Abstract
In this research, a dataset including 29 ketone-based covalent inhibitors with SARS-CoV-1 3CLpro inhibition activity was used to develop high predictive QSAR models. Twenty-two molecules were put in train set and seven molecules in test set. By using stepwise MLR method for molecules in train set, four molecular descriptors including Mor26p, Hy, GATS7p and Mor04v were selected to build QSAR models. MLR and ANN methods were used to create QSAR models for predicting the activity of molecules in both train and test sets. Both QSAR models were validated by calculating several statistical parameters. R2 values for the test set of MLR and ANN models were 0.93 and 0.95, respectively, and RMSE values for their test sets were 0.24 and 0.17, respectively. Other calculated statistical parameters (especially parameter) show that created ANN model has more predictive power with respect to developed MLR model (with four descriptor). Calculated leverages for all molecules show that predicted pIC50 (by both QSAR models) for all molecules is acceptable, and drawn residuals plots show that there is no systematic error in building both QSAR modes. Also, based on developed MLR model, used molecular descriptors were interpreted.
Keywords: QSAR, SARS-CoV-1, SARS-CoV-2, 3CLpro inhibition activity, COVID-19
Introduction
Coronavirus disease 19 (COVID-19) is a pandemic disease that has affected the health of peoples in the whole world. Until May 6, 2021, the World Health Organization (WHO) had reported 155,506,494 infected cases to COVID-19 (including 3,247,228 deaths) [1, 2]. The disease has spread from Wuhan in China (in late 2019) by a virus that has called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since some coronaviruses had been transmitted from animals to humans, probably, the similar event has happened for SARS-CoV-2 [3–6]. Before COVID-19 pandemic, two coronaviruses including severe acute respiratory syndrome coronavirus (SARS-CoV-1) and Middle East respiratory syndrome coronavirus (MERS-CoV) had been transmitted to human from animals [7, 8]. Although SARS-CoV-2 has lower mortality rate (2.3%) with respect to SARS-CoV-1 (mortality rate 10%) and MERS-CoV (mortality rate 35%), it has higher reproductive number (2.0–2.5) with respect to SARS-CoV-1 (1.7–1.9) and MERS-CoV (< 1) [9–11]. Despite the lower mortality rate of SARS-CoV-2, it has killed more people with respect to SARS-CoV-1 and MERS-CoV because of its global pandemic outbreak. SARS-CoV-2 virus is present in body fluids such as cerebrospinal fluid and blood and usually is transmitted through respiratory droplets [12, 13]. So, from the beginning of COVID-19 outbreak, social distancing and closing mask have been suggested to reduce the number of infected cases [14]. Infected people show a variety of symptoms such as fever, difficulty breathing, taste or smell loss, headache, muscle ache, sore throat, runny nose and nausea [15]. Most of the patients show mild symptoms (~ 80%), and just the smaller proportion of them (~ 5%) have severe disease [16]. There are four subfamilies of coronaviruses including α-coronaviruses, β-coronaviruses, δ-coronaviruses and γ-coronaviruses, in which α- and β-coronaviruses infect mammals. SARS-CoV-1, MERS-CoV and SARS-CoV-2 are belonging to β-coronaviruses subfamily [17–19]. SARS-CoV-2 is a positive-sense, single-stranded RNA virus (+ ssRNA) that has been packed in an envelope. Spike membrane glycoproteins in the surface of virus bind to angiotensin-converting enzyme 2 (ACE2) receptor in the membrane of human cells and enters virus to our cells [20–23]. Generally, designed drugs for COVID-19 treatment can be classified into four groups including drugs that prevent the replication and synthesis of RNA by targeting critical enzymes for the replication of the virus, drugs that block the binding of spike protein to ACE2 receptor on human cells, drugs that inhibit coronavirus virulence factors and drugs that inhibit a receptor or enzymes in human cells [24]. 3C-like cysteine protease (3CLpro) is the main protease of SARS-CoV-2 that catalyzes the cleavage of polypeptides to their effector forms and has essential enzymatic role for virus life cycle [25, 26]. So it can be considered as a target for design drugs in COVID-19 treatment [27–29]. Quantitative structure–activity relationship (QSAR) is a computer-assisted drug design method that relates the structural features of molecules to their activities. QSAR models are useful in drug design process because they predict the activity of molecules quantitatively and determine structural features that increase the activity of molecules [30]. In this research, we have used a series of new synthesized compounds including 29 ketone-based molecules as covalent inhibitors of SARS-CoV-1 3CLpro (that had been synthesized by Hoffman et al.) [31] to develop QSAR models with high predictive power for predicting their 3CLpro inhibition activities. Hoffman et al. had shown that the greatest active compound in their research (compound 4 in their published paper and compound m15 in this research) is the covalent inhibitor of 3CLpro SARS-CoV-1 (IC50: 0.004 µM) and 3CLpro SARS-CoV-2 (IC50:0.00027 µM) enzymes. The crystallographic structure of the complex of this compound with 3CLpro SARS-CoV-2 is available in protein data bank (PDB ID: 6XHM). Also, performed researches by other groups show that the derivatives of available molecules in this dataset are covalent inhibitors for the 3CLpro enzymes of MERS-CoV and SARS-CoV-2 [32–37]. Since SARS-CoV-1 and SARS-CoV-2 have high similarity in their genome [38] and the derivatives of molecules in this dataset are active against the 3CLpro enzymes of SARS-CoV-1 and SARS-CoV-2, designed and optimized inhibitors by using developed QSAR models in this research help to design new drugs for treating COVID-19.
Materials and methods
Materials
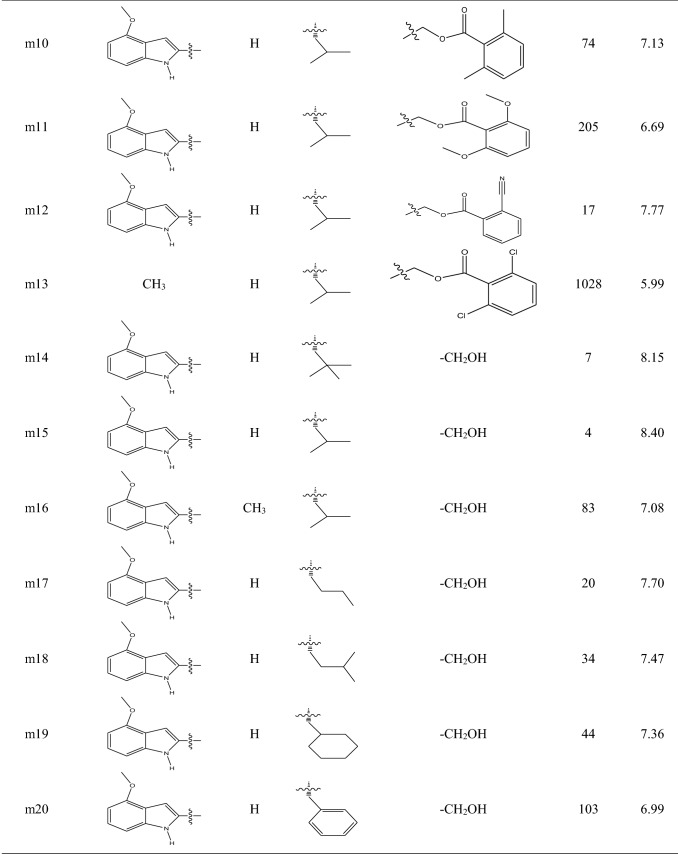
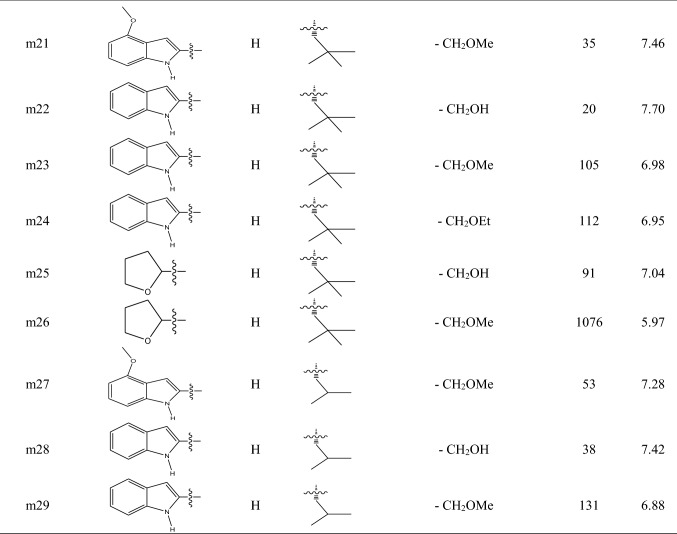
A series of molecules including 29 ketone-based covalent inhibitors of 3CLpro SARS-CoV-1 were selected from published paper by Hoffman et al. [31]. The chemical structure and activity of molecules are listed in Table 1. The activity of molecules was IC50 in nano-molar unit. In the first step, IC50 values in nano-molar unit were converted to IC50 values at molar unit and then they are converted to pIC50 by using the following equation:
| 1 |
pIC50 values had a wide range from 5.97 to 8.40. This dataset has suitable features that make it unique for developing QSAR models including the following:
Dataset has the wide range of activities (more than 2 log unit);
3CLpro SARS-CoV-1 inhibition activity in nano-molar level;
Molecule m15 in the dataset shows potent inhibition activity against 3CLpro SARS-CoV-1 (IC50: 0.004 µM) and 3CLpro SARS-CoV-2 (IC50:0.00027 µM);
Molecule m15 is a covalent inhibitor of 3CLpro SARS-CoV-2 (PDB ID: 6XHM);
Molecule m15 in the dataset shows good selectivity against other proteases [31];
Several researches have indicated that the derivatives of molecules in this dataset are covalent inhibitors of 3CLpro enzymes in SARS-CoV-1, SARS-CoV-2 and MERS-CoV [32–37], so the developed model can help to design new drugs for treating COVID-19.
Table 1.
The chemical structures and activities of molecules in dataset
To develop QSAR models, the dataset was divided into a train set containing 22 molecules for developing QSAR models and a test set including 7 molecules (molecules m3, m8, m13, m14, m17, m21 and m23) for validating them. Molecules with low, moderate and high activities were put in both train and test sets manually, and molecules with the lowest and greatest activities were put into the train set.
Programs
The three-dimensional chemical structure of all molecules was built in HyperChem (version 7.1) software and optimized by using AMBER force field (the root-mean-square gradient was set to 0.0001 kcal mol−1 Å−1) [39]. Dragon software (version 5.5) was used to calculate molecular descriptors for the optimized structures of molecules [40]. SPSS software (version 16) was used to select informative descriptors by using stepwise multiple linear regression (stepwise MLR) [41]. All other chemometrics methods for building and validating models were performed in R software (version 3.6.3) [42]. RStudio software (Version 1.1.463) was used as integrated development environment (IDE) for R programing language [43]. MLRQSAR package (version 0.1.0) was used to develop multiple linear regression (MLR) model and validate it by performing leave-one-out cross-validation and Y-randomization test on MLR model. Also, it was used to compute descriptor contribution for MLR model, calculate variance inflation factor (VIF) for descriptors, calculate several statistical parameters for validating both train and test sets of developed QSAR models and compute the applicability domain of created QSAR models based on the calculation of the leverage matrix [44, 45]. For building artificial neural network (ANN) model, h2o package (version 3.32.1.2) was used [46]. Also, ggplot2 package (version 3.3.3) was used to draw plots [47].
Methods
MLR modelling and validation
A MLR model has the following form:
| 2 |
where is constant coefficient and to are corresponding coefficients to the molecular descriptors to . Coefficients are obtained so that the sum of squared residuals (between predicted pIC50 and experimental pIC50) is minimum. Also, leave-one-out cross validation (LOOCV) and Y-randomization tests were performed on this model to indicate that the created model is robust and has not been obtained by chance [48, 49].
ANN modelling
To create an ANN model in h2o package, h2o.deeplearning option was used. Although this package is able to build both shallow feedforward ANN model (ANN model with one hidden layer) and deep feedforward ANN model (ANN model with more than one hidden layer), we built a shallow feedforward ANN model due to the small size of dataset. In deep ANN model, the number of trainable parameters increases and the small size of dataset leads to overfitting. To solve overfitting in created model, dropout technique was applied to network during its training and regularization terms were used in its cost function. Dropout removes some neurons from input and hidden layers during the training process, randomly. L1 (lasso) regularization, L2 (ridge) regularization and max_w2 (an upper limit for the (squared) sum of the incoming weights to a neuron) were added to loss function as regularization terms. The loss function in h2o.deeplearning has the following form that it is minimized for each training example j:
| 3 |
In Eq. 3, W is the collection {Wi}1:N-1, where Wi denotes the weight matrix connecting layers i and i + 1 for a network of N layers and B is the collection {bi}1:N-1, where bi denotes the column vector of biases for layer i + 1. In loss function, was set to absolute that is the sum of residuals. is the sum of all L1 norms for the weights and biases in the network, and L2 regularization is presented via that is the sum of squares of all the weights and biases in the network. and are constant variables that generally they are set to a very small value (for example 10–5). Also, maxout activation function was used for neurons in the hidden layer [50–53].
Applicability domain
The applicability domain of built QSAR models was investigated by calculating leverage matrix (H):
| 4 |
where X is descriptors matrix and the diagonal elements of H matrix are the leverages for objects (molecules). Critical leverage value was considered 3p/n, where p is the number of descriptors in model plus one and n is the number of molecules in the train set. If calculated leverage (h) for a molecule is larger than critical leverage value, its predicted activity (by created model) is not acceptable [54, 55].
Statistical parameters for validating QSAR models
For validating created QSAR models, several statistical parameters have been calculated for both train and test sets including:
| 5 |
| 6 |
| 7 |
| 8 |
| 9 |
| 10 |
| 11 |
| 12 |
| 13 |
| 14 |
| 15 |
| 16 |
| 17 |
| 18 |
| 19 |
where yi and are, respectively, the experimental and the predicted activity of molecule and and are the mean of the experimental and the predicted activities, respectively. and are the mean of the activity for train and test sets, respectively. Also, , and are the number of compounds, the number of compounds in train set and the number of compounds in test set, respectively. CCC2 is the squared concordance correlation coefficient, RMSE is the root-mean-squared error, and MAE is the mean absolute error [44, 56, 57].
Results and discussion
Model building and validation
Molecular descriptors that belong to all 22 descriptors blocks in Dragon software were calculated for all molecules. In the first step, molecular descriptors with few repeated values (fewer than 5) across samples and many zero values (with more than 10 zero values) across samples were removed. After this preprocessing step, 1203 molecular descriptor were remained. Stepwise MLR in SPSS software was used to select informative variables based on molecules in the train set. Four molecular descriptors were selected to develop QSAR models whose name and definition are listed in Table 2, and their values for all molecules are listed in Table 3. VIF values for Mor26p, Hy, GATS7p and Mor04v molecular descriptors were 1.06, 1.28, 1.12 and 1.21 which indicate that these descriptors have no collinearity and multi-collinearity problems and are suitable for creating QSAR models. Mor26p has the largest correlation with the activities of molecules (R2 = 0.59), but a predictive model cannot be created just by using this descriptor, so Hy descriptor was added by stepwise MLR and the following model was created:
| 20 |
R2 and RMSE values for the train set of this model were 0.77 and 0.22, respectively, and those for the test set were 0.79 and 0.32, respectively. R2 value for LOO-CV on the train set was 0.72 which indicates that the created model is robust, and the maximum value of R2 for ten runs of Y-randomization test was 0.17 which shows that the created model has not been obtained by chance. By adding another descriptor (GATS7p), a model with three descriptors was built:
| 21 |
Table 2.
The definition of selected descriptors by stepwise MLR
| Descriptor | Type | Descriptor block | Definition |
|---|---|---|---|
| Mor26p | 3D | 3D-MoRSE descriptors | 3D-MoRSE—signal 26/weighted by atomic polarizability |
| Hy | Others | Molecular properties | Hydrophilic factor |
| GATS7p | 2D | 2D autocorrelations | Geary autocorrelation-lag 7/weighted by atomic polarizability |
| Mor04v | 3D | 3D-MoRSE descriptors | 3D-MoRSE—signal 04/weighted by atomic van der Waals volume |
Table 3.
Experimental and predicted pIC50, descriptors values and leverage values for molecules (critical leverage value is 0.68)
| Train set | ||||||||
|---|---|---|---|---|---|---|---|---|
| Predicted pIC50 by | Descriptor values | |||||||
| Molecule | Experimental pIC50 | MLR model | ANN model | Mor26p | Hy | GATS7p | Mor04v | Leverage |
| m1 | 6.66 | 6.80 | 6.66 | 0.231 | 1.525 | 0.96 | − 0.759 | 0.0419 |
| m2 | 6.74 | 6.94 | 6.76 | 0.198 | 1.478 | 0.973 | − 0.671 | 0.0460 |
| m4 | 7.07 | 7.13 | 7.09 | 0.155 | 1.415 | 1.038 | − 1.095 | 0.0770 |
| m5 | 7.10 | 7.14 | 7.11 | 0.199 | 1.399 | 1.078 | − 0.833 | 0.0766 |
| m6 | 7.06 | 6.94 | 6.97 | 0.224 | 1.395 | 1.04 | − 0.841 | 0.0592 |
| m7 | 7.28 | 7.36 | 7.27 | 0.061 | 1.399 | 1.012 | − 1.166 | 0.1362 |
| m9 | 7.01 | 7.22 | 7.05 | 0.11 | 1.418 | 0.992 | − 0.826 | 0.0807 |
| m10 | 7.13 | 6.92 | 7.11 | 0.25 | 1.377 | 1.102 | − 1.235 | 0.1022 |
| m11 | 6.69 | 6.52 | 6.63 | 0.229 | 1.385 | 0.957 | − 1.457 | 0.1103 |
| m12 | 7.77 | 7.76 | 7.65 | 0.002 | 1.399 | 1.069 | − 1.049 | 0.2476 |
| m15 | 8.40 | 8.12 | 8.22 | 0.022 | 2.336 | 0.968 | − 0.962 | 0.1625 |
| m16 | 7.08 | 7.11 | 6.94 | 0.193 | 1.548 | 0.987 | − 0.431 | 0.0783 |
| m18 | 7.47 | 7.55 | 7.37 | 0.224 | 2.304 | 1.036 | − 1.108 | 0.1097 |
| m19 | 7.36 | 7.42 | 7.32 | 0.281 | 2.243 | 1.049 | − 0.754 | 0.1550 |
| m20 | 6.99 | 7.03 | 6.90 | 0.246 | 2.243 | 1.049 | − 2.854 | 0.5575 |
| m22 | 7.70 | 7.63 | 7.59 | 0.129 | 2.341 | 0.9 | − 0.606 | 0.2084 |
| m24 | 6.95 | 6.90 | 6.87 | 0.131 | 1.52 | 0.896 | − 1.006 | 0.0330 |
| m25 | 7.04 | 6.88 | 6.91 | 0.298 | 1.697 | 1.001 | − 0.493 | 0.1029 |
| m26 | 5.97 | 6.00 | 5.97 | 0.379 | 0.933 | 0.995 | − 0.469 | 0.1676 |
| m27 | 7.28 | 7.36 | 7.16 | 0.18 | 2.336 | 0.968 | − 1.777 | 0.1678 |
| m28 | 7.42 | 7.55 | 7.37 | 0.118 | 2.376 | 0.926 | − 1.544 | 0.1414 |
| m29 | 6.88 | 6.74 | 6.80 | 0.185 | 1.573 | 0.933 | − 1.464 | 0.0791 |
| Test set | ||||||||
|---|---|---|---|---|---|---|---|---|
| Predicted pIC50 by | Descriptor values | |||||||
| Molecule | Experimental pIC50 | MLR model | ANN model | Mor26p | Hy | GATS7p | Mor04v | Leverage |
| m3 | 6.64 | 6.95 | 6.79 | 0.193 | 1.456 | 0.981 | − 0.745 | 0.0444 |
| m8 | 7.09 | 7.06 | 6.98 | 0.123 | 1.418 | 0.972 | − 1.054 | 0.0644 |
| m13 | 5.99 | 5.54 | 5.85 | 0.601 | 0.899 | 1.06 | 0.184 | 0.5221 |
| m14 | 8.15 | 8.19 | 8.40 | − 0.012 | 2.304 | 0.939 | − 0.741 | 0.2222 |
| m17 | 7.70 | 7.77 | 7.50 | 0.146 | 2.336 | 1.02 | − 1.244 | 0.0943 |
| m21 | 7.46 | 7.25 | 7.29 | 0.066 | 1.522 | 0.943 | − 1.074 | 0.0762 |
| m23 | 6.98 | 7.00 | 6.97 | 0.123 | 1.546 | 0.91 | − 0.943 | 0.0357 |
R2 and RMSE values for the train set of this model were 0.85 and 0.17, respectively, and those for the test set were 0.82 and 0.28, respectively. R2 value for LOO-CV on the train set was 0.84 which indicates that the created model is robust, and the maximum value of R2 for ten runs of Y-randomization test was 0.29 which shows that the created model has not been obtained by chance. As seen, adding GATS7p has increased the predictive power of QSAR model. For increasing the predictive power of model, another descriptor (Mor04v) was added to the model, and according to Topliss and Costello rule (the ratio of molecules in train set to used descriptors for building model should be at least 5 to 1) [58], this is the last descriptor that we can use for developing QSAR models. By using all four descriptors, the following equation was obtained in R software:
| 22 |
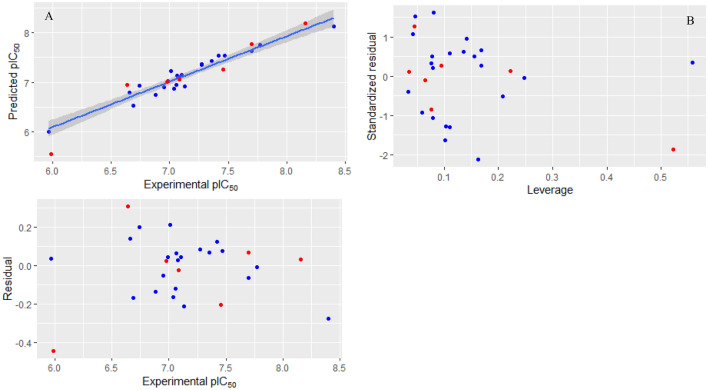
R2 values for the train and test sets of this model were 0.92 and 0.93, respectively, and RMSE values for the train and test sets were 0.13 and 0.24, respectively. R2 value for LOO-CV was 0.90 which shows that the created model is robust, and the maximum R2 value for ten runs of Y-randomization test was 0.37 which indicates that the created model has not been obtained by chance. R2 and RMSE values for the test set of created MLR models show that the created MLR model with all four descriptors has the highest predictive power. For further validation of the MLR model (MLR model with four descriptors), several statistical parameters were calculated for the train and test sets that are listed in Tables 4 and 5. Calculated values for these statistical parameters show that the created model is acceptable and has high predictive power. Predicted pIC50 for all molecules (in both train and test sets) by this model (MLR model with four descriptors) is listed in Table 3. Calculated leverages for all molecules (that are listed in Table 3) are smaller than critical leverages which show that the predicted pIC50 for all molecules (by MLR model with four descriptors) is acceptable. The plot of predicted pIC50 versus experimental pIC50, William plot and residuals plot for the MLR model (MLR model with four descriptors) are shown in Fig. 1. The William plot in Fig. 1 shows that the created model has no outlier and the predicted pIC50 for all molecules (in both train and test sets) is acceptable, and the residual plot shows that there is no systematic error in creating MLR model with four descriptors. To develop more predictive power QSAR model, these four descriptors were used as input variables for training an ANN model. In the first step, a network with one hidden layer and 10 neurons was created. For optimizing the trainable parameters of ANN model, k-fold cross-validation test was used. In this method, molecules in train set were divided into three sets, and each time, both of them were used for training ANN model and other for its validation and this process was repeated for each fold. The R2 value for each fold and their mean were calculated. The activation function for neuron in the hidden layer was set to maxout activation function. By increasing the number of neurons in the hidden layer to 100 (each time, 10 neurons were added to the hidden layer of previous network architecture), the average of R2 values for all three folds was increased. Increasing the number of neurons in the hidden layer to more than 100 neurons did not increase the average of R2 values for k-fold cross-validation test, significantly, so an ANN architecture with one hundred neurons in its hidden layer was selected as the best architecture. Also, L1 and L2 regularization terms were set to 0.00001 and max_w2 was set to its default value. Dropout ratio from 0 to 0.5 was examined for both input and hidden layers, and the best results were obtained when dropout ratio for the input layer and hidden layer was set to 0.1 and 0.3, respectively. Other parameters were set to their default. So created ANN model had four neurons in its input layer and one hundred neurons in its hidden layer (with maxout activation function) and one neuron in its output layer (with linear activation function). The predicted pIC50 for all molecules (in both train and test sets) is listed in Table 3, and the calculated statistical parameters for the train and test sets are listed in Tables 4 and 5. R2 and RMSE values for the train set of ANN model were 0.99 and 0.06, respectively, and R2 and RMSE values for the test set were 0.95 and 0.17, respectively. R2 values for folds 1, 2 and 3 were 0.89, 0.69 and 0.68, respectively, and their mean was 0.75 which indicates that the created ANN model is robust. The plot of predicted pIC50 versus experimental pIC50, William plot and residuals plot for ANN model are shown in Fig. 2. Drawn residuals plot shows that there is no bias (systematic error) in creating this ANN model. William plot shows that molecule m15 is outlier, and based on this plot, predicted pIC50 by the ANN model for all molecules (in both train and test sets) is acceptable.
Table 4.
Calculated statistical parameters for validating created QSAR models
| Statistical parameters | Threshold values | MLR | ANN | ||
|---|---|---|---|---|---|
| Train set | Test set | Train set | Test set | ||
| > 0.6 | 0.92 | 0.91 | 0.96 | 0.95 | |
| > 0.6 | 0.92 | 0.93 | 0.99 | 0.95 | |
| – | 0.13 | 0.24 | 0.06 | 0.17 | |
| ≤ 1.15 and ≥ 0.85 | 1.00 | 1.00 | 1.01 | 1.00 | |
| ≤ 1.15 and ≥ 0.85 | 1.00 | 1.00 | 0.99 | 1.00 | |
| > 0.6 | 0.92 | 0.89 | 0.98 | 0.94 | |
| > 0.6 | 0.91 | 0.92 | 0.98 | 0.95 | |
| > 0.5 | 0.92 | 0.74 | 0.93 | 0.85 | |
| > 0.5 | 0.85 | 0.83 | 0.92 | 0.90 | |
| > 0.5 | 0.88 | 0.79 | 0.92 | 0.87 | |
| < 0.2 | 0.08 | 0.09 | 0.01 | 0.05 | |
| < 0.1 | 0.00 | 0.04 | 0.00 | 0.01 | |
| < 0.1 | 0.01 | 0.01 | 0.00 | 0.00 | |
| < 0.3 | 0.01 | 0.03 | 0.00 | 0.01 | |
| – | 0.00 | 0.03 | 0.06 | 0.03 |
Table 5.
Calculated Q2-based statistical parameters for validating created QSAR models
| Parameter | MLR | ANN |
|---|---|---|
| 0.89 | 0.94 | |
| 0.89 | 0.94 | |
| 0.77 | 0.88 |
Fig. 1.
Plots for created MLR model (train set with blue color and test set with red color): (A) the plot of predicted pIC50 versus experimental pIC50; (B) William plot (critical leverage is 0.68); (C) residuals plot
Fig. 2.
Plots for created ANN model (train set with blue color and test set with red color): (A) the plot of predicted pIC50 versus experimental pIC50; (B) William plot (critical leverage is 0.68); (C) residuals plot
Descriptors interpretation
The contribution of Mor26p, Hy, GATS7p and Mor04v molecular descriptors in the building of MLR model with four descriptors was 11.70%, 23.10%, 52.60% and 3.72%, respectively, and this MLR model (with four descriptors) was used for descriptors interpretation. Negative coefficient sign for Mor26p shows that smaller values (negative values) for this descriptor are favorable for increasing the activities of molecules. For example, molecules m14 and m15 which have smaller values for this descriptor have the most potent activities among others. Among all molecules, the value of this descriptor is negative only for molecule m14. Mor26p is a descriptor that belongs to 3D molecular representations of structure based on electron diffraction (3D-MoRSE) descriptors family that has been weighted by atomic polarizability. A study by Devinyak et al. [59] shows that the weighting of these descriptors by atomic polarizability decreases the effect of hydrogen significantly and diminishes the roles of nitrogen, oxygen and fluorine atoms. Also, they found that although these descriptors have information about the whole molecule, their final values are derived mostly from short-distance atomic pairs [59]. The presence of methoxy group on phenyl ring in the R1 substituent of molecule m21 has decreased its Mor26p value and increased its activity with respect to molecule m23. The comparison of molecules m21, m23 and m26 shows that R1 substituent with two fused rings is favorable for increasing the activity of molecule with respect to R1 substituent with a ring because two fused rings decrease the Mor26p descriptor value for molecule. Replacing hydrogen atom in R2 substituent with methyl group increases the Mor26p descriptor value and decreases the activity of molecule, so bulky groups in R2 substituent are not favorable. Comparing molecules m15 and m17 with m20 shows that longer and bulky groups for R3 substituent increase the value of Mor26p descriptor, so cyclic and long-chain groups for R3 substituent are not favorable for increasing the activity of molecules. The presence of nitrile group on phenyl ring in R4 substituent in molecules m7 and m12 has decreased the value of Mor26p descriptor for these two molecules, but comparing all molecules does not reveal a specific relationship between the size of R4 substituent and Mor26p descriptor values for molecules. Hy is the hydrophilic factor for molecule, and MLR model shows that larger Hy descriptor values improve the activity of molecules. The R2 value between Hy descriptor values and the activities of molecules is 0.52. Available data in Table 2 show that molecules with greater activities such as molecules m14, m15, m17 and m22 have larger Hy value. Hydrophilic groups such as hydroxyl group are favorable for increasing Hy descriptor value. Also, the presence of atoms with negative partial charge in R1 substituent and less bulky groups in R3 substituent increases the value of Hy descriptor. The developed MLR model shows that the larger value of GATS7p descriptor is favorable for increasing the activity of molecule. Mor04v descriptor belonging to 3D-MoRSE descriptors has been weighted by atomic van der Waals volume. Weighting descriptor by atomic van der Waals volume has similar effect with the weighting of 3D-MoRSE descriptor by atomic polarizability that decreases the effect of hydrogen significantly and diminishes the roles of nitrogen, oxygen and fluorine atoms [59]. In MLR model, Mor04v descriptor has a coefficient with positive sign, so larger values of this descriptor are favorable for increasing the activity of molecules. Except for molecule m13, the values of this descriptor are negative for other molecules (Table 3). Comparing molecules m1 to m14 shows that the larger value of Mor04v descriptor for molecule m13 is related to less bulky group for R1 substituent in molecule m13. This situation is seen for molecules m25 and m26. Less bulky groups for R1 substituent increase the value of Mor26p descriptor and decrease the activity of molecules. Since Mor04v descriptor has less contribution in creating model with respect to Mor26p descriptor, less bulky groups for R1 substituent are not favorable for increasing the activity of molecules. The contribution of Mor26p, Hy, GATS7p and Mor04v molecular descriptors in the building of ANN model was 26.27%, 26.09%, 25.62% and 21.99%, respectively, that show different values in comparison with the MLR model. Although GATS7p shows the largest contribution in the building of MLR model, in ANN model all four descriptors show comparable contribution in the building of model. Also, it should be considered that Mor26p has the largest correlation with the activities of molecules (R2 = 0.59).
Comparing QSAR models
Calculated statistical parameters for the train and test sets of both models in Tables 4 and 5 show both QSAR models are acceptable and have high predictive power. Calculated , , , , ,, , and Q2-based parameters (especially parameter) show that ANN model has more predictive power with respect to MLR model. William plot in Fig. 2 shows that molecule m15 is outlier in ANN model, but as seen from Table 2, ANN model has better prediction for its activity, and probably, it has happened because of the small standard deviation value of residuals for molecules in the train set of ANN model (SD = 0.06) with respect to MLR model (SD = 0.13).
Conclusions
The results of this research show the building of MLR and ANN models based on using Mor26p, Hy, GATS7p and Mor04v molecular descriptors which are suitable for predicting the SARS-CoV-1 3CLpro inhibition activity of these ketone-based molecules. Although both created models are acceptable and show high predictive power, calculated R2- and Q2-based parameters and RMSE for both train and test sets of MLR model with four descriptors and ANN model show that the ANN model has more predictive power. The interpretation of descriptors (based on the developed MLR model with four descriptors) shows that groups with two fused rings in R1 substituent are favorable for increasing the activity of molecule, bulky groups for R2 substituent are not favorable for improving the activity of molecules, and the presence of cyclic groups and long-chain groups for R3 substituent decreases the activity of molecules.
References
- 1.https://covid19.who.int/
- 2.Tuncer T, Ozyurt F, Dogan S, Subasi A. Chemometr. Intell. Lab. Syst. 2021;210:104256. doi: 10.1016/j.chemolab.2021.104256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsafar G, Reddy V. J. Iran. Chem. Soc. 2021 doi: 10.1007/s13738-021-02299-5. [DOI] [Google Scholar]
- 4.Serte S, Demirel H. Comput. Biol. Med. 2021;132:104306. doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ton AT, Gentile F, Hsing M, Ban F, Cherkasov A. Mol. Inf. 2020;39:2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Greer RA, Song Y, Praveen H, Song Y. Eur. J. Pharm. Sci. 2021;160:105771. doi: 10.1016/j.ejps.2021.105771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alves VM, Bobrowski T, Melo-Filho CC, Korn D, Auerbach S, Schmitt C, Muratov EN, Tropsha A. Mol. Inf. 2021;40:2000113. doi: 10.1002/minf.202000113. [DOI] [PubMed] [Google Scholar]
- 8.Ciotti M, Ciccozzi M, Terrinoni A, Jiang WC, Wang CB, Bernardini S. Crit. Rev. Clin. Lab. Sci. 2020;57:365–388. doi: 10.1080/10408363.2020.1783198. [DOI] [PubMed] [Google Scholar]
- 9.Duverger E, Herlem G, Picaud F. J. Mol. Graph. Model. 2021;104:107834. doi: 10.1016/j.jmgm.2021.107834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavasotto CN, Di Filippo JI. Mol. Inf. 2021;40:2000115. doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- 11.Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills S. Judic. Rev. 2020;25:71–79. doi: 10.1080/10854681.2020.1760575. [DOI] [Google Scholar]
- 13.Kabir MA, Ahmed R, Chowdhury R, Asher Iqbal SM, Paulmurugan R, Demirci U, Asghar W. Microbes Infect. 2021 doi: 10.1016/j.micinf.2021.104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartt M. Cities and Health. 2020 doi: 10.1080/23748834.2020.1788770. [DOI] [Google Scholar]
- 15.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html
- 16.Cavasotto CN, Lamas MS, Maggini J. Eur. J. Pharmacol. 2021;890:173705. doi: 10.1016/j.ejphar.2020.173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucukoglu K, Faydal N, Bul D. Med. Chem. Res. 2020;29:1935–1955. doi: 10.1007/s00044-020-02625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sattari A, Ramazani A, Aghahosseini H. J. Iran. Chem. Soc. 2021 doi: 10.1007/s13738-021-02235-7. [DOI] [Google Scholar]
- 20.Wrobel AG, Benton DJ, Hussain S, Harvey R, Martin SR, Roustan C, Rosenthal PB, Skehel JJ, Gamblin SJ. Nat. Commun. 2020;11:5337. doi: 10.1038/s41467-020-19146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yousefi R, Moosavi-Movahedi A. J. Iran. Chem. Soc. 2020;17:1257–1258. doi: 10.1007/s13738-020-01939-6. [DOI] [Google Scholar]
- 22.Barge S, Jade D, Gosavi G, Talukdar NC, Borah J. Eur. J. Pharm. Sci. 2021;162:105820. doi: 10.1016/j.ejps.2021.105820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 24.Muhammed Y. Biosaf. Health. 2020;2:210–216. doi: 10.1016/j.bsheal.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh K, Abdul Amin S, Gayen S, Jha T. J. Mol. Struct. 2021;1237:130366. doi: 10.1016/j.molstruc.2021.130366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Krumberger M, Morris MA, Marie C, Parrocha T, Kreutzer AG, Nowick JS. Eur. J. Med. Chem. 2021;218:113390. doi: 10.1016/j.ejmech.2021.113390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chellapandi P, Saranya S. Med. Chem. Res. 2020;29:1777–1791. doi: 10.1007/s00044-020-02610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CK, Lin SM, Satange R, Lin SC, Sun SC, Wu HY, Kehn-Hall K, Hou MH. Comput. Struct. Biotechnol. J. 2021;19:2246–2255. doi: 10.1016/j.csbj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirtaleb MS, Mirtaleb AH, Nosrati H, Heshmatnia J, Falak R, Zolfaghari Emameh R. Biomed. Pharmacother. 2021;138:111518. doi: 10.1016/j.biopha.2021.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmadi R, Sepehri B, Ghavami R. J. Recept. Signal Transduct. 2019;39:264–275. doi: 10.1080/10799893.2019.1660898. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman RL, Kania RS, Brothers MA, Davies JF, Ferre RA, Gajiwala KS, He M, Hogan RJ, Kozminski K, Li LY, Lockner JW, Lou J, Marra MT, Mitchell LJ, Jr, Murray BW, Nieman JA, Noell S, Planken SP, Rowe T, Ryan K, Smith GJ, III, Solowiej JE, Steppan CM, Taggart B. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J, Liu X, Xiao G, Jiang H, Rao Z, Zhang LK, Xu Y, Yang H, Liu H. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomar S, Johnston ML, John SES, Osswald HL, Nyalapatla PR, Paul LN, Ghosh AK, Denison MR, Mesecar AD. J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai W, Jochmans D, Xie H, Yang H, Li J, Su H, Chang D, Wang J, Peng J, Zhu L, Nian Y, Hilgenfeld R, Jiang H, Chen K, Zhang L, Xu Y, Neyts J, Liu H. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c02258. [DOI] [PubMed] [Google Scholar]
- 36.Bai B, Belovodskiy A, Hena M, Kandadai AS, Joyce MA, Saffran HA, Shields JA, Khan MB, Arutyunova E, Lu J, Bajwa SK, Hockman D, Fischer C, Lamer T, Vuong W, van Belkum MJ, Gu Z, Lin F, Du Y, Xu J, Rahim M, Young HS, Vederas JC, Tyrrell DL, Lemieux MJ, Nieman JA. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, Saffran HA, McKay RT, van Belkum MJ, Joyce MA, Young HS, Tyrrell DL, Vederas JC, Lemieux MJ. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Boon SS, Wang MH, Chan RWY, Chan PKS. J. Virol. Methods. 2021;289:114032. doi: 10.1016/j.jviromet.2020.114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.HyperChem 7.1. Gainesville, USA: Hypercube, Inc. Available from: http://www.hyper.com
- 40.Milano chemometrics and QSAR research group, 2007. Available from http://www.talete.mi.it/dragon.htm
- 41.http://www.spss.com
- 42.https://www.r-project.org/
- 43.https://rstudio.com/
- 44.Sepehri B, Ghavami R, Farahbakhsh S, Ahmadi R. Int. J. Environ. Sci. Technol. 2021 doi: 10.1007/s13762-021-03271-9. [DOI] [Google Scholar]
- 45.https://www.researchgate.net/publication/350459619_MLRQSAR_package_version_010_for_R_programming_language
- 46.https://cloud.r-project.org/web/packages/h2o/index.html
- 47.https://cran.r-project.org/web/packages/ggplot2/index.html
- 48.Ghavami R, Sepehri B. J. Iran. Chem. Soc. 2016;13:519–529. doi: 10.1007/s13738-015-0761-2. [DOI] [Google Scholar]
- 49.Ghavami R, Sepehri B. J. Chromatogr. A. 2012;1233:116–125. doi: 10.1016/j.chroma.2012.01.047. [DOI] [PubMed] [Google Scholar]
- 50.Phil K. Matlab Deep Learning: With Machine Learning, Neural Networks and Artificial Intelligence. New York: Apress; 2017. [Google Scholar]
- 51.Cook D. Practical Machine Learning with H2O. Massachusetts: O’Reilly Media Inc; 2017. [Google Scholar]
- 52.J. Moolayil, Learn Keras for deep neural networks, (Jojo Moolayil, 2019)
- 53.A. Candel, E. LeDell, Deep learning with H2O, (H2O.ai, Inc, 2020)
- 54.Sepehri B, Ghavami R. Med. Chem. 2018;14:439–450. doi: 10.2174/1573406414666180321151029. [DOI] [PubMed] [Google Scholar]
- 55.Sepehri B, Ghavami R. J. Mol. Struct. 2017;1130:922–928. doi: 10.1016/j.molstruc.2016.10.079. [DOI] [Google Scholar]
- 56.Sepehri B, Rasouli Z, Hassanzadeh Z, Ghavami R. Med. Chem. Res. 2016;25:2895–2905. doi: 10.1007/s00044-016-1686-8. [DOI] [Google Scholar]
- 57.Sepehri B, Ghavami R. SAR QSAR Environ. Res. 2019;30:21–38. doi: 10.1080/1062936X.2018.1545695. [DOI] [PubMed] [Google Scholar]
- 58.Sepehri B. J. Mol. Liq. 2020;297:112013. doi: 10.1016/j.molliq.2019.112013. [DOI] [Google Scholar]
- 59.Devinyak O, Havrylyuk D, Lesyk R. J. Mol. Graph. Model. 2014;54:194–203. doi: 10.1016/j.jmgm.2014.10.006. [DOI] [PubMed] [Google Scholar]