Abstract
Background
Coronavirus disease 2019 is characterized by the elevation of a broad spectrum of inflammatory mediators associated with poor disease outcomes. We aimed at an in-silico analysis of regulatory microRNA and their transcription factors (TF) for these inflammatory genes that may help to devise potential therapeutic strategies in the future.
Methods
The cytokine regulating immune-expressed genes (CRIEG) were sorted from literature and the GEO microarray dataset. Their co-differentially expressed miRNA and transcription factors were predicted from publicly available databases. Enrichment analysis was done through mienturnet, MiEAA, Gene Ontology, and pathways predicted by KEGG and Reactome pathways. Finally, the functional and regulatory features were analyzed and visualized through Cytoscape.
Results
Sixteen CRIEG were observed to have a significant protein-protein interaction network. The ontological analysis revealed significantly enriched pathways for biological processes, molecular functions, and cellular components. The search performed in the miRNA database yielded ten miRNAs that are significantly involved in regulating these genes and their transcription factors.
Conclusion
An in-silico representation of a network involving miRNAs, CRIEGs, and TF, which take part in the inflammatory response in COVID-19, has been elucidated. Thus, these regulatory factors may have potentially critical roles in the inflammatory response in COVID-19 and may be explored further to develop targeted therapeutic strategies and mechanistic validation.
Keywords: Cytokine storm; Immuno-interactomics; COVID-19; Cytokines; MicroRNA, SARS-CoV-2
Abbreviation: AHR, Aryl hydrocarbon receptor; ARDS, acute respiratory distress syndrome; BAL, Bronchoalveolar Lavage; CC, Cellular components; CCL2, C-C motif chemokine 2; CCL3, C-C motif chemokine 3; CCL4, C-C motif chemokine 4; CCL, Chemokine (C-C motif) ligands; CCR, CC chemokine receptor; CEBPA, CCAAT/enhancer-binding protein alpha; COVID-19, Coronavirus Disease 2019; CREM, cAMP responsive element modulator; CRIEGs, Cytokine regulating immune expressed genes; CSF2, Granulocyte-macrophage colony-stimulating factor; CSF3, Granulocyte colony-stimulating factor; CXCL2, Chemokine (C-X-C motif) ligand 2; CXCL8, Interleukin-8; CXCL10, C-X-C motif chemokine 10; CXCR, C-X-C chemokine receptor; DDIT3, DNA damage-inducible transcript 3 protein; DEGs, Differentially expressed genes; E2F1, Transcription factor E2F1; EGR1, Early growth response protein 1; EP300, Histone acetyltransferase p300; ESR1, Estrogen receptor, Nuclear hormone receptor; ETS2, Protein C-ets-2; FOXP3, Forkhead box protein P3; GO, Gene Ontology; GSEs, Gene Series Expressions; HDAC1, Histone deacetylase 1; HDAC2, Histone deacetylase 2; HSF1, Heat shock factor protein 1; IL1B, Interleukin-1; IL2, Interleukin-2; IL6, Interleukin-6; IL7, Interleukin-7; IL9, Interleukin-9; IL10, Interleukin-10; IL17A, Interleukin-17A; IL-6, interleukin-6; IP-10, Interferon-Inducible Protein 10; IRF1, Interferon regulatory factor 1; JAK2, Tyrosine-protein kinase JAK2; JAK-STAT, Janus kinase (JAK)-signal transducer and activator; JUN, Transcription factor AP-1; KEGG, Kyoto Encyclopedia of Genes and Genomes; KLF4, Krueppel-like factor 4; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NFAT5, Nuclear factor of activated T-cells 5; NFKB1, Nuclear factor NF-kappa-B p105 subunit; NFKBIA, NF-kappa-B inhibitor alpha; NR1I2, Nuclear receptor subfamily 1 group I member 2; PDM, peripheral blood mononuclear cell; REL, Proto-oncogene c-Rel; RELA, Transcription factor p65; RUNX1, Runt-related transcription factor 1; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; SIRT1, NAD-dependent protein deacetylase sirtuin-1; SP1, Transcription factor Sp1; SPI1, Transcription factor PU.1; STAT1, Signal transducer and activator of transcription 1-alpha/beta; STAT3, Signal transducer and activator of transcription 3; TLR3, Toll-like receptor 3 (TLR3); TNF, Tumor necrosis factor; TNF-α, Tumor Necrosis Factor-Alpha; VDR, Vitamin D3 receptor; XBP1, X-box-binding protein 1; ZFP36, mRNA decay activator protein ZFP36; ZNF300, Zinc finger protein 300, heme oxygenase-1 (HO-1); miEAA, miRNA Enrichment Analysis and Annotation t
1. Introduction
Cytokine storm in severe or critically ill coronavirus disease 2019 (COVID-19) patients is characterized by the elevation of a wide spectrum of inflammatory mediators. These include cytokines and chemokines originating from airway epithelial cells and various immune cells and act as independent risk factors for disease severity and mortality (Liu et al., 2020, p. 19).
Various cytokines and chemokines have been observed to play dominant roles in different stages of the COVID-19 disease. Association of COVID-19 severity and mortality with higher levels of interleukin-6 (IL-6) have been corroborated in various studies (Cummings et al., 2020; Hajifathalian et al., 2020; Ruan et al., 2020). However, depending upon the stage in the natural history of COVID-19 disease, SARS CoV-2 has the menacing feature of longer persistence in the environment and various inanimate surfaces (Khokhar et al., 2020b) and different inflammatory mediators have been observed to play a dominant role in Acute kidney injury pathophysiology (Khokhar et al., 2020a). Control of this disease by newer diagnostic tools, based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas(CRISPR-Cas) system, is used for better diagnostic accuracy. (Gadwal et al., 2021, p. 19) In the initial stage, when clinical symptoms are mild, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replicates rapidly in the blood (Chen et al., 2020b). Chemokines are the inflammatory mediators that characterize this initial stage. Chemokine (C-C motif) ligands (CCL), namely CCL8, CCL9, and CCL2 expression, were found to be increased in the initial phase (Blanco-Melo et al., 2020, p. 19). In severe COVID-19 patients, serum CCL5 levels were elevated even before the occurrence of an IL-6 peak (Zhao et al., 2020). Further, in bronchoalveolar lavage (BAL) fluid of COVID-19 patients, a chemokine-rich signature was observed characterized by the expression of CCL2, CCL3, CCL4, CCL7, CCL8, chemokine (C-X-C motif) ligand 2 (CXCL2), CXCL8, CXCL17, and interferon-inducible protein 10 (IP-10) (Lu et al., 2020; Xiong et al., 2020).
During the amplification phase, inflammatory immune responses get aggravated and the disease progresses rapidly to severe/critical illness. The chemokines secreted in the initial phase recruit inflammatory innate and adaptive immune cells resulting in an exaggerated inflammatory immune response. As a result, peripheral levels of inflammatory mediators including IL-2, IL-6, IL-7, IL-10, tumor necrosis factor-alpha (TNF-α), CCL-2, and CCL-3 were highly elevated in this phase (Chen et al., 2020a; Huang et al., 2020). The unchecked elevation of inflammatory mediators leads to vascular leakage, complement cascade activation, and cytokine storm in this consummation phase (Wang et al., 2020; Yang et al., 2020; Zhou et al., 2020). Apart from the inflammatory mediators, it had also been observed that the mRNA expression levels of inflammatory genes peaked as the respiratory function deteriorated (Ong et al., 2020). Further, COVID-19 patients had also shown the possibility of alterations in transcription factors, affecting both cytokines as well as immune cells, adding another layer of dimension to the regulation and release of cytokines in a cytokine storm (Claverie, 2020, p. 19; De Biasi et al., 2020).
This study aimed to search the various databases for miRNAs that can affect the genetic expression of these inflammatory mediators and the transcription factors that regulate the expression. We identified the cytokine regulating immune expressed genes (CRIEGs) responsible for inflammation and cytokine storm in SARS-CoV from human bronchial epithelial cells. The insight into the regulation of the expression of these cytokine genes by miRNAs and transcription factors will help in devising better-targeted therapies to address the complications in severe COVID-19 disease due to cytokine storm.
2. Methodology
2.1. Identification of cytokines responsible for inflammation and cytokine storm in SARS-CoV-2
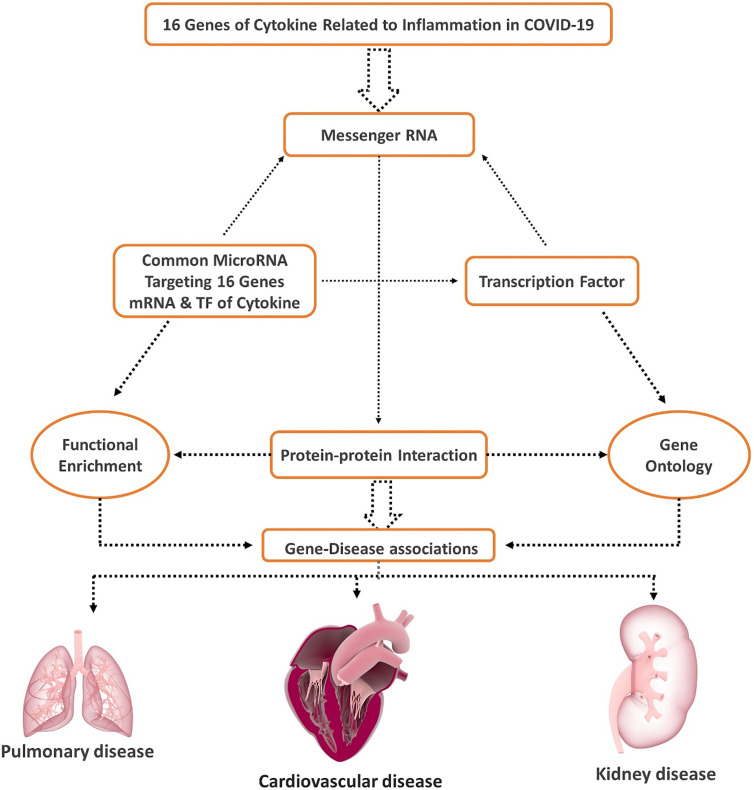
Several keywords including “Inflammation”, “Immunity”, “Immunogenetics”, “Cytokine storm”, “Acute respiratory distress syndrome”, “ARDS”, “COVID-19”, “cytokines”, “Coronavirus disease”, “SARS-CoV-2” and “Severe Acute Respiratory Syndrome” and “1999/01/01 to 2020/07/06” were searched in PubMed to identify cytokines responsible for inflammation and cytokine storm in SARS-CoV-2. (Supplementary Table 1) Fig. 1 summarizes in a flow chart the steps of data processing and analysis performed in the study. (Fig. 1).
Fig. 1.
Flow Chart of the data processing and Analysis.
2.2. Microarray data collection
We have searched in the GEO database several keywords including “SARS”, “Corona Virus”, “Blood”, “Homo sapiens”, “Expression profiling by array”, “bronchial epithelial cells” from 01/01/2012 to 17/12/2020. We found the GSE17400 contains bronchial epithelial cells of 9 (nine) samples.
2.3. Identification of cytokine regulating immune expressed genes (CRIEGs) responsible for inflammation and cytokine storm from SARS-CoV
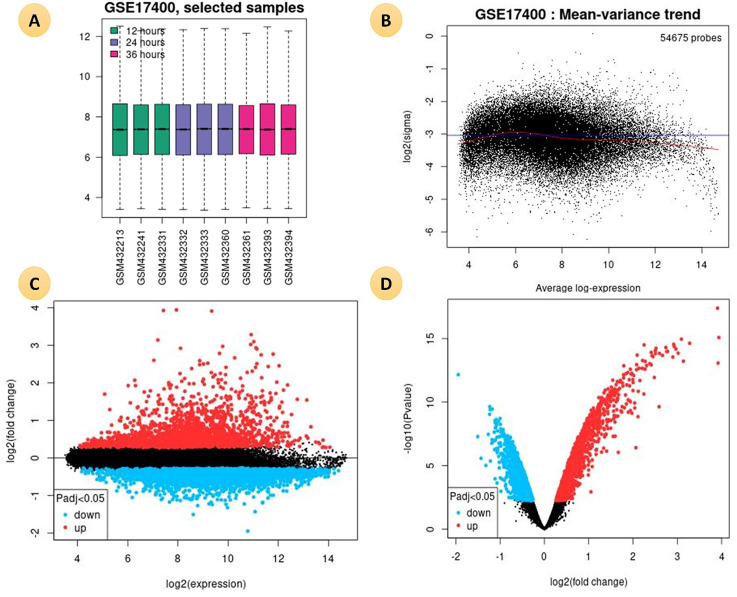
GEO2R is an online interactive web tool used to compare two groups of samples in a GEO Series to identify differentially expressed genes across experimental conditions. We obtained differentially expressed genes (DEGs) from GSE17400 dataset for innate immune responses of human bronchial epithelial cells against SARS-CoV with the help of GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) with the cut-off criteria of p < 0.05. (Fig. 2 )
Fig. 2.
(A). Boxplot is representing the distribution of the values of the selected Samples. The gene expression profile after 12 h (Green), after 24 h (violet) after 36 h (pink) in Human Bronchial Epithelial Cells (2B4 cells) infected with SARS-CoV. (B). A mean-variance trend plot is applicable to check the mean-variance relationship of the DEGs data after fitting a linear model. (C). A mean difference (MD) plot displays log2 fold change versus average log2 expression values of DEGs. (D) A volcano plot shows statistical significance (−log10 P-value) versus magnitude of change (log2 fold change) DEGs.
2.4. Identification of common transcription factors (TFs) related to cytokine regulating immune expressed genes (CRIEGs)
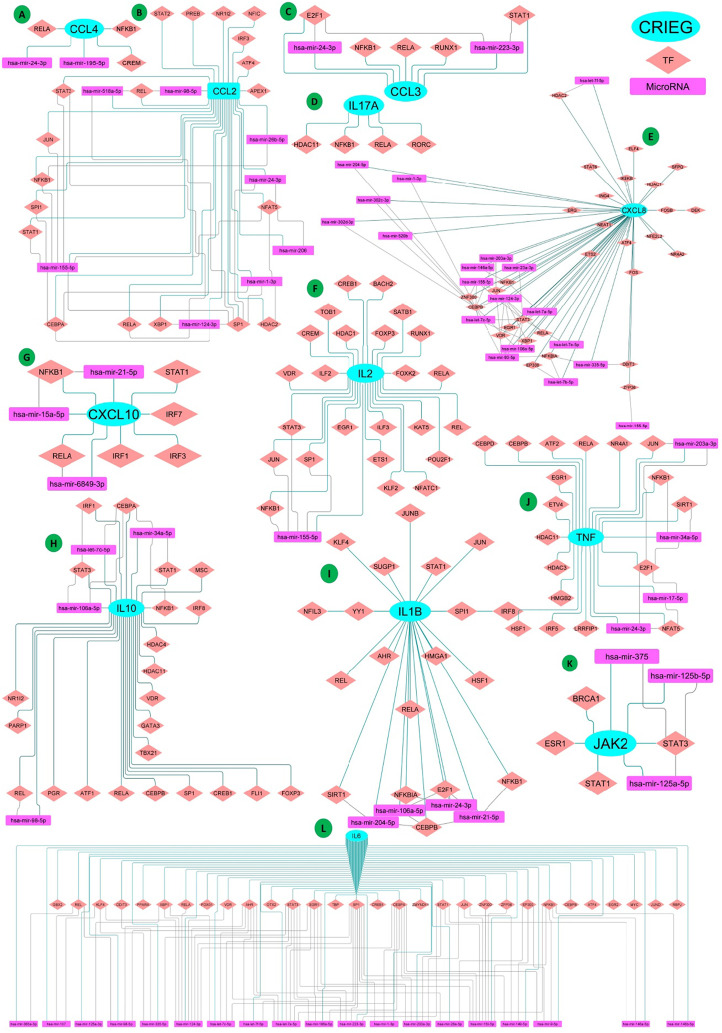
Cytokines are mostly regulated at the transcriptional level by specific transcription factors (TFs) that recruit transcriptional machinery including cofactors (Carrasco Pro et al., 2018). We identified those TFs regulating CRIEGs that commonly appeared in all five different databases of transcription factor viz., − TRRUST, RegNetwork, ENCODE, JASPAR, and CHEA. The identification of TF–target regulatory relationships is a key step for revealing functions of TFs and their regulations on CRIEG expression. We Identified more than 32 common TFs of 16 CRIEGs from well-established microRNAs and TFs target prediction database miRNet Version 2 (Chang et al., 2020). We have created an interaction network between CRIEGs and its transcription factors through the Cytoscape software [24,25]. (Fig. 3 , Supplementary Table 2).
Fig. 3.
Each CRIEG and their Transcription factor and common targeting MicroRNAs interaction network (A)CCL4; (B)CCL2; (C)CCL3; (D)IL17A; (E)CXCL8; (F)IL2; (G)CXCL10; (H)IL10; (I) IL1B; (J)TNF; (K)JAK2; (L) IL6. (Cyan colored Ellipse shaped Node: CRIEGs; Brick colored diamond shaped node: Transcription factor of CRIEGs; Pink colored rectangle shaped Node: MicroRNAs; Each node inter-connected with another node by the edges).
2.5. Identification and assortment of common regulated miRNAs of CRIEGs and transcription factors of CRIEGs
Evolutionary conserved small non-coding RNA or MicroRNA affect the gene expression by binding to specific mRNAs and regulate cell growth, differentiation, and death. miRNAs regulate multiple functions of T-cell subsets through immune homeostasis and immune tolerance that control the development, survival, and activation (Garavelli et al., 2018). The miRNA that targeted cytokine and its transcription factor genes were predicted by different well-established miRNA target prediction databases miRDB, miRBase, miRNet Version 2, and TargetScan (Chang et al., 2020). We have created an interactomics network of CRIEGs and their transcription factors with target microRNAs through the Cytoscape software [24,25] (Fig. 3, Supplementary Table 2).
2.6. Protein-protein interaction, functional enrichment and KEGG pathway analysis of CRIEGs and transcription factors of CRIEGs
The Search Tool for the Retrieval of Interacting Genes/Protein [STRING] (http://string-db.org/) was used to construct a protein-protein interaction (PPI) network using only overlapped DEGs and greater than 0.4 confidence score cut-off. The interaction networks for all 16 cytokines were constructed by Cytoscape (Otasek et al., 2019; Shannon, 2003).
Analysis of the functional and regulatory features was carried out through gene ontology (GO), KEGG pathways through DAVID (the database for annotation, visualization and integrated discovery) and STRING (functional protein association networks) biological databases.
2.7. Functional enrichment and disease relationship of common regulated miRNAs of CRIEGs and transcription factors of CRIEGs
Cytokine regulating immune expressed genes (CRIEGs) and transcription factors of CRIEGs are regulated by ten (10) common microRNAs. We identified all ten microRNAs by enrichment of two databases MIENTURNET and MiEAA (Kern et al., 2020; Licursi et al., 2019). MIENTURNET and miEAA (miRNA Enrichment Analysis and Annotation tool) perform both statistical and network-based analyses of pathways and disease-related activity in cellular processes.
3. Results
3.1. Identification of cytokine regulating immune expressed genes (CRIEGs) responsible for inflammation and cytokine storm in SARS-CoV
We compared the genes and their transcription factors obtained from the literature search with that of SARS-CoV dataset (GSE17400) and found that majority of the identified genes and transcription factors were expressed in the SARS-CoV dataset. (Table 1 ). We found 16 CRIEGs (IL-1b, IL-2, IL-7, IL-8, IL-9, IL-10, IL-17, G-CSF, GM-CSF, IFN-γ, TNF-α, CXCL10, MCP1, MIP1A, MIP1B, and IL-6) from literature which is responsible for acute respiratory distress syndrome (ARDS) in COVID-19. Further, our analysis of CRIEGs from the SARS-CoV infected human bronchial epithelial cells dataset demonstrated that the over-expression of these genes in a short time (interval of 12 h, 24 h, and 36 h) increases the severity of the disease (Yoshikawa et al., 2010).
Table 1.
List of CRIEGs and there regulating transcription factors expressed in GSE17400 data sets.
| Cytokine Strom Gene |
Transcription Factors of Cytokine Gene |
||||
|---|---|---|---|---|---|
| Gene Symbol | P Value | Fa | Gene Symbol | P Value | Fa |
| CCL2 | 0.01 | 6.07 | AHR | 0.03 | 4.25 |
| CCL4 | 0.03 | 4.59 | CREM | 0.05 | 3.75 |
| CXCL10 | 0.00 | 98.60 | DDIT3 | 0.00 | 52.60 |
| CXCL8 | 0.00 | 56.40 | E2F1 | 0.00 | 10.50 |
| IL6 | 0.00 | 139.00 | EGR1 | 0.00 | 75.60 |
| IL7 | 0.00 | 24.60 | EP300 | 0.01 | 6.13 |
| JAK2 | 0.00 | 16.40 | ESR1 | 0.00 | 7.64 |
| TNF | 0.02 | 5.26 | ETS2 | 0.00 | 82.10 |
| HDAC1 | 0.03 | 4.49 | |||
| HDAC2 | 0.00 | 9.29 | |||
| IRF1 | 0.00 | 132.00 | |||
| JUN | 0.00 | 45.20 | |||
| KLF4 | 0.00 | 58.10 | |||
| NFAT5 | 0.03 | 4.46 | |||
| NFKB1 | 0.00 | 12.70 | |||
| NFKBIA | 0.00 | 81.60 | |||
| REL | 0.00 | 14.20 | |||
| RUNX1 | 0.04 | 4.10 | |||
| SIRT1 | 0.00 | 26.40 | |||
| SP100 | 0.00 | 111.00 | |||
| SP140L | 0.00 | 53.80 | |||
| STAT1 | 0.00 | 80.60 | |||
| XBP1 | 0.02 | 4.81 | |||
| ZFP36 | 0.00 | 14.40 | |||
F:Moderated F-statistic combines the t-statistics for all the pair-wise comparisons into an overall test of significance for that gene (only available when more than two groups of samples are defined).
3.2. Identification of cytokine regulating immune expressed genes and construction of PPI network
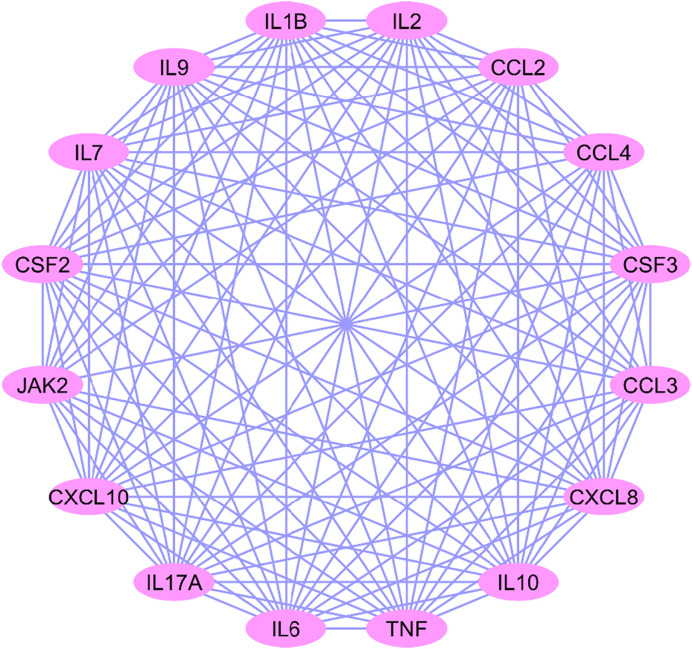
All 16 CRIEGs show interactions among themselves, based on the STRING database. The average node degree is calculated based on a number of how many interactions (at the score threshold) that a protein have on the average in the network. The local clustering coefficient is use for connection of nodes in the network. Highly connected networks have high values. A minimum significant PPI enrichment p-value indicate that the nodes are not random and that the observed number of edges is significant. In this study, PPI network consisted of 16 nodes and 117 edges, the average local clustering coefficient was 0.977, and PPI enrichment p-value was highly significant (p < 0.001) (Fig. 4 ).
Fig. 4.
Protein-Protein Interaction between Cytokine storm genes.
3.3. Identification of common transcription factors regulating CRIEGs
We identified the common transcription factors of CRIEGs through TRRUST, RegNetwork, ENCODE, JASPAR and CHEA databases. During this identification process, we found a total of 32 transcription regulators. All transcription regulators viz., AHR, CEBPA, CREM, DDIT3, E2F1, EGR1, EP300, ESR1, ETS2, FOXP3, HDAC1, HDAC2, HSF1, IRF1, JUN, KLF4, NFAT5, NFKB1, NFKBIA, NR1I2, REL, RELA, RUNX1, SIRT1, SP1, SPI1, STAT1, STAT3, VDR, XBP1, ZFP36, ZNF300 commonly regulated the transcription of 16 CRIEGs (Table 2 ).
Table 2.
List of Cytokine Strom genes, transcription factors and common targeting microRNAs.
| Cytokine Gene | Transcription Factor | MicroRNAs |
|---|---|---|
| IL1B | AHR | hsa-miR-106a-5p |
| IL2 | CEBPA | hsa-miR-155-5p |
| IL7 | CREM | hsa-miR-98-5p |
| CXCL8 | DDIT3 | hsa-miR-24-3p |
| IL9 | E2F1 | hsa-miR-204-5p |
| IL10 | EGR1 | hsa-miR-124-3p |
| IL17A | EP300 | hsa-miR-203a-3p |
| CSF3 | ESR1 | hsa-miR-335-5p |
| CSF2 | ETS2 | hsa-let-7c-5p |
| JAK2 | FOXP3 | hsa-miR-1-3p |
| TNF | HDAC1 | |
| CXCL10 | HDAC2 | |
| CCL2 | HSF1 | |
| CCL3 | IRF1 | |
| CCL4 | JUN | |
| IL6 | KLF4 | |
| NFAT5 | ||
| NFKB1 | ||
| NFKBIA | ||
| NR1I2 | ||
| REL | ||
| RELA | ||
| RUNX1 | ||
| SIRT1 | ||
| SP1 | ||
| SPI1 | ||
| STAT1 | ||
| STAT3 | ||
| VDR | ||
| XBP1 | ||
| ZFP36 | ||
| ZNF300 |
3.4. Identification of common MicroRNAs targeting CRIEGs and TFs of CRIEGs
We identified the MicroRNAs targeting CRIEGs and its TFs from various microRNA databases like miRNet, TargetScan, miRDB, miRanda, miRWalk. In our analysis, we identified 10 multi-targeting miRNAs viz.,hsa-miR-106a-5p, hsa-miR-155-5p, hsa-miR-98-5p, hsa-miR-24-3p, hsa-miR-204-5p, hsa-miR-124-3p, hsa-miR-203a-3p, hsa-miR-335-5p, hsa-let-7c-5p, and hsa-miR-1-3p. Almost all of the above identified microRNAs targeted both CRIEGs and their transcription factors simultaneously. Interestingly, microRNA targeting CCL4 was an exception. All the ten (10) microRNAs were observed to target more than one CRIEGs and its TFs (Fig. 3).
3.5. Common Pathway enrichment analysis of CRIEGs, transcription factors of CRIEGs and common targeting MicroRNAs
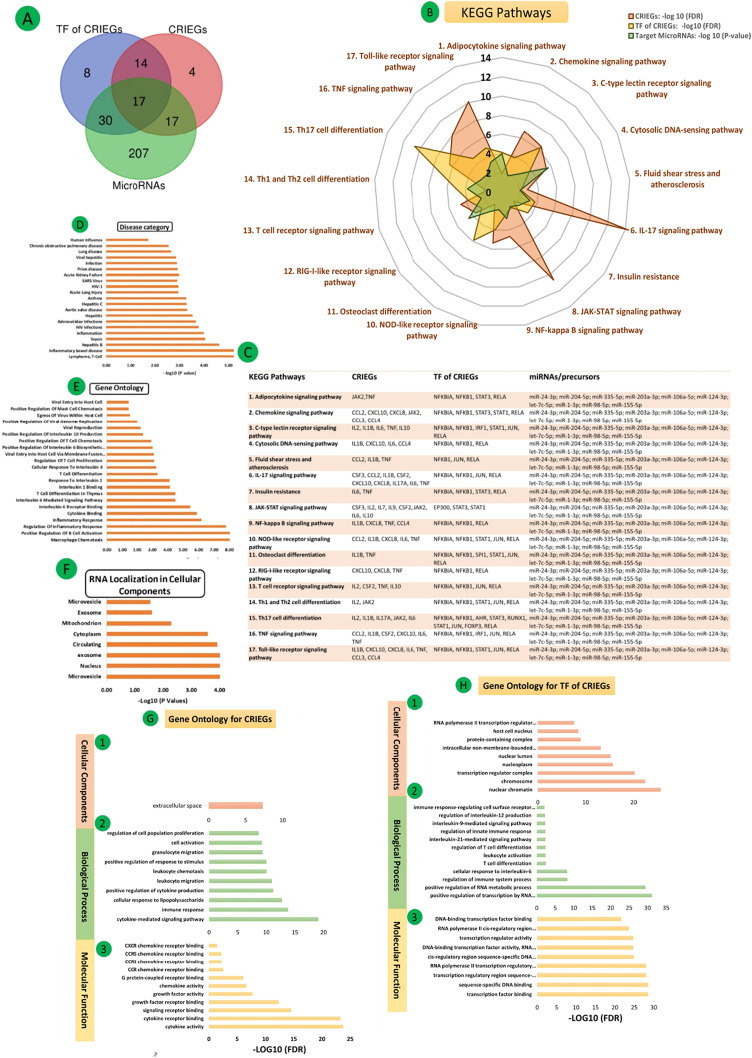
STRING and DAVID were assessed to acquire KEGG pathways enriched by CRIEGs and their TFs, common Pathways of MicroRNAs by enrichment of two databases MIENTURNET and MiEAA. (Supplementary Fig. 1 & Supplementary Table 3). All databases were selected for preferred and significant (p < 0.05) common pathways. The total 17 common pathways are involved in cytokine storm regulatory mechanism like Adipocytokine signaling pathway, Chemokine signaling pathway, C-type lectin receptor signaling pathway, Cytosolic DNA-sensing pathway, Fluid shear stress and atherosclerosis, IL-17 signaling pathway, Insulin resistance, JAK-STAT signaling pathway, NF-kappa B signaling pathway, NOD-like receptor signaling pathway, Osteoclast differentiation, RIG-I-like receptor signaling pathway, T cell receptor signaling pathway, Th1 and Th2 cell differentiation, Th17 cell differentiation, TNF signaling pathway, Toll-like receptor signaling pathway (Fig. 5 (A-C) & Supplementary Table 4).
Fig. 5.
(A). Venn diagram of common KEGG pathways involved in CRIEGs, TF of CRIEGs and MicoRNAs; (B) Common 17 KEGG pathways; (C) KEGG Pathways regulating CRIEGs, TF of CRIEGs and MicoRNAs; (D) Disease Category (DC) for MicroRNAs enrichments; (E) Gene Ontology (GO) for MicroRNAs enrichments; (F) RNA localization in cellular components; (G) Gene Ontology (GO) for CRIEGs (1) Cellular Component; (2) Biological Process (3) Molecular function; (H) Gene Ontology (GO) for TFs of CRIEGs (1) Cellular Component; (2) Biological Process (3) Molecular function.
3.6. Ontological analysis of CRIEGs
To get insights into the biomolecular significance of the identified CRIEGs, we performed gene ontology analysis by various databases and obtained enriched GO terms. STRING and DAVID were used to conduct the gene ontology analysis for CRIEGs within three categories: biological process, molecular function, and cellular component. Common statistically significant (p < 0.05) ontological processes have been identified through DAVID.
Molecular functions cytokine activity, cytokine receptor binding, signaling receptor binding, growth factor receptor binding, growth factor activity, chemokine activity, G protein-coupled receptor binding, CCR/CCR1/CCR5/CXCR5 chemokine receptor binding. Some crucial biological processes such as cytokine-mediated signaling pathways, immune responses, cellular responses to lipopolysaccharide, positive regulation of cytokine production, leukocyte migration, leukocyte chemotaxis, positive regulation of response to stimuli, granulocyte migration, cell activation, regulation of cell population proliferation are regulated by CRIEGs. Cellular components (CC) found only extracellular space [Fig. 5{G (1–3)} & Supplementary Tables 5-7].
3.7. Ontological analysis of common transcription factors
To get deep insights into the biomolecular significance of the identified common TFs, we performed gene ontology analysis by various databases and obtained enriched GO terms. STRING and DAVID have been used to conduct the GO analysis of common TFs within three categories: BP, MF, and CC. Common statistically significant (p < 0.05) ontological processes have been identified through DAVID.
Important biological processes like positive regulation of transcription by RNA polymerase II, positive regulation of RNA metabolic process, regulation of immune system process, cellular response to interleukin-6, T cell differentiation, leukocyte activation, regulation of T cell differentiation, interleukin-21-mediated signaling pathway, regulation of innate immune response, interleukin-9-mediated signaling pathway, regulation of interleukin-12 production, immune response-regulating cell surface receptor signaling pathway are involved in the metabolic regulatory process of TFs.
Most of the TFs play significant roles in different MF like transcription factor binding, sequence-specific DNA binding, transcription regulatory region sequence-specific DNA binding, RNA polymerase II transcription regulatory region sequence-specific DNA binding, cis-regulatory region sequence-specific DNA binding, DNA-binding transcription factor activity, RNA polymerase II-specific, transcription regulator activity, RNA polymerase II cis-regulatory region sequence-specific DNA binding, DNA-binding transcription factor binding.
Various transcription factors localization in multiple CC like nuclear chromatin, chromosome, transcription regulator complex, nucleoplasm, nuclear lumen, intracellular non-membrane-bounded organelle, protein-containing complex, host cell nucleus, RNA polymerase II transcription regulator complex. [Fig. 5{H (1–3)} & Supplementary Tables 8-10].
3.8. Disease category, RNA localization and Ontological analysis of frequent targeting MicroRNAs
We identified the microRNA enrichment analysis from two different databases MIENTURNET and miEA. We found out the localization of cellular components, miRNA-disease relationship and ontological functions of these important microRNAs. These ten MicroRNAs are found in different parts of the cell, such as microvesicle, nucleus, exosome, cytoplasm, and mitochondrion. These miRNAs also correlate in many diseases such as SARS, lymphoma, inflammatory bowel disease, hepatitis B, hepatitis C, asthma, Acute Lung Injury, sepsis, HIV, Adenoviridae infections, aortic valve disease, Acute Kidney Failure, prion disease, chronic obstructive pulmonary disease, and human influenza.
Involvement of miRNA in various GO terms such as macrophage chemotaxis, positive regulation of B-cell activation, regulation of inflammatory response cytokine binding, IL-6 receptor binding, IL-6 Mediated Signaling Pathway, T-cell differentiation in thymus, IL-1 binding, response To IL-1, cellular response to IL-4, regulation of T-cell proliferation, viral entry into host cell via membrane fusion with the plasma membrane, positive regulation of IL-6 biosynthetic process, T-cell chemotaxis, IL-10 production, mast cell chemotaxis, viral reproduction, positive regulation of viral genome replication, viral entry into host cell. [Fig. 5(D-F) & Supplementary Tables 11-13].
3.9. Interactomical network of CRIEGs, TFs of CRIEGs and common targeting MicroRNAs
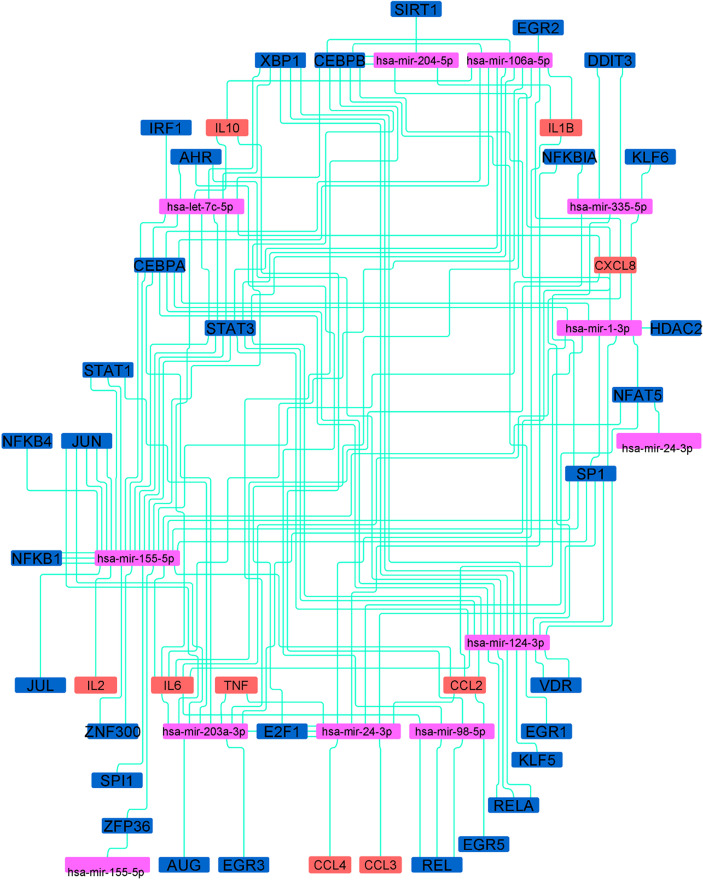
With the help of Cytoscape, we created a network of common targeting miRNAs of all the 16 CRIEGs and their 32 TFs. These interactions influence the host response to virus and contributes to the severity of the disease. The interactomical network has been depicted in Supplementary Figs. 2, 3, Figs. 3, and 6 . Supplementary Fig. 2 depicts the interaction network of CRIEGs and the TFs that regulate the expression of CRIEGs. Supplementary Fig. 3 depicts the network of interaction between CRIEGs MicroRNAs that regulate the expression of CRIEGs. Fig. 3 demonstrate the network of interaction between CRIEG, their TFs and the corresponding MicroRNAs for each of the individual CRIEGs. Lastly Fig. 6 depicts a wholesome interactome picture of various CRIEGs, their TFs and regulating MicroRNAs. (Supplementary Figs. 2, 3, Figs. 3, and 6.)
Fig. 6.
The CRIEGs and Transcription factors of CRIEG and MicroRNAs interaction network. (Brick colour node: CRIEGs; Dark blue colour node: Transcription factor of CRIEGs; Pink colour node: CRIEGs and their transcription factor targeting microRNAs). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
MicroRNAs post-transcriptionally regulate the expression of target mRNA. RNA viruses are known to utilize the host miRNA machinery for their benefit. Hence, various studies have identified miRNAs as key players in the pathogenesis and therapeutics of viral diseases. Also, miRNA scan target viral genes as well as the host inflammatory machinery, as part of the host-pathogen interactions, to counter-act the impairing effects of infection (Ghosh et al., 2008). Demirci et al. have identified 67 different human miRNA that targets the spike protein of the SARS-CoV-2 virus (Saçar Demirci and Adan, 2020). The inflammatory cascades involved in the pathophysiological pathways of COVID-19 are crucial in the development of complications in COVID-19. These pathogenetic pathways constitute but are not limited to, receptor tyrosine kinases, the JAK/STAT pathway, TNF-α receptor and toll-like receptors, IL-6 and IFN-γ, cytokine storm, and macrophage activation (Yarmohammadi et al., 2020). Hence, exploring the regulatory networks of these inflammatory markers have the two-fold advantages of discovering the interconnected nature of these dysregulated pathways and unlocking the potential of novel mechanistic-based treatment strategies. However, a clear understanding of the miRNA response in SARS-CoV-2 is still elusive. Here, we have identified the miRNAs and transcription factors of the target mRNAs which provide the necessary insight into the genetic regulation of the inflammatory response in COVID-19.
Our in-silico analysis revealed ten miRNAs involved in the regulation of the common inflammatory genes and their transcription factors.
The miR-155-5p has been widely studied in viral inflammatory pathways. It is a regulator of the HCV-induced TLR3/NF-κB pathway mediated inflammatory response. Further, elevated circulating levels of miR-155 were also observed in HBV infection (Bala et al., 2012; Wang et al., 2015). The role of miR-155-5p in the cytokine response through the TLR4/NF-κB/miR-155-5p/SOCS-1 axis in monocyte-derived macrophages has been demonstrated in dengue (Arboleda et al., 2019). It has further been observed to be upregulated in B cells in EBV infection and in PBMC of HIV-1 infected patients (Dey et al., 2016; Gao et al., 2015). In JHMV-infected (a coronavirus) mice models, miR-155 enhanced the T-cell trafficking, cytokine secretion, and cellular effectors functions (Dickey et al., 2016). Woods et al. studied 1908 mature murine miRNA expressions in influenza A virus (IAV)-infected type II alveolar cells and miR-155-5p was showed to have the highest expression (Woods et al., 2020). In FeAE cells, miR-155-5p expression induced IL-6 and IP-10 production, responsible for the recruitment of leukocytes(McAdams et al., 2015). It also regulates the NF-κB and MAPK signaling pathways (Shi et al., 2020). Our analysis found miR-155-5p to be one of the ten identified miRNAs that possibly regulate cytokine expression and triggers an inflammatory response in COVID-19. Further, miR-155-5p was found to affect the expression of multiple transcription factors, including CEBPA, JUN, NFAT5, NFKB1, SP1, SPI1, STAT1, STAT3, CEBPB, ZP36, ZNF300, ZFP36.
Another targeting miRNA identified in this study, miR-124-3p, was observed to be downregulated in JEV-infected human neural stem/progenitor cells (Mukherjee et al., 2019). A mice model showed downregulation in miR-124-3p expression in ARDS. Treatment with miR-124-3p agomir attenuated the pulmonary injury and the levels of pro-inflammatory cytokines IL-6 and TNF-α by directly targeting p65, thus showing promise in in-vitro management of pulmonary injury (Liang et al., 2020, p. 65).
Yet another miR-203a has been demonstrated to have an antagonistic role in foot-and-mouth disease virus (FMDV) infection (Gutkoska et al., 2017). This miRNA was further studied in IAV infection where an upregulated miR-203a modulated the antiviral response by targeting DR1 gene (Zhang et al., 2018, p. 1). However, further studies are needed to consolidate its role in corona-virus infections. Our in-silico analysis showed miR-203a targets transcription factors NFkB1, RELA, CREBPB, ATF4, ETS1 and 2.
miR-335-5p had the most predicted targets in the response against the Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) of alveolar macrophages. Contrastingly, Dhorne-Pollet et al. observed no effects on cytokine expression in their study (Dhorne-Pollet et al., 2019). This may be attributed to the low level of expression of miR-335-5p in most tissues, which renders its effect to be negligible despite the abundant number of predicted targets(The FANTOM Consortium et al., 2017). However, miR-24-3p facilitated PRRSV replication via suppression of heme oxygenase-1 (HO-1) (Xiao et al., 2015), and HO-1 has been reported to play role in anti-viral activity in several viral infections including HIV, hepatitis C virus, hepatitis B virus, enterovirus 71, influenza virus, respiratory syncytial virus, dengue virus, and Ebola virus(Espinoza et al., 2017, p. 1). Thus a high expression of miR-24-3p may be pathognomic for the worsening of viral infection.
Another targeting miRNA identified in this study, hsa-let-7c-5p, directly affects ACE2 and TMPRSS2; two key players in the SARS-CoV-2 infection (Chauhan et al., 2020).In rhabdomyosarcoma cells, hsa-let-7c-5p promoted the replication of enterovirus 71 (EV71) by inhibiting the MAPK4K expression (Zhou et al., 2017). Overexpression of miR-let-7c attenuated the replication of HCV through HO-1 induction (Chen et al., 2019). A differential expression of hsa-let-7 in-silico as a CRIEG indicates it has a role in immunomodulation in COVID-19.
Collectively, these studies demonstrate that mainly ten microRNAs (hsa-miR-106a-5p, hsa-miR-155-5p, hsa-miR-98-5p, hsa-miR-24-3p, hsa -miR-204-5p, hsa-miR-124-3p, hsa-miR-203a-3p, hsa-miR-335-5p, hsa-let-7c-5p, hsa-miR-1-3p) regulate the role of inflammatory mechanism in viral infection. Our in-silico analysis points towards a similar potential regulatory role of miRNA in SARS-CoV-2 mediated inflammatory cascades. Many of the target miRNA found in this study, namely miR-106a-5p, miR-1-3p, miR-98-5p, miR-24-3p, and miR-204-5p have been observed to orchestrate the gene expression of IL-1β, IL-6, IL-10, IFNγ, IL-2, and IL-17 through TFs such as ERK, STAT1, and STAT3 (Shen et al., 2019; Srivastava et al., 2017; Xiu et al., 2020; Ye et al., 2014).
Although most of these data are derived from cancer and transplantation studies, a similar regulation mechanism may be functional in the case of COVID-19. The disease severity in COVID-19 had been associated with an influx of innate immune cells and inflammatory cytokines (Hadjadj et al., 2020; Yale IMPACT Team et al., 2020, p. 19). The cytokine storm in COVID-19 leads to lung injury, multiple organ failure, and poor prognosis (Jose and Manuel, 2020; Mehta et al., 2020). TNF-α and IFN-γ together had shown to incite the cells to PANoptosis, inflammatory cell death involving the components of pyroptosis, apoptosis, and necroptosis. Further, JAK/STAT1/IRF1 axis was also involved in the regulation of inflammatory cell death due to PANoptosis (Karki et al., 2020). Further, IL-6 has also been shown to activate the Janus kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway leading to immune activation (Luo et al., 2020, p. 19).
Apart from the above-mentioned transcription factors, NF-κB also plays a crucial role in the poor prognosis of severe COVID-19 disease. NF-κB leads to an accelerated inflammatory response with increased secretion of TNF-α and IL-6. This auto-amplified pro-inflammatory loop with impaired type I IFN response culminates in Viral replication within the lungs and tissue damage (Hadjadj et al., 2020). Recent studies have demonstrated the up-regulation of various miRNAs in COVID-19 patients, thus confirming our prediction. In comparison to healthy controls, miRNAs, including miR-21, miR-155, miR-208a and miR-499, had been demonstrated to be up-regulated in the COVID-19 patients (Garg et al., 2021; Mahesh and Biswas, 2019). The varying functional significance and organ specificity of these upregulated miRNAs, i.e. miR-155 (inflammatory miRNA), miR-208a (heart-muscle specific) and miR-499 (muscle function) and miR-21 (fibrosis-associated) denotes the involvement of multiple pathways and organs in the pathophysiology of COVID-19 (Thum et al., 2008; van Rooij et al., 2009; van Rooij et al., 2007). MicroRNAs such as miR-125b, miR-138, miR-199a and miR-21 are also responsible for cytokine storms in acute respiratory distress syndrome and COPD(Guterres et al., 2020). In addition, miR-26a-5p, miR-29b-3p, and miR-34a-5p have been shown to be involved in endothelial dysfunction and inflammatory response in patients with SARS–CoV-2 infection (Centa et al., 2021). Further, the upregulation miRNAs in post-COVID-19 complications such as (miR-21, miR-155, miR-208a and miR-499) in chronic myocardial damage and inflammation (Garg et al., 2021), (let-7b-3p, miR-29a-3p, miR-146a-3p and miR-155-5p) in the post-acute COVID-19 phase; also indicates the extent of the alteration of these pathways in Post-COVID-19 sequelae. The downregulation of these miRNAs may be targeted to improve acute symptoms and distress by regulating the production of pro-inflammatory cytokines and apoptotic proteins (Guterres et al., 2020). Apart from the effect on host cellular pathways, miRNAs can also inhibit the viral infectivity by different ways like blocking the viral replication, cellular receptors and the function of viral proteins in SARS-CoV-2 (Fani et al., 2021). MicroRNAs such as miR-21-3p, miR-195-5p, miR-16-5p, miR-3065-5p, miR-424-5p and miR-421 potentially regulate the infectivity of viruses belonging to human coronavirus family through direct binding to the viral genome (Chan et al., 2020). Interestingly, newer miRNA-based therapy including antimiR-18 and antimir-125b, which potentially targets ACE2-related genes, have been proposed for nephropathy associated with COVID-19 (Widiasta et al., 2020).
We have identified other target CRIEGs of transcription factors STAT1, IRF1 &NF-κ Band the miRNAs regulating their expression. This would help better understand the cross talk and regulation of various cytokines in COVID-19 and the possible role of in regulating inflammatory cell death, leading to multiple organ failure. The main limitation of this study is that the data is derived from publicly available databases and needs experimental substantiation to prove its clinical efficacy. Moreover, our study highlights the interaction and the pathways concerning the miRNA, immune-expressed and TFs. Such expression data of all these three entities together are not available in COVID-19 patients or in-vitro models, which can establish a better understanding of the mechanisms involved.
5. Conclusion
The present study identifies an in-silico representation of a network involving miRNAs (hsa-miR-106a-5p, hsa-miR-155-5p, hsa-miR-98-5p, hsa-miR-24-3p, hsa -miR-204-5p, hsa-miR-124-3p, hsa-miR-203a-3p, hsa-miR-335-5p, hsa-let-7c-5p, hsa-miR-1-3p), CRIEGs (CCL2, CCL4, CXCL10, CXCL8, IL6, IL7, JAK2, TNF), and TF (AHR, CREM, DDIT3, E2F1, EGR1, EP300, ESR1, ETS2, HDAC1, HDAC2, IRF1, JUN, KLF4, NFAT5, NFKB1, NFKBIA, REL, RUNX1, SIRT1, SP100, SP140L, STAT1, XBP1, ZFP36) which take part in the inflammatory response in COVID-19. This study has identified the CRIEGs and miRNA, the interactions between them, which are potentially critical and can be studied further to develop targeted therapeutic strategies. The data can also be used in exploring novel pathways, which occur following SARS-CoV-2 infection. However, the data needs to be experimentally validated in vitro and in vivo.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thanks to Dr.Dipayan Roy (MD Biochemistry) for language editing and review of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mgene.2021.100990.
Appendix A. Supplementary data
Supplementary material
References
- Arboleda J.F., Fernandez G.J., Urcuqui-Inchima S. Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: Implications for the cytokine response. Infect. Genet. Evol. 2019;69:12–21. doi: 10.1016/j.meegid.2018.12.033. [DOI] [PubMed] [Google Scholar]
- Bala S., Tilahun Y., Taha O., Alao H., Kodys K., Catalano D., Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Pro S., Dafonte Imedio A., Santoso C.S., Gan K.A., Sewell J.A., Martinez M., Sereda R., Mehta S., Fuxman Bass J.I. Global landscape of mouse and human cytokine transcriptional regulation. Nucleic Acids Res. 2018;46:9321–9337. doi: 10.1093/nar/gky787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centa A., Fonseca A.S., da Silva Ferreira S.G., Azevedo M.L.V., de Paula C.B.V., Nagashima S., Machado-Souza C., dos Santos Miggiolaro A.F.R., Pellegrino Baena C., de Noronha L., Cavalli L.R. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Phys. Lung Cell. Mol. Phys. 2021;320:L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.P., Choi Y., Schork N.J. Conserved genomic terminals of SARS-CoV-2 as coevolving functional elements and potential therapeutic targets. mSphere. 2020;5 doi: 10.1128/mSphere.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020:gkaa467. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., Jaggi M., Chauhan S.C., Yallapu M.M. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2020;1–9 doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Wei C., Lee J. Micro RNA -let-7c suppresses hepatitis C virus replication by targeting Bach1 for induction of haem oxygenase-1 expression. J. Viral Hepat. 2019;26:655–665. doi: 10.1111/jvh.13072. [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang Xiaoyun, Chen H., Yu H., Zhang Xiaoping, Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Lan Y., Yuan X., Deng Xilong, Li Y., Cai X., Li L., He R., Tan Y., Deng Xizi, Gao M., Tang G., Zhao L., Wang J., Fan Q., Wen C., Tong Y., Tang Y., Hu F., Li F., Tang X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microb. Infect. 2020;9:469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J.-M. A putative role of de-Mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12:646. doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., Hochman B.R., Salazar-Schicchi J., Yip N.H., Brodie D., O’Donnell M.R. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., Paolini A., Menozzi M., Milić J., Franceschi G., Fantini R., Tonelli R., Sita M., Sarti M., Trenti T., Brugioni L., Cicchetti L., Facchinetti F., Pietrangelo A., Clini E., Girardis M., Guaraldi G., Mussini C., Cossarizza A. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R., Soni K., Saravanan S., Balakrishnan P., Kumar V., Boobalan J., Solomon S.S., Scaria V., Solomon S., Brahmachari S.K., Pillai B. Anti-HIV microRNA expression in a novel Indian cohort. Sci. Rep. 2016;6:28279. doi: 10.1038/srep28279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhorne-Pollet S., Crisci E., Mach N., Renson P., Jaffrézic F., Marot G., Maroilley T., Moroldo M., Lecardonnel J., Blanc F., Bertho N., Bourry O., Giuffra E. The miRNA-targeted transcriptome of porcine alveolar macrophages upon infection with porcine reproductive and respiratory syndrome virus. Sci. Rep. 2019;9:3160. doi: 10.1038/s41598-019-39220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey L.L., Worne C.L., Glover J.L., Lane T.E., O’Connell R.M. MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J. Neuroinflammation. 2016;13:240. doi: 10.1186/s12974-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza J.A., González P.A., Kalergis A.M. Modulation of antiviral immunity by heme oxygenase-1. Am. J. Pathol. 2017;187:487–493. doi: 10.1016/j.ajpath.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Fani M., Zandi M., Ebrahimi S., Soltani S., Abbasi S. The role of miRNAs in COVID-19 disease. Futur. Virol. 2021;16:301–306. doi: 10.2217/fvl-2020-0389. [DOI] [Google Scholar]
- Gadwal A., Roy D., Khokhar M., Modi A., Sharma P., Purohit P. CRISPR/Cas-new molecular scissors in diagnostics and therapeutics of COVID-19. Indian J. Clin. Biochem. 2021 doi: 10.1007/s12291-021-00977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Ai J., Xie Z., Zhou C., Liu C., Zhang H., Shen K. Dynamic expression of viral and cellular microRNAs in infectious mononucleosis caused by primary Epstein-Barr virus infection in children. Virol. J. 2015;12:208. doi: 10.1186/s12985-015-0441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavelli S., De Rosa V., de Candia P. The multifaceted interface between cytokines and microRNAs: An ancient mechanism to regulate the good and the bad of inflammation. Front. Immunol. 2018;9:3012. doi: 10.3389/fimmu.2018.03012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., Sonnenschein K., Pink I., Hoeper M.M., Welte T., Bauersachs J., David S., Bär C., Thum T. Circulating cardiovascular microRNAs in critically ill COVID -19 patients. Eur. J. Heart Fail. 2021;23:468–475. doi: 10.1002/ejhf.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z., Mallick B., Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2008;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterres A., de Azeredo Lima C.H., Miranda R.L., Gadelha M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkoska J., LaRocco M., Ramirez-Medina E., de los Santos T., Lawrence P. Host microRNA-203a Is antagonistic to the progression of foot-and-mouth disease virus infection. Virology. 2017;504:52–62. doi: 10.1016/j.virol.2017.01.019. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.-A., Smith N., Merkling S., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients (preprint) Infect. Dis. (Except HIV/AIDS). 2020 doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajifathalian K., Kumar S., Newberry C., Shah S., Fortune B., Krisko T., Ortiz-Pujols S., Zhou X.K., Dannenberg A.J., Kumar R., Sharaiha R.Z. Obesity is associated with worse outcomes in COVID-19: analysis of early data from New York City. Obesity. 2020 doi: 10.1002/oby.22923. oby.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., Schreiner P., Neale G., Vogel P., Webby R., Jonsson C.B., Kanneganti T.-D. COVID-19 cytokines and the hyperactive immune response: Synergism of TNF-α and IFN-γ in triggering inflammation, tissue damage, and death (preprint) Immunology. 2020 doi: 10.1101/2020.10.29.361048. [DOI] [Google Scholar]
- Kern F., Fehlmann T., Solomon J., Schwed L., Grammes N., Backes C., Van Keuren-Jensen K., Craig D.W., Meese E., Keller A. miEAA 2.0: integrating multi-species microRNA enrichment analysis and workflow management systems. Nucleic Acids Res. 2020;48:W521–W528. doi: 10.1093/nar/gkaa309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhar M., Purohit P., Roy D., Tomo S., Gadwal A., Modi A., Banerjee M., Sharma P. Acute kidney injury in COVID 19 – an update on pathophysiology and management modalities. Arch. Physiol. Biochem. 2020;1–14 doi: 10.1080/13813455.2020.1856141. [DOI] [PubMed] [Google Scholar]
- Khokhar M., Roy D., Purohit P., Goyal M., Setia P. Viricidal treatments for prevention of coronavirus infection. Pathog. Global Health. 2020;1–11 doi: 10.1080/20477724.2020.1807177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Xie J., Che D., Zhang C., Lin Y., Feng L., Chen Jinlu, Chen Jie, Chen L., Wu Z. MiR-124-3p helps to protect against acute respiratory distress syndrome by targeting p65. Biosci. Rep. 2020;40 doi: 10.1042/BSR20192132. BSR20192132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics. 2019;20:545. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Li J., Chen D., Gao R., Zeng W., Chen S., Huang Y., Huang J., Long W., Li M., Guo L., Wang X., Wu X. Dynamic interleukin-6 level changes as a prognostic indicator in patients With COVID-19. Front. Pharmacol. 2020;11:1093. doi: 10.3389/fphar.2020.01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zhang H., Zhan M., Jiang J., Yin H., Dauphars D.J., Li S.-Y., Li Y., He Y.-W. Preventing mortality in COVID-19 patients: which cytokine to target in a raging storm? Front. Cell Dev. Biol. 2020;8:677. doi: 10.3389/fcell.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Li Y.-X., Jiang L.-J., Chen Q., Wang T., Ye D.-W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesh G., Biswas R. MicroRNA-155: A master regulator of inflammation. J. Interf. Cytokine Res. 2019;39:321–330. doi: 10.1089/jir.2018.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams R.M., Bierle C.J., Boldenow E., Weed S., Tsai J., Beyer R.P., MacDonald J.W., Bammler T.K., Liggitt H.D., Farin F.M., Vanderhoeven J., Rajagopal L., Adams Waldorf K.M. Choriodecidual group B streptococcal infection induces miR-155-5p in the fetal lung in Macaca nemestrina. Infect. Immun. 2015;83:3909–3917. doi: 10.1128/IAI.00695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Akbar I., Bhagat R., Hazra B., Bhattacharyya A., Seth P., Roy D., Basu A. Identification and classification of hubs in microRNA target gene networks in human neural stem/progenitor cells following Japanese encephalitis virus Infection. mSphere. 2019;4 doi: 10.1128/mSphere.00588-19. e00588-19, /msphere/4/5/mSphere588-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E., Low J.G.H. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–882.e2. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D., Morris J.H., Bouças J., Pico A.R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saçar Demirci M.D., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8 doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Lu J., Wei J., Zhao J., Wang Meihui, Wang Ming, Shen X., Lü X., Zhou B., Zhao Y., Fu G. Investigation of the underlying hub genes and mechanisms of reperfusion injury in patients undergoing coronary artery bypass graft surgery by integrated bioinformatic analyses. Ann. Transl. Med. 2019;7 doi: 10.21037/atm.2019.10.43. 664–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Li K., Xu K., Liu Q.-H. MiR-155-5p accelerates cerebral ischemia-reperfusion injury via targeting DUSP14 by regulating NF-κB and MAPKs signaling pathways. Eur. Rev. Med. Pharmacol. Sci. 2020;24:1408–1419. doi: 10.26355/eurrev_202002_20198. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Dixit A.B., Paul D., Tripathi M., Sarkar C., Chandra P.S., Banerjee J. Comparative analysis of cytokine/chemokine regulatory networks in patients with hippocampal sclerosis (HS) and focal cortical dysplasia (FCD) Sci. Rep. 2017;7:15904. doi: 10.1038/s41598-017-16041-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium, de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., Åström G., Babina M., Bertin N., Burroughs A.M., Carlisle A.J., Daub C.O., Detmar M., Deviatiiarov R., Fort A., Gebhard C., Goldowitz D., Guhl S., Ha T.J., Harshbarger J., Hasegawa A., Hashimoto K., Herlyn M., Heutink P., Hitchens K.J., Hon C.C., Huang E., Ishizu Y., Kai C., Kasukawa T., Klinken P., Lassmann T., Lecellier C.-H., Lee W., Lizio M., Makeev V., Mathelier A., Medvedeva Y.A., Mejhert N., Mungall C.J., Noma S., Ohshima M., Okada-Hatakeyama M., Persson H., Rizzu P., Roudnicky F., Sætrom P., Sato H., Severin J., Shin J.W., Swoboda R.K., Tarui H., Toyoda H., Vitting-Seerup K., Winteringham L., Yamaguchi Y., Yasuzawa K., Yoneda M., Yumoto N., Zabierowski S., Zhang P.G., Wells C.A., Summers K.M., Kawaji H., Sandelin A., Rehli M., Hayashizaki Y., Carninci P., Forrest A.R.R., de Hoon M.J.L. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017;35:872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M.A., Licht J.D., Pena J.T.R., Rouhanifard S.H., Muckenthaler M.U., Tuschl T., Martin G.R., Bauersachs J., Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a MicroRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Quiat D., Johnson B.A., Sutherland L.B., Qi X., Richardson J.A., Kelm R.J., Olson E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cai J., Zeng X., Chen Y., Yan W., Ouyang Y., Xiao D., Zeng Z., Huang L., Liu A. Downregulation of toll-like receptor 4 induces suppressive effects on hepatitis B virus-related hepatocellular carcinoma via ERK1/2 signaling. BMC Cancer. 2015;15:821. doi: 10.1186/s12885-015-1866-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. JAMA. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-Coding RNA Res. 2020;5:153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods P.S., Doolittle L.M., Rosas L.E., Nana-Sinkam S.P., Tili E., Davis I.C. Increased expression of microRNA-155-5p by alveolar type II cells contributes to development of lethal ARDS in H1N1 influenza A virus-infected mice. Virology. 2020;545:40–52. doi: 10.1016/j.virol.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Wang X., Ni H., Li N., Zhang A., Liu H., Pu F., Xu L., Gao J., Zhao Q., Mu Y., Wang C., Sun Y., Du T., Xu X., Zhang G., Hiscox J.A., Goodfellow I.G., Zhou E.-M. MicroRNA miR-24-3p promotes porcine reproductive and respiratory syndrome virus replication through suppression of heme oxygenase-1 expression. J. Virol. 2015;89:4494–4503. doi: 10.1128/JVI.02810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Yuan, Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Yingle, Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu M., Liu Z., Tang J. Screening and identification of key regulatory connections and immune cell infiltration characteristics for lung transplant rejection using mucosal biopsies. Int. Immunopharmacol. 2020;87 doi: 10.1016/j.intimp.2020.106827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale IMPACT Team, Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmohammadi A., Yarmohammadi M., Fakhri S., Khan H. Targeting pivotal inflammatory pathways in COVID-19: A mechanistic review. Eur. J. Pharmacol. 2020;173620 doi: 10.1016/j.ejphar.2020.173620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Li Z.-L., Luo D., Huang B., Chen Y.-S., Zhang X., Cui J., Zeng Y., Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T.E., Yoshikawa N., Popov V.L., Galindo C.L., Garner H.R., Peters C.J., Tseng C.-T. Kent. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Li Jing, Li Junfeng, Yang Y., Kang X., Li Y., Wu X., Zhu Q., Zhou Y., Hu Y. Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by targeting DR1. Sci. Rep. 2018;8:6797. doi: 10.1038/s41598-018-25073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., Xu B., Dai Y., Li X., Zhang C., Peng Y., Feng Y., Li A., Hu Z., Xiang H., Ogg G., Ho L.-P., McMichael A., Jin R., Knight J.C., Dong T., Zhang Y. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Chu M., Xu S., Chen X., Liu Y., Wang Z., Zhang F., Han S., Yin J., Peng B., He X., Liu W. Hsa-let-7c-5p augments enterovirus 71 replication through viral subversion of cell signaling in rhabdomyosarcoma cells. Cell Biosci. 2017;7:7. doi: 10.1186/s13578-017-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material