Abstract
The transcriptional coactivators p300 and CREB binding protein (CBP) are important regulators of the cell cycle, differentiation, and tumorigenesis. Both p300 and CBP are targeted by viral oncoproteins, are mutated in certain forms of cancer, are phosphorylated in a cell cycle-dependent manner, interact with transcription factors such as p53 and E2F, and can be found complexed with cyclinE-Cdk2 in vivo. Moreover, p300-deficient cells show defects in proliferation. Here we demonstrate that transcriptional activation by both p300 and CBP is stimulated by coexpression of the cyclin-dependent kinase inhibitor p21WAF/CIP1. Significantly this stimulation is independent of both the inherent histone acetyltransferase (HAT) activity of p300 and CBP and of the previously reported carboxyl-terminal binding site for cyclinE-Cdk2. Rather, we describe a previously uncharacterized transcriptional repression domain (CRD1) within p300. p300 transactivation is stimulated through derepression of CRD1 by p21. Significantly p21 regulation of CRD1 is dependent on the nature of the core promoter. We suggest that CRD1 provides a novel mechanism through which p300 and CBP can switch activities between the promoters of genes that stimulate growth and those that enhance cell cycle arrest.
The p300 and CREB binding protein (CBP) transcriptional coactivators are important regulators of many cellular processes. Both proteins are highly homologous (2), contain histone acetyltransferase (HAT) domains (5, 33), and interact with a wide range of DNA binding proteins, including p53, the RelA (p65) NF-κB subunit, E2F, MyoD, AP-1, nuclear receptors, and many others (13, 14, 37, 38, 42). Interaction with p300 and CBP provides an additional level of regulation for certain transcription factors. Single-allele knockouts of both p300 and CBP have demonstrated that both are present at limiting concentrations relative to many DNA binding proteins (40, 50), which can result in cross talk between many classes of transcriptional activators as they compete for binding to the coactivator complexes (4, 19–21, 25, 45). Furthermore, and providing additional complexity, the transcriptional activities of p300 and CBP are themselves directly regulated. A number of signaling pathways, including cyclic AMP-activated protein kinase A, nerve growth factor activation of the p42/44 mitogen-activated protein kinase pathway, insulin-activated pp90rsk, and nuclear calcium signaling, have been shown to modulate p300 and CBP function (8, 27, 31, 46). In addition, the binding of other cellular coactivators such as P/CAF, P/CIP, Mdm2, and Src1 (43, 48, 49), many of which also have HAT activity, and the interaction with viral oncoproteins, such as Tax, T antigen, and E1A (3, 7, 10, 17, 24, 32), have profound effects on p300 and CBP function.
That p300 and CBP are both targets for viral oncoproteins is also an indication of the central role that both proteins play in the regulation of the cell cycle and tumorigenesis (38). In addition to interacting with cell cycle-regulating transcription factors such as p53 (4, 16, 26, 39) and E2F (25, 44), both p300 and CBP are phosphorylated in a cell cycle-dependent manner (1, 47), while cells derived from p300 knockout mice show profound proliferative defects (50). Moreover, mutations in p300 have been found to be associated with colon and gastric carcinomas, while CBP is found to be translocated in a number of leukemias (12, 30).
Previously it has been demonstrated that the cyclin-dependent kinase (CDK) inhibitor p21 strongly enhanced transactivation by the RelA (p65) NF-κB subunit (34). This was shown to correlate with inhibition of a cyclinE-Cdk2 complex bound to the carboxy termini of p300 and CBP (34). More recently it has been shown that phosphorylation of CBP by cyclinE-Cdk2 stimulates its inherent HAT activity (1). In that report it was proposed that stimulation of p300 and CBP HAT activity by cyclinE-Cdk2 promoted entry into S phase. That p21 can inhibit p300- and CBP-bound cyclinE-Cdk2 activity would thus be consistent with repression of their inherent HAT activity being associated with cell cycle arrest. This does not explain how p21 was able to stimulate RelA transactivation (34), however, or how p300 and CBP are required for the activity of transcription factors known to stimulate cell cycle arrest or differentiation through induction of p21, such as p53 and MyoD (reviewed in reference 38).
In this report we have investigated the mechanisms through which p21 regulates p300 and CBP transcriptional activity. We find that p21 stimulates transactivation by both p300 and CBP. Significantly we demonstrate that p21 induction of p300 results from the activity of a discreet domain in the amino-terminal half of the protein which functions to repress transcription. The activity of this domain is dependent on the promoter context, however, and we propose a model in which p300 and CBP activity might be switched between promoters following p21-induced cell cycle arrest.
MATERIALS AND METHODS
Plasmids.
All Gal4 fusions were constructed using the pVR-1012 Gal4 expression plasmid. pVR-1012 is a cytomegalovirus-based expression plasmid, kindly provided by agreement with Vical Inc., and has been described previously (18). pVR-1012 Gal4 was constructed by insertion of a HindIII/BamHI fragment containing the Gal4 DNA binding domain from pSG 424 (36) into the polylinker of pVR-1012. Additional restriction sites were then inserted into the polylinker using a double-stranded oligonucleotide.
Gal4 p300 (full length) was created using a SalI/NotI fragment from pBluescript p300 (34). Gal4 p300 (1239-2414) was created using a BamHI fragment from pVR 1012 p300 (34). Gal4 p300 (1-1301) was created using an XbaI fragment from pBluescript p300. Gal4 CBP (full length) was created using a BamHI fragment from RSV HA CBP (containing the murine CBP cDNA; provided by Richard Goodman, Vollum Institute, Portland, Oreg.). Gal4 CBP (1-1097) was created using an XbaI fragment from the Gal4 CBP (full length) coding sequence. Gal4 CBP (1098-2441) was created by religation of the Gal4 CBP (full-length) plasmid following the XbaI digestion used to create Gal4 CBP (1-1097). All additional plasmids encoding the amino and carboxy termini of p300 and CBP were created by PCR using Pfu Turbo (Stratagene). PCR products were engineered to have an XbaI site at the coding sequence for the amino terminus and a BglII site at the coding sequence for the carboxy terminus, which were subsequently used for subcloning into pVR 1012 Gal4. Gal4 p300 (Δ851 to 1045) and (Δ1004 to 1045) were generated in two stages. The sequence encoding the amino terminal fragment was first generated by PCR and engineered to have a SalI site at the segment encoding the amino terminus and a NotI site at the segment encoding the carboxy terminus. This fragment was then subcloned into pVR 1012 Gal4 using these restriction sites. A PCR fragment encoding a carboxy-terminal fragment (from amino acid 1045 to 2414) was then generated with NotI sites at either end, which were used to create the full-length, final plasmid with an internal deletion. pVR-1012 p300 (Δ1004 to 1045) was created from the equivalent Gal4 fusion protein by removal of Gal4 by digestion of the coding sequence with SalI.
Gal4 Sp1, Gal4 p53 (1-42), Gal4 E1B CAT, Gal4 E4 CAT, Gal4 (−31) HIV CAT, G5 TK CAT, and G0 TK CAT were supplied by Stefan Roberts (University of Dundee). The construction of Bax CAT has been described previously (45).
Calcium phosphate transient transfections and reporter gene assays.
Cells were transfected using calcium phosphate as previously described (45). In all experiments cells were harvested 24 h after transfection and chloramphenicol acetyltransferase (CAT) activity was assayed with 10 to 100 μg of protein from whole-cell extracts. All results shown are representative of at least three separate experiments.
Western blots.
Whole-cell lysates were prepared, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride before incubation with anti-Gal4 antibody (sc-510; Santa Cruz Biotechnology).
RESULTS
p300 and CBP transactivation is induced by p21 and can occur through the amino terminus.
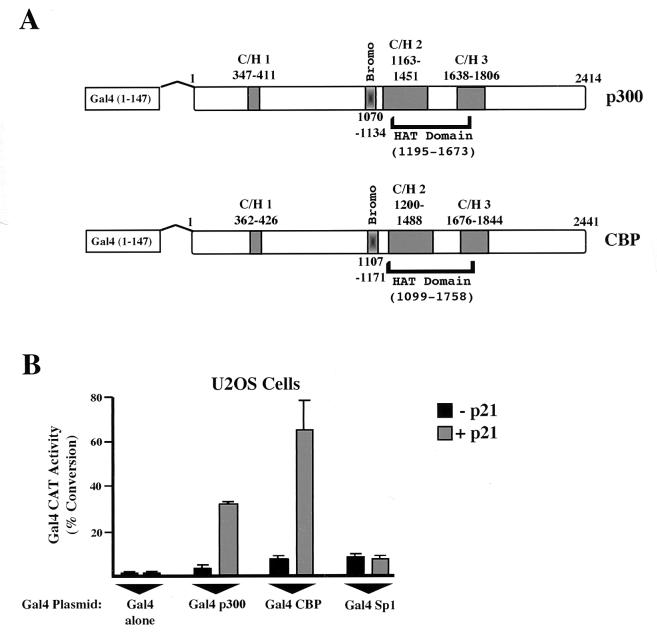
Previously it has been shown that the ability of p300 to coactivate the RelA (p65) NF-κB subunit is strongly enhanced by cotransfection of a p21 expression plasmid (34). These experiments were complicated by the need to recruit p300 to the promoter through RelA, which itself can be a regulated event (52), and thus it could not be concluded that the effect of p21 was directly on p300 itself. Similarly, other transcription factor interactions, such as those with CREB and p53, have been shown to be regulated by posttranslational modifications or the interactions of other cellular proteins (9, 15). To overcome this problem and to enable analysis of p300 and CBP directly, both proteins were fused to the Gal4 DNA binding domain (Fig. 1A). Targeting to the promoter through Gal4 also overcomes any effects from endogenous, wild-type p300 and CBP. Both Gal4 p300 and Gal4 CBP were then cotransfected with the Gal4 E1B CAT reporter plasmid into U2OS cells. As expected, both Gal4 p300 and Gal4 CBP stimulated CAT activity (Fig. 1B). Interestingly, the additional cotransfection of p21 resulted in a strong enhancement of both p300 and CBP transactivation, while Gal4 alone and Gal4 Sp1 were unaffected (Fig. 1B). Similar results were observed with 293 cells, COS cells, and human foreskin fibroblasts (data not shown and Fig. 2). These results confirmed that the transcriptional activities of both p300 and CBP are specifically induced by p21.
FIG. 1.
Transactivation by p300 and CBP is stimulated by p21. (A) Schematic diagrams of Gal4 p300 and Gal4 CBP showing the locations of the three cysteine- and histidine-rich domains (C/H), the bromodomain (Bromo), and the HAT domain. (B) Induction of Gal4 p300 and Gal4 CBP activity in U2OS cells. U2OS cells were transfected with 5 μg of the Gal4 E1B CAT reporter plasmid and expression plasmids encoding either Gal4 alone (100 ng), Gal4 Sp1 (5 ng), Gal4 p300 (full length) (100 ng) or Gal4 CBP (full length) (100 ng) as indicated. Four micrograms of Rous sarcoma virus (RSV) p21 or an RSV control (−p21) was included as indicated. Results are the means of at least three separate experiments (with standard deviations).
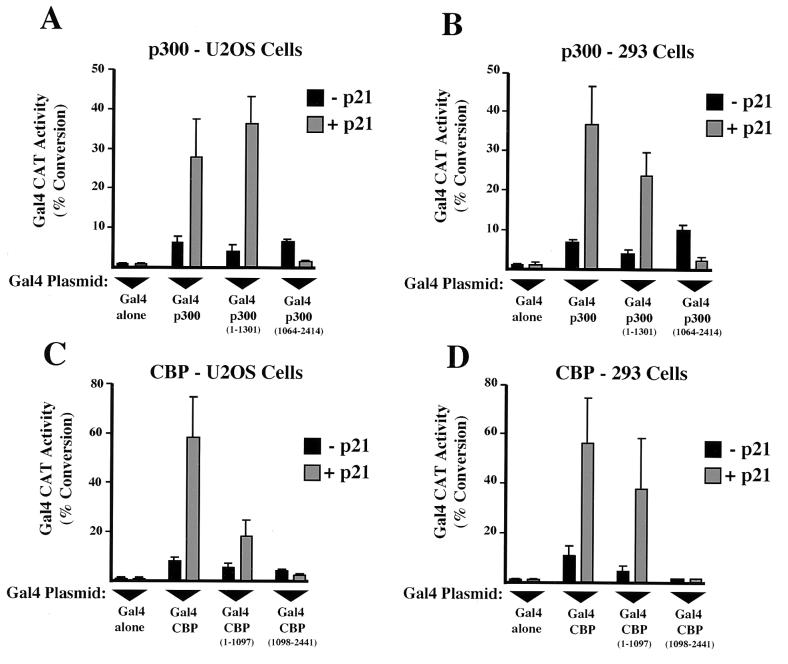
FIG. 2.
p21 stimulates transactivation by the amino termini of p300 and CBP. Transactivation by the amino-terminal domains of p300 and CBP is induced by p21 in U2OS and 293 cells. U2OS and 293 cells were transfected as for Fig. 1 with the indicated Gal4 p300 and Gal4 CBP expression plasmids. The levels of each Gal4 plasmid used were 50 ng (A); 25 ng (B); 100, 25, 12.5, and 100 ng for Gal4 alone, Gal4 CBP, Gal4 CBP (1-1097), and Gal4 CBP (1098-2441), respectively (C); and 25, 10, 10, and 50 ng for Gal4 alone, Gal4 CBP, Gal4 CBP (1-1097), and Gal4 CBP (1098-2441), respectively (D). Results are the means of at least three separate experiments (with standard deviations).
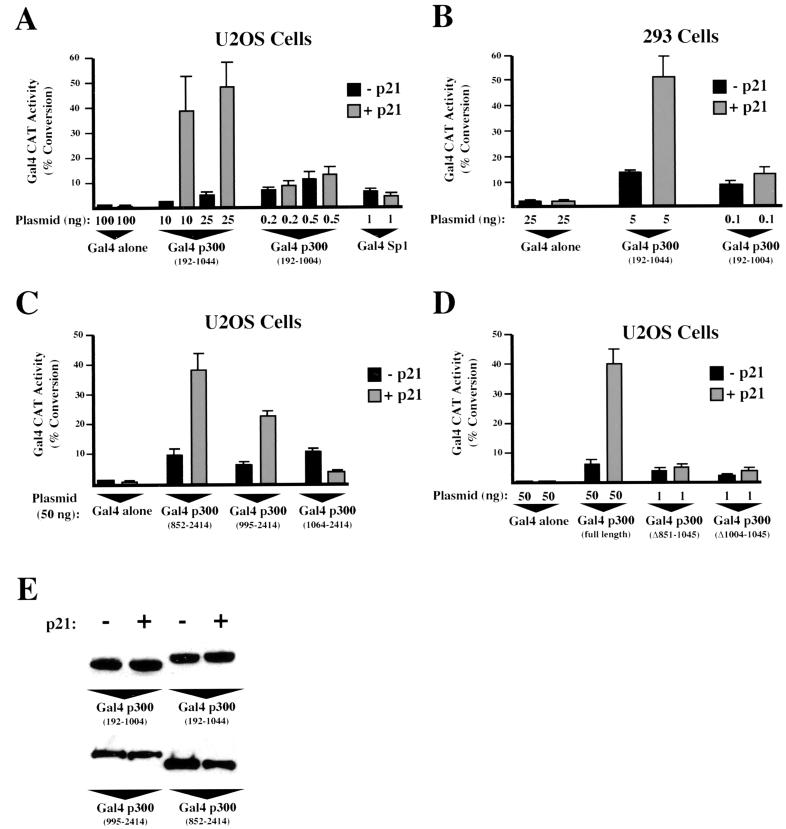
It has been shown previously that p300 and CBP bind a cyclinE-Cdk2 complex through a carboxy-terminal domain (1, 11, 34) and that the activity of this kinase is inhibited by p21 (34). To determine whether p21-induced transactivation occurred through modulation of this complex, additional Gal4 fusion plasmids encoding either the amino- or carboxy-terminal domains of p300 and CBP were constructed. Surprisingly, when these plasmids were cotransfected with Gal4 E1B CAT into either U2OS or 293 cells, p21 inducibility was only observed through the amino-terminal constructs (Fig. 2), indicating that this effect occurs independently of the previously characterized cyclinE-Cdk2 complex (11, 34) and the inherent HAT activity of p300 and CBP (5, 33). Interestingly, the carboxy-terminal p300 construct (amino acids 1064 to 2414) was repressed by coexpression of p21 (Fig. 2A and B).
Identification of a transcriptional repression domain within p300.
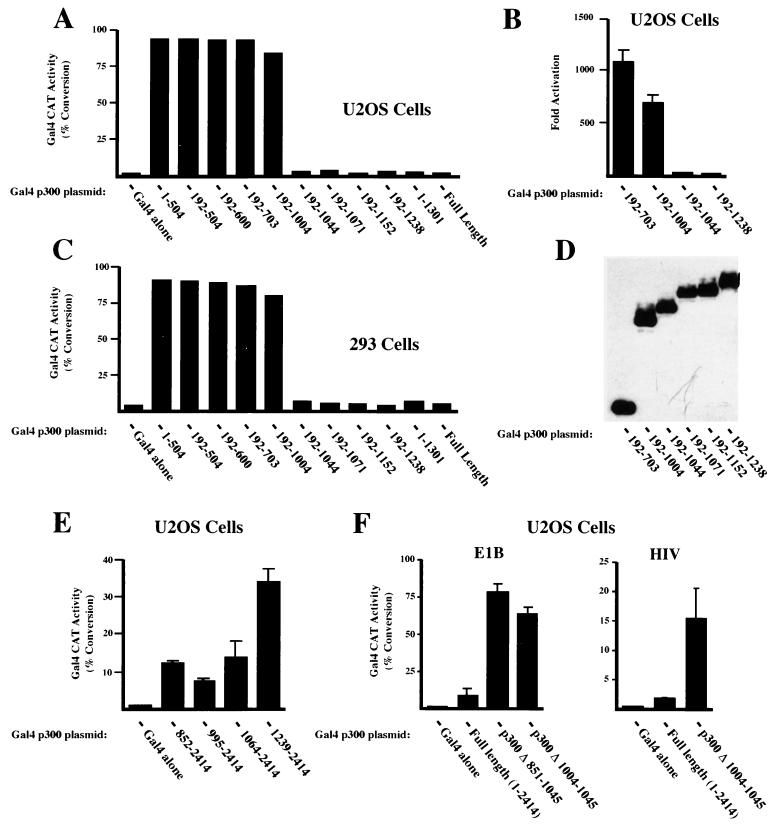
Since both p300 and CBP are similarly regulated by p21, it was decided to concentrate on p300 to further characterize this effect. A series of additional mutants with deletions from the carboxy terminus of p300 were then constructed and fused to Gal4. Cotransfection with Gal4 E1B CAT produced a striking result. While longer p300 constructs were relatively transcriptionally inactive, deletion from amino acid 1044 to 1004 resulted in a very strong increase in transactivation in both U2OS and 293 cells (Fig. 3A to C). Mutants with further deletions from the carboxy terminus were similarly transcriptionally active. Additional experiments using a range of protein concentrations in the CAT assay revealed that this represented an 85-fold stimulation of transcriptional activity in U2OS cells (Fig. 3B). Western blot analysis demonstrated that no significant differences in expression level could be observed with these proteins (Fig. 3D).
FIG. 3.
p300 transactivation is regulated by a domain located between amino acids 995 and 1044. U2OS (A, B, E, and F) or 293 (C) cells were transfected with 5 μg of the Gal4 E1B CAT reporter plasmid and 10 (A), 100 (B), 5 (C), 50 (E), or 50 ng (F) of the indicated Gal4 p300 expression plasmids. In panel F 5 μg of the Gal4 HIV CAT reporter plasmid was used where indicated. Results are the means of at least three separate experiments (with standard deviations). (D) 293 cells were transfected with 100 ng of the indicated Gal4 p300 fusion proteins. Whole-cell lysates were prepared, resolved by SDS-PAGE, and immunoblotted with anti-Gal4 antibody.
To further examine the function of this domain, and since the Gal4 fusion protein previously used to analyze the function of the carboxy terminus of p300 (Fig. 2) did not extend to the region of amino acids 1004 to 1044, additional plasmids were constructed. In contrast to the results seen with the amino terminus of p300, transactivation by both Gal4 p300 (852-2414) and Gal4 p300 (995-2414) was similar to transactivation by Gal4 p300 (1064-2414) in U2OS cells (Fig. 3E). Upon a further deletion to amino acid 1239, an increase in transcriptional activity was observed, but this was much less than that seen in Fig. 3A. Again no significant differences in expression level could be observed (data not shown). Importantly, however, deletion of amino acids 850 to 1045 and 1004 to 1045 within the context of full-length p300 also resulted in a strong increase in transactivation relative to that for the wild-type protein in U2OS cells (Fig. 3F). These experiments utilized the Gal4 E1B reporter plasmid, which contains the TATA box from the adenovirus E1B promoter. To test whether the same effect was observed with other reporter plasmids, a Gal4 HIV CAT reporter, in which Gal4 sites have been inserted immediately upstream of the human immunodeficiency virus type 1 TATA box was used. A similar increase in transactivation was observed using this reporter plasmid upon deletion of amino acids 1004 to 1045 within full-length Gal4 p300 (Fig. 3F). Western blot analysis demonstrated that these fusions were expressed as full-length proteins (data not shown).
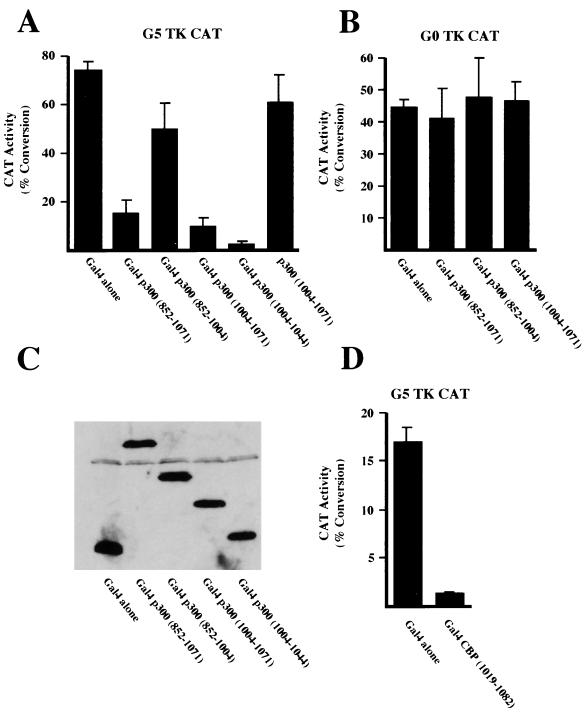
The results shown above are consistent with p300 containing a transcriptional repression domain, the activity of which is abolished by deletion of the region of amino acids 1004 to 1044. Moreover, this domain appeared to specifically repress the activity of the amino-terminal transactivation domain of p300 (Fig. 3). To test whether this domain was capable of repressing transcription in isolation or whether its activity could only function in the context of the amino terminus of p300, Gal4 fusion proteins containing the amino acid 1004 to 1044 domain alone were cotransfected with a G5 TK CAT reporter plasmid, which contains Gal4 DNA binding sites upstream of the thymidine kinase (TK) promoter and thus has a high intrinsic level of expression. Interestingly, p300 constructs containing amino acids 1004 to 1044 were strong repressors of the TK promoter while the adjacent region from amino acids 852 to 1004 had no suppressive activity (Fig. 4A). This effect required targeting to the promoter since a plasmid lacking Gal4 DNA binding sites (G0 TK CAT) was not inhibited (Fig. 4B) while p300 (1004-1071) not fused to Gal4 did not significantly affect the transcriptional activity of G5 TK CAT (Fig. 4A). Similar results were obtained with 293 cells (data not shown). Western blot analysis demonstrated that the levels of expression of these Gal4 p300 fusion proteins were equivalent (Fig. 4C). Taken together, these results demonstrate that p300 contains a domain located between amino acids 1004 and 1044 that can function as a transcriptional repressor. Interestingly, the equivalent domain of CBP could also repress transcription: a Gal4 fusion of CBP containing amino acids 1019 to 1082 was also capable of strongly repressing G5 TK CAT activity (Fig. 4D).
FIG. 4.
p300 contains a transcriptional repression domain. (A and B) U2OS cells were transfected with 5 μg of either the G5 TK CAT or G0 TK CAT reporter plasmids and 250 ng of the indicated Gal4 p300 expression plasmids. (C) 293 cells were transfected with 100 ng of the indicated Gal4 p300 fusion proteins. Whole-cell lysates were prepared, resolved by SDS-PAGE, and immunoblotted with anti-Gal4 antibody. (D) U2OS cells were transfected with 5 μg of the G5 TK CAT reporter plasmids and 250 ng of the indicated Gal4 or Gal4 CBP (1019-1082) expression plasmids. Results are the means of at least three separate experiments (with standard deviations).
Identification of the p21-regulated domain.
To determine whether p21 inducibility of p300 transactivation resulted from the action of this transcriptional repression domain, further experiments using these Gal4 fusion proteins were performed. Cotransfection of p21 with Gal4 p300 (192-1044) demonstrated that this construct retained strong p21 inducibility (Fig. 5A and B). In contrast, Gal4 p300 (192-1004) was not induced by p21. In the latter case, much lower levels of plasmid had to be used in this experiment in order to obtain results in the linear range of the CAT assay. All other amino-terminal p300 Gal4 fusion proteins containing the region from amino acid 1004 to 1044 shown in Fig. 3 were similarly strongly induced by p21, while those lacking this domain were not (data not shown). Western blot analysis demonstrated that induction of p300 transactivation did not result from increases in the expression levels of these plasmids (Fig. 5E). p21 inducibility of these plasmids was also dependent on the presence of an amino-terminal transactivation domain (51) since neither the plasmid encoding Gal4 p300 (600-1071) nor other plasmids encoding proteins lacking the amino-terminal 600 amino acids are transcriptionally active or stimulated by cotransfection of p21 (data not shown).
FIG. 5.
p300 contains a p21-responsive transcriptional repression domain. (A to D) Deletion of the p300 transcriptional repression domain abolishes p21 inducibility. U2OS (A, C, and D) and 293 (B) cells were transfected with 5 μg of the Gal4 E1B CAT reporter plasmid and the indicated Gal4, Gal4 Sp1, or Gal4 p300 expression plasmids at the levels shown. Four micrograms of Rous sarcoma virus (RSV) p21 or an RSV control (−p21) was included as indicated. Results are the means of at least three separate experiments (with standard deviations). (E) Western blot analysis of the indicated Gal4 p300 fusion proteins expressed in 293 cells. One hundred nanograms of each Gal4 p300 expression plasmid was transfected, and 4 μg of RSV p21 or an RSV control was included as indicated.
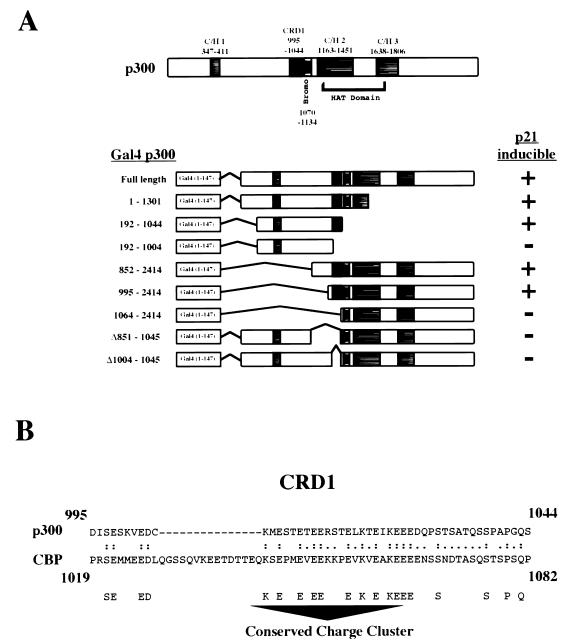
Surprisingly, although the region of amino acids 1004 to 1044 did not actively repress transactivation by the carboxy terminus of p300, it did regulate p21 inducibility. Similar to what was found for the amino terminus, Gal4 fusions of the carboxy terminus of p300 that contained the region of amino acids 1004 to 1044 were inducible by cotransfected p21 using the Gal4 E1B reporter plasmid. Deletion of this domain abolished this effect, resulting in a construct that was now repressed by p21 (Fig. 5C). Underlining the importance of this domain, internal deletion of the region of amino acids 1004 to 1044 also abolished p21 induction of transactivation of full-length p300 (Fig. 5D). These results are summarized in Fig. 6A. Western blot analysis again confirmed that p21 induction did not result from an increase in the expression levels of these proteins (Fig. 5E).
FIG. 6.
(A) Schematic diagram showing the location of the p21-inducible transcriptional repression domain (CRD1) in relationship to those of other defined motifs in p300 and summary of the deletion data in Fig. 5A to D. C/H, cysteine- and histidine-rich domain; Bromo, bromodomain. (B) Diagram showing the amino acid sequence of CRD1 in p300 and the corresponding sequence in murine CBP. Identical residues, including a conserved charge cluster domain, are indicated underneath. Sequence alignment was performed using a FASTA3 search at the European Bioinformatics Institute website (http://www2.ebi.ac.uk/fasta3/).
Thus, p300 contains a p21-regulated transcriptional repression domain from amino acid 1004 to 1044, which we have termed cell cycle regulatory domain 1 (CRD1) (Fig. 6B).
p21 regulation of p300 is dependent on the core promoter.
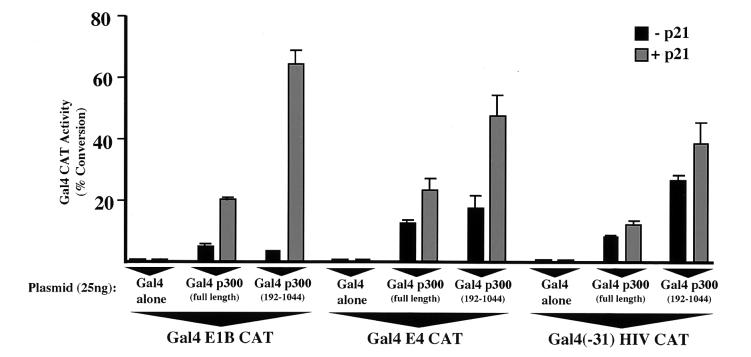
A number of laboratories have shown that the requirement for different domains of p300 and CBP can vary depending on the context within which they are functioning (22, 23, 28, 46). Since most of our previous experiments (Fig. 1 to 3 and 5) utilized the Gal4 E1B reporter plasmid, we decided to determine whether the nature of the core promoter could influence the ability of p21 to regulate p300 transcriptional activity. When a Gal4 E4 CAT reporter plasmid containing the adenovirus E4 core promoter was used, both Gal4 p300 (full length) and Gal4 p300 (192-1044) were still p21 inducible, although less so than with the Gal4 E1B CAT reporter (Fig. 7). Surprisingly, however, with the Gal4 HIV CAT reporter p300 was no longer significantly p21 inducible (Fig. 7). Although the repression function of CRD1 still functions with the Gal4 HIV CAT reporter (Fig. 3F), with both the E4 and human immunodeficiency virus core promoters, p300 displayed a higher intrinsic level of transactivation than with the Gal4 E1B CAT reporter. With lower levels of the p300 plasmid, where less transactivation was seen, these reporters were still unresponsive to p21 stimulation (data not shown).
FIG. 7.
The activity of CRD1 is dependent on the core promoter. U2OS cells were transfected with 5 μg of the indicated Gal4 CAT reporter plasmids and 25 ng of either Gal4, Gal4 p300, or Gal4 p300 (192-1044) expression plasmids. Four micrograms of Rous sarcoma virus (RSV) p21 or an RSV control (−p21) was included as indicated. Results are the averages of three separate experiments (with standard deviations).
CRD1 still represses transcription when p300 is not fused to Gal4.
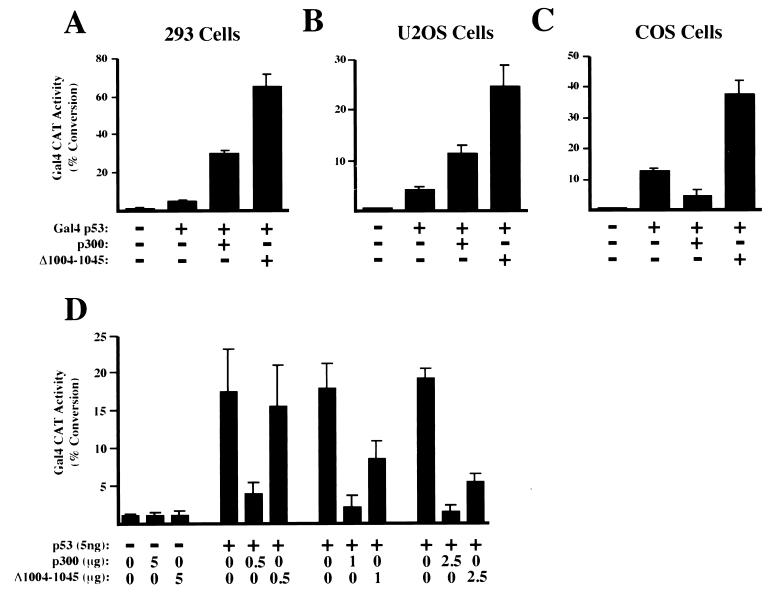
Although CRD1 clearly exerted a strong regulatory function on p300 as a Gal4 fusion, it was important to demonstrate similar behavior when p300 was recruited to DNA through an interaction with a heterologous DNA binding protein. To determine if this was the case, p300 not fused to Gal4 was cotransfected with a Gal4 p53 activation domain fusion protein and the Gal4 E1B CAT reporter plasmid in 293 cells. As expected, wild-type p300 strongly stimulated transactivation through Gal4 p53 while no effect was seen with the reporter plasmid alone (Fig. 8A and data not shown). Cotransfection of p300 (Δ1004 to 1045) consistently produced a significantly higher level of coactivation than cotransfection of wild-type p300 (Fig. 8A). Similar results were observed with U2OS cells (Fig. 8B). Surprisingly, in COS cells, wild-type p300 repressed Gal4 p53, while p300 (Δ1004 to 1045) still strongly functioned as a coactivator (Fig. 8C). To investigate this further, experiments were performed with full-length p53 and the promoter of the Bax gene, a known p53 target gene which is induced during apoptosis (29). p53 strongly stimulated transcription from the Bax CAT reporter plasmid. Wild-type p300 again strongly repressed p53 transactivation, and, although repression was still seen with p300 (Δ1004 to 1045), it was strongly reduced (Fig. 8D). Similar results were observed with the Bax promoter in U2OS cells and with p53 transactivation of p21WAF1/CIP1 CAT and PG13 CAT (which contains multimerized p53 binding sites) in COS cells (data not shown). Thus, although p300 can have diverse effects on transcription, CRD1 can function as a repression domain when not fused to Gal4.
FIG. 8.
Analysis of CRD1 function in non-Gal4-fused p300. 293 (A), U2OS (B), and COS (C) cells were transfected with 5 μg of the Gal4 E1B CAT reporter plasmid, 2.5 ng (A and B) or 5 ng (C) of the Gal4 p53 expression plasmid, and 2.5 μg of the p300 or p300(Δ1004-1045) expression plasmid as indicated. (D) COS cells were transfected with 5 μg of the Bax CAT reporter plasmid together with the indicated quantities of p53, p300, or p300(Δ1004-1045) expression plasmids. Results are the means of at least three separate experiments (with standard deviations).
DISCUSSION
In this report we have characterized the regulation of p300 and CBP by the CDK inhibitor p21. We have shown that p300 contains a previously undescribed and novel transcriptional repression domain located between amino acids 1004 and 1044, which we have designated CRD1 (Fig. 6). CRD1 can function to repress transcription both in isolation (Fig. 4) and within the context of p300 (Fig. 3 and 8). p21 stimulation of p300 transcriptional activity is abolished by deletion of this domain (Fig. 5). Importantly, we have also demonstrated that the activity of CRD1 is dependent on the structure of the core promoter (Fig. 7). This promoter specificity is reminiscent of previous reports examining the domains within p300 and CBP that are utilized. For example, STAT1 has been shown to require the CH3 domain of CBP while the retinoic acid receptor (RAR) does not (23). Furthermore, while CREB and STAT1 require CBP HAT activity, the RAR does not and utilizes P/CAF instead (22). Similarly, the requirement for amino-terminal versus carboxy-terminal activities and the inherent HAT activity of p300 and CBP vary according to the cellular stimuli for Pit1-mediated transactivation (46). It is possible that this differential usage of p300 domains and coactivators is responsible for the divergent results seen upon p300 cotransfection in Fig. 8. While these experiments clearly demonstrated the repressive function of CRD1 when not fused to Gal4, inhibition of transactivation by p300 itself should be viewed with some caution. For example, should COS cells have low levels of an essential p300-interacting protein such as P/CAF, overexpression of transiently transfected p300 would have the effect of diluting out these transcriptionally active complexes, leading to an apparent repression of transcription. We were unable to observe stimulation of transcription by lower levels of wild-type p300 in COS cells (data not shown), however, and the repressive effects seen are consistent with CRD1 having a dominant effect on transcriptional activation under some circumstances. The cell type, transcription factor, and promoter specificity of these p300-repressive effects will be the subject of future investigations.
The region of CBP equivalent to p300 CRD1 also represses transcription, demonstrating that this effect is functionally conserved between both proteins (Fig. 4). A comparison of these domains in p300 and CBP reveals a conserved sequence of highly charged amino acids (Fig. 6B). While many transactivation and repression domains contain highly charged domains (6, 41), we have not so far determined the significance of this motif with respect to p300 function. It is unclear at present through what mechanism CRD1 represses transcription and mediates p21 stimulation of p300. We have also found that a mutant transdominant-negative CDK2 expression plasmid has effects on CRD1 similar to those of p21 in 293 cells (data not shown). Furthermore, p21 does not directly bind CRD1 (data not shown), suggesting that regulation of CRD1 is indirect and arises from cell cycle arrest. Although CRD1 functions to repress transcription at all promoters tested, p21 stimulation through this domain appears to be highly selective and specific (Fig. 7). We have found that in addition to not stimulating p300 transactivation with the Gal4 HIV CAT reporter plasmid, p21 is unable to reverse repression of G5 TK CAT by Gal4-fused CRD1 (data not shown). Thus, while it is probable that a cosuppressor complex interacts directly with CRD1, it does not appear likely that its interaction with p300 or its intrinsic function is cell cycle regulated. Rather, it is likely that cell cycle-regulated components of the basal transcription complex or pol II holoenzyme are selectively required at different promoters and function as the target for CRD1. The elucidation of the precise mechanism through which CRD1 functions will be the subject of future investigations.
Previously, it has been shown that a cyclinE-Cdk2 complex binds the carboxy terminus of CBP and stimulates its intrinsic HAT activity (1). With the reporter plasmids and cell lines used in this study, we have not been able to observe stimulation of transcription by the HAT domain of p300 alone (data not shown). We observed, however, that in the absence of CRD1, p21 represses the transcriptional activity of the carboxy terminus of p300 (Fig. 2 and 5). Further mapping of this effect has demonstrated that this results from repression of a transactivation domain at the very carboxy terminus of p300 and occurs independently of the HAT domain (data not shown). While this effect correlates with the previously described binding site for cyclinE-Cdk2 within p300 (11), we have not yet precisely mapped the effect. We cannot rule out, however, the possibility that cell cycle regulation of the HAT activity of p300 does occur, but given the differential usage of this HAT domain (22, 46) we suggest that the effect of this is also promoter context dependent.
p300 and CBP are required both for E2F activity, which promotes transition from G1 to S phase (25, 35, 44), and for p53 and MyoD function, which promotes cell cycle arrest (reviewed in reference 38). The presence of multiple cell cycle-regulated domains within p300 suggests a model for how this coactivator might function as a regulator of both growth arrest and cellular proliferation. Promoters requiring the inherent HAT activity or other transactivation functions contained within the carboxy termini of p300 and CBP will be suppressed following cell cycle arrest through the p21-mediated inhibition of the cyclinE-Cdk2 activity binding the carboxy termini of p300 and CBP. In contrast, at other promoters where this inherent HAT or carboxy-terminal activity is not required, possibly as a result of the involvement of P/CAF or other proteins interacting with p300 and CBP, p21 can stimulate p300 and CBP activity through CRD1. Thus, we speculate that p300 and CBP activity will have a central regulatory role in the response to signals inducing growth or differentiation and will direct the switch from using growth-promoting genes to using those required following cell cycle arrest.
ACKNOWLEDGMENTS
We thank Neil Chapman, Louise Copeland, David Gregory, and Pavel Cabart for their help and assistance on this project, Stefan Roberts, Gary Nabel, and Richard Goodman for providing invaluable reagents, and members of the Division of Gene Regulation and Expression at the University of Dundee for their helpful comments, support, and critical reading of the manuscript.
This work was funded through a grant from the BBSRC Integrated Cellular Responses Initiative (L.A.A.) and a BBSRC studentship (A.W.S.). Additional funding was provided from the MRC (G.A.W.), Tenovus, and a Royal Society University Fellowship (N.D.P.).
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1A oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 4.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Brown H J, Sutherland J A, Cook A, Bannister A J, Kouzarides T. An inhibitor domain In C-Fos regulates activation domains containing the Hob1 motif. EMBO J. 1995;14:124–131. doi: 10.1002/j.1460-2075.1995.tb06982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H W, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 8.Chawla S, Hardingham G E, Quinn D R, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felzien L K, Farrell S, Betts J C, Mosavin R, Nabel G J. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol Cell Biol. 1999;19:4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giles R H, Peters D J M, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 13.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 14.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–120. [PubMed] [Google Scholar]
- 15.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 17.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y J, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 18.Hartikka J, Sawdey M, Cornefert-Jensen F, Margalith M, Barnhart K, Nolasco M, Vahlsing H L, Meek J, Marquet M, Hobart P, Norman J, Manthorpe M. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum Gene Ther. 1996;7:1205–1217. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- 19.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hottiger M O, Felzien L K, Nabel G J. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S, Heymann R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 22.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 24.Kwok R P S, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriot S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the coactivator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee C W, Sorensen T S, Shikama N, La Thangue N B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 26.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y Z, Chrivia J C, Latchman D S. Nerve growth factor up-regulates the transcriptional activity CBP through activation of the p42/p44(MAPK) cascade. J Biol Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita T, Reed J C. Tumor-suppressor p53 is a direct transcriptional activator of the human Bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 30.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong J, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 31.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90rsk. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 32.Nemethova M, Wintersberger E. Polyomavirus large T antigen binds the transcriptional coactivator protein p300. J Virol. 1999;73:1734–1739. doi: 10.1128/jvi.73.2.1734-1739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 34.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 35.Qin X Q, Livingston D M, Kaelin W G, Adams P D. Deregulated transcription factor E2f-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadowski I, Ptashne M. A vector for expressing Gal4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Curr Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 38.Snowden A W, Perkins N D. Cell cycle regulation of the transcriptional coactivators p300 and CREB binding protein. Biochem Pharmacol. 1998;55:1947–1954. doi: 10.1016/s0006-2952(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 39.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 42.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 43.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional coactivator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 44.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster G A, Perkins N D. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal A K, Rose D W, Glass C K, Rosenfeld M G. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 47.Yakiuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kilodalton product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 49.Yao T, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao T P, Oh S P, Fuchs M, Zhou N D, Chng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida E, Nakajima T, Murakami K, Fukamizu A. Identification of N-terminal minimal transactivation domain of CBP, p300 and Caenorhabditis elegans homologues. Gene. 1998;208:307–314. doi: 10.1016/s0378-1119(98)00008-0. [DOI] [PubMed] [Google Scholar]
- 52.Zhong H H, Voll R E, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]