Graphical abstract
Abstract
mRNA Lipid nanoparticles (LNPs) have recently been propelled onto the center stage of therapeutic platforms due to the success of the SARS-CoV-2 mRNA LNP vaccines (mRNA-1273 and BNT162b2), with billions of mRNA vaccine doses already shipped worldwide. While mRNA vaccines seem like an overnight success to some, they are in fact a result of decades of scientific research. The advantage of mRNA-LNP vaccines lies in the modularity of the platform and the rapid manufacturing capabilities. However, there is a multitude of choices to be made when designing an optimal mRNA-LNP vaccine regarding efficacy, stability and toxicity. Herein, we provide a brief on what we consider to be the most important aspects to cover when designing mRNA-LNPs from what is currently known and how to optimize them. Lastly, we give our perspective on which of these aspects is most crucial and what we believe are the next steps required to advance the field.
Current Opinion in Biotechnology 2022, 73:329–336
This review comes from a themed issue on Nanobiotechnology
Edited by Peter M Tessier and Ravi S Kane
For complete overview of the section, please refer to the article collection, “Nanobiotechnology”
Available online 26th October 2021
https://doi.org/10.1016/j.copbio.2021.09.016
0958-1669/© 2021 Elsevier Ltd. All rights reserved.
Introduction
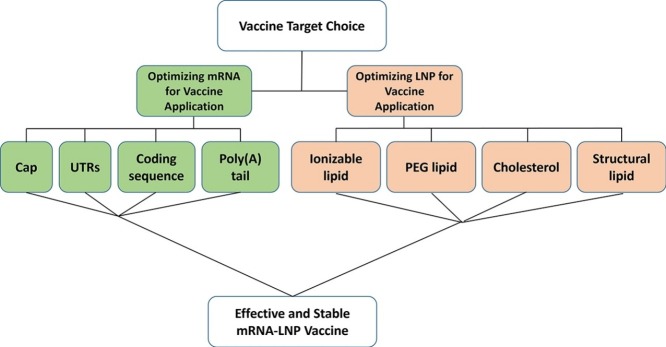
Generally, mRNA-LNP design is comprised of several elements: (1) mRNA sequence design and nucleotide modification choice, (2) Optimization of a LNP formulation to encapsulate and deliver mRNA and (3) long-term storage. Each one of these phases entails a plethora of choices and considerations to make. Currently, mRNA-LNPs are evaluated based on their ability to express encoded mRNA, their immunogenicity due to recognition of mRNA as a foreign entity by intracellular RNA sensors, LNP formulation stability and toxicity. All these are examined in light of the route of administration and therapeutic goal. Importantly, the design of an optimal therapeutic mRNA-LNP for systemic administration differs from an optimal intra-muscularly administered mRNA-LNP vaccine. We will focus on the optimal design of efficacious mRNA-LNP vaccines regarding expression, stability, and toxicity. We will summarize what is currently known from the literature, from our own experience and from comparison of the two recently greenlighted mRNA vaccines for SARS-CoV-2, Moderna’s mRNA-1273 and Pfizer/BioNTech’s BNT162b2. While many elaborative reviews covering the theories behind each component exist, we aim to provide a brief on what is currently known and what we deem to be most important.
Optimizing mRNA sequence and modifications for mRNA-LNP vaccines
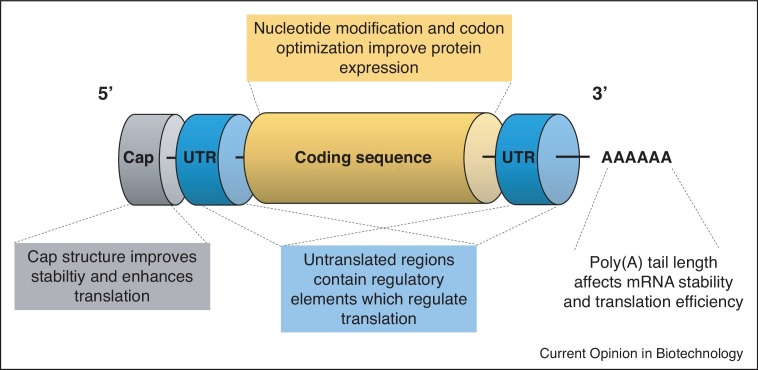
The main considerations for mRNA sequence and modification selection are: (1) expression of encoded mRNA, (2) immunogenicity due to recognition of mRNA as a foreign entity by intracellular RNA sensors and (3) mRNA stability. These are controlled by designing mRNA with modified nucleotides, sequence modifications and mRNA capping modalities (Figure 1 ).
Figure 1.
Optimizing mRNA sequence and modifications for mRNA-LNP vaccines.
A scheme representing the mRNA construct elements to consider when designing an mRNA-LNP vaccine.
Nucleotide modifications
Nucleotide modifications are considered the most important breakthrough that initiated the mRNA therapeutics field. Non-modified mRNA molecules are recognized by cellular RNA sensors which result in innate immunity activation [1,2]. While activation of innate immune pathways can be advantageous in the context of vaccination by serving as an adjuvant, these innate immune responses may also be detrimental to mRNA therapy, since they dramatically reduce mRNA translation [3,4]. In 2005, a groundbreaking study by Kariko and colleagues demonstrated that incorporation of naturally occurring, chemically modified nucleosides such as pseudouridine (Ψ), thiouridine (s2U), and 5-methylcytidine (m5C) resulted in a significant reduction in the immunogenicity of the mRNA [5]. Most importantly, several studies demonstrated that incorporation of modified nucleosides resulted in enhanced stability of the RNA molecule, and increased protein translation [6, 7, 8,9•], including the N1-methylpseudouridine modification which has been employed in both mRNA-1273 and BNT162b2 [10,11]. Contrastingly, CureVac's sequence-optimized candidate, CVnCoV, is exclusively composed of unmodified nucleosides. This approach implements sequence-engineered, unmodified mRNA, to provide a robust and balanced immune response. However, preliminary phase 2b/3 data indicates that the vaccine candidate was substantially less effective than the other two leading mRNA vaccines by Pfizer/BioNTech and Moderna) (NCT04652102) [12,13]. While the inferior efficacy of CureVac's vaccine candidate could be attributed to the inclusion of unmodified nucleosides, several other dissimilarities between the candidates should be taken into account, such as differences in non-coding elements and storage conditions (see Table 1 ). Therefore, when considering nucleotide modifications, there is a tradeoff to consider between potential innate adjuvant responses driven by unmodified nucleosides and enhanced protein expression attributed to modified nucleosides. Currently, both FDA authorized mRNA vaccines incorporate modified nucleotides.
Table 1.
Comparison of mRNA and LNP formulation elements of three SARS-CoV-2 mRNA LNP vaccines
| BNT162b2 (Pfizer/BioNTech) | mRNA-1273 (Moderna) | CVnCoV (CureVac) | |
|---|---|---|---|
| Approval Status [48, 49, 50] | FDA Approved August 23, 2021 | Emergency Use Authorization (EUA) December 18, 2020 | Phase 2b/3 (NCT04652102) |
| Submitted for full Approval August 25, 2021 | |||
| Lipid Formulation (% Molar ratio of Ionizable: Cholesterol: Neutral lipid: PEGylated lipid) [9•] | 46.3:42.7:9.4:1.6 | 50:38.5:10:1.5 | 50:38.5:10:1.5 |
| N:P Molar Ratio [9•] | 6 | ||
| Cap [51, 52, 53] | Cap-1 | ||
| 5′-UTR [54] | 5′ -untranslated region derived from human alpha-globin RNA with an optimized Kozak sequence | Undisclosed | 5′ UTR: Artifacts from restriction and transcription site, plus Kozak sequence |
| S protein antigen [53,55] | Codon-optimized sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P | Codon-optimized sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P | Codon-optimized and engineered sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P |
| Modified nucleotides [54] | N1-methyl-pseudouridine | N1-methyl-pseudouridine | Unmodified nucleotides |
| 3′-UTR [54] | 3′ UTR comprising two sequence elements derived from the amino-terminal enhancer of split (AES) mRNA and the mitochondrial encoded 12S ribosomal RNA | Undisclosed | 3′ UTR comprising human alpha-globin 3′ UTR sequence element |
| Poly(A) Tail [52,53] | A 110-nucleotide poly(A)-tail consisting of a stretch of 30 adenosine residues, followed by a 10-nucleotide linker sequence and another 70 adenosine residues. | Undisclosed | 64 Poly (A) tail |
| Storage [9•,56] | −60°C to −80°C for six months −15°C to −25°C for two weeks 2°C–8°C for 5 days | −15°C to −20°C for six months 2°C–8°C for 30 days | 2°C–8°C for three months |
| Buffer [9•] | PBS | Tris | Undisclosed |
| Cryoprotectant [9•] | Sucrose | Sucrose | Undisclosed |
mRNA capping
mRNA capping dramatically increases translation efficiency and intra-cellular mRNA stability by binding to the eukaryotic translation initiation factor 4E (eIF4E) [14,15]. Capping of in vitro transcribed (IVT) mRNA is frequently performed using a cap analog, which can be added during the IVT process or post-transcription. However, the mRNA can be reversely capped which results in rapid degradation and poor translation. To avoid reverse 5′ cap incorporation, anti-reverse cap analogs (ARCA) have been developed, which ensure correct capping orientation [16]. Further modifications were developed over the years to improve the performance of ARCA, with the recently developed ‘CleanCap’ analog (TriLink Biotechnologies, San Diego, CA, USA) most frequently used nowadays in mRNA companies [17]. Therefore, stable, correctly oriented caps need to be incorporated for an effective mRNA-LNP vaccine.
UTR selection
UTR selection also needs to be considered, since they greatly affect mRNA decay and translational efficiency. 5′ UTR features such as start codons and secondary structures may compromise ribosome recruitment, scanning and start codon recognition, and thus should be avoided. Grossly, the 5′ UTR sequences are critical for protein expression while the 3′ UTRs are more likely to affect mRNA half-life [18, 19, 20]. For example, the β globin 3′ UTR and a duplication of the β-globin 3′ UTR stabilize are widely used to stabilize mRNA [21].
ORF design
ORF sequence design also carries a critical impact on translation efficiency and immunogenicity of the mRNA due to recognition by cellular sensors. In addition to the nucleotide modifications mentioned, codon optimization has been demonstrated to significantly enhance protein expression by incorporating frequent codons and/or codons with higher tRNA abundances [22]. Another form of sequence optimization is enrichment of guanine-cytosine (GC) content, which has been shown to increase steady-state mRNA levels in vitro and protein expression in vivo, and has been employed by CureVac in its recent SARS-CoV-2 mRNA vaccine candidate, CVnCoV (NCT04652102) [22,23]. Furthermore, the mRNA sequence dictates the secondary structure. Interestingly, this secondary structure can influence mRNA degradation by hydrolysis. Recently, specially designed algorithms that design optimal mRNA sequences for maximal base stacking regions have been reported to significantly improve mRNA stability [24•].
Poly(A) tail
The poly(A) tail also contributes to mRNA translation and stability by reducing RNA exonuclease activity. In addition, the poly(A) tail binds to poly(A)-binding proteins (PABP), which recruit eIF4G and eIF4E, increasing the affinity to the mRNA cap and promoting a circular mRNA structure and efficient translation. Contrastingly, PABP has also been shown to participate in microRNA-mediated inhibition of translation [25]. When considering the length of the poly(A) tails, there is no consensus. While it is suggested that longer poly(A) tails (120–150 nucleotides) increase mRNA stability, the dual effect of PABP-mediated inhibition of translation warrants further optimization. For example, Lima et al. demonstrated that mRNAs with high translation efficiency actually had short poly(A) sequences (∼33−34 nucleotides) [25]. mRNA created by IVT can be polyadenylated in two ways: either by encoding the poly(A) on the DNA template used or by enzymatic addition by poly(A) polymerase to the mRNA after IVT. Encoding the poly(A) tail on the DNA plasmid ensures the production of a defined poly(A) tail length, whereas enzymatic polyadenylation of the mRNA produces varying lengths of poly(A) tails and is therefore, less favorable.
LNP formulation considerations
LNPs, the mRNA vehicles, both protect mRNA payloads from degradation and enable their efficient delivery into target cells. Generally, LNPs are a lipid formulation comprised of various ratios of a neutral structural lipid, cholesterol, a PEG-lipid and the ionizable lipid.
Ionizable lipid
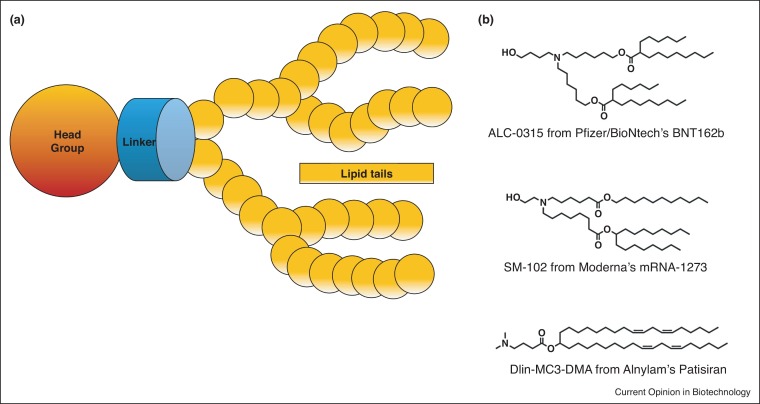
The ionizable lipid is considered the most important component and generally sets apart different mRNA-LNPs. Libraries of ionizable lipids are constantly screened to find the optimal lipid to enhance expression and provide better immune responses in mRNA-LNP vaccines while retaining low toxicity profiles [26,27]. Ionizable lipids are comprised of: (1) an ionizable head group, (2) a linker region and (3) the hydrocarbon chains (Figure 2 a). While ionizable lipids structures vary, some common grounds for effective ionizable lipids for mRNA-LNP vaccines have emerged.
Figure 2.
LNP formulation considerations.
(a) A scheme representing the elements of the ionizable lipid. (b) Chemical structures of the three clinically approved ionizable lipids. ALC-0315 (Pfizer/BioNtech’s BNT162b), SM-102 (Moderna’s SM-102) and Dlin-MC3-DMA (siRNA LNPs — Alnylam’s Patisiran) [34].
The ionizable head group: mRNA-1273 and BNT162b2 vaccines each comprise a different ionizable lipid, however, they both contain an amino-alcohol head group with pKas of 6.75 and 6.09, respectively [28]. A study by Moderna claimed that the optimal range of ionizable lipid pKa for eliciting an adaptive immune response following an mRNA vaccine is 6.6–6.9, whereas a pKa range of 6.2–6.6 has been shown to be optimal for protein expression following IV delivery [27]. Interestingly, this is consistent with mRNA-1273 ionizable lipid’s pKa, yet not with BNT162b2.
The linker region connects the head group with the lipid hydrocarbon tails, also gains attention for its effect on the in-vivo activity of RNA-LNPs. It is currently hypothesized that the linker region contributes to head group pKa and RNA-LNP endosomal escape potential [29]. Research into linker region optimization is an ongoing process. Ionizable lipid libraries with various linker structures have been designed and screened for their ability to delivery of RNA-LNPs more efficiently [30].
The lipid tails of the ionizable lipids are also to be considered. Lipid structure (and hence LNP structure) affects expression efficiency, by altering endosomal escape, stability during storage and toxicity [31]. Interestingly, studies have claimed correlations between various unsaturation degrees and symmetry of hydrophobic tails to more efficient and stable LNPs. For example, branched hydrocarbon lipid tails may create a more cone-shaped structure that enhances endosomal escape [32•]. Furthermore, ester linkages are introduced into lipid hydrocarbon chains to improve the ionizable lipid’s pharmacokinetic properties. The first FDA-approved ionizable lipid DLin-MC3-DMA (MC3) raised some concerns due to its long tissue half-life post-administration. This propelled the development of biodegradable ionizable lipids to improve lipid metabolism and prevent toxicities. Introducing one or more ester linkages, in the hydrocarbon tails as well as the linker region, leads to in-vivo cleavage by esterases, which dramatically improves the pharmacokinetic properties of the lipids, and reduces toxicities [26]. Studies constantly attempt to find the optimal ester bond position in the chains. It is assumed that placing these bonds too close to the head group-linker region reduces overall LNP efficacy by altering head group pKa. Therefore, it is preferable to locate them further downstream in the lipid tails [26,29]. Currently, both mRNA-1273 and BNT162b2 contain branched hydrocarbon tails with biodegradable linkages (Figure 2b).
Polyethylene glycol (PEG)-lipid
Polyethylene glycol (PEG) lipids generally comprise <2.5% of the total formulation. Nevertheless, they play a key role in controlling particle size and elongate shelf-life stability by preventing aggregation and fusion of LNPs. However, PEG-lipids need to be balanced since at high concentrations they prevent the delivery of RNA into cells. Recently, there have been some reports of immune responses to PEG molecules which have prompted the search for safe and potent PEG alternatives for mRNA LNPs, such as polysarcosine [33••]. Both mRNA-1273 and BNT162b2 contain 1.5 and 1.6% PEG-lipid in formulation, respectively [9].
Long-term preservation
The long-term storage and stability of mRNA-LNPs is a major consideration in the design of the formulations, and one which hasn’t been explored thoroughly, given its high clinical relevance and effect on vaccine distribution and ultimately, price. It's astonishing to look at the speed of development of Moderna’s mRNA-1273, the first mRNA vaccine for SARS-CoV-2 to reach clinical trials. From the first viral genome publication, it took a mere two days to finalize the vaccine sequence, 25 days to manufacture the first dose and 63 days until the first participant was dosed in the Phase 1 clinical study (NCT04283461) [35]. However, scaling up to supply a global vaccine demand is challenging [36]. The ultra-low storage requirement of currently approved mRNA vaccines (−80°C for BNT162b2 and −20°C for mRNA-1273) due to mRNA-LNP instability at room temperature is a system constraint. Several studies have been carried out with the purpose of finding optimal LNP freezing conditions. Currently, it is accepted that to freeze LNPs, cryoprotectants such as sucrose or trehalose should be added after mRNA-LNP formulation [37]. Zhao et al. investigated the long-term storage conditions of lipid-like nanoparticles (LLNs). They report that the addition of 5% sucrose or trehalose to LLNs-mRNA formulations stored in liquid nitrogen, preserved the physicochemical properties of the particles, and allowed maintenance of in vivo delivery efficacy for three months [38].
Lyophilization of mRNA LNPs can also be accomplished by the addition of lyoprotectants. For example, lyophilization provides Moderna’s phase II mRNA-LNP CMV vaccine (mRNA-1647) stability for at least 18 months at 5°C, however, lyophilization is an expensive and time-consuming process [39,40].
Currently, both mRNA-1273 and BNT162b contain 10% final concentration of sucrose in the final product, pre-dilution [9•]. Of note, although sucrose is currently used as a cryoprotectant in both authorized COVID-19 mRNA vaccines, initial temperature requirements were different, with Moderna’s mRNA-1273 stored at −15°C to −20°C and Pfizer/BioNTech’s BNT162b2 at −60°C to −80°C. Recent stability data has prompted an approval by the European Medicines Agency (EMA) to store the BNT162b2 vaccine at −15°C to −25°C for two weeks [34]. While distribution and storage requirements have been solved from an engineering perspective by designing temperature-controlled shipping containers, biotechnological advances to improve stability at non-frozen temperatures are still in demand.
A current hypothesis claims that freezing the LNPs protects the mRNA strands within the LNPs rather than stabilizes the LNPs themselves. LNPs may degrade over time by aggregation, fusion or leakage of mRNA from the LNPs, however, aggregation and fusion can be solved by the addition of stabilizing lipids such as PEG and mRNA leakage hasn't been previously reported [9•] Therefore, while LNP stability is important, it is not the limiting factor for short-term non-frozen LNP stability. mRNA degradation within LNPs occurs at a faster rate and dictates the storage time and temperature. This occurs mainly via chemical degradation by hydrolysis of the nucleic acid phosphodiester backbone and oxidation in presence of water or acids/bases. Initially, LNPs encapsulating smaller RNA payloads such as siRNA contained a single phospholipid outer layer with ionizable lipids inverted internally with little to no water entrapped. However, recent reports suggest that LNP-encapsulated mRNA contain within their core ionizable lipids, cholesterol and water. Some studies indicate that up to 24% of the LNP core is made of water, which affects mRNA stability within the LNP [41]. This can perhaps explain why FDA-approved siRNA LNPs (Patisiran) can be stored at 2−8°C for 24 months [42] while current mRNA-LNP COVID vaccines can be stored for one month at this temperature pre-dilution [43,44].
Conclusion and General guidelines
The successful design of effective mRNA-LNPs encompasses multiple considerations and choices which need to be made in accordance with the therapeutic purpose. While there is no single recipe for a successful mRNA-LNP, some common grounds do emerge when analyzing what is currently known on successful mRNA vaccines (Table 1).
In terms of mRNA sequence, modified and mRNA optimized sequences are required for maximal protein expression. Regarding formulation, currently approved mRNA LNP vaccines contain similar ratios of lipids with a varying ionizable lipid and PEG-Lipid. Even though the ionizable lipid is not identical in both vaccines, they are somewhat structurally similar regarding the amino-alcohol head group and branched hydrocarbon lipid tails with ester bonds.
Interestingly, while studies are constantly attempting to enhance protein expression from mRNA-LNPs [45], it seems that for vaccine purposes there is a weak correlation between the formulation’s protein expression level and the ability to elicit adaptive immune responses [27,46•]. Therefore, the endpoint for screening formulations should be the ability to elicit an adaptive immune response and not the overall protein expression.
Stability and long-term storage are crucial for mRNA-LNP formulations. From an analysis of the global health innovation center at duke university, an estimated 4.1 Billion mRNA-LNP vaccine doses are planned to be manufactured during 2021, note that the entire SARS-CoV-2 vaccine landscape is projected at 12 Billion doses for this year [47••]. The requirement to store these vaccines at ultra-low temperatures is challenging. A current hypothesis claims that the critical point is the mRNA strand stability within the LNP rather than the vehicle stability. Therefore, while algorithmic optimization of mRNA structure and modifications can improve stability, the best current vaccines should be formulated with cryoprotectants such as sucrose and novel solutions for LNP stability at non-frozen temperatures are still in demand.
To sum up, mRNA-LNP vaccines are an effective modular vaccine platform that enables rapid manufacturing of new vaccines and should be optimally designed considering the therapeutic target.
Conflict of interest statement
The authors declare the following competing financial interest(s): D.P. receives licensing fees (to patents on which he is an inventor) from, invested in, consults (or on scientific advisory boards or boards of directors) for, lectured (and received a fee) or conducts sponsored research at TAU for the following entities: ART Biosciences, BioNtech RNA Pharmaceuticals, Centricus, Diagnostear Ltd., EPM Inc., Earli Inc., lmpetis Biosciences, Kernal Biologics, Newphase Ltd., NLC Pharma Ltd., NanoGhosts Ltd., Roche, SirTLabs Corporation, and Teva Pharmaceuticals Inc.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
CRediT authorship contribution statement
Edo Kon: Conceptualization, Writing - original draft. Uri Elia: Conceptualization, Writing - original draft. Dan Peer: Conceptualization, Writing - original draft.
Acknowledgement
EK thanks the Yoran Institute for Human Genome Research for their support.
References
- 1.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 2.Tatematsu M., Funami K., Seya T., Matsumoto M. Extracellular RNA sensing by pattern recognition receptors. J Innate Immun. 2018;10:398–406. doi: 10.1159/000494034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S., et al. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit Cytolytic T cell responses. Mol Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., et al. Type i IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 9•.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review that discusses proposed structures of mRNA-LNPs, factors that impact mRNA-LNP stability and strategies to optimize mRNA-LNP product stability.
- 10.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 12.Kremsner P.G., Guerrero R.A.A., Arana E. Efficacy and safety of the CVnCoV SARS-CoV-2 mRNA vaccine candidate: results from herald, a phase 2b/3, randomised, observer-blinded, placebo-controlled clinical trial in ten countries in Europe and Latin America. Lancet. 2021 doi: 10.1016/S1473-3099(21)00677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolgin E. COVID vacine flop spotlights mRNA design challenges. Nature. 2021;594 doi: 10.1038/d41586-021-01661-0. [DOI] [PubMed] [Google Scholar]
- 14.Xu S., Yang K., Li R., Zhang L. mRNA vaccine era—mechanisms, drug platform and clinical prospection. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissman D. mRNA transcript therapy. Expert Rev Vaccines. 2014;14:265–281. doi: 10.1586/14760584.2015.973859. [DOI] [PubMed] [Google Scholar]
- 16.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics-developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 17.Pascolo S. Synthetic messenger RNA-based vaccines: from scorn to hype. Viruses. 2021;13:1–12. doi: 10.3390/v13020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gergen J., Petsch B. mRNA-based vaccines and mode of action. Nat Rev Drug Discov. 2021;17:261–279. [Google Scholar]
- 19.Asrani K.H., Farelli J.D., Stahley M.R., Miller R.L., Cheng C.J., Subramanian R.R., Brown J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018;15:756–762. doi: 10.1080/15476286.2018.1450054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T., et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlake T., Thess A., Thran M., Jordan I. mRNA as novel technology for passive immunotherapy. Cell Mol Life Sci. 2019;76:301–328. doi: 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:0933–0942. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Wayment-Steele H.K., Kim D.S., Choe C.A., Nicol J.J., Wellington-Oguri R., Sperberg R.A.P., Huang P.-S., Das R. Theoretical basis for stabilizing messenger RNA through secondary structure design. bioRxiv Prepr Serv Biol. 2020 doi: 10.1101/2020.08.22.262931. [DOI] [Google Scholar]; An interesting study that presents simple calculations for estimating RNA stability against hydrolysis, and a model that links the average unpaired probability of an mRNA to its overall hydrolysis rate.
- 25.Lima S.A., Chipman L.B., Nicholson A.L., Chen Y.H., Yee B.A., Yeo G.W., Coller J., Pasquinelli A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol. 2017;24:1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Ther Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:1–30. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramishetti S., Hazan-Halevy I., Palakuri R., Chatterjee S., Naidu Gonna S., Dammes N., Freilich I., Kolik Shmuel L., Danino D., Peer D. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv Mater. 2020;32:1–8. doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 31.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J Control Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 32•.Zhang Y., Sun C., Wang C., Jankovic K.E., Dong Y. Lipids and lipid derivatives for RNA delivery. Chem Rev. 2021 doi: 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive review covering decades of advances in research of lipids for RNA delivery from conception to approval of the three LNP-RNA formulations. The review entails an exhaustive account of lipid chemical structures, characterization, formulation methods and structure activity relationships.
- 33••.Nogueira S.S., Schlegel A., Maxeiner K., Weber B., Barz M., Schroer M.A., Blanchet C.E., Svergun D.I., Ramishetti S., Peer D., et al. Polysarcosine-functionalized lipid nanoparticles for therapeutic mRNA delivery. ACS Appl Nano Mater. 2020;3:10634–10645. [Google Scholar]; A study that demonstrates the potential of replacing the PEG lipid in an mRNA formulation with Polysarcosine, a polymer made of repetitive units of an endogenous amino acid. The study demonstrates that LNPs formed with polysarcosine yield higher protein secretion with reduced immunostimulatory response compared to PEG-lipid based LNPs.
- 34.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021 doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moderna . 2021. Moderna's Work on our COVID-19 Vaccine. [Google Scholar]
- 36.Irwin A. What it will take to vaccinate the world against COVID-19. Nature. 2021 doi: 10.1038/d41586-021-00727-3. [DOI] [PubMed] [Google Scholar]
- 37.Lball R., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomedicine. 2017;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karve S. vol 1. 2018. (Process of Preparing mRNA - Loaded Lipid Nanoparticles). [Google Scholar]
- 40.Moderna . 2021. Cytomegalovirus (CMV) Overview mRNA 1647. [Google Scholar]
- 41.Arteta M.Y., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc Natl Acad Sci U S A. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CDER . 2017. Product Quality Review (S) NDA 210922 ONPATTRO (Patisiran) Lipid Complex Injection Addendum to Drug Product Quality Review. [Google Scholar]
- 43.2021. Bnt162b - Fact Sheet for Healthcare Providers Administering Vaccine (Vaccination Providers) [Google Scholar]
- 44.2020. Emergency Use Authorization (EUA) Application for mRNA-1273. [Google Scholar]
- 45.Kon E., Hazan-Halevy I., Rosenblum D., Cohen N., Chatterjee S., Veiga N., Raanani P., Bairey O., Benjamini O., Nagler A., et al. Resveratrol enhances mRNA and siRNA lipid nanoparticles primary CLL cell transfection. Pharmaceutics. 2020;12:1–14. doi: 10.3390/pharmaceutics12060520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Elia U., Ramishetti S., Rosenfeld R., Dammes N., Bar-Haim E., Naidu G.S., Makdasi E., Yahalom-Ronen Y., Tamir H., Paran N., et al. Design of SARS-CoV-2 hFc-conjugated receptor-binding domain mRNA vaccine delivered via lipid nanoparticles. ACS Nano. 2021;15:9627–9637. doi: 10.1021/acsnano.0c10180. [DOI] [PubMed] [Google Scholar]; A study that screens mRNA-LNP vaccine formulations for their ability to elicit an adaptive immune response. Following the initial ionizable lipid screen, the authors designed an mRNA-vaccine encapsulating a sequence encoding a SARS-CoV-2 human FC-conjugated receptor binding domain. The novel mRNA vaccine demonstrated a robust humoral response with a high level of SARS-CoV-2 neutralizing antibodies.
- 47••.Center DGHI . 2021. Launch and Scale Spedometer.https://launchandscalefaster.org/covid-19/vaccinemanufacturing [Google Scholar]; The Launch and Scale Speedometer initiative systematically analyzes, generates and shares valuable insights on the introduction and scaling of medical interventions. Among its activities, the platform provides insights and data on the COVID-19 vaccine landscape, regarding manufacturing, purchases and more. The initiative is led by the Duke Global Health Innovation Center, with support from the Bill & Melinda Gates Foundation.
- 48.FDA . 2021. Comirnaty and Pfizer-BioNTech COVID-19 Vaccine. [Google Scholar]
- 49.Moderna . 2021. Moderna Media Releases. [Google Scholar]
- 50.CureVac . 2021. CureVac News Room. [Google Scholar]
- 51.Granados-Riveron J.T., Aquino-Jarquin G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.WHO . 2020. World Health Organization: International Nonproprietary Names Programme. Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein; p. 11889. [Google Scholar]
- 53.WHO . 2020. World Health Organization: International Nonproprietary Names Programme. Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein; p. 11868. [Google Scholar]
- 54.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J Control Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nance K.D., Meier J.L. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent Sci. 2021;7:748–756. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]