Abstract
Microglia are an essential population of resident immune cells in the central nervous system (CNS) and retina. These microscopic cells possess sub-cellular processes that make them challenging to image due to limited resolution and contrast. The baseline behavior of microglial processes in the living retina has been poorly characterized, and yet are essential to understanding how these cells respond under conditions of health, development, stress and disease. Here we use in vivo adaptive optics scanning light ophthalmoscopy combined with time-lapse imaging and quantification of process motility, to reveal the detailed behavior of microglial cells in a population of healthy mice. We find microglial processes to be dynamic at all branch-levels, from primary to end-protrusions. Cell-processes remodel at average speeds of 0.6 ± 0.4 µm/min with growth and deletion bursts of 0–7.6 µm/min. Longitudinal imaging in the same mice showed cell-somas to remain stable over seconds to minutes, but show migration over days to months. In addition to characterizing in vivo process motility and Sholl analysis using a microglial reporter mouse, we also demonstrate that microglia can be imaged without fluorescent labels at all. Phase-contrast imaging using safe levels of near-infrared light successfully imaged microglia soma and process remodeling with micron-level detail noninvasively, confirmed by simultaneous imaging of fluorescent microglial cells in transgenic mice. This label-free approach provides a new opportunity to investigate CNS immune system noninvasively without requiring transgenic or antibody labeling which could have off-target effects of changing normal microglial behavior. Additionally, CNS microglia study can now be conducted without the need for cranial window surgery which have the potential to change their behavior due to local or systemic inflammation.
1. Introduction
Microglia are resident immune cells in the retina and brain. The healthy central nervous system (CNS) contains millions of microglial cells; and comprises as much as 5–12% of all of the neuro-glial cells within the brain [1]. Healthy baseline microglia have microscopic cell bodies and thin branching processes that give them a ramified appearance. In the last two decades, it was discovered that these cells undergo dynamic and frequent process remodeling as the cells interact with their neural, glial and vascular neighbors in the healthy neural parenchyma [2,3]. When considering the density, constant structural remodeling and motility of microglia, the metabolic demands of this activity must serve an essential part of development, healthy brain wiring and response to foreign bodies and inflammatory threat to the nervous system [2–5]. Studies in the brain and in vitro have now established that this constant surveillance is a part of healthy neuronal function.
Less is known, however about microglia dynamics in the living retina, an extension of the central nervous system. While there are vast similarities in these tissues, there are also notable differences. Retinal microglia exhibit a more uniformly distributed appearance laterally, yet strong segregation axially with cell somas and processes highly confined to the plexiform layers of the retina which also happen to contain the dominant retinal vascular stratifications. It is tempting to speculate that this association may be attributed to the affinity of these cells to assist with synaptic remodeling which is expectedly rich at the plexiform layers. Still other reasons may have to do with a “no room” hypothesis in which cell somas have little room to compete with the densely packed somas of neuro-glia in the outer and inner nuclear layers. There are clearly many mysteries that are yet poorly characterized in the retina, and many may be revealed only through detailed study in the living retina, where neural, glial and vascular function remain intact and at physiological homeostasis.
Study of the dynamism of retinal microglia is essential to understanding the retina not only in health, but also to monitor the response to the myriad of diseases that impact retinal vision. In retinal disease such as glaucoma, diabetic retinopathy, age-related macular degeneration, uveitis and quite possibly all retinal disease that manifests as cell loss, retinal remodeling and inflammation may have a corresponding microglial response that may help or hinder disease progression and normal neural function.
Much of our understanding of retinal microglia have been from ex vivo studies of retinal microglia that have confirmed morphological activity of the cell processes [6,7]. However, histological artifacts and removal of retinal tissue have the potential to strongly impact the native activity of these cells. In the in vivo retina, studies have followed the response of cell bodies as a whole in health and disease [8–11], using transgenic mouse lines such as the CX3CR1-GFP which contain fluorescently labelled microglia, monocytes and natural killer cells. Studying microglia in the eye has the advantage that the imaging is noninvasive and the eye’s optical transparency avoids the scattering media complications of deep tissue imaging in the brain. Despite the theoretical benefits of studying retinal microglia in vivo, microglial cell-processes have been challenging to image in the living eye due to their microscopic size and limitations in imaging contrast to render these cells’ processes visible. Moreover, the activity of these subcellular parts is uniquely interesting due to their role in immune health and disease [5]. Wave aberrations of the anterior optics of the eye severely confound the imaging of these cells in the living retina. Left uncorrected, the spatial resolution of in vivo retinal imaging is often worse than ∼10–15 µm, insufficient to image the details of cell processes of microglia. Adaptive optics technology [12–14] has overcome that barrier, giving high spatial resolution imaging to structurally resolve cells in vivo with micron-level resolution [10,15–18].
Despite the recent advances, in vivo imaging studies have reported a surprising lack of process motility in retinal microglial cells in their baseline state, using scanning laser ophthalmoscopy (SLO) [19,20]. This is a surprising finding given that both the retina and the brain display an intact blood-brain-barrier and each tissue is derived from the same ontogenetic origins [5]. Moreover, retinal microglia studied with high power microscopes in vitro also show substantial process remodeling [6]. Microglia imaged in the living brain are well documented to have motile processes in the baseline state [2,3]. There are several reasons that could explain the surprising lack of process motility reported in vivo [19,20]. The first may be attributed to limited spatio-temporal resolution of the SLO imaging which would blur dynamic cellular changes at this level. Another complication may arise from the high light levels (100–230 µW) required to excite the exogenous fluorescence of these cells that must be recorded over long periods of time to image the glacial pace of remodeling [18,19]. Such imaging approaches have the potential to create a phototoxic environment of the retina that could alter baseline kinetics.
In this study, we use a custom-built adaptive optics scanning light ophthalmoscope (AOSLO) designed for the mouse eye which provides ∼1 micron lateral resolution in vivo [21]. We used this to image microglial cells in the healthy retina to find that the cell processes are in fact dynamic and remodel over all branch orders. We quantify the process motility of microglia in a population of living mice to better characterize this remodeling under baseline conditions. To appreciate the temporal dynamics over both large and small time-epochs, we characterize microglia kinetics over seconds to months. Such imaging of baseline dynamics of single cells was facilitated by a number of critical advances important for imaging these cells unperturbed. First, for fluorescence imaging of CX3CR1-GFP, we used detailed time-lapse videography using low levels of in vivo imaging light (53 µW of 488 nm ex., and 200–500 µW of 796 nm reflectance). These low light levels in a human eye would compare to ANSI-safe light exposures [22–24]. Secondly, our dual registration approach [25–27] provided sufficient signal in the near infrared channel to facilitate micron-level registration based on retinal landmarks that do not change over months in health (nerve fiber bundles, vascular landmarks). Such registration not only allowed us to provide stable registration within the same imaging session lasting hours, but also allowed us to return to the same location with micron precision to perform imaging follow up at the scale of days, weeks and even months which revealed process motility and cell migration. Third, we present a detailed quantification of microglial process motility in a population of seven healthy mice, using a custom semi-automated image-analysis approach.
In a final advance, we report an exciting finding that microglial dynamics can be imaged and quantified without fluorescent labels at all. Using our recently developed phase-contrast AOSLO imaging [28,29], the dynamics of individual processes of CX3CR1-GFP positive cells were imaged label-free using near infrared light alone, and the process velocities were quantified. Study in the mouse allowed us to confirm that the dynamics cells imaged label-free co-localized with fluorescent protein under CX3CR1 expression, confirming that these immune cells can be imaged label-free and can be identified through morphology and kinetics alone. Additionally, other sub-cellular dynamics inside the cell could be visualized in the label-free channel alone, and were not observed with simultaneous fluorescence imaging. Combined, these advances enable a new level of study of microglia in their natural environment without many of the negative confounds of prior limits of resolution, surgery, contrast, or requirements of fluorescent agents.
2. Methods
2.1. Animals
Two strains of mice were purchased from pure stock repositories at Jackson Labs. CX3CR1-GFP homozygous knock-in mice, which express EGFP (enhanced green fluorescent protein) in microglia, monocytes and other immune cells that contain the chemokine CX3 receptor 1, and C57BL/6J (The Jackson Laboratory, stocks 005582 and 000664 respectively, Bar Harbor, Maine, USA). The progeny of paired C57BL/6J and CX3CR1-GFP homozygote mice produced litters of CX3CR1-GFP positive heterozygote mice. Seven CX3CR1-GFP+ heterozygous mice from multiple litters were imaged (4 males and 3 females; age 7.0 ± 2.9 months old, weight 29.1 ± 6.0 g (mean ± SD)). Mice were fed standard chow and water ad libitum, and housed in 12-hour light-dark cycles. Mice were treated in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Guidelines of the University Committee on Animal Resources at the University of Rochester (Rochester, NY, USA) were followed.
2.2. Animal preparation for imaging
A detailed protocol of mouse preparation is given in our previous publication [30]. Briefly, mice were anesthetized with a cocktail of ketamine-xylazine (100 mg/kg and 10 mg/kg respectively), via an intraperitoneal (IP) injection. A gas mixture of 1% (v/v) isofluorane and supplemental oxygen was used to give stable anesthesia over ∼2 hours of imaging. Body temperature was maintained at 37°C using a supplemental heat-pad. Pupil dilation was achieved with a drop of 1% tropicamide (Sandoz, Switzerland) and 2.5% phenylephrine (Akorn, Illinois, USA). Our custom AOSLO imaging system operates in free-space, meaning there is no physical contact between the system and the mouse eye – thus ensuring no disruption of the normal internal pressure of the eyeball, and therefore the physiological state of the retina. Corneal hydration was achieved with 1) a rigid contact lens with +10 D power and 1.6 mm base-curve (Advanced Vision Technologies, Colorado, USA), and 2) by applying an ophthalmic lubricant (GenTeal, Alcon Laboratories Inc., Texas, USA) on the exposed outer-rim of the cornea/sclera, every ∼20 minutes. This ensured stable imaging for >2 hours without clouding of the cornea which can lead to increased light-scattering and therefore reduced image quality. The mouse head was stabilized on a stereotaxic stage with bite-bar (Bioptigen, North Carolina, USA), with three translational and three rotational degrees of freedom to allow alignment of mouse pupil and navigation to different retinal locations, with field size ranging –5° over extended periods of time. In a subset of experiments, C57BL/6J mice were IP injected with fluorescein (AK-FLUOR 10%, 100 mg/mL, 0.1 mL of 2.5% w/v injected, Akorn, Illinois, USA), for calibration of longitudinal chromatic aberration (LCA), as described below.
2.3. AOSLO system for imaging fluorescence and label-free phase-contrast
The custom mouse AOSLO for simultaneous imaging of fluorescence and near-infrared reflectance has been described in detail previously [21,30]. The system relays a raster-scan onto the mouse retina using five 4-f afocal telescopes and two orthogonal scanners, and collects the reflected and fluorescent light in double-pass configuration. Wavefront sensing is performed using a Hartmann-Shack wavefront sensor with a 904 nm laser (QFLD-905-10S, QPhotonics, Michigan, USA), and closed-loop adaptive optics correction at 13 Hz is achieved with a membrane-based deformable mirror (DM97-15, ALPAO, France). EGFP and fluorescein fluorescence were excited using a 488 nm laser (iChrome Multi-Laser Engine, Toptica Photonics, New York, USA), and emission collected using a bandpass filter (two stacked 520Δ35 nm filters, FF01-520/35-25, Semrock, New York, USA). Simultaneous reflectance imaging was performed using a near-infrared 796Δ17 nm superluminescent diode (S790-G-I-15, Superlum, Ireland).
Light from the retina was detected by two photomultiplier tubes (PMTs), one each for visible and near-infrared (NIR) wavelengths (H7422-40 and -50 respectively, Hamamatsu, Japan). Fluorescence was detected through a confocal pinhole (4.9 Airy disk diameters, ADD). NIR reflectance was imaged both in confocal mode (2.1 ADD) and in label-free phase-contrast mode. The phase-contrast approach imaged forward refracted and multiply-scattered light from a detection plane conjugate to the RPE/choroid complex to enhance the contrast of translucent cells, as recently published by our group [28]. The raster scanning system collected videos at a frame-rate of 25 Hz, over fields of view of 3 to 5 degrees (102 to 170 µm) [31], over typically 480 × 608 pixels. Videos were desinusoided in real-time [32]. The measured in vivo imaging resolution was: lateral: 0.77 µm for 520 nm fluorescence and 1.2 µm for 796 nm reflectance, and axial: 10.5 µm and 16.1 µm for fluorescence and reflectance respectively, based on calculations from our group previously published [21,30]. This ensured near-diffraction limited imaging in the lateral dimension, limited only by the numerical aperture of the eye, and sufficient axial sectioning to isolate the fluorescence of each of the three stratified layers of microglial cells in the retina [6].
2.4. Commercial SLO imaging
Wide field imaging was performed with commercial confocal SLO (Heidelberg Spectralis HRA, Heidelberg Engineering Inc., Carlsbad, CA). AOSLO fields are super-imposed on the same in Fig. 1. For the SLO, two modalities were used: NIR confocal reflectance and blue-light fluorescence (ex: 488 ± 2 nm, em: 505 nm long-pass). Field size 55 degrees. NIR focus was positioned on structures in nerve fiber layer (NFL)/superficial vasculature. Fluorescence focus was optimized for best visual contrast of microglial cells.
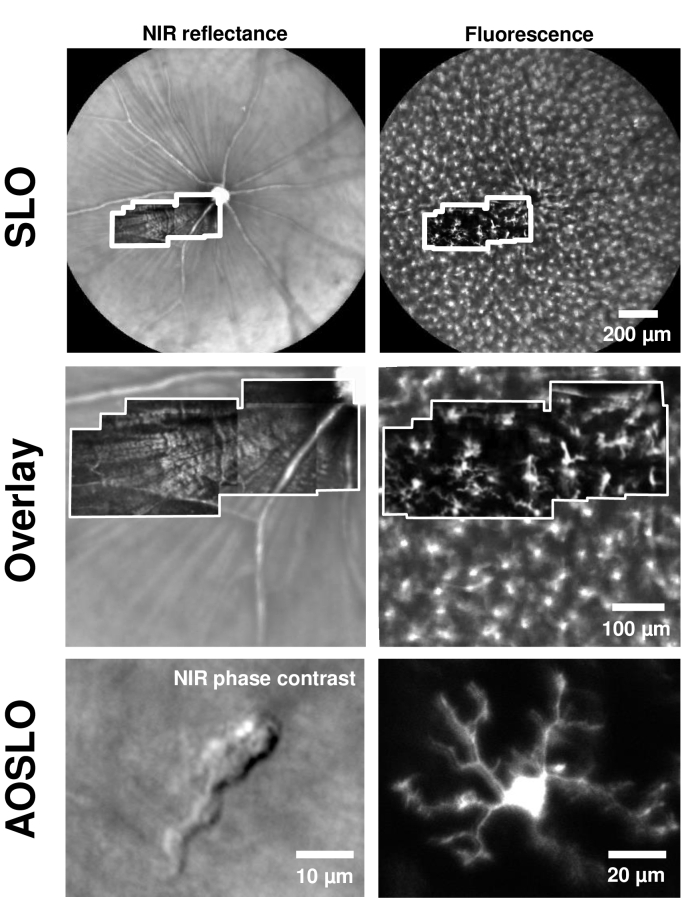
Fig. 1.
Adaptive optics (AO) resolves details of single microglial cell processes in vivo, in NIR label-free and fluorescence channels. Top: Wide-field images of the retina captured with a commercial SLO, with NIR confocal reflectance (left) and EGFP fluorescence (right) in a CX3CR1-GFP mouse. Overlaid inset (white border) shows montage of AOSLO dual-channel images from the same mouse (with simultaneous collection of confocal reflectance and fluorescence). Middle: Zoom-in of top images, to compare SLO images with overlaid AOSLO montages from the same mouse (white border). AOSLO offers much higher axial sectioning, therefore isolating fluorescence from a single stratification of microglia, unlike the SLO. Bottom: High-resolution AOSLO images (single fields) showing microglial cell soma and its processes, in both label-free NIR phase contrast (left) and fluorescence (right). (Bottom panel shows two different locations. Figure 8 shows simultaneous the CX3CR1-GFP fluorescence from same cell imaged in the label free configuration on the left.)
2.5. In vivo correction of longitudinal and transverse chromatic aberrations
Before imaging fluorescent microglia in a CX3CR1-GFP mouse, a C57BL/6J mouse of similar age was injected IP with fluorescein (details above) and imaged on the AOSLO for coarse calibration and correction of the large chromatic aberrations of the mouse eye [21], which has an optical power of ∼520 D. Because fluorescein has approximately same excitation and emission spectra as EGFP, we used this strategy to coarsely optimize the excitation plane of focus to the inner retina and align the conjugate pinhole to maximize confocal contrast and efficiency of the AOSLO. Using a C57BL/6J mouse with similar age to the target CX3CR1-GFP mouse (littermates when possible) provided best matching of axial length [31] and chromatic properties. We used known anatomical structures (retinal capillary stratifications, ∼5 µm diameters) to align focal planes of 520 nm fluorescence (blood plasma) and 796 nm reflectance. The two wavelength channels were independently focused. The visible light was focused on a laminar capillary stratification. Depending on the experiment need, the near-infrared reflectance channel was either focused on the same layer, to simultaneously image the same cell in two wavelengths, or intentionally focused on a reflective layer with high structural contrast (typically nerve fiber layer (NFL)), to help in online and offline registration of imaging location. This approach facilitated the best dual-channel registration based on cross-correlation [26,33]. The detector pinholes were placed conjugate to the illumination planes, for confocal imaging.
After the approximate chromatic calibration in a C57BL/6J mouse using fluorescein, the CX3CR1-GFP mice were subsequently imaged using the same AOSLO and reoptimized for the small LCA shifts known to exist in different mice [21]. We navigated to a 3-5° field-of-view retinal location, typically in the superficial or intermediate laminar layer of microglia [6], with 1 to 4 visible microglial cells within the imaging field. Navigation was done using only 13 µW of 488 nm light, for brief ∼10 s durations, to minimize phototoxicity and microglial activation. For fine calibration, the visible fluorescence channel was focused on the microglial cells, with the near-infrared reflectance focused either on the same layer or the NFL layer. For the former case, to know which layer the fluorescence was coming from, the channel was briefly turned into a visible reflectance channel, using 515 nm laser excitation (7 µW) which passed through the fluorescence emission filters.
After correction for LCA the following steps were taken for in vivo correction of transverse chromatic aberration (TCA). TCA is mainly impacted by change in the optical path in the pupil plane. Therefore, TCA correction was done each time we navigated to a new imaging location, which has the potential to shift the entrance pupil position at the eye. To adjust for TCA a 515 nm laser light source was turned on briefly to align a capillary or blood vessel in visible and near-infrared reflectance. Lateral alignment was done by laterally translating the mouse pupil only, without rotating the head of the mouse. Finally, for precise micrometer level correction, linescan mode (15 kHz imaging) [34] was turned on to visualize individual blood cells flowing through capillaries, which gave the imaging operator high contrast features to help align the two wavelength channels to within an error of the 3–7 µm size of a flowing red blood cell, in both the axial (LCA) and lateral (TCA) dimensions.
2.6. Time-lapse imaging of microglia
Once a field with 1 to 4 cells was selected, steps were taken to ensure that the imaging location did not change for the duration of time-lapse imaging. During imaging, we marked the computer monitor (with real-time AOSLO video-feed) with erasable markers to help the operator track and keep the field of view locked on the same region of interest. Features marked included fluorescent microglia somas and capillaries and NFL bundles in the reflectance channel. Where possible, structures with different orientations were marked, to increase sensitivity to translational movements of the mouse due to breathing or mouse-holder positional drift. The imaging protocol used for time-lapse imaging is visualized in Fig. 2(A). For the population analysis of process motility, EGFP fluorescence was continuously recorded for 40 seconds using only 53 µW of 488 nm light. Such recording was repeated every 5 minutes, for a total duration of between 35 to 95 minutes in seven CX3CR1 mice (mean=67.2 ± 18.9 minutes), depending on image quality, depth of anesthesia and animal cooperation. Continuous imaging of near-infrared reflectance helped the user visually maintain the same imaging location using small translational and rotational adjustments to the mouse head position.
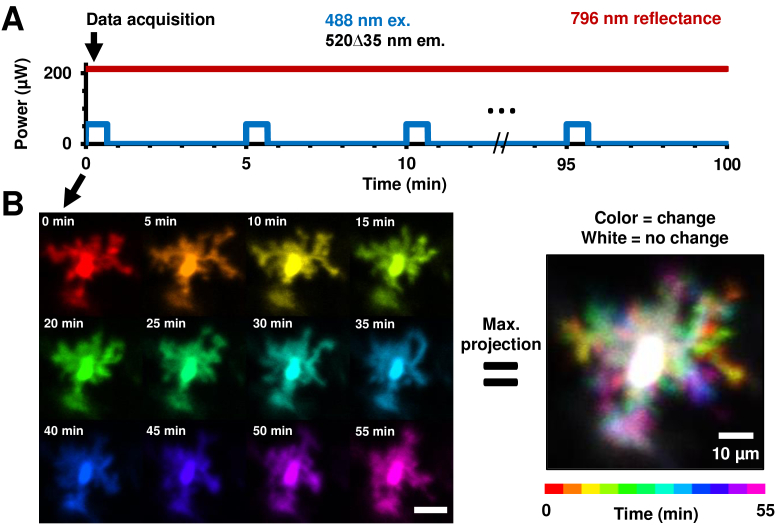
Fig. 2.
Experimental protocol for time-lapse imaging of microglia with in vivo AOSLO. A) Plot of power levels of multiple imaging wavelengths impinging on the retina during one imaging session in a mouse. The same retinal location (3–5° FOV) was imaged and tracked in 5-minute intervals. Near-infrared reflectance provided continuous registration throughout the imaging session, while fluorescence imaging for 40 seconds each (per timepoint) visualized the microglial cells. B) Images of the same microglial cell imaged at 5-minute intervals, with each timepoint given a unique color. Scale bar is 20 µm. A maximum projection image of such a stack of colored images gives the image on the right with the custom color-map at the bottom representing time. Moving parts of the cells (its processes) are thus represented in color (single or combinatorial hue) while the stationary part (soma) is represented as white.
2.7. Registration of images from multiple time-points
The near-infrared reflectance channel was used for registration of respiration and other eye motion [33] over seconds to hours. The same channel was also used to precisely register locations imaged across days to months. To account for any remaining uncorrected TCA due to small eye/pupil movements, augmented registration was performed on the binned time-lapse images of fluorescent microglial cells, with the cell somas acting as the dominant registration landmarks, providing exceptional sparse point sources for registration. This enabled determination of process displacement and speed relative to the soma. This additional step was only performed for the population process motility analysis, where the maximum imaging interval was 95 minutes.
2.8. Data visualization
The motility of the processes of microglial cells are best appreciated in videos (Visualization 1 (17MB, avi) , Visualization 2 (26MB, avi) , Visualization 3 (24MB, avi) , Visualization 4 (25.5MB, avi) , Visualization 5 (37MB, avi) , Visualization 6 (117.6MB, avi) and Visualization 7 (201.6MB, avi) ). To represent motion in a static image (Fig. 2(B)), we used a custom color-code in MATLAB (Version 9.2, MathWorks, Massachusetts, USA) that we defined previously [17] (ARVO abstract). Each timepoint was given a unique color, with each color in the map always adding to white. Thus, in a maximum projection image (Fig. 2(B), right) of the stack of time-lapse images, white pixels represented static parts of the cells, while colored pixels represented moving parts of the cells; with each color indicating the time at which the process was present at that location. This methodology was used in future figures to visualize motility in primary to tertiary end-processes of microglia (Fig. 3–Fig. 4).
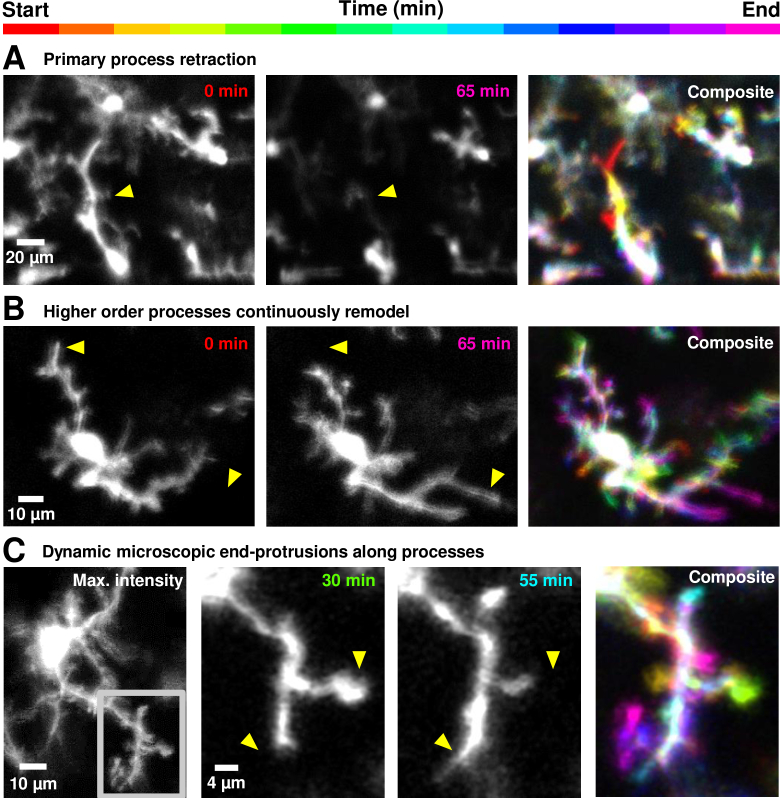
Fig. 3.
Heterogeneity in process motility in healthy retinal microglial cells of the living mouse, from primary to tertiary end-processes. A) Rare primary process retraction observed in a microglial cell (all timepoints shown in Visualization 2 (26MB, avi) ). Yellow arrow-heads mark the same retinal landmarks. B) In another cell, secondary and tertiary processes remodel (Visualization 3 (24MB, avi) ). In Fig. 5 and Fig. 6, these observations of motility are quantified at the microscopic process level, using spatio-temporal image analysis. C) Left: Stable imaging of a single microglial cell over 1.6 hours (95 minutes) (further explored in Fig. 8), with maximum intensity projection computed over the imaging time-course. Gray inset shows a single branch of the cell further investigated in subsequent panels to right. High-resolution view of a branch shown in gray inset in first panel, showing motile end-processes, only a few microns long, successfully tracked over 95 minutes (Visualization 5 (37MB, avi) ). Yellow arrow-heads mark the same retinal landmarks.
Fig. 4.
Changing directionality of ramified processes may indicate bias towards dynamic and invisible cytokine/chemokine gradients. In one microglial cell, imaged within the nerve fiber layer, processes are observed to attain a directional bias at certain timepoints (Visualization 4 (25.5MB, avi) ). Dark yellow arrow show change in directional bias of processes of one cell, over tens of minutes.
2.9. Quantification of cell-process displacement and speed
For each image location, a maximum intensity projection (MIP) image was computed from the stack of registered frames from the same focal plane. The MIP image rendered all microglial process-paths visible, regardless of their persistence. On this MIP image, processes-paths were manually traced, from soma to the tip of the process (representative traces shown in Fig. 5(A), labelled a-d). Next, for each process-path, the tip of the process was tracked at all time-points of time-lapse acquisition, typically in 5-minute intervals.
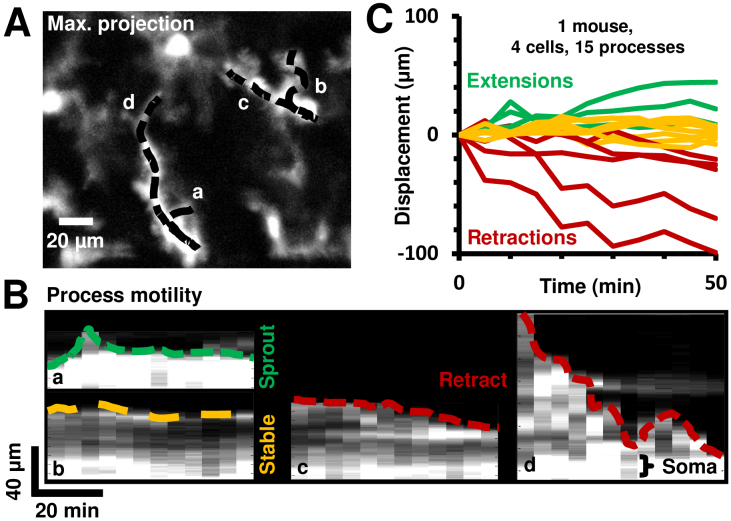
Fig. 5.
Quantification of cell-process motility and speed using space-time image analysis in a representative mouse. A) Maximum projection image of a time-lapse sequence at one AOSLO field. Process paths are traced and overlaid as shown with representative traces a-d. B) Four representative space-time images, each corresponding to a unique cell-process, showing the intensity across a process path as a function of time. The tip of the process is traced in color (green: extension, yellow: stable, red: retraction). C) Similar displacement vs time curves are plotted for all processes analyzed in one mouse.
The image intensity along each path was plotted on a vertical axis, representing space (in µm). This path intensity was plotted for each of the subsequent imaged time points, producing a space-time image of process path intensities over time, with time on the horizontal axis (Fig. 5(B)). Thus, a unique space-time image of path intensity profiles was created for every cell-process analyzed, across 129 processes in 26 cells at 9 imaging locations in 7 mice. Four representative space-time images are shown in Fig. 5(B). These images were quantitatively analyzed for process displacement and speed using methodology developed by our lab [30]. Next, the displacement of the tip of the process was plotted as a function of time, with extending processes marked in green, stable ones marked in yellow and retracting processes marked in red. Processes with instantaneous displacement less than 15 µm (the typical size of a cell soma) over 35–95 minutes were categorized as ‘stable’. All other processes were categorized as extending or retracting based on the direction of motion relative to the soma. The local slope of the displacement profile gave the instantaneous cell-process speed, while total displacement divided by total time gave the average speed. The histogram of a population of microglial process displacement magnitudes from seven mice was computed.
The net-displacement of each process is defined as the displacement of a cell-process (relative to soma) at the end of an imaging session, relative to the beginning of the time-lapse session. Conversely, the maximum displacement of each process was defined as the difference between the minimum and maximum displacement of the cell-process (relative to soma) over the entire imaging session.
2.10. In vivo Sholl analysis
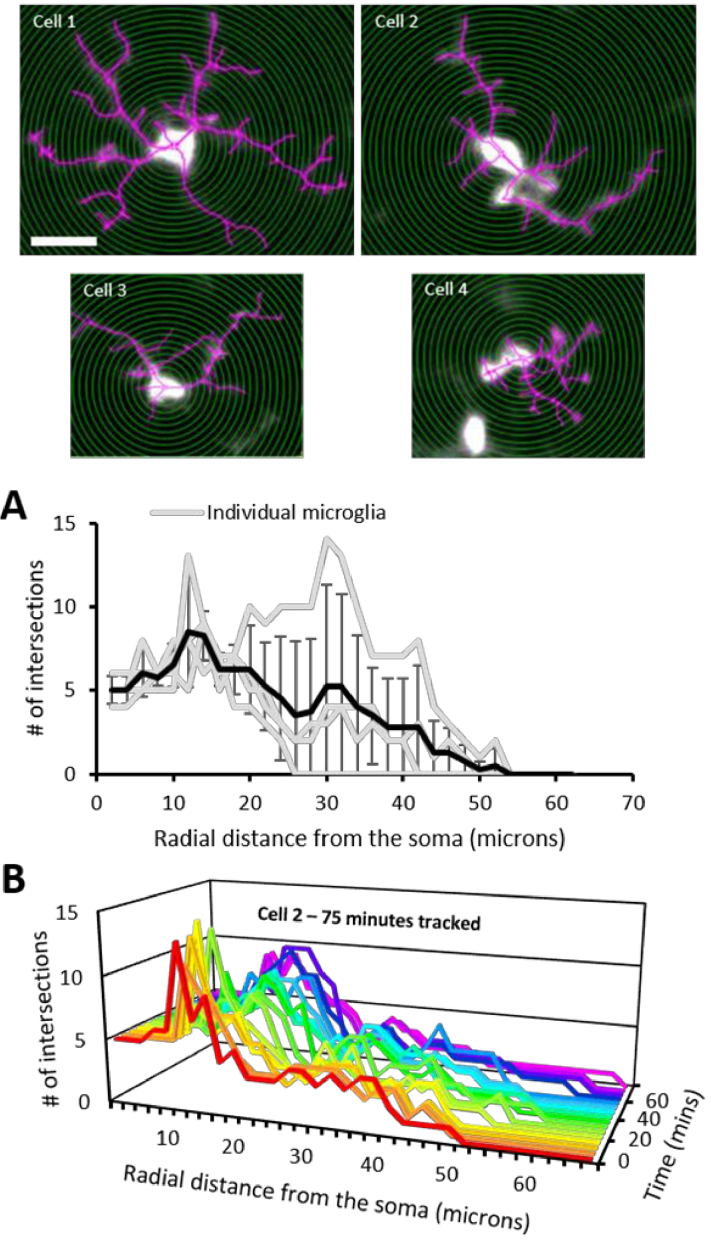
AOSLO videos of microglia (n=4 cells, chosen for their exemplary process resolution) were each averaged over 40 seconds and the resultant time-projected image was used for Sholl analysis. The ImageJ plugin, Simple Neurite Tracer (SNT v 3.2.3, [35,36]), was used for assisted tracing of microglial processes (Fig. 7, magenta) and performing Sholl analysis. For each cell, the number (#) of intersections were recorded at 2-micron radial intervals from the center of the soma. Next, to track process dynamics over time, Sholl analysis was performed for one cell (cell 2, Fig. 7, Visualization 8 (5.2MB, avi) ) over a duration of 75 minutes; this cell was chosen for its excellent resolution throughout the imaging session. For this case, one-minute videos were recorded at 5-minute intervals and the first 40 seconds were binned to the produce time-projected images (16 images total), before performing Sholl analysis.
Fig. 7.
Sholl analysis for assessing microglial complexity and dynamics. Top: Fluorescent AOSLO images (cells 1-4) used for Sholl analysis (scalebar = 20 µm). Concentric 2 µm radii (green) and process tracings (magenta) are overlayed. A. The number (#) of intersections for each cell (gray) plotted for every consecutive 2 µm distance from the center of the soma. Black line represents the mean ±1 SD. B. The number (#) of intersections vs. radial distance from the soma tracked in cell #2 for 75 minutes (5-minute intervals). Also see Visualization 8 (5.2MB, avi) for a time-lapse video of the fluorescence data and the corresponding cell tracings which enabled this time-varying Sholl analysis.
2.11. Longitudinal imaging over seconds to months
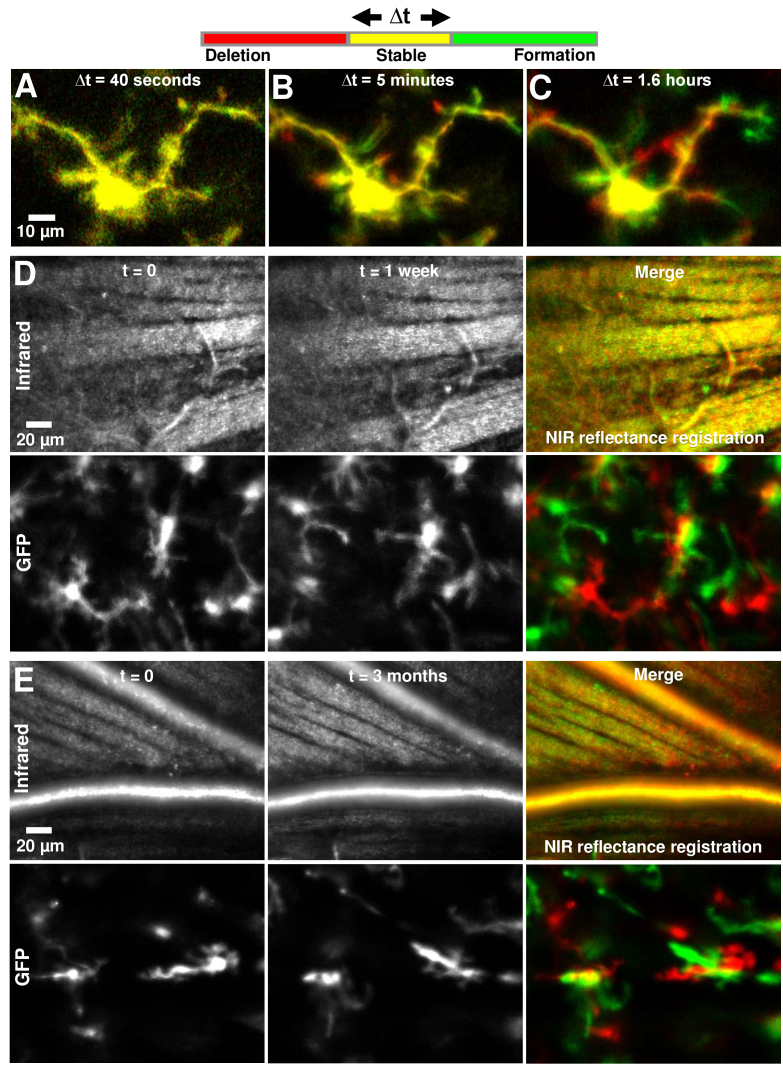
Because the imaging approach is non-destructive/noninvasive, we were able to return the mice healthy and uninjured to the cage for subsequent imaging. This allowed longitudinal imaging of the same mouse over days to months. The high spatial resolution of AOSLO imaging allowed returning to the same 3–5° retinal location with micrometer precision, using anatomical markers of vasculature and nerve fiber bundles to guide precise spatial follow-up under the presumption that these structures do not remodel in the healthy mouse over this time scale. Multiple temporal bandwidths were imaged: The same individual cell was tracked in vivo over intervals of 40 seconds, 5 minutes and 1.6 hours (Fig. 8). Separately, the same 5° retinal locations were tracked in vivo over intervals of 1 week and 3 months.
Fig. 8.
Imaging single-cell dynamics of microglia, from seconds to months. Each image is a red-green merge of two timepoints at the same imaging location, with imaging interval given by Δt. Thus, parts of the cell/image that don’t move in a certain interval appear yellow. A-C) Δt=40 seconds, 5 minutes and 1.6 hours (95 minutes) respectively. The same cell is tracked, in the same imaging session. A ‘zoom-in’ of the top-right branch of this cell is shown in Visualization 5 (37MB, avi) and Fig. 3(C). D-E) Δt=1 week and 3 months respectively. In each case, the EGFP fluorescence channel is shown on the bottom panel while the near-infrared reflectance channel is shown on the top. The reflectance channel provided precise registration of imaging location.
Next, red-green merges were used to visualize changes in cells over multiple temporal bandwidths, with t=0 labelled red and t=Δt labelled green. Thus, red represented deletion of cellular structures, while green, addition, and yellow, no change. The interval Δt was set to 40 seconds, 5 minutes, 1.6 hours (duration of typical continuous imaging session), 1 week and 3 months. The near-infrared confocal reflectance channel was used to ensure co-localized imaging over time, using anatomical markers. To visualize registration efficacy, red-green image merges were used, such that yellow image-regions would indicate exceptional structural registration.
2.12. Near-infrared label-free imaging of microglial cell process motility
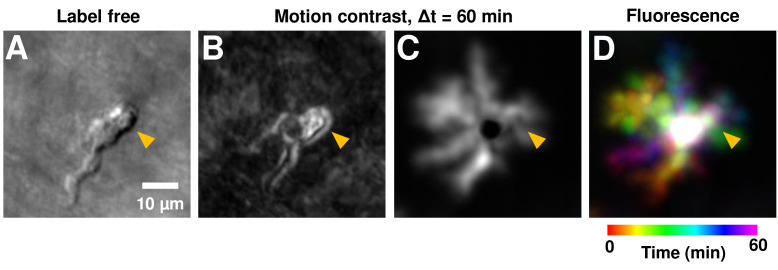
Cells were imaged simultaneously in two wavelength channels (Fig. 9, Visualization 6 (117.6MB, avi) –Visualization 7 (201.6MB, avi) ). The first collected CX3CR1 fluorescence as described above, to confirm the precise location of the EGFP+ cells. The second channel, for near infrared (796 nm) imaging, was modified to collect forward and multiply scattered light from the RPE/choroid complex, as described in our recent publication [28]. This imaging channel will be referred to as the ‘label-free’ or ‘phase contrast’ modality. It images translucent cell parts not visible in conventional confocal imaging at the same wavelengths. After a single EGFP+ cell was identified, time-lapse imaging was performed for ∼60 minutes at the same cell, with near continuous recording. Raw data was recorded as 60 videos of 1 minute duration, with <1 second gap between videos. (Note: this was different from the 5-minute interval time-lapse recording done for the population analysis of fluorescent cells in the previous sections. Time-lapse video frames were generated by averaging every 10 s (250 frames) of registered AOSLO video data. Registration of images was performed as described in Section 2.7. Process motility was tracked and quantified independently for both the label-free and fluorescence channels.
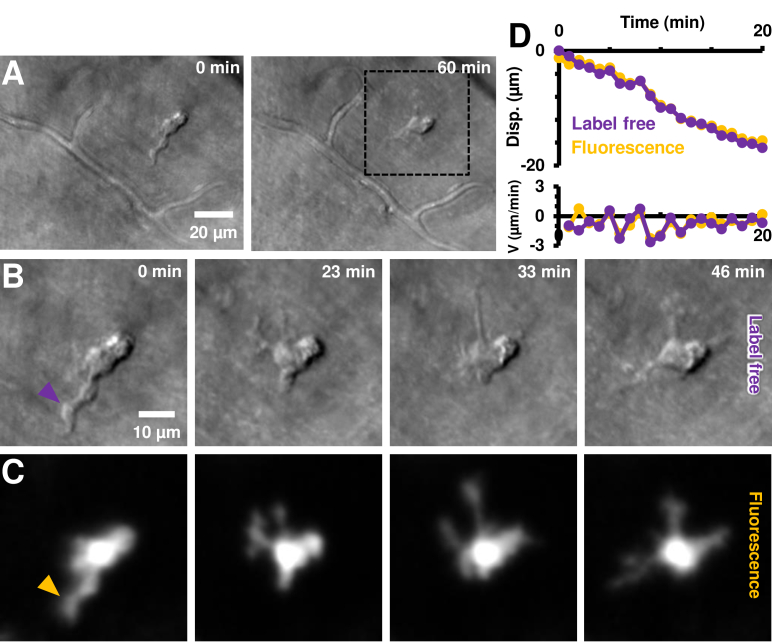
Fig. 9.
Label-free imaging of EGFP+ cell process motility. A) Label-free imaging with near-infrared light (796 nm) using phase-contrast AOSLO. An EGFP+ cell is observed in the top-right quadrant of the image; and confirmed with positive CX3CR1 labelling in C. The cell is tracked in vivo for ∼1 hour. B) Zoom-in of dotted black box region in A, shown at four representative times. Complete time-lapse (10 second resolution) shown in Visualization 7 (201.6MB, avi) . Cell soma and processes can be visualized without use of any exogenous contrast agents. C) Simultaneously imaged fluorescence showing positive CX3CR1 labelling. Cell processes visible are visible in both the infrared label-free and visible channels, showing the potential use of our label-free approach for noninvasive imaging in both mice and humans. Power levels at cornea: label-free 796 nm: 212 µW, fluorescence 488 nm ex.: 53 µW. D) Measured process displacement (disp.) as a function of time, measured independently in the label-free and fluorescence channels. The cell-process tracked is shown with arrowheads in B-C, purple (label-free) and dark-yellow (fluorescence). Instantaneous velocity (V) of the process is similarly shown on the bottom panel.
3. Results
3.1. Retinal microglial processes are motile over all branch orders, in the healthy eye in vivo
The details of individual processes of retinal microglial cells were successfully imaged in vivo with adaptive optics. All imaged cells in the study showed evidence of motility on the order of minutes. While some processes were more motile than others (discussed below), no cells showed lack of dynamism, consistent with the high turnover of cellular processes in the living animal as shown in other brain tissues. Images captured were from a mono-layer of microglia; this was attributed to the known anatomy/stratification of these cells and also to the tight axial resolution of AOSLO. AOSLO images were empirically of better quality than that of SLO, as seen in Fig. 1, yielding higher signal to background ratio as well as the detailed structure of single microglial processes. Time-lapse imaging with AOSLO revealed microglial processes to be highly motile (Fig. 2, Visualization 1 (17MB, avi) ). Processes were motile over all branch orders, from primary to tertiary end-processes (Fig. 3 and 4, Visualization 2 (26MB, avi) , Visualization 3 (24MB, avi) , Visualization 4 (25.5MB, avi) and Visualization 5 (37MB, avi) ). Over time, processes remodeled to show both new formations and deletions of extended processes. A few representative time points are shown in Fig. 3; the complete time sequences are shown in Visualization 1 (17MB, avi) , Visualization 2 (26MB, avi) , Visualization 3 (24MB, avi) , Visualization 4 (25.5MB, avi) and Visualization 5 (37MB, avi) . We show the temporal complexity of the remodeling by assigning a hue saturated value (HSV) value to each unique measurement in time. Prominent changes in a specific time epoch are thus colored in the composite image. Stable branches are manifest as ‘white’, containing all HSV values. The resultant composite images, Fig. 3 (right) show the dynamism in a single spectral image where red-yellow-orange-green represent process deletion, and cyan-blue-purple-magenta show process formation relative to the middle imaging time point. Hue combinations show a graded response in time.
The yellow arrowheads in Fig. 3 illustrate several key features that remodel over time. Microglial cells showed process motility in all branch orders imaged, from primary processes (first branch that extends from the soma) (Fig. 3(A)) to secondary and tertiary process movements (Fig. 3(B)) to the ‘bulbous tips’ or end-processes (Fig. 3(C)). This finding is new in comparison with prior reports of lack of process motility in the healthy eye [19,20] (Reference 20 is an ARVO abstract). The finding is more consistent with in vivo reports in the brain [2,3] and ex vivo reports in the retina that all processes are motile [6]. Such remodeling is presumably the product of F-actin skeleton remodeling which both forms and resolves the process skeleton over time [37].
Our imaging approach even captured the motility of microglial terminal-protrusions with bulbous tips (Fig. 3(C)). The limits to which the spatial resolution and temporal bandwidth of our imaging approach could be pushed is shown by tracking the dynamics of these smallest resolvable structures on the microglial cells over 1.6 hours. Stable image tracking of a single microglial cell was confirmed with micrometer precision (Fig. 3(C), left). This enabled investigation of motility in branches of that cell, across image areas as small as 39 × 31 µm (Fig. 3(C), gray inset in leftmost panel). Dynamic and microscopic end-protrusions along the cell processes were thus imaged (Visualization 5 (37MB, avi) shows all 20 imaged time-points, subset of time-points shown in Fig. 3(C)).
In addition to progressive remodeling that appeared directionally stochastic, we also observed one cell where there was apparent intentional directionality of ramified processes over time (Fig. 4, Visualization 4 (25.5MB, avi) ). All process limbs appeared to change direction in temporal concert, we speculate on one interpretation further in Discussion.
Process movements were often compensatory in nature, to conserve cell volume, given the sequential nature of the retraction of one process being followed by the extension of another (Visualization 3 (24MB, avi) and Visualization 5 (37MB, avi) ); such putative conservation of cell volume/area being consistent with the quantification of linear process displacements in Section 3.4. Whereas process formation and deletion were dynamic over the course of minutes, microglia somas were relatively stable over seconds to minutes (Soma activity over days to months is investigated in Section 3.6).
3.2. Measurement of microglial process motility–displacement
To complement the empirical observations of motile processes in time-lapse sequences, we also quantified process motility using in vivo space-time images of individual cell processes (Fig. 5). High-resolution imaging, coupled with stable imaging over long periods of time, allowed time-lapse registration with minimal distortions, warp or artefact to enable tracing of individual process paths (Fig. 5(A)) and the measurement of process motility relative to the cell soma (Fig. 5(B)). Such space-time analysis revealed processes to undergo extension (green), retraction (red), de novo sprout formation or remain stable (yellow) (Fig. 5(B)-(C)).
Three broad types of motile processes were identified (Fig. 5(C), representative mouse), spanning extensions, retractions and stable processes. A more detailed analysis of process displacement was then done over a population of 7 mice, with a wider spectrum of changes visualized, as described in the next section and in Fig. 6.
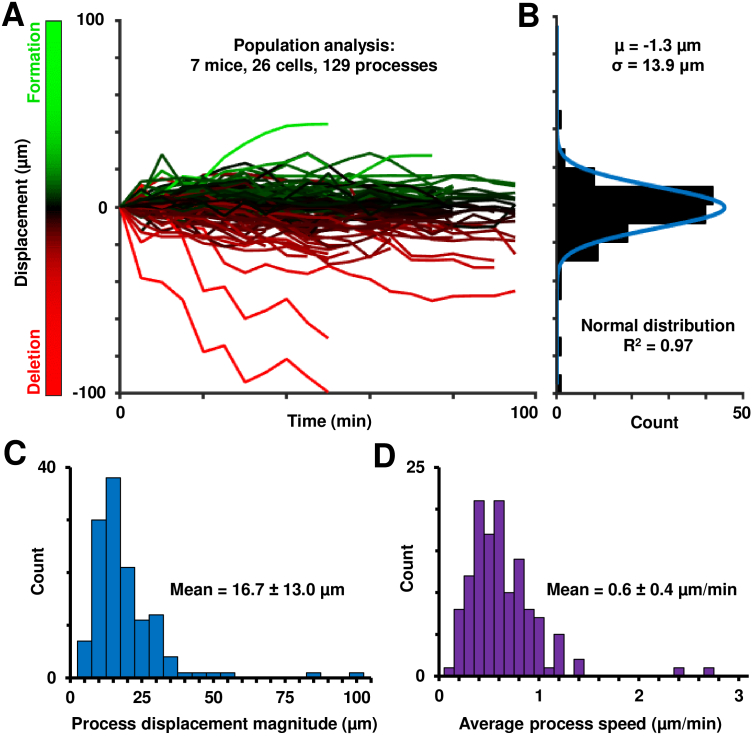
Fig. 6.
Population analysis of in vivo retinal microglial process motility. A) Space-time plots (displacement vs time) plots of 129 processes across 7 mice, with extensions-to-retractions shown in a green-red colormap. B) Histogram of net-displacements of these processes, showing overall balanced extensions and retractions of microglial processes across a population. Blue fit is a normal distribution. C) Histogram of maximum displacement magnitude per process, as defined in the Methods section 2.8. D) Histogram of the average microglial process speeds of 129 processes across the population studied.
3.3. Population analysis of microglial process displacements show that process remodeling is not impacted by our imaging approach
In 129 processes from 26 microglial cells in 7 mice, the measured process displacement magnitude ranged between 0 µm (representing no movement) and 99.1 µm (representing a rare primary process retraction), over 35–95 minutes of time-lapse imaging (Fig. 6(A), extensions-to-retractions represented in a green-to-red colormap). The maximum process extension measured was 44.4 µm. The histogram of net displacements showed an even distribution of process extensions and retractions across the population over time (Fig. 6(B)). This was quantified in the following way. A normal distribution fit well to the histogram (R2=0.97); the mean of the fit distribution was near zero (−1.3 µm). The raw mean histogram was also similarly small, at −4.3 µm). This result indicates a relative balance between process extractions and retractions and indicates a state of homeostasis in the healthy retina [6], and no perturbation of process geometry due to either the imaging light or other animal preparation factors. While two processes showed substantial net retraction of their processes (of −99.1 and −70.3 µm respectively), they were outliers compared to the 129 processes analyzed, as shown by the above population analysis. This net change close to zero indicates that cells are not becoming progressively more ameboid in shape which would indicate a net process retraction.
Despite an overall net zero formation/deletion ratio, quantification of process motility shows dynamic process remodeling in the healthy retina. The magnitude of the maximum displacement per process (Fig. 6(C)) across the population was 16.7 ± 13.0 µm (mean ± 1 SD), representing motile processes in the healthy retinal microglial cell.
3.4. Measurement of microglial process velocity
The displacement-time profiles above were analyzed to determine the velocities of cell-processes. For instantaneous speed, the first derivative of the position was computed, over 5-minute imaging intervals. Over the population of seven mice, the instantaneous speed ranged from 0 µm/min (representing no motion) to a maximum instantaneous speed of 7.6 µm/min. We did not observe faster speeds in our dataset, even when examining finer temporal intervals. Traced processes were not biased to only the moving elements because maximum projection images were used for process path determination. This presumably included all visible processes regardless of their motion. Next, for each process, we computed the mean of all instantaneous velocities to reveal the average remodeling speed per process. This revealed a robust distribution of the average speed per cell-process (Fig. 6(D)), with a population average of 0.6 ± 0.4 µm/min (mean ± 1 SD, n=129 processes). These measurements are much lower than those reported in the in vivo brain and ex vivo retina; we contrast these differences in the Discussion. For completeness, a histogram of all measured instantaneous speeds (n=1718 instances) is also shown, in Supplement 1 (1.4MB, pdf) Fig. S1.
3.5. In vivo Sholl analysis
Sholl analysis was performed for four retinal microglial cells (Fig. 7) to assess in vivo process arbor complexity. For these cells, the maximum mean number of intersections occurred at 12 µm from the soma with a max radial distance of 52 µm from the soma (Fig. 7(A)). Some cells exhibited a large, broad process arbor (cell 1; total intersections detected = 189, maximum number of intersections at 30 µm from the soma, maximum radial distance = 52 µm) and some were small and compact (cell 4; total intersections detected = 65, maximum number of intersections at 12 µm from the soma, maximum radial distance = 24 µm).
In vivo AOSLO allowed for tracking the same microglia for >1 hour. Sholl analysis was performed on cell 2 for images acquired at 5-minute intervals for 75 minutes. The dynamic nature of this microglia was captured (with respect to distance from the soma) and the change over time is seen in Fig. 7(B) and Visualization 8 (5.2MB, avi) . At t=0 minutes, the total number of intersections detected was 106. The maximum number (#) of intersections was at 12 µm from the soma and the maximum radial distance was 50 µm. At t=75 minutes, the total number of intersections detected was 109, maximum number of intersections was at 14 µm from the soma and the maximum radial distance was 66 µm.
3.6. Imaging single-cell microglial changes in vivo, from seconds to months
Microglial cell dynamics were imaged in temporal epochs of seconds, minutes, hours, weeks and months, and represented as red-green merges of the two furthest timepoints (Fig. 8). In an imaging interval of 40 seconds, no substantial changes to a microglial cell’s structure were observed (Fig. 8(A)), represented by the mostly yellow soma and processes. In an imaging interval of 5 minutes in the same cell, end-processes can be clearly seen to be motile, with green colored formations and red deletions (Fig. 8(B)). Figure 8(C) shows the longest we ever tracked a single cell in the same imaging session: 1.6 hours. The yellow color of the soma indicates stability of imaging location, with red and green remodeled processes surrounding it.
To examine microglial activity beyond a single imaging session we also longitudinally tracked the same retinal locations over weeks-months. The same location was navigated to using anatomical features (capillaries and nerve fiber bundles) imaged in the near-infrared confocal reflectance channel. This allowed us to image microglial cell dynamics at the same location over a very large temporal bandwidth of seconds to months, representing about five orders of magnitude in time. The same mice and same locations were successfully imaged one week (Fig. 8(D)) and three months apart (Fig. 8(E)). To achieve this, image registration not only needed to account for respiration motion and biological eye motion within a single imaging session, but also needed to register to the same location across repeat sessions which often yielded slightly different head orientations, native field orientations due to new gaze locations and optical stability over months. Despite these challenges, we achieved exceptional registration using the near-infrared channel to lock onto the same anatomical landmarks over time. Notably, retinal nerve fiber bundles and capillary maps were relatively unchanged over this time epoch in the healthy mouse and facilitated good cross-correlational registration [33]. The grayscale images of the NIR reflectance channel, and the yellow features in the red-green merges thereof, show the successful registration of imaging location (Fig. 8(D)-(E), top panels). Having confirmed correct registration of imaging location, the EGFP fluorescence channel showed that microglial somas are displaced over these time periods (Fig. 8(D)-(E), bottom panels), showing that somas migrate at least locally over these temporal scales. Further characteristics of somas could be explored: in the three-month example, a putative perivascular microglia which was oriented along one major blood vessel disappeared, while either the same or a different cell appeared three months later, now oriented along the neighboring vessel (Fig. 8(E)).
3.7. Label-free (phase-contrast) imaging of process motility of EGFP+ immune cells using NIR light in vivo
Having characterized and confirmed process motility in fluorescently tagged microglial cells, we also applied recent advances in NIR phase-contrast AOSLO [28]. In this paradigm, no fluorescence is needed to provide contrast of translucent cells, instead using scattered or cellular-refracted light alone. Using this approach, we successfully imaged an EGFP+ cell and the motility of its individual processes without any exogenous contrast (Fig. 9(A)); the cell was located in the nerve fiber layer. Process remodeling was observed in the label-free channel (Visualization 6 (117.6MB, avi) −7). Despite eye motion and drift over the continuous recording of 1 hour, our image registration approach provided complete stability of the background structures including a stationary capillary network over the hour-long recording (Fig. 9(A), Visualization 6 (117.6MB, avi) ). To confirm that this cell was indeed either a microglia, hyalocyte or perivascular macrophage, we performed simultaneous label-free and EGFP fluorescence imaging. Dynamic remodeling was observed in both imaging modalities (Fig. 9(B)-(C), Visualization 7 (201.6MB, avi) ). Cell process motility can be clearly seen in both channels in the time-lapse sequences. The visualizations show much finer temporal resolution of 10 seconds per video-frame, to show the detail of motion. This fine temporal resolution was enabled by the exceptionally high SNR of the phase contrast imaging that uses NIR light rather than the comparatively low SNR fluorescence imaging (Supplement 1 (1.4MB, pdf) Fig. S2).
Given the anatomy and location, the cell is most likely a microglial cell rather than a hyalocyte or perivascular macrophage, which may also express CX3CR1-GFP. We discuss this further in Section 4.5.
Next, process motility was measured independently in the two channels for the same cell-process, marked by arrowheads in Fig. 9(B)-(C): purple (label-free) and dark-yellow (fluorescence). We found that process motility can be successfully quantified using label-free imaging (Fig. 9(D), purple plot), with an average process speed of 0.8 µm/min and a total displacement of 16.3 µm over the analyzed time of 20 mins (displacement measured every one minute). An independent measurement using only the fluorescence channel (dark-yellow plot) showed that the measurements in the two channels matched, with an error less than the optical resolution limit (mean absolute difference in measured displacements in two channels = 0.60 ± 0.41 µm, mean±1SD, n=20 time instances). Furthermore, a subset of the measurements were repeated; the measurement variability in process displacement (in a single channel) was found to be 0.56 ± 0.33 µm (mean±1SD, n=15 measurements). While the same cell was imaged, not all processes visible in the fluorescence channel yielded high contrast in the NIR image. Conversely, subcellular features visible in the phase-contrast modality were not visible in fluorescence, as detailed in the next section.
3.8. Label-free imaging reveals subcellular dynamics missed in fluorescence
Having confirmed that process displacement and speed can be quantified even with label-free imaging, we next sought out differences between the fluorescence and label-free imaging of microglia. In the cell in Fig. 9 and Visualization 6 (117.6MB, avi) –Visualization 7 (201.6MB, avi) , we observed putative subcellular features and their dynamics were visible in the label-free channel which were not visualized with fluorescence. These putative sub-cellular features were seen to rapidly move in the label-free time-lapse sequence, especially in the top-right portion (orange arrowhead, Fig. 10) of the putative cell soma, whereas such subcellular motility was not visible in fluorescence (Visualization 7 (201.6MB, avi) and Fig. 10(B)). This is possibly due to the higher SNR of the phase contrast approach, but it is also possible that visualization of such subcellular motility is not dependent on the surface/volumetric expression of transgenic mice that have fluorophores tagged to the surface protein. In addition to videos, such subcellular dynamics can also be visualized by computing the motion contrast (standard deviation of pixel values in time dimension) of the above video stack (Fig. 10(B)-(C)). The top-right part of the soma (orange arrowhead) is very bright in label-free motion contrast (Fig. 10(B)), representative of the high motility of subcellular structures here (visible in Visualization 7 (201.6MB, avi) , left channel). In contrast, the fluorescence motion contrast (Fig. 10(C)) misses this high activity happening in this top-right region of the putative soma (orange arrowhead), showing only a dark region corresponding to the central part of the soma which is relatively stable/immobile. (Note: one may wonder why the other processes are not readily visible in the label-free motion contrast (Fig. 10(B)). This is due to limits of image dynamic range: the changes in pixel intensity of the putative filopodia are much higher than that of the other processes. However, those processes are still readily visible in the label-free intensity images of the same cell shown in Fig. 9(B).)
Fig. 10.
Label-free imaging reveals subcellular dynamics which may be missed in fluorescence. A) An intensity image of near-infrared label-free imaging of a microglia at a single time-point (same as in Fig. 9(B)). B) A motion contrast image of the label-free channel, computed from 60 minutes of time-lapse imaging. The bright area (orange arrowhead) in the top-right of the putative soma shows high motility of subcellular features, likely filopodia. C) Motion contrast image of the EGFP fluorescence channel, computed over 60 minutes of imaging. All process paths are visible as bright areas. The central soma is dark, due to relative immobility. D) Color-coded maximum intensity projection of the fluorescence time-lapse sequence, shown for consistency with how microglial motility is represented in previous figures.
4. Discussion
4.1. Summary
Microglial dynamics are essential to understand the composition of the healthy and diseased retinal phenotype. Here we have focused our attention on the aspects of the healthy phenotype to identify the baseline characteristics from which disease can be better understood. Moreover, this provides a foundation from which normal microglial activity may facilitate normal homeostasis in the steady state [38], with a renewed focus on retinal microglia which have been less studied than cerebral microglia. In pursuit of this goal, we have developed the imaging strategy to the point where the detailed complexity as well as process and cell soma dynamics are visualized from seconds to months in the same animals in vivo using time-lapse adaptive optics ophthalmoscopy. We found process motility to be highly active and measured substantial process remodeling from primary through terminal branches over time. This is in stark contrast to recent in vivo reports in the retina which found a surprising lack of process motility in ∼95% of GFP+ cells in the normal retina [19]. The simultaneous capture of two imaging modalities (NIR and fluorescence) allowed us to achieve micron-level registration to create stable time-lapse images not only over hours in the same animals, but also facilitated return to the same retinal area with micron-level accuracy months later (Fig. 8).
The motility of subcellular features was tracked from primary branches to the smallest micron-sized end (terminal) processes. A space-time image analysis approach was developed to quantify the displacement and velocity of cell-processes. Process motility was analyzed over a population of microglia, across 7 mice. Twenty-six cells were identified and AOSLO imaging was optimized so that the imaging field was positioned over the cells and focus optimized for the soma and process arbor. Data was recorded as long as the mouse preparation remained stable. All data was used for the quantification analysis so long as a cell could be imaged continuously for >35 minutes, and the complete cell remained within the FOV after post-processing. In total 129 processes were tracked in all data collected. Process deletions and formations were in balance indicating at least two essential findings critical for the interpretation of this study. First, the relative conservation of mass is indicative of a remodeling process that both forms and deletes branches in a way that preserves cellular resources. For example, remodeling of the f-actin cytoskeleton and its components appear to remain roughly conserved [37]. Secondly, this net-zero formation/deletion balance indicates that the imaging light we employ is non-destructive. It is expected that phototoxic light levels would induce a cytokine or chemotaxic gradient that would tip microglia from a ramified to more ameboid state [2,39]. Instead, we found dynamic activity with balanced formation and deletion suggestive of a safe imaging modality that leaves microglia in their native state.
Moreover, interesting dynamics were observed within the process arbors of single immune cells (Fig. 4) such that local processes from a single cell would form a polarized geometry as though a local invisible gradient were driving process formation and deletion in concert. Here, we have no direct evidence from our experimental protocol, yet posit that local chemokine gradients may drive this apparent motion at the microscopic scale (as has been demonstrated in the brain). Further work is needed to determine whether this directional polarization reveals invisible gradients that may be tracked within small locations in the retinal tissue by tracking this behavior in microglia in the living retina.
Having benchmarked these functions in healthy CX3CR1 mice with EGFP labelled microglia, we were able to apply recent advances in phase-contrast AO imaging to image EGFP+ immune cells and their dynamic processes label-free with near-infrared light alone. Process motility was quantified in label-free phase-contrast imaging, and shown to match measured motility in fluorescence. In addition to confirming the non-destructive nature of imaging, the NIR imaging provides the exciting possibility of future noninvasive imaging of microglia in humans. Finally, immune cell motility was tracked over a large temporal bandwidth, of seconds to months, by registering to stable vascular and retinal landmarks with near-infrared imaging. While beneficial in this study, we expect that such registration may be challenging in longer-duration studies in diseases where vascular and retinal anatomy is also undergoing remodeling over long intervals.
4.2. High resolution AOSLO allowed quantitative imaging of process motility previously missed by SLO
A required discussion item is the discrepancy of our findings with recent studies that have reported a surprising lack of process motility in ramified microglia [19,20], using SLO alone (without adaptive optics). Our findings are quite the opposite. We found remarkable remodeling of ramified microglia, from primary to terminal branches.
The following may explain the difference in findings. It is possible that the limited spatio-temporal resolution of SLO imaging restricted the prior studies to visualizing only the most prominent branches which would lead to an underestimation of the branch turnover due to focus on only the brightest processes. We believe therefore that the uncorrected optical aberrations of the eye prevented such studies from visualizing the native process motility of retinal microglia due to optical blur and the resulting less-precise spatial registration. Our approach, using adaptive optics and space-time image quantification, now enables visualizing and quantifying this motility in the in vivo retina. Our findings are more similar to the reports of microglial dynamism in the in vivo brain [2,3] and ex vivo retina [6]. For comparison and as a resource, we summarize the relevant studies and our own findings in Table 1.
Table 1. Comparison of reports which studied microglial process motility in the healthy retina and brain. Notes: Measured velocities are mean ± standard deviation, except for Nimmerjahn et al. 2005, which reported standard error of mean. ‘Inst.’ is instantaneous.
| Study | Species | Organ | Technique | Conclusion about process motility | Measured velocities (µm/min) | Inst. velocity range (µm/min) | λ (nm) | Power levels (µW) | Sample size |
|---|---|---|---|---|---|---|---|---|---|
| Current Study | Mouse | Retina (in vivo) | AOSLO | Motile | 0.6 ± 0.4 | 0 - 7.6 | 488 | 53 | 7 mice, 26 cells, 129 processes |
| 796 | 212 | ||||||||
| Nimmerjahn et al. 2005 | Mouse | Brain (in vivo) | Two-photon imaging | Motile | 1.5 ± 0.1 | 0.4 - 3.8 | 840 - 930 | 9000 - 18000 | 8 mice, 14 cells, 242 measures |
| Lee et al. 2008 | Mouse | Retina (ex vivo) | Microscopy retinal explant | Motile | 5.4 ± 2.3 | - | - | - | 3 mice, 37 cells, 367 processes |
| Alt et al. 2010 | Mouse | Retina (in vivo) | AOSLO | Motile | Not quantified | - | 491 | 500 - 1000 | - |
| Alt and Lin 2012 | Mouse | Retina (in vivo) | SLO | Motile | 7.8 ± 2.1 | - | 491 | - | - |
| Wahl et al. 2019 | Mouse | Retina (in vivo) | AOSLO | Motile | 1.3 and 4.8 | - | 488 | 100 - 230 | 1 cell, 2 processes |
| Miller et al. 2019 | Mouse | Retina (in vivo) | SLO | Not motile | Not motile | - | 488 | 100 | - |
| Hammer et al. 2020 | Human | Retina (in vivo) | AO-OCT | Motile | 15.9 ± 2.0 | - | - | - | 6 subjects, 36 cells, 75 processes |
In our study, we find that while the process displacement over the population may have an average near zero, there are visibly highly dynamic segments that rapidly remodel. This begs another scientific question: what signaling at the genetic, structural or chemotactic level signals some processes to rapidly remodel while others (extensions of the same cell) remain stationary? Further study is needed to unveil this remarkable behavior.
4.3. Comparison with process motility reported in brain and ex vivo retina
One of the leading questions is how similar are retinal and brain microglia? If it were possible to identify key similarities, imaging retinal microglia could help infer the activity of these immune cells in the brain. In general, we found that the motility of retinal microglia we imaged were roughly on-par with the motility observed in the brain (Table 1). A clear confound in doing the direct comparison is that each study performed velocimetry in a unique way. Moreover, average velocities can be biased by the selection process of the chosen microglial processes which is usually grader-facilitated. For example, there may be a tendency to mark and measure only the most dynamic processes which would lead to an overestimation of the population velocity by overlooking the more stationary and “uninteresting” branches. The converse is also true. Therefore, future work on the heels of imaging innovations will need to build more robust and automated quantification strategies that are independent of human bias.
Recognizing this, we marked and measured all visible branches in imaged microglial cells. We measured the average retinal microglial process velocity in mice to be 0.6 ± 0.4 µm/min (mean ± 1 SD, n=129 processes from 26 cells in 7 mice), averaged across 35 to 95 minutes of time-lapse imaging per process. The instantaneous process velocity (measured in 5-minute intervals) ranged from 0 µm/min (representing no motion) to a maximum of 7.6 µm/min. Our measured process velocities are similar to that reported in the in vivo mouse brain (1.47 ± 0.09 µm/min, mean ± 1 SEM, 242 measurements in 14 cells across 8 mice) [2]. Comparably, microglia in the brain showed a range of 0.4 to 3.8 µm/min.
At the detailed level, our measured mean velocity is ∼2.5x less than that reported in the brain and may represent a real or limited sample comparison. However, the observed range of velocities is well within the same order of magnitude in both retina and cortex which is noteworthy, especially given that data arises from different laboratories, using different imaging modalities, anesthetics and in different CNS tissues. Further investigations are warranted to tease apart the nuanced differences between retinal and cortical microglia. Because AOSLO is non-invasive, we are excited to think that a future experiment may choose to image microglia motility in the retina first, and then perform cranial window two-photon imaging of brain microglia in the same mouse that would further reduce biological variability in this crucial measurement.
Another interpretation of the subtle differences in the brain and retina findings is that brain imaging of microglia inherently requires surgery in order to visualize this cell population [2,3], whereas our AOSLO approach is completely non-invasive. Despite the care of the surgical preparation of a craniotomy or thinned-skull preparation, it is possible that the surgical intervention may alter the global or local baseline immune response in brain studies. Furthermore, a common technique used for brain imaging employs near infrared light (in vivo two-photon microscopy). While the wavelength is beneficial to mitigate scatter, this approach uses very high light-levels (9–18 mW of 840–930 nm two-photon excitation [2]). At these power levels and wavelength, there is also potential for changes in tissue due to thermal heating which may alter baseline microglial process motility dynamics. By comparison, our approach uses 42 to 84 times less NIR light power, thus having fewer confounds, including mitigation of thermal or photochemical alterations of the tissue homeostasis. We used 212 µW of 796 nm light in our AOSLO imaging, which we have found can be used to image continuously without damage in the retina. This strict imaging light-budget is required as the retina is one of the most light-sensitive tissues in the body. Towards clinical application, similar power levels used over similar radiant exposure areas in the in vivo human eye correspond to ANSI-safe light levels and is regularly used in human imaging without damage [22,23]. For fluorescence imaging, we used limited periods (40 s each) of 53 µW of 488 nm excitation, which is the lowest light level reported to date for microglia fluorescence imaging.
It is worthwhile mentioning that recent evidence in 2020 has suggested that the founder stock of the CX3CR1 homozygous mice may contain a mixed C57BL/6J and C57BL/6N lineage (as performed by a single nucleotide polymorphism panel analysis, Jackson Labs) which could produce a fraction of mice with the Rd8 retinal degeneration mutation. As our mice are backcrossed onto the pure C57BL/6J, it is unlikely the imaged mice contain the Rd8 phenotype [40]. While we do not have genotypic data to rule this out, we do have phenotypic evidence (Fig. 1(A)) that our imaged mice did not have geographical lesions in their fundus after that is characteristic of the Rd8 retinal degeneration phenotype that appears as early as 6 weeks of age [40]. Our mice much older (7 ± 2.9 months) when imaged and lack of fundus phenotype indicated no underlying degeneration. We therefore deem it unlikely that mice had underlying retinal degeneration that would skew our microglial measurements from normal.
In the ex vivo mouse retina, process velocity has been reported to be 5.44 ± 2.33 µm/min (mean ± 1 SD, n=367 processes from 37 cells in 3 mice) [6]. While this early seminal work advanced our understanding of microglia process dynamics in the retina, the approach is understandably limited by the ex vivo preparation, which can induce inflammatory cues and release of cytokines and chemokines, which are among the key drivers of process remodeling. Thus, there is always the concern that ex vivo microglia behavior may not accurately represent the baseline state of these cells in vivo. Our in vivo measurements are consistent with this, with average process velocities measured to be around 9 times smaller than that reported in ex vivo microglia. For reader convenience, we provide Table 1 as a helpful summary of these different studies in the retina and brain, compared with our own.
4.4. Sholl analysis reveals process turnover complexity
Sholl analysis provides a measure of microglial process arbor complexity. Recently, 2-photon imaging in the murine brain has been used to image microglia in vivo and perform Sholl analysis [4,41–43], but to our knowledge, there is no such analysis in the living retina. Sholl analysis for cell 1 (Fig. 7) exhibited higher complexity at radial distances further from the soma when compared with to cell 2 and 3. This shows the heterogeneity of microglial complexity.
Recent studies using Sholl analysis in mice/post-mortem human ex vivo tissue reported larger numbers of intersections at radial distances further from the soma [44,45] compared with our study. This may be due to the reported homo/hetro-zygous differences in microglial density [19], differences in species or the possibility that the in vivo preparation potentially may miss the finest of processes that go unmissed in ex vivo preparations. Regardless, we demonstrate that not only can Sholl analysis be performed on microglia within the living retina, but the dynamism of process turnover can be revealed through the same analysis performed over time. Figure 7(B) and Visualization 8 (5.2MB, avi) show that the arbor complexity changes over time. Such analyses may serve to characterize microglial process turnover in health and disease.
4.5. Label-free imaging of EGFP+ immune cell dynamics: intracellular study and clinical translation potential
An exciting finding of our study is that EGFP+ immune cell dynamics can be readily imaged and quantified in the healthy retina using label-free imaging with NIR light alone, using phase-contrast AOSLO [28,46–48]. Cell-type was confirmed using simultaneous imaging of CX3CR1 EGFP fluorescence. Beyond just process dynamics and somatic migration, the phase-contrast approach revealed what appears to be putative sub-cellular constituents of the cytosol and organelle movement (Fig. 9,10, Visualization 7 (201.6MB, avi) ). In particular, the cellular contents of microglia are an essential target to image as one of the primary functions of this immune cell population is to engulf, digest and remove debris through lysosomal breakdown [49]. To our knowledge, such tentative sub-cellular study has not been possible using single or multi-photon approaches due either to limitations of imaging, or lack of sub-cellular specificity of current fluorescent markers to components within the immune/microglial cell. While further work is required to test whether these are features of the surface of the cell or true intracellular organelles, the dynamic videos shown here in addition to evidence from Joseph et al. 2020 (Video 6) [29] are suggestive of internal cellular features, since the boundary geometry of the cell is changing while the putative internal contents are moving at a different rate. Future work may combine organelle fluorescence with phase contrast images to establish the potential of imaging this putative label-free organelle motility.. Sub-cellular pH labels or other markers may also facilitate confirmation of this finding. Regardless, imaging the contents of any cell population has utility and we expect further study using the phase-contrast approach will reveal yet more about the in vivo behaviors of these remarkable cells.
Given the anatomy and location, the EGFP+ cell imaged label-free in Fig. 9,10 and Visualization 6 (117.6MB, avi) –Visualization 7 (201.6MB, avi) is most likely a microglial cell rather than a hyalocyte or perivascular macrophage, which may also express CX3CR1-GFP. Hyalocytes are most commonly found over 50 microns above the vitreo-retinal surface [50]. A cell 50 microns above the ganglion/RNFL plane would not be resolved due to substantial optical defocus blur in the AOSLO which has a narrow depth of field. Moreover, the identity of a perivascular macrophage is unlikely as these cells are typically in close abluminal contact with vessels whereas the cell in Fig. 9 is 20–30 micrometers away from the nearest vessel and far from the glia limitans where they are found in the central nervous system [51]. Thus, the combination of CX3CR1 expression, axial location within the ganglion cell/NFL and distance far from the vessel make this imaged cell most consistent with a retinal microglial cell.
In the translational domain, our imaging with ANSI-safe levels of NIR light [22–24] opens the door to imaging the same in human beings, using phase-contrast AOSLO systems for the human eye [22,46,47], and builds upon reports of imaging immune cells in the human eye using other modalities [52–55]. The two main challenges we see to the implementation of our AOSLO approach in human beings is the ∼4x poorer axial resolution due to the ∼2x smaller numerical aperture of the human eye [21], and having good image registration due to the more pronounced eye movements in the awake human compared to the anesthetized mouse, both of which are not insurmountable in the near future. In a recent advance [55], a population of putative macrophage dynamics could be imaged in the living human retina, through the use of optical coherence tomography. This study showed that through a different modality, cell dynamism can be imaged and tracked using safe levels of light in a clinical setting. This finding is synergistic with our current study. Our study in the mouse affords the important advantage that we can both image cells label-free, and confirm their identity using positive protein fluorescence confirmation with transgenic mice. In this way, mice may provide the critical intermediary to confirm cell identity to aid in clinical interpretation and application. With such confirmations, our study offers structural and functional biomarkers of these cells that can now be tracked in the human eye without interpretive dispute. We expect further improvements from machine learning approaches as well to facilitate quantification of such dynamism.
Acknowledgements
The authors thank Qiang Yang, Jie Zhang and Jennifer Strazzeri, for their technical contributions to this work. We also thank Colin Chu and Robin Sharma for critical feedback on the manuscript.
Typographical corrections were made to Figs. 1–10.
Funding
National Institutes of Health10.13039/100000002 (P30 EY001319, R01 EY028293); Dana Foundation10.13039/100001068 (David Mahoney Neuroimaging Award); Research to Prevent Blindness10.13039/100001818 (Career Development Award, Stein Award, Unrestricted grant to Department of Ophthalmology).
Disclosures
Aby Joseph (F,P) and Jesse Schallek (F,P) hold patents at the University of Rochester and were also supported by a research grant from Hoffman-LaRoche.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (1.4MB, pdf) for supporting content.
References
- 1.Lawson L. J., Perry V. H., Dri P., Gordon S., “Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain,” Neuroscience 39(1), 151–170 (1990). 10.1016/0306-4522(90)90229-W [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn A., Kirchhoff F., Helmchen F., “Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo,” Science 308(5726), 1314–1318 (2005). 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- 3.Davalos D., Grutzendler J., Yang G., Kim J. V., Zuo Y., Jung S., Littman D. R., Dustin M. L., Gan W.-B., “ATP mediates rapid microglial response to local brain injury in vivo,” Nat Neurosci 8(6), 752–758 (2005). 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- 4.Stowell R. D., Sipe G. O., Dawes R. P., Batchelor H. N., Lordy K. A., Whitelaw B. S., Stoessel M. B., Bidlack J. M., Brown E., Sur M., Majewska A. K., “Noradrenergic signaling in the wakeful state inhibits microglial surveillance and synaptic plasticity in the mouse visual cortex,” Nat Neurosci 22(11), 1782–1792 (2019). 10.1038/s41593-019-0514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman S. M., Wong W. T., “Microglia in the retina: roles in development, maturity, and disease,” Annu. Rev. Vis. Sci. 4(1), 45–77 (2018). 10.1146/annurev-vision-091517-034425 [DOI] [PubMed] [Google Scholar]
- 6.Lee J. E., Liang K. J., Fariss R. N., Wong W. T., “Ex vivo dynamic imaging of retinal microglia using time-lapse confocal microscopy,” Invest. Ophthalmol. Vis. Sci. 49(9), 4169–4176 (2008). 10.1167/iovs.08-2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang K. J., Lee J. E., Wang Y. D., Ma W., Fontainhas A. M., Fariss R. N., Wong W. T., “Regulation of dynamic behavior of retinal microglia by CX3CR1 signaling,” Invest. Ophthalmol. Vis. Sci. 50(9), 4444–4451 (2009). 10.1167/iovs.08-3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paques M., Simonutti M., Augustin S., Goupille O., El Mathari B., Sahel J.-A., “In vivo observation of the locomotion of microglial cells in the retina,” Glia 58(14), 1663–1668 (2010). 10.1002/glia.21037 [DOI] [PubMed] [Google Scholar]
- 9.Alt C., Lin C. P., “In vivo quantification of microglia dynamics with a scanning laser ophthalmoscope in a mouse model of focal laser injury,” in (2012), Vol. 8209, pp. 820907–820907–9. [Google Scholar]
- 10.Zawadzki R. J., Zhang P., Zam A., Miller E. B., Goswami M., Wang X., Jonnal R. S., Lee S.-H., Kim D. Y., Flannery J. G., Werner J. S., Burns M. E., Pugh E. N., “Adaptive-optics SLO imaging combined with widefield OCT and SLO enables precise 3D localization of fluorescent cells in the mouse retina,” Biomed. Opt. Express 6(6), 2191–2210 (2015). 10.1364/BOE.6.002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P., Zam A., Jian Y., Wang X., Li Y., Lam K. S., Burns M. E., Sarunic M. V., Pugh E. N., Zawadzki R. J., “In vivo wide-field multispectral scanning laser ophthalmoscopy–optical coherence tomography mouse retinal imager: longitudinal imaging of ganglion cells, microglia, and Müller glia, and mapping of the mouse retinal and choroidal vasculature,” J. Biomed. Opt 20(12), 126005 (2015). 10.1117/1.JBO.20.12.126005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang J., Williams D. R., Miller D. T., “Supernormal vision and high-resolution retinal imaging through adaptive optics,” J. Opt. Soc. Am. A 14(11), 2884 (1997). 10.1364/JOSAA.14.002884 [DOI] [PubMed] [Google Scholar]
- 13.Williams D. R., “Imaging single cells in the living retina,” Vision Res. 51(13), 1379–1396 (2011). 10.1016/j.visres.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roorda A., Duncan J. L., “Adaptive optics ophthalmoscopy,” Annu. Rev. Vis. Sci. 1(1), 19–50 (2015). 10.1146/annurev-vision-082114-035357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biss D. P., Sumorok D., Burns S. A., Webb R. H., Zhou Y., Bifano T. G., Côté D., Veilleux I., Zamiri P., Lin C. P., “In vivo fluorescent imaging of the mouse retina using adaptive optics,” Opt. Lett. 32(6), 659–661 (2007). 10.1364/OL.32.000659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alt C., Biss D. P., Tajouri N., Jakobs T. C., Lin C. P., “An adaptive-optics scanning laser ophthalmoscope for imaging murine retinal microstructure,” in (2010), Vol. 7550, pp. 755019–755019–11. [Google Scholar]
- 17.Schallek J. B., Joseph A., “Time-lapse imaging of retinal microglia in vivo show dynamic process motility at rest,” Invest. Ophthalmol. Vis. Sci. 58, 316 (2017). [Google Scholar]
- 18.Wahl D. J., Ng R., Ju M. J., Jian Y., Sarunic M. V., “Sensorless adaptive optics multimodal en-face small animal retinal imaging,” Biomed. Opt. Express 10(1), 252–267 (2019). 10.1364/BOE.10.000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller E. B., Zhang P., Ching K., Pugh E. N., Burns M. E., “In vivo imaging reveals transient microglia recruitment and functional recovery of photoreceptor signaling after injury,” PNAS 201903336 (2019). [DOI] [PMC free article] [PubMed]
- 20.Miller E., Zhang P., Goswami M., Zawadzki R. J., Pugh E. N., Burns M. E., “Missing motility: high-resolution in vivo imaging of retinal microglia reveals stationary ramified cells.,” Invest. Ophthalmol. Vis. Sci. 57, (ARVO 2016 Annual Meeting Abstract) (2016).
- 21.Geng Y., Dubra A., Yin L., Merigan W. H., Sharma R., Libby R. T., Williams D. R., “Adaptive optics retinal imaging in the living mouse eye,” Biomed. Opt. Express 3(4), 715–734 (2012). 10.1364/BOE.3.000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi E. A., Granger C. E., Sharma R., Yang Q., Saito K., Schwarz C., Walters S., Nozato K., Zhang J., Kawakami T., Fischer W., Latchney L. R., Hunter J. J., Chung M. M., Williams D. R., “Imaging individual neurons in the retinal ganglion cell layer of the living eye,” Proc Natl Acad Sci USA 114(3), 586–591 (2017). 10.1073/pnas.1613445114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T., Jung H., Liu J., Droettboom M., Tam J., “Noninvasive near infrared autofluorescence imaging of retinal pigment epithelial cells in the human retina using adaptive optics,” Biomed. Opt. Express 8(10), 4348–4360 (2017). 10.1364/BOE.8.004348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delori F. C., Webb R. H., Sliney D. H., “Maximum permissible exposures for ocular safety (ANSI 2000), with emphasis on ophthalmic devices,” J. Opt. Soc. Am. A 24(5), 1250–1265 (2007). 10.1364/JOSAA.24.001250 [DOI] [PubMed] [Google Scholar]
- 25.Gray D. C., Merigan W., Wolfing J. I., Gee B. P., Porter J., Dubra A., Twietmeyer T. H., Ahmad K., Tumbar R., Reinholz F., Williams D. R., “In vivo fluorescence imaging of primate retinal ganglion cells and retinal pigment epithelial cells,” Opt. Express 14(16), 7144–7158 (2006). 10.1364/OE.14.007144 [DOI] [PubMed] [Google Scholar]
- 26.Morgan J. I. W., Dubra A., Wolfe R., Merigan W. H., Williams D. R., “In vivo autofluorescence imaging of the human and macaque retinal pigment epithelial cell mosaic,” Invest. Ophthalmol. Vis. Sci. 50(3), 1350–1359 (2009). 10.1167/iovs.08-2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granger C. E., Yang Q., Song H., Saito K., Nozato K., Latchney L. R., Leonard B. T., Chung M. M., Williams D. R., Rossi E. A., “Human retinal pigment epithelium: in vivo cell morphometry, multispectral autofluorescence, and relationship to cone mosaic,” Invest. Ophthalmol. Vis. Sci. 59(15), 5705–5716 (2018). 10.1167/iovs.18-24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guevara-Torres A., Williams D. R., Schallek J. B., “Origin of cell contrast in offset aperture adaptive optics ophthalmoscopy,” Opt. Lett. 45(4), 840–843 (2020). 10.1364/OL.382589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph A., Chu C. J., Feng G., Dholakia K., Schallek J., “Label-free imaging of immune cell dynamics in the living retina using adaptive optics,” eLife 9, e60547 (2020). 10.7554/eLife.60547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph A., Guevara-Torres A., Schallek J., “Imaging single-cell blood flow in the smallest to largest vessels in the living retina,” eLife 8, e45077 (2019). 10.7554/eLife.45077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmucker C., Schaeffel F., “A paraxial schematic eye model for the growing C57BL/6 mouse,” Vision Res. 44(16), 1857–1867 (2004). 10.1016/j.visres.2004.03.011 [DOI] [PubMed] [Google Scholar]
- 32.Yang Q., Yin L., Nozato K., Zhang J., Saito K., Merigan W. H., Williams D. R., Rossi E. A., “Calibration-free sinusoidal rectification and uniform retinal irradiance in scanning light ophthalmoscopy,” Opt. Lett. 40(1), 85–88 (2015). 10.1364/OL.40.000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubra A., Harvey Z., “Registration of 2D images from fast scanning ophthalmic instruments,” in Biomedical Image Registration, Lecture Notes in Computer Science (Springer, Berlin, Heidelberg, 2010), pp. 60–71. [Google Scholar]
- 34.Guevara-Torres A., Joseph A., Schallek J. B., “Label free measurement of retinal blood cell flux, velocity, hematocrit and capillary width in the living mouse eye,” Biomed. Opt. Express 7(10), 4228–4249 (2016). 10.1364/BOE.7.004228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira T. A., Blackman A. V., Oyrer J., Jayabal S., Chung A. J., Watt A. J., Sjöström P. J., van Meyel D. J., “Neuronal morphometry directly from bitmap images,” Nat Methods 11(10), 982–984 (2014). 10.1038/nmeth.3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arshadi C., Günther U., Eddison M., Harrington K. I. S., Ferreira T. A., “SNT: a unifying toolbox for quantification of neuronal anatomy,” Nat Methods 18(4), 374–377 (2021). 10.1038/s41592-021-01105-7 [DOI] [PubMed] [Google Scholar]
- 37.Franco-Bocanegra D. K., McAuley C., Nicoll J. A. R., Boche D., “Molecular mechanisms of microglial motility: changes in ageing and Alzheimer’s disease,” Cells 8(6), 639 (2019). 10.3390/cells8060639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kierdorf K., Prinz M., “Microglia in steady state,” J. Clin. Invest. 127(9), 3201–3209 (2017). 10.1172/JCI90602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giulian D., “Ameboid microglia as effectors of inflammation in the central nervous system,” J. Neurosci. Res. 18(1), 155–171 (1987). 10.1002/jnr.490180123 [DOI] [PubMed] [Google Scholar]
- 40.Mattapallil M. J., Wawrousek E. F., Chan C.-C., Zhao H., Roychoudhury J., Ferguson T. A., Caspi R. R., “The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes,” Invest. Ophthalmol. Vis. Sci. 53(6), 2921–2927 (2012). 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grier B. D., Belluscio L., Cheetham C. E. J., “Olfactory sensory activity modulates microglial-neuronal interactions during dopaminergic cell loss in the olfactory bulb,” Front. Cell. Neurosci. 10, 178 (2016). 10.3389/fncel.2016.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madry C., Kyrargyri V., Arancibia-Cárcamo I. L., Jolivet R., Kohsaka S., Bryan R. M., Attwell D., “Microglial ramification, surveillance, and Interleukin-1β release are regulated by the two-pore domain K+ Channel THIK-1,” Neuron 97(2), 299–312.e6 (2018). 10.1016/j.neuron.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendes M. S., Le L., Atlas J., Brehm Z., Ladron-de-Guevara A., Matei E., Lamantia C., McCall M. N., Majewska A. K., “The role of P2Y12 in the kinetics of microglial self-renewal and maturation in the adult visual cortex in vivo,” eLife 10, e61173 (2021). 10.7554/eLife.61173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda A., Shinozaki Y., Kashiwagi K., Ohno N., Eto K., Wake H., Nabekura J., Koizumi S., “Microglia mediate non-cell-autonomous cell death of retinal ganglion cells,” Glia 66(11), 2366–2384 (2018). 10.1002/glia.23475 [DOI] [PubMed] [Google Scholar]
- 45.Aires I. D., Boia R., Rodrigues-Neves A. C., Madeira M. H., Marques C., Ambrósio A. F., Santiago A. R., “Blockade of microglial adenosine A2A receptor suppresses elevated pressure-induced inflammation, oxidative stress, and cell death in retinal cells,” Glia 67(5), 896–914 (2019). 10.1002/glia.23579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chui T. Y. P., VanNasdale D. A., Burns S. A., “The use of forward scatter to improve retinal vascular imaging with an adaptive optics scanning laser ophthalmoscope,” Biomed. Opt. Express 3(10), 2537 (2012). 10.1364/BOE.3.002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scoles D., Sulai Y. N., Langlo C. S., Fishman G. A., Curcio C. A., Carroll J., Dubra A., “In vivo imaging of human cone photoreceptor inner segments,” Invest. Ophthalmol. Vis. Sci. 55(7), 4244–4251 (2014). 10.1167/iovs.14-14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sulai Y. N., Scoles D., Harvey Z., Dubra A., “Visualization of retinal vascular structure and perfusion with a nonconfocal adaptive optics scanning light ophthalmoscope,” J. Opt. Soc. Am. A 31(3), 569–579 (2014). 10.1364/JOSAA.31.000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majumdar A., Cruz D., Asamoah N., Buxbaum A., Sohar I., Lobel P., Maxfield F. R., “Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils,” MBoC 18(4), 1490–1496 (2007). 10.1091/mbc.e06-10-0975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiao H., Hisatomi T., Sonoda K.-H., Kura S., Sassa Y., Kinoshita S., Nakamura T., Sakamoto T., Ishibashi T., “The characterisation of hyalocytes: the origin, phenotype, and turnover,” Br J Ophthalmol 89(4), 513–517 (2005). 10.1136/bjo.2004.050658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faraco G., Park L., Anrather J., Iadecola C., “Brain perivascular macrophages: characterization and functional roles in health and disease,” J Mol Med 95(11), 1143–1152 (2017). 10.1007/s00109-017-1573-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castanos M. V., Zhou D. B., Linderman R. E., Allison R., Milman T., Carroll J., Migacz J., Rosen R. B., Chui T. Y. P., “Imaging of macrophage-like cells in living human retina using clinical OCT,” Invest. Ophthalmol. Vis. Sci. 61(6), 48 (2020). 10.1167/iovs.61.6.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurokawa K., Liu Z., Crowell J., Zhang F., Miller D. T., “Method to investigate temporal dynamics of ganglion and other retinal cells in the living human eye,” in Ophthalmic Technologies XXVIII (International Society for Optics and Photonics, 2018), Vol. 10474, p. 104740W. [Google Scholar]
- 54.Liu Z., Tam J., Saeedi O., Hammer D. X., “Trans-retinal cellular imaging with multimodal adaptive optics,” Biomed. Opt. Express 9(9), 4246–4262 (2018). 10.1364/BOE.9.004246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer D. X., Agrawal A., Villanueva R., Saeedi O., Liu Z., “Label-free adaptive optics imaging of human retinal macrophage distribution and dynamics,” Proc Natl Acad Sci USA 117(48), 30661–30669 (2020). 10.1073/pnas.2010943117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.