ABSTRACT
Over the past 20 years, humankind has encountered three severe coronavirus outbreaks. Currently ongoing, COVID-19 (coronavirus disease 2019) was declared a pandemic due to its massive impact on global health and the economy. Numerous scientists are working to identify efficacious therapeutic agents for COVID-19, although treatment ability has yet to be demonstrated. The SUMO (small ubiquitin-like modifier) system has diverse roles in viral manipulation, but the function of SUMO in coronaviruses is still unknown. The objective of this review article is to present recently published data suggesting contributions of the host SUMO system to coronavirus infection. These findings underscore the potential of SUMO as a novel target for anti-coronavirus therapy, and the need for a deeper understanding of coronavirus pathology to prepare and prevail against the current and emerging coronavirus outbreaks.
KEYWORDS: SUMO/coronavirus/COVID-19/Therapy
Introduction
Coronavirus disease 2019 (COVID-19) is a newly emerging human infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. COVID-19 has globally spread, with initial cases detected in Wuhan, China and major outbreaks in the United States, Brazil, Russia, India, and Europe [2]. As of 12 March 2021, the number of COVID-19 patients reached 117,799,584 with 2,615,018 deaths across over 250 countries and territories (WHO COVID-19 Dashboard, https://who.sprinklr.com/). About 80% of COVID-19 patients recovered without special treatment, whereas the remaining 14% and 6% of patients have severe disease and critical illness, respectively [3]. In particular, people who are elderly or have chronic medical conditions, such as lung or cardiovascular disease, cancer, diabetes, or immune disorders, are more likely to develop severe pneumonia with acute respiratory distress syndrome [4]. Currently, there is no well-established therapy for COVID-19, and we still face the challenge of developing safe and effective vaccines and antiviral therapeutics.
In the past two decades, three coronaviruses causing severe illness in humans, SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 have led sequentially to enormous public health and economic crises [1]. These coronaviruses belong to the Betacoronavirus genus in the Coronaviridae family [5]. They are pleomorphic RNA viruses consisting of a positive-sense, single-stranded RNA genome 27 to 32 kb in length (Figure 1). Two-thirds of the viral RNA genome is translated into two large polyproteins, which are further cleaved into 16 nonstructural proteins involved in transcription and replication, while the remaining viral genome encodes the structural proteins, such as spike (S), envelope (E), membrane (M), and nucleocapsid (N), and several accessory proteins [6,7]. RNA viruses generally have higher evolutionary rates than any other viruses due to their high polymerase error rate and shuffling by recombination and reassortment [8]. The SARS-CoV-2 genome has a sequence homology of 77.5% with SARS-CoV and 50% with MERS-CoV [9]. While SARS and MERS affected 8,422 people with a 11% morality rate and 2,494 people with a 34% morality rate, respectively, COVID-19 has been spreading more efficiently with a lower mortality rate of 6% [10].
Figure 1.
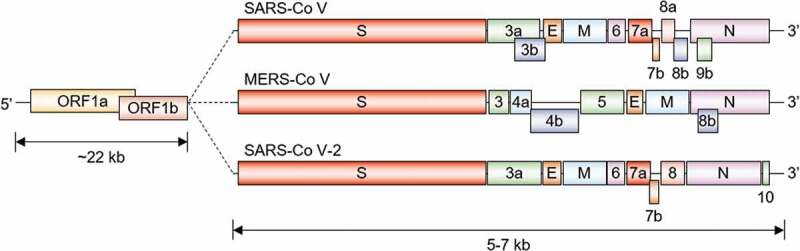
Schematic representation of the genome architecture of the three coronaviruses
SARS-CoV, MERS-CoV, and SARS-CoV-2 have a single-stranded RNA genome, which comprises ~22 kb of two large genes, ORF1a and ORF1b, and 5–7 kb of structural genes encoding structural proteins, spike (S), envelope (E), membrane (M), nucleocapsid (N), and accessory proteins. Different coronaviruses contain unique characteristics in terms of the number, genomic organization, and sequence of accessory genes.
While it is unclear when the COVID-19 pandemic will end, it is noteworthy to mention that the 1918 flu pandemic lasted from January 1918 to December 1920 [11]. Kassa and colleagues’ mathematical model estimated the possibility of circulating COVID-19 outbreaks via resurgence in infection [12]. Furthermore, because various coronaviruses persist in wildlife reservoirs, it is almost impossible to completely prevent future outbreaks of new coronaviruses [7]. Therefore, preparing for future coronavirus epidemics and pandemics is essential. The systemic changes in prevention, diagnosis, and treatment are necessary for mounting a more efficient and effective response to the next epidemic or pandemic [13]. Thus, studying the diverse mechanisms governing coronavirus biology is essential to establish effective antiviral therapeutic strategies.
Viruses have gradually evolved and developed many strategies to exploit host cellular mechanisms to counteract antiviral responses and maximize survival and reproduction [14]. Among such cellular pathways, post-translational modification (PTM) by small ubiquitin-related modifier (SUMO) is an obvious control strategy that contributes to replication and propagation of a wide range of DNA and RNA viruses [15]. Numerous viral proteins are not only substrates of this reversible PTM but also modulators of the host SUMO pathway. It is clear from numerous studies that the SUMO machinery is important for both adaptation of virus to host cells and resistance to virus infection; however, only few reports have presented the correlation between SUMO and coronaviruses [16]. The human E2 SUMO-conjugating enzyme Ubc9 directly binds and links covalently SUMO to SARS-CoV nucleocapsid protein that is involved in viral RNP assembly and replication [17,18]. Recent proteomics analyses of SARS-CoV-2-infected cells have also revealed a significant decrease in the expression of E3 SUMO ligase RanBP2, which acts as a regulator of the retrovirus restriction factor TRIM5α, implying that TRIM5α-mediated antiviral activity may be regulated by SUMO upon SARS-CoV-2 infection [19,20].
This review presents an overview of the diverse roles of the SUMO system in viral infections, especially that of coronavirus, from the perspective of the host cell and the virus. In addition, SUMO is proposed as a novel target for anti-coronavirus therapy.
The SUMO system
The SUMO protein post-translationally modifies a diverse array of substrates that play important roles in various cellular processes, including transcription, DNA replication, cell cycle progression, nucleo-cytoplasmic transport, apoptosis, genome integrity, and stability maintenance, and cellular stress responses [21]. Humans have four genes that encode SUMO proteins, SUMO-1, −2, −3, and −4. SUMO-2 and SUMO-3 are almost identical and are hereafter referred to as SUMO-2/3; SUMO-2/3 shares about 50% similarity with SUMO-1 [22].
Their paralogs are first translated as precursors with a C-terminal extension. The precursors then undergo specific proteolytic cleavage to yield mature proteins with a pair of Gly residues. However, it is unclear whether SUMO-4’s product can be covalently coupled to other proteins, because its precursor is not likely to be processed in vivo [23]. The mature form of SUMO is conjugated to lysine (K) side chains of substrate proteins via an enzyme cascade similar to that used for ubiquitin-protein conjugation [24,25] (Figure 2). A heterodimeric E1 SUMO-activating enzyme (SAE1/SAE2) first forms a thioester linkage through its active-site cysteine to the carboxyl terminus of SUMO, and then the SUMO moiety is transferred to the active-site cysteine of a E2 SUMO-conjugating enzyme (Ubc9). The SUMO is finally conjugated to the lysine side chain(s) of a target protein with the aid of one of several E3 SUMO ligases. Site-specific proteases can disassemble SUMO chains on substrates [26].
Figure 2.
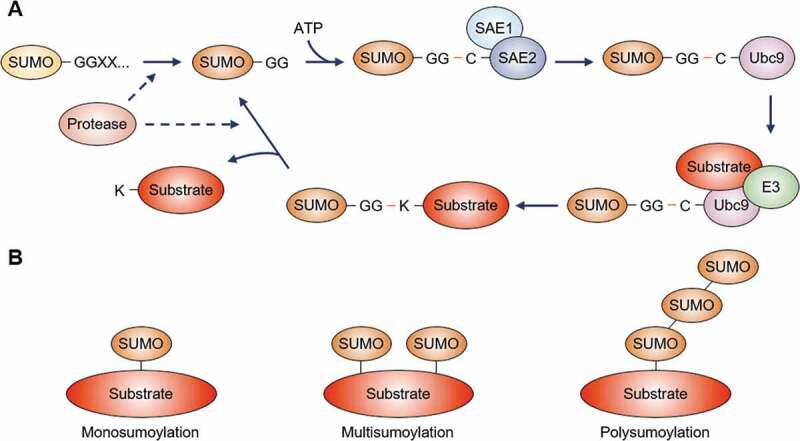
The SUMO pathway
(A) The precursor form of SUMO is processed by a SUMO-specific protease, creating a C-terminal Gly-Gly (GG) motif. In the presence of ATP, mature SUMO is activated by the heterodimeric E1 SUMO-activating enzymes, SAE1 and SAE2, through the cysteine (C) residue of SAE2. Subsequently, SUMO transferred to the cysteine residue of the E2 SUMO-conjugating enzyme (Ubc9) is finally conjugated to the lysine (K) residues of substrate proteins by aid of an E3 SUMO ligase. The SUMO protease deconjugates SUMO from proteins or edits SUMO chains, and then SUMO is recycled through the conjugation system.(B) SUMO is covalently linked to a single lysine residue of a protein (monosumoylation) or multiple lysine residues of a single protein (multisumoylation). Repeated sumoylation cycles build SUMO chain(s) attached on a substrate (polysumoylation).
Functions of the SUMO pathway in viral infection
In eukaryotes, SUMO is an essential regulator of homeostasis when cells respond to stresses, such as osmotic shock, hypoxia, heat, oxidative stress, nutrient deprivation, genotoxic stresses, or viral infection [27], and protein sumoylation levels by SUMO-1 and/or SUMO-2/3 are increased sharply in response to such stimuli [28]. There are two aspects of SUMO function during infection [16]. First, sumoylation of specific immune response factors mediates an antiviral effect, thus preventing viral load and contributing to viral elimination. Second, numerous viruses can exploit SUMO modification to invade and replicate within host cells, resulting in several viral proteins and their targets becoming SUMO substrates.
Many viruses benefit from protein sumoylation. Latent membrane protein 1 (LMP1), a viral oncoprotein, directly binds and modulates Ubc9 to increase SUMO conjugation in Epstein–Barr virus (EBV) infected cells [29,30]. LMP1-induced protein sumoylation regulates nuclear accumulation, stability, and function of interferon regulatory factor 7 (IRF7), a master regulator of the innate immune response, during latent infections [31]. Also, IRF family transcription factors undergo diverse post-translational modification leading to regulation of their properties. For example, whereas phosphorylation of IRF3 and IRF7 stimulates transcription of type I interferon (IFN) [32], their sumoylation negatively regulate IFN gene expression [33]. Infection of types A and B influenza viruses (IAV and IBV) triggers not only sumoylation of influenza viral proteins but also global reprogramming of host protein sumoylation [34,35]. To note, 76 SUMO1 substrates and 117 SUMO2 substrates demonstrated increased sumoylation states upon IAV infection, and IAV RNA polymerase activity in the nucleus contributed to remodeling of the host SUMO system [34]. Viral proteins are also substrates of sumoylation during viral infection. For example, Ubc9 binding and subsequent sumoylation of UL44, a human cytomegalovirus (HCMV) DNA polymerase subunit, results in its decreased localization in viral DNA replication compartments via altering intranuclear distribution and conversely causing a positive effect on viral replication and production [36]. Similarly, the hantavirus nucleocapsid protein interacts with Ubc9 during infection, and its sumoylation mediates assembly of structural proteins with the viral genome and localization at the perinuclear regions for viral replication [37,38]. Sumoylation of IAV nonstructural protein NS1 accelerates viral replication via enhancing NS1’s stability [39]. In addition, human papillomavirus (HPV) E2 and EBV Rta proteins serve as targets of Ubc9-mediated sumoylation, which specifically stimulate the transcription of their target genes, completely contributing to viral replication [40,41]. Furthermore, several E3 ligases, PIAS1, PIASxβ, and RanBPM, also promote sumoylation of the Rta protein [42–44].
Conversely, some viruses benefit from impaired sumoylation. The CELO adenovirus Gam1 protein promotes degradation of Ubc9 or SAE1/SAE2 in a proteasome-dependent manner [45,46], which is crucial for viral replication and transcriptional regulation of specific genes via inactivating histone deacetylase 1 (HDAC1) [47,48]. HPV-16/18 E6 oncoprotein mediates proteasomal degradation of Ubc9, resulting in decreased global sumoylation levels and perhaps subsequent development of cervical cancer [49], and inhibits E3 SUMO ligase PIASy-induced senescence and sumoylation of its substrates [50]. In addition, in HeLa cells, coxsackievirus B5 protein alters Ubc9’s cellular localization and eventually contributes to decreased sumoylation status of specific substrates [51]. Widespread reduction of cellular sumoylated protein levels, including antiviral proteins promyelocytic leukemia and Sp100, is induced by herpes simplex virus type 1 (HSV-1) ubiquitin ligase ICP0 in a proteasome-dependent way and is critical to suppressing intrinsic immunity [52,53]. The EBV protein kinase BGLF4 functions to suppress global SUMO levels, and its SUMO binding capacity is required to induce the cellular DNA damage response and to facilitate lytic EBV replication [54].
Sumoylation of coronavirus proteins
Although protein sumoylation is a critical factor in virus-induced pathogenesis, the relationship between sumoylation and coronaviruses has yet to be established. There are, however, a limited number of reports suggesting a potential role of sumoylation in SARS-CoV infection. During the SARS infection, the nucleocapsid protein binds to the viral RNA genome to form the ribonucleoprotein core, which is able to interact with a number of host proteins [55,56]. Since the nucleocapsid is expressed abundantly in infected cells, it has often been investigated as a target of SARS rapid diagnosis kits [57,58]. The SARS-CoV nucleocapsid protein is 422 amino acids long, consisting of the N-terminal structural domain (NTD) and C-terminal structural domain (CTD) sandwiched between three intrinsically disordered regions (IDRs) [59,60]. The flexible linker region (LKR) between NTD and CTD has a Ser/Arg-rich region that contains a number of potential phosphorylation sites directly related to nucleocapsid oligomerization [61–65].
In addition to phosphorylation, biochemical characterization revealed that the nucleocapsid has a motif for binding hUbc9 and is post-translationally modified by SUMO, in which the major site is the K62 within NTD and strongly promotes homo-oligomerization of the protein [17,18]. Moreover, this modification is required for normal subcellular localization of nucleocapsid and interference of host cell division [18]. Therefore, this lysine sumoylation of nucleocapsid may not be a result of the antiviral host defense system but instead is likely due to viral hijacking of the cellular SUMO system to enhance viral replication and pathogenesis [66]. Coronaviruses do not encode SUMO conjugation enzymes; however, evolution of coronaviruses may have conferred the means for these coronaviruses to hijack this modification in the host to promote survival. Although the sumoylation site of SARS-CoV-2’s nucleoscapsid has yet to be determined, two distinct bioinformatics tools, SUMOplot and GPS-SUMO, predict the same SUMO conjugation site at the K124 as SARS-CoV along with two additional sites, at the K15 and K19 lysine positions. These sites need to be further explored and understood to elucidate the role of SUMO modification in SARS-CoV-2 infection.
Alterations of SUMO profile in coronavirus-infected cells
Similar to the changes in the SUMO proteome in response to various cellular stresses, there is a dramatic change in the SUMO proteome in virus-infected host cells [15]. However, to our knowledge, alteration in SUMO conjugation patterns has not been determined in cells infected with coronaviruses, even though a transcriptome analysis revealed that COVID19 patients exhibited changes in the expression of genes regulating ubiquitin or SUMO conjugation [67]. Instead of a SUMO-focused analysis, Bojkova and colleagues carried out proteomic studies to monitor the protein levels in human Caco-2 cells upon infection by SARS-CoV-2 [20]. They revealed the reestablishment of central pathways, including translation, splicing, carbon metabolism, and nucleic acid metabolism. Such research results provoked further study, and Ortea and Bock assessed host cell proteomic data upon SARS-CoV-2 infection [19]. This study additionally suggested a new network of proteins involved in the inflammatory response or mitotic chromosome segregation in the host proteome altered by SARS-CoV-2. Particularly, the nuclear pore complex protein RanBP2 (E3 SUMO protein ligase) is placed at the center of this identified network [19]. The RanBP2/SUMO-modified RanGAP1/Ubc9 complex directly interacts with TRIM5α, a cytoplasmic protein that recognizes specific retroviral capsids and prevents its invasion into the host cell cytoplasm [68], and regulates its antiretroviral activity and localization by sumoylation [69] (Figure 3). Moreover, some COVID-19 patients have mutations in RANBP2 [70] and TRIM5 is positively correlated with the proliferation of natural killer cell response in COVID-19 [71]. Missense mutations in RANBP2 were identified as a major cause of familial and recurrent acute necrotizing encephalopathy [72], suggesting that such mutations may also contribute to the acute necrotizing encephalopathy reported in some COVID-19 patients [73–75]. Since TRIM5α promotes the premature disassembly of HIV-1 capsid in the cytoplasm of the host cell and the replication and propagation of coronavirus are also mediated in the cytoplasm [68], the role of RanBP2-mediated TRIM5α sumoylation in coronavirus infection deserves further investigation.
Figure 3.
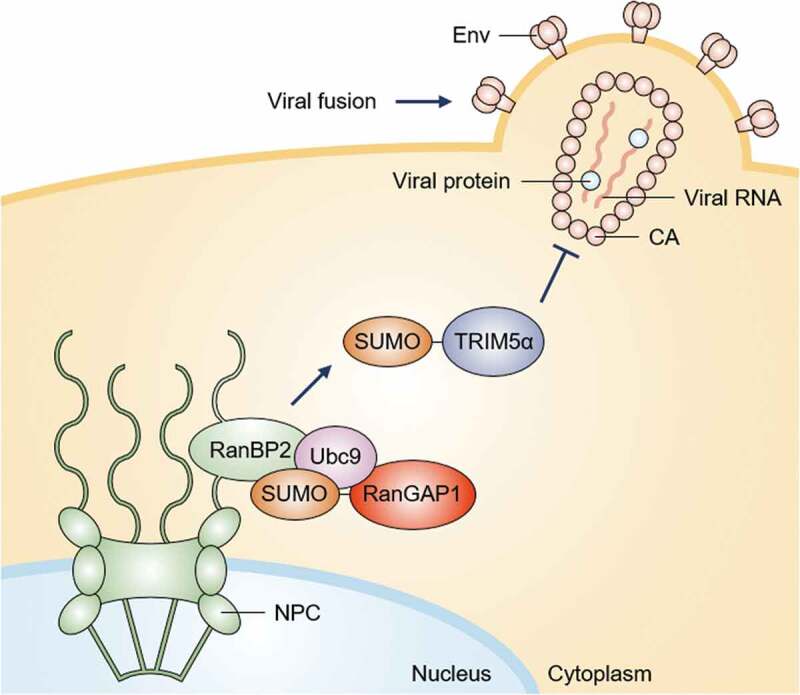
The function of RanBP2-mediated sumoylation of TRIM5α in HIV-1 infection
The envelope glycoprotein (Env) mediates human immunodeficiency virus type 1 (HIV-1) fusion with the cell membrane. The HIV-1 core consists of the capsid (CA) protein and its contents. The capsid proteins surround two copies of the RNA genome and proteins, such as reverse transcriptase and integrase. Before or during capsid disassembly, which is necessary for nuclear import of the viral genome, TRIM5α blocks retroviral infection by intercepting capsids before they reach the nucleus and promotes premature disassembly. The E3 SUMO ligase RanBP2 is associated with Ubc9 and sumoylated RanGAP1 at cytoplasmic filaments of nuclear pore complexes (NPCs). RanBP2 promotes sumoylation of TRIM5α in the cytoplasm, resulting in its proper localization and antiviral activity.
The sumoylation of TRIM28 typically suppresses the transcription of endogenous retroviral (ERV) genes. IAV infection induces the loss of SUMO-modified TRIM28, a transcriptional repressor, thus promoting the expression of endogenous retroviral (ERV) RNAs that are sensed as non-self by host pattern recognition receptors (PRRs) [76,77] (Figure 4). Consequently, the derepression of ERVs transcription induces the subsequent activation of IFN-mediated antiviral response via the RIG-I-, MAVS-, TBK1-, and JAK1-dependent pathway [76]. TRIM28 may play a critical role in SARS-CoV-2’s entry into human cells [78]. It was recently reported that angiotensin-converting enzyme 2 (ACE2), which is co-expressed with TRIM28 in type II pneumocytes, is the cellular receptor protein for SARS-CoV-2 [79,80]. Also, the knockdown of TRIM28 stimulates ACE2 expression via IFN-γ dependent immune response [78]. Although the SUMO modification of TRIM28 has not been studied in coronavirus-infected cells, studying SUMO function in coronaviruses may provide clues to developing new therapies and life-saving vaccines for coronavirus diseases.
Figure 4.
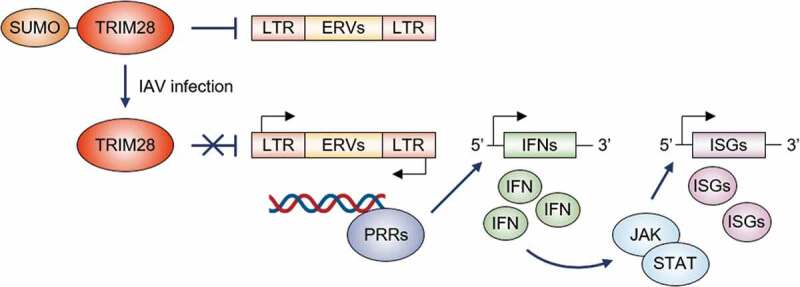
The role of sumoylation of TRIM28 in influenza A virus infection
The sumoylation of TRIM28 typically suppresses the transcription of endogenous retroviral (ERV) genes. Upon the infection of the influenza A virus (IAV), the decreased level of SUMO-modified TRIM28 derepresses the expression of ERV-encoded double-stranded RNAs that are recognized as non-self by pattern recognition receptors (PRRs), and then PRRs trigger the expression and secretion of interferons (IFNs). The IFN-mediated stimulation of the JAK/STAT signaling pathway subsequently induces the activation of hundreds of IFN-stimulated genes (ISGs) encoding), which encode proteins with diverse antiviral functions.
Conclusions and perspectives
Coronaviruses are well-known zoonotic pathogens that infect many vertebrate species, including humans, and can cause respiratory, gastrointestinal, hepatic, and neurological disorders [81]. Coronaviruses possess unstable RNA genomes that mutate continuously and undergo genetic reassortments, result in high-frequency polymerase error and RNA recombination. Therefore, developing therapeutic agents against coronavirus proteins is considerably more difficult due to the changes in viral proteins resulting from the genome’s mutations [82]. Three different coronaviruses, SARS-CoV, MERS-CoV, and SARS-CoV-2, have accounted for considerable outbreaks, in the last two decades, with SARS-CoV-2 currently causing a pandemic that has taken a toll on human life and the global economy [1]. As of now, we do not know when this pandemic will end, and furthermore, future SARS-CoV-2 outbreaks may trigger pronounced crises. Therefore, there is an impetus to understand coronavirus infection and spread and develop antiviral therapies. Herein, we have described recent observations of how the SUMO system may be involved in coronavirus replication and pathogenesis. Protein sumoylation contributes to certain pathological conditions and has the capability to both benefit and harm in viral infections [16]. Studying SUMO function in coronaviruses may provide clues for developing new therapies and life-saving vaccines for the current and upcoming coronavirus diseases.
Funding Statement
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (no. 2020R1C1C1009367). English language editing was provided by Enago (https://www.enago.com/)
Disclosure statement
The author declares no conflict of interest.
References
- [1].Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern (vol 395, pg 470, 2020). Lancet. 2020a;395(10223):496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hui DS, Azhar EI, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Anderson RM, Heesterbeek H, Klinkenberg D, et al. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395(10228):931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen Y, Liu QY, Guo DY.. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Graham RL, Baric RS. Recombination, Reservoirs, and the Modular Spike: mechanisms of Coronavirus Cross-Species Transmission. J Virol. 2010;84(7):3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim JM, Chung YS, Jo HJ, et al. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Singhal T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediatr. 2020;87(4):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kassa SM, Njagarah JBH, Terefe YA. Analysis of the mitigation strategies for COVID-19: from mathematical modelling perspective. Chaos Solitons Fractals. 2020;138:109968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gates B. Responding to Covid-19 - A Once-in-a-Century Pandemic? N Engl J Med. 2020;382(18):1677–1679. [DOI] [PubMed] [Google Scholar]
- [14].Nathanson N, Mcgann KA, Wilesmith J, et al. The Evolution of Virus Diseases - Their Emergence, Epidemicity, and Control. Virus Res. 1993;29(1):3–20. [DOI] [PubMed] [Google Scholar]
- [15].Lowrey AJ, Cramblet W, Bentz GL. Viral manipulation of the cellular sumoylation machinery. Cell Commun Signal. 2017;15(27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilson VG. Viral Interplay with the Host Sumoylation System. In: Sumo Regulation of Cellular Processes. 2nd ed. Vol. 963. 2017. p. 359–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fan Z, Zhuo Y, Tan XY, et al. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J Med Virol. 2006;78(11):1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li FQ, Xiao H, Tam JP, et al. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579(11):2387–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bock JO, Ortea I. Re-analysis of SARS-CoV-2-infected host cell proteomics time-course data by impact pathway analysis and network analysis: a potential link with inflammatory response. Aging (Albany NY). 2020;12(12):11277–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82(1):357–385. [DOI] [PubMed] [Google Scholar]
- [22].Muller S, Hoege C, Pyrowolakis G, et al. SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell Biol. 2001;2(3):202–210. [DOI] [PubMed] [Google Scholar]
- [23].Owerbach D, McKay EM, Yeh ETH, et al. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337(2):517–520. [DOI] [PubMed] [Google Scholar]
- [24].Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17(9):581–595. [DOI] [PubMed] [Google Scholar]
- [25].Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73(1):355–382. [DOI] [PubMed] [Google Scholar]
- [26].Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13(12):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guo C, Henley JM. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. Iubmb Life. 2014;66(2):71–77. [DOI] [PubMed] [Google Scholar]
- [28].Enserink JM. Sumo and the cellular stress response. Cell Div. 2015;10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bentz GL, Moss CR, Whitehurst CB, et al. LMP1-Induced Sumoylation Influences the Maintenance of Epstein-Barr Virus Latency through KAP1. J Virol. 2015;89(15):7465–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bentz GL, Whitehurst CB, Pagano JS. Epstein-Barr Virus Latent Membrane Protein 1 (LMP1) C-Terminal-Activating Region 3 Contributes to LMP1-Mediated Cellular Migration via Its Interaction with Ubc9. J Virol. 2011;85(19):10144–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bentz GL, Shackelford J, Pagano JS. Epstein-Barr Virus Latent Membrane Protein 1 Regulates the Function of Interferon Regulatory Factor 7 by Inducing Its Sumoylation. J Virol. 2012;86(22):12251–12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. [DOI] [PubMed] [Google Scholar]
- [33].Kubota T, Matsuoka M, Chang TH, et al. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283(37):25660–25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Domingues P, Golebiowski F, Tatham MH, et al. Global Reprogramming of Host SUMOylation during Influenza Virus Infection. Cell Rep. 2015;13(7):1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pal S, Santos A, Rosas JM, et al. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 2011;158(1–2):12–27. [DOI] [PubMed] [Google Scholar]
- [36].Sinigalia E, Alvisi G, Segre CV, et al. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PLoS One. 2012;7(11):e49630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Alfadhli A, Love Z, Arvidson B, et al. Hantavirus nucleocapsid protein oligomerization. J Virol. 2001;75(4):2019–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Maeda A, Lee BH, Yoshimatsu K, et al. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology. 2003;305(2):288–297. [DOI] [PubMed] [Google Scholar]
- [39].Xu K, Klenk C, Liu B, et al. Modification of Nonstructural Protein 1 of Influenza A Virus by SUMO1. J Virol. 2011;85(2):1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Miller G, El-Guindy A, Countryman J, et al. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv Cancer Res. 2007;97:81. [DOI] [PubMed] [Google Scholar]
- [41].Wu YC, Roark AA, Bian XL, et al. Modification of papillomavirus E2 proteins by the small ubiquitin-like modifier family members (SUMOs). Virology. 2008;378(2):329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chang LK, Lee YH, Cheng TS, et al. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J Biol Chem. 2004;279(37):38803–38812. [DOI] [PubMed] [Google Scholar]
- [43].Chang LK, Liu ST, Kuo CW, et al. Enhancement of transactivation activity of Rta of Epstein-Barr virus by RanBPM. J Mol Biol. 2008;379(2):231–242. [DOI] [PubMed] [Google Scholar]
- [44].Liu ST, Wang WH, Hong YR, et al. Sumoylation of Rta of Epstein-Barr virus is preferentially enhanced by PIASx beta. Virus Res. 2006;13(2):163–170. [DOI] [PubMed] [Google Scholar]
- [45].Boggio R, Colombo R, Hay RT, et al. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16(4):549–561. [DOI] [PubMed] [Google Scholar]
- [46].Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases - A viral strategy to counteract sumoylation. J Biol Chem. 2007;282(21):15376–15382. [DOI] [PubMed] [Google Scholar]
- [47].Colombo R, Boggio R, Seiser C, et al. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3(11):1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Glotzer JB, Saltik M, Chiocca S, et al. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature. 2000;407(6801):207–211. [DOI] [PubMed] [Google Scholar]
- [49].Heaton PR, Deyrieux AF, Bian XL, et al. HPV E6 proteins target Ubc9, the SUMO conjugating enzyme. Virus Res. 2011;158(1–2):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bischof O, Schwamborn K, Martin N, et al. RETRACTED: the E3 SUMO Ligase PIASy Is a Regulator of Cellular Senescence and Apoptosis. Mol Cell. 2006;22(6):783–794. [DOI] [PubMed] [Google Scholar]
- [51].Gomes R, Guerra-Sa R, Arruda E. Coxsackievirus B5 induced apoptosis of HeLa cells: effects on p53 and SUMO. Virology. 2010;396(2):256–263. [DOI] [PubMed] [Google Scholar]
- [52].Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76(2):841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sloan E, Tatham MH, Groslambert M, et al. Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 2015;11(7):e1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li RW, Wang LY, Liao GL, et al. SUMO Binding by the Epstein-Barr Virus Protein Kinase BGLF4 Is Crucial for BGLF4 Function. J Virol. 2012;86(10):5412–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ng ML, Tan SH, See EE, et al. Early events of SARS coronavirus infection in Vero cells. J Med Virol. 2003;71(3):323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Risco C, Anton IM, Enjuanes L, et al. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70(7):4773–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].He QG, Chong KH, Chng HH, et al. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin Diagn Lab Immun. 2004a;13(2):417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lin Y, Shen X, Yang RF, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13(3):141–145. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chang CK, Sue SC, Yu TH, et al. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13(1):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huang QL, Yu LP, Petros AM, et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry-Us. 2004;43(20):6059–6063. [DOI] [PubMed] [Google Scholar]
- [61].He RT, Dobie F, Ballantine M, et al. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2004b;316(2):476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Luo H, Ye F, Chen K, et al. SR-Rich Motif Plays a Pivotal Role in Recombinant SARS Coronavirus Nucleocapsid Protein Multimerization. Biochemistry-Us. 2005;44(46):15351–15358. [DOI] [PubMed] [Google Scholar]
- [63].Peng TY, Lee KR, Tarn WY. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. Febs J. 2008;275(16):4152–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Surjit M, Kumar R, Mishra RN, et al. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol. 2005;79(17):11476–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wu CH, Yeh SH, Tsay YG, et al. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem. 2009;284(8):5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].El Motiam A, Vidal S, Seoane R, et al. SUMO and Cytoplasmic RNA Viruses: from Enemies to Best Friends. 2020;1233:263–277. Proteostasis and Disease: From Basic Mechanisms to Clinics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gill S, Dos Santos CC, O’Gorman DB, et al. (2020). Transcriptional Profiling of Leukocytes in Critically Ill COVID19 Patients: implications for Interferon Response and Coagulation. [DOI] [PMC free article] [PubMed]
- [68].Van Gent M, Sparrer KMJ, Gack MU. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu Rev Virol. 2018;5(1):385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Maarifi G, Fernandez J, Portilho DM, et al. RanBP2 regulates the anti-retroviral activity of TRIM5 alpha by SUMOylation at a predicted phosphorylated SUMOylation motif. Commun Biol. 2018;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Koralnik IJ, Tyler KL. COVID-19: a Global Threat to the Nervous System. Ann Neurol. 2020;88(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Maucourant C, Filipovic I, Ponzetta A, et al. Natural killer cell activation related to clinical outcome of COVID-19. medRxiv; 2020. [Google Scholar]
- [72]. Singh RR, Sedani S, Lim M, et al. RANBP2 mutation and acute necrotizing encephalopathy: 2 cases and a literature review of the expanding clinico-radiological phenotype. Eur J Paediatr Neuro. 2015;19(2):106–113. [DOI] [PubMed] [Google Scholar]
- [73].Dixon L, Varley J, Gontsarova A, et al. COVID-19-related acute necrotizing encephalopathy with brain stem involvement in a patient with aplastic anemia. Neurol-Neuroimmunol. 2020;7(5):e789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Elkady A, Rabinstein AA. Acute necrotizing encephalopathy and myocarditis in a young patient with COVID-19. Neurol-Neuroimmunol. 2020;7(5). [Google Scholar]
- [75].Poyiadji N, Shahin G, Noujaim D, et al. COVID-19-associated Acute Hemorrhagic Necrotizing Encephalopathy: imaging Features. Radiology. 2020;296(2):E119–E120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schmidt N, Domingues P, Golebiowski F, et al. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc Natl Acad Sci U S A. 2019;116(35):17399–17408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Xu XX, Wan H, Nie L, et al. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell. 2018;9(3):246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang Y, Fan Y, Huang Y, et al. TRIM28 regulates SARS-CoV-2 cell entry by targeting ACE2. bioRxiv; 2020b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li WH, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(5):1016-+. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yin YD, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Woo PCY, Huang Y, Lau SKP, et al. Coronavirus Genomics and Bioinformatics Analysis. Viruses-Basel. 2010;2(8):1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]


