Abstract
We have investigated how the upstream sequence element (USE) of the lamin B2 poly(A) signal mediates efficient 3′-end formation. In vitro analysis demonstrates that this USE increases both the efficiency of 3′-end cleavage and the processivity of poly(A) addition. Cross-linking using selectively labeled synthetic RNAs confirms that cleavage stimulation factor interacts with the sequences downstream of the cleavage site, while electrophoresis mobility shift assays demonstrate that the USE directly stabilizes the binding of the cleavage and polyadenylation specificity factor to the poly(A) signal. Thus in common with other poly(A) signals, the lamin B2 USE directly enhances the binding of basal poly(A) factors. In addition, a novel 55-kDa protein binds to the USE and the core poly(A) signal and appears to inhibit cleavage. The binding activity of this factor appears to change during the cell cycle, being greatest in S phase, when the lamin B2 gene is transcribed.
The 3′ ends of mRNAs transcribed by RNA polymerase II are generated by cleavage of the nascent transcript. Predominantly, this cleavage event occurs at a polyadenylation [poly(A)] site and is followed by the template-independent addition of a ∼250-nucleotide (nt) poly(A) tail (for a recent review, see reference 56). The poly(A) tail appears to be required for efficient export to the cytoplasm (25), efficient translation, and mRNA stability (reviewed in reference 48). The minimal mammalian poly(A) signal comprises a highly conserved AAUAAA element (44, 59) 8 to 31 nt upstream of the site of transcript cleavage (9) and a poorly conserved GU- or U-rich element downstream. These downstream sequence elements (DSEs) were first shown to be important for processing of the rabbit β globin (20) and adenovirus E2A (33) poly(A) signals and are now known to be a general requirement for both efficient and accurate 3′-end formation. Cleavage usually occurs after an A residue, with a preference for a CA dinucleotide (50).
Sequences upstream of the AAUAAA element are also important for the efficiency of certain poly(A) signals. Early examples of these upstream sequence elements (USEs) were found in the poly(A) signals of viral genes, such as the human immunodeficiency virus type 1 (HIV-1) (10, 22), simian virus 40 (SV40) late (8), and adenovirus L1 (15), L3 (42, 43), and L4 (51) poly(A) signals. Recently we have also identified USEs in the poly(A) signals of the human complement C2 (36, 37) and lamin B2 (7) genes.
Despite the apparent simplicity of the 3′-end formation reactions, a considerable number of proteins are required for cleavage and poly(A) addition (reviewed in reference 14), perhaps reflecting the importance of this process in the regulation of gene expression (45). The cleavage and polyadenylation specificity factor (CPSF) is a tetrameric factor comprising subunits of 160, 100, 73, and 30 kDa (5, 27) that is required for both steps of 3′-end formation. CPSF binds specifically to the AAUAAA element (3), via the 160-kDa subunit (27). The trimeric cleavage stimulation factor (CstF, comprising 77-, 64-, and 50-kDa subunits [53]) is normally required only for efficient cleavage and binds to the DSE (21, 31) via its 64-kDa subunit. CPSF and CstF bind to RNA cooperatively (21, 38), with the 77-kDa subunit of CstF contacting the 160-kDa component of CPSF (39). Two additional factors, CFIm and CFIIm, are also required only for the cleavage reaction (52); one or both of these are believed to be the endonucleases responsible for cleaving the RNA. CFIIm has yet to be fully characterized, while four CFIm subunits (of 72, 68, 59, and 25 kDa) have been purified (46, 47). Preliminary evidence suggests that CFIm forms three dimers, all sharing the 25-kDa subunit. The final factor required for cleavage is poly(A) polymerase (PAP), which interacts with CPSF (also via the 160-kDa subunit) and stabilizes its binding to RNA (6, 39). The involvement of PAP in the cleavage reaction effects the tight coupling of cleavage and polyadenylation in vivo.
Addition of the poly(A) tail proceeds in a biphasic reaction (49, 54): initial addition of adenylate residues is slow and distributive, becoming rapid and processive when a critical length of approximately 10 residues is reached. The 49-kDa poly(A) binding protein II (PABII) binds to the growing poly(A) tail and increases the stability and processivity of the PAP-CPSF complex (6). Addition of poly(A) to precursors with tails of around 200 nt is very slow (49), implying both a return to distributive addition and the existence of a mechanism to regulate the length of the added tail (6, 55). It is not yet clear how this length control manifests, although the conformation of the poly(A) tail–PABII complex may be important (28).
Surprisingly, it appears that different USEs influence 3′-end formation in different ways. The simplest mechanism is that the USE mediates additional interactions with the basal poly(A) factors, as is seen with the HIV-1 (23) and equine infectious anemia virus (24) USEs, which enhance the binding of CPSF. Alternatively, a trans-acting factor bound to the USE may recruit the basal poly(A) factors. For example, the USE of the SV40 late poly(A) signal has been found to interact with the U1A protein of U1 snRNP, which may contact CPSF and so stabilize its binding (30). Finally, USEs can act by a combination of these mechanisms. Previously we have shown that the binding of the polypyrimidine tract binding protein (PTB) to the USE of the human complement C2 gene activates cleavage, while interaction of CstF with the USE is also required for the efficiency of both cleavage and polyadenylation (37). We have now turned our attention to the USE of the human lamin B2 poly(A) signal (7). This USE (which overlaps an origin of replication [see Fig. 1A]) is required for efficient cleavage and poly(A) addition. Several proteins cross-link to this poly(A) signal in nuclear extract, including CstF, hnRNP C, and an unidentified protein of ∼55 kDa. Interestingly, the 55-kDa protein inhibits cleavage in vitro, and its binding activity appears to be regulated by the cell cycle. Finally, we demonstrate that recruitment of CPSF is the main function of the lamin B2 USE and the cause of enhanced 3′-end formation.
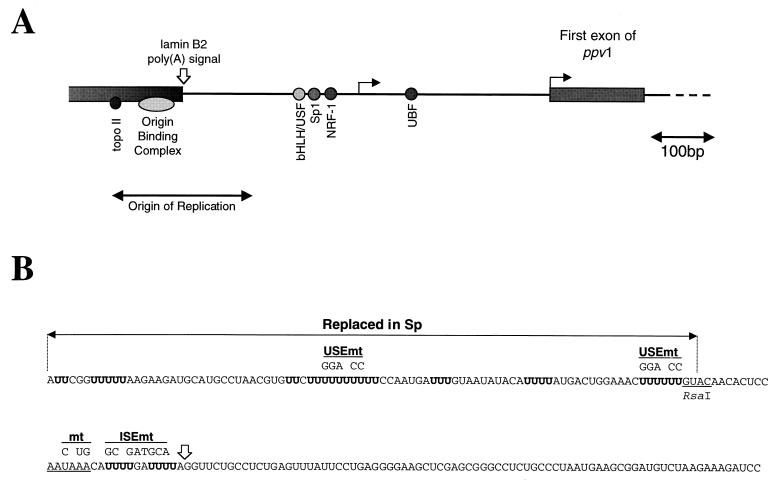
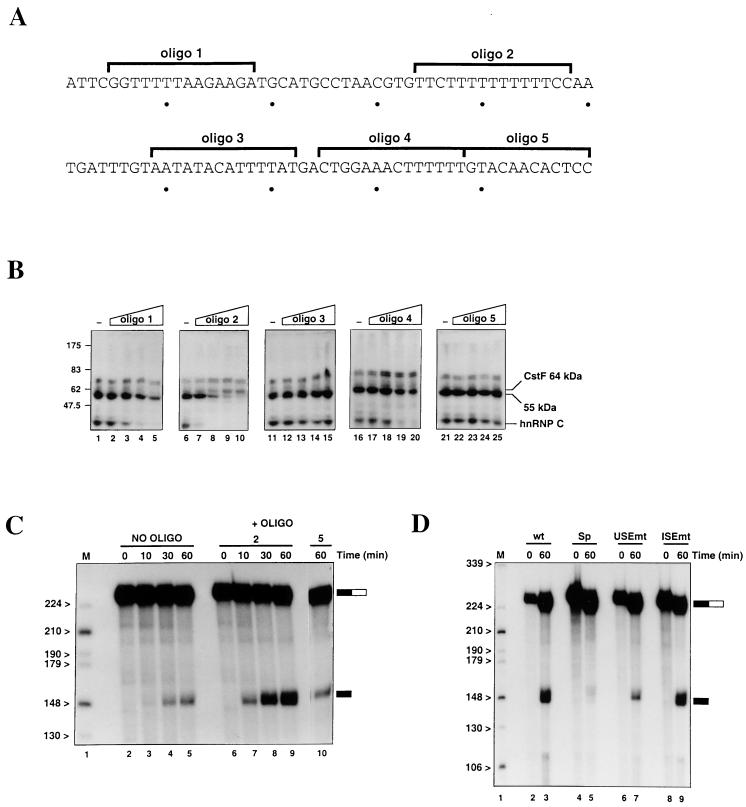
FIG. 1.
Location and sequence of the lamin B2 poly(A) signal. (A) Schematic representation of the short intergenic region separating the 3′ UTR of the lamin B2 gene from the start of the downstream gene (ppv1). The open arrow indicates the lamin B2 poly(A) site, and the two filled arrows denote the two clusters of start sites mapped for ppv1 (7). The positions of transcription factor binding sites, and the location of the origin overlapping with the 3′ UTR of the lamin B2 gene, are also shown (4, 17). (B) Sequence of the wild-type 199-bp lamin B2 poly(A) signal fragment, showing the AAUAAA (underlined) and the U tracts in the USE (boldfaced). The mtRNA contains three point mutations that inactivate the AAUAAA, the USEmt RNA contains point mutations in two U tracts, and the ISEmt RNA contains changes in the sequence between the AAUAAA and the cleavage site. The other RNAs are as follows. Sp has the first 90 nt of the USE replaced by spacer sequence; USE contains the 100 nt of the USE; and the three precleaved RNAs (pre-wt, pre-mt, and pre-Sp) are identical to the full-length wt, mt, and Sp RNAs, respectively, except that they end at the normal cleavage site.
MATERIALS AND METHODS
Plasmid constructions.
All lamin B2 constructs used were based on pGEM4 (Promega), with poly(A) signal fragments inserted at the HincII site. The wild-type (wt) lamin B2 RNA is transcribed from plasmid pG4Lwt, which carries a 199-bp poly(A) signal fragment, and PCR was used to introduce the various changes to this sequence (Fig. 1B). Templates were generated using EcoRI, except for the 5′ ligation fragment template, which was linearized upstream of the AATAAA with RsaI, and those for the precleaved RNAs [linearized at the poly(A) addition site using a FokI site engineered downstream]. The template for the Sp RNA contains a 90-bp fragment of lacZ sequence inserted at the HindIII site upstream of a 109-bp RsaI-BstYI poly(A) signal fragment. pG4USE+ contains a 100-bp PCR fragment containing the sequence from upstream of the AAUAAA.
In vitro transcription.
RNAs were transcribed and purified as previously described (37), except for the 3′ fragments used for RNA ligations. These were primed with 500 μM ApC (Sigma) (phosphorylated using T4 polynucleotide kinase) in the presence of a reduced concentration of ATP (50 μM). Because AC corresponds to the first two incorporated nucleotides, this priming does not introduce an additional base at the 5′ end of the RNA.
In vitro cleavage and poly(A) addition reactions, UV cross-linking, and immunoprecipitations were all performed as described previously (37), with the following alterations: samples were UV irradiated on ice in a Stratalinker 1800, and protein G (rather than protein A)-Sepharose was used in the immunoprecipitations. Twenty microliters of polyclonal (rabbit) anti-PTB serum, 100 μl of anti-CstF/64kDa hybridoma medium (53), and 5 μl of anti-hnRNP C ascites fluid (12) were used.
RNA electrophoresis mobility shifts.
Reaction mixtures (25 μl) contained 83 mM KCl, 0.5 μg of tRNA, 1% (wt/vol) polyvinyl alcohol, 0.016% (vol/vol) Igepal CA-630, 16.5 mM HEPES (pH 7.9), 8.3% (vol/vol) glycerol, 0.17 mM EDTA, 0.08 mM dithiothreitol, 45 ng of purified CPSF, and 150 fmol of synthetic precleaved RNA. Reaction mixtures were incubated at 30°C for 10 min and a 5-μl aliquot was removed for the zero time point. Forty picomoles of unlabeled precleaved wt (pre-wt) RNA was added to the remainder of the reaction mixture, and 5.2-μl aliquots were removed at various times. All aliquots were placed on ice and treated with 10 μg of tRNA. For the oligonucleotide competitions, a 30-μl reaction mixture was assembled and 5-μl aliquots were added to 100 pmol of the oligonucleotide. These reaction mixtures were incubated at 30°C for 10 min and then treated as described above. Samples were run on 3% native polyacrylamide gels (acrylamide-bis-acrylamide, 37.5:1) at 200 V for 2 h at 4°C in 0.25× Tris-borate-EDTA.
Oligonucleotide-mediated ligation of RNA.
Ten-microliter annealing reaction mixtures in 1× ligase buffer containing the DNA bridging oligonucleotide (AC CTAAAATCAAAATGTTTATTGGAGTGTTGTACAAAAAAGTTTCCAG TCATAAAATGTATATT) and the two RNA fragments (final concentration for each, 2 mM) were held at 70°C for 5 min and then allowed to cool to 30°C over the course of an hour. The reaction volume was increased to 20 μl with the addition of 4,000 cohesive-end units of T4 DNA ligase (New England Biolabs), 50 U of human placental RNase inhibitor (Amersham), and ATP (to a concentration of 2 mM), and incubation at 30°C continued for 16 h. The ligated RNA was gel purified following extraction against acidic phenol-chloroform and precipitation with 2.5 volumes of ethanol (in the presence of ammonium acetate).
Cell culture and nuclear run-on analysis.
Growth and synchronization of NT2/D1 cells and nuclear run-on analysis were all carried out as previously described (7). Adherent HeLa cells were synchronized as follows. Thymidine was added at a concentration of 2.5 mM for 24 h in order to accumulate cells at the G1/S-phase border. Cells were released by three washes with fresh medium and were allowed to recover for 4 h in the presence of 50 μM deoxycytidine before being exposed to nocodazole (1 μg/ml for 8 to 16 h). Mitotic cells were recovered by “shake-off” and either used for extract preparation or allowed to recover for either 3 h (and then harvested as G1 cells) or 6 h (and then arrested with aphidicolin for 2 to 6 h). Cells were released as before and allowed to recover for either 2 h (and then harvested as S-phase cells) or 12 h (and then harvested as G2 cells). Whole-cell extracts were prepared by the method of Naka and Brownlee (40).
RESULTS
The sequences of the lamin B2 poly(A) signal, and the various synthetic RNAs used to characterize the lamin B2 USE are shown in Fig. 1B.
The lamin B2 USE is required for efficient in vitro cleavage.
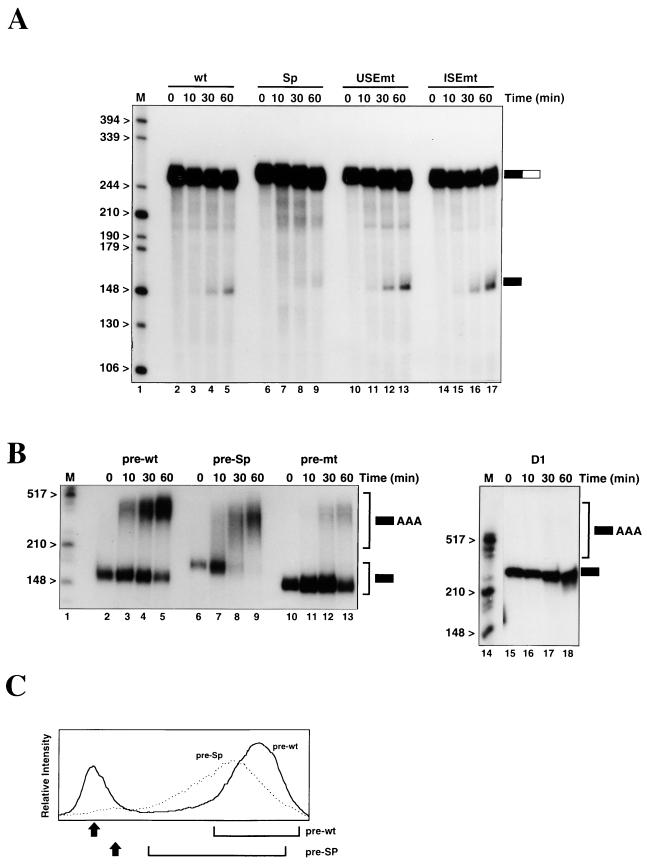
Cleavage assays were performed in nuclear extract in the presence of 3′ dATP to block poly(A) addition (Fig. 2A). Approximately 8% of the wt RNA (Fig. 2A, lanes 2 to 5) is cleaved in 60 min. As expected, mutation of the AAUAAA sequence prevents appearance of the cleaved fragment, confirming that the products observed are genuine (data not shown). Replacement of all but the last 10 nt of the upstream sequence with unrelated sequence (Sp; Fig. 2A, lanes 6 to 9) results in only 1% of the RNA being cleaved. Thus, the upstream sequence is required for efficient cleavage at the lamin B2 poly(A) signal in vitro. Previously, we characterized a number of mutations in the U tracts of the USE which resulted in reduced use of the lamin B2 poly(A) signal in vivo (7). To investigate these mutations in vitro, we used the USEmt RNA (Fig. 2A, lanes 10 to 13), which contains 10 point mutations. Surprisingly, the effects of these changes in vitro were small and somewhat variable. In the experiment for which results are shown in Fig. 2A, cleavage of the USEmt and wt RNAs was equivalent, although in other batches of extract a twofold reduction in cleavage of USEmt has been observed. The reason for this variable effect will be discussed below.
FIG. 2.
The lamin B2 USE is required for efficient cleavage and poly(A) addition in vitro. (A) Cleavage assays were performed in nuclear extract using wt, Sp, USEmt, or ISEmt RNAs. The positions of the input RNAs (solid- and -open box) and the 5′ cleavage products (solid box) are indicated to the right of the gel. Note that for the Sp RNA (lanes 6 to 9), both the input and the 5′ fragment are slightly longer than those for the other RNAs. (B) Poly(A) addition reactions in nuclear extract using the pre-wt, pre-Sp, pre-mt, and D1 RNAs. [The D1 RNA has no poly(A) signal sequences.] The positions of the input and poly(A)+ RNAs are indicated. (C) Poly(A) addition is qualitatively different for the pre-wt and pre-Sp substrates. The graphs represent quantitation of lanes 5 and 9 of panel B—the 60-min time points. Arrows indicate the positions of the input RNA, and brackets show the extent of the heterogeneous poly(A)+ band.
U-rich sequences between the AAUAAA and the cleavage site enhance the processing of the adenovirus L3 (43) and complement C2 (36) poly(A) signals. For the lamin B2 poly(A) signal, however, mutation of the U-rich internal sequence (ISEmt) resulted in a threefold increase in the efficiency of cleavage compared to that of the wt RNA (Fig. 2A, lanes 14 to 17). This suggests that the wild-type sequence represents a suboptimal environment for the AAUAAA, a phenomenon previously reported for the adenovirus L1 (43) and HIV-1 (23) poly(A) signals. Using in vivo poly(A) signal competition assays, however, we observed no difference between the wild-type and ISEmt poly(A) signals (unpublished data). The reason for the discrepancy between these in vivo and in vitro results will be addressed below.
The lamin B2 USE is required for efficient polyadenylation.
Incubation of synthetic “precleaved” RNA in nuclear extract in the presence of ATP allows addition of a poly(A) tail, resulting in conversion of the RNA from a discrete band to a slower-migrating heterogeneous population. For the pre-wt RNA, ∼75% of the input material is polyadenylated after 60 min (Fig. 2B, lanes 2 to 5), with the majority of the RNA receiving a tail of ∼200 nt. Replacement of most of the USE by the spacer sequence (pre-Sp; Fig. 2B, lanes 6 to 9) does not reduce the proportion of the input RNA polyadenylated (∼80% of input RNA is modified in lane 9). In contrast to pre-wt RNA, however, the poly(A)+ pre-Sp RNA runs as a much broader band, reflecting a larger variation in the length of the poly(A) tail added (Fig. 2C). A similar effect is seen with a pre-USEmt mutation (data not shown), implying that this is a specific consequence of altering the USE, rather than an artifact of the spacer used. Thus, the USE is not required for the overall level of polyadenylation but is required for the production of full-length tails.
Polyadenylation of the pre-mt RNA (in which the AAUAAA has been mutated) (Fig. 2B, lanes 10 to 13) is significantly less efficient than that seen for the wt RNA, although it is greater than the level of nonspecific polyadenylation seen with the extract used. Thus, an RNA lacking any poly(A) signal sequences (D1 RNA) gives no detectable poly(A) addition under identical conditions (Fig. 2B, lanes 15 to 18). This suggests that the USE of the lamin B2 poly(A) signal is able to partially compensate for the AAUAAA sequence during the polyadenylation reaction, although it is absolutely required for cleavage.
A number of proteins can be cross-linked to the lamin B2 poly(A) signal.
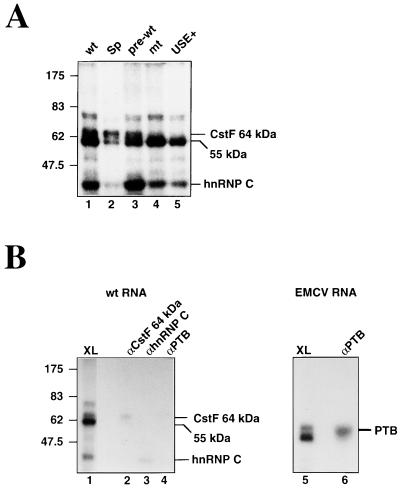
We used UV cross-linking (34) to investigate the proteins binding to the lamin B2 RNA. For the wt RNA, four main cross-linked species, of approximately 75, 64, 55, and 40 kDa, are produced in nuclear extract (Fig. 3A, lane 1). The 40-kDa species represents hnRNP C1 or C2 (both of which preferentially cross-link to U-rich RNA [18]), as shown by the fact that this band can be immunoprecipitated from the cross-linking reactions using the anti-hnRNP C 4F4 antibody (Fig. 3B, lane 3). The 64-kDa protein can be precipitated from the nuclear extract (Fig. 3B, lane 2) using antibodies specific for the RNA binding subunit of CstF. Consistent with this, mutation of the hexanucleotide (Fig. 3A, lane 4) virtually abolishes the 64-kDa cross-linking signal. The undiminished cross-linking of the other proteins to this RNA suggests that they are able to bind to RNA in the absence of a functional poly(A) signal. Replacement of the USE with the spacer sequence (Fig. 3A, lane 2) results in a relative decrease in the intensity of the 55-kDa protein, suggesting that this protein binds to the USE. Given the extremely poor cleavage of this RNA in nuclear extract, the undiminished cross-linking of the 64-kDa subunit of CstF to this RNA presumably reflects the nonquantitative nature of this assay. Interestingly, the cross-linking profile of the pre-wt RNA (Fig. 3A, lane 3) is very similar to that of the wt RNA. This is somewhat surprising given that the presumed binding site for CstF lies downstream of the cleavage site, a region absent from this RNA. The final RNA tested in these cross-linking reactions (Fig. 3A, lane 5) contains only the 100 nt of the USE. This RNA also gives 55-kDa and hnRNP C signals, confirming that these proteins cross-link to the USE.
FIG. 3.
UV cross-linking of proteins to the lamin B2 poly(A) signal. (A) Cross-linking of wt, Sp, pre-wt, mt, and USE+. The migration of prestained molecular weight markers is indicated to the left. (B) Immunoprecipitation of cross-linked proteins using antibodies specific for the 64-kDa RNA-binding subunit of CstF (lane 2), hnRNP C (lane 3), and PTB (lane 4). Lane 1 shows half of the input material used in each case. Lane 6 shows precipitation of PTB cross-linked to the D1 RNA, with half of the input material shown in lane 5.
We have previously reported that PTB, an abundant nuclear protein of approximately 55 kDa, cross-links to the USE of the human complement C2 gene (37). The 55-kDa protein that cross-links to the lamin B2 poly(A) signal protein is not precipitated with polyclonal serum raised against PTB (Fig. 3B, lane 4), while this antiserum efficiently precipitates PTB cross-linked to domain 1 of the encephalomyocarditis virus internal ribosome entry site (26) (Fig. 3B, lanes 5 and 6). We conclude that the 55-kDa protein observed here is not PTB.
Direct mapping of CstF binding.
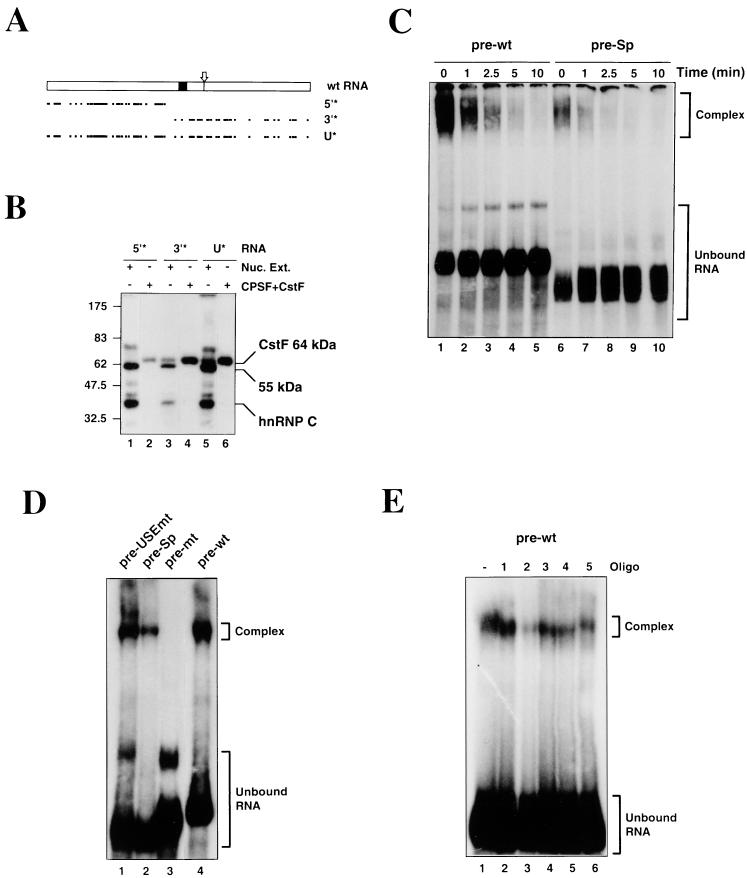
We have observed that the 64-kDa subunit of CstF cross-links efficiently to pre-wt RNA, suggesting that CstF interacts with the USE of the lamin B2 poly(A) signal. To map the binding site of CstF, we used selectively labeled synthetic RNAs produced by the oligonucleotide-mediated ligation of RNA segments with T4 DNA ligase (35). Figure 4A shows the extent of labeling in the two selectively labeled RNAs (5′* and 3′*), as well as in uniformly labeled wt RNA (U*). The cross-linking of identical amounts of these RNAs in both nuclear extract and highly purified CstF and CPSF is shown in Fig. 4B. As before, strong signals are seen for the 64-kDa subunit of CstF, hnRNP C, and the 55-kDa protein when the full-length labeled RNA is used in nuclear extract (Fig. 4B, lane 5). In contrast, when label is restricted to the USE (Fig. 4B, lane 1), cross-linking of the 64-kDa subunit of CstF is no longer seen, although both the 55-kDa protein and hnRNP C give clear signals. Cross-linking of the CstF 64-kDa subunit is still observed when label is restricted to the “core” poly(A) signal (Fig. 4B, lane 3), although the hnRNP C and 55-kDa signals (both resulting from the labeled U-rich internal sequence) are greatly reduced. The results obtained when the cross-linking is performed in purified CstF and CPSF are consistent with this: both the uniformly labeled (Fig. 4B, lane 6) and 3′-labeled (lane 4) RNAs give very strong CstF 64-kDa cross-linking signals, while the signal seen with the 5′ label is greatly reduced (lane 2). These results strongly suggest that the predominant CstF binding site lies downstream of the AAUAAA.
FIG. 4.
Interaction of CstF and CPSF with the lamin B2 poly(A) signal. (A) Schematic of the distribution of label in the 5′* and 3′* selectively labeled RNAs and the uniformly labeled (U*) wt RNA. Solid box, AAUAAA element; arrow, cleavage site; dots, individual labeled U residues; dashes, labeled U tracts. (B) Cross-linking of the uniformly and selectively labeled RNAs in nuclear extract (Nuc. Ext.; lanes 1, 3, and 5) or purified CPSF and CstF (lanes 2, 4, and 6). (C) Stability of CPSF interacting with the pre-wt (lanes 1 to 5) and pre-Sp (lanes 6 to 10) RNAs. The CPSF-RNA complexes were separated from the free RNA following the addition of an excess of unlabeled pre-wt RNA. (D) Electrophoresis mobility shift assay using purified CPSF and pre-USEmt (lane 1), pre-Sp (lane 2), pre-mt (lane 3), and pre-wt (lane 4) RNAs. (E) Effects of DNA oligonucleotide competitors (Oligo) on the binding of CPSF to the pre-wt RNA.
The stability of the CPSF-RNA complex is enhanced by the USE.
Since the above data show that the USE does not bind CstF, we investigated whether the USE interacts with CPSF. As we have never observed cross-linking of CPSF to the lamin B2 poly(A) signal, we used electrophoretic mobility shift assays to examine the stability of CPSF binding to the pre-wt and pre-Sp RNAs. Purified CPSF was incubated with the RNAs at 30°C for 10 min, and then aliquots were withdrawn at time zero. Excess cold pre-wt RNA was added to the remainder of the reaction mixture, incubation was continued, and aliquots were withdrawn at the times indicated. RNA-protein complexes were resolved on 3% native polyacrylamide gels. It can be seen that for the zero time points (Fig. 4C, lanes 1 and 6) significantly more complex is formed with pre-wt than with pre-Sp RNA. Following the addition of the unlabeled competitor RNA, both the pre-wt and pre-Sp complexes dissociate rapidly, confirming that these are genuine complexes formed with CPSF. Thus, we conclude that the USE directly stabilizes the binding of CPSF to the lamin B2 poly(A) signal.
It should be noted that although pre-Sp differs substantially from pre-wt RNA, replacement of the USE (rather than specific mutation) is required to overcome the redundancy seen when mutations are made to this sequence (7). Thus, the pre-USEmt RNA (the precleaved version of the USEmt RNA) and the pre-wt RNA give similar amounts of complex (Fig. 4D; compare lanes 1 and 4). In contrast, no stable complex is formed between CPSF and the pre-mt RNA containing the mutation to the AAUAAA (Fig. 4D, lane 3), despite some specific polyadenylation of this RNA in nuclear extract (see Fig. 2).
Cross-linking in the presence of competitor oligonucleotides.
We previously found that sense DNA oligonucleotides can be used as specific competitors in UV cross-linking reactions, to determine protein binding sites (37). We now used such oligonucleotides to investigate the binding of the various proteins observed in the cross-linkings. Figure 5A shows the sequence of the lamin B2 USE and the positions of the five sense oligonucleotides used, while the effects of increasing amounts of each oligonucleotide (0, 1, 10, 50, and 100 pmol) on the cross-linking profile of the wt RNA are shown in Fig. 5B. It should be noted that the changes in cross-linking profiles observed in these experiments do not result from RNase H-mediated cleavage; RNA recovered from these reactions prior to cross-linking is no more degraded than RNA incubated in extract alone (data not shown).
FIG. 5.
A negative factor binds to the lamin B2 poly(A) signal and inhibits cleavage in nuclear extract. (A) Sequence of the USE showing the locations of the five DNA oligonucleotides used as competitors in the cross-linking and cleavage reactions. (B) Cross-linking of wt RNA in nuclear extract in the presence of increasing amounts (0, 1, 10, 50, and 100 pmol) of the different DNA oligonucleotides. (C) Effects of the oligonucleotide competitors on cleavage of wt RNA. Time courses of cleavage in the absence (lanes 2 to 5) or presence (lanes 6 to 9) of 100 pmol of oligonucleotide 2 are shown, as is cleavage at 60 min in the presence of oligonucleotide 5 (lane 10). (D) Cleavage of the wt (lanes 2 and 3), Sp (lanes 4 and 5), USEmt (lanes 6 and 7), and ISEmt (lanes 8 and 9) RNAs in nuclear extract, all in the presence of 100 pmol of oligonucleotide 2.
Oligonucleotides 3 (AATATACATTTTAT) and 5 (GTACAACACTCC), which both lie in the 3′ half of the USE, appear to have no significant effects on protein cross-linking at any concentration. Oligonucleotides 1 (GGTTTTTAAGAAGA) and 4 (CTGGAAACTTTTTT) are similar to each other in their effects, significantly reducing the amount of hnRNP C cross-linking (beginning at 10 and 50 pmol, respectively) and slightly reducing the cross-linking of the 55-kDa protein at the highest concentrations used. The most striking effects, however, are seen with oligonucleotide 2 (TTCTTTTTTTTTTCC). One picomole of this oligonucleotide virtually abolishes the cross-linking of hnRNP C, while in the presence of 50 to 100 pmol, cross-linking of the 55-kDa protein also disappears. This confirms that hnRNP C and the 55-kDa protein are binding to the U tracts in the poly(A) signal, either in the USE or in the sequence between the hexanucleotide and the cleavage site. Interestingly, there appears to be an increase in the cross-linking of the 64-kDa subunit of CstF to the RNA as the binding of the 55-kDa protein and hnRNP C are reduced. This suggests that the 55-kDa protein and/or hnRNP C bind to the USE at the expense of CPSF, thereby reducing the cross-linking of CstF. We have also examined the effects of these competitor oligonucleotides on the binding of CPSF. As shown in Fig. 4E, only oligonucleotide 2 (lane 3) had any significant effect on the amount of complex formed (although it should be noted that this lane is slightly underloaded). This is consistent with CPSF interacting with the U-rich sequences in the USE.
Inhibition of the lamin B2 poly(A) signal.
The competition experiments described above suggest that the binding of the 55-kDa protein and/or hnRNP C to the USE prevents the binding of the 3′-end formation machinery. Therefore, we have investigated the effects of the oligonucleotide 2 competitor on cleavage of the lamin B2 poly(A) signal in nuclear extract. As shown in Fig. 5C, enhanced cleavage of the wt RNA is seen after 60 min when the oligonucleotide is present (lanes 6 to 9) compared with the standard processing conditions (lanes 2 to 5). Thus, one or more of the proteins removed from the RNA by this competitor reduces the efficiency of cleavage of this poly(A) signal in nuclear extract. Importantly, the use of nonspecific DNA oligonucleotides does not affect the cleavage of the wt RNA, confirming that this is a specific effect of the competitor used (for example, Fig. 5C, lane 10, shows cleavage after 60 min in the presence of oligonucleotide 5, which had little effect on the cross-linking of proteins).
The presence of competitor oligonucleotide 2 also results in equal cleavage (∼30% of input) of the wt (Fig. 5D, lanes 2 and 3) and ISEmt (lanes 8 and 9) RNAs. Presumably, under equilibrium conditions, mutation of the U-rich internal sequence disrupts some of the binding of the inhibitory factor, relieving the repressive effect and allowing more-efficient cleavage of this RNA compared to that of the wt. In addition, the USEmt RNA (Fig. 5D, lanes 6 and 7) is consistently cleaved threefold less efficiently than the wt RNA in the presence of the competitor. In the absence of the competitor, mutation of the U-rich USE will have two effects: (i) removal of some of the binding sites for the inhibitory factor, which will increase the efficiency of processing, and (ii) disruption of the USE, which will decrease the efficiency of processing. Differences in the level of the 55-kDa protein between independent preparations of nuclear extract presumably account for the variable effects of the USEmt mutation in the absence of the competitor. Finally, as expected, processing of the Sp RNA (Fig. 5D, lanes 4 and 5) is still very inefficient, with only 3% of the input RNA cleaved in the assay shown.
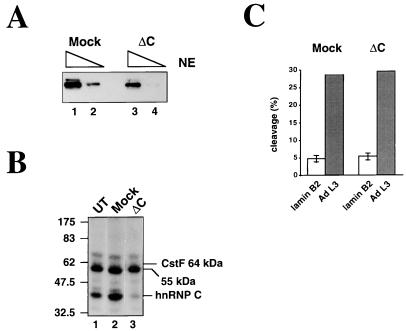
Using monoclonal antibody 4F4, we have managed to partially deplete nuclear extract of hnRNP C (Fig. 6A). Although not a complete depletion, we note that this decrease in concentration is sufficient to significantly reduce the amount of hnRNP C cross-linking to the lamin B2 poly(A) signal in the depleted (ΔC) extract (Fig. 6B, lane 3). Furthermore, we observe no difference between the depleted and mock-depleted extracts when cleavage of either the lamin B2 wt RNA or an adenovirus L3 poly(A) signal RNA is assayed (Fig. 6C). (Note that the depletion protocol has resulted in a slight decrease in the efficiency of lamin B2 cleavage in both the mock and ΔC extracts compared with that in Fig. 2A.) Additionally, oligonucleotide competitors 1 and 4 (which both significantly reduced the cross-linking only of hnRNP C and not of the 55-kDa protein [Fig. 5B]) activate cleavage of the wt lamin B2 RNA to a lesser degree than oligonucleotide 2 (data not shown). Taken together, these results suggest that inhibition of lamin B2 processing is not caused by the binding of hnRNP C. This is consistent with previous studies that failed to find a role for hnRNP C in 3′-end formation (13). Although we cannot rule out the possibility that inhibition is due to a protein not detected by cross-linking, it seems most likely that the inhibitory effect is due to the binding of the 55-kDa protein.
FIG. 6.
hnRNP C does not inhibit the lamin B2 poly(A) signal. (A) Western blot of 1-μl (lanes 1 and 3) and 0.2-μl (lanes 2 and 4) equivalents of mock- and hnRNP C-depleted (ΔC) nuclear extracts (NE). (B) Cross-linking of the wt RNA in untreated (UT; lane 1), mock-depleted (lane 2), or hnRNP C-depleted (ΔC; lane 3) extracts. (C) Quantitation of cleavage of the wt lamin B2 RNA and an adenovirus L3 poly(A) signal RNA in the mock-depleted and ΔC extracts. Error bars indicate standard deviations for three assays.
A role for the cell cycle in lamin B2 3′-end formation?
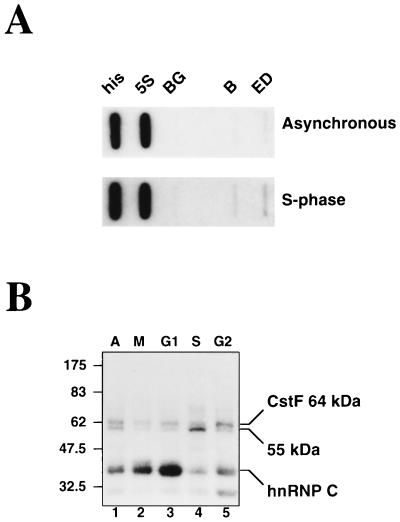
It has previously been reported that the lamin B2 gene is expressed only in S phase (4). Transcription of the lamin B2 gene as assessed by nuclear run-on analysis is approximately 2.5-fold higher (relative to the 5S rRNA control probe) for cells in S phase than for an asynchronous population (Fig. 7A). We were interested, therefore, to determine if the inhibitory effect of the 55-kDa protein could play a role in the regulation of the expression of the lamin B2 gene. To this end, we prepared whole-cell extracts from adherent HeLa cells in different stages of the cell cycle and used cross-linking to examine the amount of 55-kDa protein present. Cells in M, G1, S, and G2 were obtained by sequential treatment with thymidine, nocodazole, and aphidicolin, as described in Materials and Methods. Although complex, this protocol was used to minimize any differences that may be caused by the exposure to the different inhibitors, rather than as a result of progression through the cell cycle.
FIG. 7.
Cell cycle-dependent changes in the proteins cross-linking to the lamin B2 poly(A) signal. (A) Nuclear run-on analysis of the lamin B2 gene using asynchronous (top) and S-phase (bottom) cells. The probes used detect RNA from histone H4 (his), 5S rRNA (5S), and the lamin B2 gene (B and ED). Probe BG controls for background hybridization. (B) Cross-linking of the wt RNA in whole-cell extracts prepared from asynchronous (A; lane 1) or synchronized (lanes 2 to 5) cells.
As with the nuclear extracts, three main cross-linking signals are seen with the wt RNA in the unsynchronized extract (Fig. 7B, lane 1): the 64-kDa subunit of CstF, the 55-kDa protein, and hnRNP C (although the intensities of these cross-linking signals are much more equal than those seen previously in nuclear extract). Extracts from M phase give an altered cross-linking profile, with increased levels of hnRNP C and the appearance of additional bands representing the M-phase-specific phosphorylated forms of hnRNP C (the so-called CS proteins [41]). G1 extracts also appear to contain further increased levels of hnRNP C and reduced levels of the 55-kDa protein. Interestingly, in S phase, cross-linking of the 55-kDa protein is greatly increased, while that of hnRNP C is greatly reduced. By G2, the level of 55-kDa protein cross-linking has decreased, and the relative signals for both CstF and hnRNP C have increased. Thus, it would appear that the amount and/or binding activity of the 55-kDa protein is at a maximum in S phase, when the lamin B2 gene is being transcribed. We note that the G2 extract also shows increased cross-linking of a protein of approximately 32 kDa that is also faintly detectable in the other whole-cell extracts. Cross-linking of this species is occasionally seen in nuclear extract (data not shown) but varies to the extent that it is not even consistently seen in different batches of the same extract.
DISCUSSION
We have investigated the mechanism of activation of mRNA 3′-end formation by the USE of the lamin B2 poly(A) signal. We find that the USE enhances cleavage and is required for the processivity of poly(A) polymerase. The results presented in Fig. 4C show that the USE stabilizes the binding of CPSF, which accounts for the observed effects on cleavage and poly(A) addition. The reduced poly(A) tail lengths observed when the USE is replaced is likely to result from dissociation of a less-stable CPSF-poly(A) polymerase complex from the pre-Sp RNA prior to completion of a full-length poly(A) tail. The fact that we do not observe a quantitative effect of the USE on the overall amount of polyadenylated RNA produced (Fig. 2B) may reflect the different kinetics of the two uncoupled steps in vitro. Specifically, since poly(A) addition requires significantly fewer factors than cleavage, it may be expected that the lag between initiation of complex assembly and processing will be longer for cleavage. Therefore, instability of CPSF may have a greater effect on cleavage. The role of the USE in stabilizing CPSF binding is also consistent with the relatively high polyadenylation efficiency of the pre-mt RNA (which lacks the AAUAAA) compared with the control D1 RNA [which lacks any recognizable poly(A) signal elements]. Thus, it would appear that the USE can at least partially compensate for mutation of the hexanucleotide during poly(A) addition, but not during cleavage. This may reflect a potentially shorter lag period with the poly(A) addition complex, or perhaps stricter conformational limits imposed by the larger cleavage complex. Given the observed polyadenylation, however, it is somewhat surprising that we are unable to detect any stable binding of CPSF to the pre-mt RNA in the electrophoretic mobility shift assays.
Activation of processing by upstream sequences.
Although all of the USEs reported to date are U rich, they apparently use different mechanisms to enhance 3′-end formation. It is striking, however, that a number of splicing factors have been implicated in USE function. For example, the USE of the SV40 late poly(A) signal has been shown to interact with U1 snRNP by base pairing (57) and through the binding of the U1A protein (29). Additionally, we have previously shown that PTB binds to the USE of the human complement C2 gene and activates cleavage (37). Initially, we predicted that the 55-kDa protein cross-linking to the lamin B2 poly(A) signal was PTB and that the two human USEs would function in the same way. However, the work reported here demonstrates that the complement C2 and lamin B2 USEs employ different mechanisms, presumably reflecting the different architectures of the two poly(A) signals. Thus, while the lamin B2 poly(A) signal possesses both upstream and downstream sequence elements (7), the complement C2 poly(A) signal lacks the conventional DSE (36). The absence of a conventional CstF binding site in the C2 poly(A) signal appears to have resulted in evolution of the USE as a novel CstF binding site, allowing efficient 3′-end formation. Although our initial cross-linking results hinted that CstF might also bind to the lamin B2 USE (see Fig. 3), we have demonstrated, using selectively labeled synthetic RNAs, that CstF cross-links to the DSE of this poly(A) signal. Thus, the cross-linking of CstF to the pre-wt RNA is an artifact, perhaps resulting from the stabilization of CPSF by the USE allowing aberrant binding of CstF to the U-rich sequences immediately downstream of the AAUAAA.
The work reported here is consistent with studies on the USEs of both the HIV-1 (23) and EIAV (24) poly(A) signals, which also act by stabilizing the binding of CPSF. These retroviral USEs are required to allow differential use of the poly(A) signals in the duplicated 5′ and 3′ long terminal repeats (LTRs). As the USE lies upstream of the transcription start site, it will be included in the poly(A) signal only in the 3′ LTR, allowing for efficient utilization of this signal. Inefficient use of the poly(A) signal of the 5′ LTR results from proximity to the promoter (11), absence of the USE (16), and inhibition by the major splice donor site (1). Although no such regulation appears to be in operation for the lamin B2 gene, we note that transcription of this gene is very low. Indeed, the amount of nascent RNA hybridizing to probe ED in Fig. 7A is significantly smaller than that hybridizing to the control probes (histone H4 and 5S rRNA), despite the fact that ED is approximately threefold longer than these probes. Nascent RNA from the lamin B2 gene will, therefore, represent only a tiny fraction of the total nascent RNA in the nucleus. Given that the efficiency of a poly(A) signal correlates with the resulting levels of cytoplasmic mRNA (58), it is likely that the presence of the USE in the lamin B2 poly(A) signal is essential to ensure optimal expression of this gene.
The observation that the unrelated retroviral and lamin B2 USEs use a common mechanism suggests that CPSF recruitment has evolved as a general strategy to enhance 3′-end formation, and it will be of interest to determine if this mechanism is also used by the other known USEs. In addition, it remains to be determined exactly how the CPSF interacts with the RNA upstream of its classical binding site. Consistent with its binding to the U-rich USE, CPSF has previously been purified over a poly(U) column (38). Furthermore, the 30-kDa subunit can be cross-linked to RNA (27) and shows a distinct preference for poly(U) RNA in vitro (2), suggesting that it may be the USE-binding subunit.
Cell cycle changes in RNA binding.
The results in Fig. 7B demonstrate that the activities of both hnRNP C and the 55-kDa protein change during the cell cycle, although the mechanisms driving these changes are unknown. Cell cycle-dependent phosphorylation of hnRNP C has been reported previously (41), but this is apparently distinct from the phosphorylation that has been reported to affect RNA binding activity (32). At the moment we do not know the significance of the altered cross-linking profiles we observed, but it is striking that hnRNP C cross-linking peaks in G1, while the binding of the 55-kDa protein appears to be maximal in S phase. This reciprocal behavior may be driven by competition between the two proteins for the same sequences on the RNA and raises the possibility that the binding of only one may be directly affected, with a “knock-on” effect on the binding of the other. The fact that we do observe efficient cross-linking of both hnRNP C and the 55-kDa protein in both asynchronous whole-cell and nuclear extracts, however, suggests that this is unlikely.
The 55-kDa protein and the inhibition of lamin B2 3′-end formation.
Inhibition of processing caused by the 55-kDa protein presumably results from masking of the poly(A) signal when the RNA is bound by this factor. It should be emphasized that the 55-kDa protein binds not only to the USE but also to the U-rich internal sequences and so would be predicted to interfere with the interaction of CPSF with both the USE and the AAUAAA. At present we do not know the significance of this inhibition. However, it has been reported that the mouse lamin B2 homologue is alternatively polyadenylated (19). Although the complete structure of the human lamin B2 gene has yet to be described, a possible role of the 55-kDa protein could be occluding one poly(A) signal, allowing efficient use of an alternative signal. The apparent cell cycle regulation of the binding activity of the 55-kDa protein provides a second link between this USE and S phase: as discussed in the introduction, this poly(A) signal overlaps with a replication origin, and it is tempting to speculate that the two may in some way be connected. Work is under way to identify the 55-kDa protein and to determine the significance of the cell cycle variation in the binding activity of this factor.
ACKNOWLEDGMENTS
We are grateful to members of the N.J.P. laboratory for many helpful comments and discussions throughout these studies. Thanks go to Shona Murphy for nuclear extract, Jim Manley and Yoshio Takagaki for the CstF 64-kDa antibody and purified processing factors, Gideon Dreyfuss for the hnRNP C antibody, and Chris Smith and Matthew Wollerton for the PTB antibody. We also thank David Pritlove for suggesting the use of dinucleotide primers with T7 RNA polymerase, Dean Jackson for advice on cell synchronization, and Andy Newman for advice on the ligation of RNA.
These studies were supported by a Wellcome Program Grant to N.J.P. (no. 032773/Z/G5).
REFERENCES
- 1.Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 2.Barabino S M, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1706. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell V J, Wickens M, Bienroth S, Keller W, Sproat B S, Lamond A I. Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell. 1991;65:125–133. doi: 10.1016/0092-8674(91)90414-t. [DOI] [PubMed] [Google Scholar]
- 4.Biamonti G, Giacca M, Perini G, Contreas G, Zentilin L, Weighardt H, Guerra M, Della Valle G, Saccone S, Riva S, Falaschi A. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S phase. Mol Cell Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing on messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- 6.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in an unusually short human intergenic region. Nucleic Acids Res. 1997;25:2326–2335. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, MacDonald C C, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherrington J, Ganem D. Regulation of polyadenylation in human immunodeficiency virus (HIV): contributions of promoter proximity and upstream sequences. EMBO J. 1992;11:1513–1524. doi: 10.1002/j.1460-2075.1992.tb05196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherrington J, Russnak R, Ganem D. Upstream sequences and cap proximity in the regulation of polyadenylation in ground squirrel hepatitis virus. J Virol. 1992;66:7589–7596. doi: 10.1128/jvi.66.12.7589-7596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi D Y, Dreyfuss G. Monoclonal antibody characterization of the C-proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou Z-F, Chen F, Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation sites. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 15.DeZazzo J D, Imperiale M J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeZazzo J D, Scott J M, Imperiale M J. Relative roles of signals upstream of AAUAAA and promoter proximity in regulation of human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol. 1992;12:5555–5562. doi: 10.1128/mcb.12.12.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimitrova D S, Giacca M, Demarchi F, Biamonti G, Riva S, Falaschi A. In vivo protein-DNA interactions at a human DNA replication origin. Proc Natl Acad Sci USA. 1996;93:1498–1503. doi: 10.1073/pnas.93.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of messenger-RNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa K, Hotta K. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J. 1993;12:97–106. doi: 10.1002/j.1460-2075.1993.tb05635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil A, Proudfoot N J. A sequence downstream of AAUAAA is required for rabbit β globin mRNA 3′ end formation. Nature. 1984;312:473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 21.Gilmartin G M, Nevins J R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2189. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 22.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-1 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 24.Graveley B R, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Carmichael C C. Role of polyadenylation in nucleoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding-protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 27.Keller W, Bienroth S, Lang K M, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller W. No end yet to messenger RNA 3′ processing. Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 29.Lutz C S, Alwine J C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 30.Lutz C S, Murthy K G K, Schek N, O'Connor J P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayrand S H, Dwen T, Pedersen T. Serine/threonine phosphorylation regulates binding of C hnRNP proteins to mRNA. Proc Natl Acad Sci USA. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDevitt M A, Imperiale M J, Ali H, Nevins J R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984;37:993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 34.Moore C L, Chen J, Whoriskey J. Two proteins cross-linked to RNA containing the adenovirus L3 poly(A) site require the AAUAAA sequence for binding. EMBO J. 1988;7:3159–3169. doi: 10.1002/j.1460-2075.1988.tb03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore M J, Sharp P A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 36.Moreira A, Wollerton M, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates polyadenylation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murthy K G K, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 39.Murthy K G K, Manley J L. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 40.Naka H, Brownlee G G. Transcriptional regulation of the human factor IX promoter by the orphan receptor superfamily factors, HNF4, ARP1 and COUP/Ear3. Br J Haematol. 1996;92:231–240. doi: 10.1046/j.1365-2141.1995.269804.x. [DOI] [PubMed] [Google Scholar]
- 41.Piñol-Roma S, Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol Cell Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prescott J C, Falck-Pedersen E. Varied poly(A) site efficiency in the adenovirus major late transcription unit. J Biol Chem. 1992;267:8175–8181. [PubMed] [Google Scholar]
- 43.Prescott J C, Falck-Pedersen E. Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proudfoot N J, Brownlee G G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 45.Proudfoot N. Ending the message is not so simple. Cell. 1996;87:779–781. doi: 10.1016/s0092-8674(00)81982-0. [DOI] [PubMed] [Google Scholar]
- 46.Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 47.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 48.Sachs A, Wahle E. Poly(A) tail metabolism and function in eukaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 49.Sheets M D, Wickens M. Two phases in the addition of a poly(A) tail. Genes Dev. 1989;3:1401–1412. doi: 10.1101/gad.3.9.1401. [DOI] [PubMed] [Google Scholar]
- 50.Sheets M D, Ogg S C, Wickens M P. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sittler A, Gallinaro H, Jacob M. Upstream and downstream cis-acting elements for cleavage at the L4 polyadenylation site of adenovirus-2. Nucleic Acids Res. 1994;22:222–231. doi: 10.1093/nar/22.2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takagaki Y, Ryner L C, Manley J L. Four factors are required for 3′-end formation of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 53.Takagaki Y, Manley J L, MacDonald C C, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 54.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 55.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 56.Wahle E, Ruegsegger U. 3′ end processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 57.Wasserman K M, Steitz J A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 58.Whitelaw E, Proudfoot N J. Alpha thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilusz J, Pettine S M, Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]