Abstract
Herein, a novel molecularly imprinted polymer (MIP) based electrochemical sensor for the determination of the receptor-binding domain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2-RBD) has been developed. For this purpose, first, a macroporous gold screen-printed electrode (MP-Au-SPE) has been fabricated. The MIP was then synthesized on the surface of the MP-Au-SPE through the electro-polymerization of ortho-phenylenediamine in the presence of SARS-CoV-2-RBD molecules as matrix polymer, and template molecules, respectively. During the fabrication process, the SARS-CoV-2-RBD molecules were embedded in the polymer matrix. Subsequently, the template molecules were removed from the electrode by using alkaline ethanol. The template molecules removal was studied using cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), scanning electron microscope (SEM), energy-dispersive X-ray spectroscopy (EDX), and attenuated total reflectance spectroscopy (ATR). The fabricated MIP film acted as an artificial recognition element for the measurement of SARS-CoV-2-RBD. The EIS technique was used for the measurement of the SARS-CoV-2-RBD in the saliva solution. The electron transfer resistance (Ret) of the MIP-based sensor in a ferri/ferrocyanide solution increased as the SARS-CoV-2-RBD concentration increased due to the occupation of the imprinted cavities by the SARS-CoV-2-RBD. The MIP-based sensor exhibited a good response to the SARS-CoV-2-RBD in the concentration range between 2.0 and 40.0 pg mL-1 with a limit of detection of 0.7 pg mL-1. The obtained results showed that the fabricated MIP sensor has high selectivity sensitivity, and stability.
Keywords: Molecularly imprinted polymer, SARS-CoV-2-RBD, Macroporous gold screen-printed electrode, Electrochemical sensor
Graphical abstract
1. Introduction
Nowadays, the rapid spread of coronavirus disease 2019 (COVID-19) is causing public health and economic crises worldwide. International Monetary Fund estimates that the crisis from COVID-19 can lead to a global loss of $28 trillion by 2025 (Popoola, 2020). Therefore, researchers need to find a way to reduce the global cost of COVID-19. The fabrication of cheap and fast response diagnostic tools can be one of these attempts.
Current methods applied to determine COVID-19 rely on the interaction of a bio-recognizer element such as antibodies (Mojsoska et al., 2021; Seo et al., 2020; Yakoh et al., 2021), and aptamer (Song et al., 2020; Tabrizi et al., 2021) with a biomolecule related to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The short fragment of the nucleotide chains (Morales-Narváez and Dincer, 2020), the spike protein (Zhang et al., 2021), the SARS-CoV-2-nucleo-protein (Tian et al., 2021; Zhang et al., 2020), and the SARS-CoV-2-receptor binding domain (SARS-CoV-2-RBD) (Suh et al., 2021) are the most relevant biomarkers related to the SARS-CoV-2. Although these diagnostic tools are sensitive and selective, however, they have several drawbacks such as short shelf-life, high cost, and should be stored in a refrigerator.
Therefore, it is a crucial demand to fabricate a cheap, selective, sensitive, and portable diagnostic tool to facilitate rapid testing for COVID-19.
In recent decades, several research groups have worked on the fabrication of the molecularly imprinted polymer (MIP) that acts as the artificial recognizer to detect various disease biomarkers such as the SARS-CoV-2-nucleo-protein (Raziq et al., 2021), Troponin T (Karimian et al., 2013), epithelial ovarian cancer antigen (Wang et al., 2010), the dual detection of epidermal growth factor receptor and vascular endothelial growth factor (Johari-Ahar et al., 2018), dual detection of prostate-specific antigen and myoglobin (Karami et al., 2019), and cancer biomarker-prostate specific antigen (Jolly et al., 2016).
During the fabrication of a MIP, the target molecule acting as a template is trapped in the polymer matrix during the polymerization of monomers. As the template molecule is removed from the structure of the polymer, several cavities (binding sites) are generated in the polymer. The binding sites are complementary to the target molecule in size, shape, and orientation of functional groups. Then, the fabricated MIP can selectively rebind with its target (Lowdon et al., 2020).
The combination of the MIP with a transducer allowed us to develop an advanced sensing device for the detection of biomarkers related to Covid-19.
Among the various transducers that have been used for the fabrication of MIP-based sensors, the electrochemical transducer has several advantages such as portability, high sensitivity, low cost, easiness to use, and integrability with nano/macro materials (Karimian et al., 2020; Khanmohammadi et al., 2020; Lahcen and Amine, 2019; Tajik et al., 2019).
Up to now, several nano/macro materials have been applied for the fabrication of electrochemical MIP-based sensors such as gold nanoparticles (Motia et al., 2020), magnetic nanoparticles (Yang et al., 2020), graphene (Liang et al., 2017; Liu et al., 2019), carbon nanotubes (Meng et al., 2019), and Three-dimensional-ensembles of gold nanowires (Beluomini et al., 2019).
Three-dimensional electroactive materials that have porous structures are good candidates for the fabrication of electrochemical-based sensors with high analytical performances because of their high electroactive surface area, conductivity, chemical, and thermal stability (Lang et al., 2013; Ma et al., 2021; Qiu et al., 2014). In this sense, the use of porous gold has recently become a favored material to be incorporated in the electrode because of its easy fabrication, and high analytical performance for the detection of the wide range of biomolecules (Rong et al., 2018; Silva et al., 2019; Sukeri et al., 2015; Xiao et al., 2016).
To the best of our knowledge, the use of a macroporous gold screen-printed electrode (MP-Au-SPE) for the fabrication of a MIP-based electrochemical sensor has not been reported yet. In this research work, the SARS-CoV-2-RBD was selected as a model analyte for the MIP-based measurement. The SARS-CoV-2-RBD is a part of the spike protein that binds to the angiotensin-converting enzyme-2 on the membrane of cells to gain entry into the cell, leading to infection (Lan et al., 2020).
For this purpose, first, we have converted the gold screen-printed electrode (Au-SPE) to the MP-Au-SPE using two simple electrochemical and chemical steps. After that, the MIP polymer was synthesized on the surface of the MP-Au-SPE by electrochemical polymerization of ortho-phenylenediamine (oPD) in the presence of SARS-CoV-2-RBD as the matrix polymer and the template molecule, respectively. The obtained results showed that the proposed MIP/MP-Au-SPE for measurement of the SARS-CoV-2-RBD in human saliva samples has high sensitivity, selectivity, reproducibility, and fast response time.
2. Experimental section
2.1. Reagents and chemicals
All chemicals were of analytical reagent grade and used without further purification. Double deionized (DI) water (18.6 MΩ) was used throughout the research work. Phosphoric acid (H3PO4), oPD, potassium hydroxide (KOH), potassium ferricyanide (Fe(CN)6 3−), potassium ferrocyanide (Fe(CN)6 4−), and ascorbic acid (AA) were obtained from Alfa Aesar. SARS-CoV-2-RBD (∼35 kDa), human serum albumin (HSA), human immunoglobulin A (HIgA), and human immunoglobulin G (HIgG) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Human SARS-CoV-2-RBD ELISA Kit was obtained from Thermo Fisher Scientific. The Au-SPEs (Ref 250 AT) have been obtained from Metrohm-DropSens.
2.2. Preparation of the MP-Au-SPE
The MP-Au-SPE was prepared in three steps: cleaning the Au-SPE, electrochemical oxidation of Au-SPE, and chemical reduction. The Au-SPE was first washed with ethanol/DI water (50:50) and then cleaned with piranha solution to remove all the organic components on the surface of the Au-SPE electrode. After that, the Au-SPE was washed with DI water several times. Subsequently, the Au-SPE was immersed in 2 M H2SO4 and a voltage of +1.85 V was applied for 200 s to oxidize the working electrode of the Au-SPE. The amperogram of the fabrication process is shown in Fig. S1. As it can be seen, in the beginning, the intensity of the current was high but after 130 s, the current dropped to 120 μA, dramatically. During the oxidation process, the color of the working electrode changed from yellow to brown (Fig. S2). After that, the electrode was washed with DI water several times and subsequently 100 μL of 0.1 M AA (a reducing agent) was dropped on the surface of the oxidized working gold electrode. The color of the working gold electrode was changed immediately from brown to black. The fabricated MP-Au-SPE was finally washed with DI water. The cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), and the related electrochemical properties of the Au-SPE and MP-Au-SPE are included in the supplementary data section. The roughness factor (RF) for the MP-Au-SPE and Au-SPE were found to be 49.28, and 0.77, respectively. The heterogeneous electron transfer constant (k0) for the Au-SPE and MP-Au-SPE in a ferri/ferrocyanide (Fe(CN)6 3−/4−) solution were also found to be 0.058 cm s-1 and 0.39 cm s-1. The obtained results indicated that the electrochemical properties of the MP-Au-SPE were significantly better than Au-SPE (Fig. S3, Fig. S4, and Fig. S5).
The surface roughness of the Au-SPE and MP-Au-SPE were also studied with atomic force microscopy (AFM) (Fig. S6). As it can be seen, the surface of the MP-Au-SPE was porous in comparison with the Au-SPE. The statistical analysis of the data obtained from the AFM images are summarized in Table S1. The results indicated that the maximum peak to valley height (Rt) of the roughness profile and the root mean square deviation (Rq) for the Au-SPE were 10.7 nm and 4.2 nm, respectively. In contrast, these values for the MP-Au-SPE were 30.6 nm and 12.6 nm.
2.2.1. Preparation of the MIP electrode
The MIP/MP-Au-SPE was prepared in two steps: electrochemical fabrication of the MIP, removal of the molecular templates from the MIP.
To fabricate the MIP-based sensor, the MP-Au-SPE was first immersed in an acetate buffer (0.5 M, pH 5.2) containing 7.5 mM oPD and 1.5 μg mL-1 of SARS-CoV-2-RBD. The electro-polymerization of oPD on the MP-Au-SPE was carried out by the CV method by sweeping the potential 20 times between -0.2 V and 1.0 V with a scan rate of 50 mV s-1 (Fig. S7). During the oPD polymerization process to poly-oPD (PoPD), the SARS-CoV-2-RBD template molecules were entrapped into the polymer. After that, the electrode was washed with acetate buffer (0.2 mol.L-1, pH = 4.8) to remove loosely attached non-polymerized oPD. The MIP-based electrode was then washed with alkaline ethanol (ethanol/0.25 M NaOH, (2:1 v/v)) for 15 min at 50 °C, under stirring to remove the SARS-CoV-2-RBD template molecules. During this process, several functional monomers were orderly assembled with their functional groups, preparing fitting sites in the cavities. Finally, the sensor was washed with 0.1 M phosphate-buffered saline (PBS, pH 7.4) and stored at room temperature.
Fig. S8 shows the schematic fabrication of the MIP/MP-Au-SPE.
The non-imprinted polymer (NIP) sensor (NIP/MP-Au-SPE) was fabricated in the same condition that was used for the MIP/MP-Au-SPE but the template molecule (SARS-CoV-2-RBD) was not added to the solution. The NIP/MP-Au-SPE is the control electrode to examine the accuracy of the analytical performance of the MIP/MP-Au-SPE.
2.3. The response mechanism of the MIP/MP-Au-SPE
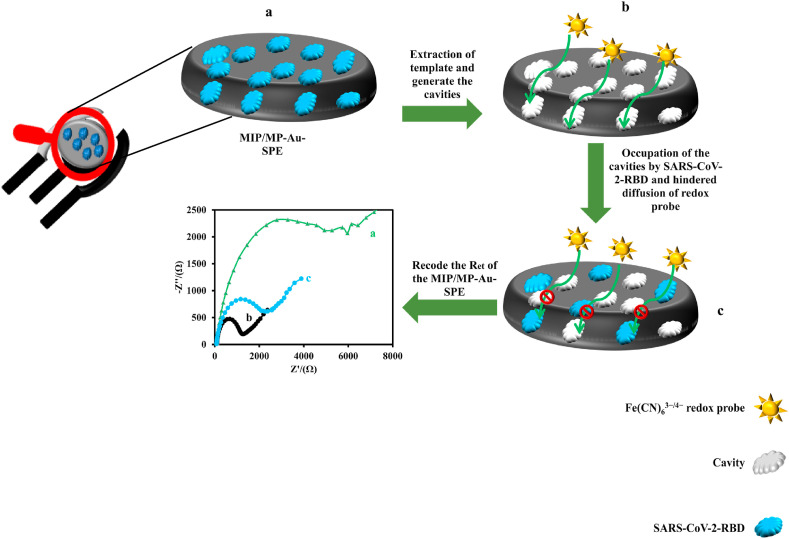
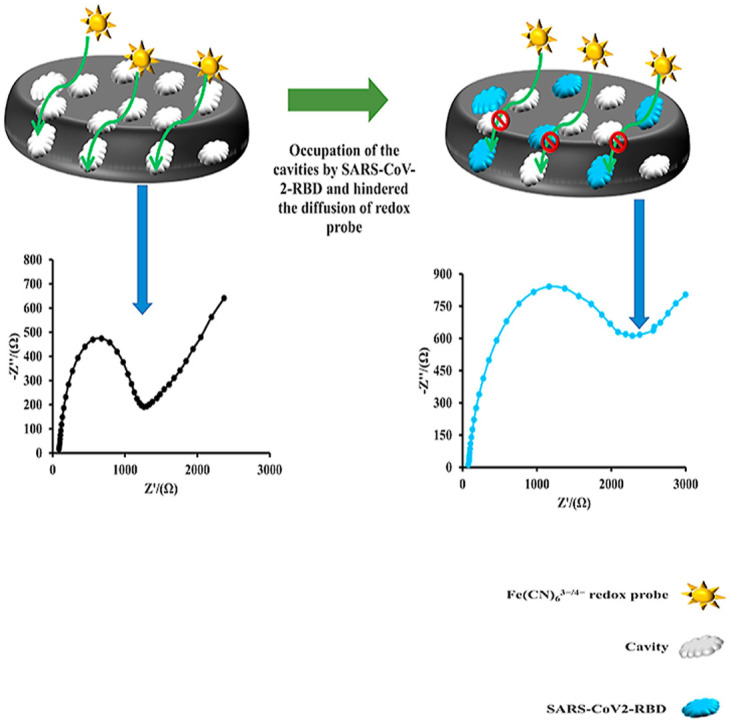
Fig. 1 presents the schematic diagram of the response mechanism of the proposed MIP-based electrochemical sensor. As the SARS-CoV-2-RBD (template molecule) was removed from the MIP, several specific polymer-based cavities (binding sites) were generated for the guest analyte. Therefore, these imprinted binding sites are complementary to the SARS-CoV-2-RBD molecule in shape, size, and chemical functionality. Since there were several cavities on the surface of the electrode, the Fe(CN)6 3−/4− redox probe could pass through the polymer matrix and reach the surface of the MP-Au-SPE. Therefore, the electron transfer resistance (Ret) of the electrode before the incubation of the guest molecules was low. When the SARS-CoV-2-RBD solution was dropped on the surface of the MIP/MP-Au-SPE, the SARS-CoV-2-RBD molecules were selectively bound to the binding sites and occupied them. Therefore, the permeability of the MIP to the Fe(CN)6 3−/4− probe decreased. Hence, the Ret of the MIP/MP-Au-SPE increased. The increase in the Ret of the electrode had a linear logarithmic relationship with the SARS-CoV-2-RBD concentration.
Fig. 1.
Schematic illustration of the response mechanism of the MIP sensor to the SARS-CoV-2-RBD.
2.4. Measurement process
During the measurement process, 50.0 μL of saliva sample was mixed with 50.0 μL of phosphate-buffered saline (PBS, 0.2 M, pH 7.4) containing a fixed concentration of the SARS-CoV-2-RBD. The mixture was then dropped on the surface of the MIP/MP-Au-SPE to react for 20 min. After that, the electrode was rinsed thoroughly with 0.1 M PBS. Finally, 100 μL of Fe(CN)6 3−/4− solution (0.1 M PBS, pH 7.4) was then dropped on the MIP/MP-Au-SPE surface to record the signal.
3. Experimental
3.1. Morphology characterization of the electrodes
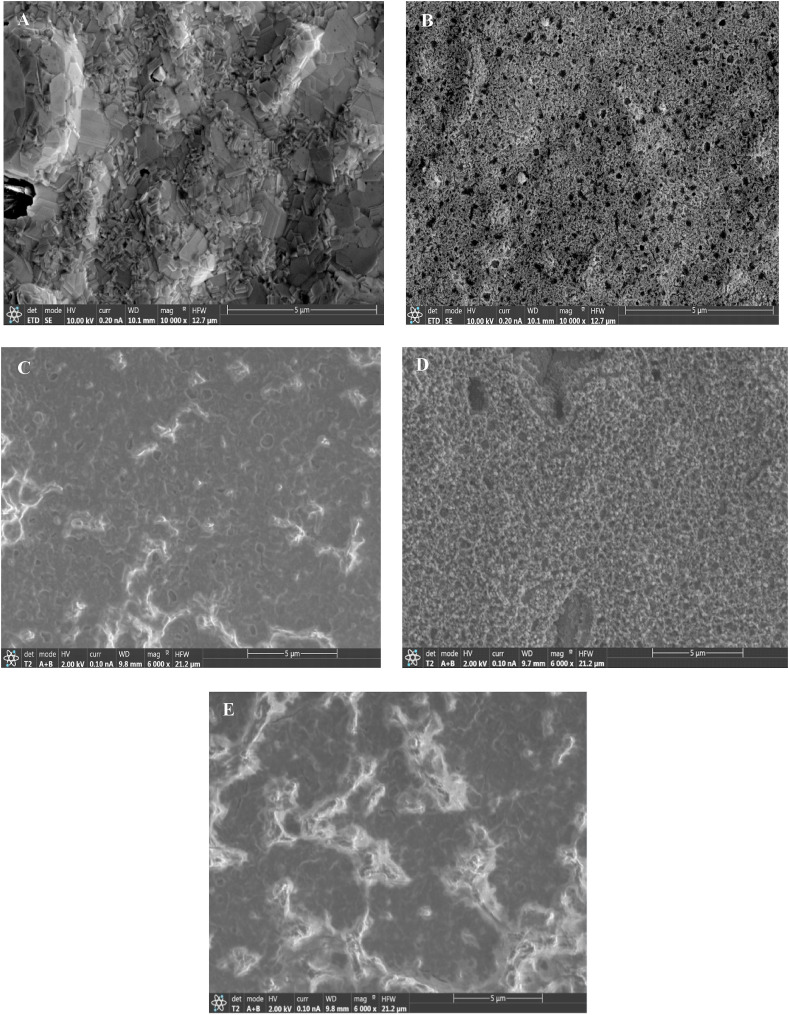
Fig. 2 shows the SEM images of an Au-SPE (A), a MP-Au-SPE (B), a MIP/MP-Au-SPE before template removal (C), a MIP/MP-Au-SPE after template removal (D), and a NIP/MP-Au-SPE (E), respectively. As it can be seen, the surface morphology of the Au-SPE changed dramatically after the electrochemical oxidation and chemical reduction treatment. The MP-Au-SPE has a three-dimensional macroporous structure compared to the Au-SPE. After the MIP fabrication, the morphology of the electrode surface changed. As it can be seen, the surface of the porous electrode was coated with a layer of polymer (Fig. 2C). After the extraction of the template molecules (SARS-CoV-2-RBD) (Fig. 2D), the porosity of the electrode increased. The reasonable explanation is that the elution of the template molecules formed several cavities on the structure of the polymer. The SEM image of the NIP/MP-Au-SPE is shown in Fig. 2E. As it can be seen, the surface morphology of the NIP/MP-Au-SPE was similar to that of the MIP/MP-Au-SPE before the template extraction because the same monomer and method were used for the fabrication of them.
Fig. 2.
SEM images of a Au-SPE (A), a MP-Au-SPE (B), a MIP/MP-Au-SPE before removal of the SARS-CoV-2-RBD (C), a MIP/MP-Au-SPE after removal of the SARS-CoV-2-RBD (D), and a NIP/MP-Au-SPE (E).
The energy-dispersive X-ray spectroscopy (EDX) and attenuated total reflectance spectroscopy (ATR) analysis of a MIP/MP-Au-SPE before (a) and after (b) the extraction of template molecules are shown in Fig. S9. The results indicated that the template molecule was extracted from the MIP film, successfully. The details are mentioned in the supplementary data section.
3.2. Electrochemical behavior of the MIP/MP-Au-SPE
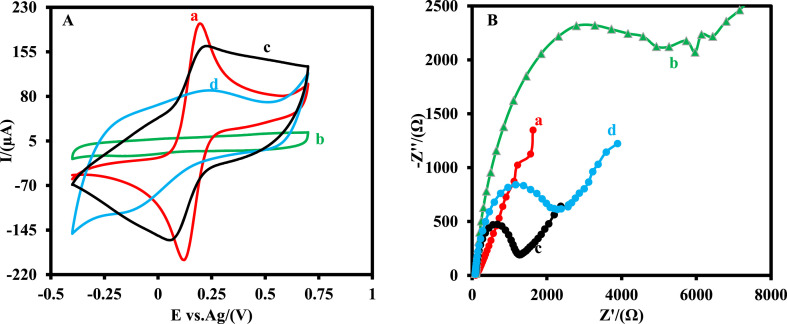
The EIS and CV methods were applied to study the step-by-step fabrication process of the MIP/MP-Au-SPE in 0.5 mM Fe(CN)6 3−/4− solution (0.1M PBS, pH 7.4) (Fig. 3 ).
Fig. 3.
(A) CV and (B) EIS measurements of the MP-Au-SPE (a), the MIP/MP-Au-SPE before removal of SARS-CoV-2-RBD (b), the MIP/MP-Au-SPE after removal of the SARS-CoV-2-RBD (c), the MIP/MP-Au-SPE after the occupation the cavities with 10.0 pg mL-1 SARS-CoV-2-RBD (d). CV measurements were done at a scan rate of 0.05 V s−1 in a 0.5 mM Fe(CN)63−/4− solution. EIS measurements were done by applying an alternating current (AC) potential of 10 mV, and a direct current (DC) potential of 0.15 V in the frequency range of 100 kHz-0.1 Hz in a 0.5 mM Fe(CN)63−/4− solution.
Fig. 3A shows the CV of the MP-Au-SPE (a), the MIP/MP-Au-SPE before extraction of the template molecules (b), the MIP/MP-Au-SPE after extraction of the template molecules (c), and the MIP/MP-Au-SPE after the incubation with 10.0 pg mL-1 SARS-CoV-2-RBD (d). As it can be seen, a couple of well-defined and reversible redox peaks (anodic peak current (Ipa) = 193.4 μA, cathodic peak current (Ipc) = -190.4 μA, peak-to-peak separation (ΔE) = 0.044 V) were observed for the Fe(CN)6 3−/4− probe at the MP-Au-SPE (a). These redox peaks of the probe disappeared as oPD was polymerized in the presence of SARS-CoV-2-RBD on the MP-Au-SPE surface as a polymer matrix and a template molecule, respectively (b). After the template molecules removal (c), a pair redox peak of the Fe(CN)6 3−/4− probe was observed, indicating that several cavities were generated on the surface of the sensor. Therefore, the Fe(CN)6 3−/4− probe can go through the MIP structure and reach the MP-Au-SPE. However, the recorded signal intensities of the Fe(CN)6 3−/4− probe with the MIP/MP-Au-SPE was lower (Ipa = 158.9 μA, Ipc = -156.3 μA) and ΔE was higher (ΔE = 0.12 V) than those values that were recorded with the MP-Au-SPE due to the high Ret of the polymer matrix (PoPD).
As the fabricated MIP/MP-Au-SPE was incubated with 10.0 pg mL-1 SARS-CoV-2-RBD (d), some of the cavities in the polymer matrix were occupied with SARS-CoV-2-RBD. Therefore, the Fe(CN)6 3−/4− probe could not pass through the MIP and reach the MP-Au-SPE. Hence, the intensity of the signal (Ipa = 89.4 μA, Ipc = -110.7 μA) decreased and an in the ΔE increased (ΔE = 0.37 V).
The electrochemical behavior of the electrodes was also studied with the EIS method. Fig. 3B shows the Nyquist plots of the impedance spectra for the same electrode. It can be seen that the Ret of the MP-Au-SPE dramatically increased from 87 Ω (a) to 8100 Ω (b) as the PoPD was fabricated in the presence of template molecules. After the extraction of the template molecules (c), the Ret of the electrode decreased to 1290 Ω. The Ret of the electrode was increased to 2550 Ω as the electrode was incubated with SARS-CoV-2-RBD (d). The reasonable explanation is that the SARS-CoV-2-RBD re-bound to the MIP and blocked the mass transfer pathways between the MP-Au-SPE and the Fe(CN)6 3−/4− probe.
The electrochemical properties of the NIP/MP-Au-SPE before (a) and after (b) washing with the alkaline ethanol were also studied (Fig. S10). As it can be seen, the CVs (A) and EIS (B) of the NIP/MP-Au-SPE did not change. The reasonable explanation is that the cavities did not generate in the PoPD structure and hence, the Fe(CN)6 3−/4− probe could not penetrate into the PoPD. Due to the high ability of the EIS technique to probe the interfacial properties at the electrode surface, further confirmation of the analytical performance of the MIP/MP-Au-SPE was studied with this method.
3.3. Optimization of parameters
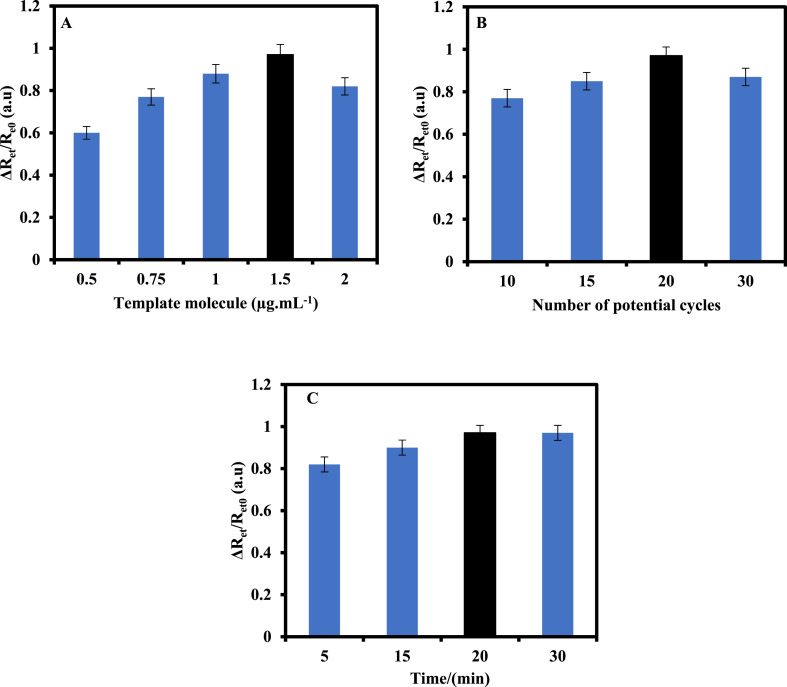
The effect of the template molecule concentration in the response of the sensor was investigated. For this purpose, the MIP film was prepared in the presence of various SARS-CoV-2-RBD concentrations (0.5–2.0 μg mL-1) as template molecules. The fabricated sensor was then used for the measurement of SARS-CoV-2-RBD (10.0 pg mL-1). As can be seen in Fig. 4 A, the response of the MIP/MP-Au-SPE films to SARS-CoV-2-RBD increased as template molecules concentration increased up to 1.5 μg mL-1, suggesting that the number of cavities was increased in the polymer. As the concentration of the template molecule was increased to 2 μg mL-1, the signal of the MIP/MP-Au-SPE to the SARS-CoV-2-RBD decreased due to the surface aggregation of template molecule in the high concentrations of the template molecule, leading to a decrease in the number of cavities of the polymer matrix.
Fig. 4.
(A) Effect of the template molecule concentration (B) the number of potential cycles and (C) the incubation time on the response of MIP/MP-Au-SPE to 10 pg mL-1 SARS-CoV-2-RBD. The standard deviation error bars were obtained by using four different sensors.
The effect of the amount of the deposited polymer on the response of the MIP/MP-Au-SPE was also investigated varying the polymerization cycles from 10 to 30.
As shown in Fig. 4B, the normalized response of the MIP/MP-Au-SPE to 10.0 pg mL-1 SARS-CoV-2-RBD was dependent on the number of potential cycles which is related to the thickness of the polymer matrix. As it can be seen, the response of the MIP sensor to 10.0 pg mL-1 SARS-CoV-2-RBD increased as the number of cycles increased from 10 to 20. During the process, the number of binding sites (cavities) increased, and therefore, the sensitivity of the MIP/MP-Au-SPE increased. However, the response of the MIP-based sensor decreased as the number of cycles increased to 30. The reasonable explanation is that the permeability of the Fe(CN)6 3−/4− to the surface of electrode decreased as the thickness of the non-conductive MIP film increased.
The influence of the incubation time between target molecules (SARS-CoV-2-RBD) and binding sites (cavities) on the response of the MIP/MP-Au-SPE was also studied (Fig. 4C). The response of the MIP/MP-Au-SPE to 10.0 pg mL-1 SARS-CoV-2-RBD increased as the incubation time increased from 5 to 20 min and then remained unchanged at longer incubation times, suggesting that the formation of SARS-CoV-2-RBD/cavities bind reached a saturation level. Therefore, 1.5 μg mL-1 SARS-CoV-2-RBD as template molecule concentration and 20 times scanning potential were selected for the MIP fabrication as the optimum fabrication conditions. Also, 20 min was applied for the interaction of the target molecule (SARS-CoV-2-RBD) with the MIP/MP-Au-SPE throughout this work as the optimum interaction time.
3.4. Analytical performance of the MIP/MP-Au-SPE
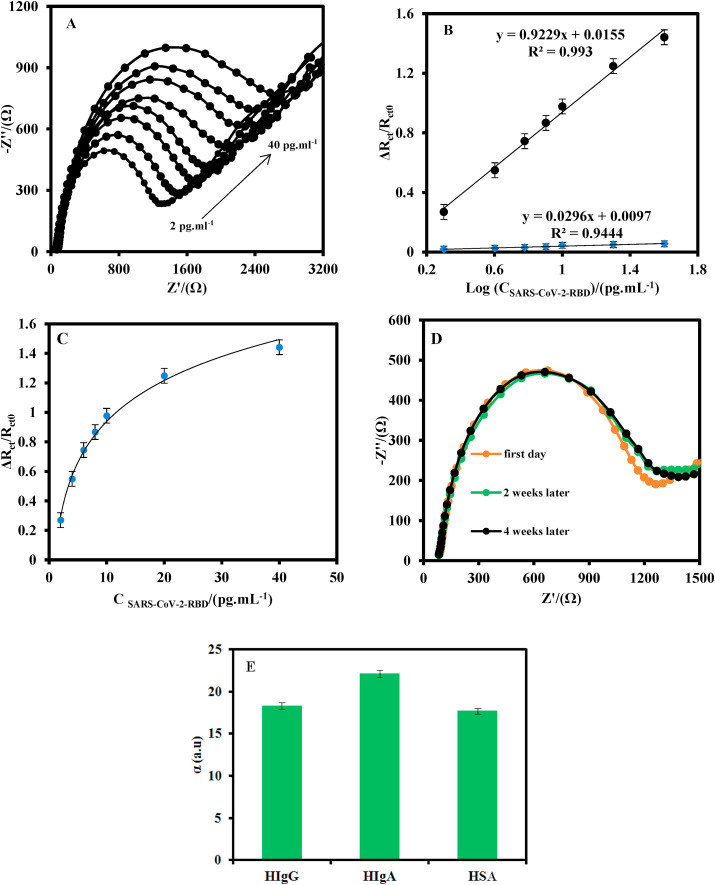
Fig. 5A shows the EIS plots of the MIP/MP-Au-SPE in a 0.5 mM Fe(CN)6 3-/4- solution after the incubation with saliva sample containing different concentrations of SARS-CoV-2-RBD. As it can be seen, the Ret of the MIP sensor increased as the SARS-CoV-2-RBD concentration was increased. The reasonable explanation is that the SARS-CoV-2-RBD re-bound with the cavities in the MIP and hindered the diffusion of Fe(CN)6 3-/4- to the surface of the electrode. Also, Fig. 5B shows the plot of the normalized response (ΔRet = Ret Cx-Ret0)/Ret0 versus the logarithm of the SARS-CoV-2-RBD concentration, where Ret0 and Ret Cx are the electron transfer resistance in the absence of analyte and in the presence of the fixed concentration of analyte, respectively. As shown, the normalized response has a linear relationship with the logarithm of the SARS-CoV-2-RBD concentration in the dynamic linear range of 2.0 to 40.0 pg mL-1 with a regression equation (eq) (1):
| (1) |
Fig. 5.
(A) EIS of the MIP/MP-Au-SPE at the optimum operating conditions for different concentrations of SARS-CoV-2-RBD (2, 4, 6, 8, 10, 20, and 40 pg mL-1) at 5.0 mM Fe(CN)63−/4− solution (0.1 M PBS, pH 7.4). (B) Linear-logarithmic relationship between the normalized signal and SARS-CoV-2-RBD concentration was recorded by MIP/MP-Au-SPE (a) and NIP/MP-Au-SPE (b). (C) Binding isotherm of SARS-CoV-2-RBD in MIP/MP-Au-SPE. (D) The EIS of the MIP/MP-Au-SPE at 5.0 mM Fe(CN)63−/4− solution (0.1 M PBS, pH 7.4) during 4 weeks. (E) Plot of the selectivity factor of the MIP/MP-Au-SPE to the SARS-CoV-2-RBD compare to HIgG, HIgA, and HSA as interfering agents. The standard deviation error bars were obtained by using four different sensors. The EIS measurements were done by applying an AC potential of 10 mV, and a DC potential of 0.15 V in the frequency range of 100 kHz-0.1 Hz.
The limit of detection (LOD) was estimated to be 0.7 pg mL-1 (20 fM) at (3σ/S), where σ is the standard deviation of the absolute values of the Ret of the blank solution for four different MIP/MP-Au-SPE, and S is the slope of the calibration curve. The LOD of the proposed MIP-based sensor is lower than the photoelectrochemical-based aptasensor (Tabrizi et al., 2021), and fluorescence-based aptamer assay (Song et al., 2020).
According to the literature, each SARS-CoV-2 has between 25 and 40 spike proteins on its surface (Highfield, 2020). As each spike protein of the SARS-CoV-2 has an RBD, therefore, the LOD of the proposed MIP based sensor was found to be 3-4.8 × 102 Copies/μL by using eq (2):
| (2) |
The comparisons of the analytical performance of the MIP/MP-Au-SPE with other electrochemical-based MIP sensors for proteins are summarized in Table S1. It can be seen that the analytical performance of the MIP/MP-Au-SPE is better than the other sensor in most cases.
A Langmuir isotherm model (eq (3)) was applied to Fig. 5C to calculate the apparent dissociation constant (KD), and maximum binding capacity (Bmax). In this equation, ΔRet is the normalized signal, C is the SARS-CoV-2-RBD concentration, Ret max is the maximum signal observed and it is equal to the maximum binding capacity (Bmax), and KD is the apparent dissociation constant which it is equal to the SARS-CoV-2-RBD concentration required to provide half of the maximum response produced.(Moreira et al., 2014)
| (3) |
The value of KD and B max were calculated to be 2.7 pg mL-1 and, 3.35 kΩ, respectively. Also, the Gibbs free energy for desorption bind of the MIP/SARS-CoV-2-RBD was established to be -65.96 kJ mol−1 by using eq (4): (Borea et al., 1998)
| (4) |
where R is the universal gas constant of 8.31 J.K−1.mol−1, and T is the temperature in Kelvin (298.15 K).
The stability of the MIP/MP-Au-SPE was investigated (Fig. 5D). The Ret of MIP/MP-Au-SPE changed by approximately 9.7% after 4 weeks, indicating that the proposed sensor had a good stability.
The selectivity of the MIP/MP-Au-SPE was also investigated (Fig. 5E). For this purpose, different MIP/MP-Au-SPE were fabricated to measure 10 pg mL-1 SARS-CoV-2-RBD and the interfering agents (0.5 ng mL-1) such as HIgG, HIgA, and HSA antibodies, each one with a different MIP/MP-Au-SPE. The selectivity factor (α) was estimated by eq (5) (Zhang et al., 2018)
| (5) |
where (ΔRet/Ret0)TM is the normalized response of the MIP/MP-Au-SPE to the template molecule and (ΔRet/Ret0)IA is the normalized response of the MIP/MP-Au-SPE to the interfering agent. To calculate the α, first, the (ΔRet/Ret0)TM of the MIP/MP-Au-SPE to 10.0 pg mL-1 SARS-CoV-2-RBD was measured for one of the MIP/MP-Au-SPE. Then, the (ΔRet/Ret0)IA of the MIP/MP-Au-SPE for the interfering agents (0.5 ng mL-1) using a new electrode for each interfering agent. The values of α for the MIP/MP-Au-SPE to SARS-CoV-2-RBD in the presence of HIgG, HIgA, and HSA antibodies were 18.3, 22.1, and 17.7, respectively. As it can be seen, the values of α for all the electrodes were greater than 1, indicating the high selectivity of the MIP/MP-Au-SPE to SARS-CoV-2RBD. To avoid the memory effect of the interfering agents in the response of MIP/MP-Au-SPE, the electrode was washed thoroughly with 0.1 M PBS after each measurement.
The reproducibility was also evaluated for the determination of 10 pg mL-1 SARS-CoV-2-RBD by using five different MIP-based sensors. The relative standard deviation (RSD) of the response was calculated as 4.3%.
The responses of the proposed MIP-based sensor to two different concentrations of the SARS-CoV-2RBD in saliva samples were compared with the enzyme-linked immunosorbent assay (ELISA) method (Table S3). As it can be seen, there were no statistically significant difference between the responses of the MIP/MP-Au-SPE and those by the ELISA. It indicates that the MIP/MP-Au-SPE can be used as a reliable technique for the determination of SARS-CoV-2RBD in saliva serum samples.
4. Conclusions
In this work, we have designed a MIP-based sensor for the selective detection of the SARS-CoV-2-RBD by using the MP-Au-SPE and PoPD. The MP-Au-SPE that had a larger electroactive surface area and low electron transfer resistance improved the sensitivity to Fe(CN)6 3−/4− probe. Additionally, the fabrication of the MIP that acted as an artificial recognizer membrane improved the selectivity to the SARS-CoV-2-RBD. The EIS and CV methods were used to study both template removal and target attaching on the MIP. As the SARS-CoV-2-RBD re-bounded to cavities, the Fe(CN)6 3−/4− probe could not pass through the MIP and therefore the Ret of the sensor was increased. The proposed sensor was applied to detect the SARS-CoV-2-RBD in the range of 2.0 to 40.0 pg mL-1. The LOD and KD were found to be 0.7 pg mL-1, and 2.68 pg mL-1, respectively. Although the proposed MIP sensor for SARS-CoV-2-RBD has a good potential to be a point-of-care tool due to its easy fabrication, fast response, low cost, good sensitivity and selectivity, disposability, however, it has some disadvantages. First, the EIS-based measurement is an expensive method in comparison with the potentiostatic methods. Second, the proposed sensor cannot be used for the fabrication of the wearable sensor due to not being a flexible sensor.
CRediT authorship contribution statement
Mahmoud Amouzadeh Tabrizi: Supervision, Project administration, Funding acquisition, Conceptualization, Methodology, Data curation, Formal analysis, Validation, Investigation, Visualization, Writing – original draft. Juan P. Fernández-Blázquez: Writing – review & editing. Dahiana Mojena Medina: Writing – review & editing. Pablo Acedo: Supervision, Writing – review & editing, Resources, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 801538. The authors would like to thank the characterization services of the ICTP-CSIC for the AFM measurements of the sample.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113729.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Beluomini M.A., Karimian N., Stradiotto N.R., Ugo P. Sensor. Actuator. B Chem. 2019;284:250–257. [Google Scholar]
- Borea P.A., Varani K., Gessi S., Gilli P., Dalpiaz A. Farmaco. 1998;53(4):249–254. doi: 10.1016/s0014-827x(98)00017-2. [DOI] [PubMed] [Google Scholar]
- Johari-Ahar M., Karami P., Ghanei M., Afkhami A., Bagheri H. Biosens. Bioelectron. 2018;107:26–33. doi: 10.1016/j.bios.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Jolly P., Tamboli V., Harniman R.L., Estrela P., Allender C.J., Bowen J.L. Biosens. Bioelectron. 2016;75:188–195. doi: 10.1016/j.bios.2015.08.043. [DOI] [PubMed] [Google Scholar]
- Karami P., Bagheri H., Johari-Ahar M., Khoshsafar H., Arduini F., Afkhami A. Talanta. 2019;202:111–122. doi: 10.1016/j.talanta.2019.04.061. [DOI] [PubMed] [Google Scholar]
- Karimian, N., Vagin, M., Zavar, M.H.A., Chamsaz, M., Turner, A.P.F., Tiwari, A., 2013. 50, 492-498. [DOI] [PubMed]
- Karimian N., Hashemi P., Khanmohammadi A., Afkhami A., Bagheri H. Anal. Bioanal. Chem. Res. 2020;7(3):281–301. [Google Scholar]
- Khanmohammadi A., Jalili Ghazizadeh A., Hashemi P., Afkhami A., Arduini F., Bagheri H. J. Iran. Chem. Soc. 2020;17(10):2429–2447. [Google Scholar]
- Lahcen A.A., Amine A. Electroanalysis. 2019;31(2):188–201. [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lang X.-Y., Fu H.-Y., Hou C., Han G.-F., Yang P., Liu Y.-B., Jiang Q. Nat. Commun. 2013;4(1):2169–2177. doi: 10.1038/ncomms3169. [DOI] [PubMed] [Google Scholar]
- Liang Y., Yu L., Yang R., Li X., Qu L., Li J. Sensor. Actuator. B Chem. 2017;240:1330–1335. [Google Scholar]
- Liu Y., Liang Y., Yang R., Li J., Qu L. Talanta. 2019;195:691–698. doi: 10.1016/j.talanta.2018.11.052. [DOI] [PubMed] [Google Scholar]
- Lowdon J.W., Diliën H., Singla P., Peeters M., Cleij T.J., van Grinsven B., Eersels K. Sensor. Actuator. B Chem. 2020;325 doi: 10.1016/j.snb.2020.128973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Liu N., Xu Z., Wang J., Luo X. Microchem. J. 2021;161 [Google Scholar]
- Meng X., Xiao Z., Scott S.K. Propellants explos. Pyrotech. 2019;44(10):1337–1346. [Google Scholar]
- Mojsoska B., Larsen S., Olsen D.A., Madsen J.S., Brandslund I., Alatraktchi F.A. Sensors. 2021;21(2):390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. Biosens. Bioelectron. 2020;163 doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira F.T.C., Sharma S., Dutra R.A.F., Noronha J.P.C., Cass A.E.G., Sales M.G.F. Sensor. Actuator. B Chem. 2014;196:123–132. [Google Scholar]
- Motia S., Bouchikhi B., Llobet E. El Bari, N. 2020;216 doi: 10.1016/j.talanta.2020.120953. Talanta. [DOI] [PubMed] [Google Scholar]
- Qiu H.J., Li X., Xu H.-T., Zhang H.-J., Wang Y. J. Mater. Chem. C. 2014;2(46):9788–9799. [Google Scholar]
- Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Biosens. Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong K., Huang L., Zhang H., Zhai J., Fang Y., Dong S. Chem. Commun. 2018;54(64):8853–8856. doi: 10.1039/c8cc04454f. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Silva T.A., Khan M.R.K., Fatibello-Filho O., Collinson M.M. J. Electroanal. Chem. 2019;846 [Google Scholar]
- Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Anal. Chem. 2020;92(14):9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- Suh J.-S., Kim H.-S., Kim T.-J. Sensor. Actuator. B Chem. 2021;334 doi: 10.1016/j.snb.2021.129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukeri A., Saravia L.P.H., Bertotti M. Phys. Chem. Chem. Phys. 2015;17(43):28510–28514. doi: 10.1039/c5cp05220c. [DOI] [PubMed] [Google Scholar]
- Tabrizi M.A., Nazari L., Acedo P. Sensor. Actuator. B Chem. 2021 doi: 10.1016/j.snb.2021.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik S., Beitollahi H., Aflatoonian M.R. J. Electrochem. Sci. Eng. 2019;9(3):187–195. [Google Scholar]
- Tian J., Liang Z., Hu O., He Q., Sun D., Chen Z. Electrochim. Acta. 2021;387 [Google Scholar]
- Wang Y., Zhang Z., Jain V., Yi J., Mueller S., Sokolov J., Liu Z., Levon K., Rigas B., Rafailovich M.H. Sensor. Actuator. B Chem. 2010;146(1):381–387. [Google Scholar]
- Xiao X., Si P., Magner E. Bioelectrochemistry. 2016;109:117–126. doi: 10.1016/j.bioelechem.2015.12.008. [DOI] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yan W., Guo C., Zhang J., Yu L., Zhang G., Wang X., Fang G., Sun D. Anal. Chim. Acta. 2020;1106:1–21. doi: 10.1016/j.aca.2020.01.044. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang G., Xiong C., Zheng L., He J., Ding Y., Lu H., Zhang G., Cho K., Qiu L. Biosens. Bioelectron. 2018;105:121–128. doi: 10.1016/j.bios.2018.01.035. [DOI] [PubMed] [Google Scholar]
- Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Chem. Commun. 2020;56(70):10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- Zhang M., Li X., Pan J., Zhang Y., Zhang L., Wang C., Yan X., Liu X., Lu G. Biosens. Bioelectron. 2021;190 doi: 10.1016/j.bios.2021.113421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web references
- Highfield R. 2020. https://www.sciencemuseumgroup.org.uk/blog/coronavirus-the-spike/
- Popoola N. 2020. https://punchng.com/imf-projects-28tn-covid-19-losses-in-five-years/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.