Abstract
Thyroid hormone (T3) nuclear receptors (TR) are ligand-dependent transcription factors which regulate growth, differentiation, and development. One emerging hypothesis suggests that TR mediate these diverse effects via a large network of coregulators. Recently, we found that TR-mediated transcriptional responses varied in six cell lines derived from different tissues. We therefore used human TR subtype β1 (TRβ1) as bait to search for coregulators in human colon carcinoma RKO cells with a yeast two-hybrid system. RKO cells exhibited T3-dependent and -independent transcriptional activation. One of the three positive clones was identified as Ear-2, which is a distant member of the chick ovalbumin upstream promoter-transcription factors of the orphan nuclear receptor family. The physical interaction between Ear-2 and TRβ1 was further confirmed by specific binding of Ear-2 to glutathione S-transferase–TRβ1. In addition, Ear-2 was found to associate with TRβ1 in cells. As a result of this physical interaction, binding of TRβ1 to the T3 response elements was inhibited. Using reporter systems, we found that both the basal activation and the T3-dependent activation mediated by TRβ1 were repressed by Ear-2 in CV1 cells. In RKO cells, however, the T3-independent transcriptional activity was more sensitive to the repression effect of Ear-2 than the T3-dependent transcriptional activity. The repression effect of Ear-2 was reversed by steroid hormone receptor coactivator 1. These results suggest that TR-mediated responses reflect a balance of corepressors and coactivators in cells. These findings further strengthen the hypothesis that the diverse activities of TR are achieved via a large network of coregulators that includes Ear-2.
Thyroid hormone, 3,3′,5-triiodo-l-thyronine (T3), is important for the growth, development, and differentiation of vertebrates. It is also essential for maintaining metabolic-energetic homeostasis. These biological activities of T3 are mediated by T3 nuclear receptors (TR), which are members of the steroid hormone and retinoic acid receptor superfamily (28). Like other nuclear receptors, TR consists of modular domains which include the amino-terminal A/B domain, the central DNA binding domain (domain C), and the hormone binding domains (domains D and E). Two TR genes, α and β, located on chromosomes 17 and 3, respectively, give rise to receptor isoforms (α1, α2, β1, and β2) by alternative splicing. TR mediates the action of T3 by binding to the cis elements on the promoter regions of target genes. TR binds to specific sequences known as T3 response elements (TRE), containing the consensus AGGTCA half-site binding motifs arranged in an inverted repeat, everted repeat, or direct repeat separated by 4 nucleotides (DR4) (4). The transcriptional activity of TR depends not only on the types of TRE but also on T3. In the absence of T3, TR acts as a repressor. Binding of T3 leads TR to function as a transcriptional activator (11).
While the mechanism(s) underlying the repression and activation functions of TR is not fully understood, recent studies indicate that these two functions could be mediated via the interaction of TR with a network of coregulatory proteins (20). It is known that in the absence of T3, TR is bound to corepressors, such as N-CoR, leading to transcription silencing (11). Binding of T3 leads to the dissociation of N-CoR from TR and allows TR to recruit coactivators, including steroid hormone nuclear receptor coactivator 1 (SRC-1) (26), CREB binding protein (CBP; also called p300) (13), and CBP-associated factors (14). These coactivators bridge the DNA-bound receptors to proteins in the preinitiation complex, thereby enhancing transcription. Some of the coactivators are histone acetyltransferases which act to modify the chromatin structure to facilitate easy accessibility for the binding of other transcription factors (1, 25, 29).
Recently, Xu et al. reported the establishment of SRC-1 knockout mice (34). The relatively minor phenotype changes and the up-regulation of the coactivator transcription initiation factor 2 (TIF2) in these mice suggest compensation of its function by other, redundant coactivators (34). Furthermore, it has also been shown that the expression of some coregulatory proteins, i.e., SRC-1, CBP, and N-CoR, is tissue specific (22). Thus, it is reasonable to postulate that there are other coregulatory proteins yet to be identified. This possibility was further supported by Fondell et al. (10), who isolated a complex of proteins associated with the hormone-bound TR from HeLa cells. None of the associated proteins was identified as the previously isolated coactivators, such as SRC-1 and CBP (10).
Recently, we observed that with the same TRE, the transcriptional activities of TR subtype β1 (TRβ1) differed in six cell lines derived from different tissues. We postulated that some tissue-dependent factors could play a role in modulating the transcriptional responses and are reflective of the cellular context. We therefore used a yeast two-hybrid system to search for TR-interacting proteins in human colon carcinoma RKO cells. In RKO cells, TRβ1 not only mediated T3-dependent transcriptional activity but also mediated potent T3-independent transcriptional activity. Using intact human TRβ1 as bait, we identified Ear-2, an orphan nuclear receptor, as one of the interacting proteins. Ear-2 is a distant member of the chick ovalbumin upstream promoter-transcription factors (COUP-TF) (32). Biochemical characterization indicated that Ear-2 interacted with the C-terminal region of TRβ1. Furthermore, we showed that Ear-2 was a strong negative regulator for the T3-dependent and T3-independent transcriptional activities of TR. The repression effect of Ear-2 could be reversed by SRC-1, indicating that the action of TR is based on the balance of modulation by coactivators and corepressors.
MATERIALS AND METHODS
Cell culture.
Monkey kidney CV1 cells, rat pituitary growth hormone-producing GC cells, human cervical carcinoma HeLa cells, epidermoid carcinoma A431 cells, breast carcinoma MCF-7 cells, human hepatocellular carcinoma SK-Hep-1 cells, and colon carcinoma RKO cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Quality Biological Inc., Gaithersburg, Md.).
Preparation of plasmids.
To construct the expression vector, Ear-2 cDNA was released from pGEM3Zf(−)Ear-2 (a generous gift from Todashi Yamamoto, University of Tokyo) (23) with NcoI-BamHI and ligated to a vector containing both T7 and cytomegalovirus (CMV) promoters (Invitrogen, Carlsbad, Calif.). To prepare mutant Ear-2, the cDNA for Ear-2 was cloned into the HindIII and BamHI sites of pBluescript SK(+); the resulting construct was called pBS-Ear-2. Single-stranded pBS-Ear-2 was prepared according to the manufacturer's instructions for the Muta-Gene Phagemid in vitro mutagenesis kit (Bio-Rad Laboratories, Hercules, Calif.) with host strain CJ236. The primer sequence for mutagenesis was 5′-CATTACGGTGTCTTCACCTGCGGATCCTGCAAGGTCTTTTCAAGCAACGATCCGC-3′, which changed the amino acid sequence from 73CysGluGlyCysLysSer80 (wild-type receptor sequence) to 73CysGlySerCysLysVal80 (mutated receptor sequence). The primer was annealed to the single-stranded template, and the second strand was synthesized. After transformation, the mutant plasmid was isolated and identified by restriction site mapping and DNA sequencing.
To construct the glutathione S-transferase (GST)–Ear-2 expression plasmid, pGEM3Zf(−)Ear-2 was digested with NcoI and XhoI and cloned into pGEX-6P-1 (Pharmacia Biotech, Piscataway, N.J.).
Yeast two-hybrid system.
A human colon carcinoma RKO cell cDNA library in pGAD10 (Clontech Laboratories, Inc., Palo Alto, Calif.) was constructed with a cDNA construction kit according to the manufacturer's instructions. In this library, the cDNAs were fused to the Gal4 activation domain downstream. The TRβ1 cDNA (NdeI-EcoRI-restricted DNA fragment from pCJ3) (16) was inserted into vector pAS2-1 to yield fusion protein Gal4DNA-BD-TRβ1 in yeast Y190. The cDNAs from the pGAD10 library and pGal4-DNA-BD-TRβ1 were electroporated into yeast Y190. The transformants were plated onto agar plates containing synthetic complete medium without histidine, leucine, and tryptophan. The pGAD10 cDNAs from the positive colonies were purified and sequenced.
Northern blotting.
Total RNAs were isolated from CV1, RKO, GC, and SK-Hep-1 cells with an RNA miniprep kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). An equal amount of total RNA from each cell line (10 μg) was analyzed on a 1% agarose gel containing formaldehyde (2.2 M) and transferred to a Hybond nylon membrane (Amersham Pharmacia Biotech UK Ltd., Piscataway, N.J.). A human poly(A) RNA-blotted membrane, MTN Blots (Clontech), was used for Northern blot analysis. The membrane was hybridized with human Ear-2 or TRβ1 cDNA labeled with [32P]dCTP by random priming and was subsequently washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.05% sodium dodecyl sulfate (SDS) at room temperature for 30 min and then 0.1× SSC–0.1% SDS at 50°C for 30 min. The blot was autoradiographed.
GST pull-down assay.
The GST-glutathione (GSH) binding system was used to assess the physical interaction of Ear-2 with TRβ1, glucocorticoid receptor (GR), estrogen receptor (ER), or SRC-1. The binding of 35S-labeled Ear-2 to TRβ1 was carried out basically as described previously (35). Briefly, Escherichia coli-expressed GST (2 μg) or GST-TRβ1 (2 μg) noncovalently bound to GSH-Sepharose beads was incubated with increasing amounts of in vitro-translated 35S-labeled Ear-2 (5, 10, and 20 μl) synthesized by use of a TNT kit (Promega) in 500 μl of buffer (20 mM Tris-Cl [pH 7.5], 100 mM NaCl, 2 mM EDTA, 0.1% Lubrol, 2 mM dithiothreitol, 0.05% bovine serum albumin, 5% glycerol). The beads were washed and boiled in SDS sample buffer. The proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). The gel was dried and autoradiographed.
In other experiments, 35S-labeled TRβ1, GR, ER, or SRC-1 (10 μl) was incubated with GST (2 μg) or GST–Ear-2 (2 μg) bound to GSH-Sepharose beads basically as described above. Detection and analysis of bound 35S-labeled TRβ1, GR, ER, or SRC-1 were done as described above.
Transient transfection assays.
Cells (4 × 105 to 8 × 105 cells/60-mm dish) were plated 24 h before transfection in DMEM containing 10% fetal bovine serum. The cells were transfected with the appropriate plasmids (0.2 μg of pTK28m, 0.2 μg of pCDM-TRβ1 [36], 0.2 μg of pCH110, and 0.025 to 1 μg of pCDM-Ear-2) by use of Lipofectamine (GIBCO-BRL, Gaithersburg, Md.). pTK28m is a chloramphenicol acetyltransferase (CAT) reporter plasmid containing two copies of TRE-Pal in tandem (a generous gift from G. Brent). pCH110 is a mammalian expression plasmid for β-galactosidase. After 24 h, the medium was replaced with fresh DMEM containing 10% T3-depleted serum. Twenty-four hours before the cells were harvested, T3 (100 nM) was added to the appropriate dishes. Cells were lysed, and CAT activity was determined as described previously (2, 15). The success of transfection was monitored by β-galactosidase activity. The CAT activity was normalized by using equal amounts of lysate proteins.
The transactivation activities of ER and GR were determined basically as described above. For the former, CV1 cells were transfected with the ER expression plasmid (HEGO; 0.2 μg), the luciferase reporter plasmid (pTK-ERE-Luc; 0.2 μg), and pCH110 (0.2 μg) in the absence or presence of pCDM-Ear-2 (0.2 μg). The total plasmids transfected were normalized to 3 μg. Estradiol (E2; 100 nM) was added to the appropriate dishes 24 h before cells were harvested. For the determination of the transcriptional activity of GR, CV1 cells were cotransfected with the CAT reporter plasmid (pMSG-CAT; 0.2 μg), pCMV-GR (0.2 μg), and pCH110 (0.2 μg) in the absence or presence of pCDM-Ear-2. The total plasmids transfected were normalized to 3 μg. Twenty-four hours before cells were harvested, dexamethasone (Dex; 100 nM) was added to the appropriate dishes. Determination of CAT activity was carried out basically as described above.
Primary rat hepatocytes were also used to analyze the effect of Ear-2 on the human S14 promoter. Hepatocytes were prepared and cultured as described previously (27). Six hours after plating (1.3 × 106 cells/35-mm petri dish), the hepatocytes were transfected with 0.6 μg of the human S14 promoter (4 kb) ligated to a luciferase reporter plasmid, 0.1 μg of the rat TRβ1 expression plasmid (or the control expression vector pCMV4), and 0.1 μg of the Ear-2 expression plasmid (or the control expression vector CMV4). Transfection was performed as described by Ota et al. (27). After 48 h, the cells were lysed and luciferase activity was measured. Data were analyzed by analysis of variance. When necessary, the data were log transformed to provide homogeneity of variance between the groups. Differences between the groups were determined by the Bonferroni technique.
Development of anti–Ear-2 antibodies.
Anti–Ear-2 antibodies in mice were developed by the DNA immunization method (5, 18). Briefly, 4-month-old female BALB/c mice were injected intradermally with 15 μg of pCDM-Ear-2. This injection was repeated once every 2 weeks for 6 months. Anti–Ear-2 antibodies were screened by immunoprecipitation of the [35S]methionine-labeled Ear-2 (3 μl) prepared by in vitro transcription-translation. Preimmune sera from the same mice were used as a control. Immunoprecipitation was carried out basically as described previously (31).
Coimmunoprecipitation of TRβ1 with Ear-2 in cells.
CV1 cells were transfected with pCLC51 (15) and pCDM-Ear-2 basically as described above. After the cells were cultured for an additional 24 h, the medium was replaced with Met- and Cys-free medium for 1 h. [35S]Met-Cys (Amersham; 40 μCi) was added and incubated with the cells for 3 h. Cells were lysed with lysis buffer (50 mM Tris [pH 7.4], 0.25 M NaCl, 5 mM EDTA, 100 mM NaF, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of aprotinin per ml, 10 μg of leupeptin per ml). Cellular extracts were immunoprecipitated with anti-TRβ1 monoclonal antibody (MAb) C4 or PC-9 (anti–Ear-2 sera). After being washed, the immunoprecipitates were analyzed by SDS-PAGE. After being dried, the gel was autoradiographed.
Determination of Ear-2 and Ear-2 mutant proteins by Western blot analysis.
Western blot analysis was carried out basically as described previously (36). Briefly, the cellular lysates mentioned above were analyzed by SDS–10% PAGE, and the proteins were transferred to nitrocellulose (Pharmacia Biotech). Blots were blocked with 10% nonfat dry milk (Bio-Rad) in 50 mM Tris-HCl (pH 7.4)–150 mM NaCl (TBS) at 25°C for 2 h and incubated with anti–Ear-2 sera (PC-9; 1:2,000 dilution) in 5% nonfat dry milk in TBS at 4°C overnight. After being washed, the blots were incubated with a 1:2,000 dilution of peroxidase-linked secondary antibody (Amersham) in 5% nonfat dry milk in TBS for 1 h at room temperature. Ear-2 proteins were detected with the ECL detection system (Amersham).
EMSA.
Two complementary oligonucleotides containing chicken lysozyme TRE (Lys), DR4, or Pal sequences were annealed, and the recessed 3′ end was filled with DNA polymerase (Klenow fragment) in the presence of [α-32P]dCTP as described by Meier et al. (21). In vitro-translated TRβ1 and/or Ear-2 was incubated with the labeled oligonucleotides in binding buffer (18, 21) in the presence or absence of antibody. For TRβ1, anti-TRβ1 MAb C4 (36) was used; for Ear-2, PC-9 was used. The electrophoretic gel mobility shift assay (EMSA) was carried out basically as described previously (18, 21).
RESULTS
TRβ1-mediated transcriptional activity is cell type dependent.
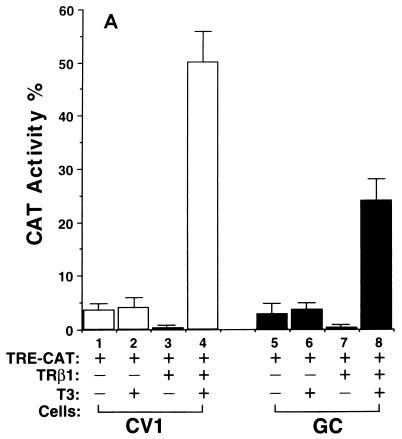
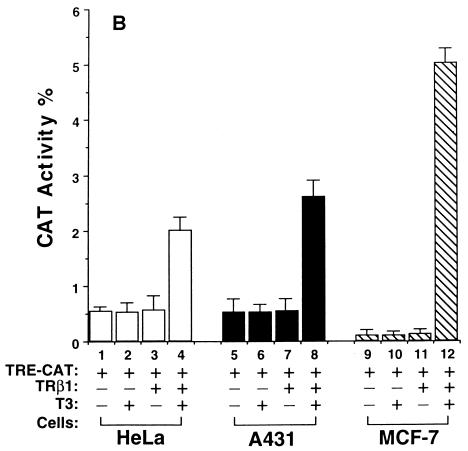
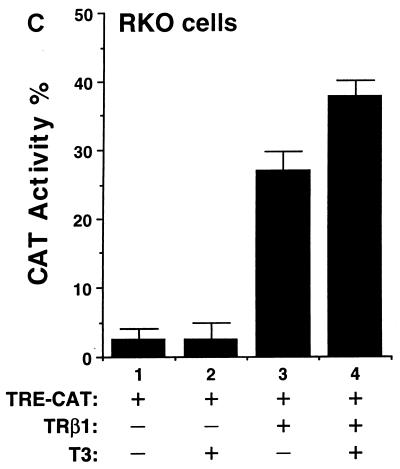
We hypothesized that the diverse effects of T3 could be mediated by the interaction of TR with a network of tissue-dependent factors and that the diverse effects reflect the cellular context. To test this hypothesis, we evaluated the transactivational responses of TRβ1 in six cultured cell lines which were derived from different tissues. They were monkey kidney cells, rat pituitary growth hormone-producing GC cells, human cervical carcinoma HeLa cells, human epidermoid carcinoma A431 cells, human breast cancer MCF-7 cells, and human colon carcinoma RKO cells. Based on the T3-independent TRβ1-mediated transcriptional activities, these six cell lines can be grouped into three classes. CV1 and GC cells belonged to the first class in that TRβ1 was a strong repressor in the absence of T3; as shown in Fig. 1A, about 90% of the basal transcriptional activity was repressed (bars 3 and 7 versus bars 1 and 5, respectively). HeLa, A431, and MCF-7 cells belonged to the second class in that TRβ1 had no effect on basal transcriptional activity in the absence of T3 (Fig. 1B, bars 3, 7, and 11 versus bars 1, 5, and 9, respectively). RKO cells belonged to the third class, in which strong T3-independent transactivation was seen (Fig. 1C, bar 3 versus bar 1).
FIG. 1.
Cell type-dependent transcriptional activity of TRβ1. Cells (4 × 105 to 8 × 105/60-mm dish) were plated 24 h before transfection in DMEM containing 10% fetal bovine serum. The cells were transfected with the appropriate plasmids by use of Lipofectamine (pTK28m, 0.2 μg; pCH110 and pCDM-TRβ1, 0.2 μg). After 24 h, all media were replaced with fresh DMEM containing 10% T3-depleted serum in the presence or absence of T3 (100 nM), and the dishes were incubated for an additional 24 h. The cells were harvested and lysed, and CAT activity was determined. The data shown are the results of three independent experiments, each with duplicates (mean ± standard deviation; n = 3). (A) CV1 cells and GC cells. (B) HeLa, A431, and MCF-7 cells. (C) RKO cells.
In all these cells, however, the addition of T3 led to transcriptional activation (bar 4 versus bar 3 and bar 8 versus bar 7 in Fig. 1A; bar 4 versus bar 3, bar 8 versus bar 7, and bar 12 versus bar 11 in Fig. 1B; and bar 4 versus bar 3 in Fig. 1C). Fold T3 activation, however, varied with cell type in that the highest activation was seen in CV1 cells (∼40-fold) and the lowest was detected in RKO cells (Fig. 1). These results indicate that the transcriptional activity of TRβ1 was cell type dependent.
Identification of Ear-2 as a TR-interacting protein and its expression in tissues.
The dramatic differences in the T3-independent transcriptional activities between RKO cells and other cell lines prompted us to search for TR-interacting proteins in RKO cells using a yeast two-hybrid system. Intact TRβ1 was fused to the GAL4 DNA binding domain to be used as bait to search the cDNA library prepared from RKO cells for interacting proteins. After repeated screening, three positive clones were identified. One of them was identified as the orphan nuclear receptor, Ear-2, by DNA sequencing. Human Ear-2 was cloned from human embryo fibroblasts by Miyajima et al. (23). It shares 59% sequence identity with TRβ1 in the DNA binding domain and 26 and 35% identities in regions II and III of the hormone binding domain, respectively (32). However, the Ear-2 gene has not been very well studied, and its functions are yet to be elucidated.
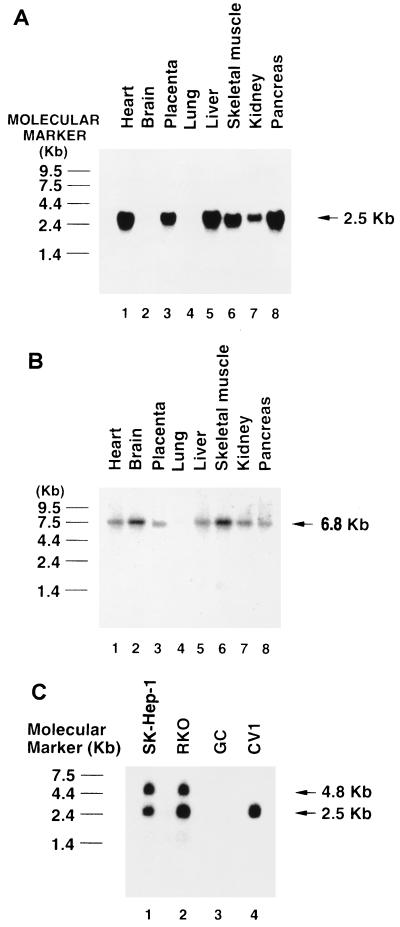
We first examined Ear-2 expression in human tissues and cultured cells by Northern blot analysis. In human tissues, Ear-2 was expressed as a single mRNA species of 2.5 kb (Fig. 2A). It was, however, differentially expressed in the heart, placenta, liver, skeletal muscle, kidney, and pancreas, with the most abundant message being found in the heart, liver, and pancreas. It was not detectable in the brain and lung. Using the same human tissue blot (Fig. 2B), we found that TRβ1 mRNA was expressed in the heart, brain, placenta, liver, skeletal muscle, kidney, and pancreas. A comparison of the expression patterns in Fig. 2A and B indicates that except for the brain, Ear-2 and TRβ were coexpressed in the same tissues.
FIG. 2.
Expression of Ear-2 in tissues and cultured cells, as determined by Northern blot analysis. (A and B) An RNA human tissue blot was purchased from Clontech and probed with human Ear-2 cDNA (A) and TRβ1 cDNA (B) labeled with [32P]dCTP. (C) Ten micrograms of total RNAs prepared from SK-Hep-1, RKO, GC, and CV1 cells was loaded onto a 1.2% agarose gel, and the blot was prepared as described in Materials and Methods. The blot was hybridized with 32P-labeled Ear-2 cDNA.
We also examined the expression of Ear-2 mRNA in several cultured cell lines (Fig. 2C). Lanes 1 and 2 of Fig. 2C show that in addition to the 2.5-kb mRNA, a second mRNA species, of 4.8 kb, was detected in human hepatocellular carcinoma SK-Hep-1 and RKO cells. In CV1 cells, however, only a single mRNA of the same size as that seen in the tissues was detected (Fig. 2C, lane 4). Ear-2 was not expressed in GC cells (Fig. 2C, lane 3). These results indicate that additional splicing products of mRNA were present in SK-Hep-1 and RKO cells. However, the nature of the protein encoded is unknown.
The hormone binding domain of TRβ1 is the binding site for Ear-2.
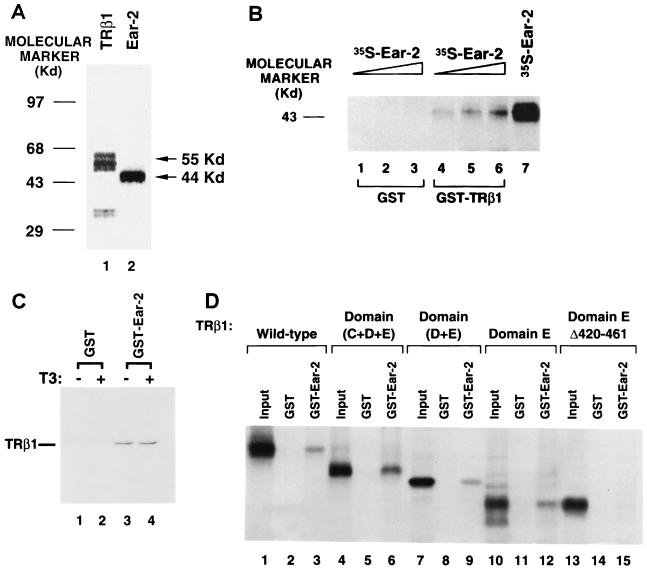
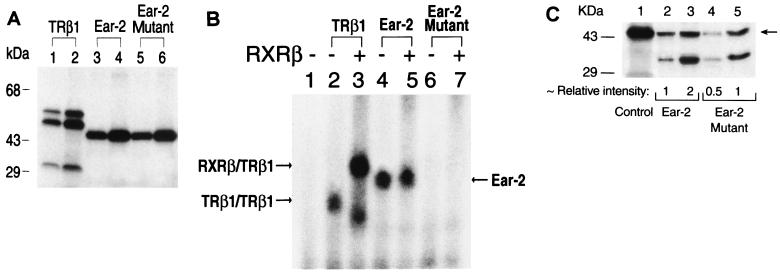
Ear-2 was shown to interact with intact TRβ1 by the yeast two-hybrid system. To identify the domain of TRβ1 to which Ear-2 bound, we used a GST pull-down assay. 35S-labeled Ear-2 was prepared by in vitro transcription-translation. Analysis by SDS-PAGE showed that it had an apparent molecular mass of 44 kDa (Fig. 3A, lane 2). For comparison, lane 1 of Fig. 3A shows the size of TRβ1 prepared by in vitro transcription-translation. Increasing concentrations of 35S-labeled Ear-2 were incubated with a constant amount of GST-TRβ1 or GST alone. As shown in Fig. 3B, 35S-labeled Ear-2 bound to GST-TRβ1 in a concentration-dependent manner (lanes 4, 5, and 6), whereas no 35S-labeled Ear-2 at the corresponding concentrations bound to the control (GST) (lanes 1, 2, and 3). The amounts of GST used in lanes 1, 2, and 3 corresponded to those used in lanes 4, 5, and 6, respectively. Lane 7 shows the input of 35S-labeled Ear-2 for comparison. These results provided additional evidence that Ear-2 physically interacted with TRβ1.
FIG. 3.
The C-terminal region of TRβ1 is critical for the interaction of TRβ1 with Ear-2. (A) Molecular size of Ear-2. Ear-2 (lane 2) and TRβ1 (lane 1) were prepared by in vitro transcription-translation in the presence of [35S]methionine, and 5 μl of the programmed lysates was analyzed by SDS-PAGE (10% gel). The dried gel was autoradiographed. (B) Binding of TRβ1 to intact Ear-2. The binding of 35S-labeled Ear-2 to GST-TRβ1 was carried out as described in Materials and Methods. E. coli-expressed GST (2 μg) or GST-TRβ1 (2 μg) noncovalently bound to GSH-Sepharose beads was incubated with increasing concentrations of in vitro-translated 35S-labeled Ear-2 (5, 10, and 20 μl [lanes 1 and 4, 2 and 5, and 3 and 6, respectively]). The bound proteins were analyzed by SDS-PAGE. Lane 7 shows the input from 10 μl of 35S-labeled Ear-2. (C) T3-independent binding of Ear-2 to TRβ1. GSH-Sepharose-bound GST–Ear-2 (10 μg) or GST (10 μg) was incubated with 35S-labeled TRβ1 in the presence or absence of T3 (100 nM) as described in Materials and Methods. After washing was done, bound 35S-labeled TRβ1 was detected as described above. Lanes 1 and 3, without T3; lanes 2 and 4, with T3; lanes 1 and 2 were from the binding of 35S-labeled TRβ1 to GST alone; lanes 3 and 4 were from the binding of 35S-labeled TRβ1 to GST–Ear-2. (D) Mapping of the region of TRβ1 binding to Ear-2. 35S-labeled wild-type TRβ1 (lanes 1 to 3), domains C, D, and E (lanes 4 to 6), domains D and E (lanes 7 to 9), domain E (lanes 10 to 12), and domain E with a deletion of amino acids 420 to 461 (EΔ420-461) were prepared from the corresponding T7 expression plasmids, pCJ3, pJL08, pJL05, pCJ4, and pCJ7, respectively (11). The 35S-labeled proteins were incubated with GST alone (lanes 2, 5, 8, 11, and 14) or GST–Ear-2 (3, 6, 9, 12, and 15) basically as described in panel A. One-tenth the inputs are shown as markers in lanes 1, 4, 7, 10, and 13.
To understand whether the interaction of Ear-2 with TRβ1 was T3 dependent, we carried out a GST pull-down assay in which Ear-2 was fused to GST (Fig. 3C). Consistent with the results shown in Fig. 3B, we found that 35S-labeled TRβ1 bound to GST–Ear-2 (Fig. 3C, lanes 3 and 4) but not to the control (GST) (lanes 1 and 2). The binding, however, was T3 independent, because no differences in the amounts of 35S-labeled TRβ1 bound were detected with T3 or without T3 (Fig. 3C, lane 3 versus lane 4).
TRβ1 consists of the amino-terminal A/B domain, the DNA binding domain (domain C), and the hormone binding domains (domains D and E). To identify the domain to which Ear-2 bound, we used truncated TRβ1 in which domain A/B, domains A/B and C, domains A/B, C, and D, and domains A/B, C, and D and amino acids 420 to 461 were deleted (16). We prepared various 35S-labeled truncated TRβ1 species and determined their binding to GST–Ear-2 (Fig. 3D). We found that domains C, D, and E (Fig. 3D, lane 6), domains D and E (lane 9), and domain E (lane 12) bound to GST–Ear-2 but not to the control (GST) (lanes 5, 8, and 11, respectively), indicating that domain E of TRβ1 was the binding site for Ear-2. Lanes 4, 7, and 10 of Fig. 3D show 1/10 the inputs of 35S-labeled domains C, D, and E, domains D and E, and domain E used in the binding experiments, respectively.
We next examined whether the binding site of TRβ1 for Ear-2 was localized at the C-terminal region of domain E. We therefore further truncated domain E of TRβ1 by deleting all of helix 12 and most of helix 11 (amino acids 420 to 461) (33). Lane 15 of Fig. 3D shows that upon deletion of this region, no binding to GST–Ear-2 was detected, suggesting the important role of amino acids 420 to 461 in the binding of Ear-2 to the hormone binding domain of TRβ1.
Development of anti–Ear-2 antibodies and detection of the interaction of Ear-2 with TRβ1 in cells by coimmunoprecipitation.
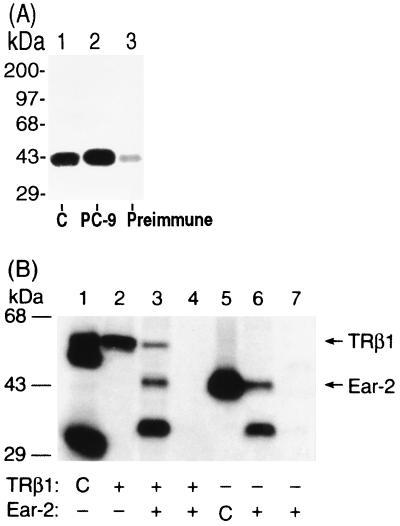
We prepared antibodies to human Ear-2 with the use of the DNA immunization technique, in which an antigen-specific immune response is elicited by injection of nonreplicative transcription units (30). We adopted the protocol described by Chowdhury et al. (5), using the cytomegalovirus promoter to drive the expression in mice of the human Ear-2 protein (pCDM-Ear-2), which is processed by the immune system to elicit the immune response (8). All five mice injected with pCDM-Ear-2 developed anti–Ear-2 antibodies after 6 to 8 weeks. A representative result from the screening of the mouse antisera is shown in Fig. 4A. Lane 1 shows 35S-labeled Ear-2 prepared by in vitro transcription-translation as a marker. Lane 2 shows that 35S-labeled Ear-2 was immunoprecipitated by anti–Ear-2 sera designated PC-9. Lane 3 shows that only a minor background signal was detected when the preimmune sera from the same mouse were used in the immunoprecipitation. These results demonstrate that the injection of mice with pCDM-Ear-2 led to the successful production of anti–human Ear-2 antibodies.
FIG. 4.
TRβ1 interacts with Ear-2 in cells. (A) Preparation of anti-Ear-2 antibody PC-9. PC-9 was prepared by injection of mice with expression plasmid pCDM-Ear-2 as described in Materials and Methods. Antisera (1 μl; lane 2) and preimmune sera (1 μl; lane 3) were screened by immunoprecipitation with in vitro-translated 35S-labeled Ear-2 (3 μl). The immunoprecipitates were analyzed by SDS–10% PAGE. The dried gel was autoradiographed. Lane 1 shows 35S-labeled Ear-2 as a marker (control [C]). (B) Coimmunoprecipitation of TRβ1 and Ear-2. CV1 cells (4 × 105/60-mm dish) were plated 1 day before transfection. The cells were transfected with 2 μg of pCLC51 and/or 2 μg of pCDM-Ear2. After being incubated overnight, the cells were metabolically labeled with [35S]Met-Cys (40 μCi) for 3 h at 37°C. Cellular extracts (500 μg) were immunoprecipitated with anti-TRβ1 antibodies J51 and J52 (2.5 μg each) (17) (lane 2) or PC-9 (5 μl of sera; lanes 3 and 6) or with antibody MOPC (5 μg; lanes 4 and 7). The immunoprecipitates were analyzed by SDS–10% PAGE. Lanes 1 and 5 show in vitro-translated 35S-labeled TRβ1 and Ear-2, respectively, as markers (control [C]). The dried gel was autoradiographed.
Using the yeast two-hybrid system and the GST pull-down assay, we have already shown that Ear-2 physically interacts with TRβ1 (see above). To demonstrate their physical interaction in cells, we expressed both proteins in CV1 cells by transfection with their expression plasmids. After metabolically labeling the cells with 35S-labeled methionine, we immunoprecipitated Ear-2 with PC-9. As shown in Fig. 4B, lane 3, three bands with molecular masses of 55, 44, and 35 kDa were immunoprecipitated with PC-9. The immunoprecipitation of these three proteins was specific because, as shown in Fig. 4B, lane 4, when an unrelated antibody (MOPC) was used, no immunoprecipitable bands were detected. The 55-kDa band was TRβ1, because when cells were transfected only with a TRβ1 expression plasmid and subsequently immunoprecipitated with an anti-TRβ1 MAb (C4) (36), 35S-labeled TRβ1 was detected (Fig. 4B, lane 2). Lane 1 of Fig. 4B shows the control, in which in vitro-translated 35S-labeled TRβ1 was directly loaded as a marker (17). The 44-kDa band observed in Fig. 4B, lane 3, represented Ear-2, because it had the same molecular mass as the control, in which in vitro-translated 35S-labeled Ear-2 was similarly immunoprecipitated with PC-9 (lane 5). However, the nature of the 35-kDa band was unclear. It could represent a degradation product of Ear-2. As shown in lane 6 of Fig. 4B, the same 44- and 35-kDa proteins were observed when the cells were transfected only with an Ear-2 expression plasmid and immunoprecipitated with PC-9. The two proteins immunoprecipitated in lane 6 of Fig. 4B were specific because, when an unrelated antibody (MOPC) was used (lane 7), no such proteins were immunoprecipitated. These results indicate that TRβ1 was coimmunoprecipitated with Ear-2 and thus provide additional evidence to demonstrate the interaction of these two receptor proteins in cells.
The binding of TRβ1 to TRE is inhibited by Ear-2.
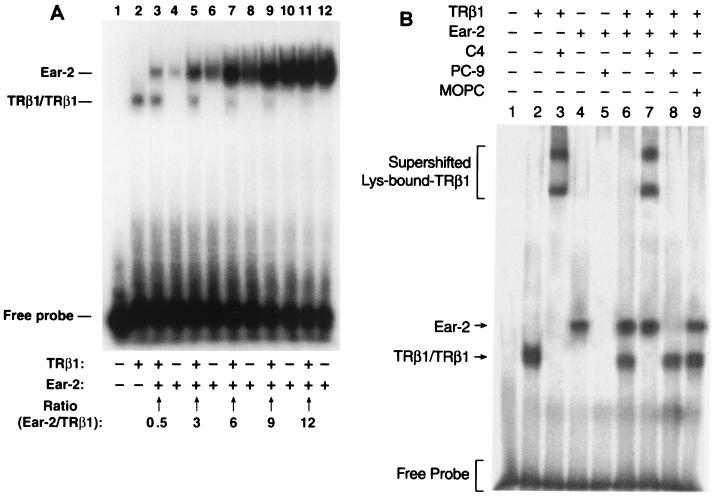
To understand the functional consequences of the physical interaction between TRβ1 and Ear-2, we first evaluated whether the binding of TRβ1 to TRE was affected by Ear-2. We carried out an EMSA with constant amounts of Lys-TRE and TRβ1 in the presence of increasing concentrations of Ear-2. Lane 1 of Fig. 5A shows the control, in which unprogrammed lysates were used. No signals were detected. Lane 2 of Fig. 5A shows the binding of TRβ1 to Lys-TRE in the absence of Ear-2. In the presence of Ear-2, however, the binding of TRβ1 to Lys-TRE was reduced (Fig. 5A, lane 3). The extent of reduction was Ear-2 dependent, because in the presence of increasing concentrations of Ear-2, the intensities of Lys-TRE-bound TRβ1 were gradually decreased (band intensity in Fig. 5A lanes: 3 > 5 > 7 > 9). Furthermore, the binding of TRβ1 to Lys-TRE was completely inhibited when the ratio of Ear-2 to TRβ1 reached 12 (Fig. 5A, lane 11).
FIG. 5.
The binding of TRβ1 or Ear-2 to TRE is modulated by TRβ1 or Ear-2. 35S-labeled Lys-TRE, TRβ1, and Ear-2 receptors were prepared as described in Materials and Methods. (A) A constant amount of 32P-labeled Lys-TRE was incubated with a constant amount of TRβ1 (2 μl of programmed lysates) in the presence of increasing concentrations of the Ear-2 protein. Identical amounts of Ear-2 were used in lane pairs 3 and 4, 5 and 6, 7 and 8, 9 and 10, and 11 and 12. The ratios of Ear-2 to TRβ1 are indicated. After electrophoresis, the gel was dried and autoradiographed. (B) Interaction of Lys-bound Ear-2 and TRβ1 with antibodies. A constant amount of 32P-labeled Lys-TRE was incubated with 2 μl of in vitro-translated TRβ1 (lanes 2 and 3), Ear-2 (1 μl; lanes 4 and 5), or 2 μl of TRβ1 and 1 μl of Ear-2 (lanes 6 to 9). Lys-bound TRβ1 was supershifted by MAb C4 (1 μg; lanes 3 and 7), and the binding of Ear-2 to Lys-TRE was blocked by PC-9 (1 μl; lanes 5 and 8). In lane 9, a control antibody (MOPC) (1 μg) was used. (C) The binding of TRβ1-RXRβ heterodimers to Lys-TRE is inhibited by the binding of Ear-2. A constant amount (105 cpm) of 32P-labeled Lys-TRE was incubated with 2 μl of in vitro-translated TRβ1 and/or 1 μl of Ear-2 in the absence or presence of 1 μg of MAb C4, MOPC, and/or RXRβ, as indicated. After gel electrophoresis, the dried gel was autoradiographed.
Interestingly, Ear-2 was found to bind to Lys-TRE (Fig. 5A, lane 4). However, in contrast to the effect of Ear-2 on the binding of TRβ1 to Lys-TRE, the binding of Ear-2 to Lys-TRE was enhanced by TRβ1. Equal amounts of Ear-2 were present in lane pairs 3-4, 5-6, 7-8, 9-10, and 11-12 of Fig. 5A. However, the TRE-bound Ear-2 band was stronger when TRβ1 was present (intensity of Ear-2 band in Fig. 5A lanes: 3 > 4, 5 > 6, 7 > 8, 9 > 10). Ear-2 was also found to bind to Pal-TRE and DR4-TRE, and the binding of TRβ1 to these TRE was inhibited by Ear-2 (data not shown). These results indicate that the physical interaction between Ear-2 and TRβ1 affected their binding to DNA. These data further indicate that Ear-2 and TRβ1 competed for binding to TRE.
That the slower-mobility bands shown in Fig. 5A were TRE-bound Ear-2 was further confirmed by supershift experiments (Fig. 5B). Lane 2 of Fig. 5B shows Lys-bound TRβ1, which was supershifted by anti-TRβ1 MAb C4 (lane 3). The binding of Ear-2 to Lys-TRE (Fig. 5B, lane 4), however, was blocked by anti–Ear-2 sera designated PC-9 (compare lane 5 with lane 4). Lane 6 of Fig. 5B shows that both Ear-2 and TRβ1 bound to Lys-TRE. In the presence of MAb C4, Lys-bound TRβ1 was supershifted, whereas Ear-2 was unaffected (Fig. 5B, compare lane 7 with lane 6). When PC-9 was present, consistent with the results shown in Fig. 5B, lane 5, the binding of Ear-2 was blocked, whereas no effect on the binding of TRβ1 was detected (compare lane 8 with lane 6). Lane 9 of Fig. 5B is a control used to indicate that the presence of an unrelated antibody (MOPC) did not affect the gel mobility of Lys-bound Ear-2 or TRβ1 (compare lane 9 with lane 6). These results clearly indicate that the slower-mobility bands shown in Fig. 5A were Lys-bound Ear-2.
Whether the binding of Ear-2 to TRβ1 affected the formation of TRβ1-retinoid X receptor (RXR) heterodimers is addressed in the experiments shown in Fig. 5C. Lanes 2 and 3 are the controls used to show that TRβ1 bound to Lys-TRE as a homodimer and as a heterodimer with RXR subtype β (RXRβ), respectively. Lanes 5 and 6 show Lys-bound Ear-2 in the absence and presence of RXRβ, respectively, demonstrating that Ear-2 did not form a heterodimer with RXRβ. Lane 7 shows that in the presence of both Ear-2 and TRβ1, consistent with the results shown in Fig. 5A, the binding of TRβ1 to Lys was reduced (cf. lane 2) and the binding of Ear-2 to Lys was intensified (cf. lanes 5 and 6). However, whether the binding of TRβ1 with RXRβ as a heterodimer was also inhibited by Ear-2 could not be discerned in lane 8, because TRβ1-RXRβ migrated in a position similar to that of Lys-bound Ear-2 (compare the Lys-bound Ear-2 migration position in lane 5 with the position of the TRβ1-RXRβ heterodimer in lane 3). We therefore supershifted the TRβ1-RXRβ heterodimer with MAb C4 to a more retarded position, as shown in lane 9. In lane 9, Lys-bound Ear-2 was not affected by MAb C4; however, the intensity of the supershifted TRβ1-RXRβ complex was clearly lower than that in lane 4, which was supershifted TRβ1-RXRβ complex in the absence of Ear-2. These results indicate that the binding of TRβ1 to Lys-TRE as a TRβ1-RXRβ heterodimer was inhibited by Ear-2. Lane 10 shows the negative control, in which a control antibody (MOPC) was used and no supershifted TRβ1-RXRβ complex was detected. Taken together, these results indicate that the binding of TRβ1 to Lys-TRE both as a homodimer and as a heterodimer with RXRβ was inhibited by its physical interaction with Ear-2. Similar results were observed when the TRE were DR4 and Pal (data not shown).
Hormone-dependent differential repression of TRβ1 transcriptional activity by Ear-2.
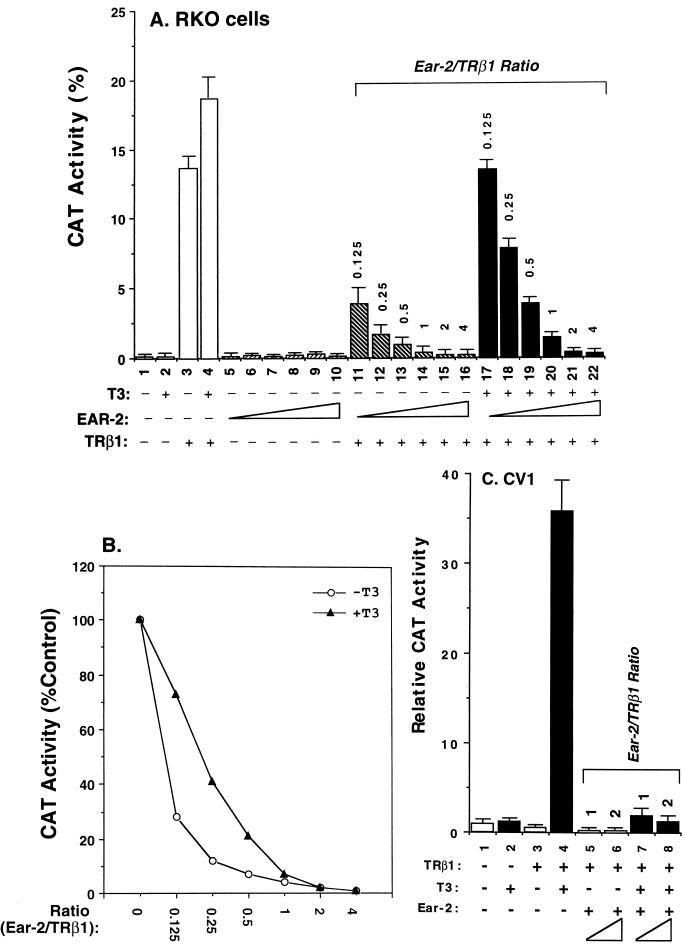
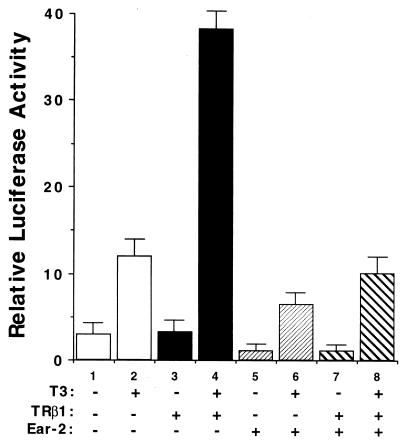
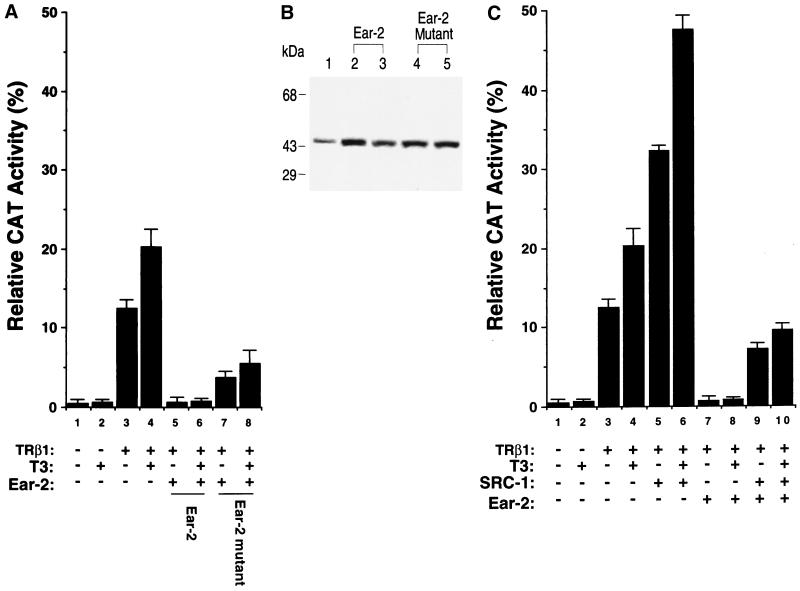
The above results prompted us to further evaluate the effect of Ear-2 on the transcriptional activity of TRβ1. Figure 6A shows that in RKO cells, both T3-independent (lanes 11 to 16 versus lane 3) and T3-dependent (lanes 17 to 22 versus lane 4) transcriptional activities were repressed in the presence of increasing concentrations of Ear-2. Lanes 5 to 10 of Fig. 6A were controls used to indicate that Ear-2 could not mediate transcription via TRE. Quantitation of the extent of the repression caused by Ear-2 shows that T3-independent activation was more sensitive to the repression effect of Ear-2 than T3-dependent activation at Ear-2/TRβ1 ratios of 0.125 to 0.5 (Fig. 6B). At higher ratios, i.e., 1 to 4, both transcriptional activities were completely repressed by Ear-2 (Fig. 6A and B).
FIG. 6.
The differential transcriptional repression by Ear-2 is cell type and T3 dependent. (A) RKO cells were transfected with the CAT reporter plasmid (pTK28m; 0.2 μg) and the TRβ1 expression plasmid (pCDM-TRβ1; 0.2 μg) in the absence (lanes 1 to 4) or the presence of increasing concentrations of pCDM-Ear-2 (lanes 5 to 10, 11 to 16, and 17 to 22). The ratios of Ear-2 to TRβ1 plasmids used in the transfection are shown (pCDM-TRβ1, 0.2 μg; pCDM-Ear-2, 0.025, 0.05, 0.1, 0.2, 0.4, and 0.8 μg for the groups in lanes 11 to 16 and 17 to 22, respectively). Lanes 5 to 10 were transfected with the same increasing concentrations of pCDM-Ear-2 but without pCDM-TRβ1. CAT activity was determined as described in Materials and Methods. (B) T3-dependent transcriptional repression by Ear-2 in RKO cells. The CAT activity in panel A was quantified and plotted. (C) CV1 cells were transfected with the CAT reporter plasmid (pTK28m; 0.2 μg) and the TRβ1 expression plasmid (pCDM-TRβ1; 0.2 μg) in the absence (lanes 1 to 4) or the presence of increasing concentrations of pCDM-Ear-2 (0.2 and 0.4 μg for the groups in lanes 5 and 6 and lanes 7 and 8, respectively). CAT activity was determined as described in Materials and Methods. Data are averages of three independent experiments, each with duplicates (mean ± standard deviation; n = 3).
To assess whether the basal repression effect of TRβ1 was also affected by Ear-2, we used CV1 cells. In these experiments, the repression of basal activity by unliganded TRβ1 was ∼70% (Fig. 6C, bar 3 versus bar 1). Figure 6C shows not only that T3-dependent activation was repressed by Ear-2 (lanes 7 and 8 versus lane 4) but also that the basal repression effect of TRβ1 was further affected (lanes 5 and 6 versus lane 3). These results indicate that Ear-2 was a potent repressor of TR-mediated transcriptional activity.
Ear-2 represses the promoter activity of the S14 T3-targeted gene.
To establish that Ear-2 plays a role in TR-mediated signaling pathways, we examined whether Ear-2 modulated the promoter activity of a T3-targeted gene. We chose to use the S14 gene because it is an important T3-targeted gene in the lipid metabolism pathway of liver and its TRE have been extensively characterized (27). Using a promoter-luciferase reporter system, we found that the promoter activity of S14 was also repressed by Ear-2 (Fig. 7). Bars 2 and 4 of Fig. 7 show the extent of T3-dependent activation mediated by the endogenous and transfected TR in a primary liver culture, respectively. These activities were repressed by Ear-2 (Fig. 7, bar 6 versus bar 2 and bar 8 versus bar 4). Consistent with the results obtained with CV1 cells, the extent of the basal repression effect of TR was also further enhanced by Ear-2 (Fig. 7, bar 5 versus bar 1 and bar 7 versus bar 3). These results indicate that Ear-2 could play an important role in TR-mediated signaling pathways.
FIG. 7.
Ear-2 suppresses basal and T3-induced responses of the human S14 promoter. The data are from a representative experiment that was repeated three times. Each bar is the mean + standard deviation of three plates with the indicated treatment. Bars 1, 3, 5, and 7 were hepatocytes maintained at 5.5 mM glucose and no T3; bars 2, 4, 6, and 8 were cells maintained at 27.5 mM glucose and 500 nM T3. As indicated below the bars, TRβ1 and Ear-2 were cotransfected as indicated in Materials and Methods. At each condition, the addition of T3 led to a significant (P, <0.05) increase in luciferase activity. At each condition, the cotransfection of Ear-2 led to a significant (P, <0.05) decrease in luciferase activity.
Mutation of the DNA binding domain of Ear-2 leads to a loss of TRE binding.
The above results indicated that Ear-2 repressed TR-mediated transcriptional activity. Since Ear-2 also bound to TRE (Fig. 5), the repression could be the result of the competition of Ear-2 with TRβ1 for binding to TRE. To understand if this was the case, we mutated the DNA binding domain (domain C) of Ear-2 from 73CysGluGlyCysLysSer80 (wild-type Ear-2) to 73CysGlySerCysLysVal80 (Ear-2 mutant). This mutant (1 and 2 μl of lysates were loaded in lanes 5 and 6 of Fig. 8A, respectively) had the same molecular mass as wild-type Ear-2 (1 and 2 μl of lysates were loaded in lanes 3 and 4 of Fig. 8A, respectively), as indicated by the proteins prepared with an in vitro transcription-translation system. This mutant, however, had lost the ability to bind to Lys-TRE in either the absence or the presence of RXRβ (lanes 6 and 7 versus lanes 4 and 5, respectively, of Fig. 8B). Lanes 2 and 3 of Fig. 8B were controls used to indicate that TRβ1 bound to Lys-TRE as a homodimer and as a heterodimer with RXRβ, respectively. Lanes 4 and 5 of Fig. 8B were also controls used to show that Ear-2 bound to Lys-TRE but did not form a heterodimer with RXRβ. The lack of binding of the mutant to Lys-TRE was not a result of smaller amounts of the Ear-2 mutant being used in the binding experiments. Equal amounts of Ear-2 and Ear-2 mutant proteins were used in the experiments.
FIG. 8.
Lack of binding of the Ear-2 mutant to TRE and its partial repression of TRβ1-mediated transcriptional activity. (A) The Ear-2 mutant has the same molecular size as wild-type Ear-2. 35S-labeled TRβ1 (lanes 1 and 2), Ear-2 (lanes 3 and 4), and mutant Ear-2 (lanes 5 and 6) proteins were prepared by in vitro transcription-translation in the presence of [35S]Met-Cys with a TNT kit. One (lanes 1, 3, and 5) or 2 (lanes 2, 4, and 6) μl from each programmed lysate was analyzed by SDS–10% PAGE. The dried gel was exposed overnight. (B) The Ear-2 mutant does not bind to TRE either as a homodimer or as a heterodimer with RXR. Two microliters of TRβ1 (lanes 2 and 3), 1 μl of Ear-2 (lanes 4 and 5), and 1 μl of Ear-2 mutant (lanes 6 and 7) prepared by in vitro transcription-translation were incubated with 32P-labeled Lys-TRE in the absence (lanes 2, 4, and 6) or the presence (lanes 3, 5, and 7) of 1 μg of RXRβ. (C) Relative protein expression of Ear-2 and the Ear-2 mutant. Cellular lysates (50 μg) of transfected CV1 cells (1 and 2 μg of pCDM-Ear-2 in lanes 2 and 3, respectively, or 1 and 2 μg of the pCDM-Ear-2 mutant in lanes 4 and 5, respectively) were analyzed by Western blot analysis with an anti–Ear-2 antibody (1:2,000 dilution of PC-9) as described in Materials and Methods. The arrow indicates Ear-2. (D) Partial repression of the T3-dependent transcriptional activity of TRβ1 by mutant Ear-2. CV1 cells were transfected with the CAT reporter plasmid (pTK28m; 0.2 μg) and the TRβ1 expression plasmid (pCDM-TRβ1; 0.2 μg) in the absence (lanes 1 to 4) or the presence of increasing concentrations of pCDM-Ear-2 (1 and 2 μg for lanes 5 and 6, respectively) or the pCDM-Ear-2 mutant (1 and 2 μg for lanes 7 and 8, respectively). The data were from three independent experiments, each with duplicates (mean ± standard deviation; n = 3). The ratios of Ear-2 and Ear-2 mutant proteins were determined from the results of Western blot analysis in panel C (see above).
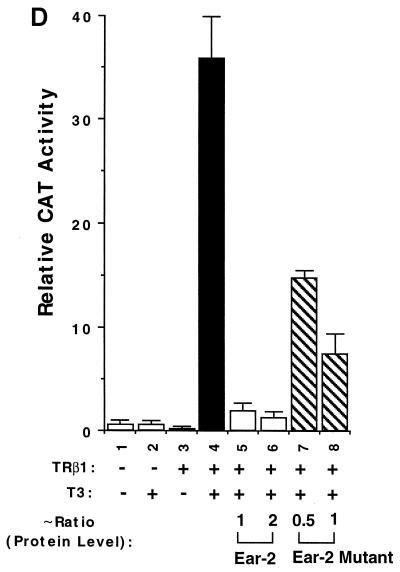
We further evaluated whether this Ear-2 mutant, which had lost DNA binding activity, could act as a repressor in CV1 cells. We transfected a constant amount of TRβ1 expression plasmid in the presence of increasing concentrations of wild-type Ear-2 or mutant Ear-2 plasmids. We concurrently determined both the CAT activities and the levels of expression of wild-type Ear-2 and mutant Ear-2 proteins by Western blotting. Figure 8C shows the levels of expression of wild-type Ear-2 and mutant Ear-2 proteins in cellular lysates, as determined by Western blotting with antibody PC-9. Lane 1 shows Ear-2 from the in vitro transcription-translation as a marker. Lanes 2 and 3 show the protein expression levels from the transfection of 1 and 2 μg of Ear-2 expression plasmid, respectively. Lanes 4 and 5 show the protein expression levels from the transfection of 1 and 2 μg of Ear-2 mutant expression plasmid, respectively. Quantitative comparison of the intensities of the bands indicated that relative ratios for the protein expression levels of the Ear-2 bands in lanes 2, 3, 4, and 5 were ∼1, 2, 0.5, and 1. Since the expression of both proteins was driven by the same CMV promoter, it is unclear why the expression of the Ear-2 mutant was lower than that of wild-type Ear-2 when the same amounts of plasmids were transfected (e.g., lane 2 versus lane 4 and lane 3 versus lane 5). The lower expression was not due to the lower reactivity of PC-9 with the Ear-2 mutant because, when in vitro-translated receptors were used, PC-9 reacted with wild-type Ear-2 and mutant Ear-2 with similar avidities (data not shown).
Figure 8D compares the repression effect of wild-type Ear-2 and mutant Ear-2 on TRβ1-mediated transcriptional activity. Bars 5 and 6 of Fig. 8D show that when 1 and 2 μg of wild-type Ear-2 expression plasmid were cotransfected with a constant concentration of TRβ1 expression plasmid, 92 and 95% of T3-dependent transcriptional activities were repressed, respectively. When 1 and 2 μg of mutant Ear-2 expression plasmid were cotransfected with TRβ1 (Fig. 8D, bars 7 and 8), ∼57 and ∼80% of the transcriptional activities of TRβ1 were repressed, respectively. The repression effect of wild-type Ear-2 in bar 5 of Fig. 8D and that of mutant Ear-2 in bar 8 were mediated by similar levels of receptor proteins (see lanes 2 and 5 of Fig. 8C). Therefore, the extents of repression mediated by wild-type Ear-2 and mutant Ear-2 could be compared. Because mutant Ear-2 had lost the ability to bind to TRE, the repression seen for mutant Ear-2 was unlikely to be mediated by competition with TRβ1 for binding to TRE. These results suggest that the repression of the transcriptional activity of TRβ1 by wild-type Ear-2 was a combination of a protein-protein interaction and competition for binding to TRE.
The repression of the T3-dependent transcriptional activity of TRβ1 by Ear-2 was reversed by SRC-1 in CV1 cells.
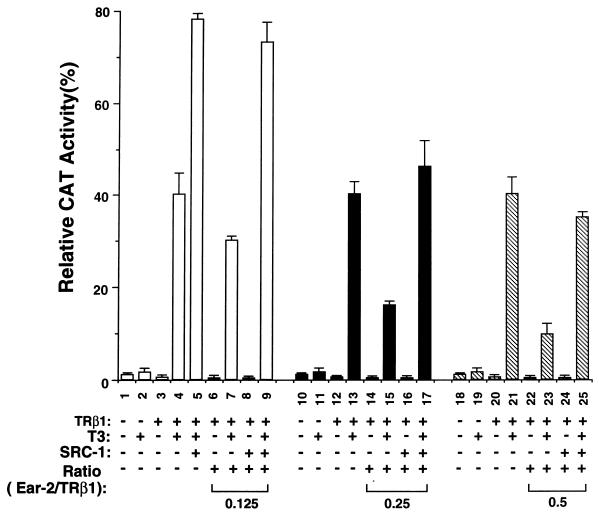
SRC-1 is a coactivator for many members of the receptor superfamily, including TR (9, 26). It interacts with the activation function 2 (AF2) region of the receptors via its LXXLL motifs (9, 19, 24). We transfected SRC-1 into CV1 cells to determine if SRC-1 could lead to a reversal of the repression caused by Ear-2 (Fig. 9). Bar 5 of Fig. 9 was a control used to indicate that SRC-1 enhanced T3-dependent TRβ1-mediated transcriptional activity. Under the experimental conditions, the enhancement was about twofold (compare bars 5 and 4 of Fig. 9). Compared to bar 4, bars 7, 15, and 23 of Fig. 9 show the Ear-2 concentration-dependent repression of TRβ1-mediated transcription. When a small amount of Ear-2 expression plasmid (Ear-2/TRβ1 ratio, 0.125) was transfected, SRC-1 not only derepressed the repression effect of Ear-2 but also enhanced TRβ1-mediated transcriptional activity (Fig. 9, bar 9 versus bars 7 and 5). When more Ear-2 expression plasmid (Ear-2/TRβ1 ratio, 0.25 or 0.5) was transfected into cells, SRC-1 was sufficient to compete with Ear-2 for TRβ1 to reverse the repression (Fig. 9, bar 17 versus bar 15 and bar 25 versus bar 23) but not sufficient for additional activation (bars 17 and 25 versus bar 5). In addition, when an Ear-2/TRβ1 ratio of >2 was used, SRC-1 was not able to reverse the repression effect of Ear-2 (data not shown).
FIG. 9.
Reversal of the Ear-2 repression of the T3-dependent transcriptional activity of TRβ1 by SRC-1 in CV1 cells. CV1 cells were cotransfected with the CAT reporter plasmid (pTK28m; 0.2 μg), the TRβ1 expression plasmid (pCDM-TRβ1; 0.2 μg), the β-galactosidase expression vector (pCH110; 0.2 μg), and the SRC-1 expression vector (pCR3.1 hSRC-1A; 1 μg) in the absence (lanes 1 to 4, 9 to 12, and 17 to 20) or the presence of increasing concentrations of pCDM-Ear-2 (lanes 6 to 9, 0.025 μg; lanes 14 to 17, 0.05 μg; lanes 22 to 25, 0.1 μg) as described in Materials and Methods. The total plasmids transfected were normalized to 3 μg by using the vector control plasmid, pCR3.1. The final ratios of Ear-2 to TRβ1 are indicated. The data were from three independent experiments, each with duplicates (mean ± standard deviation; n = 3).
The repression of the T3-independent transcriptional activity of TRβ1 by Ear-2 was reversed by SRC-1 in RKO cells.
In RKO cells, both T3-dependent and T3-independent transcriptional activities were repressed by Ear-2 (Fig. 6A and B). We first examined whether the Ear-2 mutant could repress both transcriptional activities in these cells. Figure 10A shows that, as in CV1 cells, the Ear-2 mutant was able to repress both T3-independent (bar 7 versus bar 3) and T3-dependent (bar 8 versus bar 4) transcriptional activities. Figure 10B shows that transfection of equal amounts of expression plasmids for wild-type Ear-2 and mutant Ear-2 in RKO cells led to the expression of the same levels of Ear-2 proteins (duplicates in lanes 2 and 3 versus lanes 4 and 5 of Fig. 10B). Unlike the results obtained for CV1 cells, no 35-kDa protein band was detected in RKO cells (Fig. 8C). The reasons for such differences are not clear. Thus, the extents of repression shown in bars 5 and 6 and in bars 7 and 8 of Fig. 10C were mediated by the same levels of wild-type Ear-2 and mutant Ear-2, respectively. The finding that the Ear-2 mutant retained the ability to repress TRβ1-mediated transcriptional activity suggested that competition for DNA binding was not the only mechanism for the repression effect of Ear-2 in RKO cells.
FIG. 10.
Repression of the T3-independent and T3-dependent transcriptional activities of TRβ1 by wild-type Ear-2 and mutant Ear-2 and its reversal by SRC-1 in RKO cells. (A) Repression by wild-type Ear-2 and mutant Ear-2. RKO cells (8 × 105 cells/60 mm-dish) were transfected with the CAT reporter plasmid (pTK28m; 0.5 μg), the β-galactosidase expression vector (pCH110; 0.2 μg), and pCDM-TRβ1 (0.2 μg) in the absence of Ear-2 (lanes 1 to 4), the presence of Ear-2 (pCDM-Ear-2, 0.4 μg for lanes 5 and 6), or the presence of mutant Ear-2 (pCDM-Ear-2 mutant, 0.4 μg for lanes 7 and 8). (B) Relative expression of Ear-2 and Ear-2 mutant proteins. Cellular lysates of transfected RKO cells (50 μg) were analyzed by Western blot analysis with PC-9 (1:2,000 dilution) as described in Materials and Methods. Lanes 2 and 3 and lanes 4 and 5 are duplicates. (C) Reversal of the repression effect of Ear-2 by SRC-1. RKO cells were transfected as described in Materials and Methods, except in the absence of SRC-1 (bars 1 to 4, 7, and 8) or the presence of SRC-1 (bars 5, 6, 9, and 10; pCR3.1 hSRC-1A; 1 μg). The total DNA concentration in all dishes was kept at 3 μg by supplementation with the vector control plasmid, pCR3.1. CAT activity in panels A and C was determined as described in Materials and Methods. The data in panels A and C were from three independent experiments, each with duplicates (mean ± standard deviation; n = 3).
We also ascertained if the repression effect of Ear-2 in RKO cells could be reversed by SRC-1. SRC-1 enhanced both T3-independent and T3-dependent transcriptional activities 2.6- and 2.3-fold, respectively (compare bars 5 and 3 and bars 6 and 4 in Fig. 10C). In the presence of Ear-2 (plasmid Ear-2/TRβ1 ratio, 2), both T3-independent and T3-dependent transcriptional activities were completely repressed (Fig. 10C, bar 7 versus bar 3 and bar 8 versus bar 4). In the presence of SRC-1, ∼60 and ∼50% of T3-independent (bar 9 versus bar 3) and T3-dependent (bar 10 versus bar 4) transcriptional activities were restored, respectively. These results indicate that SRC-1 was able to reverse the repression mediated by Ear-2 in RKO cells.
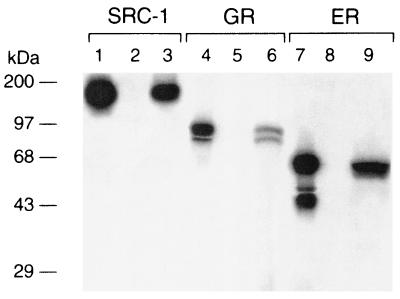
The reversal of the Ear-2-mediated repression of the transcriptional activity of TRβ1 by SRC-1 could be due to competition of TRβ1 with SRC-1 for binding to Ear-2. To test this possibility, we carried out a GST pull-down assay. As shown in Fig. 11, lane 3, 35S-labeled SRC-1 bound to GST–Ear-2, whereas no 35S-labeled SRC-1 was detected when GST alone was used (lane 2). Lane 1 of Fig. 11 shows the input of SRC-1 as a marker. These results indicated that Ear-2 physically interacted with SRC-1. This interaction raises the possibility that the reversal of the Ear-2 repression effect by SRC-1 could be due to the recruitment of Ear-2 by SRC-1, leading to the reduction of TRβ1–Ear-2 complexes.
FIG. 11.
Ear-2 binds to SRC-1, GR, and ER. E. coli-expressed GST (4 μg; lanes 2, 5, and 8) or GST–Ear-2 (4 μg; lanes 3, 6, and 9) noncovalently bound to GSH-Sepharose beads was incubated with 10 μl of in vitro-translated 35S-labeled SRC-1 (lanes 2 and 3), GR (lanes 5 and 6), or ER (lanes 8 and 9) for 2 h at 4°C. After washing was done, the bound proteins were analyzed by SDS–10% PAGE. The gel was dried and autoradiographed. Lanes 1, 4, and 7 show 10% inputs of 35S-labeled SRC-1, GR, and ER, respectively.
Ear-2 is also a negative regulator for the transcriptional activities of GR and ER.
To address the question of whether Ear-2 also functions as a negative coregulator for other members of the steroid hormone receptors, we first evaluated whether Ear-2 physically interacted with GR or ER in a GST pull-down assay. Lanes 6 and 9 of Fig. 11 show that GR and ER, respectively, bound to GST–Ear-2. Lanes 5 and 8 of Fig. 11 were the corresponding negative controls used to indicate that GR and ER, respectively, did not bind to GST alone. Lanes 4 and 7 of Fig. 11 show the 10% inputs of GR and ER, respectively. These results indicated that Ear-2 also physically interacted with other members of the steroid hormone receptors.
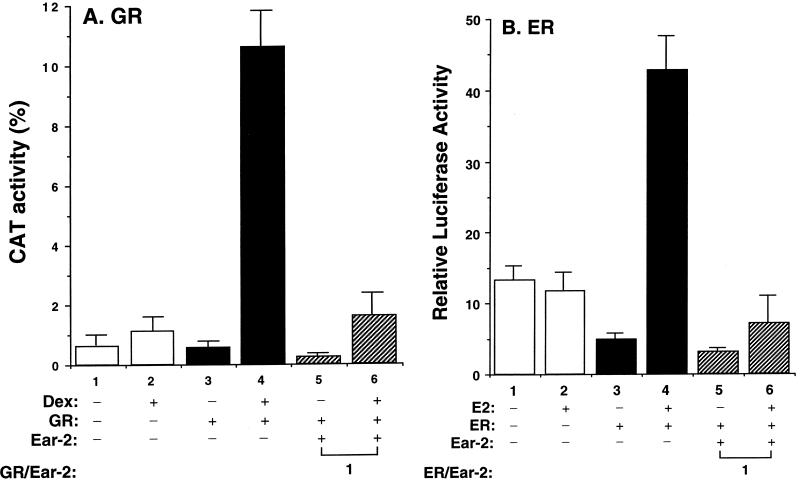
We further evaluated whether Ear-2 functions as a negative regulator in GR- or ER-mediated transactivation activity (Fig. 12). Like that of TRβ1, the Dex-dependent transactivation activity mediated by GR was repressed by Ear-2 (Fig. 12A, lane 6 versus lane 4). At a plasmid Ear-2/GR ratio of 1, the extent of repression of the Dex-dependent transactivation activity was 85%. Lanes 1 and 2 of Fig. 12A were the controls used to show the basal transcriptional activities in the absence and presence of Dex, respectively. Ear-2 also was a negative regulator for the transcriptional activity mediated by ER. A comparison of bars 6 and 4 of Fig. 12B indicates that the estrogen (E2)-dependent transactivation of ER was repressed 85% by Ear-2. Therefore, Ear-2 is a negative regulator not only for the transactivation activity of TR but also for that of other members of the steroid hormone receptors.
FIG. 12.
Ear-2 represses the hormone-dependent transcriptional activity of GR and ER. (A) CV1 cells (4 × 105 to 8 × 105/60-mm dish) were cotransfected with the CAT reporter plasmid (pMSG-CAT; 0.2 μg), pCMV-GR (0.2 μg), and pCH110 (0.2 μg) in the absence (bars 1 to 4) or the presence (bars 5 and 6) of pCDM-Ear-2. The total plasmids transfected were normalized to 3 μg. Twenty-four hours before cells were harvested, Dex (100 nM) was added to the dishes (bars 2, 4, and 6). The success of transfection was monitored by β-galactosidase activity. The CAT activity was normalized by using equal amounts of lysate proteins. The final GR/Ear-2 ratio is shown. The data were from three independent experiments, each with duplicates (mean ± standard deviation; n = 3). (B) CV1 cells were cotransfected with the luciferase reporter plasmid (pTK-ERE-Luc; 0.2 μg), the ER expression plasmid (HEGO; 0.2 μg), and pCH110 (0.2 μg) in the absence (bars 1 to 4) or the presence (bars 5 and 6) of pCMV-Ear-2. The total plasmids transfected were normalized to 3 μg. E2 (100 nM) was added to the dishes (bars 2, 4, and 6) 24 h before cells were harvested. The luciferase activity was normalized by using equal amounts of lysate proteins. The final ER/Ear-2 ratio is shown. The data were from three independent experiments, each with duplicates (mean ± standard deviation; n = 3).
DISCUSSION
Using intact TRβ1, we identified Ear-2 as one of its interacting proteins by a yeast two-hybrid system. We further demonstrated their physical interaction by using a GST pull-down assay and showed that in cells, TRβ1 was associated with Ear-2. Functional characterization indicated that Ear-2 was a negative regulator for TRβ1-mediated transcriptional activity. However, the sensitivity and type of repression depended on the cell type. In CV1 cells, Ear-2 repressed not only T3-dependent activation but also TRβ1-mediated basal repression. In RKO cells, both T3-independent activation and T3-dependent activation were repressed. However, T3-independent activation was more sensitive to the repression effect of Ear-2 than T3-dependent activation. These observations suggest that cellular context plays an important role not only in TRβ1-mediated transcription but also in the repression effect of Ear-2.
To understand how Ear-2 repressed the T3-dependent activation of TRβ1 in CV1 cells, we considered the following possibilities: (i) formation of inactive TRβ1–Ear-2 heterodimers on DNA; (ii) competition of TRβ1 and Ear-2 for DNA binding sites; and (iii) interference with TRβ1 for binding to limited amounts of coactivators. These possibilities, however, may not be mutually exclusive. Similar to the results observed for Ear-3–COUP-TF (12, 32), we found that Ear-2 bound to TRE with the half-site binding motifs arranged in three different orientations. However, the formation of TRβ1–Ear-2 heterodimers was not detected. These results are similar to those of Butler and Parker, who reported that the formation of COUP-TF homodimers and RXRβ-TRβ heterodimers is favored over that of COUP-TF–TRβ1 heterodimers (3). Therefore, the formation of inactive TRβ1–Ear-2 heterodimers on DNA is not a likely mechanism to account for the repression of TRβ1 transcriptional activity by Ear-2. We also found that Ear-2 cannot form heterodimers with RXR. Thus, the competition of Ear-2 with TRβ1 for RXR may not be a viable mechanism to account for the repression effect of Ear-2 on the transcriptional activity of TR. We did find, however, that the binding of either TRβ1 or Ear-2 to TRE was affected by the other protein through the physical interaction of the two (Fig. 5). The binding of Ear-2 to TRβ1 inhibited the binding of TRβ1 to TRE not only as a homodimer but also as a heterodimer. The binding of Ear-2 to TRE was intensified by its physical association with TRβ1, but the contrary was detected for TRβ1. Thus, the physical interaction of TRβ1 with Ear-2 enabled the latter to become a more efficient TRE binder favorably competing with TRβ1 for binding to TRE. Furthermore, the finding that competition for binding to DNA is one of the mechanisms to account for the repression of TR-mediated transcription by Ear-2 has been shown for the estrogen-stimulated transcriptional activity of the human oxytocin gene by Ear-2 (6, 7). Ear-2 binds to a site that overlaps the estrogen response element on the promoter region of the oxytocin gene (6, 7).
Using a GST pull-down assay, we found that Ear-2 also interacted with GR and ER. We further showed that Ear-2 was a negative regulator for the transcriptional activities mediated by both GR and ER. Therefore, Ear-2 is a negative regulator of transcriptional activity not only for TR but also for other members of the steroid hormone receptors. Interestingly, we also found that Ear-2 physically interacted with SRC-1. This observation raises the possibility that SRC-1 could recruit Ear-2 and compete with TRβ1 for binding to Ear-2, thereby releasing Ear-2 from the Ear-2–TRβ1 complex. This could be one of the mechanisms to account for the reversal of the Ear-2-mediated repression of the transcriptional activity of TRβ1.
We found that an Ear-2 mutant which had totally lost TRE binding activity still retained ∼60% of repression activity. This finding suggested that competition for binding to TRE is not the only mechanism for the repression effect of Ear-2 on TRβ1-mediated transcription in CV1 cells. Additional mechanisms to account for the repression effect exist. Using a series of truncated TRβ1 molecules, we found that when the region of amino acids 420 to 461 of TRβ1 was deleted, no binding to Ear-2 could be detected, suggesting the involvement of this region in the binding of TRβ1 to Ear-2.
Based on these observations, we considered another possible mechanism to account for the reversal of the Ear-2-mediated repression effect by SRC-1. The C-terminal region of TRβ1 contains the interaction domain for binding to SRC-1 (9, 19, 24). Therefore, it is reasonable to postulate that the binding of Ear-2 to this region may directly or indirectly prevent the interaction of TRβ1 with SRC-1. A model has been proposed for the interaction of SRC-1 with TRα1 (9) in which one of the several α-helical LXXLL motifs of SRC-1 is bound in a hydrophobic cleft on the surface of the receptor ligand binding domain. The cleft is formed by residues from helices 3, 5, and 12 of the ligand binding domain. Computer analysis with the GCG software package (University of Wisconsin, Madison) failed to reveal any sequence homology between SRC-1 and Ear-2. The latter protein also lacks the LXXLL motif. However, four regions of Ear-2 which are predicted to be helical in structure contain HXXHH motifs, where H is L, F, or I. These side chains could be accommodated by the hydrophobic cleft (9), providing binding sites which could compete with SRC-1 for binding to TR. Alternatively, Ear-2 may bind to another site on the C-terminal region of TRβ1 and interfere with the binding of SRC-1 by a steric mechanism. Therefore, the repression of T3-dependent TRβ1 transcriptional activity may be due at least in part to the combination of competition for binding to TRE and interference with the binding of TRβ1 to SRC-1. However, at present, we cannot rule out other possibilities.
In RKO cells, however, the molecular mechanisms underlying the repression of Ear-2 are clearly more complex. The proposed two mechanisms for the repression of TRβ1-mediated transcriptional activity could reasonably account for the observed inhibition of T3-dependent activation by Ear-2. At present, because very little is known about the molecular events leading to T3-independent activation, it is unclear how Ear-2 repressed this activity. In the beginning, we had hoped to identify specific coregulators which mediated T3-independent transcriptional activity in RKO cells. However, none of the positive clones obtained fulfilled this role. While we still do not understand how TRβ1 mediates T3-independent activation, we found that the physical interaction of Ear-2 with TRβ1 was T3 independent. It is reasonable to propose that competition for binding to TRE could be one of the possible mechanisms. However, the elucidation of other possible mechanisms to account for the repression of TR-mediated transcriptional activity by Ear-2 in RKO cells awaits future studies.
Ear-2 is a distant member of the COUP-TF group, which belongs to a distinct subfamily within the steroid-TR superfamily (32). While much progress has been made in understanding the tissue distribution, biochemical characterization, and in vivo functions of other COUP-TF members, very limited information is available for Ear-2. The ligand for Ear-2 has not been identified, nor has its physiological role. The present study, however, has clearly identified one function of Ear-2, which is to negatively regulate TR actions. This negative regulation has been demonstrated not only by using TRE reporter genes but also by using the promoter of a T3-targeted gene, S14. Our results suggest that the extent of negative regulation most likely depends on the levels of expression of Ear-2, TR, and coactivators. Thus, a very complex picture of this regulatory network emerges. Our findings in the present study have further strengthened the hypothesis that the diverse functions of TR are mediated by a large network of coregulatory proteins.
ACKNOWLEDGMENTS
We thank G. Brent and M. J. Tsai for providing pTK28m and pCR-SRC-1a, respectively. We also thank Wilfred Vieira for assistance with the production of anti-Ear-2 antibodies.
Qihong Zhu and Cary N. Mariash were supported by NIH grant P30 DK50456.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CPB co-activator is a histone acetyltransferase. Nature. 1996;38:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Bhat M K, Ashizawa K, Cheng S-Y. Phosphorylation enhances the target gene sequence-dependent dimerization of thyroid hormone receptor with the retinoid X receptor. Proc Natl Acad Sci USA. 1994;91:7927–7931. doi: 10.1073/pnas.91.17.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler A J, Parker M G. COUP-TF II homodimers are formed in preference to heterodimers with RXR alpha or TR beta in intact cells. Nucleic Acids Res. 1995;23:4143–4150. doi: 10.1093/nar/23.20.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng S-Y. New insights into the structure and function of the thyroid hormone receptor. J Biomed Sci. 1995;2:77–89. doi: 10.1007/BF02253060. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury P S, Viner J L, Beers R, Pastan I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proc Natl Acad Sci USA. 1998;95:669–674. doi: 10.1073/pnas.95.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu K, Boutin J M, Breton C, Zingg H H. Nuclear orphan receptors COUP-TFII and Ear-2: presence in oxytocin-producing uterine cells and functional interaction with the oxytocin gene promoter. Mol Cell Endocrinol. 1998;137:145–154. doi: 10.1016/s0303-7207(97)00241-4. [DOI] [PubMed] [Google Scholar]
- 7.Chu K, Zingg H H. The nuclear orphan receptors COUP-TFII and Ear-2 act as silencers of the human oxytocin gene promoter. Mol Endocrinol. 1997;19:163–172. doi: 10.1677/jme.0.0190163. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly J J, Ulmer J B, Liu M A. Immunization with DNA. J Immunol Methods. 1994;176:145–152. doi: 10.1016/0022-1759(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 9.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 10.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 12.Kadowaki Y, Toyoshima K, Yamamoto T. Ear3/COUP-TF binds most tightly to a response element with tandem repeats separated by one nucleotide. Biochem Biophys Res Commun. 1992;183:492–498. doi: 10.1016/0006-291x(92)90509-j. [DOI] [PubMed] [Google Scholar]
- 13.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CPB integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 14.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M J, O'Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 15.Lin K H, Ashizawa K, Cheng S-Y. Phosphorylation stimulates the transcriptional activity of the human β1 thyroid hormone nuclear receptor. Proc Natl Acad Sci USA. 1992;89:7737–7741. doi: 10.1073/pnas.89.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin K H, Parkison C, McPhie P, Cheng S-Y. An essential role of domain D in the hormone-binding activity of human β1 thyroid hormone nuclear receptor. Mol Endocrinol. 1991;5:485–492. doi: 10.1210/mend-5-4-485. [DOI] [PubMed] [Google Scholar]
- 17.Lin K-H, Willingham M C, Liang C-M, Cheng S-Y. Intracellular distribution of the endogenous and transfected β form of thyroid hormone nuclear receptor visualized by the use of domain-specific monoclonal antibodies. Endocrinology. 1991;128:2601–2609. doi: 10.1210/endo-128-5-2601. [DOI] [PubMed] [Google Scholar]
- 18.Lin K H, Zhu X-G, Shieh H Y, Hsu H C, Chen S T, McPhie P, Cheng S Y. Identification of naturally occurring dominant negative mutants of thyroid hormone α1 and β1 receptors in a human hepatocellular carcinoma cell line. Endocrinology. 1996;137:4073–4081. doi: 10.1210/endo.137.10.8828459. [DOI] [PubMed] [Google Scholar]
- 19.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 21.Meier C A, Parkison C, Chen A, Ashizawa K, Muchmore P, Meier-Heusler S C, Cheng S-Y, Weintraub B D. Interaction of human β1 thyroid hormone receptor and its mutants with DNA and RXR. T3 response element-dependent dominant negative potency. J Clin Investig. 1993;92:1986–1993. doi: 10.1172/JCI116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misiti S, Schomburg L, Yen P M, Chin W W. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology. 1998;139:2493–2500. doi: 10.1210/endo.139.5.5971. [DOI] [PubMed] [Google Scholar]
- 23.Miyajima N, Kadowaki Y, Fukushige S, Shimizu S, Semba K, Yamanashi Y, Matsubara K, Toyoshima K, Yamamoto T. Identification of two novel members of erbA superfamily by molecular cloning: the gene products of the two are highly related to each other. Nucleic Acids Res. 1988;16:11057–11074. doi: 10.1093/nar/16.23.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 25.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CPB are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 26.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 27.Ota Y, Mariash A, Wagner J L, Mariash C N. Cloning, expression and regulation of the human S14 gene. Mol Cell Endocrinol. 1997;126:75–81. doi: 10.1016/s0303-7207(96)03971-8. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro R C, Apriletti J W, Wagner R L, West B L, Feng W, Huber R, Kushner P J, Nilsson S, Scanlan T, Fletterick R J, Schaufele F, Baxter J D. Mechanisms of thyroid hormone action: insights from X-ray crystallographic and functional studies. Recent Prog Horm Res. 1998;53:351–392. [PubMed] [Google Scholar]
- 29.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 30.Tang D C, DeVit M, Johnston S A. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 31.Ting Y T, Wong R, Cheng S-Y. Tissue-specific stabilization of the thyroid hormone β1 nuclear receptor by phosphorylation. J Biol Chem. 1997;272:4129–4134. doi: 10.1074/jbc.272.7.4129. [DOI] [PubMed] [Google Scholar]
- 32.Tsai S Y, Tsai M J. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocrinol Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- 33.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O'Malley B W. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 35.Yap N, Yu C-L, Cheng S-Y. Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc Natl Acad Sci USA. 1996;93:4273–4277. doi: 10.1073/pnas.93.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X-G, McPhie P, Lin K-H, Cheng S-Y. The differential hormone-dependent transcriptional activity of thyroid hormone receptor isoforms is mediated by interplay of their domains. J Biol Chem. 1997;272:9048–9054. doi: 10.1074/jbc.272.14.9048. [DOI] [PubMed] [Google Scholar]