Abstract
Background
Clinically, evidence shows that uterine corpus endometrial carcinoma (UCEC) patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) may have a higher death-rate. However, current anti-UCEC/coronavirus disease 2019 (COVID-19) treatment is lacking. Plumbagin (PLB), a pharmacologically active alkaloid, is an emerging anti-cancer inhibitor. Accordingly, the current report was designed to identify and characterize the anti-UCEC function and mechanism of PLB in the treatment of patients infected with SARS-CoV-2 via integrated in silico analysis.
Methods
The clinical analyses of UCEC and COVID-19 in patients were conducted using online-accessible tools. Meanwhile, in silico methods including network pharmacology and biological molecular docking aimed to screen and characterize the anti-UCEC/COVID-19 functions, bio targets, and mechanisms of the action of PLB.
Results
The bioinformatics data uncovered the clinical characteristics of UCEC patients infected with SARS-CoV-2, including specific genes, health risk, survival rate, and prognostic index. Network pharmacology findings disclosed that PLB-exerted anti-UCEC/COVID-19 effects were achieved through anti-proliferation, inducing cytotoxicity and apoptosis, anti-inflammation, immunomodulation, and modulation of some of the key molecular pathways associated with anti-inflammatory and immunomodulating actions. Following molecular docking analysis, in silico investigation helped identify the anti-UCEC/COVID-19 pharmacological bio targets of PLB, including mitogen-activated protein kinase 3 (MAPK3), tumor necrosis factor (TNF), and urokinase-type plasminogen activator (PLAU).
Conclusions
Based on the present bioinformatic and in silico findings, the clinical characterization of UCEC/COVID-19 patients was revealed. The candidate, core bio targets, and molecular pathways of PLB action in the potential treatment of UCEC/COVID-19 were identified accordingly.
Keywords: cancer, clinical, pharmacologic (drug) therapy, target, mechanism and characterization
Highlights
Clinical characteristics of UCEC/COVID-19 patients, including specific genes, health risk, survival rate, and prognosis, were determined.
All candidates, core targets of PLB in treatment for UCEC/COVID-19, were screened out and identified.
Biological functions and molecular pathways of PLB in treatment for UCEC/COVID-19 were revealed.
Biological docking findings elucidated that PLB mediated effective molecular affinity with COVID-19.
Introduction
Coronavirus disease, caused by SARS-CoV-2, is continuously evolving and spreading around the world in 2020, especially in India (1). In western countries, including the United States, increasing reports indicate that the disease incidence, death rate, and infection rate of COVID-19 are elevating sharply owing to limited management (2). While the vaccine against coronavirus is still developing, there is still no clinical medicine to treat COVID-19 because of the current evolving situation (3). In addition to adjuvant therapy, some common drugs, such as hydrocortisone, may alternatively be prescribed to patients infected with SARS-CoV-2; however, the clinical effectiveness against COVID-19 remains limited (4). Taken together, mankind needs to screen and develop some pharmacologically bioactive compounds for the treatment of COVID-19, an emerging and evolving epidemic. In clinical practice, it has been reported that hospitalized patients with cancer seem to be highly susceptible to infection with coronavirus, resulting in an additional death toll (5). Uterine corpus endometrial carcinoma (UCEC) is a gynecological cancer characterized by biological invasiveness and potent metastasis (6). Epidemiologically, it is estimated that the incidence and mortality of UCEC are increasing in recent decades, especially in less-resourced nations, and it is still a major public health problem worldwide (7). In China, other reporting data show that the death rate of young women with UCEC is increasing yearly, especially in urban areas (8). Accordingly, cancer patients, who may be hospitalized during clinical treatment, may have a higher risk of exposure to SARS-CoV-2 infection during the early stages of COVID-19 as the novel coronavirus is transmitting rapidly, and it is not detectable during an outbreak (9). Theoretically, it could be reasoned that some of the UCEC patients that were hospitalized appear to have a high risk of infection with SARS-CoV-2. Thus, the treatment of UCEC/COVID-19 patients will become more challenging as there are no current medicines to treat UCEC and COVID-19, and the virus is still evolving in the world. Accordingly, anti-UCEC/COVID-19 medicine is urgent to be screened and developed in the current situation.
Plumbagin (PLB), a bioactive naphthoquinone, exerts potent pharmacological properties against some of the chronic diseases, such as obesity and fatty liver (10). In particular, PLB functions as an anti-cancer compound inducing cytotoxicity and suppressing cancer cells (11). The preliminary anti-tumor mechanism exerted by PLB would be through targeting the Wnt/β-catenin and AKT signaling pathways (12, 13). Interestingly, our previous findings suggest that PLB plays a potent anti-cancer effect against pancreatic cancer and hepatocellular carcinoma (14, 15). Furthermore, preclinical data indicate that PLB exerts antiproliferative activity against cervical cancer cells (16, 17). However, it has been reported that PLB has effective anti-inflammatory effects via the suppression of nuclear factor-κB activity (18). Moreover, it is also reported that PLB can possess antibacterial effects, including action against Staphylococcus aureus and Bacillus subtilis (19, 20). However, the potential relevance of PLB in UCEC has not been assessed. Moreover, the anti-UCEC/COVID-19 functions and mechanisms achieved by PLB remain uninvestigated. The evolving methodology of in silico analysis, which includes network pharmacology and molecular docking analysis, can be used for unmasking the functions and mechanism of phytocompounds to treat a medical disease before clinical investigation (21, 22). The present study was designed to determine the clinical characteristics of UCEC/COVID-19, to find an anti-UCEC/COVID-19 bio target, and to determine the mechanism exerted by PLB through network pharmacology and biological molecular docking analysis.
Materials and Methods
Selection of UCEC/COVID-19-Functional Genes
In order to select and define UCEC/COVID-19-functional genes, we used the Cancer Genome Atlas (TCGA) portal (https://portal.gdc.cancer.gov/) to download the data of Gene Expression Quantification (GEQ) in UCEC’s transcriptome profiling, accessed on September 14, 2020. Using Bioconductor’s “limma” package in R-language to screen GEQ, the criteria were set as false discovery rate (FDR) and |logfold change| > 1 for obtaining the differential genes of UCEC. In addition, we used different gene-functional modules from the Genecard database, Online Mendelian Inheritance in Man (OMIM) database, Therapeutic Target Database (TTD), and National Center for Biotechnology Information (NCBI). Next, the differential genes of UCEC and COVID-19 targets were subjected to a map through the online bioinformatic Venn diagram tool, in order to identify all the UCEC/COVID-19 shared targets (22, 23).
Clinical Information Determination of UCEC/COVID-19-Functional Genes
After processing the harvested clinical data downloaded from the TCGA database, the UCEC/COVID-19-functional genes were obtained similarly. The survival prognosis for a UCEC/COVID-19 case was assessed using the “survival” package in R-language. The prognostic determination was conducted through univariate Cox proportional hazards regression analysis. Meanwhile, the clinical characterization of UCEC/COVID-19 in patients was determined with the multivariate Cox proportional hazards regression model. Following the average risk score, patients were grouped into low and high-risk populations (24, 25).
Analysis of Candidate and Shared Genes in UCSC/COVID-19
We identified and harvested the functional genes of PLB through different online platforms: Swiss Target Prediction, Batman, Genecards, and SuperPred webserver. After gene correction, the candidate genes of PLB and UCSC/COVID-19 were further mapped using the online Venn diagram to harvest all intersection targets (26, 27).
Gene Ontology and Signaling Pathway Enrichment Analyses of Intersection Targets
We used R-language package settings, including “ClusterProfiler,” “ReactomePA,” “org.Hs.eg.Db,” and “GOplot,” to conduct the gene analysis with all intersection targets, as reported elsewhere (28). Next, to analyze the biological processes, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was determined and visualized. Gene annotation information harvested from “org.Hs.eg.Db”, p-value Cutoff = 0.05, q-value Cutoff = 0.05 during enrichment, contributing to output the corresponding histogram.
Screening Core Targets of Plumbagin in the Treatment of COVID-19/UCEC
All interaction targets of PLB and COVID-19/UCEC were loaded into the STRING tool (version 11.0) for functional protein association networks. Then the network interaction relationship between the target and target function-related proteins was obtained. Accordingly, target interaction in the PPI network diagram and the tsv data were produced and collected. We applied the NetworkAnalyzer in Cytoscape_v3.7.1 to analyze the topological parameters, such as the median and maximum degrees of freedom in the network. All core targets were screened according to degree value, and the upper limit of the filtering range was the maximum degree value in the topology data, whereas the lower limit was the median degree of freedom (29).
Construction of Network-Connected Visualization
Further, we used Cytoscape_v3.7.1 to construct a drug-target gene ontology biological process pathway disease visualization graph based on the results of PLB intervention in COVID-19/UCEC’s GO-based biological process and pathway enrichment, as described previously (30).
Molecular Docking Assessment
Further, we used molecular docking determination to predict and identify the binding capacity and interaction force between associated proteins, including MAPK3, TNF, PLAU, and the PLB molecule. Accordingly, the compound structures of PLB and Darunavir were obtained from PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and all investigated proteins for docking were gained from Protein Data Bank database (https://www.rcsb.org/). The PLB compound structure was optimized the minimum energy 2 (MM2) through 3D Draw module in the Chem Bio Office software. Ligand-associated PDBQT structure file format was necessary file for virtual screening. The PDBQT file of PLB compound structure was generated by using Raccoon software. And the investigated proteins/targets were processed through MGLTools (1.5.6 version), a supporting tool of Autodock Vina software, followed with hydrogenation, Gasteiger charge calculation, non-polar hydrogen combination. The original pdb file format was converted to recognized format by the Autodock Vina setting, providing ligand basis for chemically and structurally docking. Docking active center, including surrounding residues centered on the original ligand, was set by using Grid box function setting. The rationality of docking parameter settings in PLB and proteins/targets were determined according to the size of root mean square deviation (RMSD) between the docked and original ligand molecules. It was generally reasoned that RMSD ≤ 4 Å was the threshold for the conformation of ligand to match the original ligand docked. More details of the procedures were reported previously (31, 32). Additionally, two-dimensional diagrams of PLB and Darunavir with known 3D structure were created according to chemical drawing conventions in PoseView tool. And generation of structure diagrams and layout modifications were plotted using the library 2Ddraw tool.
Results
Identifying Candidate Genes of COVID-19 and UCEC
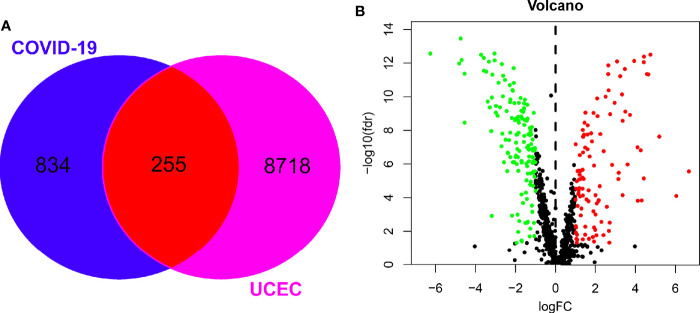
Using the Genecard and OMIM online tools, we identified a group of 1089 genes of COVID-19. A set of 8973 associated UCEC genes were determined using the TCGA database. Following the online Venn diagram assay, we collected a total of 255 mutual genes of COVID-19 and UCEC ( Figure 1A ). Finally, the volcano map of the mutual genes was obtained by R-language analysis, in which 108 genes were up-regulated and 147 others were down-regulated ( Figure 1B ).
Figure 1.
Screening of mutual genes between UCEC and COVID-19. (A), candidate, mutual genes of UCEC and COVID-19 in a Venn diagram. (B), differentially expressed genes from mutual genes shown in a volcano-plot map.
Clinical and Medical Characteristics in COVID-19/UCEC
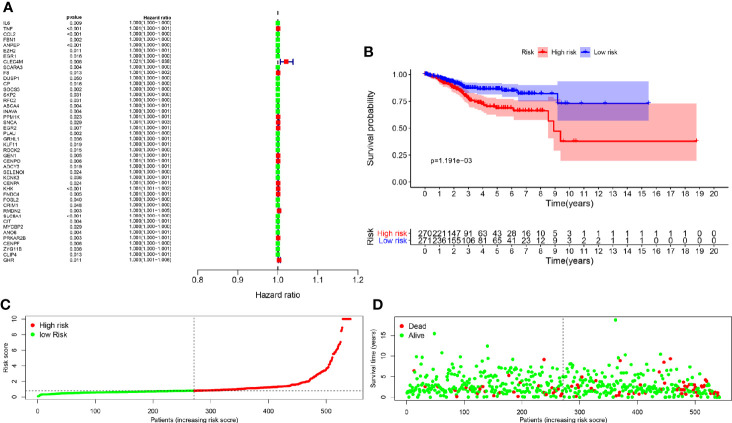
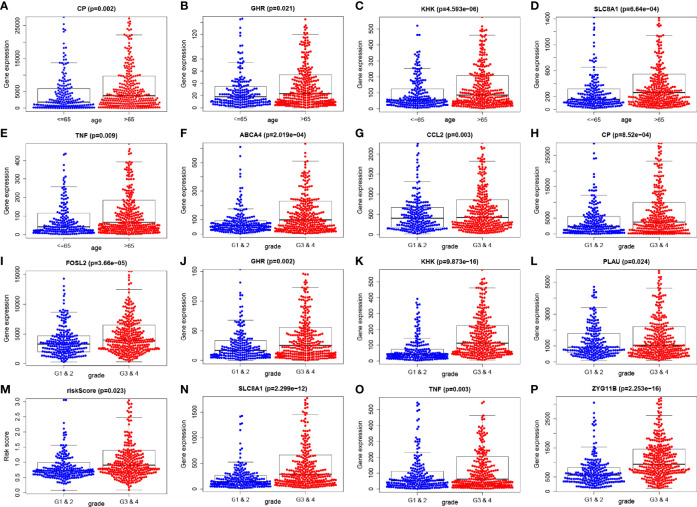
There were 44 genes with P < 0.05 in the univariate Cox analysis among the 255 differentially expressed genes, as detailed in Figure 2A and Table 1 . Further, we screened out the other 13 genes from the 44 specific genes through multivariate Cox analysis performed with hazard regression analysis, including CCL2, ANPEP, CLEC4M, SCARA3, CP, ABCA4, KHK, SLC8A1, ZYG11B, GHR, TNF, FOSL2, and PLAU ( Table 2 ). Based on the logistic regression coefficient (coef) value for patient risk evaluation, the risk value was the sum of the expression of each gene multiplied by the coef value, thereby dividing patients into high-risk and low-risk groups. As shown in the survival analysis, we found that the high- and low-risk groups related to 13 genes had a statistical significance on overall survival ( Figure 2B ); the greater the patient’s risk value, the higher the patient’s risk score ( Figure 2C ). Likewise, a higher mortality and lower overall survival were observed ( Figure 2D ). We also carried out single factor and multivariate independent prognostic analyses of the 13 associated genes. As a result, age factor had a significant difference in the single factor analysis P < 0.05, while grade and risk value had significant differences in both single factor and multivariate independent prognostic analyses. Similarly, the hazard ratio was greater than 1. The result showed the higher the risk value, the greater the prognostic risk, and it can be used as an independent prognostic analysis factor for COVID-19/UCEC ( Table 3 ). Clinical correlation analysis of these 13 genes was further performed, and the results showed that each gene had no correlation with clinical single factors, as detailed in Table 4 and Figure 3 .
Figure 2.
Clinical features of UCEC/COVID-19-related genes. (A), Univariate Cox determination was conducted to identify 44 candidate genes. (B), Multivariate Cox analysis identified 13 specific genes, including CCL2, ANPEP, CLEC4M, SCARA3, CP, ABCA4, KHK, SLC8A1, ZYG11B, GHR, TNF, FOSL2, and PLAU. The survival analysis suggested statistical significance associated with 13 specific genes in the overall survival between high- and low-risk groups. (C, D), Further analysis showed the greater the patient’s risk value, the higher the patient’s risk score; similarly, higher mortality and lower survival.
Table 1.
Univariate cox proportional hazards regression analysis of COVID-19_UCEC gene.
| Genes | HR | HR.95L | HR.95H | p-value |
|---|---|---|---|---|
| IL6 | 1.000207554 | 1.000052316 | 1.000362816 | 0.008778943 |
| TNF | 1.000753076 | 1.000353788 | 1.001152523 | 0.000217956 |
| CCL2 | 1.000273964 | 1.000143442 | 1.000404503 | 3.88E-05 |
| FBN1 | 1.000213328 | 1.000078541 | 1.000348132 | 0.001921055 |
| ANPEP | 1.000020478 | 1.000009404 | 1.000031554 | 0.000289922 |
| EZH2 | 1.000291755 | 1.000066441 | 1.00051712 | 0.011148452 |
| EGR1 | 1.000024347 | 1.000004519 | 1.000044175 | 0.016100539 |
| CLEC4M | 1.021467315 | 1.005536209 | 1.037650824 | 0.008088622 |
| SCARA3 | 1.000071903 | 1.000022389 | 1.00012142 | 0.004423996 |
| F8 | 1.001312422 | 1.000281938 | 1.002343967 | 0.012540539 |
| DUSP1 | 1.000042068 | 1.000000058 | 1.00008408 | 0.049682011 |
| CP | 1.00000685 | 1.000001293 | 1.000012407 | 0.015698793 |
| SOCS3 | 1.000066755 | 1.000023813 | 1.000109699 | 0.002312255 |
| SKP2 | 1.000236732 | 1.000021935 | 1.000451576 | 0.030762207 |
| RFC2 | 1.000238896 | 1.000022052 | 1.000455787 | 0.03082664 |
| ABCA4 | 1.000393186 | 1.000123976 | 1.000662468 | 0.004199901 |
| INAVA | 1.000351216 | 1.000111279 | 1.00059121 | 0.004116141 |
| PPM1K | 1.00050373 | 1.000069526 | 1.000938122 | 0.022973265 |
| SNCA | 1.001338861 | 1.000133987 | 1.002545187 | 0.029401172 |
| EGR2 | 1.000738441 | 1.000204013 | 1.001273155 | 0.00676005 |
| PLAU | 1.000078444 | 1.000029525 | 1.000127365 | 0.001672441 |
| GRHL1 | 1.000423737 | 1.000027483 | 1.000820148 | 0.036088497 |
| KLF11 | 1.000478342 | 1.000078569 | 1.000878274 | 0.019013985 |
| ROCK2 | 1.00024856 | 1.000047953 | 1.000449207 | 0.015159967 |
| GEN1 | 1.000503887 | 1.000155033 | 1.000852863 | 0.004637324 |
| CENPO | 1.000660487 | 1.000186113 | 1.001135086 | 0.006349271 |
| ADCY3 | 1.000405922 | 1.000066913 | 1.000745045 | 0.018930544 |
| SELENOI | 1.000198502 | 1.000026436 | 1.000370597 | 0.023751878 |
| KCNK3 | 1.000291018 | 1.000019436 | 1.000562674 | 0.0357067 |
| CENPA | 1.000547601 | 1.000071496 | 1.001023932 | 0.024172924 |
| KHK | 1.001353007 | 1.000581099 | 1.002125511 | 0.000589413 |
| FNDC4 | 1.000803206 | 1.00024437 | 1.001362354 | 0.004841882 |
| FOSL2 | 1.000052958 | 1.000002379 | 1.000103539 | 0.040155414 |
| CRIM1 | 1.000193521 | 1.00000154 | 1.000385539 | 0.048189646 |
| RMDN2 | 1.003267988 | 1.001117153 | 1.005423445 | 0.002885881 |
| SLC8A1 | 1.000303821 | 1.000142034 | 1.000465635 | 0.000232414 |
| CIT | 1.000309325 | 1.000099374 | 1.00051932 | 0.003879488 |
| MYCBP2 | 1.000211944 | 1.000021642 | 1.000402281 | 0.029044014 |
| ANO6 | 1.000306132 | 1.000100554 | 1.000511752 | 0.003513997 |
| PRKAR2B | 1.00051216 | 1.000170903 | 1.000853534 | 0.00326338 |
| CENPF | 1.000063814 | 1.000018577 | 1.000109054 | 0.005694564 |
| ZYG11B | 1.000328864 | 1.00001874 | 1.000639084 | 0.037670898 |
| CLIP4 | 1.000466365 | 1.000099814 | 1.000833051 | 0.012638528 |
| GHR | 1.00338401 | 1.000762567 | 1.006012319 | 0.011371407 |
Table 2.
Multivariate ox proportional hazards regression analysis.
| Genes | coef | HR | HR.95L | HR.95H | p-value |
|---|---|---|---|---|---|
| CCL2 | 0.000307056 | 1.000307103 | 1.000155931 | 1.000458298 | 6.84E-05 |
| ANPEP | 2.78E-05 | 1.000027785 | 1.000015648 | 1.000039923 | 7.22E-06 |
| CLEC4M | 0.01810549 | 1.018270388 | 1.000379462 | 1.036481279 | 0.045294101 |
| SCARA3 | 6.89E-05 | 1.000068931 | 1.000013251 | 1.000124614 | 0.015249326 |
| CP | 1.10E-05 | 1.000011016 | 1.000001055 | 1.000020976 | 0.030194505 |
| ABCA4 | 0.000426783 | 1.000426874 | 1.000059006 | 1.000794878 | 0.02294149 |
| KHK | 0.001078914 | 1.001079496 | 1.00003703 | 1.002123048 | 0.042394865 |
| SLC8A1 | 0.000344485 | 1.000344545 | 1.000107186 | 1.000581959 | 0.004438439 |
| ZYG11B | -0.000579096 | 0.999421071 | 0.998842689 | 0.999999788 | 0.049916327 |
| GHR | 0.003522729 | 1.003528941 | 1.000308451 | 1.0067598 | 0.031712525 |
| TNF | 0.000456603 | 1.000456708 | 0.999959582 | 1.000954081 | 0.071769993 |
| FOSL2 | -7.33E-05 | 0.999926709 | 0.999851297 | 1.000002126 | 0.056816556 |
| PLAU | 6.46E-05 | 1.000064581 | 0.999997251 | 1.000131916 | 0.060117016 |
Table 3.
Univariate and multivariate analysis of the correlation of 13 gene expression values with OS among the patients.
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| age | 1.0323 | 1.0100-1.055 | 4.32E-03 | 1.0166 | 0.9946-1.0391 | 1.41E-01 |
| Grade (G1-G4) | 2.5350 | 1.7539-3.6639 | 7.45E-07 | 2.5078 | 1.7553-3.5830 | 4.40E-07 |
| riskScore | 1.1632 | 1.1275-1.2000 | 1.98E-21 | 1.1743 | 1.1318-1.2184 | 1.23E-17 |
Table 4.
Clinical correlation analysis.
| Genes | Age (≤65 vs >65) | Grade (G1 & 2 vs G3 & 4) |
|---|---|---|
| CCL2 | 0.681 (0.496) | -2.983 (0.003) |
| ANPEP | -1.442 (0.150) | 0.992 (0.322) |
| CLEC4M | -1.444 (0.149) | -0.509 (0.611) |
| SCARA3 | 0.474 (0.636) | -0.799 (0.425) |
| CP | -3.088 (0.002) | -3.358 (8.52e-04) |
| ABCA4 | 0.437 (0.662) | -3.757 (2.019e-04) |
| KHK | -4.633 (4.593e-06) | -8.329 (9.873e-16) |
| SLC8A1 | -3.424 (6.64e-04) | -7.242 (2.299e-12) |
| ZYG11B | -1.934 (0.054) | -8.476 (2.253e-16) |
| GHR | -2.31 (0.021) | -3.085 (0.002) |
| TNF | -2.605 (0.009) | -2.976 (0.003) |
| FOSL2 | -0.29 (0.772) | -4.163 (3.66e-05) |
| PLAU | 0.169 (0.866) | -2.261 (0.024) |
| riskScore | -0.617 (0.537) | -2.277 (0.023) |
Figure 3.
Clinical correlation analysis of 13 genes was carried out and assessed; the results showed that each gene has no correlation with clinical single factors (A–P).
Identification of Candidate and Mutual Targets of PLB and COVID-19/UCEC
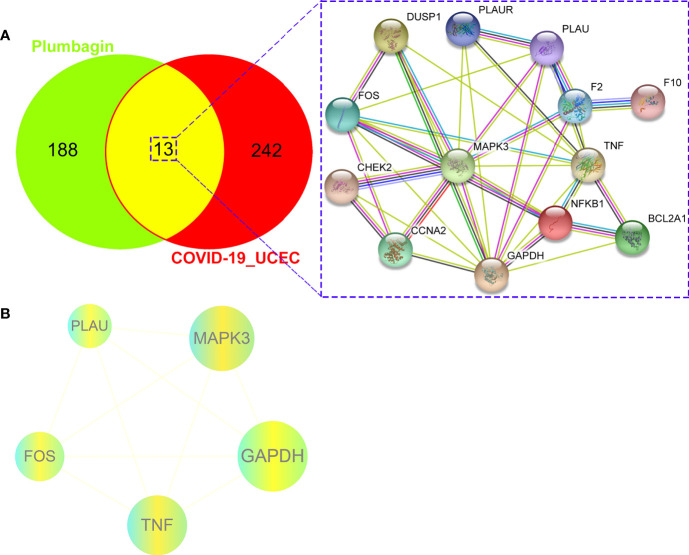
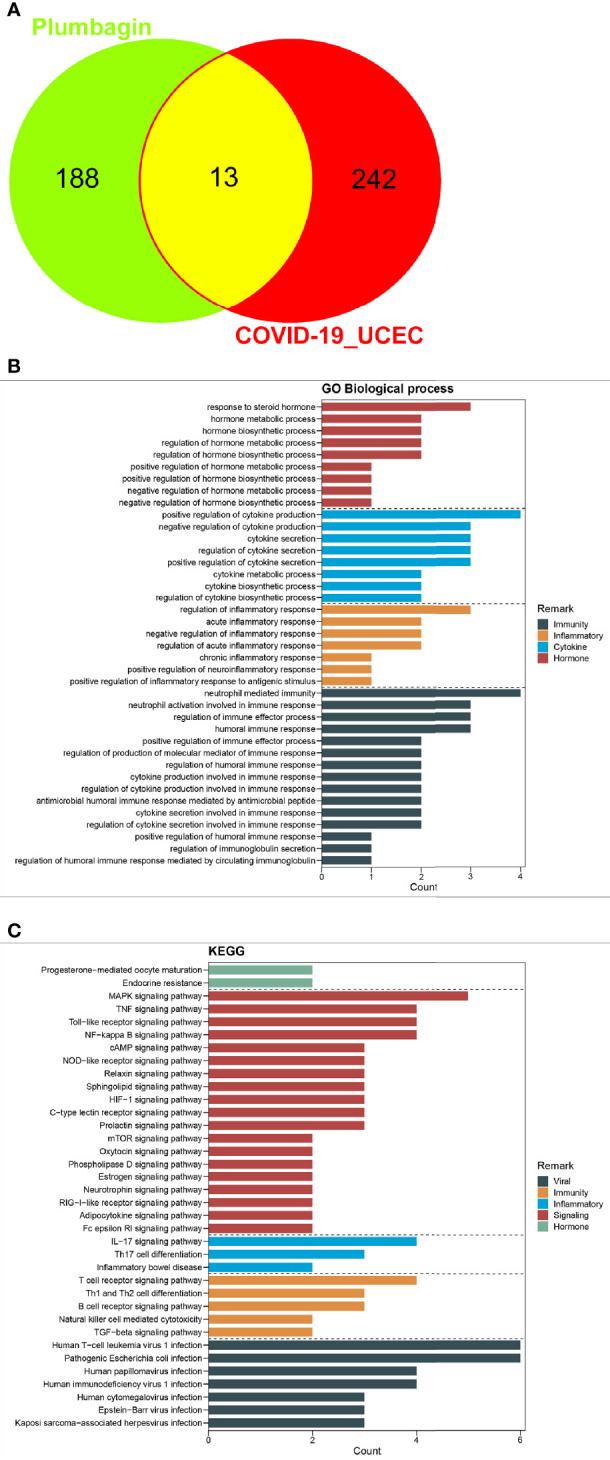
Following with the Swiss Target Prediction, TCMSP databases for screening drug targets, a series of 201 PLB-pharmacological genes were identified after correcting by the Uniprot database and removing duplicates. As a result, the 13 mutual genes of PLB and COVID-19/UCEC were screened out through an online bioinformatics platform ( Figure 4A ; more details shown in Supplemental Table 1 ).
Figure 4.
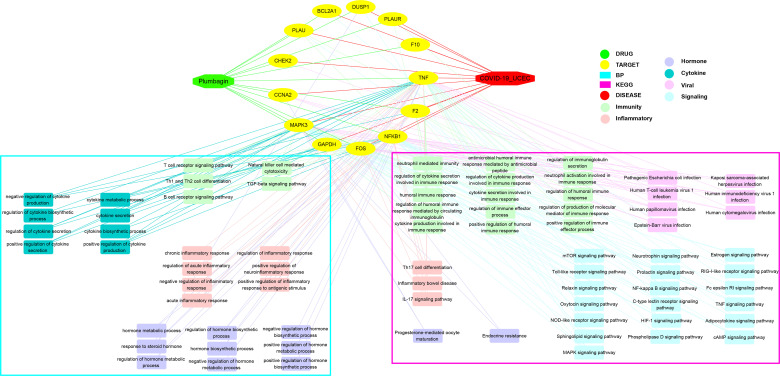
Preliminary findings of network pharmacology. (A), The candidate, mutual genes of PLB and UCEC/COVID-19 in a Venn diagram. (B, C), GO-based biologic process and KEGG-based signaling pathway of PLB against UCEC/COVID-19 following enrichment analysis.
Enrichment Outcomes of Gene Ontology and Molecular Pathway
All mutual genes were further used for GO and KEGG enrichment assays. The findings indicated the detailed GO-assayed biological processes ( Figure 4B ) and KEGG signaling pathways ( Figure 4C ) of PLB in the treatment of COVID-19/UCEC. The biological processes were mainly the regulation of cytokine secretion involved in immune response, cytokine secretion involved in immune response, neutrophil-mediated immunity, antimicrobial humoral immune response mediated by antimicrobial peptide, regulation of cytokine production involved in immune response, humoral immune response, cytokine production involved in immune response, regulation of immune effector process, regulation of humoral immune response, regulation of production of molecular mediator of immune response, neutrophil activation involved in immune response, regulation of humoral immune response mediated by circulating immunoglobulin, positive regulation of immune effector process, regulation of immunoglobulin secretion, positive regulation of humoral immune response, regulation of inflammatory response, regulation of acute inflammatory response, negative regulation of inflammatory response, positive regulation of inflammatory response to antigenic stimulus, and acute inflammatory response ( Supplemental Table 2 ). As highlighted in the pathway enrichment determination, a total of 81 KEGG signaling pathways were identified accordingly via P-adjust <0.05. The computational data principally included pathogenic Escherichia coli infection, Human T-cell leukemia virus 1 infection, Human immunodeficiency virus 1 infection, Human papillomavirus infection, Kaposi sarcoma-associated herpesvirus infection, Epstein-Barr virus infection, Human cytomegalovirus infection, Endocrine resistance, Progesterone-mediated oocyte maturation, T cell receptor signaling pathway, B cell receptor signaling pathway, Th1 and Th2 cell differentiation, TGF-beta signaling pathway, Natural killer cell-mediated cytotoxicity, IL-17 signaling pathway, Th17 cell differentiation, Inflammatory bowel disease, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, and TNF signaling pathway ( Supplemental Table 3 ).
Protein-Protein Interaction Network, Core Targets of PLB in the Treatment of COVID-19/UCEC
We identified a PPI network of PLB in the treatment of COVID-19/UCEC from 13 mutual targets through the STRING database, as shown in Figure 5A . Further, all mutual genes were submitted to the Cytoscape 3.7.1 software to determine the topological parameters of the PPI network related to the core targets of PLB in the treatment of COVID-19/UCEC. As a result, we screened and identified 5 final core targets, namely GAPDH, MAPK3, TNF, FOS, and PLAU ( Figure 5B ).
Figure 5.
Bioinformatics characteristics of PLB against UCEC/COVID-19. (A), a gene-assayed network connection of PLB against UCEC/COVID-19. (B), 5 identified predictive core bio targets of PLB in the potential treatment of UCEC/COVID-19.
Integrative Network Connection
Collectively, we used the Cytoscape 3.7.1 software to integrate the bioinformatic findings. As a result, a network connection of PLB-target-BP-KEGG-COVID-19/UCEC was optimized ( Figure 6 ).
Figure 6.
Interaction network connection of candidates, core bio targets, BP, functions, and molecular pathways of PLB in the treatment of UCEC/COVID-19 by using Cytoscape analysis.
Biological Molecular Docking Findings
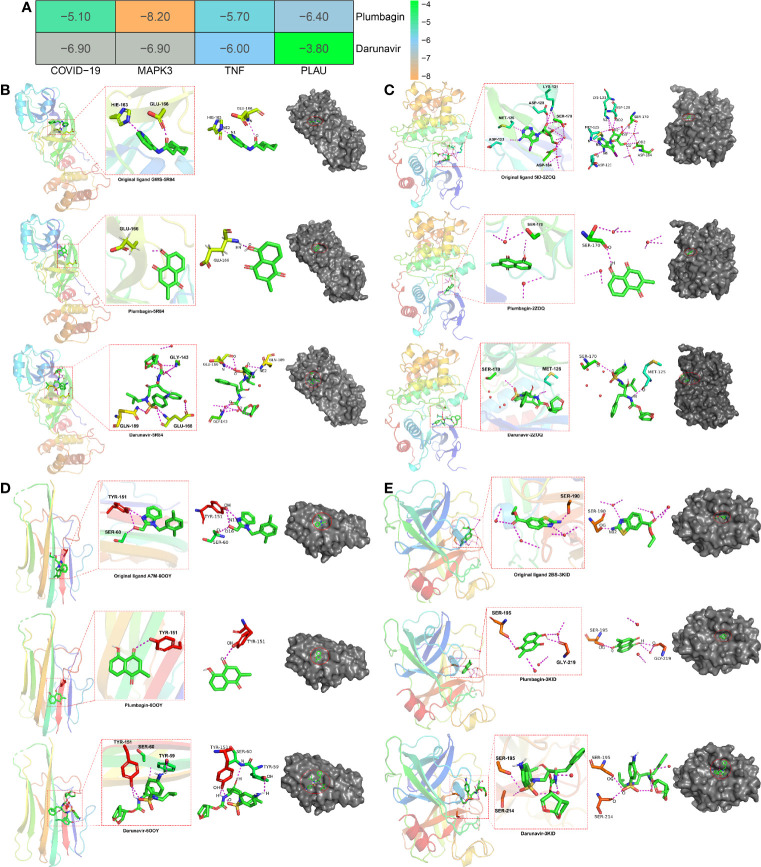
Using bioinformatics and computational biology, we determined the active cavities and binding affinity in MAPK3, TNF, and PLAU docked with PLB using the pymol 2.3 software, more docking parameter detail showed in Supplemental material ( Supplemental Tables 4–7 ) ( Figure 7A ). In COVID-19 (PDB ID: 5R84) (http://www.rcsb.org/structure/5R84), the root mean squared error (RMSD) of the original ligand GWS was 0.593 Å, and its hydrogen bond with the 5R84 protein acted on the protonation state of HIS-163 -HIE-163 (2.7 Å), and the amino acid residue GLU-166 (3.0 Å). PLB formed a hydrogen bond with the amino acid residue GLU-166 (2.1 Å), and in the surface model, PLB occupied the same active cavity as the original ligand, indicating that it exerted better binding characteristics with the 5R84 protein. Darunavir formed a hydrogen bond with amino acid residues GLY-143 (2.8 Å), GLU-166 (2.8 Å), GLN-189 (2.9 Å) ( Figure 7B ). In MAPK3 (PDB ID: 2ZOQ), the RMSD of the original ligand 5ID was 1.421 Å, which hydrogen-bonded with the ZQOQ protein to act on the amino acid residues ASP-123 (2.6), MET-125 (2.9), ASP- 128 (2.9), LYS-131 (3.3), SER-170 (3.0), and ASP-184 (3.2). PLB formed a hydrogen bond with amino acid residue SER-170 (2.2). In the surface model, plumbagin occupied the same active cavity as the original ligand, and the affinity between PLB and MAPK3 was small, indicating that PLB and the 2ZOQ protein had good binding properties. Darunavir forms a hydrogen bond with amino acid residues MET-125 (2.2 Å) and SER-170 (3.3 Å) ( Figure 7C ). In TNF (PDB ID: 6OOY), the RMSD of the original ligand A7M was 2.980 Å, and its hydrogen bond with the 6OOY protein acted on amino acid residues SER-60 (2.9) and TYR-151 (2.8). PLB formed a hydrogen bond with the amino acid residue TYR-151 (2.8), and in the surface model, PLB occupied the same active cavity as the original ligand, indicating that PLB played better binding characteristics with the 6OOY protein. Darunavir formed a hydrogen bond with amino acid residues TYR-59 (2.5 Å), SER-60 (3.1 Å), TYR-151 (2.5 Å) ( Figure 7D ). In PLAU (PDB ID: 3KID) (http://www.rcsb.org/structure/3KID), the RMSD of the original ligand 2BS was 1.730 Å, and its hydrogen bond with the 3KID protein acted on the amino acid residue SER-190 (2.3). PLB formed hydrogen bonds with amino acid residues SER-195 (2.7) and GLY-219 (2.2). In the surface model, PLB occupied the same active cavity as the original ligand, and the affinity of PLB to PLAU was smaller, indicating that PLB and the 3KID protein had better binding characteristics. Darunavir forms a hydrogen bond with amino acid residues TYR-59 (2.5 Å), SER-195 (3.0 Å), SER-214 (2.5 Å) ( Figure 7E ). In two-dimensional model, more detailed data were presented in Supplemental Figure 1 .
Figure 7.
Biological molecular docking findings of PLB against UCEC/COVID-19. (A), molecular affinity and the binding energy of PLB docked with core proteins, including MAPK3, TNF, and PLAU. (B–E), in silico characteristics of PLB, docked with core proteins.
Discussion
COVID-19, induced by the newly evolved coronavirus, becomes a cosmopolitical challenge due to SARS-CoV-2 severely threatening human life in many countries (33). Globally, SARS-CoV-2 is evolving and transmitting widespread, leaving a continual increment of death toll, as the specific medicine is absent (34). Fortunately, great efforts are being made to develop a targeting vaccine to suppress SARS-CoV-2 in some countries. However, there is an unknown period of time before the new specific vaccine is completed (35). Statistically, a growing number of people are living with chronic diseases in modern life, such as cardiovascular disorders and cancer. Also, the incidence of these diseases is mounting yearly in the world, especially in western countries (36). In addition, patients suffering from cancer may suffer immunological suppression and dysfunction of immunity, being potentiality prone to hospital-acquired infection (37). In the current grim situation, as in the global outbreak of COVID-19, there is no effective management for handling this problem, producing an increment of the death toll (38). Accordingly, during the early outbreak of COVID-19, hospital-originated infections of this virus were high as it was potentially undetected (2). Mounting epidemiological evidence shows that the cases of UCEC, characterized by malignant metastasis, are increasing around the world (39). As a result, numerous hospitalized cancer patients might be at an increased risk of infection with the new coronavirus, especially during the early outbreak. More significantly, the current treatment against UCEC shows reduced pharmacological effectiveness when over infection with SARS-CoV-2, causing unwanted increment in mortality.
Reportedly, PLB, a naturally occurring naphthoquinone, has been found to possess pharmaceutical anticancer properties via cytotoxic action against human cancer cell lines (11). In preliminary mechanism studies, the anti-cervical carcinoma activity of PLB was linked to the induction of apoptosis. In addition, based on the marked anti-inflammatory benefits of PLB, we preliminarily hypothesized that PLB may have effective pharmacological activities in patients with CC/COVID-19. Following an in silico investigation, all candidates, 255 mutual and 13 specific genes of COVID-19/UCEC, were screened out. The DGE determination resulted in 108 up-regulated and 147 down-regulated genes in COVID-19/UCEC patients. Accordingly, these DGE-assayed findings are likely to characterize the UCEC cases infected with SARS-CoV-2. In further independent prognostic and survival assays, some of the key differentially expressed genes, such as CCL2, ANPEP, CLEC4M, SCARA3, CP, ABCA4, KHK, SLC8A1, ZYG11B, GHR, TNF, FOSL2, and PLAU, may be used as molecular markers to detect and identify and stage UCEC patients infected with the novel coronavirus. Overall, current clinical investigations demonstrate that 255 mutual genes in COVID-19/UCEC patients are likely to function as candidate therapeutical bio targets. By the use of network pharmacology analysis, we screened out and determined a total of 13 mutual genes with PLB in the treatment COVID-19/UCEC before core target identification. Further analysis identified 5 core targets, including GAPDH, MAPK3, TNF, FOS, and PLAU. The current evidence highlighted that the core genes may be the pharmacologically active bio targets of PLB in the treatment of COVID-19/UCEC. Following an integrative enrichment assay, the bioinformatic data revealed that the anti-COVID-19/UCEC function and mechanism mediated by PLB might effectively be achieved by cytotoxicity, anti-proliferation, inducing apoptosis, anti-inflammation, immunomodulation, and modulation of some of key molecular pathways, such as Human T-cell leukemia virus 1 infection, Human immunodeficiency virus 1 infection, Human cytomegalovirus infection, T cell receptor signaling pathway, B cell receptor signaling pathway, Th1 and Th2 cell differentiation, TGF-beta signaling pathway, Natural killer cell-mediated cytotoxicity, IL-17 signaling pathway, Th17 cell differentiation, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, and TNF signaling pathway. Based on the biological molecular docking method, the anti-COVID-19/UCEC effect of PLB could be achieved by some of the core genes, including MAPK3, TNF, and PLAU, as PLB exerted better active cavities and binding affinity in MAPK3, TNF, and PLAU when docking. In view of these results, we hypothesize that PLB is a likely candidate to be used for the potential treatment of UCEC patients infected with SARS-CoV-2 in the current evolving situation before future clinical trials.
Conclusions
The current bioinformatic and computational findings reveal the anti-COVID-19/UCEC pharmacological functions and mechanisms achieved by PLB. Moreover, all core targets of PLB treatment in COVID-19/UCEC were identified, indicating potential pharmacological significance. Interestingly, biological molecular docking data indicate that PLB is a likely candidate to be applied clinically in the therapy of UCEC patients infected with SARS-CoV-2.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author Contributions
RL and MS conceived and designed the study. YML and SY performed the data analysis and data interpretation. YML and XL conducted the bioinformatics and statistical analyses. RL, YML, and SY prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the National Natural Science Foundation of China (No. 81660091) and the National Natural Science Foundation of Guangxi (No. 2019GXNSFBA185015, 2018GXNSFAA281242).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.714909/full#supplementary-material
Abbreviations
UCEC, uterine corpus endometrial carcinoma; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TCGA, The Cancer Genome Atlas; GO, Gene Ontology; BP, Biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; RMSD, root mean square deviation; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform. Plumbagin, PLB; GEQ, Expression Quantification; FDR, false discovery rate; OMIM, Online Mendelian Inheritance in Man; TTD, Therapeutic Target database; NCBI, National Center for Biotechnology Information; PPI, Protein-protein interaction.
References
- 1. Kakar A, Nundy S. COVID-19 in India. J R Soc Med (2020) 113(6):232–3. doi: 10.1177/0141076820927668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moghadas SM, Shoukat A, Fitzpatrick MC, Wells CR, Sah P, Pandey A, et al. Projecting Hospital Utilization During the COVID-19 Outbreaks in the United States. Proc Natl Acad Sci USA (2020) 117:9122–6. doi: 10.1073/pnas.2004064117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esposito S, Noviello S, Pagliano P. Update on Treatment of COVID-19: Ongoing Studies Between Promising and Disappointing Results. Infez Med (2020) 28:198–211. [PubMed] [Google Scholar]
- 4. Russell CD, Millar JE, Baillie JK. Clinical Evidence Does Not Support Corticosteroid Treatment for 2019-Ncov Lung Injury. Lancet (2020) 395:473–5. doi: 10.1016/S0140-6736(20)30317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harky A, Chiu CM, Yau THL, Lai SHD. Cancer Patient Care During COVID-19. Cancer Cell (2020) 37:749–50. doi: 10.1016/j.ccell.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. MacKintosh ML, Crosbie EJ. Prevention Strategies in Endometrial Carcinoma. Curr Oncol Rep (2018) 20:101. doi: 10.1007/s11912-018-0747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I. Endometrial Cancer. Lancet (2005) 366:491–505. doi: 10.1016/S0140-6736(05)67063-8 [DOI] [PubMed] [Google Scholar]
- 8. Zhou JY, Zhang L, Wei LH, Wang JL. Endometrial Carcinoma-Related Genetic Factors: Application to Research and Clinical Practice in China. BJOG (2016) 123:90–6. doi: 10.1111/1471-0528.14007 [DOI] [PubMed] [Google Scholar]
- 9. Wee LE, Conceicao EP, Sim XYJ, Aung MK, Tan KY, Wong HM, et al. Minimizing Intra-Hospital Transmission of COVID-19: The Role of Social Distancing. J Hosp Infect (2020) 105:113–5. doi: 10.1016/j.jhin.2020.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pai SA, Munshi RP, Panchal FH, Gaur IS, Mestry SN, Gursahani MS, et al. Plumbagin Reduces Obesity and Nonalcoholic Fatty Liver Disease Induced by Fructose in Rats Through Regulation of Lipid Metabolism, Inflammation and Oxidative Stress. BioMed Pharmacother (2019) 111:686–94. doi: 10.1016/j.biopha.2018.12.139 [DOI] [PubMed] [Google Scholar]
- 11. Tripathi SK, Panda M, Biswal BK. Emerging Role of Plumbagin: Cytotoxic Potential and Pharmaceutical Relevance Towards Cancer Therapy. Food Chem Toxicol (2019) 125:566–82. doi: 10.1016/j.fct.2019.01.018 [DOI] [PubMed] [Google Scholar]
- 12. Sakunrangsit N, Ketchart W. Plumbagin Inhibited AKT Signaling Pathway in HER-2 Overexpressed-Endocrine Resistant Breast Cancer Cells. Eur J Pharmacol (2020) 868:172878. doi: 10.1016/j.ejphar.2019.172878 [DOI] [PubMed] [Google Scholar]
- 13. Sakunrangsit N, Ketchart W. Plumbagin Inhibits Cancer Stem-Like Cells, Angiogenesis and Suppresses Cell Proliferation and Invasion by Targeting Wnt/β-Catenin Pathway in Endocrine Resistant Breast Cancer. Pharmacol Res (2019) 150:104517. doi: 10.1016/j.phrs.2019.104517 [DOI] [PubMed] [Google Scholar]
- 14. Pan Q, Zhou R, Su M, Li R. The Effects of Plumbagin on Pancreatic Cancer: A Mechanistic Network Pharmacology Approach. Med Sci Monit (2019) 25:4648–54. doi: 10.12659/MSM.917240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou R, Wu K, Su M, Li R. Bioinformatic and Experimental Data Decipher the Pharmacological Targets and Mechanisms of Plumbagin Against Hepatocellular Carcinoma. Environ Toxicol Pharmacol (2019) 70:103200. doi: 10.1016/j.etap.2019.103200 [DOI] [PubMed] [Google Scholar]
- 16. Srinivas P, Gopinath G, Banerji A, Dinakar A, Srinivas G. Plumbagin Induces Reactive Oxygen Species, Which Mediate Apoptosis in Human Cervical Cancer Cells. Mol Carcinog (2004) 40:201–11. doi: 10.1002/mc.20031 [DOI] [PubMed] [Google Scholar]
- 17. Nair S, Nair RR, Srinivas P, Srinivas G, Pillai MR. Radiosensitizing Effects of Plumbagin in Cervical Cancer Cells Is Through Modulation of Apoptotic Pathway. Mol Carcinog (2008) 47:22–33. doi: 10.1002/mc.20359 [DOI] [PubMed] [Google Scholar]
- 18. Luo P, Wong YF, Ge L, Zhang ZF, Liu Y, Liu L, et al. Anti-Inflammatory and Analgesic Effect of Plumbagin Through Inhibition of Nuclear Factor-κb Activation. J Pharmacol Exp Ther (2010) 335:735–42. doi: 10.1124/jpet.110.170852 [DOI] [PubMed] [Google Scholar]
- 19. Bhattacharya A, Jindal B, Singh P, Datta A, Panda D. Plumbagin Inhibits Cytokinesis in Bacillus Subtilis by Inhibiting FtsZ Assembly–A Mechanistic Study of Its Antibacterial Activity. FEBS J (2013) 280:4585–99. doi: 10.1111/febs.12429 [DOI] [PubMed] [Google Scholar]
- 20. Periasamy H, Iswarya S, Pavithra N, Senthilnathan S, Gnanamani A. In Vitro Antibacterial Activity of Plumbagin Isolated From Plumbago Zeylanica L. Against Methicillin-Resistant Staphylococcus Aureus. Lett Appl Microbiol (2019) 69:41–9. doi: 10.1111/lam.13160 [DOI] [PubMed] [Google Scholar]
- 21. Wu K, Wei P, Liu M, Liang X, Su M. To Reveal Pharmacological Targets and Molecular Mechanisms of Curcumol Against Interstitial Cystitis. J Adv Res (2019) 20:43–50. doi: 10.1016/j.jare.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li R, Ma XY, Song YQ, Zhang YY, Xiong WB, Li L, et al. Anti-Colorectal Cancer Targets of Resveratrol and Biological Molecular Mechanism: Analyses of Network Pharmacology, Human and Experimental Data. J Cell Biochem (2019) 120:11265–73. doi: 10.1002/jcb.28404 [DOI] [PubMed] [Google Scholar]
- 23. Li R, Guo C, Li Y, Liang X, Yang L, Huang W. Therapeutic Target and Molecular Mechanism of Vitamin C-Treated Pneumonia: A Systematic Study of Network Pharmacology. Food Funct (2020) 11:4765–72. doi: 10.1039/D0FO00421A [DOI] [PubMed] [Google Scholar]
- 24. Li R, Song Y, Ji Z, Li L, Zhou L. Pharmacological Biotargets and the Molecular Mechanisms of Oxyresveratrol Treating Colorectalcancer: Network and Experimental Analyses. Biofactors (2020) 46:158–67. doi: 10.1002/biof.1583 [DOI] [PubMed] [Google Scholar]
- 25. Liang Y, Zhou R, Liang X, Kong X, Yang B. Pharmacological Targets and Molecular Mechanisms of Plumbagin to Treat Colorectal Cancer: A Systematic Pharmacology Study. Eur J Pharmacol (2020) 881:173227. doi: 10.1016/j.ejphar.2020.173227 [DOI] [PubMed] [Google Scholar]
- 26. Su M, Guo C, Liu M, Liang X, Yang B. Therapeutic Targets of Vitamin C on Liver Injury and Associated Biological Mechanisms: A Study of Network Pharmacology. Int Immunopharmacol (2019) 66:383–7. doi: 10.1016/j.intimp.2018.11.048 [DOI] [PubMed] [Google Scholar]
- 27. Li R, Li Y, Liang X, Yang L, Su M, Lai KP. Network Pharmacology and Bioinformatics Analyses Identify Intersection Genes of Niacin and COVID-19 as Potential Therapeutic Targets. Brief Bioinform (2021) 22:1279–90. doi: 10.1093/bib/bbaa300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li R, Wu K, Li Y, Liang X, Tse WKF, Yang L, et al. Revealing the Targets and Mechanisms of Vitamin A in the Treatment of COVID-19. Aging (Albany NY) (2020) 12:15784–96. doi: 10.18632/aging.103888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu F, Pan Q, Wang L, Yi S, Liu P, Huang W. Anticancer Targets and Mechanisms of Calycosin to Treat Nasopharyngeal Carcinoma. Biofactors (2020) 46:675–84. doi: 10.1002/biof.1639 [DOI] [PubMed] [Google Scholar]
- 30. Li R, Guo C, Li Y, Qin ZQ, Huang WJ. Therapeutic Targets and Signaling Mechanisms of Vitamin C Activity Against Sepsis: A Bioinformatics Study. Brief Bioinform (2021) 22(3):bbaa079. doi: 10.1093/bib/bbaa079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li R, Guo C, Li Y, Liang X, Su M. Functional Benefit and Molecular Mechanism of Vitamin C Against Perfluorooctanesulfonate-Associated Leukemia. Chemosphere (2021) 263:128242. doi: 10.1016/j.chemosphere.2020.128242 [DOI] [PubMed] [Google Scholar]
- 32. Li R, Wu K, Li Y, Liang X, Lai KP, Chen J. Integrative Pharmacological Mechanism of Vitamin C Combined With Glycyrrhizic Acid Against COVID-19: Findings of Bioinformatics Analyses. Brief Bioinform (2021) 22:1161–74. doi: 10.1093/bib/bbaa141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int J Antimicrob Agents (2020) 55:105924. doi: 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate Drugs Against SARS-CoV-2 and COVID-19. Pharmacol Res (2020) 157:104859. doi: 10.1016/j.phrs.2020.104859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dror AA, Eisenbach N, Taiber S, Morozov NG, Mizrachi M, Zigron A, et al. Vaccine Hesitancy: The Next Challenge in the Fight Against COVID-19. Eur J Epidemiol (2020) 35:775–9. doi: 10.1007/s10654-020-00671-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cockerham WC, Hamby BW, Oates GR. The Social Determinants of Chronic Disease. Am J Prev Med (2017) 52:5–12. doi: 10.1016/j.amepre.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ariza-Heredia EJ, Chemaly RF. Update on Infection Control Practices in Cancer Hospitals. CA Cancer J Clin (2018) 68:340–55. doi: 10.3322/caac.21462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weinberger DM, Cohen T, Crawford FW, Mostashari F, Olson D, Pitzer VE, et al. Estimating the Early Death Toll of COVID-19 in the United States. bioRxiv (2020). doi: 10.1101/2020.04.15.20066431 [DOI] [Google Scholar]
- 39. Sorosky JI. Endometrial Cancer. Obstet Gynecol (2012) 120:383–97. doi: 10.1097/AOG.0b013e3182605bf1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.