Abstract
Angiogenesis-related gene expression is associated with the efficacy of anti-VEGF therapy. We tested whether intratumoral mRNA expression levels of genes involved in vascular morphogenesis and early vessel maturation predict response, recurrence-free survival (RFS), and overall survival (OS) in a unique cohort of patients with colorectal liver metastases (CLM) treated with bevacizumab-based chemotherapy followed by curative liver resection. Intratumoral mRNA was isolated from resected bevacizumab-pretreated CLM from 125 patients. In 42 patients, a matching primary tumor sample collected before bevacizumab treatment was available. Relative mRNA levels of 9 genes (ACVRL1, EGFL7, EPHB4, HIF1A, VEGFA, VEGFB, VEGFC, FLT1, and KDR) were analyzed by RT-PCR and evaluated for associations with response, RFS, and OS. P values for the associations between the individual dichotomized expression level and RFS were adjusted for choosing the optimal cut-off value. In CLM, high expression of VEGFB, VEGFC, HIF1A, and KDR and low expression of EGFL7 were associated with favorable RFS in multivariable analysis (P < 0.05). High ACVRL1 levels predicted favorable 3-year OS (P = 0.041) and radiologic response (PR = 1.093, SD = 0.539, P = 0.002). In primary tumors, low VEGFA and high EGFL7 were associated with radiologic and histologic response (P < 0.05). High VEGFA expression predicted shorter RFS (10.1 vs. 22.6 months; HR = 2.83, P = 0.038). High VEGFB (46% vs. 85%; HR = 5.75, P = 0.009) and low FLT1 (55% vs. 100%; P = 0.031) predicted lower 3-year OS rates. Our data suggest that intratumoral mRNA expression of genes involved in vascular morphogenesis and early vessel maturation may be promising predictive and/or prognostic biomarkers.
Introduction
Bevacizumab in combination with chemotherapy has improved the survival of patients with metastatic colorectal cancer, and in 25% of those with liver-limited disease, surgical resection of colorectal liver metastases (CLM) combined with perioperative chemotherapy affords a chance for cure (1, 2). Although randomized controlled trials are missing, the addition of bevacizumab to perioperative chemotherapy has been associated with improved radiologic and histologic response rates, which translates into improved survival (3–5). However, the majority of patients recurs and succumbs to metastatic disease. It is therefore critical to identify new biomarkers to select patients most likely to benefit from resection of CLM and achieve a durable remission, and those who may better undergo other treatment concepts.
Intratumoral expression of genes involved in angiogenesis have shown promise as predictive and prognostic biomarkers for bevacizumab efficacy in metastastic colorectal cancer (6). However, there is a paucity of data regarding the role of genes involved in vascular morphogenesis and early vessel maturation in predicting outcomes in metastatic colorectal cancer, which may serve as novel biomarkers for efficacy of bevacizumab-based chemotherapy. Identifying such biomarkers may guide patient selection for liver resection as well as offer insight on antiangiogenic resistance mechanisms and targets for novel drug development. We investigated whether relative mRNA levels of genes involved in vascular morphogenesis and early vessel maturation can predict response, probability of cure, and survival in a unique cohort of patients undergoing bevacizumab-based perioperative chemotherapy followed by liver resection with curative intent. Specifically, we examined mRNA levels of genes involved in blood vessel sprouting and lymphangiogenesis [HIF1A (hypoxia inducible factor 1, alpha), VEGFA (encoding vascular endothelial growth factor A), VEGFB, VEGFC, FLT1 (fms-related tyrosine kinase 1 encoding VEGF receptor 1), KDR (kinase insert domain receptor encoding VEGFR 2)], tubulogenesis [EGFL7 (EGF-like domain 7)], and early vessel maturation [EPHB4 (ephrin receptor b4), ACVRL1 (activin A receptor type II-like 1 encoding ALK1)].
Materials and Methods
We investigated 125 consecutive patients with resectable or borderline resectable CLM who received 3 months of neoadjuvant and 3 months of adjuvant bevacizumab-based combination chemotherapy and underwent liver resection in curative intent (2005–2011). Patients’ clinical data were obtained from a prospectively maintained database. Patient characteristics are given in Table 1. The median age of patients was 62 years (range, 30–80 years), of which 59.2% (n = 74) were male. The median follow-up was 3.8 years (range, 0.02–7.6 years). One patient with short follow-up (0.02 years) was included as response data were available. The study was approved by the institutional review board.
Table 1.
Baseline characteristics among patients with resected liver metastases from colorectal cancer
|
N
|
% |
With CLM available |
With primary tumor available |
|||
|---|---|---|---|---|---|---|
| n = 149 | % | n = 125 | % | n = 42 | % | |
| Age | ||||||
| Median (range) | 62 (30–80) | 62 (30–80) | 64 (47–80) | |||
| <65 years | 90 | 60.4 | 75 | 60.0 | 23 | 54.8 |
| ≥65 years | 59 | 39.6 | 50 | 40.0 | 19 | 45.2 |
| Sex | ||||||
| Male | 87 | 58.4 | 74 | 59.2 | 28 | 66.7 |
| Female | 62 | 41.6 | 51 | 40.8 | 14 | 33.3 |
| Timing of metastases | ||||||
| Metachronous | 55 | 36.9 | 48 | 38.4 | 16 | 38.1 |
| Synchronous | 94 | 63.1 | 77 | 61.6 | 26 | 61.9 |
| Number of metastasesa | ||||||
| 1–2 | 88 | 59.5 | 73 | 58.4 | 16 | 39.0 |
| >2 | 60 | 40.5 | 52 | 41.6 | 25 | 61.0 |
| Size of metastasesa | ||||||
| 1–50 mm | 127 | 85.8 | 107 | 85.6 | 36 | 87.8 |
| >50 mm | 21 | 14.2 | 18 | 14.4 | 5 | 12.2 |
| Distribution of metastases | ||||||
| Unilobar | 76 | 51.0 | 62 | 49.6 | 16 | 38.1 |
| Bilobar | 73 | 49.0 | 63 | 50.4 | 26 | 61.9 |
| Primary tumor sitea | ||||||
| Right colon | 39 | 26.4 | 32 | 25.8 | 8 | 19.0 |
| Left colon | 60 | 40.5 | 51 | 41.1 | 21 | 50.0 |
| Rectum | 49 | 33.1 | 41 | 33.1 | 13 | 31.0 |
One patient had missing information.
Radiologic response was assessed according to RECIST (7). Patients with disease progression under neoadjuvant treatment (<5%), which is considered a contraindication, did not undergo liver resection and were not included in this study (8). Tissue samples were not available in these patients. Histologic response was assessed according to the classification proposed by Rubbia-Brandt and colleagues (9). As described for this classification, tumor regression grade 1 and 2 were classified as major histologic response, tumor regression grade 3 as partial histologic response, and tumor regression grade 4 and 5 as no histologic response.
After microdissection, intratumoral mRNA was isolated from resected formalin-fixed paraffin-embedded, bevacizumab-pretreated CLM. In 42 of these patients, a matching primary tumor (PT) sample collected before bevacizumab treatment was available. Relative mRNA levels were calculated as ratios of the target gene and a reference gene [ACTB (β-actin)]. Relative mRNA levels of 9 genes (ACVRL1, EGFL7, EPHB4, HIF1A, VEGFA, VEGFB, VEGFC, FLT1, KDR) were analyzed by RT-PCR in CLM and primary tumor samples and evaluated for associations with response, recurrence-free survival (RFS), and overall survival (OS). Because of the limited patient number, the number of selected genes was restricted to the 9 most promising according to previous studies. In CLM, histologic response was not correlated with gene expression due to a high rate of missing gene expression data caused by tumor destruction in the major histologic response group. Laser capture microdissection, isolation of mRNA, and RT-PCR was performed as previously described (10).
Statistical analyses
The primary endpoint was RFS, which was calculated from the date of liver resection until the first observation of recurrence or death from any cause. If a patient had not recurred or died, RFS was censored at the date of the last follow-up. OS was calculated as the time from the date of surgery until death from any cause or until the date of the last date known to be alive. OS, radiographic [partial response (PR), stable disease (SD)], and histologic responses (major, partial, none) were the secondary outcome measures.
Reported mRNA expression values were quantified as ratios between two absolute measurements: The gene of interest versus an internal housekeeping gene [ACTB (β-actin)]. No transformation was conducted for mRNA expression levels that were continuous variables. To assess the associations between gene expression and RFS, the expression levels were dichotomized into a low and a high subgroup at the optimal cut-off value if the expression level was measureable. The adapted maximal χ2 method of Miller and Siegmund (1982) and Halpern (1982) was used to determine which cut-off value best separated patients into poor prognosis and good prognosis subgroups, in terms of likelihood of recurrence. We compared RFS by three groups: no expression, low expression, and high expression. Patients who had no gene expression measureable had complete histologic response (CR) and no tumor tissue detected in the resected CLM. The corresponding P value was adjusted using 2,000 bootstrap-like simulations. The optimal cut-off values selected in analyzing RFS were applied for analyses of OS. The multivariable Cox regression model was used to evaluate the independent effects of gene expression levels on RFS and OS, when adjusting for age at surgery, number of metastases, presence of bilobar hepatic disease, and time to metastases (metachronous versus synchronous). The associations between gene expression levels and radiologic response were examined using the two-sample Wilcoxon test. The changes in the gene expression levels between the primary tumor and corresponding liver metastases were examined using the paired sign test.
In addition, recursive partitioning analyses were performed to identify patterns of gene expression levels associated with RFS, OS, and radiologic and histologic response. A mixture cure model was used to assess the cure rate by expression levels of each gene (11). Two recurrence patterns, intrahepatic only and extrahepatic were examined using cumulative incidence of recurrence in the competing risks model.
No adjustment for multiple comparisons was performed because this study was exploratory and hypothesis generating. No sample size or power calculation was performed because of the retrospective name of the study. All tests were two-sided with an α significance of 0.05. All analyses were performed using the SAS statistical package version 9.4 (SAS Institute), and R package (R Foundation for Statistical Computing).
Results
Gene expression in CLM and association with RFS, OS, and radiologic response
Thirteen (10.4%) patients achieved a complete histologic response (CR) and had a median RFS of 29.1 months. Using these patients as a reference group, VEGFB, VEGFC, HIF1A, KDR, and EPHB4 expression predicted RFS in univariable analysis. Specifically, high VEGFB [not reached vs. 8.3 months (low); HR = 0.84 and HR = 3.36, P = 0.001], VEGFC [not reached vs. 10.3 months (low); HR = 0.60 and HR = 2.34, P = 0.034], HIF1A [not reached vs. 9.5 months (low); HR = 0.67 and HR = 2.62, P = 0.017], and KDR expression [17.8 vs. 9.5 months (low); HR = 1.24 and HR = 2.70, P = 0.039] were associated with significantly longer RFS. Conversely, low EPHB4 expression [13.0 vs. 7.7 months (high); HR = 1.88 and HR = 4.67, P = 0.043] was associated with improved RFS. In multivariable analysis, the associations between VEGFB (HR = 0.71 and HR = 3.29, P < 0.001), VEGFC (HR = 0.66 and HR = 2.07, P = 0.025), HIF1A (HR = 0.60 and HR = 2.26, P = 0.002), and KDR (HR = 1.16 and HR = 2.29, P = 0.015) and RFS remained significant. In addition, low EGFL7 expression was associated with improved RFS (16.0 months vs. 8.4 months (high); HR = 1.46 and HR = 2.45, P = 0.044). Kaplan–Meier curves for RFS are given in Supplementary Figs. S1–S6. With regard to OS, the 3-year OS rate of the 13 patients with a complete histologic response was 75%. High VEGFB (91% vs. 57% (low); HR = 0.54 and HR = 2.12, P = 0.013) and ACVRL1 [62% vs. 36% (low); HR = 1.25 and HR = 3.37, P = 0.034)] mRNA levels predicted significantly outcomes in univariable analysis (Supplementary Figs. S7 and S8). In multivariable analysis, only the association between ACVRL1 expression and 3-year OS remained significant (HR = 0.82 and HR = 2.46, P = 0.041). Data on RFS and OS are given in Table 2. Radiologic response was associated with high ACVRL1 [PR = 1.01 (range, 0.00–3.49), SD = 0.52 (range, 0.00–1.93), P = 0.026] and low VEGFA mRNA levels [PR = 4.58 (range, 0.00–3.23), SD = 6.22 (range, 0.00–15.51), P = 0.035]. Expression levels of the other investigated genes were not associated with radiologic response (data not shown).
Table 2.
Association of intratumoral gene expression with RFS and OS in CLM
| RFS |
OS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | n | Median (95% CI) | Uni HR (95% CI) | P a | Multi HR (95% CI) | P b | 3-yr rate ± SE | Uni HR (95% CI) | P b | Multi HR (95% CI) | P b |
| ACVRL1 | 0.59 | 0.11 | 0.034 | 0.041 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 0.390 | 12 | 9.5 (4.1–16.5) | 2.86 (1.06–7.73) | 2.95 (1.07–8.12) | 0.36 ± 0.16 | 3.37 (0.98–11.56) | 2.46 (0.69–8.71) | ||||
| > 0.390 | 95 | 11.7 (9.2–14.4) | 1.97 (0.91–4.30) | 1.73 (0.78–3.82) | 0.71 ± 0.05 | 1.25 (0.44–3.56) | 0.82 (0.27–2.46) | ||||
| EGFL7 | 0.052 | 0.044 | 0.51 | 0.88 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| 0.001 | 63 | 16.0 (10.1–20.3) | 1.58 (0.71–3.51) | 1.46 (0.64–3.32) | 0.70 ± 0.07 | 1.30 (0.44–3.81) | 0.91 (0.30–2.80) | ||||
| > 0.001 | 42 | 8.4 (5.4–11.7) | 3.05 (1.34–6.91) | 2.45 (1.06–5.67) | 0.62 ± 0.09 | 1.76 (0.58–5.30) | 1.11 (0.34–3.61) | ||||
| EPHB4 | 0.043 | 0.16 | 0.20 | 0.66 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 0.286 | 99 | 13.0 (10.1–16.3) | 1.88 (0.87–4.06) | 1.75 (0.79–3.88) | 0.69 ± 0.05 | 1.38 (0.49–3.89) | 1.00 (0.34–2.93) | ||||
| > 0.286 | 12 | 7.7 (4.5–10.8) | 4.67 (1.67–13.06) | 2.78 (0.97–8.00) | 0.32 ± 0.18 | 2.89 (0.77–10.90) | 1.59 (0.39–6.59) | ||||
| HIF1A | 0.017 | 0.002 | 0.27 | 0.61 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 1.725 | 82 | 9.5 (6.5–11.1) | 2.62 (1.21–5.70) | 2.26 (1.01–5.02) | 0.62 ± 0.06 | 1.74 (0.61–4.94) | 1.20 (0.40–3.62) | ||||
| > 1.725 | 20 | 91.1+ | 0.67 (0.23–1.96) | 0.60 (0.20–1.82) | 0.81 ± 0.10 | 0.87 (0.22–3.49) | 0.71 (0.17–2.91) | ||||
| VEGFA | 0.36 | 0.063 | 0.26 | 0.65 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 6.573 | 65 | 13.0 (9.2–16.3) | 1.96 (0.88–4.33) | 1.65 (0.73–3.73) | 0.73 ± 0.06 | 1.28 (0.43–3.74) | 0.94 (0.30–2.94) | ||||
| > 6.573 | 29 | 9.5 (4.9–11.1) | 2.89 (1.23–6.79) | 2.68 (1.11–6.47) | 0.53 ± 0.12 | 2.12 (0.68–6.61) | 1.39 (0.40–4.75) | ||||
| VEGFB | 0.001 | <.0001 | 0.013 | 0.065 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 4.135 | 65 | 8.3 (5.6–10.3) | 3.36 (1.55–7.32) | 3.29 (1.45–7.46) | 0.57 ± 0.07 | 2.12 (0.74–6.04) | 1.56 (0.50–4.82) | ||||
| > 4.135 | 28 | 91.1+ | 0.84 (0.33–2.10) | 0.71 (0.27–1.87) | 0.91 ± 0.06 | 0.54 (0.14–2.16) | 0.44 (0.11–1.83) | ||||
| VEGFC | 0.034 | 0.025 | 0.10 | 0.34 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 0.481 | 98 | 10.3 (8.3–13.5) | 2.34 (1.08–5.07) | 2.07 (0.93–4.61) | 0.62 ± 0.06 | 1.65 (0.59–4.64) | 1.16 (0.39–3.43) | ||||
| > 0.481 | 13 | 72.7+ | 0.60 (0.18–2.02) | 0.66 (0.19–2.26) | 1.00 ± 0.00 | 0.29 (0.03–2.58) | 0.26 (0.03–2.36) | ||||
| FLT1 | 0.23 | 0.27 | 0.62 | 1.00 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 1.403 | 101 | 10.5 (8.3–14.3) | 2.16 (0.99–4.70) | 1.86 (0.84–4.11) | 0.66 ± 0.05 | 1.50 (0.53–4.23) | 1.01 (0.34–2.99) | ||||
| > 1.403 | 11 | 18.5 (10.3–54.4) | 1.20 (0.40–3.55) | 1.40 (0.46–4.28) | 0.72 ± 0.18 | 0.94 (0.17–5.13) | 1.02 (0.18–5.82) | ||||
| KDR | 0.039 | 0.015 | 0.27 | 0.63 | |||||||
| 0 | 13 | 29.1 (10.3–81.2) | 1 (reference) | 1 (reference) | 0.75 ± 0.13 | 1 (reference) | 1 (reference) | ||||
| ≤ 0.691 | 65 | 9.5 (5.8–12.8) | 2.70 (1.23–5.95) | 2.29 (1.02–5.13) | 0.58 ± 0.07 | 1.70 (0.59–4.93) | 1.04 (0.34–3.19) | ||||
| > 0.691 | 39 | 17.8 (11.7–91.1) | 1.24 (0.53–2.91) | 1.16 (0.49–2.79) | 0.82 ± 0.07 | 0.99 (0.31–3.15) | 0.72 (0.21–2.40) | ||||
mRNA levels of gene were dichotomized on the optimal cut-off value using the maximal χ2 method on RFS. P value for univariable analysis of RFS was adjusted for considering potential multiple cut-off value.
P values were based on log-rank test in the univariate analysis and Wald test in the multivariate analysis within Cox regression model. Multivariate Cox regression model was adjusted for age (<65 vs. ≥65 yrs), number of mets (1–2 Mets vs. >2 Mets), timing to metastases (metachronous vs. synchronous), and bilobar (no vs. yes).
Recursive partitioning analyses in CLM for RFS, OS, and radiologic response
Recursive partitioning analyses in CLM showed that VEGFB expression was the most important predictor of RFS, and EGFL7 and VEGFB levels further defined prognostic subgroups (Fig. 1). Similarly, VEGFB level was the primary prognostic marker of OS, whereas EGFL7 and VEGFA levels were relevant in subgroups (Fig. 2). No recursive partitioning tree was created for radiologic response.
Figure 1.

A, recursive partitioning tree for RFS in CLM. B, Kaplan–Meier curves for terminal nodes for RFS (NR, not reached).
Figure 2.
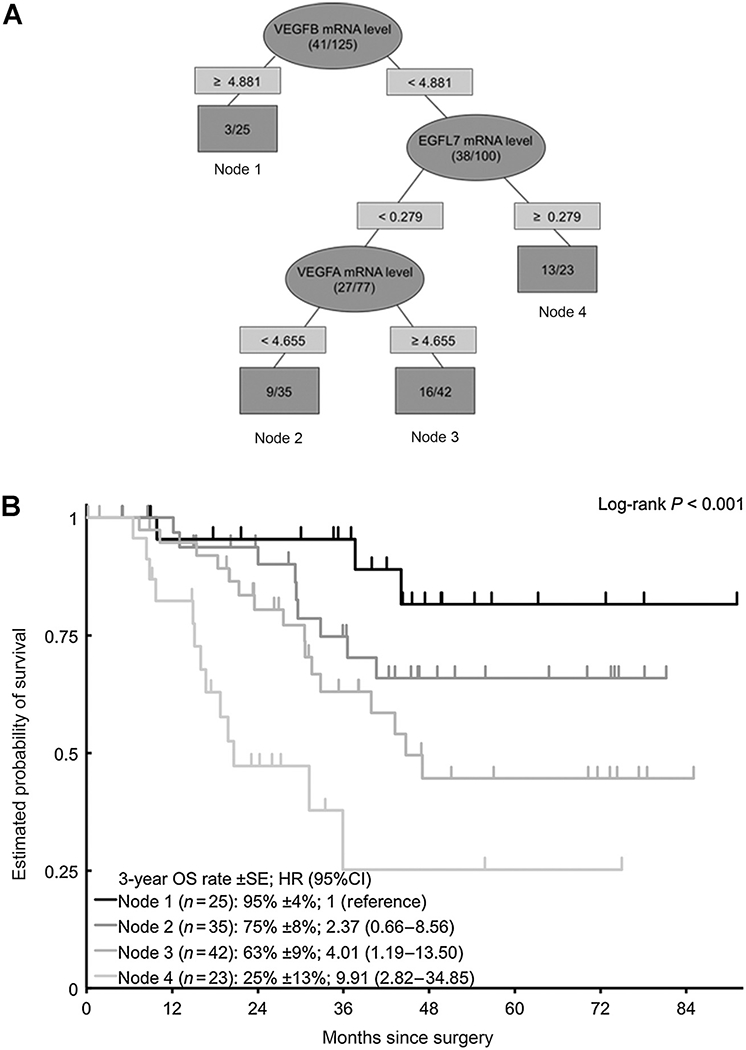
A, recursive partitioning tree for OS in CLM. B, Kaplan–Meier curves for terminal nodes for OS.
Gene expression in CLM and probability of cure
In CLM, EPHB4 and EGFL7 mRNA levels were associated with probability of cure. Patients with high EPHB4 [OR = 3.15; 95% confidence interval (CI), 1.15–8.61; P = 0.025] or EGFL7 (OR = 2.40; 95% CI, 1.19–4.84; P = 0.015) expression had lower probability of cure. Expression levels of the other investigated genes were not associated with probability of cure (data not shown). Probability of cure was not assessed in primary tumors due to the limited sample size.
Gene expression in CLM and pattern of recurrence
High expression of VEGFB [49% (SE ±11%) vs. 71% (±6%; low) vs. 40% (±15%; CR), P = 0.005], VEGFC [23% (±12%) vs. 69% (±5%; low) vs. 40% (±15%; CR), P = 0.008], and KDR [44% (±9%) vs. 72% (±6%; low)vs. 40% (±15%; CR), P = 0.003] were associated with significantly lower 2-year extrahepatic recurrence rates. Expression levels of the other investigated genes were not associated with patterns of recurrence (data not shown).
Expression of VEGFB and VEGFC in CLM and association with hypoxia
HIF1A expression was measured as a marker for hypoxia. High expressions of VEGFB and VEGFC were significantly associated with high HIF1A expression (P < 0.001 and P = 0.002, respectively).
Gene expression in primary tumors and association with RFS, OS, and response
In primary tumors, high VEGFA levels predicted significantly shorter RFS (10.1 vs. 22.6 months; HR = 2.83, P = 0.038). In terms of 3-year OS rates, high VEGFB (46% vs. 85%; HR = 5.75, P = 0.009) and low FLT1 (55% vs. 100%; P = 0.031) were associated with inferior outcomes. Low VEGFA levels were associated with radiologic response (PR = 3.53; SD = 5.65; P = 0.032). High EGFL7 predicted histologic response (major histologic response = 0.32; partial histologic response = 0.001; no histologic response = 0.001; P = 0.043). Expression levels of the other investigated genes were not associated with RFS, OS, radiologic, and histologic response (data not shown).
Correlation between gene expression in primary tumors and CLM
Low expression of VEGFB in primary tumors was associated with increased expression in CLM, whereas high expression in primary tumors was associated with reduced expression in CLM (r = −0.67, P < 0.001).
Discussion
Our data show for the first time that intratumoral mRNA expression levels of genes involved in vascular morphogenesis and early vessel maturation predict response, survival, and probability of cure in patients with liver-limited metastatic colorectal cancer undergoing bevacizumab-based perioperative chemotherapy and curative liver resection. Furthermore, our results suggest that perioperative bevacizumab-based therapy may induce intratumoral gene expression changes in a manner that affects clinical outcomes.
Previous studies have shown high levels of concordance between VEGF mRNA expression in primary tumors and corresponding liver metastases, supporting the notion that primary tumor gene expression can serve as a reliable surrogate of bevacizumab efficacy in CLM (12). Interestingly, our data demonstrated that certain genes may have divergent prognostic effects depending on previous treatment. Among patients in whom bevacizumab-naïve primary tumor tissue was available for analysis, high VEGFA expression was associated with lower radiologic response in the resected CLM and shorter RFS, while high VEGFB expression was associated with decreased 3-year OS rates. These findings are consistent with previous studies showing these markers to be negative prognosticators (13, 14). VEGFA, the target of bevacizumab, binds FLT1 and KDR, and is considered a main driver of angiogenesis (15). Conversely, VEGFB binds only to FLT1 and has been shown to potentiate, rather than initiate angiogenesis, and also plays a role in tissue protection (15). In contrast with VEGFB, high primary tumor FLT1 expression was associated with increased 3-year OS. Current evidence suggests that the clinical relevance of FLT1 expression in colorectal cancer depends on disease stage and therapy rendered. In the adjuvant setting, high FLT1 expression has been associated with shorter time-to-recurrence in patients with stage II and III colorectal cancer (16). However, in patients with metastatic colorectal cancer receiving chemotherapy and a VEGFR inhibitor, a more recent biomarker study showed that high FLT1 expression to be associated with higher radiologic response and longer progression-free survival (PFS), which supports the finding of the current study (10).
In contrast with our findings in primary tumor tissue, high VEGFB and VEGFC expression in CLM predicted longer RFS and OS as well as lower extrahepatic recurrence rates. One hypothesis that may reconcile these findings is that although increased VEGF signaling in primary tumors may reflect an intrinsically resistant phenotype, effective VEGFA inhibition may cause compensatory VEGFB and VEGFC upregulation in CLM as an attempted escape mechanism, and may therefore serve as a marker of bevacizumab efficacy or sensitivity (17). This hypothesis is supported by the fact that prognostic beneficial low VEGFB expression in primary tumors was associated with an increase in expression levels in CLM. On the basis of our findings for VEGFB and VEGFC, we hypothesized that those patients with the best treatment effect had a compensatory upregulation of HIF1A expression. We tested that hypothesis and showed that high expression of HIF1A predicted favorable clinical outcome and was associated with high VEGFB and VEGFC expression. Indeed, preclinical and clinical data demonstrate that bevacizumab induces intratumoral hypoxia and subsequent HIF1A expression, and that VEGF/KDR/HIF1A signaling can be variably affected by bevacizumab (18).
Furthermore, recursive partitioning analyses demonstrated that the pattern of genes for RFS and OS was similar with high VEGFB expression identifying patients with the most favorable clinical outcome. This consistent observation supports the idea of VEGFB expression being a clinically relevant biomarker for bevacizumab efficacy.
High KDR expression in CLM was associated with significantly increased RFS and 3-year OS. Recent studies investigating KDR expression in patients with metastatic colorectal cancer receiving bevacizumab-based chemotherapy have demonstrated similar associations (19, 20). With regard to EGFL7, high expression in CLM predicted shorter RFS and lower probability of cure, whereas in primary tumors high expression was associated with major histologic response. The implications of these contradictory observations are unclear and require further investigation. EGFL7 is expressed by endothelial cells and regulates tubulogenesis in addition to endothelial cell proliferation, migration, and invasion as well as extracellular matrix remodeling (21, 22). EGFL7 is overexpressed in various tumor types promoting tumor angiogenesis and irregular vessel formation (23–25). Moreover, in a murine hepatocellular carcinoma model, EGFL7 has been shown to promote tumor growth via EGFR signaling, and knockout of EGLF7 results in smaller tumors with reduced vessel density (24). In patients with metastatic colorectal cancer receiving first-line bevacizumab-based chemotherapy, low EGFL7 expression has been associated with better radiologic response and longer PFS (26). Anti-EGFL7 in combination with anti-VEGF therapy appears to be a promising treatment approach, and the anti-EGFL7 antibody MEGF0444A is currently being investigated in patients with metastatic colorectal cancer receiving bevacizumab-based chemotherapy (NCT01399684; ref. 27).
High expression of ACVRL1 in CLM, which encodes a coreceptor within the TGFB pathway and is a member of the TGFB receptor 1 (TGFBR1) family, was associated with increased radiologic response and longer OS. This is consistent with preclinical data demonstrating that ACVRL1 is associated with VEGFA downregulation, and therefore improved efficacy of bevacizumab-based chemotherapy (28). In conjunction with TGFBR2, ACVRL1 binds the ligands TGFB1 and TGFB3 to mediate endothelial cell migration and proliferation and early vessel maturation (29). Lack or mutation of ACVRL1 is associated with vascular malformation (30). Little is currently known about the role of ACRVL1 expression in tumors. However, consistent with our findings, one study showed low expression to be associated with poor prognosis in patients with nasopharyngeal tumors (31).
Finally, high EPHB4 expression in CLM was associated with shorter RFS and lower probability of cure. After binding of its ligand, EFNB2, this tyrosine kinase receptor promotes angiogenesis and vessel maturation in a synergistic manner with VEGFA. Our findings regarding EPHB4 expression is in line with that of a previous study showing high EPHB4 expression to be predictive of nonresponse and worse survival in metastatic colorectal cancer patients receiving bevacizumab (32).
Certain limitations of our study should be acknowledged, including the limited sample size and retrospective design. Another limitation is that gene expression levels in CLM could be measured in only a subset of patients who had a major histologic response and was therefore not correlated to histologic response. Furthermore, samples of primary tumors were only available in about a third of patients, which also limits the strength of our conclusions. In addition, 13 of 125 (10%) patients had a complete pathologic response after bevacizumab-based chemotherapy. Analyses were therefore only performed in patients who were not the best responders. Comparing gene expression profiles in CLM at diagnosis and after resection would shed important insight on bevacizumab-induced changes on tumor biology, concordance with primary lesions, and further define the clinical relevance of these biomarkers. Moreover, prospective studies investigating corresponding tissue samples of CLM collected before and after bevacizumab-based therapy would clarify the relevance of expression changes as prognosticators, treatment effects, and biological mechanisms.
In conclusion, this study shows for the first time that expression of genes involved in vascular morphogenesis and early vessel maturation pathways may be promising predictive and/or prognostic biomarkers in metastatic colorectal cancer patients with resected CLM treated with bevacizumab-based chemotherapy. These novel biomarkers may help to identify who do not benefit from this invasive treatment approach and should better undergo other treatment concepts. Future prospective studies are warranted to validate these findings.
Supplementary Material
Acknowledgments
Grant Support
S. Stremitzer is a recipient of an Erwin Schrödinger fellowship of the Austrian Science Fund (J3501-B13). T. Gruenberger receives financial support by Roche, Merck-Serono, Sanofi-Aventis, Bayer, and Amgen. H.-J. Lenz receives financial support from the NIH (P30CA014089), the Gloria Borges Wonderglo Foundation, and the Daniel Butler Research Fund.
Disclosure of Potential Conflicts of Interest
T. Gruenberger reports receiving a commercial research grant from Merck; has received speakers bureau honoraria from Roche, Merck, Amgen, and Eli Lilly; and is a consultant/advisory board member for Roche, Merck, and Amgen. H.-J. Lenz is a consultant/advisory board member for Bristol-Myers Squibb, Merck KG, and Genentech. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
References
- 1.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004;350:2335–42. [DOI] [PubMed] [Google Scholar]
- 2.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol 2013;14:1208–15. [DOI] [PubMed] [Google Scholar]
- 3.Gruenberger B, Tamandl D, Schueller J, Scheithauer W, Zielinski C, Herbst F, et al. Bevacizumab, capecitabine, and oxaliplatin as neoadjuvant therapy for patients with potentially curable metastatic colorectal cancer. J Clin Oncol 2008;26:1830–5. [DOI] [PubMed] [Google Scholar]
- 4.Cui CH, Huang SX, Qi J, Zhu HJ, Huang ZH, Yu JL. Neoadjuvant chemotherapy (NCT) plus targeted agents versus NCT alone in colorectal liver metastases patients: a systematic review and meta-analysis. Oncotarget 2015;6:44005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klinger M, Tamandl D, Eipeldauer S, Hacker S, Herberger B, Kaczirek K, et al. Bevacizumab improves pathological response of colorectal cancer liver metastases treated with XELOX/FOLFOX. Ann Surg Oncol 2010;17:2059–65. [DOI] [PubMed] [Google Scholar]
- 6.Lambrechts D, Lenz HJ, de Haas S, Carmeliet P, Scherer SJ. Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol 2013;31:1219–30. [DOI] [PubMed] [Google Scholar]
- 7.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 8.Adam R, Pascal G, Castaing D, Azoulay D, Delvart V, Paule B, et al. Tumor progression while on chemotherapy: a contraindication to liver resection for multiple colorectal metastases? Ann Surg 2004;240:1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubbia-Brandt L, Giostra E, Brezault C, Roth AD, Andres A, Audard V, et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neoadjuvant chemotherapy followed by liver surgery. Ann Oncol 2007;18:299–304. [DOI] [PubMed] [Google Scholar]
- 10.Wilson PM, Yang D, Azuma M, Shi MM, Danenberg KD, Lebwohl D, et al. Intratumoral expression profiling of genes involved in angiogenesis in colorectal cancer patients treated with chemotherapy plus the VEGFR inhibitor PTK787/ZK 222584 (vatalanib). Pharmacogenomics J 2013; 13:410–6. [DOI] [PubMed] [Google Scholar]
- 11.Corbiere F,Joly P.ASAS macro for parametric and semiparametric mixture cure models. Comput Methods Programs Biomed 2007;85:173–80. [DOI] [PubMed] [Google Scholar]
- 12.Kuramochi H, Hayashi K,Uchida K, Miyakura S, Shimizu D,Vallbohmer D, et al. Vascular endothelial growth factor messenger RNA expression level is preserved in liver metastases compared with corresponding primary colorectal cancer. Clin Cancer Res 2006;12:29–33. [DOI] [PubMed] [Google Scholar]
- 13.Des Guetz G, Uzzan B, Nicolas P, Cucherat M, Morere JF, Benamouzig R, et al. Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br J Cancer 2006;94:1823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayasinghe C, Simiantonaki N, Kirkpatrick CJ. VEGF-B expression in colorectal carcinomas and its relevance for tumor progression. Histol Histopathol 2013;28:647–53. [DOI] [PubMed] [Google Scholar]
- 15.Bry M, Kivela R, Leppanen VM, Alitalo K. Vascular endothelial growth factor-B in physiology and disease. Physiol Rev 2014;94:779–94. [DOI] [PubMed] [Google Scholar]
- 16.Ning Y, Lurje G, Danenberg K, Cooc J, Yang D, Pohl A, et al. VEGF and VEGFR1 gene expression levels and tumor recurrence in adjuvant colon cancer. J Clin Oncol 27:15s, 2009. (suppl; abstr 4040). [Google Scholar]
- 17.Mesange P, Poindessous V, Sabbah M, Escargueil AE, de Gramont A, Larsen AK. Intrinsic bevacizumab resistance is associated with prolonged activation of autocrine VEGF signaling and hypoxia tolerance in colorectal cancer cells and can be overcome by nintedanib, a small molecule angiokinase inhibitor. Oncotarget 2014;5:4709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvani M, Trisciuoglio D, Bergamaschi C, Shoemaker RH, Melillo G. Differential involvement of vascular endothelial growth factor in the survival of hypoxic colon cancer cells. Cancer Res 2008;68:285–91. [DOI] [PubMed] [Google Scholar]
- 19.Weickhardt AJ, Williams C, Lee C, Simes J, Murone C, Wilson K, et al. Vascular endothelial growth factors (VEGF) and VEGF receptor expression as predictive biomarkers for benefit with bevacizumab in metastatic colorectal cancer (mCRC): analysis of the phase III MAX study. J Clin Oncol 29: 2011. (suppl; abstr3531). [Google Scholar]
- 20.El-Khoueiry AB, Pohl A, Danenberg K, Cooc J, Zhang W, Yang D, et al. Wt Kras and gene expression levels of VEGFR2, EGFR, and ERCC-1 associated with progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC) treated with first-line 5-FU or capecitabine with oxaliplatin and bevacizumab (FOLFOX/BV or XELOX/BV). J Clin Oncol 27:15s, 2009(suppl; abstr 4056). [Google Scholar]
- 21.Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature 2004;428:754–8. [DOI] [PubMed] [Google Scholar]
- 22.Nichol D, Stuhlmann H. EGFL7: a unique angiogenic signaling factor in vascular development and disease. Blood 2012;119:1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Pena C, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer 2008;47:794–802. [DOI] [PubMed] [Google Scholar]
- 24.Wu F, Yang LY, Li YF, Ou DP, Chen DP, Fan C. Novel role for epidermal growth factor-like domain 7 in metastasis of human hepatocellular carcinoma. Hepatology 2009;50:1839–50. [DOI] [PubMed] [Google Scholar]
- 25.Nichol D, Shawber C, Fitch MJ, Bambino K, Sharma A, Kitajewski J, et al. Impaired angiogenesis and altered Notch signaling in mice overexpressing endothelial Egfl7. Blood 2010;116:6133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen TF, Nielsen BS, Sorensen FB, Johnsson A, Jakobsen A. Epidermal growth factor-like domain 7 predicts response to first-line chemotherapy and bevacizumab in patients with metastatic colorectal cancer. Mol Cancer Ther 2014;13:2238–45. [DOI] [PubMed] [Google Scholar]
- 27.Johnson L, Huseni M, Smyczek T, Lima A, Yeung S, Cheng JH, et al. Anti-EGFL7 antibodies enhance stress-induced endothelial cell death and anti-VEGF efficacy. J Clin Invest 2013;123:3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, et al. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci U S A 2000;97:2626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawinkels LJ, GarciadeVinuesa A,TenDijke P. Activin receptor-like kinase 1 as a target for anti-angiogenesis therapy. Expert Opin Investig Drugs 2013;22:1371–83. [DOI] [PubMed] [Google Scholar]
- 30.Urness LD, Sorensen LK, Li DY. Arteriovenous malformations in mice lacking activin receptor-like kinase-1. Nat Genet 2000;26: 328–31. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Zeng Z, Fan S, Wang J, Yang J, Zhou Y, et al. Evaluation of the prognostic value of TGF-beta superfamily type I receptor and TGF-beta type II receptor expression in nasopharyngeal carcinoma using high-throughput tissue microarrays. J Mol Histol 2012;43:297–306. [DOI] [PubMed] [Google Scholar]
- 32.Guijarro-Munoz I, Sanchez A, Martinez-Martinez E, Garcia JM, Salas C, Provencio M, et al. Gene expression profiling identifies EPHB4 as a potential predictive biomarker in colorectal cancer patients treated with bevacizumab. Med Oncol 2013;30:572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


