Abstract
Hedysarum polybotrys var. alaschanicum is an important medicinal plant and is widely used in traditional Chinese medicine. The complete chloroplast genome of H. polybotrys var. alaschanicum was assembled from Illumina pair-end sequence reads. The whole chloroplast genome is 122,933 bp in length and encodes a total of 110 genes, including 76 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The overall GC content of the cp genome is 35.3%. A maximum likelihood (ML) phylogenetic analysis revealed that H. polybotrys var. alaschanicum was close to Hedysarum semenovii.
Keywords: Hedysarum polybotrys var. alaschanicum, Chloroplast genome, phylogenetic analysis
Hedysarum polybotrys Hand.-Mazz. var. alaschanicum (B. Fedtsch.) H. C. Fu et Z. Y. Chu, a perennial leguminous herb, is widely used in traditional Chinese medicine to strengthen the body (Hu et al. 2010; Dong et al. 2013). The genus Hedysarum L. is economically and medicinally important and widely distributed in the northern hemisphere (Choi and Ohashi 1998). Lots of work has been completed to infer the phylogeny of this genus relying on multilocus (both plastid and nuclear markers) (e.g. Liu et al. 2017; Nafisi et al. 2019), however, the phylogenetic relationship is not well known (Yurkevich et al. 2021). In this study, we report and characterize the complete chloroplast (cp) genome of H. polybotrys var. alaschanicum (GenBank accession number: MZ327727) to provide genomic data for future research.
An individual of H. polybotrys var. alaschanicum was collected in Chunkun Mountain, Baotou, Inner Mongolia, China (E110°44′27″,N 41°17′23″). The specimen was deposited at the herbarium of Baotou Teachers’ College (contact person: Yang Yanci, email: yycjyl@163.com) under the voucher number CKS-2020-07-B06KYYHQ. Total genomic DNA was isolated from silica-dried leaf material using modified CTAB method (Doyle 1987), and then sequenced using an Illumina Hiseq 2500 platform at Biomarker Technologies Inc. (Beijing, China). A total of 707,801 reads were assembled to generate the chloroplast (cp) genome of H. polybotrys var. alaschanicum. Reference-guided assembly was used to reconstruct the cp genomes with the programs MIRA 4.0.2 (Chevreux et al. 2004) and MITObim v1.7 (Hahn et al. 2013). In the process, cp genome of Hedysarum petrovii (MT120797) was used as reference genome. The complete cp genome was annotated in GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand), and then manually corrected by comparing with the published complete cp genomes (GenBank accession numbers: NC_046493, NC_047344) of Hedysarum species using GENEIOUS R11.
The complete cp genome of H. polybotrys var. alaschanicum is 122,933 bp in length. Different from typical quadripartite structure of most angiosperm cp genomes, the cp genome of this species lacks inverted repeat region. The cp genome encodes a total of 110 genes, consisting of 76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The overall GC content of the cp genome is 35.3%.
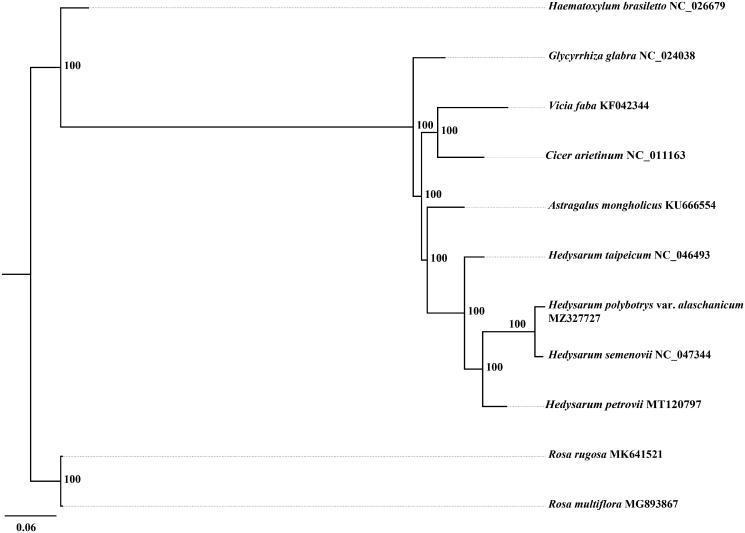
The phylogenetic tree was constructed based on nine complete cp genomes of Fabaceae plants and two complete cp genomes of Rosaceae plants (outgroups). All of the 11 complete cp genome sequences were aligned using MAFFT (Katoh and Standley 2013) with default parameter. To reconstruct the phylogenetic relations of these species we used the GTR + G model with maximum likelihood (ML) analysis. A ML tree was constructed with RAxML v8 (Stamatakis 2014). The branch support values were assessed using a rapid bootstrapping analysis with 1000 replicates. The phylogenetic tree indicated that H. polybotrys var. alaschanicum was close to Hedysarum semenovii with high support (Figure 1). The cp genome of H. polybotrys var. alaschanicum will provide useful genetic information for further study on genetic diversity and phylogeny of Hedysarum species.
Figure 1.
Maximum-likelihood phylogenetic tree for H. polybotrys var. alaschanicum based on 11 complete chloroplast genome sequences. The number on each node indicates the bootstrap value.
Funding Statement
This research was financially supported by the Scientific Research Project of Higher Education Institutions of Inner Mongolia under Grant number NJZZ20360 and the Initial Scientific Research Fund of Baotou Teachers’ College under Grant number BTTCRCQD2018-01.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ327727. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA749549, SRP329995, and SAMN20376023, SAMN20376024, respectively.
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S.. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH, Ohashi H.. 1998. Proposal to conserve the name Hedysarum (Leguminosae: Papilionoideae) with a conserved type. Taxon. 47(4):877–877. [Google Scholar]
- Dong YM, Tang DY, Zhang N, Li Y, Zhang CH, Li L, Li MH.. 2013. Phytochemicals and biological studies of plants in genus Hedysarum. Chem Cent J. 7(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FD, Li XD, Zhao LG, Feng SL, Wang CM.. 2010. Antidiabetic properties of purified polysaccharide from Hedysarum polybotrys. Can J Physiol Pharmacol. 88(1):64–72. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PL, Wen J, Duan L, Arslan E, Ertuğrul K, Chang ZY.. 2017. Hedysarum L. (Fabaceae: Hedysareae) is not monophyletic – evidence from phylogenetic analyses based on five nuclear and five plastid sequences. PLoS One. 12(1):e0170596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi H, Kazempour-Osaloo S, Mozaffarian V, Schneeweiss GM.. 2019. Molecular phylogeny and divergence times of the genus Hedysarum (Fabaceae) with special reference to section Multicaulia in Southwest Asia. Plant Syst Evol. 305(10):1001–1017. [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkevich OY, Samatadze TE, Selyutina IY, Romashkina SI, Zoshchuk SA, Amosova AV, Muravenko OV.. 2021. Molecular cytogenetics of eurasian species of the genus Hedysarum L. (Fabaceae). Plants. 10(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ327727. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA749549, SRP329995, and SAMN20376023, SAMN20376024, respectively.