8.
HDAC activity is required for Blimp-1 repression on the c-myc promoter. Ten micrograms of Blimp-1 expression plasmid or a control with Blimp-1 cDNA inserted in reverse orientation were transfected into 18-81 cells with 1 μg of the c-myc promoter (−1100/+580) driving a luciferase reporter. Immediately after transfection, cells were split into two parts and were subjected to either treatment with 100 ng of TSA/ml or no treatment before being harvested 16 h later. Data shown are the averages of nine independent transfections, and error bars show 1 standard deviation from the mean.
Blimp-1 expression causes deacetylation of histone H3 bound to the c-myc promoter.
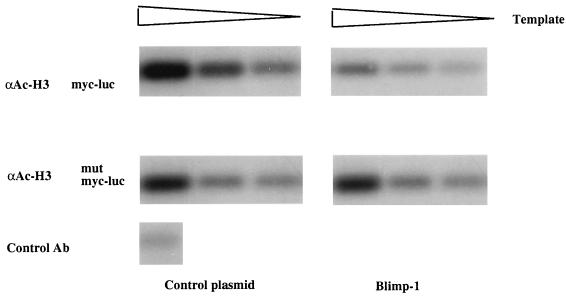
To test further the hypothesis that histone deacetylation is involved in Blimp-1-dependent repression of c-myc transcription, we employed the recently developed ChIP approach (43). In this assay, histones in chromatin are cross-linked to DNA and acetylated histone H3 in chromatin is immunoprecipitated; DNA sequences of interest are then amplified by PCR. Under these conditions, a decrease in the amount of the PCR product will reflect a decrease in acetylated histone H3 bound to the amplified sequence. 18-81 cells were cotransfected with luciferase reporters dependent on either a wild-type c-myc promoter or a c-myc promoter containing a mutation in the Blimp-1 binding site (27) and with an expression plasmid for Blimp-1 or a control plasmid. As previously reported (40), luciferase assays showed that Blimp-1 repressed the c-myc promoter approximately fivefold (data not shown). Transfected cells were treated with formaldehyde to covalently cross-link histones to DNA, and chromatin was isolated, fragmented, and immunoprecipitated with antibodies to acetylated histone H3, as previously described (43). Acetylated chromatin in the immunoprecipitates was purified, and following removal of the cross-links, c-myc promoter DNA sequences in the immunoprecipitates were detected by PCR using specific primers for the transfected promoter. The linearity of the PCR was obtained by amplification of twofold serial dilutions of the DNA samples, and PCR products were quantitated by a phosphorimager. c-myc sequences were readily amplified after the ChIP assay was performed on cells cotransfected with the c-myc promoter and a control plasmid; in contrast, expression of Blimp-1 caused an approximately 70% decrease in the amount of c-myc promoter sequences associated with acetylated histone H3 (Fig. 9, top panels). However, expression of Blimp-1 did not alter the amount of acetylated histone H3 associated with the c-myc promoter lacking a Blimp-1 binding site (Fig. 9, lower panels), thus showing that the observed changes in histone H3 acetylation depend on binding of Blimp-1 to its cognate site in the c-myc promoter.
FIG. 9.
Blimp-1 expression causes deacetylation of histone H3 bound to the c-myc promoter. Top, a luciferase reporter driven by the c-myc promoter was cotransfected into 18-81 pre-B cells along with control (left) or Blimp-1 expression (right) plasmids. Bottom, a luciferase reporter driven by the c-myc promoter containing a mutation in the Blimp-1 site was cotransfected into 18-81 pre-B cells along with control (left) or Blimp-1 expression (right) plasmids. Transfected cells were cross-linked and subjected to the ChIP assay. The chromosomal immunoprecipitations were performed with either anti-acetylated histone H3 (αAc-H3) or control antibody (Ab) (as indicated). Twofold serial dilutions of DNA recovered after immunoprecipitation with the indicated antibody were amplified by PCR assay using primers specific for the c-myc promoter. PCR products were analyzed by Southern blot hybridization with an internal probe recognizing the amplified c-myc promoter.
We conclude that Blimp-1 causes deacetylation of histone H3 associated with the c-myc promoter in vivo. These data are consistent with the TSA-dependent inhibition of Blimp-1 repression shown in Fig. 7 and provide additional support for the idea that recruitment of HDAC is important for Blimp-1-dependent repression of the c-myc promoter in vivo.
DISCUSSION
Blimp-1 is a transcriptional repressor that is capable of initiating a program of terminal differentiation in BCL1 lymphoma cells (62). The c-myc gene is a physiologically important target for Blimp-1 in B cells (40), and it is therefore reasonable to study the mechanism of Blimp-1 repression in the context of c-myc transcription. We have presented data showing that Blimp-1 is an active repressor of the c-myc promoter and that its activity is independent of an adjacently bound activator, YY1. We identified two regions of the Blimp-1 protein that associate with HDACs and showed that endogenous Blimp-1 and HDAC associate and that the complexes bind DNA. Finally, we showed that HDAC activity is necessary for Blimp-1-dependent repression and that Blimp-1 expression causes deacetylation of histone H3 associated with the c-myc promoter. Thus, our study shows that recruitment of HDACs is one mechanism by which Blimp-1 represses transcription.
Active repression by Blimp-1.
Several lines of evidence indicate that Blimp-1 is an active repressor that does not mediate repression simply by interfering with the binding or function of an activator. First, even though Blimp-1 binds very close to the activator YY1 on the c-myc promoter, Blimp-1 and YY1 act independently of one another. Blimp-1 repression does not depend on binding or transcriptional activating properties of YY1 (Fig. 1). Secondly, truncation mutants of Blimp-1 that retain the ability to bind DNA are unable to repress the c-myc promoter (Fig. 2), showing that DNA binding is not sufficient to cause transcriptional repression. Finally, both full-length Blimp-1 and an isolated domain of Blimp-1 (aa 557 to 714) are sufficient to repress transcription in the Gal4-fusion protein assay (Fig. 3), demonstrating that repression is independent of binding to a Blimp-1 site and can occur on a synthetic promoter where YY1 and other c-myc transcriptional activators do not bind.
However, these data do not rule out the possibility that Blimp-1 may also repress transcription on some promoters by interfering with the binding of a transcriptional activator. Blimp-1 binding sites in both the c-myc and INF-β promoters are very similar to interferon-stimulated response element sites (11, 27, 31, 34) and Blimp-1 might displace or compete with interferon regulatory factor 1 (IRF-1), interferon-stimulated gene factor 3, or other activators which recognize the same sites. Our previous studies failed to detect any protein other than Blimp-1 binding to the Blimp-1 site in the c-myc promoter in B-cell lines (27, 40), so this mechanism does not appear to play a role for c-myc repression. However, the human homolog of Blimp-1, PRD1-BF1, binds to a site on the IFN-β promoter that is also recognized by the activators IRF-1 and IRF-3 (38, 56). It has been suggested that Blimp-1 displaces positive regulatory proteins to limit the expression of IFN-β following viral infection. It is possible that in other Blimp-1 target genes yet to be identified, the Blimp-1 site can be recognized by IRF family activators as well as by Blimp-1. In these cases, Blimp-1 would repress transcription by a combination of active and passive mechanisms.
Blimp-1 repression domains.
In the context of the c-myc promoter, multiple domains of Blimp-1 appear to be required for repression (Fig. 2). As discussed below, a role for HDAC activity is established by the inhibitor studies and the ChIP assay. However, recruitment of HDAC was not sufficient for Blimp-1 to repress the c-myc promoter since Blimp-1 mutants H1 and H2 contain one or both domains which associate with HDAC but are unable to repress transcription (Fig. 2). It may be that HDAC is the only protein which needs to associate with Blimp-1 for it to act as a repressor and the truncation and deletion mutants which retain HDAC association domains have an abnormal three-dimensional conformation which does not allow efficient recruitment of HDAC on the c-myc promoter. This is consistent with the finding that some domains required for repression, such as the N-terminal acidic region, were unable or only partially able to repress transcription independently in the Gal4-fusion protein assay, whereas one HDAC association domain of Blimp-1 (aa 557 to 714) was sufficient to repress in this assay (Fig. 7). However, it is not consistent with our demonstration that mutant forms of Blimp-1 associate with HDAC in the GST assay in vitro (Fig. 5) and in the coimmunoprecipitation assay in vivo (data not shown). These data indicate that the truncation and deletion mutants of Blimp-1 do retain the ability to associate with HDAC.
Therefore, we favor an alternate explanation, which is that mechanisms in addition to recruitment of HDAC are required for Blimp-1-dependent repression in the context of the c-myc promoter. The c-myc promoter has binding sites for multiple transcriptional activators and is subject to complex regulation (44) and is thus more complicated than that of the synthetic (Gal4)4-tk promoter, where recruitment of HDAC appears to be sufficient for transcriptional repression (Fig. 7). The fact that TSA treatment did not completely inhibit Blimp-1 repression on the c-myc promoter (Fig. 8) also supports the notion that Blimp-1 has more than one repression mechanism. Additional proteins may associate with Blimp-1 to mediate other mechanisms of repression in the context of the c-myc promoter. Consistent with this suggestion, a yeast two-hybrid screen has recently shown that the proline-rich region of Blimp-1 associates with the murine homolog of Groucho (50). We do not currently know how association with Groucho may affect the ability of Blimp-1 to associate with HDAC, but since Blimp-1 contains two domains that can associate with HDAC, it seems likely that Groucho and HDAC could associate simultaneously with Blimp-1. There is precedent for proteins mediating transcriptional repression to associate with HDAC and other proteins since the corepressor SMRT has been shown to recruit both HDAC and TFIIB (65).
HDAC recruitment and Blimp-1 repression.
Our work shows that for Blimp-1, like several other transcriptional repressors (16, 48, 64), HDAC activity is important for transcriptional repression. Most currently described transcription factors that repress transcription via HDACs require corepressors such as Sin3, SMRT/NcoR, RbAp-46, RbSp-48, Ski, SAP18, or SAP30 (1, 8, 9, 20, 21, 32, 33, 36, 46, 47, 69). These corepressors act as a bridge between the DNA binding protein and HDACs. However, Blimp-1 appears to associate directly with HDAC1 and HDAC2 (Fig. 5), and we have been unable to detect association between Blimp-1 and Sin3 or NcoR. Thus, Blimp-1 is similar to proteins such as the Rb family proteins, YY1, PLZF, and Bcl-6, that have been reported to associate directly with HDAC. Our EMSAs further show that Blimp-1 and HDAC, present at endogenous levels in plasmacytoma nuclear extracts, associate with one another (Fig. 6). Similar studies have been used to show the association of NF-κB p65 with CBP/p300 (70). These data provide direct evidence that Blimp-1 recruits HDAC to DNA.
The two HDAC association domains on Blimp-1, a proline-rich region and a Zn finger region, show no obvious homology with the other currently identified HDAC association motifs, such as the LXCXE-like motif on Rb family proteins (12), a 30-amino-acid glycine-rich region on YY1 (66), or the POZ domains on Bcl6 and PLZF (9, 15, 39). Different protein association surfaces of HDACs may be involved in association with different partners, or the association motifs on the partners may have a common three-dimensional structure which is not readily apparent by inspection of their primary sequences. Alternatively, a bridging protein might be involved since for Blimp-1, as well as other proteins such as Rb and YY1, the possible involvement of a bridging protein has not been definitively ruled out. Further analyses will be necessary to define the precise protein-protein interactions and domains involved between HDAC and its partners.
The HDAC inhibitor TSA inhibits Blimp-1's ability to repress transcription, both in a Gal4 assay and when assayed with the natural c-myc promoter (Fig. 8). In addition, our ChIP studies show that expression of Blimp-1 leads to deacetylation of histone H3 bound to the c-myc promoter and to concommitant repression of c-myc promoter activity (Fig. 9). Both histone deacetylation and transcriptional repression of this promoter depend on the presence of the Blimp-1 binding site. Thus, the functional importance of HDAC recruitment by Blimp-1 is supported by two different but complementary experimental approaches.
ACKNOWLEDGMENTS
We are grateful to Gerald Siu and Xiaoming Zou for critically reading the manuscript and to Dimitrios Thanos and members of the Calame laboratory for many helpful discussions. We thank Wen-Ming Yang and Edward Seto for HDAC2 cDNA, Christian Hassig and Stuart Schrieber for HDAC1 cDNA, and Leila Alland and Ron DePinho for testing the interaction between Blimp-1 and mSin3. We are grateful to D. Thanos and T. Maniatis and members of their laboratories for advice on the ChIP assay and to S. Ghosh and members of his lab for advice on EMSA protocol.
This work was supported by USPHS grant RO1 AI 43576 to K.C.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Artandi S, Calame K. Association of DNA-binding transcription activators in solution. In: Adolph K, editor. Methods in molecular genetics. I. New York, N.Y: Academic Press; 1993. pp. 267–279. [Google Scholar]
- 3.Blackman M A, Tigges M A, Minie M E, Koshland M E. A model system for peptide hormone action in differentiation: interleukin 2 induces a B lymphoma to transcribe the J chain gene. Cell. 1986;47:609–617. doi: 10.1016/0092-8674(86)90625-2. [DOI] [PubMed] [Google Scholar]
- 4.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 5.Buyse I M, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppola J A, Cole M D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 7.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 8.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 9.Dhordain P, Lin R J, Quief S, Lantoine D, Kerckaert J P, Evans R M, Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dmitrovsky E, Kuehl W M, Hollis G F, Kirsch I R, Bender T P, Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986;322:748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- 11.Fan C M, Maniatis T. Two different virus-inducible elements are required for human beta-interferon gene regulation. EMBO J. 1989;8:101–110. doi: 10.1002/j.1460-2075.1989.tb03353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher A L, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 14.Geraudie J, Hourdry J, Vriz S, Singer M, Mechali M. Enhanced c-myc gene expression during forelimb regenerative outgrowth in the young Xenopus laevis. Proc Natl Acad Sci USA. 1990;87:3797–3801. doi: 10.1073/pnas.87.10.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 18.Hagman J, Belanger C, Travis A, Turck C W, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 19.Hansen U, Tenen D G, Livingston D M, Sharp P A. T antigen repression of SV40 early transcription from two promoters. Cell. 1981;27:603–613. doi: 10.1016/0092-8674(81)90402-5. [DOI] [PubMed] [Google Scholar]
- 20.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 21.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 22.Herschbach B M, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman L B, Liebermann D A. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991;6:903–909. [PubMed] [Google Scholar]
- 24.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 25.Johnson F B, Krasnow M A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- 26.Kakkis E, Calame K. A plasmacytoma-specific factor binds the c-myc promoter region. Proc Natl Acad Sci USA. 1987;84:7031–7035. doi: 10.1073/pnas.84.20.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakkis E, Riggs K J, Gillespie W, Calame K. A transcriptional repressor of c-myc. Nature. 1989;339:718–721. doi: 10.1038/339718a0. [DOI] [PubMed] [Google Scholar]
- 28.Kang T, Martins T, Sadowski I. Wild type GAL4 binds cooperatively to the GAL1-10 UASG in vitro. J Biol Chem. 1993;268:9629–9635. [PubMed] [Google Scholar]
- 29.Kaplan J, Calame K. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 1997;25:1108–1116. doi: 10.1093/nar/25.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato G J, Dang C V. Function of the c-Myc oncoprotein. FASEB J. 1992;6:3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- 31.Keller A D, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 32.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 33.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 34.Levy D E, Kessler D S, Pine R, Reich N, Darnell J E., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Manley J L. Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Leo C, Schroen D J, Chen J D. Characterization of receptor interaction and transcriptional repression by the corepressor SMRT. Mol Endocrinol. 1997;11:2025–2037. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- 37.Liebermann D A, Hoffman B. Differentiation primary response genes and proto-oncogenes as positive and negative regulators of terminal hematopoietic cell differentiation. Stem Cells (Dayton) 1994;12:352–369. doi: 10.1002/stem.5530120402. [DOI] [PubMed] [Google Scholar]
- 38.Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 39.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 40.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 41.Littlewood T D, Evan G I. The role of myc oncogenes in cell growth and differentiation. Adv Dent Res. 1990;4:69–79. doi: 10.1177/08959374900040011001. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Yang N, Teng C T. COUP-TF acts as a competitive repressor for estrogen receptor-mediated activation of the mouse lactoferrin gene. Mol Cell Biol. 1993;13:1836–1846. doi: 10.1128/mcb.13.3.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 44.Marcu K B, Bossone S A, Patel A J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 45.Matsui K, Nakanishi K, Cohen D I, Hada T, Furuyama J, Hamaoka T, Higashino K. B cell response pathways regulated by IL-5 and IL-2. Secretory microH chain-mRNA and J chain mRNA expression are separately controlled events. J Immunol. 1989;142:2918–2923. [PubMed] [Google Scholar]
- 46.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 47.Nomura T, Khan M M, Kaul S C, Dong H D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;15:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 49.Prochownik E V, Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature. 1986;322:848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- 50.Ren B, Chee K J, Kim T H, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renkawitz R. Transcriptional repression in eukaryotes. Trends Genet. 1990;6:192–197. doi: 10.1016/0168-9525(90)90176-7. [DOI] [PubMed] [Google Scholar]
- 52.Riggs K J, Merrell K T, Wilson G, Calame K. Common factor 1 is a transcriptional activator which binds in the c-myc promoter, the skeletal alpha-actin promoter, and the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1991;11:1765–1769. doi: 10.1128/mcb.11.3.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riggs K J, Saleque S, Wong K K, Merrell K T, Lee J S, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rinkenberger J L, Wallin J J, Johnson K W, Koshland M E. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity. 1996;5:377–386. doi: 10.1016/s1074-7613(00)80263-0. [DOI] [PubMed] [Google Scholar]
- 55.Ruzek M C, Mathur A. Plasma cells tumors decrease CD23 mRNA expression in vivo in murine splenic B cells. Eur J Immunol. 1995;25:2228–2233. doi: 10.1002/eji.1830250817. [DOI] [PubMed] [Google Scholar]
- 56.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 57.Schmid P, Schulz W A, Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989;243:226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- 58.Schule R, Umesono K, Mangelsdorf D J, Bolado J, Pike J W, Evans R M. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 59.Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 61.Tedder T F, Tuscano J, Sato S, Kehrl J H. CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu Rev Immunol. 1997;15:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- 62.Turner C A, Jr, Mack D H, Davis M M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 63.Vincent S, Marty L, Le Gallic L, Jeanteur P, Fort P. Characterization of late response genes sequentially expressed during renewed growth of fibroblastic cells. Oncogene. 1993;8:1603–1610. [PubMed] [Google Scholar]
- 64.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 65.Wong C W, Privalsky M L. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 69.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 70.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]