Abstract
Two core small nucleolar RNP (snoRNP) proteins, Nop1p (fibrillarin in vertebrates) and Nop58p (also known as Nop5p) have previously been reported to be specifically associated with the box C+D class of small nucleolar RNAs (snoRNAs). Here we report that Nop56p, a protein related in sequence to Nop58p, is a bona fide box C+D snoRNP component; all tested box C+D snoRNAs were coprecipitated with protein A-tagged Nop56p. Analysis of in vivo snoRNP assembly indicated that Nop56p was stably associated with the snoRNAs only in the presence of Nop1p. In contrast, Nop58p and Nop1p associate independently with the snoRNAs. Genetic depletion of Nop56p resulted in inhibition of early pre-rRNA processing events at sites A0, A1, and A2 and mild depletion of 18S rRNA. However, Nop56p depletion did not lead to codepletion of the box C+D snoRNAs. This is in contrast to Nop58p, which was required for the accumulation of all tested box C+D snoRNAs. Unexpectedly, we found that Nop1p was specifically required for the synthesis and accumulation of box C+D snoRNAs processed from pre-mRNA introns and polycistronic transcripts.
The nucleolus is often referred to as a ribosome factory, as most steps in the production of cytoplasmic ribosomes (synthesis of pre-rRNAs, pre-rRNA processing and modification, and ribosomal assembly) occur in this subnuclear compartment (reviewed in reference 38). The nucleolus also functions in the synthesis of other ribonucleoprotein (RNP) particles (signal recognition particle, telomerase, and the U6 RNP), as well as pre-tRNA processing (reviewed in reference 33). More surprisingly, the nucleolus has been shown to function in cell cycle control and the regulation of gene expression by the sequestration and release of regulatory complexes (reviewed in references 3 and 14).
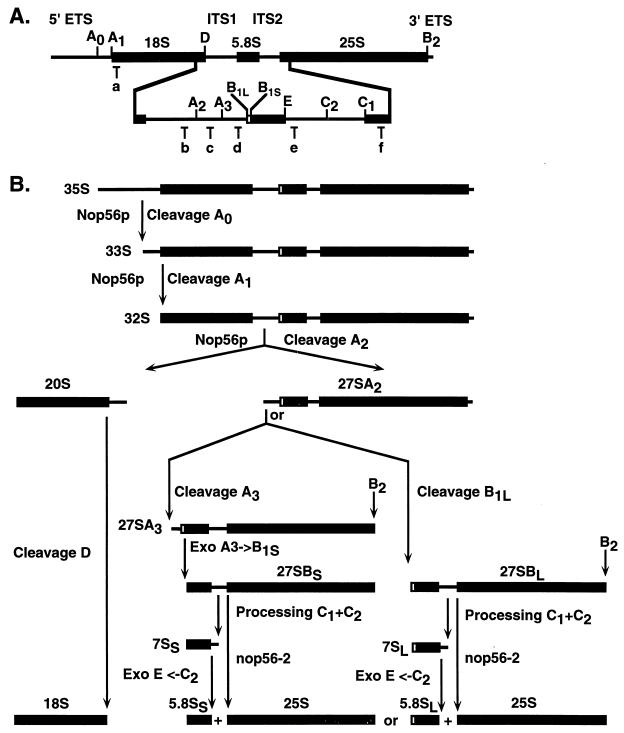
Three classes of small nucleolar RNAs (snoRNAs) have been defined on the basis of conserved features in their primary and secondary structures (reviewed in references 24 and 41). The vast majority of the box C+D and box H+ACA classes of snoRNAs function in the selection of sites of 2′-O methylation and pseudouridylation in the pre-rRNAs, respectively. Human box C+D snoRNAs also direct modification of the spliceosomal snRNA U6 (45). The third class of snoRNAs is defined by its sole known member, the RNA component of the endonuclease RNase MRP, which is closely related to RNase P (8). All tested box C+D snoRNAs are dispensable for normal growth in yeast, with the exception of U3 and U14. Depletion of these snoRNAs inhibits pre-rRNA cleavage at sites A0, A1, and A2 and strongly impairs 18S rRNA synthesis (Fig. 1).
FIG. 1.
Structure of the ribosomal DNA and pre-rRNA-processing pathway in yeast. (A) In the 35S primary transcript, the sequences of the mature 18S, 5.8S, and 25S pre-rRNAs are flanked by the external transcribed spacers (5′ and 3′ ETS) and separated by the internal transcribed spacers (ITS1 and ITS2). The cleavage sites are indicated by uppercase letters (A0 to E), and the oligonucleotide probes used are indicated by lowercase letters (a to f). (B) Successive cleavage of the 35S pre-rRNA at sites A0 and A1 generates the 33S and 32S pre-rRNAs. Cleavage of the 32S pre-rRNA at site A2 then generates the 20S and 27SA2 pre-rRNAs, which are precursors to the RNA components of the small and large ribosomal subunits, respectively. The mature 18S rRNA is generated by cleavage of the 20S pre-rRNA at site D. The 27SA2 precursor is either cleaved at site A3 by RNase MRP, generating the 27SA3 pre-rRNA, or at site B1L to yield 27SBL pre-rRNA. The 27SA3 pre-rRNA is rapidly digested by the 5′-to-3′ exonucleases Xrn1p and Rat1p to yield the 27SBS pre-rRNA. Processing at site B2, the 3′ end of the 25S rRNA, occurs concomitantly with 27SB 5′-end formation, since a probe specific for the 10-nucleotide extension beyond site B2 detects 27SA but not 27SB (21). The 27SBS and 27SBL pre-rRNAs both follow the same pathways of processing to 25S and 5.8SS/L through cleavage at sites C1, the 5′ end of the mature 25S rRNA, and C2 in ITS2, followed by 3′-to-5′ exonucleolytic digestion of 7SS and 7SL from site C2 to E by the exosome complex. Pre-rRNA processing analysis in GAL-regulated and thermosensitive alleles of Nop56p (nop56-2) indicated that Nop56p is required for the early pre-rRNA cleavage at sites A0, A1, and A2. In addition, 27SB processing was inhibited in nop56-2 strains.
All snoRNAs are associated with specific proteins in small nucleolar RNP (snoRNP) particles. The box C+D snoRNAs are associated with Nop1p (fibrillarin in vertebrates) and Nop58p (also known as Nop5p) (26, 39, 50). The 2′-O-methyltransferase has not yet been unambiguously identified. However, Nop1p is predicted to have a binding site for S-adenosyl-methionine (the methyl donor), and a point mutation designated nop1-3 inhibited pre-rRNA methylation without affecting processing, making it a strong candidate to be the rRNA methyltransferase (32, 43). The presence of the methyltransferase as an integral component of the box C+D snoRNPs would be closely analogous to the identification of the putative rRNA Ψ synthase, Cbfp5p, as a component of the H+ACA snoRNPs (20, 25, 47).
The box C+D snoRNAs are believed to be relatively unstructured, with a terminal stem bringing together the conserved motifs C and D that are very likely to function as protein binding sites. Selection of sites of methylation occurs through the formation of a snoRNA–pre-rRNA hybrid by Watson-Crick base pairing (18, 31; reviewed in reference 40) that locates box D at a fixed distance of 5 bp from the nucleotide to be modified. In most RNA species, a second site of pre-rRNA interaction is present, together with the duplicated motifs C′ and D′ (19, 44).
In addition to their roles in RNA modification, the conserved C and D motifs are also essential for accumulation and nucleolar localization of the snoRNAs (7, 16, 27, 30, 37, 48). These functions are most likely also mediated by snoRNP proteins.
A search for mutations which were lethal in combination with conditional alleles of Nop1p identified two essential genes, NOP56 and NOP58 (15). Nop56p and Nop58p shared 45% identical amino acid residues, including a carboxy-terminal domain with a repetitive structure rich in lysine, aspartate, and glutamate residues (KKD/E domain) that is also present in the nucleolar proteins Cbf5p and Dpb3p (25, 49). Both Nop56p and Nop58p were required for ribosome synthesis and copurified biochemically with Nop1p (15, 26). Like Nop1p, Nop58p was specifically associated with the box C+D snoRNAs (26). Nop58p was isolated independently by Wu et al. as an abundant nucleolar antigen interacting with several snoRNAs and was designated Nop5p (50).
Here we report that Nop56p is a core component of the box C+D snoRNPs. An assembly pathway for the box C+D snoRNP is presented in which Nop1p and Nop58p bind independently to the snoRNAs while Nop56p associates with Nop1p on the snoRNPs. In addition, we address the involvement of Nop1p and Nop56p in the accumulation and synthesis of box C+D snoRNAs.
MATERIALS AND METHODS
Strains.
The Saccharomyces cerevisiae strains Y606 [nop56::HIS3 + pRS315(LEU2)-NOP56] and Y799 [nop56-2] were described previously (15). Strains YDL401 (22), YDL522-17 (GAL::nop58) (26), D255 (GAL::nop1) (42), nop1-TS (nop1-2, nop1-3, nop1-4, nop1-5, and nop1-7) (44), and rrp6Δ::Kl TRP1 (2) were described previously. Strains YDL527-1 and YDL527-7 (GAL::nop56) were constructed in YDL401 by use of a one-step PCR strategy (22). This resulted in the direct fusion on the chromosome of a HIS3-pGAL cassette in front of the ATG of NOP56. PCR amplification was done with plasmid pTL26 (22) and oligonucleotides LD188 and LD189. Transformants were selected on 2% raffinose–2% sucrose–2% galactose (RSG) minimal medium lacking histidine, screened for glucose sensitivity, and analyzed by PCR on yeast colonies with oligonucleotides LD154 and LD155. The RNA analyses presented in Fig. 5 and 7 were performed in duplicate on two independently isolated GAL::nop56 strains (YDL527-1 and YDL527-7). The results shown are for strain YDL527-1.
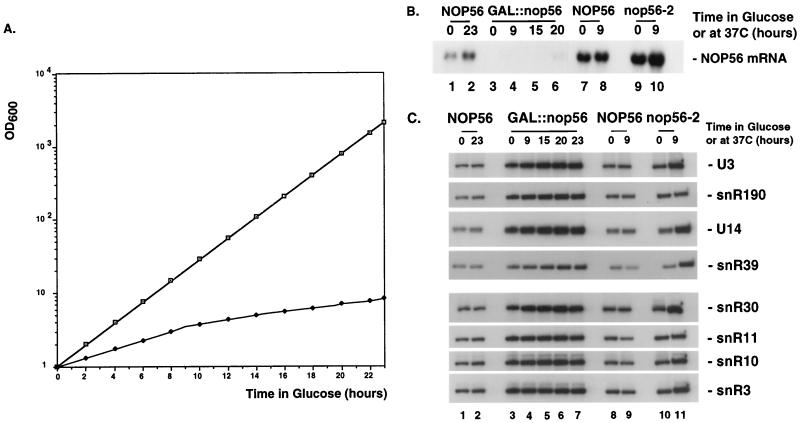
FIG. 5.
Nop56p is not required for snoRNA accumulation. (A) Growth of the GAL::nop56 (solid circles) and NOP56 (open squares) strains following transfer to glucose medium. Cell density was measured at regular intervals, and the cultures were periodically diluted to maintain exponential growth. The results are presented on an exponential scale with the OD600 values corrected for the dilution factor. (B) Northern-blot hybridization of NOP56 mRNA in GAL::nop56 and nop56-2 strains. Total RNA was extracted from GAL::nop56 and nop56-2 strains following growth under permissive conditions on RSG medium or at 23°C (0 h) and following transfer to repressive conditions on glucose medium or at 37°C for the times indicated. Isogenic control strains were used. Total RNA was resolved in a 1.2% agarose gel containing formaldehyde. (C) Steady-state levels of the snoRNAs on depletion and heat inactivation of Nop56p. Total RNA was extracted from GAL::nop56 and nop56-2 strains grown in RSG medium or at 23°C (0 h) and following transfer to glucose medium or to 37°C for the times indicated. Isogenic control strains were used. RNA was resolved in an 8% polyacrylamide gel under denaturing conditions.
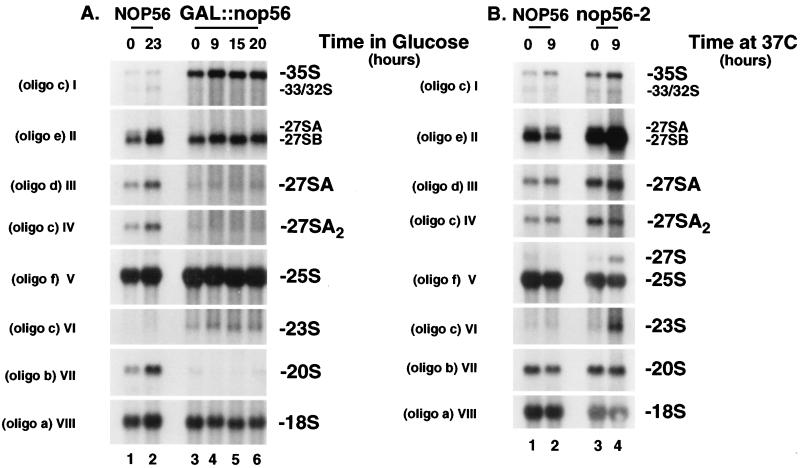
FIG. 7.
Pre-rRNA processing in GAL::nop56 and nop56-2 strains. (A) GAL::nop56 strain in permissive RSG medium (0 h) and at time points (9 to 20 h) after transfer to glucose medium. (B) nop56-2 strain grown at 23°C (0 h) and following transfer to 37°C for 9 h. Isogenic control strains were used in parallel. Total RNA was extracted and resolved in a 1.2% agarose gel containing formaldehyde. The steady-state levels of precursors and mature rRNAs were determined by hybridization with oligonucleotides a (row VIII), b (row VII), c (rows I, IV, and VI), d (row III), e (row II), and f (row V). The oligonucleotides are depicted in Fig. 1 and are described in Materials and Methods. Oligonucleotides d and e do not formally distinguish between 27SA2 and 27SA3, but 27SA3 is substantially less abundant than 27SA2, and the reduced amount of 27SA observed in rows II and III is a consequence of the reduced 27SA2 levels.
For Nop56p depletion and heat inactivation, the wild-type strains used were YDL401 and Y606, respectively. YDL401 was used as the wild-type control for Nop1p depletion and Nop1p heat inactivation.
Time courses, RNA extraction, Northern hybridization, and primer extension analysis.
For depletion of Nop56p (YDL527-1 and YDL527-7), cells growing exponentially in permissive RSG complete medium at 30°C were harvested by centrifugation, washed, and resuspended in prewarmed yeast extract peptone-dextrose (YPD) medium. During growth, the cells were diluted with prewarmed medium and constantly maintained in exponential phase.
RNA extraction, Northern hybridization, and primer extension were done as described previously (23). Standard 1.2% agarose-formaldehyde–8 and 6% polyacrylamide gels were used to analyze the processing of the high- and low-molecular-weight rRNA species and the steady-state levels of the snoRNAs. Nine micrograms of total RNA were used per lane for the Northern blot and primer extension experiments. For heat inactivation of Nop56p, strain Y799 was grown at 23°C in YPD medium and transferred to 37°C. For depletion of Nop1p, strain D255 was grown in RSG medium to mid-log phase, washed, and transferred to YPD medium. For Nop1p heat inactivation, the nop1-2 to nop1-7 strains were grown in YPD medium at 23°C and transferred to 37°C. Nop58p depletion was achieved as previously described (26). The rrp6-Δ strain was grown at 30°C in SD −trp.
Analysis of methylation levels.
The overall level of rRNA methylation was assessed by in vivo pulse-labeling of the RNAs with either [3H]uracil or [3H]methionine followed by autoradiography. A GAL::nop56 strain (YDL527-1) and the isogenic wild-type control (YDL401), transformed with a plasmid expressing the URA3 gene (pFL44S) (5), were grown at 30°C in minimal medium lacking uracil, methionine, and histidine and containing 2% galactose, 2% sucrose, and 2% raffinose. Exponentially growing cells were washed and transferred to prewarmed minimal medium lacking uracil, methionine, and histidine and containing 2% glucose. At identical optical densities at 600 nm (OD600) of 0.41 (for the GAL::nop56 strain, this corresponded to 6 h after transfer to nonpermissive conditions), wild-type and mutant cells were pulse-labeled for 5 min with 100 μCi of either [3H]uracil or [3H]methionine/ml. Aliquots (1 ml) of cultures were pelleted and snap frozen in liquid nitrogen. Total RNA was extracted and resolved on a 1.2% agarose–formaldehyde gel. The gels were transferred to GeneScreen Plus membranes (NEF-976; Dupont De Nemours), sprayed with tritium enhancer (NEF-970G; Dupont De Nemours), and exposed for autoradiography.
Western blot analysis.
For protein extraction, cells equivalent to an OD600 of 10 were harvested and resuspended in 200 μl of sodium dodecyl sulfate loading buffer with 50 μl of glass beads. The cells were vortexed for 1 min and incubated for 1 min at 95°C three times successively. The lysates were cleared by centrifugation for 10 min at 14 krpm, and supernatants equivalent to 0.375 OD600 unit of cells were loaded per lane. Samples were run on a sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis gel and blotted according to standard procedures. The blot was decorated with monoclonal mouse anti-Nop1p antibody (mA66; dilution, 1:20; kindly provided by J. Aris, University of Florida) or with polyclonal rabbit anti-Srp14p (dilution, 1:500; kindly provided by J. Brown, University of Edinburgh) and developed with the ECL detection kit (Amersham).
Oligonucleotides.
A comprehensive snoRNA database is available (http: //www.bio.umass.edu/biochem/rna-sequence/Yeast_snoRNA_Database/snoRNA _DataBase.html) (36). The probes for box C+D snoRNAs were as follows: snR38, GAGAGGTTACCTATTATTACCCATTCAGACAGGGATAACTG (255); snR39, CGACAGCATCGTCAATGACTAGTCGAATATGTATTGGG (256); snR54, CTCTACAAGATCGTTTGATCACAGTCAGTAGAACGAAAGTTATTG (257); snR59, GGTGATTAAACGACAGCATTGTCAAAGACTAGTCGA (258); and snR73, GCTCAGTACCACGCCCTGT (259). U3, U14, U18, U24, snR4, snR13, and snR190 were as described previously (26). The probes for box H+ACA snoRNAs (snR3, snR10, snR11, snR30, snR31, snR33, snR36, snR37, and snR42) and for MRP RNA were all described previously (26). The probes for spliceosomal snRNAs were U5 (AAGTTCCAAAAAATATGGCAA) and U6 (AAAACGAAATAAATCTCTTTGTAAAAC). The oligonucleotides used for pre-rRNA hybridization were oligonucleotides a, CATGGCTTAATCTTTGAGAC (008); b, CGGTTTTAATTGTCCTA (004); c, TTGTTACCTCTGGGCCC (003); d, CCAGTTACGAAAATTCTTG (005); e, GGCCAGCAATTTCAAGTTA (013); and f, CTCCGCTTATTGATATGC (007).
A probe specific to NOP56 mRNA was generated by PCR on genomic DNA with oligonucleotides LD154 and LD155 and labeled using the Prime-a-Gene labeling kit (Promega).
The following oligonucleotides were used to construct yeast strains: LD154, CATATGATGACAATGTGG; LD155, AATCAAATCCTGAGCAAG; LD188, AAGTGAAAT TT TT TTGAGTGATGAGATGGGAAATGAAAAAT TT TGTGCCTCTTGGCCTCCTCTAGT; and LD189, AAACAGTAGGTATTCAATAGGAGCCATCT TTCACTAAATACTGT TCTGCACGCTCGAAT TCCT TGAATTTTCAAA.
RESULTS
Nop56p is specifically associated with the box C+D snoRNAs.
The association of Nop56p with the snoRNAs was addressed by immunoprecipitation experiments. Epitope-tagged alleles of Nop56p were constructed in which two copies of the z domain of Staphylococcus aureus protein A (ProtA) were fused in frame with the ATG of either full-length or carboxy-terminally truncated versions of Nop56p. Both tagged alleles were found to be functional, as shown by the absence of growth defects when they were expressed in a fully deleted nop56-Δ background (reference 15 and data not shown).
Coprecipitation experiments with immunoglobulin G (IgG)-agarose beads revealed that ProtA-Nop56p is associated with all tested box C+D snoRNAs: U3, U14, U18, U24, snR4, snR13, snR39, and snR190 (Fig. 2A, lanes 7 to 12, and data not shown). No coprecipitation was observed with a nontagged Nop56p control strain (Fig. 2A, lanes 4 to 6). A ProtA-Nop58p strain was used as a control and efficiently coprecipitated the snoRNAs (Fig. 2A, lanes 1 to 3) (26). To exclude the possibility that the highly charged KKD/E domain of Nop56p led to nonspecific snoRNA binding, the association was also tested in ProtA-Nop56ΔKKE strains (see Materials and Methods). Truncation of the carboxy-terminal region of Nop56p had no effect on association with the snoRNAs (Fig. 2A, lanes 13 to 18, and data not shown).
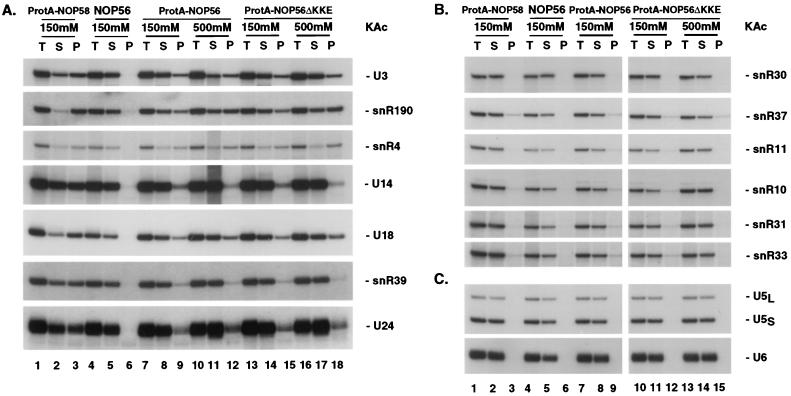
FIG. 2.
Nop56p is specifically associated with the box C+D snoRNAs. Immunoprecipitation on IgG-agarose was performed on lysates from strains expressing fusions between ProtA and Nop56p (ProtA-NOP56 or ProtA-NOP56ΔKKE). An isogenic wild-type control strain (NOP56) and a ProtA-NOP58 strain were used as controls. The concentrations of KAc used during immunoprecipitation are indicated. RNA was extracted from equivalent amounts of total (T), supernatant (S), and pellet (P) fractions, separated on an 8% polyacrylamide gel, and analyzed by Northern hybridization. (A) Probes specific for box C+D snoRNAs; (B) probes specific for box H+ACA snoRNAs; (C) probes specific for the snRNAs U5 and U6.
As previously observed for ProtA-Nop1p and ProtA-Nop58p, the efficiency of coprecipitation with ProtA-Nop56p varied for different box C+D snoRNA species, presumably reflecting differences in accessibility within the snoRNP structures. The efficiency of coprecipitation with ProtA-Nop56p was generally lower than that observed with ProtA-Nop58p. The interactions between the ProtA-tagged alleles of Nop56p and the box C+D snoRNAs appeared to be specific. Very limited amounts of the box H+ACA snoRNAs tested (snR3, snR10, snR11, snR30, snR31, snR33, snR36, snR37, and snR42) were recovered in the pellet fractions (Fig. 2B, lanes 7 to 15, and data not shown). The spliceosomal snRNAs U5 and U6 and the MRP RNA were also not precipitated (Fig. 2C and data not shown). The immunoprecipitation experiments were performed at two salt concentrations: 150 and 500 mM KAc. Nonspecific association between Nop1p and the box H+ACA snoRNAs was reported at 150 mM but was lost under the more stringent conditions of 500 mM (13). Similarly, the weak association between Nop56p and the box H+ACA snoRNAs seen at 150 mM was lost at 500 mM (Fig. 2B, compare lanes 9 and 12 with lane 15). As previously reported (26), Nop58p also coprecipitated several H+ACA species slightly above the background level at 150 mM (Fig. 2B, lanes 1 to 3; see, for example, snR37). The coprecipitation of the box C+D snoRNAs with Nop56p was not greatly affected by the salt concentration (Fig. 2A).
We conclude that Nop56p is associated with all tested box C+D snoRNAs. The interactions between Nop56p and the snoRNAs were specific, resistant to stringent immunoprecipitation conditions, and not dependent on the highly charged carboxyl KKD/E domain.
Association of Nop56p with the box C+D snoRNAs requires Nop1p.
Nop56p and Nop58p interact genetically and physically with Nop1p. The requirement for Nop1p in the association of Nop56p and Nop58p with the box C+D snoRNAs was assessed by immunoprecipitation in strains with Nop1p depleted. The chromosomal NOP1 gene was placed under the control of a GAL promoter in strains expressing either a ProtA-Nop56ΔKKE or ProtA-Nop58ΔKKE fusion. In each case, two independently isolated strains were analyzed. In these experiments carboxy-terminally truncated versions of Nop56p and Nop58p that lack the KKD/E repeat regions were used, as they were reported to be less sensitive to proteolytic degradation during immunoprecipitation (15). Deletion of these domains had no effect on interaction with the snoRNAs (Fig. 2) (15, 26, 50).
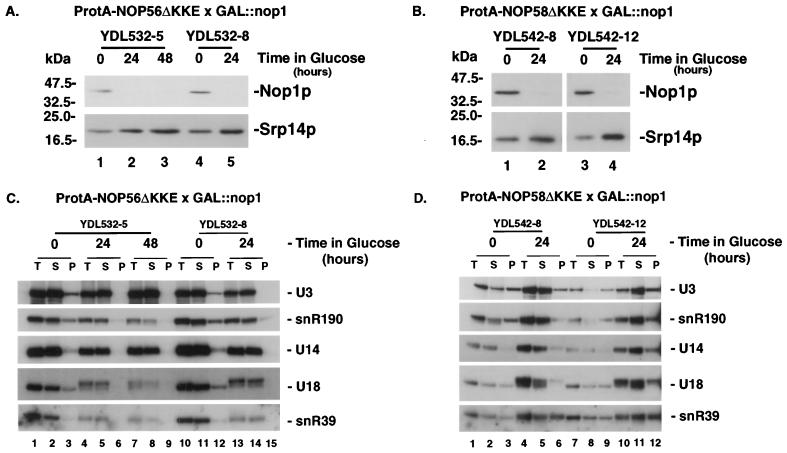
ProtA-NOP56ΔKKE × GAL::nop1 (YDL532-5 and -8) and ProtA-NOP58ΔKKE × GAL::nop1 (YDL542-8 and YDL542-12) strains were grown in galactose medium (zero-hour samples) and transferred to glucose medium for 24 or 48 h. The steady-state level of Nop1p was analyzed by Western blotting and found to be substantially reduced 24 h after transfer (Fig. 3A and B). Coprecipitation of the box C+D snoRNAs U3, U14, U18, U24, snR190, and snR39 with ProtA-Nop56p was seen in cell lysates prepared after growth on galactose medium (Fig. 3C, lanes 3 and 12, and data not shown). However, this precipitation was lost following transfer to glucose medium for 24 or 48 h (Fig. 3C, lanes 6, 9 and 15). In contrast, immunoprecipitation of the box C+D snoRNAs U3, U14, U18, U24, snR190, and snR39 with ProtA-Nop58p was seen in lysates from cells grown on either galactose (Fig. 3D, lanes 3 and 9, and data not shown) or glucose (Fig. 3D, lanes 6 and 12) medium. Western blot analysis showed that Nop1p was not required for the accumulation of the ProtA-Nop56ΔKKE and ProtA-Nop58ΔKKE fusions (data not shown).
FIG. 3.
Nop56p, but not Nop58p, is dependent on Nop1p for binding to the snoRNAs. (A and B) Western analysis of Nop1p levels in ProtA-NOP56ΔKKE × GAL::nop1 (A) and ProtA-NOP58ΔKKE × GAL::nop1 (B) strains. (C and D) Immunoprecipitation on IgG-agarose was performed on lysates from strains expressing ProtA epitope-tagged alleles of Nop56p (C) or Nop58p (D) in the presence (0-h time points) or absence (24- and 48-h time points) of Nop1p. RNA was extracted from equivalent amounts of total (T), supernatant (S), and pellet (P) fractions, separated on an 8% polyacrylamide gel, and analyzed by Northern hybridization.
We conclude that Nop1p is required for the stable association of Nop56p with the box C+D snoRNAs but is not required for the Nop58p-snoRNA association.
The requirement for Nop58p in the Nop56p-snoRNA interaction is more difficult to assess, since depletion of Nop58p results in strong codepletion of all tested box C+D snoRNAs (e.g., snR190 [Fig. 4A] [26]). However, a 3′-extended version of U24 is accumulated in strains with Nop58p depleted (Fig. 4A) (26). This species was efficiently coprecipitated with Nop56p in a GAL::nop58 strain expressing a ProtA-Nop56ΔKKE fusion (Fig. 4A, lane 6).
FIG. 4.
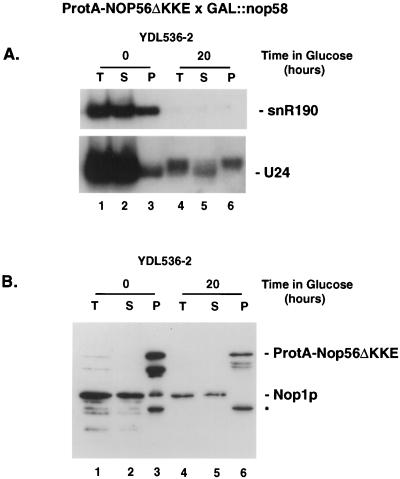
Effects of Nop58p depletion on Nop56p association with the snoRNAs and Nop1p. Immunoprecipitation on IgG-agarose was performed on lysates from a strain expressing ProtA-Nop56ΔKKE in the presence (0 h) or absence (20 h) of Nop58p. Total protein and RNA were extracted from equivalent amounts of total (T), supernatant (S), and pellet (P) fractions. (A) Northern blot hybridization with probes directed against box C+D snoRNAs. (B) Western blot decorated with anti-Nop1p antibodies (mA66); these cross-react with the ProtA tag on Nop56p. Several degradation products of ProtA-Nop56ΔKKE were observed, including one (∗) migrating slightly faster than Nop1p (15).
We conclude that, at least in the case of the 3′-extended form of U24, Nop56p associates with a box C+D snoRNA in the absence of Nop58p.
Nop1p and Nop56p were reported to be tightly associated and to coprecipitate in stoichiometric amounts (15). In order to determine whether these interactions are dependent on the snoRNAs, cell lysates were prepared from the ProtA-NOP56ΔKKE × GAL::nop58 strain, YDL536-2, following growth in permissive RSG medium and 20 h after transfer to glucose medium. Following immunoprecipitation of ProtA-Nop56ΔKKE, the proteins were analyzed by Western blotting with anti-Nop1p antibodies. Under permissive conditions, Nop1p was readily detected in the pellet fractions (Fig. 4B, lane 3) along with the ProtA-Nop56ΔKKE fusion protein that is also decorated by the antibodies due to its ProtA domain. Several ProtA-Nop56ΔKKE degradation products were also detected. Following depletion of Nop58p and the snoRNAs, Nop1p was not detectably coprecipitated with ProtA-Nop56p (Fig. 4B, lane 6).
We conclude that Nop1p and Nop56p do not associate in the absence of snoRNAs, indicating that the two proteins only interact within the snoRNPs.
Nop56p is not required for snoRNA accumulation.
All tested box C+D snoRNAs were codepleted with Nop58p (26), and we therefore tested the steady-state levels of the snoRNAs in the absence of Nop56p.
To allow depletion of Nop56p, the chromosomal copy of NOP56 was placed under the control of a regulated GAL10 promoter using a one-step, PCR-based strategy (see Materials and Methods). The growth rate of GAL::nop56 strains was already impaired on permissive RSG medium (a doubling time of 2 h for the wild type and 4 h for the GAL::nop56 strain). Following transfer to nonpermissive glucose medium, the growth rate decreased, with a major reduction in doubling time occurring between 9 and 10 h after transfer (Fig. 5A). Eighteen hours after transfer, the doubling time of GAL::nop56 strains had increased to 16 h. Total RNA was extracted at various time points after transfer to glucose medium, and the levels of NOP56 mRNA were analyzed by Northern hybridization. In the GAL::nop56 strains, the NOP56 mRNA was barely detected in RSG medium (Fig. 5B, lane 3), which was presumably the basis for the 50% growth rate inhibition. No mRNA was detected after transfer to glucose medium (Fig. 5B, lanes 4 to 6). The analyses presented in Fig. 5 (also see Fig. 7) were performed in duplicate on two independently isolated GAL::nop56 strains (YDL527-1 and YDL527-7 [see Materials and Methods]). The strains showed identical phenotypes; results are shown only for YDL527-1.
The steady-state levels of the snoRNAs were analyzed by Northern hybridization in GAL::nop56 strains and in the previously described temperature-sensitive (TS)–lethal nop56-2 strain (15) during growth under permissive conditions and following transfer to glucose medium or 37°C. All tested box C+D snoRNAs, U3, U14, U18, U24, snR4, snR13, snR39, and snR190 (Fig. 5C and data not shown), were accumulated at levels close to those of the wild type. The steady-state levels of the snoRNAs showed a mild increase in the mutants, presumably as a consequence of an overall decrease in rRNA levels (see below), since constant amounts of total RNA were loaded. Several box H+ACA snoRNAs tested, snR3, snR10, snR11, snR30, snR36, and snR42 (Fig. 5C and data not shown), also accumulated to wild-type levels.
We conclude that genetic depletion of Nop56p and heat inactivation in nop56-2 strains had no effect on the accumulation of the box C+D snoRNAs.
Nop1p is required for snoRNA accumulation.
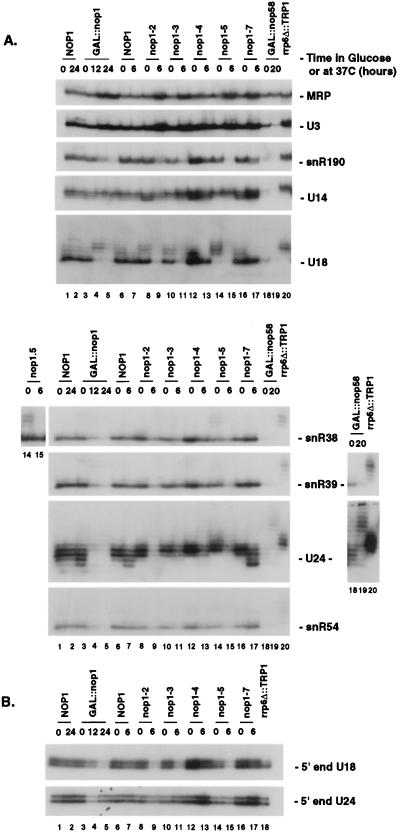
During the analysis of Nop1p depletion, we were surprised to see the appearance of slow-migrating forms of the U18 and U24 snoRNAs (Fig. 3C to D and data not shown). This observation prompted us to analyze the requirement for Nop1p in snoRNA accumulation and synthesis. A GAL::nop1 strain (42) was grown in galactose medium (zero hour) and transferred to glucose medium for 12 and 24 h. Five TS nop1 alleles (43) that do not affect Nop1p accumulation and have distinct ribosome synthesis defects were also analyzed. These were grown at 23°C (zero hour) and transferred to 37°C for 6 h.
A range of strategies have evolved for the expression of snoRNAs, which can be excised from the introns of pre-mRNAs or synthesized from their own promoter and terminator sequences, either as monocistronic or polycistronic transcription units (reviewed in reference 29); both U18 and U24 are intron encoded. All the tested intronic box C+D snoRNAs, U18, U24, snR38, snR39, snR54, and snR59, were depleted following transfer of the GAL::nop1 strain to glucose medium (Fig. 6A, compare lanes 2 and 5, and data not shown). Accumulation of snoRNAs synthesized from polycistronic transcripts was tested for species expressed from transcription units encoding two snoRNAs (U14 and snR190), three snoRNAs (snR51 and snR70), and 7 snoRNAs (snR73 and snR76) (9, 10, 34, 35). All tested snoRNAs encoded in polycistronic transcripts, except U14, were strongly codepleted with Nop1p (Fig. 6A and data not shown). U3 (Fig. 6A) and snR4 (data not shown) are expressed as monocistronic transcripts and were unaffected on Nop1p depletion, as was the MRP RNA (Fig. 6A).
FIG. 6.
Nop1p is required for the accumulation and synthesis of snoRNAs processed from introns and polycistronic transcripts. Steady-state levels of intronic and polycistronic snoRNAs on depletion and heat inactivation of Nop1p are shown. Total RNA was extracted from a GAL::nop1 strain and from nop1-2 to nop1-5 and nop1-7 strains following growth under permissive conditions on galactose or at 23°C (0 h) and transfer to a glucose-based medium or 37°C for the times indicated. For comparison, total RNA was extracted from a null allele of rrp6 (rrp6Δ::TRP1) and from a GAL-regulated NOP58 strain (GAL::nop58). (A) RNA separated on 6% polyacrylamide gels and analyzed by Northern hybridization. (B) Primer extension using internal U18 or U24 primers. Inserts show longer exposures of the lanes indicated.
Accumulation of the intronic box C+D snoRNAs was strongly reduced at 37°C in the nop1-2, nop1-3, and nop1-5 strains but was unaffected in the nop1-4 and nop1-7 strains. The nop1-4 and nop1-7 mutations, therefore, uncoupled the lethality of nop1 mutations from effects on snoRNA accumulation. The effects of different nop1 alleles on accumulation of polycistronic snoRNAs were similar to those on the intronic box C+D snoRNAs, with depletion seen in the nop1-2, nop1-3, and nop1-5 strains at 37°C.
Alterations in the lengths of several snoRNAs were seen in nop1 mutants. For U24, shorter forms accumulated in the nop1-7 strain (Fig. 6A, lane 17) while longer forms appeared in the nop1-5 strain at 23°C (Fig. 6A, lane 14). These were, however, shorter than the species detected in the GAL::nop58 strain (Fig. 6A, lane 19). Extended forms of snR38, snR51, and U18 were also seen in the nop1-5 strain (Fig. 6A, lane 14, and data not shown), and extended snR39, snR51, snR73, and U18 were seen on depletion of Nop1p (Fig. 6A, lane 4, and data not shown). The strongest accumulation of extended species was seen in the nop1-5 strain at 23°C, the permissive temperature for growth and pre-rRNA processing, and was not accompanied by a clear reduction in snoRNA levels. Nop1p was therefore required both for snoRNA accumulation and for accurate synthesis, with these functions uncoupled in the nop1-5 strain.
Effects on snoRNA length were most evident for U18 and U24. Primer extension analysis (Fig. 6B) from internal U18 and U24 primers revealed that 5′-end formation was not affected on Nop1p depletion or inactivation. The reduction in primer extension signals was in good agreement with the results of the Northern analysis. We conclude that Nop1p is required for normal 3′-end formation of these snoRNAs.
Deletion of the gene encoding Rrp6p, a component of the exosome complex of 3′→5′ exonucleases (2, 6), led to the accumulation of snoRNAs that were 3′ extended by 3 to 4 nucleotides (Fig. 6A, lane 20) (1). The 3′-extended forms of U18, snR38, snR39, snR51, and snR73 seen in the nop1 mutants comigrated with those detected in rrp6-Δ strains. This suggests that Nop1p may interact with the exosome complex to promote accurate 3′ trimming of the snoRNAs.
We conclude that Nop1p is required for the accumulation and accurate 3′-end formation of intron-encoded and polycistronic snoRNAs. It is notable that the effects on snoRNA accumulation and synthesis seen on depletion of Nop1p were not seen on depletion of Nop56p (Fig. 5C). This indicates that Nop1p associates with the snoRNAs in the absence of Nop56p.
Requirement for Nop56p in pre-rRNA processing.
During the analysis of Nop1p, distinctly different effects on ribosome and snoRNA synthesis were observed in genetically depleted strains and in TS mutants (42, 43) (see above). We therefore compared pre-rRNA processing in GAL::nop56 strains and the previously reported nop56-2 TS strain (15).
GAL::nop56 strains were grown in RSG medium and transferred to glucose medium. Total RNA was extracted 9, 15, and 20 h after transfer and analyzed by Northern hybridization (Fig. 7A [the oligonucleotide probes used are indicated, and their locations are shown in Fig. 1A]). Importantly, even in permissive RSG medium (Fig. 7A, lanes 0), the GAL::nop56 strains have a strongly reduced level of NOP56 mRNA (Fig. 5B) and are substantially impaired in growth (50% reduction).
In GAL::nop56 strains, strong accumulation of 35S pre-rRNA was seen (Fig. 7A, row I), accompanied by depletion of the 32S (Fig. 7A, row I), 20S (Fig. 7A, row VII), and 27SA2 (Fig. 7A row IV) pre-rRNAs. The 23S RNA, which extends from the 5′ end of the pre-rRNA transcript to site A3 in ITS1, was also detected in GAL::nop56 strains (Fig. 7A, row VI). Oligonucleotide c (Fig. 7A, row I) hybridizes to both the 33S and 32S pre-rRNAs (Fig. 1), which are not resolved on the gel, but the 32S pre-rRNA is much more abundant and generates most of the signal. This phenotype is highly characteristic of substantial inhibition of cleavage at sites A0, A1, and A2 (Fig. 1). GAL depletion always allows a certain degree of leakage, and in this case, the residual processing appeared to be sufficient to produce 18S rRNA at steady-state levels close to those of the wild type (Fig. 7A, row VIII). No clear alteration in the levels of the 27SB pre-rRNA (Fig. 7A, row II) or the mature 25S rRNA (Fig. 7A, row V) was observed.
In the nop56-2 strain at 37°C (Fig. 7B, lane 4), some accumulation of the 35S pre-rRNAs and 23S RNA was seen, indicating a mild inhibition of the cleavages at sites A0, A1, and A2. The effects on the 35S, 27SA2, and 20S pre-rRNAs were, however, much less marked than in the strains with Nop56p depleted. In contrast, clear accumulation of the 27SB pre-rRNA was seen in the nop56-2 strain at 37°C, accompanied by a reduced level of the mature 25S rRNA (Fig. 7B, row V, lane 4).
We conclude that cleavage at sites A0, A1, and A2 is affected in both GAL::nop56 and nop56-2 strains, but with different severities. It is notable that in neither mutant does the magnitude of the reduction in the mature rRNA levels appear sufficient to account for the severe growth inhibition observed. Moreover, little further impairment in processing was seen on transfer of the GAL::nop56 strain from RSG medium to glucose, although growth is substantially more inhibited. We conclude that the defect in pre-rRNA processing is probably not the primary cause of lethality on depletion of Nop56p.
The effects of Nop56p depletion on 2′-O methylation were assessed by metabolic labeling. A GAL::nop56 strain and the isogenic wild-type control were grown in RSG medium and transferred to glucose medium for 6 h before being pulse-labeled for 5 min with either [3H]methionine or [3H]uracil. Total RNA was extracted from the same numbers of cells, separated on 1.2% agarose–formaldehyde gels, transferred to GeneScreen membranes, and visualized by fluorography. Incorporation in the strain with Nop56p depleted was substantially lower than in the wild type, presumably due to its reduced growth rate. However, the reductions in the incorporation of tritiated methionine and tritiated uracil were not clearly different (data not shown).
We conclude that 2′-O methylation of the rRNA was more resistant to depletion of Nop56p than was pre-rRNA processing. The same phenomenon was observed on depletion of Nop58p (26) but not on depletion of Nop1p, which inhibited both pre-rRNA processing and nucleolar methylation (42), consistent with the proposal that Nop1p is the methyltransferase.
DISCUSSION
Nop56p is the third common component of the large family of box C+D snoRNPs to be identified. ProtA-tagged Nop56p coprecipitated all tested box C+D snoRNAs with no significant coprecipitation of the box H+ACA snoRNAs or other small RNAs (MRP RNA and snRNAs). The highly charged carboxy-terminal KKD/E domain of Nop56p was not required for interactions with the snoRNAs. A similar carboxy-terminal truncation in Nop58p also had no effect on interaction with the snoRNAs (26).
Previous studies indicated that Nop58p coprecipitated with all tested box C+D snoRNAs and was required for their accumulation (26), showing it to be an integral component of all box C+D snoRNPs. Similarly, Nop1p coprecipitated with all tested box C+D snoRNAs (4, 13) and was required for the accumulation of all species expressed from pre-mRNA introns and polycistronic transcripts and must also be a component of the corresponding snoRNPs. Since Nop1p coprecipitated with Nop56p in stoichiometric amounts (15) and these interactions did not take place in the absence of the snoRNAs, we conclude that Nop1p, Nop56p, and Nop58p are common components of most, and probably all, box C+D snoRNPs.
Based on our initial analysis of the in vivo assembly of the snoRNPs, we propose that Nop1p and Nop58p bind independently to the box C+D snoRNAs, with Nop56p then associating with Nop1p on the snoRNP. The association of Nop56p with the snoRNAs was only detected in the presence of Nop1p, whereas Nop58p could interact with the snoRNAs in the absence of Nop1p. Moreover, Nop1p and Nop58p are required for the stability of box C+D snoRNAs, whereas Nop56p is not, indicating that neither Nop1p nor Nop58p is dependent on Nop56p for association with the snoRNAs. This is consistent with the recent report that recombinant Xenopus fibrillarin binds directly to the U16 snoRNA (11) and indicates that Nop56p only interacts with snoRNAs that have prebound Nop1p. All tested box C+D snoRNAs were lost in strains with Nop58p depleted (26), complicating the analysis of its role in snoRNP assembly. A 3′-extended form of U24 was, however, accumulated on depletion of Nop58p and was efficiently precipitated with ProtA-Nop56p, demonstrating that the association of Nop56p with this species at least was not dependent on Nop58p.
Most box C+D snoRNAs are synthesized by posttranscriptional processing, either from introns excised from pre-mRNAs or from polycistronic pre-snoRNAs. In both cases, 5′ processing of the mature snoRNAs involves the 5′→3′ exonucleases Rat1p and Xrn1p, while 3′ processing involves the exosome complex of 3′→5′ exonucleases (1, 34, 46). All tested box C+D snoRNAs that are synthesized from introns or polycistronic transcripts were codepleted with Nop1p, with the exception of U14. Nop1p was not, however, required for normal accumulation of the U3 and snR4 snoRNAs that are encoded in monocistronic transcripts. This is distinct from the effects of Nop58p depletion, which strongly reduced the levels of all box C+D snoRNAs, including snR4, U3, and U14 (26). In contrast, the levels of all tested snoRNAs were unaffected either by depletion of Nop56p or by the conditional-lethal nop56-2 mutation.
Alterations in the position of the mature 3′ ends of several snoRNAs were seen in nop1 mutants. The mutant alleles of NOP1 uncoupled various defects in ribosome synthesis (43) and also differ in their effects on snoRNA synthesis and accumulation. The strongest effects on 3′-end formation were shown by the nop1-5 strain at the permissive temperature (23°C), at which snoRNA accumulation was unaffected. Discrete 3′-extended forms of many snoRNAs are observed in strains lacking the Rrp6p component of the exosome complex, which is responsible for the final trimming of the pre-snoRNAs (2, 6). The extended snoRNAs in the nop1 and rrp6 mutants were similar in size, suggesting that Nop1p interacts with the exosome during snoRNA 3′ maturation. The formation of longer forms of the snoRNAs on depletion or mutation of snoRNP components is slightly surprising; if anything, the loss of these factors might have been expected to allow the exosome complex further access into the mature snoRNA region. This indicates that the snoRNP proteins play a more active role than simply blocking the exosome. They may be required to promote the final trimming reaction and/or to displace other factors that protect the 3′ ends of the pre-snoRNAs.
It was surprising that Nop1p was required for the accumulation of snoRNAs synthesized from polycistronic and intronic precursors but not monocistronic snoRNAs. These appear to have highly homologous box C+D regions and 3′-terminal stems, which likely constitute the snoRNP protein binding site. The obvious difference is that the monocistronic snoRNAs have a 5′-cap structure and therefore do not undergo 5′ processing by Rat1p. It is possible that in addition to influencing 3′ processing, Nop1p is also involved in protection of the 5′ ends during pre-snoRNA processing.
What is the function of Nop56p? Depletion of Nop56p inhibited cleavage at sites A0, A1, and A2, leading to impaired synthesis of the 18S rRNA (Fig. 1). These cleavages were also inhibited in strains with Nop1p, Nop58p, or the U3 and U14 snoRNAs depleted (17, 26, 28, 42, 50), suggesting that Nop56p is required for the normal functioning of U3 and/or U14. Interestingly, the GAL::nop56 and nop56-2 strains had distinct pre-rRNA-processing phenotypes. Greater inhibition of the cleavages at sites A0, A1, and A2 was seen on depletion of Nop56p than in the nop56-2 strain. In contrast, the nop56-2 strain had additional defects in the 25S rRNA synthesis pathway that were not observed on Nop56p depletion. The effects on ribosome synthesis of genetic depletion of Nop1p and conditional-lethal point mutations were also found to be distinctly different, and some mutations in NOP1 also interfere with 25S rRNA synthesis (42, 43).
In neither the strains with Nop56p depleted nor the nop56-2 strains does the reduction in the mature rRNAs appear sufficient to account for the severe growth inhibition. This is in contrast to depletion of Nop58p or Nop1p, both of which substantially reduced 18S rRNA levels. Ribose methylation of rRNA was also not greatly affected on Nop56p depletion. As previously proposed, Nop56p could be involved in the correct assembly of ribosomal particles (15); mutations in NOP1 that lead to specific defects in ribosome assembly have been reported. Deletion of the cap binding complex proteins CBP20 and CBP80 (Gcr3p and Mud13p in yeast) was recently reported to be synthetic lethal with mutations in the snoRNP proteins Nop58p and Cbf5p (12). In the absence of Gcr3p and Mud13p, some inhibition of pre-rRNA processing was observed, and synergistic defects in ribosome synthesis were proposed as the basis for the colethality (12). Since the inhibition of rRNA synthesis does not appear to be the basis of the lethality seen on depletion of Nop56p, additional functions of the snoRNP proteins in other aspects of RNA metabolism appear possible. The cap binding complex is required for splicing commitment complex formation, and pre-mRNA splicing is therefore a possible target.
A homologue of Nop1p and a single homologue of Nop56p-Nop58p are present in Archaea, where they may be associated with methylation guide RNAs (24). We propose that Nop56p was derived from an ancestral Nop58p-like protein by gene duplication, but the proteins have clearly undergone substantial divergence in function.
ACKNOWLEDGMENTS
We thank J. Aris (University of Florida) and J. Brown (University of Edinburgh) for the anti-Nop1p and anti-Srp14p antibodies, respectively; J. Beggs (University of Edinburgh) for probes against U5 and U6 snRNAs; C. Allmang (University of Edinburgh) for the rrp6-Δ strain; E. Bertrand (CNRS, Montpellier) for critical reading of the manuscript; and E. Hurt and T. Gautier (University of Heidelberg) and A. Fatica, Carlo Presuti, and I. Bozzoni (University of Roma) for communicating results prior to publication.
D.L.J.L. was the recipient of a long-term fellowship from the European Commission (TMR). This work was supported by the Wellcome Trust.
REFERENCES
- 1.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachant J B, Elledge S J. Mitotic treasures in the nucleolus. Nature. 1999;398:757–758. doi: 10.1038/19641. [DOI] [PubMed] [Google Scholar]
- 4.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 6.Briggs M W, Burkard K T, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 7.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNA metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 10.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A, Galardi S, Altieri F, Bozzoni I. Fibrillarin binds directly and specifically to U16 box C/D snoRNA. RNA. 2000;6:88–95. doi: 10.1017/s1355838200991623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj I W. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 14.Garcia S N, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- 15.Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G M, Jarmolowski A, Struck J C, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes J M, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 19.Kiss-Laszlo Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3471. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 24.Lafontaine D L J, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 25.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H D, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 30.Narayanan A, Speckmann W, Terns R, Terns M P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicoloso M, Qu L H, Michot B, Bachellerie J P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 32.Niewmierzycka A, Clarke S. S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 33.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu L H, Henras A, Lu Y J, Zhou H, Zhou W X, Zhu Y Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samarsky D A, Fournier M J. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:161–164. doi: 10.1093/nar/27.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 39.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 41.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 42.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 44.Tycowski K T, Shu M D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 45.Tycowski K T, You Z H, Graham P J, Steitz J A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 46.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver P L, Sun C, Chang T H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]