Abstract
Inhibition of proteasome-mediated protein degradation machinery is a potent stress stimulus that causes accumulation of ubiquitinated proteins and increased expression of heat shock proteins (Hsps). Hsps play pivotal roles in homeostasis and protection in a cell, through their well-recognized properties as molecular chaperones. The inducible Hsp expression is regulated by the heat shock transcription factors (HSFs). Among mammalian HSFs, HSF1 has been shown to be important for regulation of the heat-induced stress gene expression, whereas the function of HSF2 in stress response is unclear. Recent reports have suggested that both HSF1 and HSF2 are affected during down-regulation of ubiquitin-proteasome pathway (Y. Kawazoe et al., Eur. J. Biochem. 255:356–362, 1998; A. Mathew et al., Mol. Cell. Biol. 18:5091–5098, 1998; D. Kim et al., Biochem. Biophys. Res. Commun. 254:264–268, 1999). To date, however, no unambiguous evidence has been presented as to whether a single specific HSF or multiple members of the HSF family are required for transcriptional induction of heat shock genes when proteasome activity is down-regulated. Therefore, by using loss-of-function and gain-of-function strategies, we investigated the specific roles of mammalian HSFs in regulation of the ubiquitin-proteasome-mediated stress response. Here we demonstrate that HSF1, but not HSF2, is essential and sufficient for up-regulation of Hsp70 expression during down-regulation of the ubiquitin proteolytic pathway. We propose that specificity of HSF1 could be an important therapeutic target during disease pathogenesis associated with abnormal ubiquitin-dependent proteasome function.
Regulation of protein degradation by the ubiquitin-proteasome pathway enables cells rapidly to reduce levels of defined proteins that control diverse processes, such as gene expression, cell signaling, immune responses, and stress adaptation. Therefore, proteasome-mediated degradation has to display a high degree of specificity, carried out by complex cascades of enzymes, toward its numerous substrates (6). Recently, a variety of inhibitors of the 26S proteasome have been identified. For example, the peptide aldehyde MG132 and the natural products lactacystin and its derivative clasto-lactacystin β-lactone all selectively inhibit degradation of proteins by the ubiquitin-proteasome pathway (15).
When the ubiquitin-proteasome network is down-regulated, certain heat shock proteins, such as Hsp70, and other molecular chaperones are induced (5, 13, 14, 21, 22, 45). Expression of heat shock genes is regulated at the transcriptional level by activation of the heat shock transcription factors (HSFs) (25, 42). Unlike in yeast and Drosophila, the existence of multiple HSFs in eukaryotes suggests that the various HSFs might have specialized functions in response to distinct physiological and environmental stimuli. Among the three known mammalian HSFs (HSF1, HSF2, and HSF4), HSF1 mediates the stress response induced by heat, heavy metals, and oxidants, while HSF2 has been suggested to have a role as a developmental regulator (1, 2, 23, 31, 32, 34, 35, 37, 38), and the role of HSF4 is still unknown (27). Recently, the phenotype of homozygous HSF1-deficient (hsf1−/−) mice was reported to include defects of the chorioallantoic placenta and increased prenatal lethality, growth retardation, female infertility, elimination of the classical heat shock response, and exaggerated tumor necrosis factor alpha production, which results in increased mortality after endotoxin challenge (43). These results demonstrate that, in addition to regulation of the stress response under pathological conditions, HSF1 is required for extraembryonic development and postnatal growth.
The specific functions of HSF1 and HSF2 in the regulation of heat shock gene expression in response to distinct stimuli, and especially during down-regulation of proteasome activity, are unclear. Mathew and coworkers (21) have provided evidence for the activation and accumulation of HSF2 when the ubiquitin-proteasome pathway is down-regulated, whereas all of the three avian HSFs have been shown to be activated by proteasome inhibitors; consequently, all of the major Hsps are markedly induced (13). Furthermore, Kim and coworkers (14) have suggested that treatments with MG132 and lactacystin result in altered phosphorylation state and activation of the DNA-binding ability of HSF1 in mouse cells. These findings raise the question of whether both HSF1 and HSF2 regulate heat shock gene expression in the ubiquitin proteolytic pathway.
To establish the specific roles of HSF1 and HSF2 in regulation of the ubiquitin-proteasome network, we used loss-of-function and gain-of-function strategies. In this study, by using HSF1-deficient cells, we show that the induction of heat shock response upon proteasome inhibition requires HSF1, as measured by increased Hsp70 expression, and this property cannot be substituted by other HSFs. Moreover, reintroduction of the HSF1 gene into HSF1-deficient cells containing normal HSF2 expression can restore the inducible Hsp70 expression in response to down-regulation of the ubiquitin-proteasome pathway. Our results provide compelling evidence for HSF1, not HSF2, being the critical transcription factor for heat shock gene expression following proteasome inhibition.
MATERIALS AND METHODS
Cell culture and experimental treatments.
Human K562 erythroleukemia cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and antibiotics (penicillin and streptomycin) in a humidified 5% CO2 atmosphere at 37°C. K562 cells stably overexpressing HSF2-α and HSF2-β isoforms (2α-C7 and 2β-D5, respectively [17]) were maintained in RPMI 1640 medium containing G418 (500 μg/ml; Gibco/BRL). Mouse embryonic fibroblasts (MEF cells) from wild-type and HSF1-deficient mice (24) were maintained in Dulbecco's modified Eagle's medium containing 10% FCS, 10 mM minimal essential medium nonessential amino acids, 0.96 μl of β-mercaptoethanol per 100 ml, and antibiotics (penicillin and streptomycin). For experimental treatments, cells were seeded at 5 × 106 cells per 10-cm-diameter plate (HSF2-α- and HSF2-β-overexpressing cells were plated in RPMI 1640 medium without G418). Hemin (Aldrich) was added to a final concentration of 40 μM, MG132 (Peptide Institute, Inc.) was added to 10 μM, and clasto-lactacystin β-lactone (Calbiochem) was added to 5 and 10 μM; cells were then incubated at 37°C for the indicated time periods. Heat shock was performed at 42°C in a water bath.
Plasmid constructions.
A full-length 1,974-bp cDNA coding for mouse HSF1 (clone C12A) was digested from pGEM-1 vector (34) and subcloned under the cytomegalovirus promoter into the EcoRI site of pCI vector (Promega). Flag-tagged mouse HSF1 was a kind gift from Richard Morimoto (Northwestern University, Evanston, Ill.). Flag-tagged mouse HSF2-α and HSF2-β expression plasmids were constructed by PCR using forward (5′-CGGAATTCCAACGTGCCGGCTTTC-3′) and reverse (5′-CCATCGATTCCACTTGGGAG-3′) primers. The forward primer contains an EcoRI restriction site consensus sequence (underlined), and the reverse primer contains a ClaI restriction site (underlined). The resulting EcoRI and ClaI sites were used to clone the fragment in frame with the Flag tag in pFLAG-CMV-2 (Kodak).
Transfections.
MEF cells were transfected by electroporation (975 μF, 280 V) using a Bio-Rad Gene Pulser electroporator. Cells (2 × 106) were washed, resuspended in 0.4 ml of OptiMEM (Gibco-BRL), and placed in a 0.4-cm gap electroporation cuvette (BTX). Plasmid DNA (30 μg) was added, and after a brief incubation at room temperature, the cells were subjected to a single electric pulse followed by dilution to 2 × 105 cells/ml with Dulbecco's modified Eagle's medium containing 10% FCS, 10 mM minimal essential medium nonessential amino acids, 0.96 μl of β-mercaptoethanol per 100 ml, and antibiotics (penicillin and streptomycin). Thereafter, cells were incubated at 37°C for 40 h prior to use.
Gel mobility shift assay.
Whole cell extracts were prepared from treated cells as previously described (26) and incubated (12 μg of protein) with a 32P-labeled oligonucleotide representing the proximal heat shock element (HSE) of the human hsp70 promoter. The protein-DNA complexes were analyzed on a native 4% polyacrylamide gel as described previously (26). The signal intensities of the protein-DNA complexes were quantitated with a phosphorimaging scanner (Bio-Rad).
Nuclear run-on assay.
Nuclear run-on transcription reactions were performed with nuclei isolated from MG132-, hemin-, or heat shock-treated cells in the presence of 100 μCi of [α-32P]dUTP (3,000 Ci/mmol; Amersham) as previously described (3). Radiolabeled RNA was hybridized to nitrocellulose-immobilized plasmids for the human hsp70 (pH2.3 [41]), human hsp90/89α (pUCHS801 [11]), and rat GAPDH (pGAPDH [9]) genes and a Bluescript vector (Stratagene). The hybridizations were carried out in 50% formamide–6× SSC (1× SSC is 0.15 M sodium chloride and 0.015 M sodium citrate)–10× Denhardt's solution-0.2% sodium dodecyl sulfate (SDS) at 42°C for 72 h. Filters were washed with high-stringency conditions (0.2× SSC–0.2% SDS at 65°C) and visualized by autoradiography.
Western analysis.
Whole cell extracts (12 μg of protein) were subjected to SDS–8% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose filter (Protran nitrocellulose; Schleicher & Schuell) by using a Bio-Rad semidry transfer apparatus. Proteins were detected as follows: HSF1 by a polyclonal antibody specific to mouse HSF1 (35), HSF2 by a polyclonal antibody specific to mouse HSF2 (35), the inducible form of Hsp70 by 4g4 (Affinity Bioreagents, Inc.), the mouse Hsp70 by SPA-810 (StressGen), Hsp90 by SPA-835 (StressGen), Hsc70 by SPA-815 (StressGen), Flag epitope by M2 (Sigma), and actin by monoclonal antiactin antibody N350 (Amersham). Horseradish peroxidase-conjugated secondary antibodies were purchased from Promega and Amersham. The blots were developed with an enhanced chemiluminescence method (ECL kit; Amersham).
Northern analysis.
Poly(A) mRNA was isolated from the treated K562 cells by using a poly(A) mRNA purification kit (Pharmacia). RNA was separated on a 1% agarose-formaldehyde gel, transferred to a nylon filter (Hybond-N; Amersham), and hybridized at 65°C with an [α-32P]dCTP (50 μCi, 3,000 Ci/mmol; ICN)-labeled 931-bp HindIII/PstI cDNA insert coding for human HSF2 (31) (hHSF2 cDNA was a kind gift from Robert Kingston, Harvard Medical School, Boston, Mass. [37]) and [α-32P]dCTP-labeled plasmids for human hsp70 (pH2.3 [41]) and rat GAPDH (pGAPDH [9]). Following hybridization, filters were washed with high-stringency conditions (0.1× SSC–0.1% SDS at 65°C) and visualized by autoradiography. The intensities of radioactive signals were quantitated with a phosphorimaging scanner (Bio-Rad).
RESULTS
Inhibition of the ubiquitin-proteasome pathway leads to rapid transcriptional induction of heat shock genes.
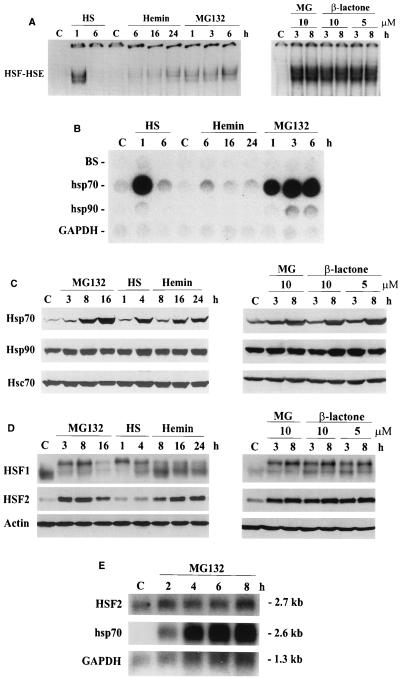
To investigate the HSF DNA-binding activity during down-regulation of the ubiquitin-proteasome pathway in human K562 erythroleukemia cells, gel mobility shift analysis was performed with an HSE-containing oligonucleotide. As shown in Fig. 1A, the HSF HSE binding induced by heat shock and MG132 was readily detectable after 1 h of treatment. In contrast to the heat-induced HSF HSE binding, which had totally disappeared after 6 h of heat shock, the MG132-induced HSF HSE binding remained constant or even increased during the 6-h period. Similar results were obtained using clasto-lactacystin β-lactone, a specific covalent inhibitor of the 26S proteasome. This confirms that the MG132-induced HSF HSE-binding activity is due to inhibition of proteasome function. In comparison to the effects of potent proteasome inhibitors, the hemin-induced HSF HSE binding was detected only after a 6-h treatment, whereafter it increased until 24 h (Fig. 1A), in agreement with earlier studies showing a slow and sustained HSF activation related to erythroid differentiation of K562 cells (38, 40).
FIG. 1.
Heat shock response caused by inhibition of the ubiquitin-mediated proteolysis (A) HSF HSE-binding activity in whole cell extracts from control (C), heat-shocked (HS; 42°C for 1 and 6 h), hemin-treated (40 μM for 6, 16, and 24 h), MG132-treated (10 μM for 1, 3, 6, and 8 h), and clasto-lactacystin β-lactone-treated (10 or 5 μM for 3 and 8 h) K562 cells was analyzed by gel mobility shift assay. Extracts (12 μg) were incubated with a 32P-labeled oligonucleotide representing the proximal HSE of the human hsp70 promoter. Protein-DNA complexes were resolved on a 4% nondenaturing polyacrylamide gel as described elsewhere (26). (B) Transcription rates of hsp70 and hsp90 genes were analyzed by nuclear run-on assay. Equal number of nuclei from control (C), heat-shocked (HS; 1 and 6 h at 42°C), hemin-treated (40 μM for 6, 16, and 24 h), and MG132-treated (10 μM for 1, 3, and 6 h) K562 cells were used for in vitro [32P]dUTP labeling of newly synthesized transcripts which were hybridized to immobilized DNA probes for human hsp70, human hsp90/89α, rat GAPDH, and a Bluescript vector (BS) as described elsewhere (3). GAPDH was used as an internal control. (C and D) For protein analysis, whole cell extracts (12 μg) isolated from control (C), MG132-treated (10 μM for 3, 8, and 16 h), heat-shocked (HS; 42°C for 1 and 4 h), hemin-treated (40 μM for 8, 16, and 24 h), and clasto-lactacystin β-lactone-treated (10 μM for 3 and 8 h) K562 cells were analyzed by SDS-PAGE and Western immunoblotting using antibodies against HSF1, HSF2, Hsp70, Hsp90, Hsc70, and actin. (E) Poly(A) mRNA from control (C) and MG132-treated (10 μM for 2, 4, 6, and 8 h) K562 cells was analyzed by Northern blotting using 32P-labeled cDNA probes for human HSF2, hsp70, and GAPDH. GAPDH was used as a control for equal loading. mRNA sizes are indicated on the right.
Since exposure to elevated temperatures is a well-known inducer of heat shock gene expression, we examined whether the proteasome inhibition-induced HSF HSE binding is accompanied by enhanced transcription of heat shock genes. The nuclear run-on experiment shown in Fig. 1B demonstrates that similarly to heat shock, the proteasome inhibitor MG132 strongly induced hsp70 and to a lesser extent hsp90 transcription after 1 h. In agreement with the HSE-binding activity (Fig. 1A), heat shock stimulated hsp70 and hsp90 transcription transiently, whereas the MG132-induced transcription persisted for at least 6 h (Fig. 1B). In contrast to the prominent transcriptional induction of hsp70 gene in response to heat stress and blocked proteasome function, hsp70 transcription was significantly less affected by hemin treatment (Fig. 1B). Next, we measured heat shock protein accumulation in response to proteasome inhibition, heat shock, and hemin treatment. Hsp70 protein levels dramatically increased by 8 h of treatment with MG132 and clasto-lactacystin β-lactone (Fig. 1C). Exposures to heat shock (4 h) and hemin (16 h) resulted in equal increases in Hsp70 levels (Fig. 1C). The amounts of Hsp90 and Hsc70 were not affected by these treatments.
To gain insight to the regulatory mechanism of heat shock response upon proteasome down-regulation, the HSF1 and HSF2 proteins were analyzed by Western blotting. As shown in Fig. 1D, no changes in HSF1 protein levels were observed in response to treatment with the proteasome inhibitors. However, heat shock, MG132, and clasto-lactacystin β-lactone, but not hemin, induced hyperphosphorylation of HSF1 (Fig. 1D), which can be detected by the appearance of slower-migrating HSF1 bands resulting from induced phosphorylation (7, 14, 35). HSF2 protein levels were dramatically increased in MG132 and clasto-lactacystin β-lactone-treated cells by 3 h, whereas heat shock had no effect on HSF2 (Fig. 1D). Hemin treatment increased HSF2 levels, but with different kinetics compared to MG132. By 8 h, the HSF2 protein levels were slightly elevated and thereafter the amounts of HSF2 continued to increase (Fig. 1D). The increase in HSF2 protein was due solely to inhibited proteasome function, as HSF2 mRNA levels did not increase in MG132-treated K562 cells (Fig. 1E). In contrast, consistent with the transcriptional induction of hsp70 gene shown in Fig. 1B, the hsp70 mRNA amounts dramatically increased after 2 h of MG132 treatment and continued to increase during the 8-h period (Fig. 1E).
Heat shock response induced by proteasome inhibition requires HSF1.
Since down-regulation of the proteasome function induced HSF HSE-binding activity and heat shock gene transcription, and both HSF1 and HSF2 were affected, it was important to establish the specific roles of these transcriptional regulators by using two strategies that abrogated HSF1 and HSF2 activities. First, we studied the effect of proteasome inhibition on HSF2 activity by using K562 cell clones stably overexpressing either the HSF2-α (2α-C7) or HSF2-β (2β-D5) isoform. Overexpression of HSF2-α has an enhancing effect on activation of HSF2, whereas overexpression of the dominant negative HSF2-β isoform prevents HSF2 activation (17). As shown in Fig. 2, both 1-h heat shock and 3-h MG132 treatments induced equivalent HSF HSE binding in cells expressing either HSF2-α or HSF2-β and in mock-transfected cells (Fig. 2). In contrast, no HSF HSE binding was detected in 2β-D5 cells upon hemin treatment (Fig. 2) (17).
FIG. 2.
Proteasome inhibition induces HSF HSE binding in the absence of HSF2 activity. Whole cell extracts from control (C), heat-shocked (HS; 42°C for 1 h), hemin-treated (He; 40 μM for 16 h), and MG132-treated (MG; 10 μM for 3 h) mock-transfected K562 cells (vector) or K562 cells overexpressing the HSF2-α (2α-C7) or HSF2-β (2β-D5) isoform were analyzed as for Fig. 1A.
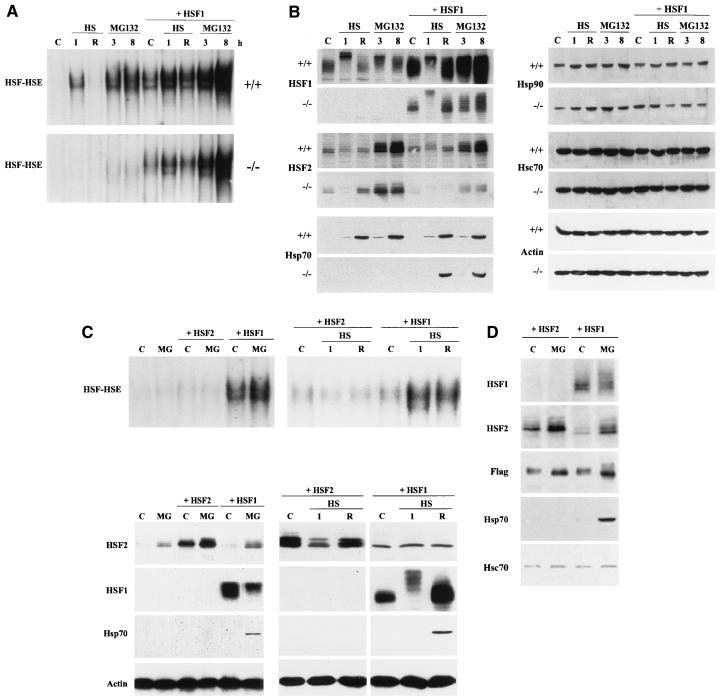
To elucidate the role of HSF1 during the heat shock response induced by proteasome inhibition, we investigated the effect of MG132 on HSF1 and HSF2 in wild-type and hsf1−/− MEF cells (24). Both heat shock and MG132 treatments strongly induced HSF HSE binding in wild-type cells, whereas in the HSF1-deficient cells, treatment with MG132 induced only a modest HSF HSE-binding activity (Fig. 3A), which was perturbed with anti-α-HSF2 antibody (data not shown). Similar results were obtained with clasto-lactacystin β-lactone (data not shown). As expected, heat shock did not activate HSF HSE binding in the hsf1−/− cells (Fig. 3A) (24).
FIG. 3.
HSF1, but not HSF2, can restore Hsp70 expression. (A) Whole cell extracts from control (C), heat-shocked (HS; 42°C for 1 h without [1] or with [R] a 3-h recovery at 37°C), and MG132-treated (10 μM for 3 and 8 h) wild-type (+/+) and hsf1−/− MEF cells, as well as wild-type and hsf1−/− cells transiently transfected with mouse HSF1 (+ HSF1), were analyzed as described for Fig. 1A. (B) The samples described above were analyzed by SDS-PAGE and immunoblotted with antibodies against HSF1, HSF2, Hsp70, Hsp90, Hsc70, and actin. (C) Whole cell extracts from control (C), heat-shocked (HS; 42°C for 1 h without [1] or with [R] a 3-h recovery at 37°C), and MG132-treated (10 μM for 8 h) MEF cells deficient for HSF1, as well as hsf1−/− cells transiently transfected with mouse HSF2 (+ HSF2) or HSF1 (+ HSF1), were subjected to gel mobility shift analysis (upper panels) and Western immunoblotting (lower panels) using antibodies against HSF2, HSF1, Hsp70, and actin. (D) Whole cell extracts from control (C) and MG132-treated (10 μM for 8 h) hsf1−/− cells transiently transfected with Flag epitope-tagged mouse HSF1 (+ HSF1) and HSF2 (+ HSF2) were subjected to Western immunoblotting using antibodies against HSF1, HSF2, Flag, Hsp70, and Hsc70.
Expression of Hsp70 in HSF1-deficient cells can be rescued by exogenous HSF1.
To further analyze whether HSF1 is the main regulator of heat shock gene expression during down-regulation of proteasome activity, mouse HSF1 was reintroduced into wild-type and hsf1−/− MEF cells by transient transfection. In both cases, overexpression of HSF1 induced a basal HSF HSE-binding activity (Fig. 3A). Heat shock for 1 h further increased the HSF HSE-binding activity, and after a 3-h recovery at normal temperature, the HSE-binding activity was decreased to the basal level. Interestingly, upon 3 h of MG132 treatment, the HSF HSE-binding activity was similarly increased when exogenous HSF1 was expressed in wild-type cells and in cells deficient for HSF1, and the HSE-binding activity was further enhanced by 8 h of MG132 treatment (Fig. 3A). As shown in Fig. 3B, both wild-type and hsf1−/− cells expressed significant amounts of the exogenous HSF1, and a 1-h heat shock and 3- to 8-h MG132 treatment resulted in an altered phosphorylation state of HSF1. However, in contrast to heat shock, which induced hyperphosphorylation of HSF1, more intermediate phosphorylation forms were detected in response to blocked proteasome function. Despite the significant increase in HSF2 protein levels after 3 h of MG132 treatment in both wild-type and HSF1-deficient MEF cells (Fig. 3B), only a minor HSF2 HSE-binding activity was detected in hsf1−/− cells (Fig. 3A). The Hsp70 protein levels were strongly increased after a 1-h heat shock followed by a 3-h recovery and after an 8-h MG132 treatment in wild-type MEF cells. In contrast, in HSF1-deficient cells, no Hsp70 protein was detected, although the endogenous HSF2 protein levels were equal in both wild-type and hsf1−/− cells (Fig. 3B).
Strikingly, reintroduction of the mouse HSF1 gene into the hsf1−/− cells fully restored the Hsp70 expression in response to heat shock and inhibition of the ubiquitin proteolytic pathway. Overexpression of HSF1 did not markedly affect Hsp70 protein levels in wild-type cells (Fig. 3B). Equivalent expression of Hsp90 and Hsc70 was present in both wild-type cells and in HSF1-deficient cells, and overexpression of HSF1 did not affect the levels of these proteins (Fig. 3B). To compare the effects of exogenous HSF1 and HSF2 on Hsp70 expression upon exposure to heat shock or inhibition of proteasome function, we transiently transfected hsf1−/− cells with the mouse HSF2. In contrast to the spontaneous DNA-binding activity in HSF1-transfected cells, overexpression of HSF2 caused neither basal nor inducible HSF HSE binding (Fig. 3C, upper panels). Despite overexpression of HSF2, Hsp70 expression could not be restored in hsf1−/− cells treated with the proteasome inhibitors or heat shock (Fig. 3C, lower panels). To compare the relative expression levels of exogenous HSF1 and HSF2, we repeated the previous experiment by using Flag epitope-tagged forms of HSF1 and HSF2. As shown in Fig. 3D, the antibody specific for the Flag epitope revealed equivalent expression levels of the transfected genes, and Hsp70 expression was rescued only by HSF1, thereby verifying the results in Fig. 3B and C.
DISCUSSION
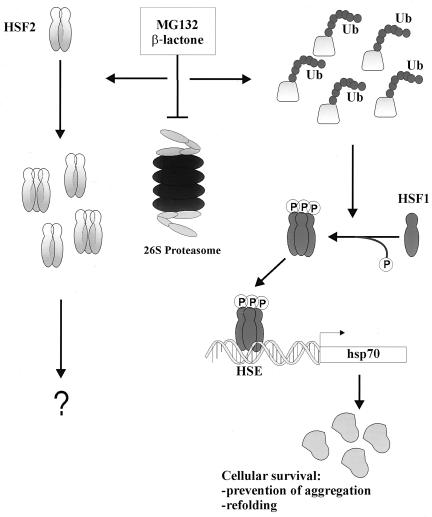
Originally identified by their increased expression following heat shock, Hsp70 proteins function as molecular chaperones that assist protein folding processes in most cellular compartments (4, 10, 20, 28). Apart from the well-characterized heat-induced stress response mediated by HSF1 (25, 42), regulation of the inducible Hsp70 expression during nonstressful conditions and certain pathophysiological states is largely unknown. Using a gain-of-function strategy in HSF1-deficient cells, we have established the absolute requirement of HSF1 in the regulation of hsp70 gene expression in the ubiquitin-proteasome network. Our findings are summarized in the following model of Hsp70 induction upon proteasome inhibition (Fig. 4). Down-regulation of the ubiquitin-proteasome pathway results in increased amounts of polyubiquitinated proteins (6), which in turn lead to HSF1 activation and HSF1-dependent hsp70 transcription (this study). According to the model, we anticipate that by associating with the aberrant proteins targeted for degradation, Hsp70 and other molecular chaperones promote the cellular survival by preventing protein aggregation and facilitating refolding. Most importantly, although accumulation and partial oligomerization of HSF2 can be observed, HSF2, or other HSFs, cannot functionally substitute for HSF1 during down-regulation of the ubiquitin-proteasome pathway. The proposed model does not exclude the possible existence of a positive regulator of HSF1 (45) or a negative regulator of HSF2 (44).
FIG. 4.
Model of HSF1-dependent Hsp70 induction in the ubiquitin (Ub)-proteasome pathway. Down-regulation of ubiquitin-mediated proteolysis by proteasome inhibitors, such as MG132 and clasto-lactacystin β-lactone, results in the accumulation of malfolded, abnormal, and short-lived proteins normally degraded by the 26S proteasome (6). Increase in the amount of polyubiquitinated proteins leads to oligomerization, phosphorylation, and acquisition of HSF1 HSE-binding activity, which in turn induces hsp70 transcription and accumulation of the Hsp70 protein, leading to enhanced molecular chaperone activity. It can be speculated that by associating with the polyubiquitinated proteins, molecular chaperones, among them Hsp70, promote the cellular survival by preventing protein aggregation and facilitating the malfolded proteins to refold. On the other hand, proteasome inhibition results in accumulation and partial oligomerization of the labile HSF2 protein, but the specific role of HSF2 under these conditions remains to be elucidated.
The signal for HSF1 activation upon inhibition of the ubiquitin-proteasome pathway most probably results from the accumulation of undegraded, unfolded, and misfolded proteins. Interestingly, this kind of protein deposition has been implicated in disease pathogenesis of a number of chronic degenerative diseases, such as cystic fibrosis, Alzheimer's disease, Cushing's disease, and amyotrophic lateral sclerosis (6, 12), all of which exhibit abnormal proteins that accumulate either inside or outside of the cell. Whether the increased levels of Hsp70 and other Hsps observed in some of the degenerative diseases are dependent on HSF1 is an important question for future studies. Another aspect of Hsp accumulation is thermotolerance, which can be acquired by induction of Hsps in response to mild heat stress (19, 29). Thermotolerance is an endogenous mechanism for cells to withstand subsequent greater thermal injury, and in addition to hyperthermia, the beneficial effects of acquired thermotolerance could protect against injury from other forms of noxious stimuli (cross-tolerance). Alternatively, pharmacological strategies such as pretreatment with the proteasome inhibitor MG132 for as little as 2 h markedly increase the survival of cells subjected to high temperatures, thus conferring cross-tolerance (5, 16). Our finding that HSF1 is indispensable for the inducible expression of Hsp70 upon treatment with MG132 and other proteasome inhibitors is likely of importance in understanding the regulation of cytoprotection and discovering its potential medical applications.
The physiological role of two structurally highly related HSFs, HSF1 and HSF2, and their specific functions in the regulation of heat shock gene expression have remained unresolved. Here we demonstrate that the increase in HSF2 expression does not correspond to the enhanced Hsp70 expression, as neither accumulation of the endogenous HSF2 protein in response to blocked proteasome function nor overexpression of the exogenous HSF2 prior to inhibition of proteasome function induces Hsp70 expression. Furthermore, HSF2 is not capable of compensating for HSF1 in the physiological heat shock response or acquired thermotolerance, as demonstrated with MEF cells derived from HSF1-deficient mice (24). It has been proposed that HSF2 might have other target genes, such as the thioredoxin gene (18, 31), and during development no correlation between activation of HSF2 and expression of heat shock genes has been observed (1, 33). Abundant expression of Hsp70 in K562 cells undergoing hemin-mediated erythroid differentiation was originally shown to be due to HSF2 activation (38, 39). Also, HSF1 has been reported to mediate hemin-induced hsp70 expression (44). However, our recent studies suggest that the antibody supershift technique used in these studies is not adequate for determining the composition of HSF-HSE complexes (30), stressing the usefulness of the knockout cells as experimental tools.
Under normal cellular as well as stressful conditions, cells must continuously use both reparative and degradative pathways to cope with the accumulation of abnormal proteins. It has been speculated that enzymes of the ubiquitin-conjugating system are assisted by Hsp70 and other molecular chaperones in their task to identify proteins with nonnative conformation (8, 36). The high cross-species conservation of the components of cellular stress response machinery, including the 26S proteasome complex, reflects the need for effective degradation of damaged proteins. It is plausible that the decreased rate of macromolecular degradation is a signal for HSF1 activation, resulting in beneficial effects on protein homeostasis by increasing the concentration of thermoprotectant molecules and molecular chaperones.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Helena Saarento and Päivi Nykänen for excellent technical assistance, and we thank John E. Eriksson and Marko Kallio for valuable comments and discussions.
This work was supported by the Academy of Finland, the Sigrid Jusélius Foundation, and the Finnish Cancer Organizations (L.S.), by the Finnish Medical Society DUODECIM (T.-P.A.), and by the Emil Aaltonen Foundation (L.P.). L.P. and T.-P.A. are supported by the Turku Graduate School of Biomedical Sciences. I.J.B. received support from the National Institutes of Health (K14 award, grant HL60667) and an Established Investigator Award from the American Heart Association.
REFERENCES
- 1.Alastalo T-P, Lönnström M, Leppä S, Kaarniranta K, Pelto-Huikko M, Sistonen L, Parvinen M. Stage-specific expression and cellular localization of the heat shock factor 2 isoforms in the rat seminiferous epithelium. Exp Cell Res. 1998;240:16–27. doi: 10.1006/excr.1997.3926. [DOI] [PubMed] [Google Scholar]
- 2.Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji S S, Theodorakis N G, Morimoto R I. Heat shock-induced translational control of HSP70 and globin synthesis in chicken reticulocytes. Mol Cell Biol. 1984;4:2437–2448. doi: 10.1128/mcb.4.11.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Bush K T, Goldberg A L, Nigam S K. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotto J J, Kline M, Morimoto R I. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. J Biol Chem. 1996;271:3355–3358. doi: 10.1074/jbc.271.7.3355. [DOI] [PubMed] [Google Scholar]
- 8.Craig E A, Baxter B K, Becker J, Halladay J, Ziegelhoffer T. Cytosolic hsp70s of Saccharomyces cerevisiae: roles in protein synthesis, protein translocation, proteolysis, and regulation. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 31–52. [Google Scholar]
- 9.Fort P, Marty L, Piechaczyk M, El Sabrouty S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 11.Hickey E, Brandon S E, Smale G, Lloyd D, Weber L A. Sequence and regulation of a gene encoding a human 89-kilodalton heat shock protein. Mol Cell Biol. 1989;9:2615–2626. doi: 10.1128/mcb.9.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman R J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 13.Kawazoe Y, Nakai A, Tanabe M, Nagata K. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur J Biochem. 1998;255:356–362. doi: 10.1046/j.1432-1327.1998.2550356.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim D, Kim S H, Li G C. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem Biophys Res Commun. 1999;254:264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 15.Lee D H, Goldberg A L. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 16.Lee D H, Goldberg A L. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leppä S, Pirkkala L, Saarento H, Sarge K D, Sistonen L. Overexpression of HSF2-β inhibits hemin-induced heat shock gene expression and erythroid differentiation in K562 cells. J Biol Chem. 1997;272:15293–15298. doi: 10.1074/jbc.272.24.15293. [DOI] [PubMed] [Google Scholar]
- 18.Leppä S, Pirkkala L, Chow S C, Eriksson J E, Sistonen L. Thioredoxin is transcriptionally induced upon activation of heat shock factor 2. J Biol Chem. 1997;272:30400–30404. doi: 10.1074/jbc.272.48.30400. [DOI] [PubMed] [Google Scholar]
- 19.Li G C, Nussenzweig A. Thermotolerance and heat shock proteins: possible involvement of Ku autoantigen in regulating HSP70 expression. In: Feige U, Morimoto R I, Yahara I, Polla B S, editors. Stress-inducible cellular responses. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 425–449. [DOI] [PubMed] [Google Scholar]
- 20.Lindquist S, Craig E A. The heat shock response. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 21.Mathew A, Mathur S K, Morimoto R I. The heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meriin A B, Gabai V L, Yaglom J, Shifrin V I, Sherman M Y. Proteasome inhibitors activate stress kinases and induce Hsp72. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 23.Mezger V, Rallu M, Morimoto R I, Morange M, Renard J-P. Heat shock factor 2-like activity in mouse blastocysts. Dev Biol. 1994;166:819–822. doi: 10.1006/dbio.1994.1361. [DOI] [PubMed] [Google Scholar]
- 24.McMillan D R, Xiao X, Shao L, Graves K, Benjamin I J. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto R I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 26.Mosser D D, Theodorakis N G, Morimoto R I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto R I, Nagata K. HSF4, a new member of the human heat shock family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 29.Parsell D A, Lindquist S. Heat shock proteins and stress tolerance. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 457–494. [Google Scholar]
- 30.Pirkkala L, Sistonen L. Antibody supershift assay is inadequate for determining HSF stoichiometry in HSE complexes. Cell Stress Chaperones. 1999;4:259–261. doi: 10.1379/1466-1268(1999)004<0259:asaiif>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirkkala L, Alastalo T-P, Nykänen P, Seppä L, Sistonen L. Differentiation lineage-specific expression of human heat shock transcription factor 2. FASEB J. 1999;13:1089–1098. doi: 10.1096/fasebj.13.9.1089. [DOI] [PubMed] [Google Scholar]
- 32.Rabindran S K, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci USA. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rallu M, Loones M T, Lallemand Y, Morimoto R I, Morange M, Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci USA. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarge K D, Zimarino V, Holm K, Wu C, Morimoto R I. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 35.Sarge K D, Murphy S P, Morimoto R I. Activation of heat shock gene transcription by HSF1 involves oligomerization, acquisition of DNA binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheffner M, Smith S, Jentsch S. The ubiquitin-conjugation system. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 65–98. [Google Scholar]
- 37.Schuetz T J, Gallo G J, Sheldon L, Tempst P, Kingston R E. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci USA. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sistonen L, Sarge K D, Phillips B, Abravaya K, Morimoto R I. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sistonen L, Sarge K D, Morimoto R I. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theodorakis N G, Zand D J, Kotzbauer P T, Williams G T, Morimoto R I. Hemin-induced transcriptional activation of the hsp70 gene during erythroid maturation in K562 cells is due to a heat shock factor-mediated stress response. Mol Cell Biol. 1989;9:3166–3173. doi: 10.1128/mcb.9.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu B, Hunt C, Morimoto R I. Structure and expression of the human gene encoding major heat shock protein HSP70. Mol Cell Biol. 1985;5:330–341. doi: 10.1128/mcb.5.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 43.Xiao X, Zuo X, Davis A A, McMillan D R, Curry B B, Richardson J A, Benjamin I J. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshima T, Yura T, Yanagi H. Heat shock factor 1 mediates hemin-induced hsp70 gene transcription in K562 erythroleukemia cells. J Biol Chem. 1998;273:25466–25471. doi: 10.1074/jbc.273.39.25466. [DOI] [PubMed] [Google Scholar]
- 45.Zhou M, Wu X, Ginsberg H N. Evidence that a rapidly turning over protein, normally degraded by proteasomes, regulates hsp72 gene transcription in HepG2 cells. J Biol Chem. 1996;271:24769–24775. doi: 10.1074/jbc.271.40.24769. [DOI] [PubMed] [Google Scholar]