Abstract
Phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF-2α) is a well-characterized mechanism regulating protein synthesis in response to environmental stresses. In the yeast Saccharomyces cerevisiae, starvation for amino acids induces phosphorylation of eIF-2α by Gcn2 protein kinase, leading to elevated translation of GCN4, a transcriptional activator of more than 50 genes. Uncharged tRNA that accumulates during amino acid limitation is proposed to activate Gcn2p by associating with Gcn2p sequences homologous to histidyl-tRNA synthetase (HisRS) enzymes. Given that eIF-2α phosphorylation in mammals is induced in response to both carbohydrate and amino acid limitations, we addressed whether activation of Gcn2p in yeast is also controlled by different nutrient deprivations. We found that starvation for glucose induces Gcn2p phosphorylation of eIF-2α and stimulates GCN4 translation. Induction of eIF-2α phosphorylation by Gcn2p during glucose limitation requires the function of the HisRS-related domain but is largely independent of the ribosome binding sequences of Gcn2p. Furthermore, Gcn20p, a factor required for Gcn2 protein kinase stimulation of GCN4 expression in response to amino acid starvation, is not essential for GCN4 translational control in response to limitation for carbohydrates. These results indicate there are differences between the mechanisms regulating Gcn2p activity in response to amino acid and carbohydrate deficiency. Gcn2p induction of GCN4 translation during carbohydrate limitation enhances storage of amino acids in the vacuoles and facilitates entry into exponential growth during a shift from low-glucose to high-glucose medium. Gcn2p function also contributes to maintenance of glycogen levels during prolonged glucose starvation, suggesting a linkage between amino acid control and glycogen metabolism.
Phosphorylation of eukaryotic initiation factor 2 (eIF-2) is an important mediator of translational control in response to environmental stresses that is conserved from yeast to mammals (11, 15, 36, 52). eIF-2 delivers initiator Met-tRNA to the translational machinery by a mechanism requiring the hydrolysis of GTP associated with eIF-2 (32). Phosphorylation of the α subunit of eIF-2 at serine-51 impedes the guanine nucleotide exchange reaction that recycles eIF-2–GDP to the functional GTP-bound form. A family of protein kinases have been described that catalyze eIF-2α phosphorylation in response to a number of different stresses, including nutrient deprivation, viral infection, heat shock, and ischemia. Among mammals, the characterized eIF-2α kinases include the RNA-dependent protein kinase PKR, which is important for an antiviral defense pathway mediated by interferon (40); the heme-regulated inhibitor kinase HRI, which reduces protein synthesis in erythroid tissues in response to iron deficiency (9); and the pancreatic eIF-2α kinase PEK, which responds to stress in the endoplasmic reticulum and has also been called PKR-like endoplasmic reticulum kinase (20, 46, 48). Phosphorylation of eIF-2α reduces protein synthesis, facilitating a coordinated strategy to remedy the stress-induced problems in mammalian cells.
In contrast to the many different eIF-2α kinases present in mammals, the yeast Saccharomyces cerevisiae has only a single eIF-2α kinase, Gcn2p (23, 52). Furthermore, Gcn2 protein kinase does not regulate total protein synthesis in yeast but rather stimulates the expression of a single species of mRNA, encoding Gcn4p, in response to amino acid starvation (24). Gcn4p is a transcriptional activator of genes involved in the synthesis of amino acids. Regulation of GCN4 translation involves four short open reading frames (ORFs) located in the 5′ noncoding portion of the GCN4 mRNA (1, 24). In cells not limiting for amino acids, the upstream ORFs block translation of the GCN4 coding sequences. During starvation for one of several different amino acids, Gcn2p phosphorylation of eIF-2α leads to reduced eIF-2–GTP levels, alleviating the inhibitory effects of the upstream ORFs and allowing for increased GCN4 translation. Elevated levels of Gcn4p stimulate the expression of enzymes required to synthesize many different amino acids (22).
Induction of Gcn2 protein kinase activity by amino acid limitation is mediated by a Gcn2p domain homologous with histidyl-tRNA synthetase (HisRS) enzymes (54). Uncharged tRNAs were shown to directly bind with the HisRS-related region, and mutations in this domain that block tRNA interaction also abolish kinase function in vitro and in vivo (56). These studies suggest that the HisRS-related domain of Gcn2p can associate with multiple species of uncharged tRNAs that accumulate in cells starved for amino acids, facilitating activation of the kinase and phosphorylation of eIF-2α. Purine starvation also enhances GCN4 expression by Gcn2p, supporting the idea that there is coordinated regulation of nucleotide and amino acid biosynthetic pathways (43). A second RNA-binding region is found in the carboxy terminus of Gcn2p. This domain was reported to be required for association of Gcn2p with ribosomes, and adjacent sequences are proposed to mediate dimerization between Gcn2p molecules (41, 42, 60). Ribosomal association of Gcn2p is required for GCN4 translational control, and this interaction may support monitoring of uncharged tRNA levels in cells. Monitoring of uncharged tRNA levels by the HisRS-related sequences of Gcn2p may also be facilitated by Gcn1p and Gcn20p, which are associated with ribosomes and are required for high levels of eIF-2α phosphorylation by Gcn2p during amino acid starvation conditions (30, 31, 51).
Given that eIF-2α phosphorylation in mammals is induced in response to both carbohydrate and amino acid limitations (11, 15, 37), we addressed whether Gcn2p control of GCN4 expression in yeast can be controlled by different nutrient limitations. We report that (i) starvation for glucose induces Gcn2p phosphorylation of eIF-2α and stimulates GCN4 translational expression and (ii) distinct mechanisms regulate Gcn2 protein kinase activity during amino acid limitation and in response to carbohydrate deficiency. Gcn2 protein kinase enhances storage of amino acids in the vacuoles and contributes to the maintenance of glycogen pools in response to glucose starvation.
MATERIALS AND METHODS
Plasmids and strains.
Plasmid p180 expresses a GCN4-lacZ fusion including the entire GCN4 5′ noncoding region with four upstream ORFs inserted into YCp50, a low-copy-number plasmid marked with URA3 (34). Plasmid p227 is a similar construct with mutations in the initiation codon of each of the four upstream ORFs of GCN4 (34). A 6-kb SalI-to-EcoRI fragment expressing GCN4-lacZ was removed from plasmid p180 and inserted between the SalI and EcoRI restriction sites of the low-copy-number TRP1-based plasmid pRS314 (47), generating plasmid pYB41. Plasmids expressing different mutant versions of GCN2 were marked by URA3 and are either high or low copy number, as indicated.
Strains used in this study are listed in Table 1. The four principal strain backgrounds (EG328-1A, JC482, H1896, and F113) were each derived in part from strain S288C. EG328-1A and JC482 are also derived from crosses with W303. All RY-designated strains were derived from EG328-1A (21). Plasmid p180 and p227 were transformed into EG328-1A and selected for by uracil prototrophy, generating strains RY124 and RY127, respectively. GCN2 was deleted in strain EG328-1A by transforming a linear fragment expressing gcn2::LEU2 derived from p500 after BamHI digestion (55). Transformed cells were assayed for sensitivity to 3-aminotriazole (3-AT), and deletion of GCN2 was confirmed by mating with strain H1069 (gcn2Δ); the resulting gcn2::LEU2 strain was designated RY139. Plasmids p180 and p227 were introduced into RY139, producing RY133 and RY134, respectively. Strain RY196 was produced by introducing pYB41 into RY139. The one-step PCR method was used to replace the entire coding regions of GCN1 with TRP1 in RY124, generating strains RY281, and the GCN20 ORF was replaced with TRP1 in RY282. The gcn1::TRP1 and gcn20::TRP1 knockout strains displayed growth sensitivity to 3-AT and did not complement the impaired general control pathway when mated with their respective mutant counterparts, H2079 (gcn1Δ) and H2512 (gcn20Δ). To delete GCN3 in RY124, a 3.7-kb EcoRI-to-PvuI restriction fragment expressing gcn3::LEU2 was purified from Ep149 (18), transformed into this strain, and selected for by leucine prototrophy. The resulting strain, RY283, was growth sensitive to 3-AT and did not complement the amino acid biosynthetic defect associated with strain H1395 (gcn3::LEU2). Strains RY284 (gcn1::TRP1), RY285 (gcn20::TRP1), and RY286 (gcn3::LEU2) contain p227. GCN4 was deleted in EG328-1A by digesting p401 expressing gcn4::LEU2 with PstI and PvuII and transforming the digested DNA into this strain, followed by selection for leucine prototrophy. The resulting strain, RY290, was severely impaired for growth in SD medium (see below) in the absence of amino acids and did not complement the strain H1716 (gcn4-Δ1). The SUI2-S51A allele was introduced into EG328-1A by “pop-in/pop-out allele replacement” (44), which involves transforming p594 linearized with BglII into RY124 and selecting for uracil prototrophy. Plasmid p594 contains SUI2-S51A inserted into pRS306, a URA3-based integrating plasmid (47). Transformants containing integrated SUI2-S51A were grown in the presence of 5-fluoroorotic acid to select for loss of URA3 and plasmid excision. The resulting strain, RY287, was growth sensitive to 3-AT; this sensitivity could be overcome by introducing wild-type SUI2 on a low-copy-number plasmid into this strain.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| EG328-1A | MATα ura3-52 leu2 trp1 | 21 |
| F113 | MATa ino1 ura3 can1 | 1 |
| H1069 | MATa ura3-52 leu2-3 leu2-112 gcn2::LEU2 | A. Hinnebusch |
| H1395 | MATa ura3-52 leu2-3 leu2-112 gcn3::LEU2 [HIS4-lacZ, ura3-52] | 19 |
| H1716 | MATa ura3-52 leu2-3 leu2-112 gcn4-Δ1 inol-13 his3-609 | A. Hinnebusch |
| H1896 | MATa ura3-52 leu2-3 leu2-112 trp1Δ63::[GCN4-lacZ, TRP1] sui2Δ gcn2Δ p1097[SUI2, LEU2] | 16 |
| H2079 | MATa ino1 ura3 gcn1Δ can1 | 30 |
| H2512 | MATa ino1 ura3 gcn20Δ can1 | 51 |
| JC482 | MATα ura3-52 leu2 his4-359 | 8 |
| RY124 | MATα ura3-52 leu2 trp1 p180[GCN4-lacZ, URA3] | This study |
| RY127 | MATα ura3-52 leu2 trp1 p227[uORFsΔ-GCN4-lacZ, URA3] | This study |
| RY133 | MATα ura3-52 leu2 trp1 gcn2::LEU2 p180[GCN4-lacZ, URA3] | This study |
| RY134 | MATα ura3-52 leu2 trp1 gcn2::LEU2 p227[uORFsΔ-GCN4-lacZ, URA3] | This study |
| RY139 | MATα ura3-52 leu2 trp1 gcn2::LEU2 | This study |
| RY196 | MATα ura3-52 leu2 trp1 gcn2::LEU2 pYB41[GCN4-lacZ, TRP1] | This study |
| RY281 | MATα ura3-52 leu2 trp1 gcn1::TRP1 p180[GCN4-lacZ, URA3] | This study |
| RY282 | MATα ura3-52 leu2 trp1 gcn20::TRP1 p180[GCN4-lacZ, URA3] | This study |
| RY283 | MATα ura3-52 leu2 trp1 gcn3::LEU2 p180[GCN4-lacZ, URA3] | This study |
| RY287 | MATα ura3-52 leu2 trp1 SUI2-S51A p180[GCN4-lacZ, URA3] | This study |
| RY290 | MATα ura3-52 leu2 trp1 gcn4::LEU2 | This study |
Growth medium and conditions.
Yeast strains were inoculated into synthetic minimal medium containing 2% glucose (SD medium) (27) with the required amino acids at 30°C with constant shaking. Cells were grown to mid-logarithmic phase with an A600 between 0.3 to 0.6 and then inoculated into fresh minimal medium containing either 2 or 0.05% glucose for nonstarvation or glucose starvation conditions, respectively. Alternatively, to elicit amino acid or purine starvation, cells were introduced into SD medium supplemented with 10 mM 3-AT or 50 μg of 8-aza-adenine (AzaA) per ml (43). Cells were harvested after 4 h of growth in nonstarvation medium or 6 h of growth under starvation conditions, unless otherwise indicated. To study the growth differences between strains RY124 and RY133, cells were grown in minimal medium supplemented with 0.05% glucose for 6 or 24 h, collected, and inoculated into SD medium. The starting cell densities of the wild-type and gcn2Δ cells in SD medium were identical; growth at 30°C was monitored by A600, and cell numbers were counted using a hemacytometer and microscopy. Furthermore, cell viability was monitored by plating onto agar medium containing 1% yeast extract, 2% peptone, and 2% glucose.
Gcn4p-LacZ enzyme assay.
Cells were grown to mid-logarithmic phase and shifted to SD medium, minimal medium containing 0.05% glucose, or SD supplemented with either 3-AT or AzaA as described above. Where indicated, all 20 amino acids were added to the minimal medium. Additionally, minimal medium supplemented with 3% glycerol or 3% ethanol was used to assess GCN4 expression in medium containing nonfermentable carbon sources. Following incubation with shaking at 30°C for 4 h in nonstarvation medium or 6 h in starvation medium, cells were chilled on ice and collected by centrifugation. Gcn4p-LacZ enzyme activity steadily increased in cells incubated in 0.05% glucose medium from 4 to 6 h. No differences in GCN4 expression was measured when cells were shifted to nonstarvation medium for 4 or 6 h. Cell pellets were resuspended in 250 μl of breaking buffer containing 0.1 M Tris-HCl (pH 8), 20% glycerol, 1 mM β-mercaptoethanol, and 100 μM phenylmethylsulfonyl fluoride in 50-ml Falcon tubes and broken by vortexing with glass beads for 2 min by 30-s intervals that were interspersed with periods of chilling on ice. Cell lysates were collected and clarified by centrifugation in a microcentrifuge at 15,000 × g. The assay for β-galactosidase enzyme activity involved mixing between 10 and 100 μl of lysate with Z buffer (100 mM sodium phosphate [pH 7.5], 10 mM KCl, 2 mM MgSO4, 4.5 mM β-mercaptoethanol) in a total volume of 1 ml. Reactions were initiated by the addition of 200 μl of ο-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml), incubated for 10 to 60 min at 30°C, and terminated by addition of freshly made 1 M Na2CO3. Sample A420 was obtained, and results were calculated as nanomoles of ONPG hydrolyzed per minute per milligram of total protein. Protein concentrations were obtained as milligrams per milliliter by the Bradford method (6).
Immunoblot analysis.
Yeast cells were grown to mid-logarithmic phase and shifted to minimal medium with 2 or 0.05% glucose or SD medium with addition of 3-AT or AzaA at 30°C. Cells were collected by centrifugation, and lysates were prepared using glass beads and vortexing. Equal amounts of protein extracts were separated by electrophoresis in a sodium dodecyl sulfate-polyacrylamide gel and transferred to nitrocellulose filters. Filters were blocked in TBS-T (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Tween 20, 4% nonfat dry milk). Phosphorylated eIF-2α was visualized by using TBS-T containing an affinity-purified antibody that specifically recognizes eIF-2α phosphorylation at serine-51, kindly provided by Gary Krause (Wayne State University) (14). Total eIF-2α in yeast lysates was detected by immunoblotting with a polyclonal antibody prepared against a polyhistidine-tagged version of yeast eIF-2α expressed and purified from Escherichia coli. Filters were washed in TBS-T, and eIF2α-antibody complex was detected using horseradish peroxidase-labeled secondary antibody and chemifluorescent substrate. The Gcn2p immunoblot analysis was similarly carried out using a polyclonal antibody that specifically recognizes Gcn2p.
Measurement of amino acid levels in the cytoplasm and vacuoles of yeast.
Strains were grown to mid-logarithmic phase and shifted to minimal medium with 2 or 0.05% glucose as described above; 5 × 108 cells were collected by centrifugation and washed twice with H2O. Amino acids levels in the cytoplasm and vacuoles were measured as previously described (35, 50). Cells were resuspended in 1.5 ml of a solution of 2.5 mM K3PO4, 0.6 M sorbitol, 10 mM glucose, and 0.2 mM CuCl2 and incubated for 20 min at 30°C (35). To determine cell density, a 10-μl aliquot of the sample suspension was assayed for A600. The remaining portion of the cell suspension was applied to a vacuum filtration apparatus, using a Millipore filter with a pore size of 0.45 μm. The filtrate was used to measure the amino acid levels in the cytoplasm as described below. To measure the vacuolar amino acid levels, permeabilized cells adhering to the filter were washed four times with 0.5 ml of a solution of 2.5 mM K3PO4 and 0.6 M sorbitol. Then the cells were resuspended into 1 ml of H2O, and a 10-μl aliquot was assayed for A600 to determine cell density. The cell suspension were boiled for 20 min and clarified by centrifugation in a microcentrifuge at 5,000 × g (35).
Total free amino acid levels in the cytoplasmic and vacuolar preparation were determined by the ninhydrin method (50). Aliquots of the samples were mixed with H2O to a final volume of 400 μl, and reactions were started by addition of 200 μl of ninhydrin reagent (Sigma) and heating at 100°C for 10 min. The reaction mixtures were cooled to room temperature, and 1 ml of 95% ethanol was added to terminate the reaction. The absorbance of the solution at 570 nm was measured, and the amounts (nanomoles) of amino acids in each sample were calculated using a standard amino acid curve. A standard curve for amino acid concentrations was prepared using a solution of 50 μM leucine in 0.05% glacial acetic acid. Measurements between 0 and 400 μl of this stock solution were characterized by the ninhydrin method. Concentrations of amino acids in each sample were normalized by cell numbers obtained from the A600 and expressed as nanomoles/A600.
Glycogen measurements.
Glycogen levels were determined as described previously (4, 28, 57). Strains RY124 and RY133 were grown in minimal medium containing either 2 or 0.05% glucose, collected by centrifugation, and lysed using glass beads and vortexing. Glycogen in lysates was digested to glucose using amyloglucosidase and α-amylase. Glucose-6-phosphate dehydrogenase and hexokinase were used to determine the released glucose level, which is proportional to the increase of NADPH measured by the extinction change at 340 nm. Different concentrations of purified glycogen were assayed to prepare a standard curve. Glycogen levels were reported as micrograms of glycogen per milligram of protein.
RESULTS
Expression of GCN4 is induced in response to starvation for glucose.
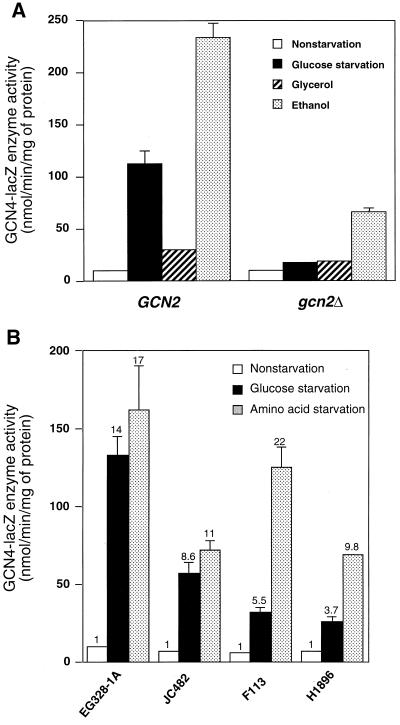
Starvation for glucose in mammalian cells leads to elevated phosphorylation of eIF-2α and altered translation initiation (37). To determine whether GCN4 control in yeast is regulated under glucose starvation conditions, we introduced plasmid p180 expressing a GCN4-lacZ fusion containing all four upstream ORFs into the wild-type strain EG328-1A and measured β-galactosidase activity under different starvation conditions. Glucose starvation was achieved by shifting cells grown to mid-logarithmic phase in SD medium into medium containing only 0.05% glucose. For experimental controls, we elicited amino acid or purine starvation by adding 3-AT, an inhibitor of the HIS3 gene product, or AzaA, a pseudo-feedback inhibitor for the first enzyme of purine biosynthesis, respectively, to SD medium. Consistent with previous studies (34, 43), expression of GCN4-lacZ was increased over 14-fold during amino acid or purine starvation compared with the nonstarved or repressed conditions (Table 2). During glucose starvation, Gcn4p-LacZ enzyme activity was also found to be increased by almost 15-fold (Table 2). To assess whether induction of GCN4 expression also occurs when cells are shifted from glucose to alternative carbon sources, we transferred cells grown in SD medium to minimal medium containing a nonfermentable carbon source, glycerol or ethanol. Incubation of GCN2 cells in minimal medium containing glycerol contributed to only a modest increase in Gcn4p-LacZ enzyme compared to nonstarved cells, while in ethanol there was a 24-fold increase (Fig. 1A). These results indicate that stimulation of GCN4 expression can occur when cells are shifted from glucose to a poor carbon source which contributes to a significantly reduced doubling time.
TABLE 2.
GCN2 protein kinase is required to stimulate GCN4 expression in response to starvation for glucose, as well as amino acid and purine limitations
| Strain/GCN2 allele | Plasmida/diagram | Gcn4p-LacZ enzyme activity (nmol/min/mg of protein)b
|
|||
|---|---|---|---|---|---|
| Glucose
|
3-AT | AzaA | |||
| 2% | 0.05% | ||||
| RY124/GCN2 | p180/□□□□ LacZ | 11 | 160 | 210* | 160* |
| RY127/GCN2 | p227/XXXX LacZ | 500 | 1,300 | 600 | 700 |
| RY133/gcn2Δ | p180/□□□□ LacZ | 9 | 16 | 15 | 13 |
| RY134/gcn2Δ | p227/XXXX LacZ | 600 | 1,200 | 600* | 600 |
Plasmid p180 encodes GCN4-lacZ with the four ORFs in 5′ noncoding region of GCN4, as illustrated by the open boxes. Plasmid p227 encodes GCN4-lacZ with substitutions destroying the function of each of the four ORFs, as denoted by the X's.
Measured in lysates prepared from the indicated isogenic strains that were cultured in minimal medium containing 2 or 0.05% glucose as described in Materials and Methods. To elicit starvation for amino acids or purines, 3-AT or AzaA, respectively, was added to SD medium. Each enzyme measurement was derived from three independent experiments. Asterisks mark samples with a standard error between 20 to 30%; all other measurements had a standard error of less than 20%.
FIG. 1.
GCN4 expression is increased in response to starvation for either glucose or amino acids. (A) RY124 (GCN2) and RY133 (gcn2Δ) cells expressing GCN4-lacZ that included the four upstream ORFs in the 5′ noncoding region of GCN4 were cultured in minimal medium with 2% glucose (nonstarvation), 0.05% glucose (glucose starvation), 3% glycerol, or 3% ethanol. Cultures were collected, and Gcn4p-LacZ enzyme activity was measured in lysate preparations as detailed in Materials and Methods. (B) Four different yeast strains expressing GCN4-lacZ, including the four upstream ORFs, were cultured in SD medium (nonstarvation), minimal medium supplemented with 0.05% glucose (glucose starvation) or SD medium supplemented with 3-AT (amino acid starvation). In the case of the histidine auxotroph JC482, sulfometuron methyl was used to elicit starvation for amino acids instead of 3-AT (56). For each strain background, the fold increase of β-galactosidase activity during amino acid or glucose limitation compared with nonstarvation conditions is listed above the histograms.
To determine whether glucose starvation induces GCN4 expression in other yeast strain backgrounds, we carried out similar starvation experiments in three additional strain backgrounds. In strain JC482, we found that starvation for glucose led to a ninefold increase in Gcn4p-LacZ enzyme activity, similar to that measured for amino acid starvation (Fig. 1B). By comparison, glucose starvation led to about a 6-fold increase in GCN4-lacZ expression in F113 background, while amino acid starvation led to more than 20-fold increase compared to the repressed growth conditions. Finally, in strain H1896, containing a chromosomally integrated GCN4-lacZ, we found only a 4-fold elevation in GCN4 expression during glucose limitation, compared to a 10-fold increase under amino acid starvation conditions (Fig. 1B). Our results demonstrate that GCN4 expression in many different yeast strains increases in response to glucose limitation, although the magnitude of induction differed between the surveyed strain backgrounds.
Glucose starvation induces translational expression of GCN4 and is dependent on Gcn2 protein kinase.
GCN4 translational expression is mediated by four upstream ORFs located in the 5′ noncoding region of the GCN4 mRNA. To investigate whether increased GCN4 expression in response to glucose starvation is mediated by these upstream ORFs, strain EG328-1A was transformed with plasmid p227 containing the GCN4-lacZ fusion with nucleotide substitutions in the initiation codons of each of the four ORFs. These mutations render the upstream ORFs nonfunctional for translation control, and any increase of Gcn4p-LacZ enzyme activity from p227 would be attributable to transcriptional control (23, 24, 34). In response to glucose limitation, there was a 2.6-fold increase of GCN4-lacZ expression from p227 (Table 2). By comparison, minimal differences in β-galactosidase activity were detected when the strain was starved for amino acids or purines compared to the nonstarved growth conditions. Given that there was nearly a 15-fold increase of GCN4 expression in the presence of the upstream ORFs in EG328-1A, these results strongly suggest that glucose starvation induces GCN4 expression primarily at the translational level, although there also appears to be a superposition of a modest transcriptional regulation.
Translational control of GCN4 requires increased phosphorylation of eIF-2α by Gcn2 protein kinase in response to amino acid or purine starvation. To determine whether induction of GCN4 expression during glucose limitation is dependent on GCN2, this gene was deleted in EG328-1A. Expression of GCN4-lacZ, which included the four upstream ORFs, during glucose starvation was dramatically reduced in the gcn2Δ cells compared to the wild-type GCN2 cells. Similar reductions in GCN4 expression were measured in the gcn2Δ strain grown in amino acid or purine starvation conditions (Table 2). There was a twofold increase in GCN4-lacZ expression from p227 under glucose starvation conditions in the gcn2Δ strain, suggesting that the previously noted transcriptional induction of GCN4 expression occurs independently of Gcn2 protein kinase. In the example of the shift to ethanol medium discussed earlier, deletion of GCN2 also contributed to a reduction of Gcn4p-LacZ activity, although there was still a significant increase compared to nonstarved cells (Fig. 1A). Together, these results indicate that Gcn2p mediates translational expression of GCN4 during glucose limitation, as well as during amino acid and purine starvation conditions.
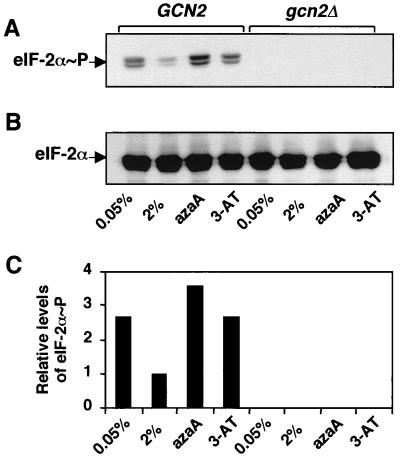
Glucose starvation stimulates phosphorylation of eIF-2α by Gcn2 protein kinase. To determine whether glucose starvation enhances phosphorylation of eIF-2α by Gcn2 protein kinase, we carried out an immunoblot analysis using a polyclonal antibody that specifically recognizes eIF-2α phosphorylated at serine-51. Consistent with earlier studies that used the alternative method of isoelectric focusing gel electrophoresis (16, 43, 56), there was increased phosphorylation of eIF-2α in response to limitation for either amino acids or purines compared with the repressed conditions (Fig. 2A). A similar increase in the levels of phosphorylated eIF-2α was observed during glucose starvation. In the absence of Gcn2 protein kinase, there was no detectable phosphorylation of eIF-2α (Fig. 2A). Levels of total eIF-2α present in the cells grown in the different growth media were unchanged, as determined by a second immunoblot analysis using a polyclonal antibody that recognizes both phosphorylated and nonphosphorylated eIF-2α (Fig. 2B). These results indicate that there is increased eIF-2α phosphorylation by Gcn2p kinase in response to limiting glucose in the medium (Fig. 2C).
FIG. 2.
Phosphorylation of eIF-2α is increased in yeast starved for glucose, amino acid, or purines. RY124 (GCN2) and RY133 (gcn2Δ) cells were cultured in minimal medium with 2% glucose (nonstarvation) or with 0.05% glucose (glucose starvation) or were grown in SD medium supplemented with 3-AT (amino acid starvation) or AzaA (purine starvation). Cell lysates were characterized by immunoblotting analysis using either a polyclonal antibody that specifically recognizes eIF-2α phosphorylated at serine-51 (A) or an antibody that recognizes both phosphorylated and nonphosphorylated forms of eIF-2α (B). The levels of phosphorylated eIF-2α in RY124 and RY133 cells cultured under each condition were measured by densitometry and presented as a histogram (C). Values are listed relative to that measured in RY124 grown under nonstarvation conditions.
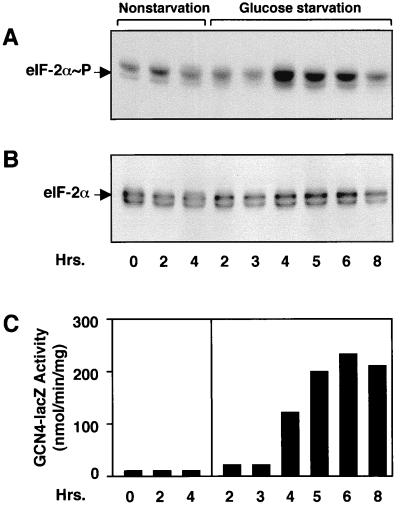
We next followed the time course of eIF-2α phosphorylation and induced GCN4 expression in response to glucose starvation conditions. Strain RY124 cells grown to mid-logarithmic phase in SD medium were collected and introduced into minimal medium containing either 2 or 0.05% glucose. Cells deprived of glucose were assayed after 2 h of incubation, followed by 1-h increments thereafter. Phosphorylation of eIF-2α as measured by immunoblotting was increased following 4 h of glucose starvation, and these elevated levels of phosphorylation were sustained up to 6 h of glucose limitation (Fig. 3A). The levels of β-galactosidase activity were also increased in cells limiting for glucose for 4 h and continued to accumulate after 6 h of growth in minimal medium containing 0.05% glucose (Fig. 3C). By 8 h of glucose starvation, phosphorylation of eIF-2α was reduced and there was no further accumulation of β-galactosidase enzyme activity (Fig. 2A and C). No increases in eIF-2α phosphorylation or Gcn4p-LacZ enzyme activity were detected during the nonstarvation conditions (Fig. 2A and C). Furthermore, the steady-state levels of total eIF-2α remained unchanged throughout the experiment (Fig. 3B). These results indicate that eIF-2α phosphorylation by Gcn2 protein kinase is increased transiently after glucose starvation. Furthermore, the increases of GCN4 expression and phosphorylation of eIF-2α by Gcn2p happen simultaneously in response to limiting glucose.
FIG. 3.
The time course of increased phosphorylation of eIF-2α is coincident with induced GCN4 expression in response to glucose starvation. Wild-type RY124 cells were grown in SD medium to mid-logarithmic phase and then shifted to fresh minimal medium with 2% glucose (nonstarvation) or with 0.05% glucose (glucose starvation). Cells were incubated with shaking at 30°C, and aliquots of the cultures were analyzed at the indicated times for eIF-2α phosphorylation or for Gcn4p-LacZ enzyme activity. Time zero represents analysis of cells collected just prior to the shift of media. (A) Immunoblot analysis using a polyclonal antibody that specifically recognizes eIF-2α phosphorylated at serine-51. (B) Measurement of eIF-2α levels using a polyclonal antibody that recognizes both phosphorylated and nonphosphorylated forms of eIF-2α. (C) Gcn4p-LacZ enzyme activity in each culture sample.
Stimulation of GCN4 expression in response to glucose starvation is dependent on HisRS-related region but not the ribosome association domain of Gcn2p.
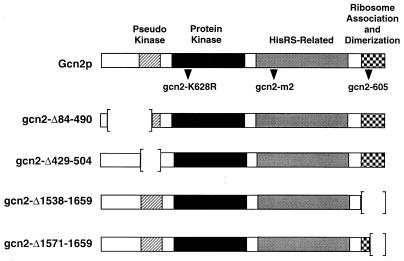
The Gcn2 protein kinase contains several domains that are required for stimulation of GCN4 translational expression during amino acid starvation conditions (Fig. 4). In addition to the kinase catalytic domain, Gcn2p function requires a pseudo-protein kinase region that has sequences homologous to only a portion of the protein kinase domain (59), the HisRS-related domain (54, 56), and the ribosomal binding region that is flanked by sequences that mediate Gcn2p dimerization (41, 42, 60). To determine whether each of these Gcn2p domains is required for the induction of GCN4 expression in response to glucose starvation, plasmids expressing different Gcn2p mutants containing defined alterations in each of these regions were introduced into strain RY196 (gcn2Δ). Expression of GCN4-lacZ was measured during glucose- or amino acid-limiting conditions. In wild-type cells containing a chromosomally encoded wild-type Gcn2p, Gcn4p-LacZ enzyme activity was increased 8-fold during glucose limitation and 10-fold in response to amino acid starvation (Table 3). With Gcn2p encoded on a plasmid, there was increased GCN4 expression during the nonstarvation conditions. Illustrating this point, cells expressing Gcn2p from a high-copy-number plasmid had 120 U of Gcn4p-LacZ enzyme activity during nonstarvation conditions, with about a twofold increase during glucose limitation. This increase in basal GCN4 expression is consistent with previous studies noting that elevated expression of Gcn2 protein kinase can lead to higher GCN4 translation in the absence of amino acid starvation (45, 55). Expression of GCN4-lacZ in cells containing each of the plasmid-encoded Gcn2p mutants was similar to that measured in the gcn2Δ cells, indicating that at least under these repressing conditions, these mutant kinases had reduced activities.
FIG. 4.
Diagram of Gcn2 protein kinase mutants containing mutations in each of the Gcn2p domains. The box depicts the sequence of Gcn2 protein kinase, including the pseudo-kinase, protein kinase, HisRS-related, and ribosome association and dimerization domains. Gcn2p is 1,659 residues in length, 69 residues longer than previously reported (54). The location of mutations in gcn2-K628R, gcn2-m2, and gcn2-605 are indicated below the Gcn2p diagram. Sequences deleted in Gcn2p mutants are illustrated by brackets.
TABLE 3.
Induced GCN4 expression during glucose limitation is dependent on the function of the HisRS-related domain of GCN2 protein kinase but does not require the ribosome binding region
| Relevant genotype | Gcn4p-LacZ enzyme activity (nmol/min/mg of protein)a
|
||
|---|---|---|---|
| Glucose
|
3-AT | ||
| 2% | 0.05% | ||
| GCN2 | 20 | 160 | 190 |
| gcn2Δ | 14 | 16 | 8 |
| GCN2 alleles encoded on a low-copy-number plasmid | |||
| GCN2 | 52 | 155 | 205 |
| gcn2-K628R | 12 | 16 | 9 |
| gcn2-Δ84-490 | 9 | 11 | 7 |
| gcn2-Δ429-504 | 13 | 16 | 11 |
| gcn2-m2 | 14 | 16 | 9 |
| gcn2-605 | 10 | 60 | 10 |
| gcn2-Δ1538-1659 | 11 | 21 | 10 |
| gcn2-Δ1571-1659 | 12 | 20 | 11 |
| GCN2 alleles encoded on a high-copy-number plasmid | |||
| GCN2 | 120 | 230 | 210 |
| gcn2-K628R | 14 | 19 | 12 |
| gcn2-Δ84-490 | 12 | 18 | 11 |
| gcn2-Δ429-504 | 15 | 19 | 12 |
| gcn2-m2 | 12 | 24 | 10 |
| gcn2-605 | 15 | 138 | 20 |
Strain RY196 (gcn2Δ) was transformed with low- or high-copy-number plasmids encoding the indicated GCN2 allele or vector alone. Enzyme activity was measured in lysates prepared from transformed cells cultured in SD medium, minimal medium supplemented with 0.05% glucose, or SD supplemented with 3-AT as described in Materials and Methods. All measurements were carried out with two or more independent transformants, and all values had a standard error of 20% or less.
In response to glucose limitation, only the gcn2-605 mutant, containing mutations that impair ribosomal binding of Gcn2p, was found to induce GCN4-lacZ expression. Characterization of Gcn2p recombinant proteins revealed that the gcn2-605 mutation does not affect the ability of carboxy-terminal portions of Gcn2p to dimerize (J. Narsimhan and R. Wek, unpublished data). β-Galactosidase enzyme activity was elevated six- or ninefold when gcn2-605 protein was expressed from a low- or high-copy-number plasmid, respectively. By comparison, gcn2-605 cells were unable to stimulate GCN4 expression when grown in amino acid-limiting conditions. Deletion of the carboxy termini in gcn2-Δ1538-1659 and gcn2-Δ1571-1659 impaired GCN4 translational control in response to both glucose and amino acid limitation, suggesting that Gcn2p dimerization facilitates activation of Gcn2 protein kinase in response to glucose starvation conditions. The steady-state levels of each of the mutant versions of Gcn2p expressed from a high-copy-number plasmid were found to be similar during the starvation and nonstarvation conditions, indicating that reduced levels of the mutant proteins is not the underlying basis for their impaired function (data not shown). These results suggest that while there are shared features between the mechanisms regulating Gcn2 protein kinase during glucose and amino acid limitation, there are differences involving the requirement of ribosomal association of Gcn2p in the activation process.
GCN20 is not essential for GCN4 translational control during glucose limitation.
It has been proposed that during amino acid starvation, uncharged tRNA in the vicinity of ribosomes induces Gcn2p phosphorylation of eIF-2α, leading to inhibition of the guanine nucleotide exchange activity catalyzed by the multisubunit protein eIF-2B. Gcn1p and Gcn20p form a complex that associates with ribosomes and is proposed to mediate activation of Gcn2 protein kinase in response to elevated levels of uncharged tRNA (31). To determine whether Gcn1p and Gcn20p are required to respond to glucose starvation, we deleted the GCN1 and GCN20 genes individually and characterized the resulting strains for GCN4 translational control. Consistent with previous reports, deletion of either gene reduced GCN4-lacZ expression in response to amino acid starvation (Table 4). Both Gcn1p and Gcn20p were also required for stimulation of GCN4-lacZ expression in response to purine starvation, with only a twofold increase in gcn1Δ and gcn20Δ cells. While Gcn1p was required for translational control in the glucose-limited cells, there was almost an eightfold induction of β-galactosidase activity in the absence of Gcn20p function. The induction of GCN4 expression by glucose limitation was also observed in gcn20Δ cells in the F113 strain background (data not shown). By comparison, deletion of GCN2 or substitution of alanine for the phosphorylation site, serine-51, in eIF-2α (SUI2-S51A) resulted in only a twofold increase in GCN4 expression during glucose limitation (Table 4). Furthermore, there was only about a twofold induction in the expression of GCN4-lacZ devoid of upstream ORFs in the gcn20Δ cells grown in glucose-limiting conditions, suggesting that this increased expression in the gcn20Δ strain resulted primarily from translational control.
TABLE 4.
Increased translational expression of GCN4 during glucose limitation is partially independent of Gcn20p function
| Strain/relevant genotype | Gcn4p-LacZ enzyme activity (nmol/min/mg)a
|
|||
|---|---|---|---|---|
| Glucose
|
3-AT | AzaA | ||
| 2% | 0.05% | |||
| Expression of GCN4-lacZ with four upstream ORFs | ||||
| RY124/wild type | 10 | 130 | 160* | 130* |
| RY281/gcn1Δ | 7 | 14 | 13 | 13 |
| RY282/gcn20Δ | 7 | 55 | 13 | 13* |
| RY283/gcn3Δ | 4 | 11 | 6 | 9 |
| RY133/gcn2Δ | 7 | 15 | 13 | 12 |
| RY287/SUI2-S51A | 9 | 14 | 16 | ND |
| Expression of GCN4-LacZ without upstream ORFs | ||||
| RY127/wild type | 420 | 1,200 | 540 | 650 |
| RY284/gcn1Δ | 310 | 690 | 430 | 420 |
| RY285/gcn20Δ | 320 | 760 | 440 | 340 |
| RY286/gcn3Δ | 300 | 870 | 320 | 320 |
The indicated strains contained either plasmid p180 encoding GCN4-lacZ with the four ORFs in the 5′ noncoding region of GCN4 or p227 containing GCN4-lacZ with each of the upstream ORFs abolished by mutations. Enzyme activity was measured in lysates prepared from cells cultured in SD medium, minimal medium supplemented with 0.05% glucose, or SD supplemented with 3-AT or AzaA as described in the Materials and Methods. Asterisks indicate values with a standard error of 25% or less; all other measurements had standard errors of 15% or less. ND, not determined.
A similar genetic analysis was carried out using cells deleted for GCN3, encoding the α subunit of eIF-2B that mediates inhibition of exchange function by phosphorylated eIF-2 (10, 38, 39). Loss of Gcn3p function blocked the induction of GCN4-lacZ expression in response to each of the three starvation conditions (Table 4), indicating that stimulation of GCN4-lacZ expression in response to glucose starvation is fully dependent on inhibition of eIF-2B function by phosphorylated eIF-2α. These results suggest that Gcn20p is not essential for the translational induction of GCN4-lacZ expression during glucose-limiting conditions and that both Gcn1p and Gcn3p are positive activators required for the induction of GCN4 expression in response to carbohydrate starvation.
Amino acid levels contribute to increased GCN4-lacZ expression during glucose starvation conditions.
The HisRS-related domain of Gcn2p is essential for stimulation of GCN4 expression in response to glucose starvation. This suggests that glucose starvation may impair aminoacylation of tRNA, contributing to the activation of Gcn2 protein kinase. To assess the contribution of amino acid levels in the translational regulation of GCN4 during glucose limitation, we measured Gcn4p-LacZ enzyme activity in the medium supplemented with all 20 amino acids (Table 5). There was over a 10-fold increase in β-galactosidase activity in response to glucose limitation with the addition of only amino acids essential for the RY124 strain and only about a twofold increase in the expression of GCN4 devoid of the upstream ORFs (Table 5). In glucose-limiting medium supplemented with all 20 amino acids, induction of GCN4 expression was partially reduced, with almost a fivefold increase of GCN4-lacZ expression compared to the nonstarvation conditions. This induction of GCN4-lacZ expression in the presence of all amino acids was largely dependent on the activity of the Gcn2 protein kinase, as GCN4 expression in the isogenic RY133 (gcn2Δ) strain grown in the presence of complete amino acids was increased only twofold in response to glucose limitation. This reduced level of induction was comparable to that measured in the gcn2Δ cells containing GCN4-lacZ without the upstream ORFs (Table 5). These results suggest that stimulation of GCN4 translation during glucose limitation is partially attributable to altered amino acid levels.
TABLE 5.
Addition of amino acids partially suppresses the induction of GCN4 expression in response to glucose limitation
| Strain/GCN2 allele | Plasmida | Gcn4p-LacZ enzyme activity (nmol/min/mg of protein)b
|
|||
|---|---|---|---|---|---|
| Glucosec
|
Glucose + amino acidsd
|
||||
| 2% | 0.05% | 2% | 0.05% | ||
| RY124/GCN2 | p180 | 11 | 120 | 10 | 47 |
| RY127/GCN2 | p227 | 470* | 1,300* | 450* | 990* |
| RY133/gcn2Δ | p180 | 10 | 18* | 7 | 16* |
| RY134/gcn2Δ | p227 | 490 | 1,100 | 530* | 1,100* |
Plasmid p180, encoding GCN4-lacZ with the upstream GCN4 ORFs, and p227, containing GCN4-lacZ without the ORFs, were transformed into the indicated GCN2 and gcn2Δ strains.
Cells were inoculated into synthetic minimal medium supplemented with 2 or 0.05% glucose and amino acids as indicated. Each value was measured in triplicate. Asterisks indicate a standard error of less than 30%; all other values had a standard error of 15% or less.
Cells were incubated in synthetic minimal medium supplemented with either 2 or 0.05% glucose plus only the required amino acid (leucine, valine, isoleucine, and tryptophan), with constant shaking at 30°C.
Cells were cultured in synthetic minimal medium supplemented with either 2 or 0.05% glucose plus all 20 amino acids.
Gcn2 protein kinase facilitates cell growth after prolonged glucose starvation.
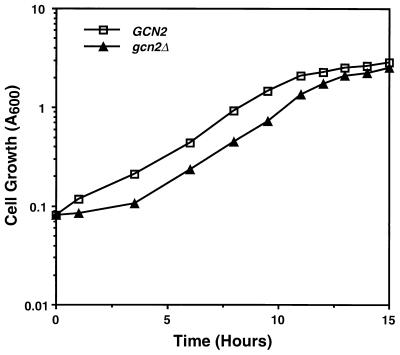
Our results show that glucose starvation induces GCN4 translational expression by activating Gcn2p phosphorylation of eIF-2α. To explore the consequences of this Gcn2p-mediated translational control, the growth levels of wild-type GCN2 and gcn2Δ cells in SD medium were compared following extended incubations in glucose-deficient medium. Strains RY124 (GCN2) and RY133 (gcn2Δ) were first incubated in minimal medium supplemented with 0.05% glucose for 6 or 24 h. No differences in cell viability between the GCN2 and gcn2Δ cells were observed after 24 h of glucose starvation. Cells were then transferred into SD medium at equal densities as judged by A600 and by counting cell numbers, and their growth was monitored. While there were no differences in growth between RY124 and RY133 cells following a 6-h incubation in minimal medium supplemented with 0.05% glucose, the gcn2Δ cells incubated for 24 h were delayed from entering exponential growth by 2.5 h compared with wild-type cells (Fig. 5). Once the strains entered the exponential phase, RY124 and RY133 cells grew at similar rates. This delay of 2.5 h between GCN2 and gcn2Δ cells was observed in three independent experiments. These results indicate that loss of Gcn2p activity in cells starved for glucose for an extended period of time impairs their ability to resume growth when shifted into medium containing 2% glucose. Addition of all 20 amino acids to the minimal medium expedited entry of the gcn2Δ cells into exponential growth, although it was still delayed compared to wild-type GCN2 cells similarly cultured in the presence of amino acids.
FIG. 5.
Impaired Gcn2p function delays growth following extended glucose starvation. RY124 (GCN2) and RY133 (gcn2Δ) cells were grown in minimal medium supplemented with 0.05% glucose for 22 h. Following this glucose-limited growth, cells were diluted into SD medium and incubated with shaking at 30°C. Cells were monitored for growth by measuring A600 and counting cell number. The gcn2Δ cells displayed a 2.5-h extension of the lag period prior to exponential growth in three independent experiments.
Vacuolar amino acid pools are diminished in gcn2Δ cells during glucose limitation.
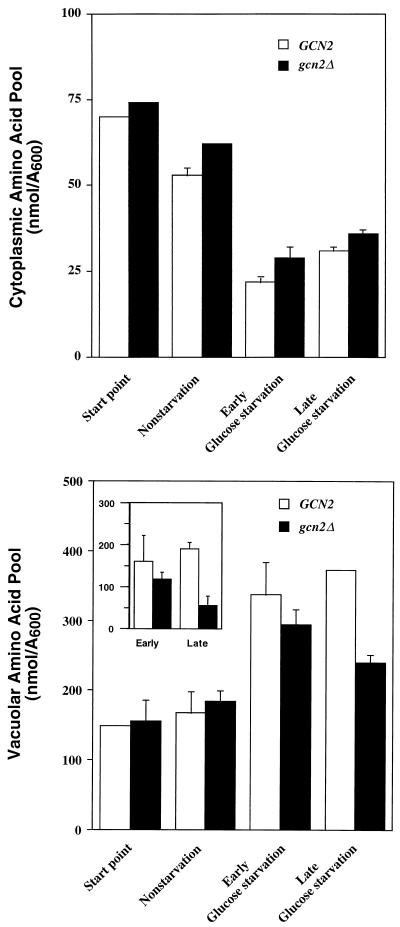
Our results show that altered levels of amino acids during glucose limitation may contribute to Gcn2p induction of GCN4 expression. Gcn4p is a transcriptional activator of genes involved in amino acid biosynthesis. Gcn2p induction of GCN4 translational expression may have physiological effects on the amino acid pools, contributing to delayed growth of gcn2Δ cells following a protracted starvation for glucose. To investigate the changes in amino acid pools, the levels of amino acids were directly measured in the cytoplasm and vacuoles, large intracellular organelles in S. cerevisiae that function as storage vesicles for amino acids (26, 33, 35). Strains RY124 and RY133 were grown to mid-logarithmic phase in SD and then shifted into fresh minimal medium supplemented with 2 or 0.05% glucose. Cells were subsequently collected at different time points following the medium shift, and amino acid levels were measured by the ninhydrin method. After 6 h of glucose limitation, there was a 2.5-fold decrease in the cytoplasmic pool of amino acids, consistent with the idea that reduced amino acid and charged tRNA levels contribute to regulation of Gcn2 protein kinase; by comparison, there was a twofold elevation in the vacuolar pool of amino acids with no statistical difference between cells with or without Gcn2p function (Fig. 6). At this starvation time point, the vacuoles contained greater than 90% of the free amino acids in the cell.
FIG. 6.
Measurement of vacuolar and cytoplasmic amino acid pools in wild-type and gcn2Δ cells cultured during nonstarvation and glucose starvation conditions. RY124 (GCN2) and RY133 (gcn2Δ) cells were grown in SD medium to mid-logarithmic phase and shifted to fresh minimal medium supplemented with either 2% glucose (nonstarvation) or 0.05% glucose (glucose starvation). Cell cultures were incubated with shaking at 30°C, and amino acid levels were measured in the cytoplasm (top) and vacuoles (bottom) by the ninhydrin method as described in Materials and Methods. Start point represents analysis of cells collected just prior to the shift of medium. Nonstarved cells were collected and analyzed after 4 h of growth in SD medium. Early and late glucose starvation represent analyses of cells collected after 6 and 20 h of incubation, respectively, in low-glucose medium. The inset represents the accumulation of vacuolar amino acid levels during early and late glucose starvation that exceeds the basal levels present at the start point.
Following incubation for 20 h in glucose-limiting medium, the vacuolar pool of amino acids in the gcn2Δ cells was reduced by 35% compared with the wild-type strain; no differences were detected in the cytoplasmic amino acid pool between the wild-type and gcn2Δ strains (Fig. 6). As illustrated in the inset to Fig. 6, to assess the contribution of increased GCN4 translational expression to the accumulation of amino acid pool in vacuoles, the basal vacuolar amino acid pool under nonstarvation conditions was subtracted from the measurements taken during glucose starvation. Following this assessment, there was a fourfold difference after the longer starvation period. The cytoplasmic amino acid pools were similar between wild-type and gcn2Δ cells (Fig. 6). These results indicate that in response to glucose limitation, there is an increase in the vacuolar amino acid pool concomitant with a reduction in the cytoplasmic amino acid level. Increased GCN4 translational expression by Gcn2p contributes to the accumulation of amino acids in vacuoles during prolonged glucose starvation. When all 20 amino acids were added to the glucose-limiting medium, the cytoplasmic amino acid pool increased by about 50% at the early glucose starvation (22 ± 1 versus 31 ± 1 nmol/A600) and twofold at the later glucose-limiting condition (31 ± 1 versus 60 ± 2 nmol/A600). Given that addition of all 20 amino acids also significantly reduced GCN4-lacZ expression (Table 5), these results are consistent with the idea that altered cytoplasmic amino acid pools can control the activity of Gcn2 protein kinase through accumulation of uncharged tRNA during glucose starvation.
To directly assess the contribution of Gcn4p in the accumulation of vacuolar amino acids in response to limiting glucose, we constructed an isogenic gcn4Δ strain, RY290. The gcn4Δ cells display a severe growth defect in SD medium that can be largely overcome by the addition of all 20 amino acids. When we carried out an experiment similar to that described in Fig. 6, we found that the vacuolar amino acid pool was elevated in both glucose-limiting and nonlimiting conditions (Fig. 7). At the late glucose starvation period, the vacuolar amino acids levels were reduced by over 30% to 310 nmol/A600, as found for the gcn2Δ cells (Fig. 6). These results suggest that cells can accumulate large vacuolar pools of amino acids independent of GCN4 function. We proposed that the reason why the gcn4Δ strain had high vacuolar amino acid levels in the 2% glucose medium is that these cells are growth limiting for amino acids. We next grew the gcn4Δ strain in SD medium supplemented with all 20 amino acids and then transferred these cells to minimal medium containing either 2 or 0.05% glucose without amino acids. While the vacuolar amino acid levels were again elevated in the glucose-limiting condition, we found about a 25% reduction in the presence of elevated glucose concentrations (Fig. 7). When cells were transferred to minimal medium containing 2% glucose and all 20 amino acids, gcn4Δ cells were able to attain a growth rate approaching the wild-type level and reduced levels of amino acids in the vacuole (224 nmol/A600) that were comparable to those for wild-type and gcn2Δ cells. Together, these experiments suggest that Gcn4p does not play a major role in the sharp increase in the vacuolar amino acid levels during early glucose starvation.
FIG. 7.
Amino acids accumulate in the vacuole of gcn4Δ cells. (A) RY124 (GCN2), RY133 (gcn2Δ), and RY290 (gcn4Δ) cells were grown in SD medium to mid-logarithmic phase and shifted to minimal medium containing either 2% glucose (nonstarvation) or 0.05% glucose (glucose starvation). Cultures were incubated with shaking at 30°C, and vacuolar amino acid levels were measured as described in Materials and Methods. (B) As for panel A except that all 20 amino acids were added to the SD medium prior to the medium shift. Two independent experiments yielding similar results were carried out for each panel.
Impaired GCN4 translational control during glucose limitation decreases glycogen storage levels.
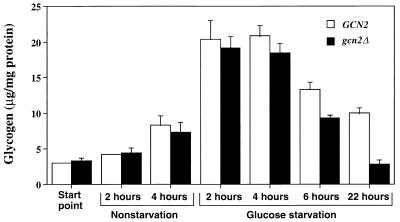
Yeast cells accumulate glycogen in response to starvation for many different nutrients, including nitrogen and carbohydrates (25). To investigate whether Gcn2 protein kinase control of GCN4 expression affects the levels of this glucose storage polymer, we cultured RY124 and RY133 in minimal medium containing 2 or 0.05% glucose, harvested cells at the indicated times, and assayed for glycogen (Fig. 8). Glycogen levels were increased sevenfold after 2 h of incubation in glucose-limiting medium, and there were no significant differences between GCN2 and gcn2Δ cells. The glycogen levels were reduced in both RY124 and RY133 following 6 h of incubation in the glucose-deficient medium, with the gcn2Δ cells having 70% of the levels measured for the wild-type cells. Following a 22-h culture period, glycogen levels in the gcn2Δ cells were further diminished, with RY133 having fourfold less glycogen than wild-type RY124. These results indicate that stimulation of GCN4 translational expression by Gcn2 protein kinase during an extended glucose starvation contributes to the maintenance of both glycogen and amino acid storage pools in response to glucose-limiting growth conditions.
FIG. 8.
Glycogen levels are reduced in gcn2Δ mutants after extended starvation for glucose. RY124 (GCN2) and RY133 (gcn2Δ) were grown in SD medium to mid-logarithmic phase and then shifted to fresh minimal medium with 2% glucose (nonstarvation) or with 0.05% glucose (glucose starvation). Cell cultures were incubated with shaking at 30°C, and glycogen levels were measured in cells sampled at the indicated times. Start point represents analysis of cells just prior to the shift of medium.
DISCUSSION
In this report, we show that Gcn2p-mediated phosphorylation of eIF-2α, and the accompanying translational control of GCN4, is induced in response to glucose limitation as well as to amino acid starvation. Activation of Gcn2 protein kinase is transient during glucose starvation, with elevated phosphorylation of eIF-2α occurring between 4 to 6 h following a shift into glucose-limiting medium (Fig. 3). The mechanisms regulating Gcn2p during carbohydrate limitation are, in part, conserved with those operating during amino acid limiting conditions (Fig. 9). In both starvation conditions, the gcn2-m2 mutant, impaired for the association of Gcn2p with uncharged tRNA in vitro, was unable to induce GCN4 translation (Table 3). These observations suggest that accumulation of uncharged tRNAs in the cell contributes to Gcn2p activation during glucose starvation as well as amino acid limitation (Fig. 9). Supporting this view, our results show that altered amino acid levels partially contributed to induced GCN4 expression (Table 5) and cytoplasmic amino acid pool levels were greatly reduced in response to glucose limitation (Fig. 6). However, there are also important differences in the regulatory mechanisms in response to amino acid and glucose limitation. First, the gcn2-605 mutant impaired for association with ribosomes effectively induced GCN4 expression during glucose limitation but was unable to stimulate translational control during amino acid starvation conditions (Table 3). A second important difference was that Gcn20p was not essential for induced GCN4 expression during glucose limitation but was required in response to amino acid starvation (51) (Table 4). Gcn20p can complex with Gcn1p and has been found to be associated with ribosomes in an ATP-stimulated process (31). These results indicate that induction of Gcn2p activity and GCN4 translational control occurs in response to a wider spectrum of nutrient deprivations than was previously thought and suggest that regulation of eIF-2α kinase activity in yeast during glucose limitation can be mediated by ribosome-independent events.
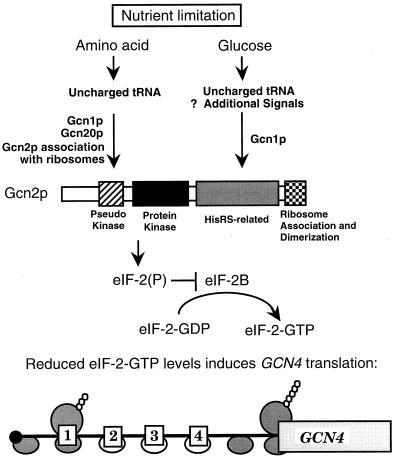
FIG. 9.
Model for regulation of Gcn2 protein kinase and GCN4 translation during amino acid or glucose limitation. Uncharged tRNAs that accumulate during amino acid starvation are proposed to associate with the HisRS-related domain of Gcn2p, leading to a conformational change in the protein and activation of eIF-2α kinase activity (23, 52, 54, 56). Gcn1p and Gcn20p are required for elevated levels of eIF-2α phosphorylation by Gcn2p and are proposed to facilitate tRNA interaction with Gcn2p in the vicinity of the ribosome (31). Ribosomal association of Gcn2p is mediated by RNA-binding sequences in the carboxy terminus of Gcn2p, and this interaction is proposed to facilitate activation of the eIF-2α kinase during amino acid starvation (42, 60). Activation of Gcn2p during glucose limitation requires the function of the HisRS-related domain, suggesting uncharged tRNAs present during carbohydrate limitation signal activation of Gcn2p. Additional signals may also participate in the activation of this eIF-2α kinase during glucose limitation. While Gcn1p is required for regulation of Gcn2p during glucose starvation, Gcn20p is in part dispensable. It has been suggested that the EF3-like domain in Gcn1p facilitates delivery of uncharged tRNA to the HisRS-related domain of Gcn2p in the vicinity of the ribosomes (31). Gcn20p associates with Gcn1p and is proposed to enhance its regulatory function. The details of this enhancement are uncertain, but our results suggest that the role of Gcn20p is not simply to facilitate Gcn1p-mediated interaction of uncharged tRNA with Gcn2 protein kinase. Furthermore, Gcn2p sequences required for association of the protein kinase with ribosomes are not required for induction of GCN4 translation in response to glucose limitation. Elevated phosphorylation of eIF-2α by Gcn2 protein kinase during nutrient limitation reduces the exchange of eIF-2-GDP for eIF-2-GTP that is catalyzed by eIF-2B (24). After translation of upstream ORF1 in the 5′ leader of the GCN4 mRNA, the reduced eIF-2-GTP levels resulting from nutrient limitation are proposed to delay subsequent reinitiation of translation. This allows for the 40S ribosome devoid of eIF-2-GTP, as illustrated by the open circles, to scan through the inhibitory upstream ORF2, ORF3, and ORF4 located in the 5′ noncoding portion of the GCN4 mRNA. In the interval between ORF4 and the GCN4 coding sequences, scanning ribosomes associate with eIF-2–GTP and initiate translation at the GCN4 coding sequences.
Physiological significance of Gcn2p induction of GCN4 translation in response to glucose limitation.
After an extended period of glucose starvation, cells lacking Gcn2p function delay entry into exponential growth in SD medium (Fig. 5). We found elevated levels of free amino acids in wild-type and gcn2Δ cells during glucose starvation, with an increase in the vacuolar amino acid pool and a concomitant reduction in the cytoplasmic amino acid levels (Fig. 6). During the early period of glucose starvation up to 6 h, the accumulation of vacuolar amino acids was independent of Gcn4p translation control, as similar levels were observed in GCN2 and gcn2Δ cells (Fig. 6). However, after longer periods of glucose limitation, vacuolar amino acid levels in gcn2Δ cells were lower than in the wild-type strain. These results suggest that Gcn2 protein kinase induction of GCN4 translational expression contributes to the storage of amino acids when carbohydrates are limiting. This reduction in the storage pool of amino acids may be one reason why there is delayed growth of gcn2Δ cells following glucose starvation. A second contributing factor may be accelerated glycogen turnover in gcn2Δ cells starved for periods longer than 6 h (Fig. 8).
In support of the idea that vacuolar storage of amino acids is triggered in cells limiting for nutrients, Messenguy et al. (33) observed that reduced assimilation of ammonia drove amino acids from the cytoplasm into vacuoles. In this study, concentrations of the individual amino acids were found to vary greatly, with glutamate constituting nearly a third of the total pool, and arginine and alanine each constituting about 10%. With reduced assimilation of ammonia, there was a further flux of amino acids into the vacuolar compartment, along with a significant elevation in the concentration of the predominant amino acids. It was rationalized that the reduced rates of protein synthesis and cell growth accompanying nitrogen limitation would trigger the cells to store amino acids preferentially in vacuoles. Our studies are consistent with this model (Fig. 6 and 7). The findings that the level of amino acid accumulation in vacuoles following nutrient limitation exceeded the net loss measured in the cytoplasm (Fig. 6) and that accumulation occurred in the absence of GCN4 function (Fig. 7) suggest that protein turnover and uptake mechanisms significantly contribute to this storage strategy.
Gcn4p contributes to the transcriptional activation of over 50 different genes in yeast (23). While the majority of these genes are directly involved in amino acid biosynthetic pathways, Gcn4p also regulates the expression of genes important for purine synthesis, aminoacylation of tRNA, membrane transport, and the formation of metabolic precursors used in amino acid synthesis (12, 23, 43, 58). The magnitude and kinetics of the Gcn4p-mediated control of these genes vary depending on the number of Gcn4p-binding sites in their promoters and the additional transcriptional factors that function in combination with Gcn4p. The transient induction of GCN4 translation during glucose starvation would indicate that increased expression of at least a subset of these genes contributes to accumulation of amino acids in the vacuolar compartment. These induced genes may represent specifically those pathways involved in the synthesis of key amino acids whose concentrations are greatly elevated in the vacuoles. The storage of amino acids during nutrient limitation would ensure ready access to nitrogen when carbohydrates again become accessible. Based on these observations, we propose that one physiological role for the induction of GCN4 translational expression during glucose limitation is to store nitrogen in the form of amino acids for future use.
The rationale for storage of glycogen in response to nutrient limitation follows a similar strategy of adaptation to different growth conditions (25). Our results indicate a significant reduction in glycogen levels in gcn2Δ cells during an extended glucose starvation, establishing a linkage between Gcn2p-mediated translational control and glycogen metabolism (Fig. 8). Currently, no genes directly involved in glycogen accumulation have been described to be under Gcn4p-mediated regulation. This suggests that reduced glycogen levels in gcn2Δ cells during an extended glucose starvation may be an indirect consequence of reduced levels of intermediary metabolites. We note that Glc7p, a type 1 serine/threonine protein phosphatase, was found to be important for both glycogen accumulation and GCN4 control of amino acid biosynthesis, suggesting that this enzyme also regulates protein phosphorylation in these two pathways (53).
Translational control by eIF-2α phosphorylation in yeast and mammals during nutrient deprivation.
There are many parallels between the translation control mediated by nutrient starvation in mammalian and yeast cells. In response to either amino acid or glucose limitation in mammalian cells, there is an increase in the phosphorylation of eIF-2α. While eIF-2α phosphorylation in mammals leads to a reduction in total translation initiation, there is evidence supporting a simultaneous induction of selected gene expression (2, 3, 7, 29). For example, Andrulis et al. (2) observed that the activity of asparagine synthetase is sharply increased in response to elevation of many different uncharged tRNA levels. Subsequently, it was shown that this enzyme induction was due to transcriptional control involving positive-acting sequences centered about 70 nucleotides upstream of the asparagine synthetase transcriptional start site that mediates amino acid control by some unknown factor(s) (17). Interestingly, glucose limitation was also shown to increase expression of asparagine synthetase mRNA (3). Many of these regulatory features are reminiscent of the amino acid control system in yeast, and we note that Gcn4p is a transcriptional activator of both ASN1 and ASN2, encoding asparagine synthetase isozymes in yeast (13).
The identity of the mammalian protein kinase(s) catalyzing phosphorylation of eIF-2α during nutrient limitation is not known. Stimulation of PEK autokinase activity has been linked to glucose limitation by a mechanism involving impaired glycosylation of proteins in the endoplasmic reticulum (20). Recent studies have also identified a mammalian Gcn2p homologue that shares the HisRS-related region juxtaposed to the kinase domain (5, 49). Mutations in either the catalytic domains or HisRS-related sequences impair mammalian Gcn2p function in an in vivo translation system (49). Given our results identifying glucose limitation as a regulator of Gcn2p in yeast, it is inviting to speculate that translational control by different nutrient limitations in mammalian cells is mediated by the newly identified Gcn2p homologue. The specific role of each of the mammalian eIF-2α kinases in translation control during starvation for individual nutrients and the role of eIF-2α phosphorylation in gene-specific regulation in mammals await further study.
ACKNOWLEDGMENTS
We thank Mark Goebl, Peter Roach, Janice Blum, Anna DePaoli-Roach, Wayne Wilson, Shuhao Zhu, Krishna Vattem, and members of the Wek laboratory for advice during the course of this work and comments on the manuscript. Additional thanks go to Alan Hinnebusch for helpful discussions and generously providing plasmids, strains and reagents, Gary Krause for eIF-2α antibody, and Robert Harris and Jinnie Garret for advice on amino acid measurements.
This work was supported in part by Public Health Service grant GM49164 from the National Institutes of Health and by American Cancer Society grant RPG MBC-87806 (R.C.W.).
REFERENCES
- 1.Abastado J P, Miller P F, Jackson B M, Hinnebusch A G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis of GCN4 translational control. Mol Cell Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrulis I L, Hatfield G W, Arfin S M. Asparaginyl-tRNA aminoacylation levels and asparagine synthetase expression in cultured Chinese hamster ovary cells. J Biol Chem. 1979;254:10629–10633. [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa-Tessmann I P, Pineda V L, Nick H S, Schuster S M, Kilberg M S. Transcriptional regulation of the human asparagine synthetase gene by carbohydrate availability. Biochem J. 1999;339:151–158. [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmeyer H U, Bernt E, Schmidt F, Stork H. d-Glucose. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Vol. 3. New York, N.Y: Academic Press, Inc.; 1974. pp. 1196–1201. [Google Scholar]
- 5.Berlanga J J, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brostrom C O, Brostrom M A. Regulation of translation initiation during cellular response to stress. Prog Nucleic Acid Res Mol Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- 8.Cannon J F, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J J, London I M. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 10.Cigan A M, Bushman J L, Boal T R, Hinnebusch A G. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc Natl Acad Sci USA. 1993;90:5350–5354. doi: 10.1073/pnas.90.11.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 139–172. [Google Scholar]
- 12.Coleman S T, Tseng E, Moye-Rowley W S. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J Biol Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 13.Dang V D, Valens M, Bolatin-Fukuhar M, Daignman-Fornier B. Cloning of the ASN1 and ASN2 genes encoding asparagine synthetases in Saccharomyces cerevisiae: differential regulation by the CCAAT-box-binding factor. Mol Microbiol. 1996;22:681–692. doi: 10.1046/j.1365-2958.1996.d01-1715.x. [DOI] [PubMed] [Google Scholar]
- 14.DeGracia D J, Sullivan J M, Neumar R W, Alousi S S, Hikade K R, Pittman J E, White B C, Rafols J A, Krause G S. Effect of brain ischemia and reperfusion on the localization of phosphorylated eukaryotic initiation factor 2α. J Cereb Blood Flow Metab. 1997;17:1291–1302. doi: 10.1097/00004647-199712000-00004. [DOI] [PubMed] [Google Scholar]
- 15.de Haro C, Mendez R, Santoyo J. The eIF-2α kinases and the control of protein synthesis. FASEB J. 1996;10:1378–1388. doi: 10.1096/fasebj.10.12.8903508. [DOI] [PubMed] [Google Scholar]
- 16.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T F, Hinnebusch A G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 17.Guerrini L, Gong S S, Mangasarian K, Basilico C. cis- and trans-acting elements involved in amino acid regulation of asparagine synthetase gene expression. Mol Cell Biol. 1993;13:3203–3212. doi: 10.1128/mcb.13.6.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannig E M, Hinnebusch A G. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol Cell Biol. 1988;8:4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannig E M, Williams N P, Wek R C, Hinnebusch A G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990;126:546–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding H P, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 21.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 22.Hinnebusch A G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- 23.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF-2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 199–244. [Google Scholar]
- 24.Hinnebusch A G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 25.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–282. [Google Scholar]
- 26.Jones E W, Fink G R. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces, metabolism and gene expression. Cold Spring HarborNew York, N.Y.: Cold Spring Harbor Laboratory; 1982. pp. 181–299. [Google Scholar]
- 27.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 207–217. [Google Scholar]
- 28.Keppler D, Decker K. Glycogen determination with amyloglucosidase. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Vol. 3. New York, N.Y: Academic Press, Inc.; 1974. pp. 1127–1131. [Google Scholar]
- 29.Kilberg M S, Hutson R G, Laine R O. Amino acid-regulated gene expression in eukaryotic cells. FASEB J. 1994;8:13–19. doi: 10.1096/fasebj.8.1.8299885. [DOI] [PubMed] [Google Scholar]
- 30.Marton M J, Crouch D, Hinnebusch A G. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993;13:3541–3556. doi: 10.1128/mcb.13.6.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marton M J, Vazquez de Aldana C R, Qui H, Chakraburtty K, Hinnebusch A G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF-2α kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 33.Messenguy F, Colin D, ten Have J P. Regulation of compartmentation of amino acid pools in Saccharomyces cerevisiae and its effects on metabolic control. Eur J Biochem. 1980;108:439–447. doi: 10.1111/j.1432-1033.1980.tb04740.x. [DOI] [PubMed] [Google Scholar]
- 34.Mueller P P, Hinnebusch A G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 35.Ohsumi Y, Kitamoto K, Anraku Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J Bacteriol. 1988;170:2676–2682. doi: 10.1128/jb.170.6.2676-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pain V M. Translational control during amino acid starvation. Biochimie. 1994;76:718–728. doi: 10.1016/0300-9084(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 37.Pain V M, Clemens M J. Adjustment of translation to special physiological conditions. In: Trachsel H, editor. Translation in eukaryotes. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 293–324. [Google Scholar]
- 38.Pavitt G D, Ramaiah K V, Kimball S R, Hinnebusch A G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavitt G D, Yang W, Hinnebusch A G. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2β mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proud C G. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:217–256. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 41.Qiu H, Garcia-Barrio M T, Hinnebusch A G. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions of the C-terminal ribosome binding region and the protein kinase domain. Mol Cell Biol. 1998;18:2697–2711. doi: 10.1128/mcb.18.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez M, Wek R C, Hinnebusch A G. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolfes R J, Hinnebusch A G. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13:5099–5111. doi: 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 45.Roussou I, Thireos G, Hauge B M. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol Cell Biol. 1988;8:2132–2139. doi: 10.1128/mcb.8.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translation control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designated for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sood R, Porter A C, Ma K, Wek R C. Pancreatic eukaryotic initiation factor-2α kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to ER stress. Biochem J. 2000;346:281–293. [PMC free article] [PubMed] [Google Scholar]
- 49.Sood R, Porter A C, Olsen D, Cavener D R, Wek R C. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spies J R. Colorimetric procedures for amino acids. Methods Enzymol. 1957;3:467–477. [Google Scholar]
- 51.Vazques de Aldana C R, Marton M J, Hinnebusch A G. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 1995;14:3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wek R C. eIF-2 kinases: regulators of general and gene-specific translation initiation. Trends Biochem Sci. 1994;19:491–496. doi: 10.1016/0968-0004(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 53.Wek R C, Cannon J F, Dever T E, Hinnebusch A G. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol Cell Biol. 1992;12:5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wek R C, Jackson B M, Hinnebusch A G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wek R C, Ramirez M, Jackson B M, Hinnebusch A G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990;10:2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wek S A, Zhu S, Wek R C. The histidyl-tRNA synthetase-related sequence in eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang R, Chun K T, Wek R C. Mitochondrial respiratory mutants in yeast inhibit glycogen accumulation by blocking activation of glycogen synthase. J Biol Chem. 1998;278:31337–31344. doi: 10.1074/jbc.273.47.31337. [DOI] [PubMed] [Google Scholar]
- 58.Zaman Z, Bowman S B, Kornfeld G D, Brown A J, Dawes I W. Transcription factor GCN4 for control of amino acid biosynthesis also regulates the expression of the gene for lipoamide dehydrogenase. Biochem J. 1999;340:855–862. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu S, Sobolev A Y, Wek R C. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]
- 60.Zhu S, Wek R C. Ribosome binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J Biol Chem. 1998;273:1808–1814. doi: 10.1074/jbc.273.3.1808. [DOI] [PubMed] [Google Scholar]